Cancer studies have historically focused on changes in the transcriptome of tumor cells to reveal the molecular pathways responsible for cellular transformation. In the last 20 years, however, it has become clear that deregulation of translation also plays a crucial role in cancer development and progression. Major players in cancer biology, such as the PI3K pathway, tumor suppressor protein p53 and MYC, can deregulate expression of components of the translation machinery and can influence the spectrum of mRNAs which are most efficiently translated in the cell. In addition, somatic lesions affecting ribosome biogenesis, translation initiation and elongation factors have been described in a wide variety of human cancers.1 The cancer promoting action of such lesions is supported by the observation that patients with congenital mutations in ribosomal proteins or ribosome biogenesis factors develop ribosomopathies, diseases with specific pathological characteristics that often lead to an increased risk of developing hematopoietic malignancies and solid tumors.2
T-cell acute lymphoblastic leukemia (T-ALL) is an aggressive malignancy caused by accumulation of multiple co-operating oncogenic lesions in developing T cells. It has been well-established that prominent roles in T-ALL pathogenesis are played by: i) ectopic expression of oncogenic transcription factors such as TAL1, TLX1 and TLX3; ii) deregulation of the CDKN2A and CDKN2B cell cycle regulators; iii) constitutive activation of NOTCH1 signaling; and iv) hyperactivation of cytokine signaling by defects, e.g. in IL7R and JAK kinases.3,4 Recent next generation sequencing based screens in T-ALL have further expanded the spectrum of defects involved in the pathogenesis of this disease. First, it became clear that epigenetic regulators like PHF6, SUZ12, EZH2, TET1 and KDM6A are also involved in T-ALL pathogenesis.5 Second, defects in essential components of the translation machinery have been revealed. We described mutations affecting the ribosomal proteins L5/uL18 (RPL5/uL18) and L10/uL16 (RPL10/uL16) in 10% of pediatric TALL patients.6 In addition, ribosomal protein L22/eL22 (RPL22/eL22) is inactivated in another 10% of T-ALLs7 and rare mutations in the ribosomal protein L11/uL5 (RPL11/uL5) gene were found in relapsed T-ALL patients (Figure 1).8 The oncogenic capacity of ribosomal protein defects in T-ALL is illustrated by the observation that loss of one copy of RPL22/eL22 accelerates tumor formation in a mouse model of T-cell lymphoma.7 Whereas acquiring mutations in ribosomal proteins is the most direct way to influence generation and function of mature ribosomes in a cell, other mutations in T-ALL may also affect these processes. For example, up to 40% of all T-ALL cases carry loss-of-function defects in the PHF6 gene.9 While the exact role of PHF6 in T-ALL is not yet understood, it has been proposed that it functions as an epigenetic regulator. PHF6 localizes in the nucleolus, inhibits rRNA synthesis, and its inter-actors, identified by mass spectrometry, include several ribosomal proteins.10,11 These observations suggest a role for PHF6 in ribosome biogenesis, and loss of PHF6 function in T-ALL could thus represent an alternative way to interfere with proper formation of ribosomes (Figure 1).
Figure 1.
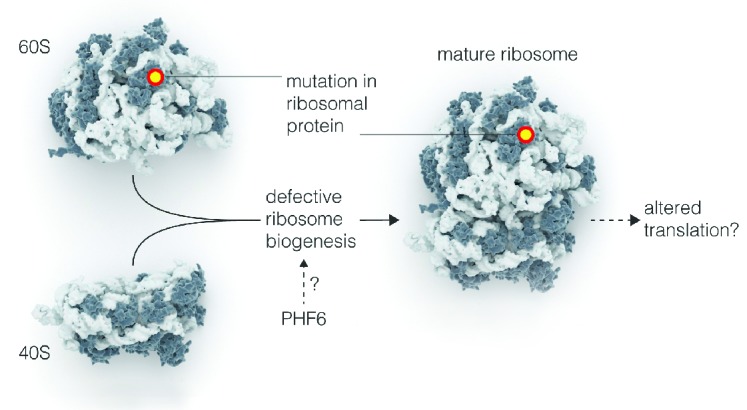
Mutations that impair maturation of the translation machinery in T-ALL. Mutations in ribosomal proteins and defective PHF6 function may impair formation of mature ribosomes (ribosome biogenesis) in T-ALL. Note that ribosome biogenesis defects have only been experimentally proven so far for mutations in ribosomal proteins, the effects of PHF6 are more speculative. Effects on altered translation are also speculative.
