Abstract
Studies of patients with congenital immunodeficiency due to mutation of the NF-κB essential modulator (NEMO) gene, have demonstrated that NEMO integrity is required for NK cell cytotoxicity. Thus, we have studied the physiology of NF-κB activation in NK cells during the cytolytic program. In resting ex vivo human NK cells or cell lines, IκB was degraded after 10 min exposure to PMA and ionomycin, or TNF and was maximally degraded by 30 min. Ligation of several NK cell activation receptors including NKp30 induced a similar response and was blocked by pretreatment with the proteosome inhibitor MG132. There was no short-term effect on p100 processing, the signature of non-canonical NF-κB activation. NK cell IκB degradation corresponded to increases in nuclear NF-κB as detected by EMSA. Supershift of stimulated NK cells and fluorescence microscopy of individual NK cells in cytolytic conjugates demonstrated that the p65/p50 heterodimer was the primary NF-κB utilized. NF-κB function was evaluated in NK92 cells transduced with a κB GFP reporter, and their conjugation with K562 cells or ligation of NKp30 ligation resulted in rapid GFP accumulation. The latter was prevented by the Syk inhibitor piceatannol. Thus, NK cell activation signaling specifically induces transcriptional activation and synthesis of new NF-κB dependent proteins during the initiation of cytotoxicity.
Keywords: NK cells, NF-κB, NEMO
Introduction
NK cells are lymphocytes of the innate immune system that are critical in host defense and immune regulation. They are activated or inhibited through the ligation of germline-encoded receptors (1) and exert their biological activity through a triad of functions: cytotoxicity, production of cytokines and chemokines and co-stimulation to other cells of the immune system (reviewed in (2)). In humans NKp46, NKp44 and NKp30, collectively termed natural cytotoxicity receptors (NCR), are major triggering receptors expressed almost exclusively on NK cells (1). NKp30 is present on the majority of resting as well as activated NK cells and acts to induce signals through CD3ζ leading to NK-mediated cytotoxicity (3). The cytolytic function of NK cells is believed to be independent of nuclear processes involving a cascade of pre-existing cytoplasmic kinases and adaptor proteins to generate calcium flux and release of perforin containing cytolytic granules.
The nuclear factor-κB (NF-κB) family of transcription factors consists of Rel A (p65), Rel B, c-Rel, NF-κB1 (p50), and NF-κB2 (p52) and participates in the regulation of many facets of innate and adaptive immunity (4). Function of the NF-κB proteins are restrained by the inhibitor of NF-κB (IκB) family of proteins that prevent the NF-κB dimers from entering the nucleus. Release of NF-κB homo- or heterodimers depends upon the tightly regulated proteosomal degradation of IκB, which is initiated by phosphorylation of IκB by the IκB kinase (IKK) complex. The IKK can contain the kinase subunits, IKKα, and IKKβ, and a regulatory subunit, NEMO (NF-κB essential modulator) or IKKγ (5). NF-κB activation using the IKK occurs through either the canonical or non-canonical pathway. In the canonical pathway, IKKβ in association with NEMO phoshorylates IκB to release NF-κB dimers. In contrast, in the non-canonical pathway, activation signaling leads to the function of NF-κB-induced kinase (NIK), a mitogen-activated protein kinase kinase kinase (MAP3K)-like enzyme that specifically phosphorylates and induces IKKα which functions independently of NEMO (5). NIK and IKKα contribute to the phosphorylation of the c-terminus of p100, thereby stimulating proteolytic processing of p100 to p52, releasing an IκB-like component, and ultimately allowing p52-containing NF-κB complexes to translocate to the nucleus and bind DNA. The relative importance of the canonical and non-canonical pathways in a given cell type depends on the specific cell surface receptors engaged and the nature of the activating stimulus received.
Although great emphasis has been placed on the role of NF-κB in immunity, few have investigated NF-κB in NK cells. Initial studies have shown that pharmacological inhibitors of NF-κB reduce NK cell cytotoxicity (6). Subsequent in vitro studies revealed that stimulation with the soluble activators IL-2, IL-18, or TLR2 ligands induces NF-κB binding activity (7, 8, 9). In addition, RelB deficiency or overexpression of a dominant negative form of IκBα (a global inhibitor of NF-κB) in mice can impair NK cell activation and functions during infection with the intracellular parasite, Toxoplasma gondii (10, 11). Finally, NK cells obtained from patients with NEMO deficiency have defective cytotoxic function associated with impaired activation of NF-κB (12). While global inhibition of NF-κB decreases NK cell activity, little is known about the specific paradigm of NF-κB activation in NK cells and the role that individual NF-κB family members may play in NK cells. The availability of different NF-κB hetero- and homodimers, whose synthesis and activation may provide important levels of selectivity in NF-κB mediated gene transcription (13). Given this variability and NK cell function defects in NK cells with defective NEMO, we wanted to understand the basic physiology of NF-κB activation relative to the cytolytic process. Here, we characterize NF-κB activation in NK cells and demonstrate that the canonical pathway of NF-κB is induced after ligation of some activating receptors including NKp30, as well as after physiologic activation by susceptible target cells. We also found that NK cell activation signaling specifically induces robust synthesis of new NF-κB dependent protein during the initiation of cytotoxicity.
Materials and Methods
NK cell lines, NK cell preparation, and cellular evaluation
The immortalized NK cell lines YTS (a subclone of the line YT) and NK92 were originally derived from patients having malignant expansions of NK cells (14) and were maintained in cell culture. MHC class-I negative K562 erythroleukemia cells were used as a susceptible target cell where indicated. An NK92 NF-κB GFP reporter cell line was generated using HIV based lentivirus packaged with κB-GFP reporter construct (System Biosciences). Cells were infected according to manufacturer recommendations and GFP expressing transductants selected by FACS and grown as pure cultures. All NK92 cell lines were grown in Myelocult-5100 culture media (StemCell Technologies) containing 100u/ml hrIL-2. Ex vivo NK cells were prepared from leukocyte-enriched blood obtained from volunteer donors by negative depletion using the human NK cell isolation kit-II (Miltenyi Biotec) using Super MACS LS separation columns according to manufacturer recommendations, on the same day that blood was taken from the donor. Enriched cells were analyzed in a flow cytometer to determine their purity and routinely contained >95% CD56+/CD3− with < 1.0% CD3+ cells, and were always used in experiments immediately after enrichment. The use of these samples was approved by the Children’s Hospital of Philadelphia Internal Review Board for the Protection of Human Subjects. Where specified, cells were incubated with 10ng/ml TNF-α, or 100ng/ml PMA + 1μg/ml Ionomycin (PMA/I) or immobilized monoclonal antibodies directed against NK cell surface receptors at 37° C. For the antibody mediated stimulation either anti-NKp30 clone Z25 (Beckman Coulter), anti-CD11a clone HI111 (BD Biosciences), anti CD28 clone 9.3 (kind gift from Dr. Jim Riley- University of Pennsylvania School of Medicine), anti-CD56 clone B159 (BD Biosciences), were used in PBS at 5μg/ml to coat polystyrene wells overnight at 4°C. Wells were then rinsed and cells incubated for the specified time at 37°C followed immediately by the addition of lysis buffer directly to the well. A zero minute timepoint was achieved by adding cells directly to a chilled well.
