Abstract
We studied 1696 patients (18 to 61 years) with acute myeloid leukemia for ASXL1 mutations and identified these mutations in 103 (6.1%) patients. ASXL1 mutations were associated with older age (P<0.0001), male sex (P=0.041), secondary acute myeloid leukemia (P<0.0001), and lower values for bone marrow (P<0.0001) and circulating (P<0.0001) blasts. ASXL1 mutations occurred in all cytogenetic risk-groups; normal karyotype (40%), other intermediate-risk cytogenetics (26%), high-risk (24%) and low-risk (10%) cytogenetics. ASXL1 mutations were associated with RUNX1 (P<0.0001) and IDH2R140 mutations (P=0.007), whereas there was an inverse correlation with NPM1 (P<0.0001), FLT3-ITD (P=0.0002), and DNMT3A (P=0.02) mutations. Patients with ASXL1 mutations had a lower complete remission rate (56% versus 74%; P=0.0002), and both inferior event-free survival (at 5 years: 15.9% versus 29.0%; P=0.02) and overall survival (at 5 years: 30.3% versus 45.7%; P=0.0004) compared to patients with wildtype ASXL1. In multivariable analyses, ASXL1 and RUNX1 mutation as a single variable did not have a significant impact on prognosis. However, we observed a significant interaction (P=0.04) for these mutations, in that patients with the genotype ASXL1mutated/RUNX1mutated had a higher risk of death (hazard ratio 1.8) compared to patients without this genotype. ASXL1 mutation, particularly in the context of a coexisting RUNX1 mutation, constitutes a strong adverse prognostic factor in acute myeloid leukemia.
Introduction
The additional sex combs-like 1 (ASXL1) gene on chromosomal band 20q111 is one of three human homologs of the additional sex combs (Asx) gene of drosophila.2 Somatic ASXL1 mutations were found in a broad variety of myeloid malignancies including chronic myelomonocytic leukemia (CMML, 45%), myelodysplastic syndromes (MDS, 16%), primary myelofibrosis (35%), and acute myeloid leukemia (AML, secondary AML 30%; de novo AML 6.5%).3 Although the exact role of ASXL1 in normal hematopoiesis and the contribution of mutated ASXL1 to the development of hematopoietic malignancies have not been fully delineated yet, there are emerging data suggesting that ASXL1 is a tumor suppressor. An animal study has shown that Asxl1 deletion or haploinsuffiency constitutes a sufficient condition for the development of myeloid neoplasia reminiscent of MDS and MDS/myeloproliferative neoplasms.4 In another study the transplantation of bone marrow cells expressing oncogeneic NRASG12D together with knocked-down Asxl1 into lethally irradiated mice promoted myeloproliferation.5 The animals with additional loss of Asxl1 showed accelerated myeloproliferation and shorter survival than animals expressing Asxl1.5 In fact, in vitro, ASXL1 mutations cause a loss of Polycomb-repressive complex 2 (PRC2)-mediated repression of leukemogeneic target genes including those from the posterior HOXA cluster.5
The first studies on ASXL1 mutations in adult AML are remarkably consistent with respect to the increasing incidence of these mutations with age and their association with distinct clinical and genetic features.6–9 In addition, ASXL1 mutations in AML appear to have an adverse impact on induction success and long-term outcome.6–10 However, there are only limited data on the prognostic relevance of ASXL1 mutations in younger patients with AML.10,11
In our study, representing the largest AML cohort (n=1696) studied for ASXL1 mutations, we focused on younger patients with AML (≤61 years) and assessed the incidence and clinical impact of these mutations in the context of other clinical and genetic factors in a well-defined population of patients intensively treated in trials of the German-Austrian AML Study Group (AMLSG).
Methods
Patients
A total of 1696 younger AML patients (18 to 61 years) were studied. Patients were enrolled in prospective treatment protocols of the AMLSG), namely AML HD98A12 (n=733; NCT00146120), AMLSG 07–0413 (n=893; NCT00151242), and APL HD9514 (n=70) for the patients with acute promyelocytic leukemia (APL). The clinical studies were approved by the local ethics review committees and all patients gave informed consent for both treatment and cryopreservation of leukemia samples according to the Declaration of Helsinki. The only criterion to include patients in our study was the availability of a pretreatment bone marrow or peripheral blood specimen for analysis of ASXL1 mutations. Cytogenetic and additional molecular analyses were performed as previously described.15–19
ASXL1 mutation analysis
A detailed description of the ASXL1 mutation analysis is provided in the Online Supplementary Material. Briefly, genomic DNA was used as a template for polymerase chain reactions to amplify several fluorescently-labeled DNA fragments covering the entire exon 12 (AMLSG 07–04) or regions within exon 12 (AML HD98A and APL HD95) identified as main mutation clusters in AML.6,20 Amplicons were screened for mutations by a GeneScan-based fragment analysis (Online Supplementary Figures S1 and S2). Samples classified as mutated after the GeneScan analysis (Online Supplementary Figure S2) were further analyzed by direct sequencing to validate the mutation and to determine the mutation type.