Besides the identification of mutations in ribosomal proteins, there are additional indications for an essential role of translational deregulation in T-ALL. Translation of most cellular mRNAs depends on a cap structure in the 5′ part of the mRNA (cap-dependent translation). However, for other mRNAs, translation is initiated by alternative structures such as an internal ribosomal entry site (cap-independent translation). It was shown that T-ALL development in mice can be accelerated by overexpression of eIF4A and eIF4E, two proteins that are part of the protein complex that initiates cap-dependent translation downstream of mTORC1.12,13 The strict dependence of T-ALL cells on cap-dependent translation is further supported by the observations that silvestrol, a drug inhibiting eIF4A, can impair proliferation of mouse and human T-ALL leukemia cells, both in vitro and in vivo,12 and that interference with eIF4E by shRNA knockdown or administration of the eIF4E inhibitor 4EGI-1 reduces the viability of T-ALL blasts in vitro.13 As far as we are aware, no specific genetic lesions in eIF4A, eIF4E or other components of the protein complex supporting cap-dependent translation have been described in T-ALL, suggesting that the role of other (known) genetic lesions in T-ALL in driving cap-dependent translation has been underestimated. An obvious candidate is the NOTCH1 pathway which is hyperactivated in 60% of T-ALL cases by mutations in the NOTCH1 receptor itself and/or by inactivation of its ubiquiting ligase FBXW7.3 Whereas hyperactive NOTCH1 signaling most likely drives T-cell transformation by influencing several pathways, NOTCH1 hyperactivates the PI3K-AKT-mTORC1 pathway and may thus support cap-dependent translation. Indeed, NOTCH1 activity in TALL cells leads to HES1 mediated downregulation of PTEN, a critical negative regulator of the PI3K pathway. Moreover, NOTCH1 can activate AKT via the LCK tyrosine kinase in T cells. Furthermore, numerous signaling molecules upstream of PI3K, including the interleukin 7 receptor alpha chain (IL7RA) and the pre-T-cell receptor alpha (PTCRA), are up-regulated upon activation of NOTCH1 signaling in T-ALL lymphoblasts.14 In addition to impairing degradation of NOTCH1, FBXW7 deletion may further enhance cap-dependent translation by directly stabilizing mTOR, another known substrate of FBXW7.15 Inactivation of the PTEN tumor suppressor, an event occurring in 20% of T-ALLs, is another way for T-ALL cells to hyperactivate the PI3K-AKT-mTORC1 axis. Finally, hyperactivation of receptor tyrosine kinases can also stimulate mTORC1 activity and cap-dependent translation (Figure 2). Interestingly, whereas constitutive activation of the receptor tyrosine kinase TRK leads to increased mTORC1 signaling in lymphoid cells, pre-leukemic cells with hyperactive TRK tend to acquire activating mutations in NOTCH1 and loss of PTEN. This leads to T-ALL during clonal evolution, resulting in even stronger activation of mTORC1 and more pronounced dependence on cap-dependent translation.13
Figure 2.