IκBα Degradation
The ability of different cell activation stimuli to induce IκBα degradation was determined by western blot analysis. Briefly, 2 × 106 YTS, NK92 or 5 × 106 ex-vivo NK cells were treated and washed in PBS and then lysed in NuPAGE LDS sample buffer (Invitrogen) and boiled for 5 min before loading. 10μg of protein per sample was separated on 4–12% Bis-Tris density gradient gels in MOPS SDS running buffer and transferred to PVDF membranes (Invitrogen), which after blocking with 3% BSA and 0.1% Tween-20 were incubated with rabbit polyclonal anti-IκBα C-21 (Santa Cruz Biotechnology). Bound antibody was detected using HRP-conjugated donkey-anti-rabbit (Amersham Biosciences) and enhanced chemiluminescence detection system (Amersham Pharmacia). Where specified, membranes were stripped in 0.2M glycine pH 2.5, 0.05% Tween-20 and 140mM NaCl in TBS at 50°C for 30 min, blocked with 3% BSA, and reprobed with rabbit anti-β-actin pAb 20–33 (Sigma). Processing of p100 (~100KD) to p52 (~52KD) was detected with mouse mAb anti-NF-κB p52 C-5 (Santa Cruz) followed by HRP-conjugated goat anti-mouse (Jackson Immunoresearch). To inhibit IκB degradation, cells were pretreated with 5μg/ml of MG132 (Biomol International).
Electrophoretic Mobility Shift Assay
Nuclear extracts were prepared by washing cells twice with 1 ml of ice-cold PBS and resuspending in 400μl of ice-cold lysis buffer containing 1M HEPES, 0.5 M EDTA, 0.1M EGTA, 2M KCl, 0.1M DTT, cocktail of Protease inhibitors at 5μg/ml (Roche), and 10% NP-40 and incubated for 15 min on ice with occasional vortexing to obtain complete cell lysis and release of nuclei. Tubes were centrifuged at 13,400 x g for 1 min, supernatant (cytoplasmic extract) was removed completely and remaining nuclei were resuspended in 25 μl of ice-cold nuclear extraction buffer containing 1M HEPES, 5M NaCl, 0.5M EDTA 0.1M DTT and cocktail of Protease inhibitors at 5μg/ml (Roche), incubated for 30 min on ice and centrifuged at 13,400 x g for 5 min. Supernatant containing the soluble nuclear proteins was aliquoted in pre-chilled tubes, snap-frozen in liquid nitrogen and stored at −80°C until use. Equal protein amounts of the extracts (10μg) as determined using detergent compatible protein assay (Bio-Rad) were utilized in experiments. DNA probes were prepared by labeling double stranded oligo (3.5pM) NF-κB (5′-AGTTGAGGGGACTTTCCCGGC-3′), or OCT1 (5′-TGTCGAATGCAAATCACTAGAA-3′) with γ32-ATP, and T4 polynucleotide kinase in buffer containing 700mM Tris-Cl (pH 7.6), 100mM MgCl2 and 50mM DTT at 37°C for 30 min (gel retardation assay kit - Promega). The reaction was stopped by adding 1μl of 0.5M EDTA, pH 8.0, on ice and the final volume was adjusted to 100 μl with addition of 89μl of buffer containing 10mM Tris-HCl (pH 8.0) and 1mM EDTA. To determine binding, nuclear extracts were incubated with 35fmole of γ32-ATP end labeled, double stranded oligonucleotide of NF-κB or OCT-1. The binding reactions were performed in gel shift binding buffer containing 50mM Tris-HCl (pH 7.5), 250mM NaCl, 2.5mM DTT, 2.5mM EDTA, 5mM MgCl2, 20% glycerol and 0.25mg/ml poly (dI-dC) at 37°C for 30 min. The specificity of the binding reaction was confirmed by a competition assay with a 100-fold molar excess of unlabelled oligonucleotide probe, which was added to the tubes 10 min prior to the addition of 32P γ-ATP labeled probe. Supershift analysis was performed by pre-incubating nuclear protein extracts with antibodies specific for p65, p52, p50, RelB or c-Rel (Santa Cruz) for 30 min prior to incubation with labeled oligonucleotide. Complexes were separated by electrophoresis on a 5% native polyacrylamide gel in Tris-borate-EDTA (45mM tris, 45mM borate, 1mM ETDA) containing 89mM Tris-HCl (pH 8.0), 89mM Boric acid, and 2mM EDTA, dried and exposed to HyBlot CL™ autoradiography film (Denville Scientific) at −80°C. Variations in the mobility of supershifted bands were due to differing electrophoresis times, while variability in band intensity or background signal were a likely feature of relative differences in NF-κB protein concentrations, differing probe intensities and exposure times.
Cell Conjugation and Immunofluorescence
To evaluate the specific NF-κB complexes migrating to the nucleus in individual NK cells, NK cells were adhered onto poly-L-Lysine coated glass slides (Sigma) for 15 min at 37°C, treated with TNF-α or media for 30 min at 37°C, then fixed and permeabilized with 4% formaldehyde, 0.1% saponin, and 0.1% TritonX-100 in PBS for 15 min. Adherent cells were then stained with DAPI (100ng/ml) and either goat anti-p65 C-20, mouse anti-p50 E-10, mouse anti-p52 C-5, rabbit anti-RelB C-19, or rabbit anti-c-Rel N-466, followed by Alexa Flour 647-conjugated chicken anti-goat, 568-conjugated anti-mouse, or 568-conjugated anti-rabbit. All incubations with antibodies were performed at room temperature for 1hr followed by at least 2 washes in PBS containing 0.1% saponin. To evaluate NF-κB complexes migrating to the nucleus in true effector-target cell conjugates, NK cells were first stained with CFSE (10μM) for 15 min 37°C, then conjugated to K562 target cells at a 2:1 ratio for 15 min at 37°C in suspension, and then adhered to slides for 15 min. After adherence, conjugates and unconjugated controls were fixed, permeabilized and stained. Slides were mounted using ProLong anti-fade reagent (Molecular Probes) and 0.15mm glass coverslips. All antibodies were used in the range of 1–20μg/ml and diluted in saponin containing buffer. Cells were imaged using a Zeiss epifluroscence microscope with a 63x objective. Each conjugated NK cell was confirmed by the presence of a CFSE stained cell bound to a non-CFSE-stained cell. DAPI only, CFSE only and isotype controls were included to set exposure times and to ensure a lack of bleed through.