Statistical analysis
Statistical analyses of clinical outcome were performed according to previous reports.16 The median follow-up for survival was calculated according to the method of Korn.21 The definition of complete remission (CR), event-free survival (EFS), relapse-free survival (RFS), and overall survival (OS) as well as cytogenetic categorization into favorable-, intermediate-, and adverse-risk groups followed recommended criteria.22 Pairwise comparisons between patients’ characteristics (covariates) were performed using the Mann-Whitney test for continuous variables and the Fisher exact test for categorical variables. The Kaplan-Meier method was used to estimate the distribution of EFS, RFS and OS.23 Estimation of confidence intervals for the survival curves was based on the Greenwood formula for standard error estimation. A logistic regression model was used to analyze associations between baseline characteristics and the achievement of CR. A Cox model was used to identify prognostic variables.24 In addition to ASXL1 mutation status, age, sex, hemoglobin level, logarithm of white blood cell count, type of AML (de novo, secondary AML, therapy-related AML), percentage of peripheral blood and bone marrow blasts, cytogenetic risk group,22 and mutational status of NPM1, FLT3 (ITD and TKD), CEBPA (CEBPA double-mutated, CEBPAdm), IDH1, IDH2 (IDH2R140, IDH2R172), RUNX1, MLL (PTD), and DNMT3A were included as explanatory variables in the regression analyses, as indicated, without further selection (full models). The full model presentation was chosen to allow estimation of the relative impact of the new marker (ASXL1 mutational status) in the concert of the already known prognostic and predictive markers. For multivariable analyses a missing value imputation technique was used as recommended for the situation termed missing at random.25 We estimated missing data for covariates by using 50 multiple imputations in chained equations that incorporated predictive mean matching.26 All statistical analyses were performed with the statistical software environment R version 2.14.0, using the R packages rms version 3.3–1, survival version 2.36–8, and cmprsk version 2.2–2.33.
Results
Demographics, clinical baseline characteristics and outcomes of the entire study population
The median age at diagnosis in the entire study cohort (n=1696) was 48.3 years (range, 18–61 years). The patients’ baseline characteristics are summarized in Online Supplementary Table S2. The CR rate was 73.1% (1230 of 1683 patients). With a median follow-up for survival of 5.6 years (95%-confidence interval, 5.4 to 5.9 years), the estimated 5-year rates for EFS, RFS and OS in the entire cohort were 28.2%, 42.6% and 40.2%, respectively (Online Supplementary Table S2). In total, 422 patients underwent allogeneic hematopoietic stem cell transplantation in first CR.
Frequency and types of ASXL1 mutations
ASXL1 mutations were detected in 103 (6.1%) of the 1696 patients. The types of mutation at the DNA and protein levels are described in Online Supplementary Table S3. The most common mutation, found in 60% (62/103) of the mutated cases, was a duplication of guanine at cDNA position 1934 (c.1934dupG). Other ASXL1 mutations found in more than one patient were c.1900_1922del (n=17) and c.1934delG (n=6). All frame shift mutations resulted in premature stop codons with consecutive loss of the c-terminal plant-homeo-domain. The wild-type allele was retained in all mutated samples. Of note, the majority (>90%) of the mutations clustered within or around a glycine-rich domain located between amino acids 642 and 685.27
Clinical and genetic characteristics of acute myeloid leukemia with ASXL1 mutations
Patients with ASXL1 mutations were older (P<0.0001), more frequently males (P=0.04), and more often had a history of MDS (P<0.0001) (Table 1). ASXL1 mutations were associated with lower values for white blood cell count (P=0.002), lactate dehydrogenase in serum (P=0.0008), proportion of bone marrow (P<0.0001) and circulating (P<0.0001) blasts (Table 1).
Table 1.
Clinical and cytogenetic characteristics according to the ASXL1 mutation status.
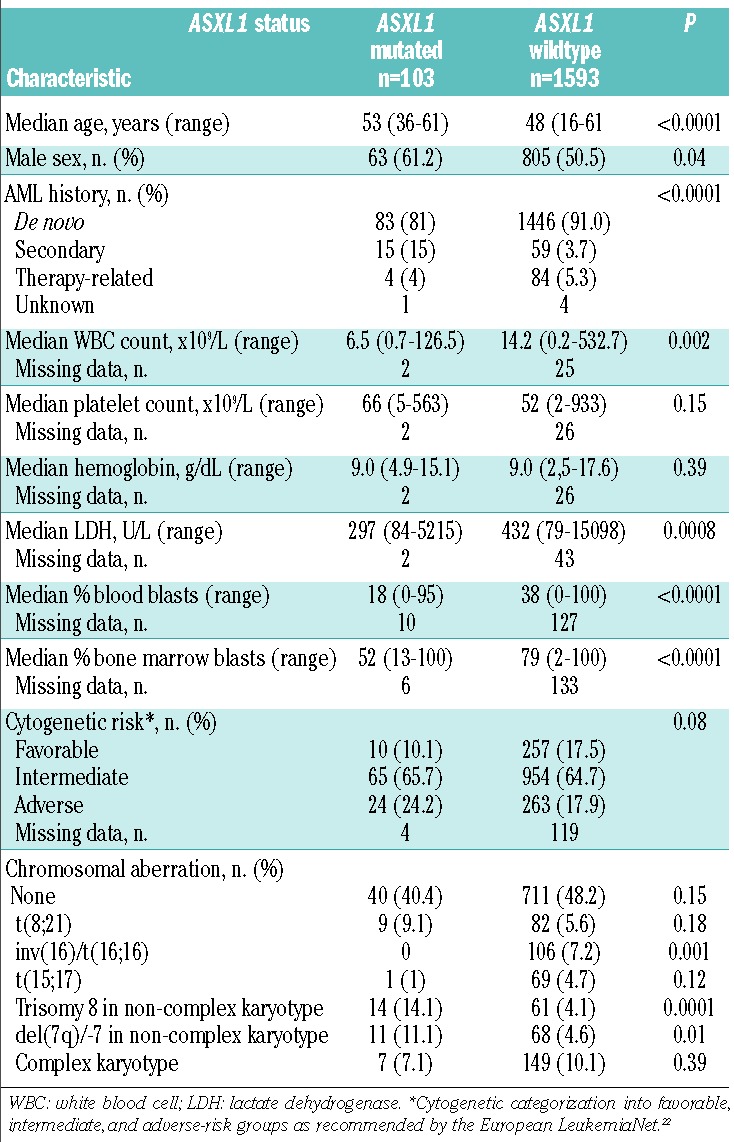
Among the ASXL1 mutated cases with informative cytogenetics (n=99), 40 (40.4%) patients had a normal karyotype; ASXL1 mutations tended (P=0.08) to be more frequent in AML with adverse-risk cytogenetics and less frequent in AML with favorable-risk cytogenetics compared to wild-type ASXL1 (Table 1); in core-binding factor-AML, ASXL1 mutations were only found in AML with t(8;21)(q22;q22) (9/91; 9.9%); none of the 106 cases with inv(16)(p13.1q22) had an ASXL1 mutation. ASXL1 mutations were further associated with trisomy 8 (P=0.0001), and with del(7q)/-7 (P=0.01) within a non-complex karyotype and in the absence of balanced translocations.
