In a recent issue of Haematologica, Matsuo et al.1 pinpoint the pejorative effect of EVI1 overexpression in 18 acute myeloid leukemias (AML) with MLL rearrangements. However, EVI1 overexpression has also been reported in patients with translocations involving chromosome 3 and the EVI1 gene.2,3 Because of the poor prognosis associated to these anomalies, it is important to investigate them at an early stage in order to adapt patient management. Indeed, previous reports4–6 and the 2008 WHO classification7 indicate that EVI1-rearranged (EVI1-r) AML display typical features, such as absence of thrombopenia, atypical megakaryocytes and multilineage dysplasia2–4 which can be detected by current diagnostic reference methods. In this line, we compared a cohort of 17 EVI1-r AML, aged between 8 and 79-years old (median 54 years) to 1822 other cases of AML diagnosed in the same laboratory over 14 years. At diagnosis, there were similar hemoglobin levels or white blood cell counts in both groups. Median platelet counts were 123×109/L, higher than 100×109/L in 53% of EVI1-r AML patients, compared to 25% in the control AML population (P=0.02). These subnormal counts were associated with platelets dysplasia (giant and hypogranular) in 57%. Bone marrow (BM) megakaryocytes were present in all EVI1-r AML cases, while they were seen in only 54% of the control cohort (P<0.0001). In EVI1-r AML, megakaryocytes were small, with monolobated or bilobated nuclei and appeared in characteristic clusters. Multilineage dysplasia was present in 75% of the EVI1-r AML cases (vs. 17.6%; P<0.001). Myeloperoxidase (MPO) cytochemistry and flow cytometry was negative in 57% of EVI1-r AML patients (23%; P=0.008). Of note, 78.5% of EVI1 patients had less than 10% MPO positive blasts, and MPO was also poorly expressed by mature neutrophils. Classification indeed showed a significant increase of AML with minimal differentiation among EVI1-r AML (31% vs. 7.5%; P=0.002).
Karyotypic examination found classical features of EVI1-r AML. Nine patients had inv(3)(q21q26.2), cryptic in a normal karyotype at diagnosis and fully disclosed at relapse in one patient. Translocations were present in 7 other cases, with different partners [(t(3;3), n=4; t(3;12), n=1; t(3;21), n=1; t(2;3), n=1)]. Monosomy 7, another classical feature of EVI1-r AML, was observed in 8 cases and del(7q) in one case.
Ten EVI1-r AML patients had de novo AML. Antecedents of myeloproliferative neoplasm (chronic myeloid leukemia n=2, essential thrombocytosis n=1, myelomonocytic leukemia n=1) were retrieved in 4, of myelodysplastic syndrome in one and of lymphoproliferative disorder in 2 (1 diffuse large B-cell lymphoma and 1 Waldenström disease). This incidence of 41% of secondary AML was significantly higher than in the reference cohort (19%; P=0.035).
Interestingly, 5 patients with secondary AML had very similar cytomorphological characteristics, yet did not carry EVI1 rearrangement. Cytogenetics showed for all a t(1;3)(p32q21), involving PRDM16. As for EVI1 patients, platelet counts were normal at diagnosis (mean 259×109/L). BM smears were characteristically rich in small, monolobated and clustered megakaryocytes (more than 50/smear). All showed multilineage dysplasia and, as for EVI1 patients, MPO was characteristically low and completely absent in 3 cases. Prognosis was dismal for both EVI1 and PRDM16 AML, with 9 months overall survival. The 14 patients who could not receive allogeneic transplantation died within 12 months.
This study consolidates the unusual base-line characteristics and clinical features of EVI1-r AML cases. Moreover, it indicates a very low rate of MPO expression in EVI1-r AML patients. It is interesting to note that relationships have been reported between EVI1 expression and MPO regulation,8,9 suggesting that the translocation could interfere with MPO production in EVI1-r AML. Moreover, a mouse model has shown a relationship between EVI1 and thrombopoiesis,10 indicating that the peculiar features of EVI1-r AML could be directly related to the abnormal expression of this gene. This report also adds the novel information that similar hematologic and morphological features can be associated to PRDM16 rearrangement, a gene closely related to EVI,11 likely to impact the same pathways. This would notably be the case in rearrangements where the RPN1 gene is translocated to either EVI1 or PRDM16.
EVI1-r AML have recently been reported to carry molecular anomalies providing them with a specific signature.12 It would be interesting to investigate whether those are also found in PRDM16-r AML.
In conclusion, these two rare but very similar entities, identifiable during the first steps of AML diagnosis, should prompt the investigation of EVI1 rearrangement, followed by that of PRDM16 if EVI1 is normal. A proposed algorithm (Figure 1) could include the association of absence of thrombocytopenia, abnormal platelets on a blood smear, micromegakaryocytes in clusters, multilineage dysplasia and low MPO-expressing blasts together with the notion of a secondary AML. Based on these features, cytogeneticians should be made aware of a possible chromosome 3q anomaly. The latter, and especially inv(3) can be tricky to detect and, when uncertain, should be confirmed by FISH analysis. The poor prognosis associated with these rare diseases should lead instigate the rapid search for a donor, with a view to allogeneic transplantation of hematopoietic stem cells.