Image analysis
Images of NK cells were analyzed using the Volocity software package classification module (Improvision). In experiments evaluating conjugates between an NK cell and target cell, a conjugate was identified as a CFSE+ NK cell adherent to a CFSE− target cell. At least 30 NK cells per condition were evaluated and images were cropped based upon CFSE fluorescence to allow specific evaluation of fluorescence within the NK cell. Analysis of translocation was performed by obtaining the mean fluorescence intensity (MFI) of all pixels in each NK cell (defined as the CSFE positive region for cells in conjugates, or the whole cell region for experiments using NK cells only) for the given NF-κB protein, as well as the MFI of the NF-κB protein for pixels contained within the nucleus (defined by the DAPI positive region). As an additional quantitative measure of nuclear NF-κB content, the ratio of nuclear MFI to total cell MFI was calculated. To express the activation-induced change in nuclear NF-κB content, the mean ratio of nuclear to total cell NF-κB MFI after activation was divided by the mean ratio of nuclear to total cell NF-κB MFI in unactivated cells. In some cases, as an additional means of demonstrating the changes in sub-cellular NF-κB localization, colocalization dot plots were generated using Volocity in which each point represents the fluorescent intensity of an individual pixel with DAPI fluorescence expressed on the x-axis and NF-κB protein fluorescence on the y-axis.
Activation-induced NFκB reporter GFP expression in NK92 cells
NK92 NF-κB GFP reporter cells were treated with TNF-α immobilized anti-NKp30 or were conjugated with K562 target cells for 8h. Where specified, cells were pretreated for 30 min with various inhibitors: 10μM helenalin (Biomol International, LP), 50μM piceatannol (Calbiochem, La Jolla, CA), 1ug/ml actinomycin-D, or 10ug/ml cycloheximide (Sigma). The first two inhibitors have been previously described to alkylate and inhibit NF-κB p65 function in the nucleus, or block Syk function, respectively (15, 16). Cells were then evaluated for GFP fluorescence using FACS and the data were analyzed with FloJo software (TreeStar). When conjugates between NK92 and K562 cells were evaluated, the conjugates were stained with PE-conjugated anti-CD56 so that GFP fluorescence could be specifically determined in the NK92 cells and the unconjugated CD56− K562 target cells could be excluded from analysis.
mRNA isolation and analysis
After experimental treatment, cell pellets were snap frozen in liquid N2 and RNA was extracted using RNeasy Kit according to the manufacturers instructions (Qiagen, Valencia, CA). Samples were subjected to on column DNase digestion. First strand cDNA was derived from each treatment group using SuperScript II and oligo(dT) according to the manufacturers instructions (Invitrogen, Carlsbad, CA).
For quantitative real-time PCR analysis of A20 and β-actin targets were amplified using the following primer pairs: β-actin, 5′-TCAGCAAGCAGGAGTATGACGAG-3′ and 5′-ATTGTGAACTTTGGGGGATGC-3′. A20. 5′-GCCAGTTTTGTCCTCAGTTTC-3′ and 5′-CCATTCATCATTCCAGTTCCGAG-3′. Targets were amplified from 100ng cDNA using Power SYBR (ABI) according to the manufacturers instructions. PCR products were generated in quadruplicate and normalized to β-actin also generated in quadruplicate using the ABI 7500 Real-Time PCR system. Analysis was performed using SDS v1.3 Software. PCR product specificity was confirmed by performing a dissociation curve at the end of each experiment.
Results
Rapid activation signal induced IκBα degradation in NK cell lines
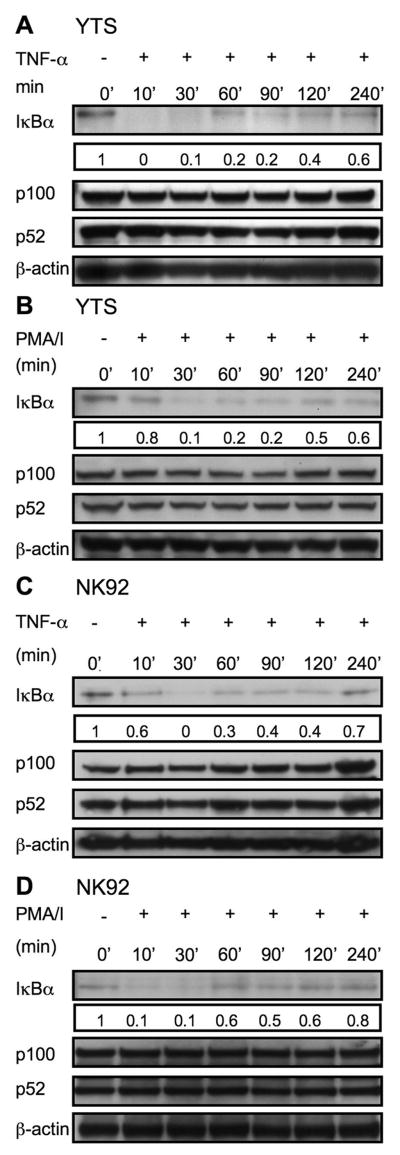
In resting cells, NF-κB is found in an inactive cytosolic form, retained by association with its inhibitor, IκB. Upon activation, IκB is rapidly phosphorylated, ubiquitinated and degraded, releasing the active NF-κB complex to translocate into the nucleus. There is, however, significant variability in activation of NF-κB in immune cell responses. For example, TNF-α signaling via TNFR1 results in rapid activation of IKK and nearly complete degradation of IκBα within 10 min, whereas signaling through TCR takes nearly 45 min (13, 17). Since activation of NF-κB in NK cells may utilize distinct receptors and may have kinetic differences from other immune cells we specifically evaluated the process in NK cells. Immortalized NK cell lines were initially used as a model. YTS and NK92 cells were chosen because they are both CD56+ and possess high cytotoxic capacity. These cell lines also provide an important functional contrast; YTS, but not NK92 cells grow independently of IL-2. To evaluate the kinetics of IκBα degradation in these NK cell lines we initially exposed the YTS and NK92 cells to different soluble stimuli for times ranging from 0 min to 4h. Western blot of cell lysates demonstrated that IκBα was degraded in YTS and NK92 cells in response to TNF-α after 10 min and persisted for 30 min (Fig. 1 A & C). PMA/I induced IκBα degradation in YTS and NK92 cells optimally at 30 min (Fig. 1 B & D). In some cases a return towards resting cell levels of IκB was noted after 60 min. Although the non-canonical pathway of NF-κB function generally takes longer than the canonical pathway to induce (18), it was also evaluated over 4h to determine if there were any predisposition towards non-canonical NF-κB activation in these cells. TNF and PMA/I stimulation in YTS and NK92 cells had no effect on processing of p100 to p52, a signature event in the non-canonical NF-κB pathway (Fig. 1A, B, C and D). Thus, soluble stimuli rapidly induce canonical NF-κB activation in NK cell lines although different stimuli possess distinct kinetics.