ASXL1 mutations showed several associations with other gene mutations (Table 2). RUNX1 (P<0.0001) and IDH2R140 mutations (P=0.007) co-occurred more frequently with ASXL1 mutations than with wild-type ASXL1. Conversely, ASXL1 mutations were only rarely detected in patients with NPM1 (P<0.0001), FLT3-ITD (P=0.0002), and DNMT3A (P=0.02) mutations. Of note, 31 of 103 (30%) patients with ASXL1 mutations had a genetic abnormality involving the RUNX1 gene on chromosome 21q22, i.e., these patients had either a somatic RUNX1 mutation (n=22) or t(8;21)(q22;q22); RUNX1-RUNX1T1 fusion (n=9).
Table 2.
Co-occurence of gene mutations in ASXL1 mutated cases.
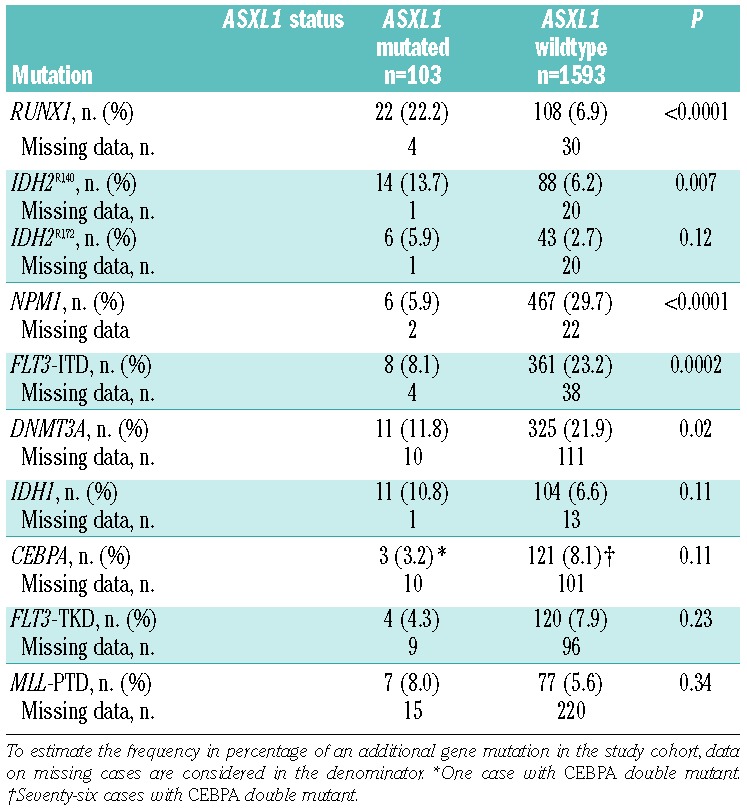
Twenty-two patients had both ASXL1 and RUNX1 mutations; among these double-mutated cases 15 presented with de novo AML, five with secondary AML, and one with therapy-related AML. The distribution of cytogenetically defined risk-groups among the ASXL1mutated/RUNX1mutated patients was as follows: 18 had intermediate-risk and three had high-risk cytogenetics; no information on cytogenetic risk group was available at diagnosis for one patient.