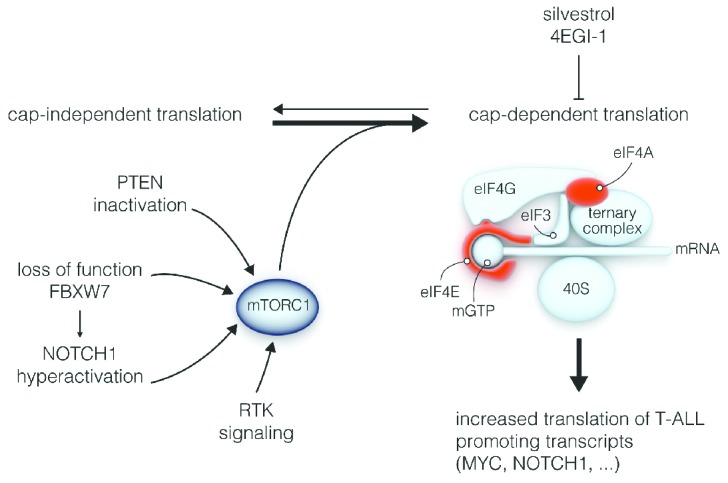
T-ALL cells depend on cap-dependent translation. Several molecular lesions in T-ALL (PTEN or FBXW7 inactivation, NOTCH1 or RTK hyperactivation) promote cap-dependent translation in T-ALL cells, resulting in more efficient translation of T-ALL promoting genes. Cap-dependent translation can be inhibited by drugs like silvestrol and 4EGI-1, that target eIF4A and eIF4E in the protein complex initiating cap-dependent translation. RTK: receptor tyrosine kinase.
How the defects in the translation machinery or cap-dependent translation described above drive T-ALL is the next important question. The Wendel laboratory recently showed that inhibition of eIF4A function with the drug silvestrol in T-ALL cells impairs the translation of components essential for T-ALL pathogenesis, such as NOTCH1, MYC, RUNX1 and BCL11B, suggesting that cells with hyperactive cap-dependent translation may shift towards a T-ALL-promoting translation program (Figure 2).12 It is not clear at this point if the defects in the ribosome are acting via a similar mechanism or one that is completely different. Whereas the RPL10/uL16 mutations impair ribosome biogenesis and translational fidelity,6,16 a thorough characterization of the effects of these ribosomal defects on the cellular translation profile in T-ALL has still not been made. Moreover, it remains to be established whether these defects are promoting leukemia by altering the translational profile of the ribosome (Figure 1) or by affecting translation-independent extra-ribosomal functions that have been assigned to these proteins.
Is the occurrence of defects in the ribosome and in the regulation of translation a specific characteristic of T-ALL? As far as we are aware, residue R98 in RPL10/uL16, a strong mutational hotspot in T-ALL, is not targeted by mutations in other cancer types, including other acute leukemias such as B-ALL or AML. At this point, we do not understand the reason for the unique occurrence of this mutation in T-ALL. In contrast, RPL22/eL22 and RPL5/uL18 were identified in the pan-cancer project as genes that are recurrently mutated in various cancer types. The dependence on cap-dependent translation is also not unique for T-ALL cells. Silvestrol and 4EGI-1 were shown to have therapeutic effects in xenograft models for various leukemias and solid tumors.17–19 It still needs to be determined, however, if this addiction to cap-dependent translation drives cell type specific translation programs in different tumor types.
In conclusion, in addition to the extensive list of defective processes and molecular aberrations already known in TALL, a central role of defective translation machinery and regulation was recently revealed in this disease. These findings offer novel opportunities for T-ALL therapy. Current treatment regimens consist of intensive schemes of chemotherapy and are associated with a multitude of long-term side-effects, while failing to induce long-term remission in 25% of pediatric and 50% of adult patients. For patients with one or several lesions driving cap-dependent translation, such as PTEN, NOTCH1 or FBXW7 mutations, drugs like 4EGI-1 or silvestrol could be considered. In contrast to what one might expect, off-target toxicity of these compounds seems limited. 4EGI-1 shows no toxicity on human CD34+ cells,20 and numerous studies showing therapeutic effects of these drugs in in vivo application in mouse tumor xenografts support the concept that there is a therapeutic window for treatment in humans.
Footnotes
Financial and other disclosures provided by the author using the ICMJE (www.icmje.org) Uniform Format for Disclosure of Competing Interests are available with the full text of this paper at www.haematologica.org.