Figure 1. Rapid activation signal induced IκBα degradation in YTS and NK92 cells.
The ability of TNF-α and PMA/I to induce IκB degradation was determined in YTS (A, B) and NK92 (C, D) cells. Cells were treated in solution for the time indicated, lysed, separated on Nu-Page gels and transferred to membranes which were probed with anti-IκBα pAb (~33KD), stripped and reprobed with anti-β-actin rabbit pAb (~38KD) (as a control for protein loading). Processing of p100 (~100KD) to p52 (~52KD) after TNF-α and PMA/I stimulation for the indicated time points was also assessed in lysates with anti-NF-κBp52 rabbit pAb. Numbers below the IκBα blots represent the ratio of the band intensity to the 0 min band intensity for IκBα. Representative results of three independent experiments are shown.
Activation receptor induced IκBα degradation in NK cell lines
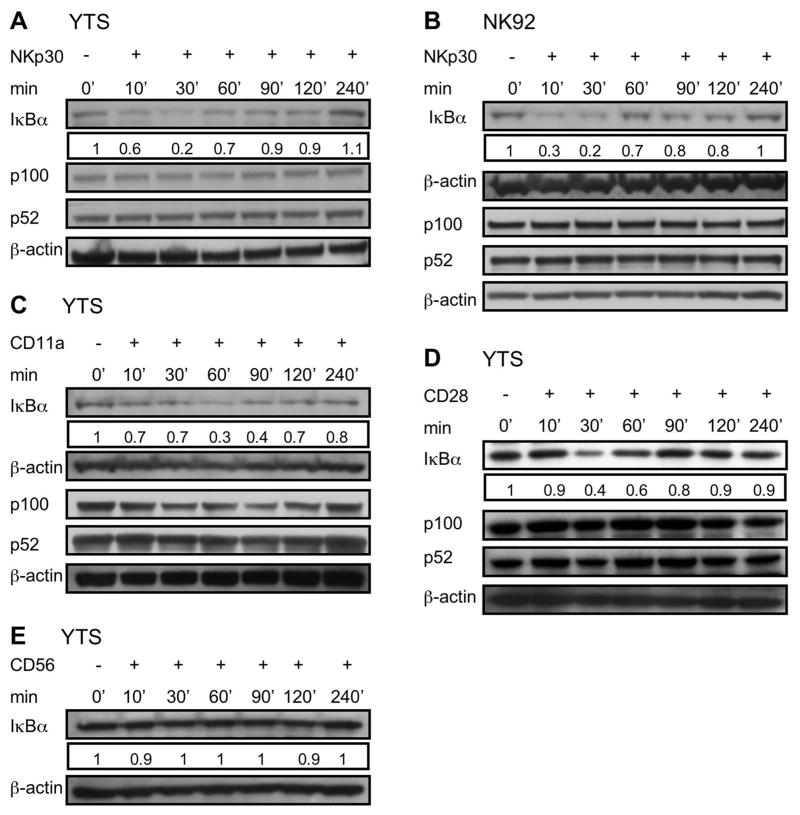
Since NK cells use the NCR family as a major receptor for inducing NK cell cytotoxic function, we next wanted to determine if triggering these receptors could induce NF-κB activation. Because NKp30 is expressed on YTS, and NK92, as well as resting and activated ex vivo NK cells, we focused on this receptor. We initially assessed the ability of NKp30 cross linking to induce IκBα degradation in YTS and NK92 cells by exposing them to immobilized anti-NKp30. Cells exposed to anti-NKp30 demonstrated IκBα degradation, which was maximal at 30 min (Fig. 2A & B). We also investigated function induced through other cell surface receptors (CD28, CD11a and CD56) in YTS cells. CD28 and CD11a were targeted since both have been shown to trigger cytotoxicity in YTS cells and CD56 was targeted as a control as it does not induce activation (19, 20). Immobilized anti-CD28 induced IκBα degradation in YTS at 30 min (Fig. 2C) while immobilized anti-CD11a induced optimal IκBα degradation at 60 min (Fig. 2D). In contrast, exposure to immobilized anti-CD56 failed to induce IκBα degradation (Fig. 2E). Stimulation of YTS and NK92 cells with anti-NKp30, anti-CD28 or anti-CD11a had no effect on processing of p100 to p52 after 4hrs (Fig. 2A–D).
Figure 2. Activating receptor specific IκBα degradation in YTS and NK92 cells.
YTS (A, C, D, E) and NK92 cells (B) were exposed to immobilized anti-NKp30 (A, B), anti-CD11a (C), anti-CD28 (D) and anti-CD56 (E) for 0, 10, 30, 60, 90, 120 and 240 min and then lysed on ice. Lysates were separated, transferred and membranes probed with anti-IκBα rabbit pAb (~33KD), stripped and reprobed with anti-β-actin rabbit pAb (~38KD) (as a control for protein loading). Processing of p100 to p52 after exposure to immobilized antibodies was also assessed for each. Numbers below IκBα blot represents the ratio of the band intensity to the 0 min band intensity for IκBα. Representative results of three independent experiments are shown.