Response to induction therapy
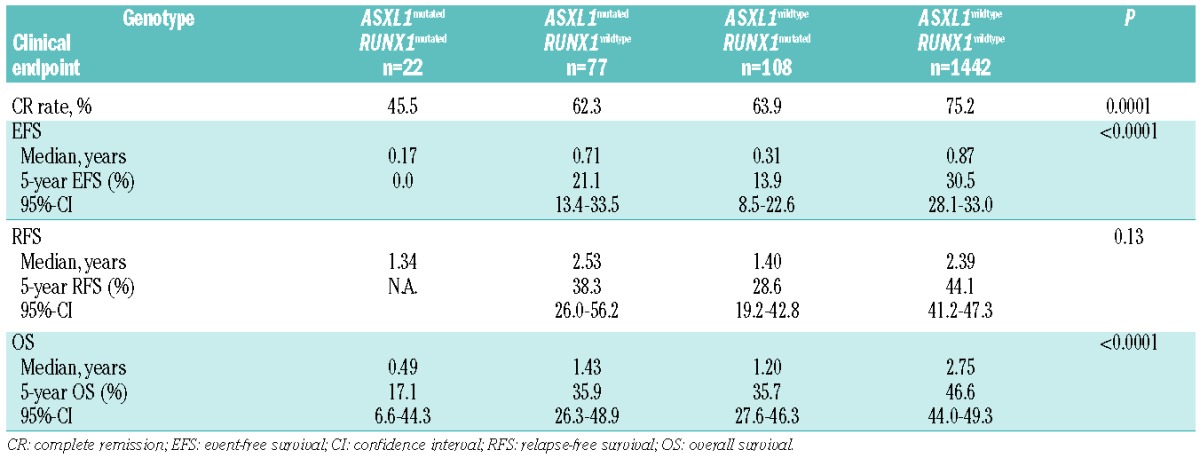
The CR rate was significantly lower in patients with ASXL1 mutations than in patients with wild-type ASXL1 (56.3% versus 74.2%; P=0.0002; Table 3); exclusion of APL patients did not change the unfavorable (P=0.0002) impact of ASXL1 mutations. The inferior CR rate in ASXL1 mutated cases was due to both a higher rate of resistant disease (29.1% versus 18.2%; P=0.03) and a higher rate of early deaths (14.6% versus 7.7%; P=0.02). Results were similar in the subset of cytogenetically normal AML (data not shown). Among the 91 AML patients with t(8;21), the CR rate did not differ between patients with (n=9) and without (n=82) ASXL1 mutation (89% versus 94%; P=0.47). Because of the association of ASXL1 with RUNX1 mutations, we assessed response according to the various ASXL1/RUNX1 genotypes: patients with the genotype ASXL1mutated/RUNX1mutated had an inferior (P=0.0001) CR rate (45.5%) compared to patients with any other ASXL1/RUNX1 genotype (Table 4). Patients with the ASXL1wildtype/RUNX1wildtype genotype had the highest CR rate (75.2%); CR rates were comparable in patients with the genotype ASXL1mutated/RUNX1wildtype (62.3%) and ASXL1wildtype/RUNX1mutated (63.9%). In multivariable analysis ASXL1 mutations were not a significant factor for achievement of CR (Online Supplementary Table S4).
Table 3.
Univariable outcome analyses according to ASXL1 mutation status.
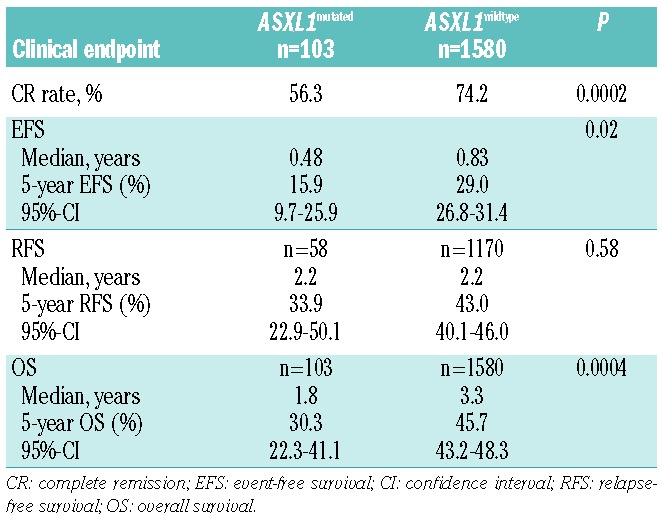
Table 4.
Univariable outcome analyses in the entire AML cohort according to combined ASXL1/RUNX1 genotype.
Survival analyses
In univariable analyses, ASXL1 mutations were associated with inferior EFS (P=0.02) and OS (P=0.0004), while there was no significant effect on RFS (P=0.58) (Table 3; Figure 1); the impact of ASXL1 mutations on outcome (EFS, P=0.028; RFS, P=0.69; OS, P=0.0006) was retained after exclusion of patients with APL. The estimated 5-year EFS and OS rates in patients with ASXL1 mutations were 15.9% and 30.3%, respectively, whereas in patients with wild-type ASXL1 they were 29.0 % and 45.7%, respectively (Table 3; Figure 1). The adverse effect of ASXL1 mutations on EFS and OS was also present in the subset of cytogenetically normal-AML (data not shown). In the subset of AML with t(8;21), none of the survival endpoints was impacted by the presence of ASXL1 mutations (EFS, P=0.77; RFS, P=0.74; OS, P=0.81). The one patient with t(15;17) and an ASXL1 mutation is still alive and in complete remission with a follow-up of 4 years. Among the 58 patients with ASXL1 mutations achieving a CR, 24 underwent allogeneic hematopoietic stem cell transplantation in first CR; Mantel-Byar analyses of allogeneic hematopoietic stem cell transplantation did not reveal a significant impact on either RFS (P=0.87) or OS (P=0.66).
Figure 1.