References
- 1.Stumpf CR, Ruggero D. The cancerous translation apparatus. Curr Opinin Genet Dev. 2011;21(4):474–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Narla A, Ebert BL. Ribosomopathies: human disorders of ribosome dysfunction. Blood. 2010;115(16):3196–3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Van Vlierberghe P, Ferrando A. The molecular basis of T cell acute lymphoblastic leukemia. J Clin Invest. 2012;122(10):3398–3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Degryse S, de Bock CE, Cox L, et al. JAK3 mutants transform hematopoietic cells through JAK1 activation, causing T-cell acute lymphoblastic leukemia in a mouse model. Blood. 2014;124(20):3092–3100. [DOI] [PubMed] [Google Scholar]
- 5.Peirs S, Van der Meulen J, Van de Walle I, et al. Epigenetics in T-cell acute lymphoblastic leukemia. Immunol Rev. 2015;263(1):50–67. [DOI] [PubMed] [Google Scholar]
- 6.De Keersmaecker K, Atak ZK, Li N, Vicente C, et al. Exome sequencing identifies mutation in CNOT3 and ribosomal genes RPL5 and RPL10 in T-cell acute lymphoblastic leukemia. Nat Genet. 2013; 45(2):186–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rao S, Lee SY, Gutierrez A, et al. Inactivation of ribosomal protein L22 promotes transformation by induction of the stemness factor, Lin28B. Blood. 2012;120(18):3764–3773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tzoneva G, Perez-Garcia A, Carpenter Z, et al. Activating mutations in the NT5C2 nucleotidase gene drive chemotherapy resistance in relapsed ALL. Nat Med. 2013;19(3):368–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Van Vlierberghe P, Palomero T, Khiabanian H, et al. PHF6 mutations in T-cell acute lymphoblastic leukemia. Nat Genet. 2010;42(4):338–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Todd MAM, Picketts DJ. PHF6 interacts with the nucleosome remodeling and deacetylation (NuRD) complex. J Proteome Res. 2012;11(8): 4326–4337. [DOI] [PubMed] [Google Scholar]
- 11.Wang J, Leung JWC, Gong Z, Feng L, Shi X, Chen J. PHF6 Regulates Cell Cycle Progression by Suppressing Ribosomal RNA Synthesis. J Biol Chem. 2013;288(5):3174–3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wolfe AL, Singh K, Zhong Y, et al. RNA G-quadruplexes cause eIF4A-dependent oncogene translation in cancer. Nature. 2014;513(7516): 65–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schwarzer A, Holtmann H, Brugman M, et al. Hyperactivation of mTORC1 and mTORC2 by multipleoncogenic events causes addiction to eIF4E-dependent mRNAtranslation in T-cell leukemia. Oncogene. 2014. September 22 [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 14.Hales EC, Taub JW, Matherly LH. New insights into Notch1 regulation of the PI3K–AKT–mTOR1 signaling axis: Targeted therapy of γ-secretase inhibitor resistant T-cell acute lymphoblastic leukemia. Cell Sign. 2014;26(1):149–161. [DOI] [PubMed] [Google Scholar]
- 15.Mao H, Kim I-J, Climent J, Kang HC, Del Rosario R, Balmain A. FBXW7 Targets mTOR for Degradation and Cooperates with PTEN in Tumor Suppression. Science. 2008;321(5895):1496–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sulima SO, Patchett S, Advani VM, De Keersmaecker K, Johnson AW, Dinman JD. Bypass of the pre-60S ribosomal quality control as a pathway to oncogenesis. Proc Natl Acad Sci USA. 2014;111(15):5640–5645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lucas DM, Edwards RB, Lozanski G, et al. The novel plant-derived agent silvestrol has B-cell selective activity in chronic lymphocytic leukemia and acute lymphoblastic leukemia in vitro and in vivo. Blood. 2009;113(19):4656–4666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kogure T, Kinghorn AD, Yan I, et al. Therapeutic potential of the translation inhibitor silvestrol in hepatocellular cancer. PLoS ONE. 2013;8(9):e76136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen L, Aktas BH, Wang Y, et al. Tumor suppression by small molecule inhibitors of translation initiation. Oncotarget. 2012;3(8):869–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tamburini J, Green AS, Bardet V, et al. Protein synthesis is resistant to rapamycin and constitutes a promising therapeutic target in acute myeloid leukemia. Blood. 2009;114(8):1618–1627. [DOI] [PubMed] [Google Scholar]