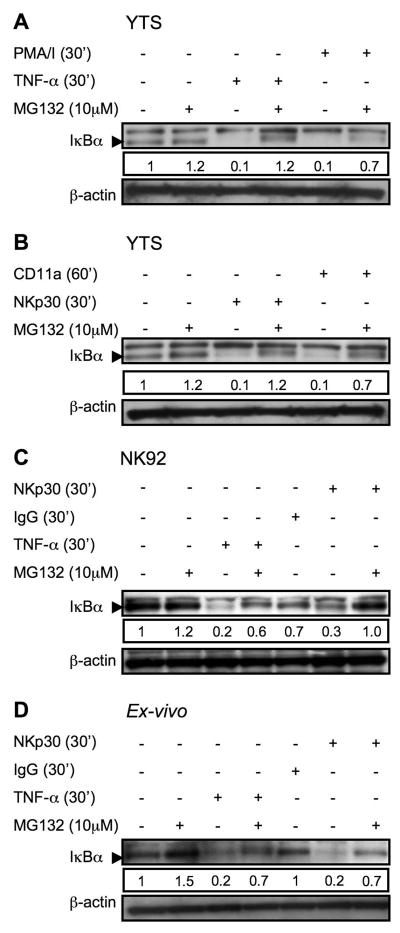
To demonstrate specificity in the induction of the NF-κB pathway in NK cells after stimulation, they were pretreated with the proteosome inhibitor MG132 to prevent degradation of IκBα that was ubiquitinated as result of activation signaling. Pretreatment of YTS cells with MG132 for 30 min effectively blocked degradation of IκBα in response to TNF-α and PMA/I (Fig. 3A). Proteosomal degradation of IκBα was also a feature of activation receptor triggering, as IκBα degradation in YTS cells after exposure to immobilized anti-NKp30 for 30 min or anti-CD11a for 60 min was blocked by MG132 (Fig. 3 B). This was not unique to YTS cells as IκBα degradation in NK92 cells stimulated with TNF-α or anti-NKp30 was also blocked by MG132 pretreatment (Fig. 3C). To ensure that the induction of this pathway was not a feature of culture propagated NK cell lines, we also evaluated ex-vivo NK cells. In freshly isolated NK cells TNF-α or immobilized anti-NKp30 induced IκBα degradation by 30 min (Fig. 3D). This induction was specific, as exposure to immobilized whole non-specific IgG molecules did not result in IκBα degradation. Furthermore, pretreatment with MG132 for 30 min also blocked degradation of IκBα in response to TNF-α or anti-NKp30 induced activation (Fig. 3D). Thus, the triggering of NK cell activation receptors induced the initial activation of the canonical pathway of NF-κB activation as evidenced by proteosomally-mediated rapid IκBα degradation.
Figure 3. Inhibition of activation induced IκBα degradation in YTS, NK92 and ex-vivo NK cells by MG132.
YTS cells were pretreated with MG132 for 30 min, restimulated with TNF-α or PMA/I (A), anti-CD11a or anti-NKp30 (B). NK 92 (C) and ex vivo NK cells (D) were pretreated with MG132 for 30 min, restimulated with TNF or anti-NKp30 for 30 min and then lysed. Lysates were separated, transferred and membranes probed with anti-IκBα rabbit pAb (~33KD), as demonstrated by the arrowhead. Membranes were then stripped and reprobed with anti-βactin rabbit pAb (~38KD) as a control for protein loading. A nonspecific band above the IκBα band was found with some lots of the pAb. Numbers below each IκBα blot represent the ratio of the band intensity to the 30 min band intensity for IκBα. Representative results of three independent experiments are shown.
NF-κB translocation in NK cells and inhibition by MG132
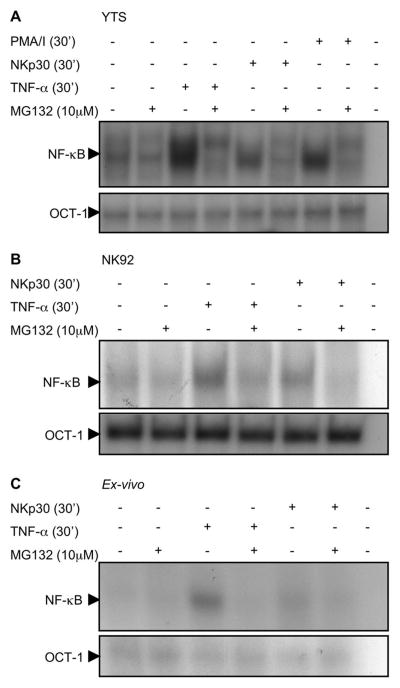
Given that NKp30 induced IκB degradation, we wanted to establish that NKp30 induces NF-κB nuclear translocation. EMSA was performed using nuclear extracts from NK cell lines and ex-vivo NK cells. Immobilized anti-NKp30 robustly induced NF-κB DNA binding activity in YTS, NK92 or ex-vivo NK cells (Fig. 4A, B & C). This was specific, as pretreatment with MG132 prevented the NKp30-induced increase in each of the cell lines as well as the ex vivo NK cells. As positive controls, YTS, NK92 and ex-vivo NK cells were stimulated with TNF-α (Fig. 4A, B & C), or PMA/I in YTS cells (Fig. 4A). Both of these signals induced NF-κB-DNA binding activity as well, which was blocked by preincubation with MG132. Equal loading was confirmed in these experiments by EMSA for Oct-1, which is not inducible. The integrity of the nuclear extracts was not altered by MG132 pretreatment, as the DNA binding activity of the ubiquitous transcription factor Oct-1 was maintained at equal levels.
Figure 4. Increased NF-κB nuclear translocation in NK cells after activation.
Nuclear protein extracts of YTS (A), NK92 (B) and ex-vivo NK cells (C) were evaluated for NF-κB (top) and Oct-1 (bottom) DNA binding activity by EMSA. Prior to lysis cells were activated with PMA/I (YTS only) or TNF-α or anti-NKp30. Cells were also pretreated with MG132. Nuclear extracts were preincubated with radiolabelled double stranded oligonucleotide NF-κB or Oct-1 probe and reaction products were analyzed on 5% non-denaturing polyacrylamide gels. Selected sections of the autoradiogram are shown. Representative results of three independent experiments are shown.
NK cell activation induces nuclear translocation of classical NF-κB heterodimers
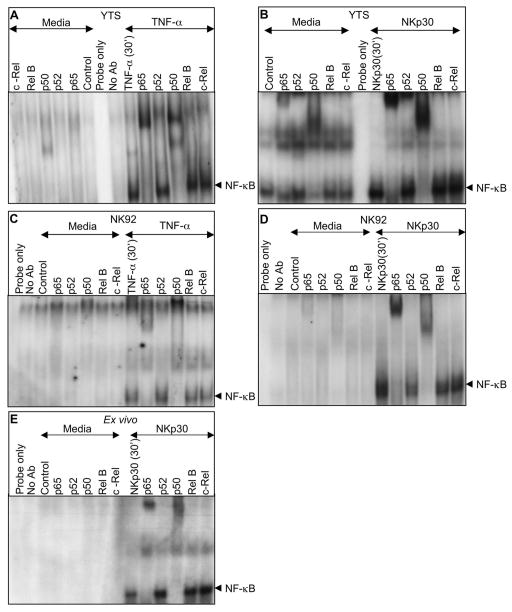
The NF-κB family of transcription factors contains five members, Rel A (p65), Rel B, c-Rel, NF-κB1 (p50), and NF-κB2 (p52). The existence of diversity among these proteins has raised the possibility that specific functions can be induced by particular heterodimers or homodimers in response to distinct stimuli. The most typical are heterodimers consisting of p65 (RelA) and p50 or c-Rel. Since the specific NF-κB dimers used in human NK cells have not been identified, we studied the NF-κB complexes activated in YTS, NK92 and ex-vivo NK cells by supershift assays. Prior to activation, basal NF-κB binding activity in YTS cell nuclei largely shifted with anti-NF-κB p50 (Fig. 5A - left) and to a much lesser extent with anti-NF-κBp65, suggesting a baseline presence of both p50 homodimers as well some p65/p50 heterodimers (Fig. 5A – left). After activation with TNF-α or immobilized anti-NKp30 (Fig. 5 A and B - right) a substantial increase in the amount of shifted p65 was found along with an increase in p50. This suggests the specific translocation of p65/p50 heterodimer after NK cell activation. A minor upshifted band of p52 is seen YTS in some cases, but is found irrespective of activation and without upshifted RelB and thus is unlikely to represent a non-canonical p52 RelB complex. To establish the consistency of this result, NK92 cells were also evaluated. As in YTS cells, basal NF-κB binding shifted with both anti anti-NF-κB p50 and to a lesser extent with anti-NF-κB p65. TNF and immobilized anti-NKp30 also resulted in substantially increased p65 and p50 bound to κB probe. To ensure that this activation paradigm was not unique to the NK cell lines, supershift assay using otherwise unactivated ex-vivo NK cells was performed. In resting cells a substantially less, but still detectable amount of basal p65 and p50 bound to κB DNA probe was detected. After activation with immobilized anti-NKp30, however there was an increase in nuclear NF-κB binding activity that shifted with anti-NF-κB p65 and anti-NF-κB p50 binding translocation in the nucleus. This suggests that there is a baseline presence of p50 in the NK cell nucleus, but that the classical heterodimer of p65/p50 is induced after NKp30 signaling in otherwise unstimulated NK cells.