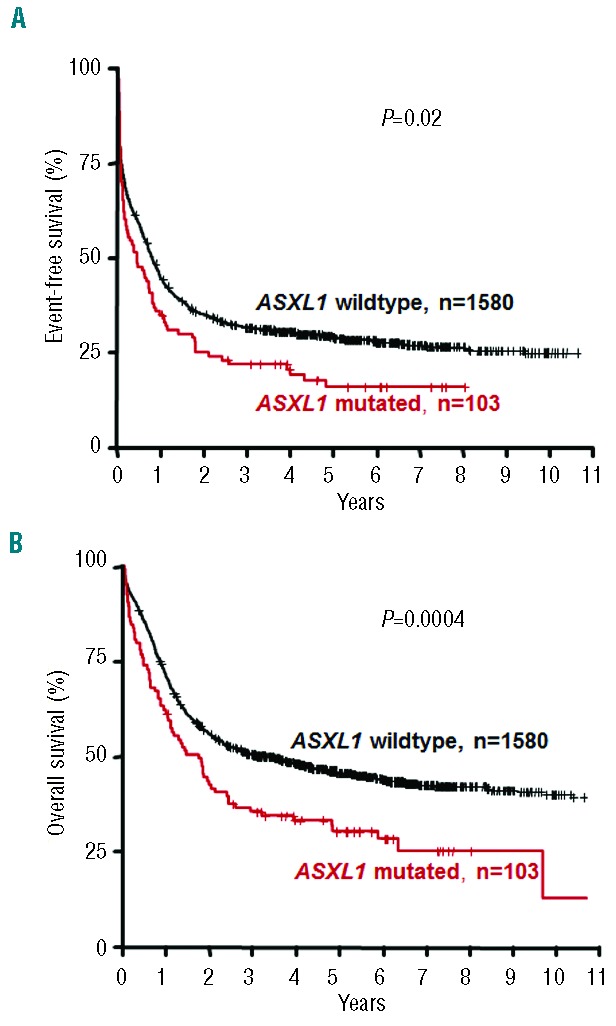
Impact of ASXL1 mutations on (A) event-free survival and (B) overall survival.
We also conducted an exploratory analysis of the composite ASXL1/RUNX1 genotypes. Patients with the genotype ASXL1mutated/RUNX1mutated had a significantly worse EFS (P<0.0001) compared to patients with any other ASXL1/RUNX1 genotype (Table 4, Figure 2). Of note, 73% of patients with the genotype ASXL1mutated/RUNX1mutated experienced an event within the first year. The dismal EFS in patients with the genotype ASXL1mutated/RUNX1mutated translated into a significantly inferior OS (P<0.0001) compared to that of patients with any other ASXL1/RUNX1 genotype (Table 4, Figure 2). Among the ten patients with the genotype ASXL1mutated/RUNX1mutated who achieved a CR, four underwent hematopoietic stem cell transplantation in first CR. The small number of patients in this molecular subgroup prevented a meaningful analysis of the impact of allogeneic stem cell transplantation in first CR.
Figure 2.
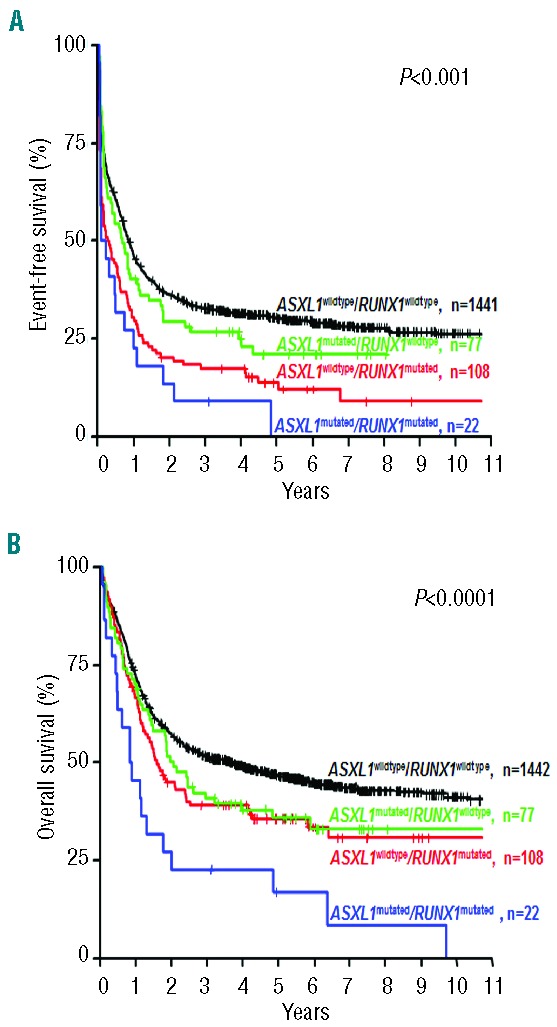
Impact of ASXL1/RUNX1 genotypes on (A) event-free survival and (B) overall survival.
Both ASXL1 and RUNX1 mutations as a single variable did not remain a significant factor for any clinical endpoint (EFS, RFS, and OS) after multivariable analyses (Online Supplementary Tables S5 and S6; Table 5). However, a significant interaction (P=0.04) for ASXL1 and RUNX1 mutations was observed in terms of OS (Table 5); the risk of death was almost twice as high in patients with the genotype ASXL1mutated/RUNX1mutated than in patients without this genotype.
Discussion
We here present the largest study assessing the incidence and clinical impact of ASXL1 mutations in younger (≤61 years) adults with newly diagnosed AML.