Figure 1.
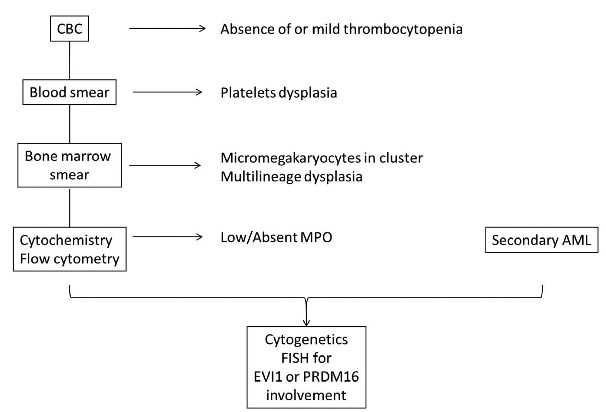
Algorithm for the suspicion of EVI1 and PRDM16 AMLs.
Footnotes
Information on authorship, contributions, and financial & other disclosures was provided by the authors and is available with the online version of this article at www.haematologica.org.
References
- 1.Matsuo H, Kajihara M, Tomizawa D, et al. EVI1 overexpression is a poor prognostic factor in pediatric patients with mixed lineage leukemia-AF9 rearranged acute myeloid leukemia. Haematologica. 2014;99(11):225–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yamazaki H, Suzuki M, Otsuki A, et al. A remote GATA2 hematopoietic enhancer drives leukemogenesis in inv(3)(q21;q26) by activating EVI1 expression. Cancer Cell. 2014;25(4):415–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gröschel S, Lugthart S, Schlenk RF, et al. High EVI1 expression predicts outcome in younger adult patients with acute myeloid leukemia and is associated with distinct cytogenetic abnormalities. J Clin Oncol. 2010;28(12):2101–2107. [DOI] [PubMed] [Google Scholar]
- 4.Grimwade D, Hills RK, Moorman A, et al. Refinement of cytogenetic classification in acute myeloid leukemia: determination of prognostic significance of rare recurring chromosomal abnormalities among 5876 younger adult patients treated in the United Kingdom Medical Research Council trials. Blood. 2010;116(3):354–365. [DOI] [PubMed] [Google Scholar]
- 5.Rogers HJ, Vardiman JW, Anastasi J, et al. Complex or monosomal karyotype and not blast percentage is associated with poor survival in acute myeloid leukemia and myelodysplastic syndrome patients with inv(3)(q21q26.2)/t(3;3)(q21;q26.2): a Bone Marrow Pathology Group study. Haematologica 2014;99(5):821–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lugthart S, Gröschel S, Beverloo H, et al. Clinical, molecular, and prognostic significance of WHO type inv(3)(q21q26.2)/t(3;3)(q21;q26.2) and various other 3q abnormalities in acute myeloid leukemia. J Clin Oncol. 2010;28(24):3890–3898. [DOI] [PubMed] [Google Scholar]
- 7.Swerdlow SH, Campo E, Harris NL, et al. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. Lyon, France: IARC Press; 2008. [Google Scholar]
- 8.Morishita K, Parganas E, Matsugi T, Ihle JN. Expression of the Evi-1 zinc finger gene in 32Dc13 myeloid cells blocks granulocytic differentiation in response to granulocyte colony-stimulating factor. Mol Cell Biol. 1992;12(1):183–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Laricchia-Robbio L, Premanand K, Rinaldi CR, Nucifora G. EVI1 impairs myelopoiesis by deregulation of PU.1 function. Cancer Res. 2009;69(4):1633–1642. [DOI] [PubMed] [Google Scholar]
- 10.Buonamici S, Chakraborty S, Senyuk V, Nucifora G. The role of EVI1 in normal and leukemic cells. Blood Cells Mol Dis. 2003;31(2):206–212. [DOI] [PubMed] [Google Scholar]
- 11.Mochizuki N, Shimizu S, Nagasawa T, et al. A novel gene, MEL1, mapped to 1p36.3 is highly homologous to the MDS1/EVI1 gene and is transcriptionally activated in t(1;3)(p36;q21)-positive leukemia cells. Blood. 2000;96(9):3209–3214. [PubMed] [Google Scholar]
- 12.Lavallée VP, Gendron P, Lemieux S, D’Angelo G, Hébert J, Sauvageau G. EVI1-rearranged acute myeloid leukemias are characterized by distinct molecular alterations. Blood. 2015;125(1):140–143. [DOI] [PMC free article] [PubMed] [Google Scholar]