Figure 5. Activation induced nuclear translocation of specific NF-κB heterodimers in NK cells.
Supershift analysis with antibodies to p65, p52, p50, Rel-B and c-Rel was performed on nuclear protein extracts of YTS (A, B), NK92 (C, D) and ex-vivo (E) NK cells. NK cells were treated with TNF-α (A, C) or anti-NKp30 (B, D, E) for 30 minutes. Nuclear extracts preincubated with NF-κB specific Abs were radiolabelled with double stranded oligonucleotide NF-κB probe and reaction products were analyzed on 5% non-denaturing polyacrylamide gels. Selected sections of the autoradiogram are shown highlighting the region of NF-κB bound probe and supershifted bands. Representative results of three independent experiments are shown.
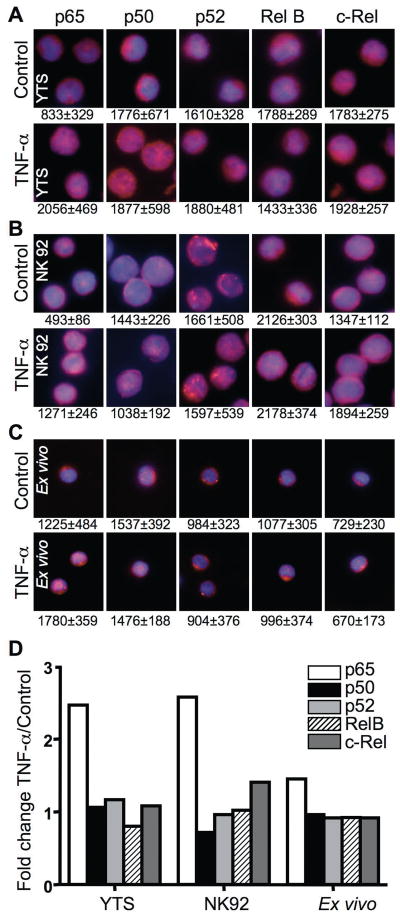
To evaluate this paradigm in individual NK cells, NF-κB nuclear translocation was measured using quantitative fluorescence microscopy and antibodies directed against different NF-κB family members. Since cell morphology was preserved using this method, the precise localization of NF-κB members could be determined and the NF-κB nuclear content quantified by measuring colocalization with nuclear stain. The localization of NF-κB in single cells after TNF-α activation was first determined using this approach in YTS, NK 92 and ex vivo NK cells (Fig. 6A–C). TNF-α treated YTS, NK92 and ex-vivo NK cells demonstrated increased nuclear p65 in comparison to untreated cells and support in part our supershift assays findings. Appreciable increases in other NF-κB family members among all NK cells studied after TNF stimulation were not found. To quantify differences, the MFI in the nucleus and the whole cell was measured in ≥50 cells per condition. The mean nuclear MFI ± SD from the ≥50 cells was calculated and is listed beneath each representative image in Fig 6. The increase in nuclear p65 MFI after TNF stimulation was significant (p<0.001) in YTS, NK92 and ex vivo NK cells. Other NF-κB proteins did not demonstrate significant increases in NF-κB nuclear MFI after TNF stimulation consistently across the different NK cells. As an additional measure of nuclear translocation after activation, the ratio of nuclear to total NF-κB MFI was calculated. The ratio in unstimulated cells was then compared to that in cells after TNF-α activation and was represented as an activation-induced fold change for each NF-κB family member (Fig. 6D). This analysis also demonstrated a selective increase in nuclear p65 after stimulation.
Figure 6. Single NK cell NF-κB nuclear translocation after activation.
Untreated (top) and TNF-α treated (bottom) YTS (A), NK92 (B) and ex-vivo (C) NK cells were fixed and stained with DAPI (blue) as well as anti-p65, anti-p50, anti-p52, anti-RelB or anti-c-Rel followed by AlexaFluor 568-conjugated secondary Ab (red). Cells were imaged using fluorescence microscopy to show a merged overlay of DAPI and different NF-κB family members. The numbers beneath each image represent the mean nuclear MFI ± SD from ≥50 cells measured using Volocity. (D) The ratio of mean fluorescence intensity in the nucleus to the whole cell was calculated using Volocity and is represented as fold change after TNF-α activation for each NF-κB family member. Each bar represents individual analysis of 50–60 cells.