ASXL1 mutations were detected in 6.1% of the patients. In line with previous studies in AML,6–9,11,27,28 we found that c.1934dupG (p.G646WfsX12) was the most frequent ASXL1 mutation, accounting for 60% of the ASXL1 mutated cases. In a prior, large study of 501 patients with unselected AML, Chou et al. found ASXL1 mutations in 10.8% of the patients.6 The incidence of ASXL1 mutations in that study increased significantly with age; ASXL1 mutations occurred in 6.4% of patients <60 years, but in 18% of patients ≥60 years.6 In accordance, a Cancer and Leukemia Group B (CALGB) study on 423 adults with cytogenetically normal-AML reported a 5-fold higher frequency of ASXL1 mutations in patients ≥60 years than in those <60 years (16.2% versus 3.2%).7 A German study on 740 unselected AML cases with a median age of 67 years identified ASXL1 mutations in 17.2% of the cases,9 whereas a study from the Dutch-Belgian Hematology-Oncology Cooperative Group (HOVON) on 882 unselected AML patients with a median age <55 years found ASXL1 mutations in 5.3% of the cases.8 Finally, the most recent study from the UK Medical Research Council (MRC) on 367 adults with AML identified ASXL1 mutations in 9% of the cases.11 In line with other studies the frequency of ASXL1 mutations in this MRC study increased with age, and was twice as high in older AML patients (≥60 years; 12%, 18/148) than in younger ones (15–59 years; 6%, 14/219).11 Based on our data and the results from previous studies,6,8,9,11 ASXL1 mutations account for less than 10% of cases in younger adults with AML, whereas in older patients the incidence increases over 10%. Base exchanges resulting in stop codons (non-sense mutations) are not detected with our screening method. However, as already reported in previous studies, which used direct sequencing for mutational analysis of ASXL1, non-sense mutations appear to be infrequent molecular alterations, at least in the younger AML population; Metzeler et al.7 did not find any nonsense mutations in 189 younger (<60 years) AML patients with a normal karyotype, and Chou et al.6 detected only one (0.3%) non-sense mutation in 312 younger (<60 years) patients with unselected AML. In addition, the overall incidence of ASXL1 mutations in our study is very much in line with the data obtained for younger AML patients in other studies. Thus, we obviously did not miss a substantial proportion of ASXL1 mutations in younger AML patients by using our screening approach.
As in other studies, we found that ASXL1 mutations were more frequent in males than in females,6,7,9 and we also observed an association of ASXL1 mutations with both lower white blood cell count7–9 and blast count,7 as well as with a prior phase of MDS.9,28 This is in line with the finding that ASXL1 mutations are frequently detected already at the stage of a MDS or CMML.20,29,30 In a recent study of 48 AML patients with myelodysplasia-related changes ASXL1 mutations were detected in one third of the cases.31
In our study ASXL1 mutations were associated with distinct genetic characteristics. More than 60% of the ASXL1 mutated cases belonged to the cytogenetically intermediate-risk group. Among patients with core-binding factor-AML ASXL1 mutations were almost exclusively detected in AML with t(8;21), a finding which is concordant with that of other studies.6,8 In line with previous reports, we found that ASXL1 mutations were frequently associated with RUNX116,9,32 and IDH29,11 mutations, whereas they were only rarely detected in patients with FLT3-ITD6–9 or NPM1 mutations.6–9,11,28 In contrast to two other studies,7,9 we and others6,11 did not observe any significant association of ASXL1 mutations with CEBPA or FLT3-TKD mutations. However, the association of ASXL1 mutations with CEBPA mutations was only reported in older patients with cytogenetically normal AML7 and for FLT3-TKD mutations in an AML cohort with a relatively high median age of 67 years.9 Thus, the selection of the AML cases and age difference might in part contribute to the differences among the results of the studies. In our study, 31 (30%) of the 103 ASXL1 mutated cases had a concurrent genetic abnormality involving the RUNX1 gene, i.e., a somatic RUNX1 mutation or t(8;21)(q22;q22); RUNX1-RUNX1T1. Similar data can be obtained from the publication by Chou et al. comprising 54 ASXL1 mutated cases.6
The presence of ASXL1 mutations in patients with a clinically preleukemic condition such as MDS or CMML and the association of ASXL1 mutations with secondary AML suggest that these mutations represent a relatively early event in leukemogenesis. One recent study backtracked ASXL1 mutations in four patients with secondary AML and demonstrated that these mutations were already present in all patients at the stage of the preceding MDS.9 Other studies provide additional evidence that ASXL1 mutations favor the development of AML from MDS or CMML. Thol et al. reported that the presence of ASXL1 frameshift mutations in patients with lower-risk MDS, i.e., those with an International Prognostic Scoring System classification of low or intermediate-1, was associated with a shorter time to AML progression,33 and a more recent study by Papaemmanuil et al. of 595 MDS cases found an inferior leukemia-free survival in patients with ASXL1 mutations (n=81).30 A study by Gelsi-Boyer et al. of 51 CMML patients identified ASXL1 mutations as an unfavorable factor for the progression to AML.34 Of note, in that study only CMML patients with an ASXL1 mutation (11/25; 44%) did progress to AML, whereas none of the patients with wild-type ASXL1 (n=26) developed AML.34
Chou et al. found in a group of 360 unselected AML cases that ASXL1 mutations had an unfavorable impact on CR and OS in univariable, but not in multivariable analyses.6 In a subsequent CALGB study in older patients with cytogenetically normal AML, ASXL1 mutations were associated with inferior CR, EFS, DFS, and OS in univariable analyses,7 but on multivariable analyses ASXL1 mutations were revealed as a relevant factor for CR, EFS, DFS, and OS only in cytogenetically normal AML classified as favorable-risk (n=220) according to the European LeukemiaNet (ELN) recommendations.7 A HOVON study of 807 AML patients <65 years showed a lower CR rate and shorter OS in patients with ASXL1 mutations than in those with wild-type ASXL1; multivariable analysis in this study identified ASXL1 mutation as a relevant factor for shorter survival.8 Schnittger et al. evaluated 481 unselected AML patients, 50% of whom were ≥60 years, and found that ASXL1 mutations were a relevant factor for a shorter survival.9 In a study by the Eastern Cooperative Oncology Group (ECOG) including younger (<60 years) AML patients (n=398), ASXL1 mutations also conferred inferior survival.10 In a recent MRC study including 367 AML cases, ASXL1 mutations were found to be an unfavorable factor with regards to the cumulative incidence of relapse and overall survival in univariable, but not in multivariable analyses.11 However, the high proportion of older patients in this study [148/367 (40%) ≥60 years] might have been a confounding factor overriding the impact of ASXL1 mutations in multivariable analyses.11 In our study, which focuses on younger patients with AML, the presence of ASXL1 mutations was associated with inferior CR, EFS, and OS. We describe for the first time an additive, prognostically unfavorable effect for the combined genotype ASXL1mutated/RUNX1mutated. While multivariable analysis did not identify ASXL1 as well as RUNX1 mutation as a single marker to be a relevant factor for long-term outcome, a significant interaction was found for these two mutations, in that patients with the genotype ASXL1mutated/RUNX1mutated had almost twice the risk of death compared to patients lacking this double mutated ASXL1/RUNX1 genotype.