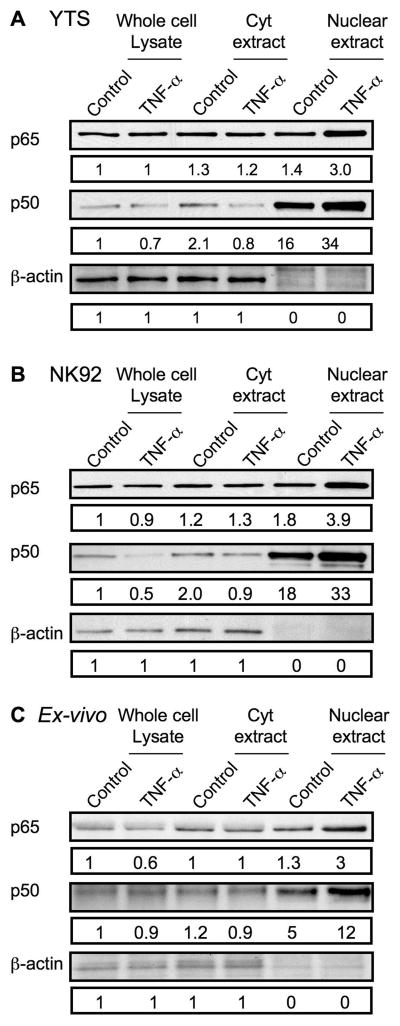
Since the classical NF-κB heterodimer of p65/p50 appeared to translocate to the nucleus after TNF stimulation of NK cells when measured by EMSA (Fig. 5), but only p65 translocation was found by microscopy (Fig. 6) we hypothesized that there was a substantial nuclear presence of NF-κB p50 in resting NK cells. To evaluate the baseline localization of p50, whole cell lysates, and cytoplasmic as well as nuclear extracts were prepared from YTS, NK92 and ex vivo NK cells that were either control- or TNF-treated. The presence of p65 and p50 in equal protein quantities from these different preparations was determined by western blot (Fig. 7A–C). β-actin is included as a control and also confirms the actin-poor nuclear fraction. Although the amount of nuclear p65 in resting cells was similar to that present in cytoplasmic extracts and whole cell lysates, there was substantially more p50 present in the nuclear fraction. TNF-treatment resulted in increased quantities of p65 in the nucleus, as well as the already elevated amount of nuclear p50 relative to p50 in cytoplasm or whole cell lysate. This demonstrates that p50 is preferentially concentrated in the nucleus of resting NK cells and would be consistent with the existence of nuclear p50 homodimers that have been previously described to serve an inhibitory role (21, 22),
Figure 7. Cytoplasmic and nuclear levels of NF-κB p65 and p50 protein in resting and activated NK cells.
(A) YTS, (B) NK92, and (C) ex vivo NK cells were treated with control media or TNF-α for 30 minutes and equal amounts of protein from whole cell lysates (left), cytoplasmic extracts (Cyt, middle), and nuclear extracts (right) were evaluated for the presence of NF-κB p65 (top), NF-κB p50 (middle), and β-actin by Western blot. Numbers below each band represent the ratio of the band intensity to that of the band for the whole cell lysate of control-treated cells.
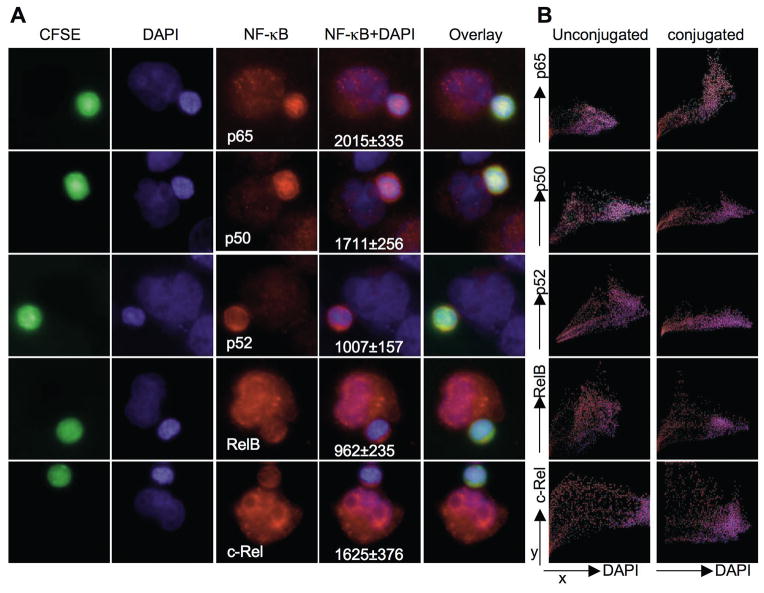
To determine the nuclear translocation of individual NF-κB family members in single ex vivo NK cells after their physiologic activation by a susceptible target cell we again used the microscopy-based approach. To facilitate the identification and analysis of ex vivo NK cells, they were first stained with CFSE, then conjugated with unlabeled K562 target cells for a total of 30 min and finally stained with antibodies directed against the individual NF-κB family members and DAPI. The entire ex vivo NK cell area was distinguished from a visibly bound unlabeled target cell using CFSE fluorescence and NF-κB fluorescence measured only within the area of CFSE fluorescence. Of the individual NF-κB family members, p65 was most consistently identified in the nucleus of conjugated NK cells (Fig. 8A) when compared to unactivated, unconjugated ex vivo NK cells (Fig. 6C). Colocalization of individual pixel fluorescent intensities within a single unconjugated NK cell or NK cell conjugated to K562 target cell was measured and plotted with DAPI and an NF-κB family member fluorescence on the x and y-axes, respectively (Fig. 8B). As pixels with low DAPI fluorescence localize to the cytoplasm, the plot for p65 fluorescence after activation was most consistent with nuclear localization (pixels having bright p65 fluorescence also having high DAPI intensity). To quantitatively analyze NF-κB nuclear translocation after conjugation of ex vivo NK cells to K562 target cells, the mean nuclear MFI of the different NF-κB family members in ex vivo NK cells conjugated to a target cell (Fig. 8A) was compared to that in unconjugated ex vivo NK cells. Only p65 nuclear MFI was significantly increased in NK cells (p<0.001) after conjugation. This increase was also confirmed using an analysis algorithm accounting for the total cell NF-κB MFI. Here the ratio of nuclear to total p65 intensity in ≥50 conjugated NK cells was increased 1.86-fold when compared to this ratio in ≥50 unconjugated NK cells. There were no such appreciable increases found for other NF-κB family members when conjugated NK cells were compared to unconjugated NK cells. Thus, the pattern of p65 fluorescence after target cell conjugation was distinguished from the other profiles, which is likely a specific characteristic of a single NK cell activated by a physiologic target cell.
Figure 8. NF-κB nuclear translocation in single NK cells after physiologic activation.
Ex vivo NK cells were stained with CFSE and conjugated with K562 cells. (A) Conjugates were fixed and stained with DAPI (blue) and anti-p65, anti-p50, anti-p52, anti-RelB or anti-c-Rel followed with Alexa Fluor 568-conjugated secondary Ab. Fluorescence for CFSE (green), DAPI (blue), NF-κB family members (red), an overlay of NF-κB and DAPI and merged overlay fluorescent intensities are shown. Representative conjugated cells are shown. Mean nuclear MFI ± SD of NF-κB family members in ≥30 ex vivo NK cells conjugated to K562 target cells are shown beneath the NF-κB+DAPI overlay image. (B) Colocalization of individual pixel fluorescent intensities within a single unconjugated NK cell (left) or NK cell conjugated to K562 target cell (right). The x-axis represents DAPI fluorescence intensity and y-axis NF-κB family member fluorescence intensity. Pixels with no DAPI fluorescence localize to the cytoplasm. Plots are representative of 10 analyses.