We report here the largest study in AML assessing the incidence and prognostic relevance of ASXL1 mutations in younger patients with AML. We confirm that ASXL1 mutations constitute a recurrent molecular alteration in AML and are associated with distinct clinical and genetic features and, most importantly, with an adverse prognosis. We extend the results of previous studies by the finding that the combined genotype ASXL1mutated/RUNX1mutated, which can be detected in up to 30% of ASXL1 mutated AML,6 is associated with a particularly dismal rate of CR achievement and long-term outcome. Thus, allogeneic hematopoietic stem cell transplantation might be evaluated prospectively as one therapeutic option in younger AML patients carrying the genotype ASXL1mutated/RUNX1mutated. In addition, since ASXL1 mutations lead to a loss of H3K27me3, in particular at the HOXA locus, pharmacological inhibition of H3K27 demethylation35 might be explored as a potential therapeutic approach in ASXL1 mutated AML.
Acknowledgments
The authors thank the AMLSG institutions and investigators who contributed to this study. They also thank Marianne Habdank for technical support with molecular analyses. This work was supported by the Deutsche Krebshilfe (grant 109675).
Footnotes
The online version of this article has a Supplementary Appendix
Authorship and Disclosures
Information on authorship, contributions, and financial & other disclosures was provided by the authors and is available with the online version of this article at www.haematologica.org.
References
- 1.Fisher CL, Berger J, Randazzo F, Brock HW. A human homolog of Additional sex combs, ADDITIONAL SEX COMBS-LIKE 1, maps to chromosome 20q11. Gene. 2003;306:115–126. [DOI] [PubMed] [Google Scholar]
- 2.Abdel-Wahab O, Dey A. The ASXL-BAP1 axis: new factors in myelopoiesis, cancer and epigenetics. Leukemia. 2013;27(1):10–15. [DOI] [PubMed] [Google Scholar]
- 3.Gelsi-Boyer V, Brecqueville M, Devillier R, Murati A, Mozziconacci MJ, Birnbaum D. Mutations in ASXL1 are associated with poor prognosis across the spectrum of malignant myeloid diseases. J Hematol Oncol. 2012;5:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang J, Li Z, He Y, et al. Loss of Asxl1 leads to myelodysplastic syndrome-like disease in mice. Blood. 2014;123(4):541–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abdel-Wahab O, Adli M, LaFave LM, et al. ASXL1 mutations promote myeloid transformation through loss of PRC2-mediated gene repression. Cancer Cell. 2012;22(2): 180–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chou WC, Huang HH, Hou HA, et al. Distinct clinical and biological features of de novo acute myeloid leukemia with additional sex comb-like 1 (ASXL1) mutations. Blood. 2010;116(20):4086–4094. [DOI] [PubMed] [Google Scholar]
- 7.Metzeler KH, Becker H, Maharry K, et al. ASXL1 mutations identify a high-risk subgroup of older patients with primary cytogenetically normal AML within the ELN favorable genetic category. Blood. 2011; 118(26):6920–6929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pratcorona M, Abbas S, Sanders MA, et al. Acquired mutations in ASXL1 in acute myeloid leukemia: prevalence and prognostic value. Haematologica. 2012;97(3):388–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schnittger S, Eder C, Jeromin S, et al. ASXL1 exon 12 mutations are frequent in AML with intermediate risk karyotype and are independently associated with an adverse outcome. Leukemia. 2013;27(1):82–91. [DOI] [PubMed] [Google Scholar]
- 10.Patel JP, Gonen M, Figueroa ME, et al. Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. N Engl J Med. 2012;366(12):1079–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.El-Sharkawi D, Ali A, Evans CM, et al. ASXL1 mutations are infrequent in young patients with primary acute myeloid leukemia and their detection has a limited role in therapeutic risk stratification. Leuk Lymphoma. 2014;55(6):1326–1331. [DOI] [PubMed] [Google Scholar]
- 12.Schlenk RF, Döhner K, Mack S, et al. Prospective evaluation of allogeneic hematopoietic stem-cell transplantation from matched related and matched unrelated donors in younger adults with high-risk acute myeloid leukemia: German-Austrian trial AMLHD98A. J Clin Oncol. 2010; 28(30):4642–4648. [DOI] [PubMed] [Google Scholar]
- 13.Schlenk RF, Döhner K, Krauter J, et al. All-trans retinoic acid improves outcome in younger adult patients with nucleophosmin-1 mutated acute myeloid leukemia – results of the AMLSG 07–04 randomized treatment trial. Blood. 2011;118(21):38–39 (abstract #80). [Google Scholar]
- 14.Schlenk RF, Germing U, Hartmann F, et al. High-dose cytarabine and mitoxantrone in consolidation therapy for acute promyelocytic leukemia. Leukemia. 2005;19(6):978–983. [DOI] [PubMed] [Google Scholar]
- 15.Gaidzik VI, Bullinger L, Schlenk RF, et al. RUNX1 mutations in acute myeloid leukemia: results from a comprehensive genetic and clinical analysis from the AML study group. J Clin Oncol. 2011;29(10): 1364–1372. [DOI] [PubMed] [Google Scholar]
- 16.Gaidzik VI, Schlenk RF, Paschka P, et al. Clinical impact of DNMT3A mutations in younger adult patients with acute myeloid leukemia: results of the AML Study Group (AMLSG). Blood. 2013;121(23):4769–4777. [DOI] [PubMed] [Google Scholar]
- 17.Schlenk RF, Döhner K, Krauter J, et al. Mutations and treatment outcome in cytogenetically normal acute myeloid leukemia. N Engl J Med. 2008;358(18):1909–1918. [DOI] [PubMed] [Google Scholar]
- 18.Paschka P, Schlenk RF, Gaidzik VI, et al. IDH1 and IDH2 mutations are frequent genetic alterations in acute myeloid leukemia and confer adverse prognosis in cytogenetically normal acute myeloid leukemia with NPM1 mutation without FLT3 internal tandem duplication. J Clin Oncol. 2010;28(22):3636–3643. [DOI] [PubMed] [Google Scholar]
- 19.Schlenk RF, Taskesen E, van Norden Y, et al. The value of allogeneic and autologous hematopoietic stem cell transplantation in prognostically favorable acute myeloid leukemia with double mutant CEBPA. Blood. 2013;122(9):1576–1582. [DOI] [PubMed] [Google Scholar]
- 20.Gelsi-Boyer V, Trouplin V, Adelaide J, et al. Mutations of polycomb-associated gene ASXL1 in myelodysplastic syndromes and chronic myelomonocytic leukaemia. Br J Haematol. 2009;145(6):788–800. [DOI] [PubMed] [Google Scholar]
- 21.Korn EL. Censoring distributions as a measure of follow-up in survival analysis. Stat Med. 1986;5(3):255–260. [DOI] [PubMed] [Google Scholar]
- 22.Döhner H, Estey EH, Amadori S, et al. Diagnosis and management of acute myeloid leukemia in adults: recommendations from an international expert panel, on behalf of the European LeukemiaNet. Blood. 2010;115(3):453–474. [DOI] [PubMed] [Google Scholar]
- 23.Kaplan E, Meier P. Nonparametric estimation from incomplete observations. J Am Stat Assoc. 1958;53:457–481. [Google Scholar]
- 24.Therneau TM, Grambusch PM. Modeling survival data: extending the Cox model. New York, NY: Springer Verlag, 2000. [Google Scholar]
- 25.Little RJ, D’Agostino R, Cohen ML, et al. The prevention and treatment of missing data in clinical trials. N Engl J Med. 2012;367(14):1355–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harrell FE. Regression modeling strategies: with applications to linear models, logistic regression, and survival analysis. New York: Springer Verlag, 2001. [Google Scholar]
- 27.Boultwood J, Perry J, Pellagatti A, et al. Frequent mutation of the polycomb-associated gene ASXL1 in the myelodysplastic syndromes and in acute myeloid leukemia. Leukemia. 2010;24(5):1062–1065. [DOI] [PubMed] [Google Scholar]
- 28.Carbuccia N, Trouplin V, Gelsi-Boyer V, et al. Mutual exclusion of ASXL1 and NPM1 mutations in a series of acute myeloid leukemias. Leukemia. 2010;24(2):469–473. [DOI] [PubMed] [Google Scholar]
- 29.Bejar R, Stevenson K, Abdel-Wahab O, et al. Clinical effect of point mutations in myelodysplastic syndromes. N Engl J Med. 2011;364(26):2496–2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Papaemmanuil E, Gerstung M, Malcovati L, et al. Clinical and biological implications of driver mutations in myelodysplastic syndromes. Blood. 2013;122(22):3616–3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Devillier R, Gelsi-Boyer V, Brecqueville M, et al. Acute myeloid leukemia with changes are characterized by a specific molecular pattern with high frequency of ASXL1 mutations. Am J Hematol. 2012;87(7):659–662. [DOI] [PubMed] [Google Scholar]
- 32.Mendler JH, Maharry K, Radmacher MD, et al. RUNX1 mutations are associated with poor outcome in younger and older patients with cytogenetically normal acute myeloid leukemia and with distinct gene and microRNA expression signatures. J Clin Oncol. 2012;30(25):3109–3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thol F, Friesen I, Damm F, et al. Prognostic significance of ASXL1 mutations in patients with myelodysplastic syndromes. J Clin Oncol. 2011;29(18):2499–2506. [DOI] [PubMed] [Google Scholar]
- 34.Gelsi-Boyer V, Trouplin V, Roquain J, et al. ASXL1 mutation is associated with poor prognosis and acute transformation in chronic myelomonocytic leukaemia. Br J Haematol. 2010;151(4):365–375. [DOI] [PubMed] [Google Scholar]
- 35.Kruidenier L, Chung CW, Cheng Z, et al. A selective jumonji H3K27 demethylase inhibitor modulates the proinflammatory macrophage response. Nature. 2012;488(7411):404–408. [DOI] [PMC free article] [PubMed] [Google Scholar]