Functional activation-induced NF-κB in NK92 cells
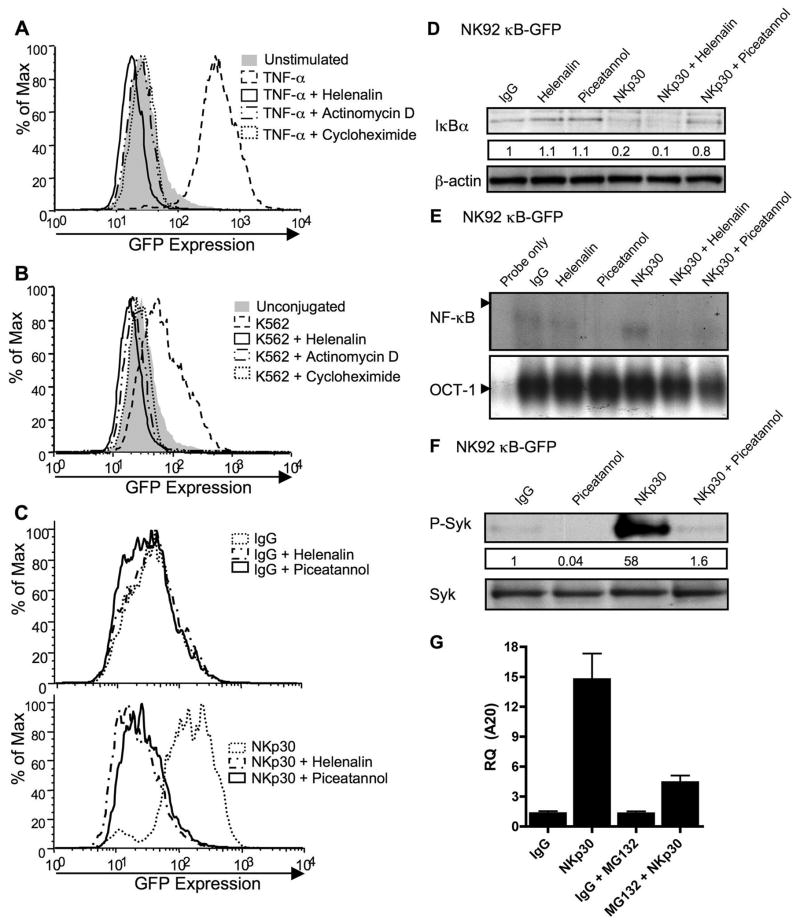
To determine if NF-κB could be functional soon after NK cell activation, NK92 cells were infected with a lentivirus containing an HIV-based vector that included a κB-GFP reporter construct. Successfully transduced cells were selected for GFP expression and grown as stable cultures, which were studied by FACS after different activation stimuli (Fig. 9A–C). To demonstrate their initial function in reporting, these cells were treated with TNF-α for 8h and analyzed for GFP expression (Fig. 9A). GFP accumulation was increased 8h after TNF-α treatment of cells. To demonstrate the specificity of NF-κB mediated GFP induction, actinomycin-D, cycloheximide, or helenalin were used to block transcription, translation and NF-κB function respectively. Helenalin has been previously demonstrated to selectively alkylate the p65 subunit of NF-κB preventing it from binding to its target sequence (15). All the three inhibitors blocked the TNF-α induced increase in GFP expression (Fig. 9A). To determine if physiologic NK cell stimulation could induce NF-κB function, reporter cells were conjugated to K562 target cells for 8h and were analyzed for fluorescence intensity of GFP by FACS. NK92 κB-GFP expressing cells incubated with K562 target cells had on average a >2-fold increase in GFP mean fluorescence intensity compared to that in unconjugated cells. Pretreatment of cells with helenalin, actinomycin-D or cycloheximide inhibited GFP expression (Fig. 9B) demonstrating that the expression was de novo and NF-κB dependent. Since a physiologic target cell could induce NF-κB function, we wanted to determine whether this rapidly induced NF-κB dependent protein synthesis could be identified after signaling by a single specific NK cell activating receptor. Thus, NK92 κB-GFP cells were stimulated with immobilized anti-NKp30 and GFP expression was analyzed relative to IgG control stimulation by FACS. 8h of stimulation with anti-NKp30 induced an increase in cell GFP content, which was dependent upon NFκB activity as the increase was inhibited by helenalin treatment (Fig. 9C). To validate the effects of helenalin on NKp30-induced NF-κB activation, IκB degradation by western blot and nuclear NF-κB DNA-binding activity by EMSA were performed. Compared to IgG control, NKp30-induced IκB degradation was not blocked by helenalin (Fig. 9D), whereas nuclear NF-κB DNA binding was (Fig. 9E). This is consistent with the previously reported effect of helenalin in preventing p65 from binding DNA, without effecting upstream functions (15). We also used the Syk-specific inhibitor piceatannol to determine the requirement for the NF-κB dependent GFP content shift upon NKp30 induced Syk signaling. As expected, NKp30-induced Syk phosphorylation, which was inhibited by preincubation with piceatannol (Fig 9F). The effect of piceatannol upstream of NF-κB activation in NKp30 signaling was demonstrated as pre-incubation with piceatannol blocked anti-NKp30-induced IκB degradation (Fig 9D), NF-κB nuclear translocation (Fig 9E) and GFP-accumulation (Fig 9C). To demonstrate that the NKp30 signaling can induce physiological NF-κB function, real-time PCR for transcripts of the NF-κB target gene A20 (23) was performed (Fig. 9G). Anti-NKp30 induced A20 gene expression, which was reduced markedly in the presence of MG132. Thus, both physiologic and NKp30 lysis receptor signaling specifically induced activation signaling leading to NF-κB function and rapid protein synthesis in NK cells.
Figure 9. Activation-induced NF-κB function in NK92 cells.
NK92 cells transduced with a κB-GFP reporter construct were stimulated with TNF-α (A), K562 target cells (B), or immobilized IgG (top) or anti-NKp30 (bottom) (C) for 8h after 30 min pretreatment with media, helenalin, actinomycin-D, cycloheximide or piceatannol. GFP expression in live cells was analyzed by FACS and representative results of five independent experiments are shown. NK92 κB-GFP cells were pretreated for 30min with media, helenalin, or piceatannol and stimulated for 30min with immobilized IgG or anti-NKp30. Whole cell lysates were evaluated for the presence of IκBα by Western blot (D) and nuclear extracts for NF-κB (top) and OCT-1 (bottom) DNA binding activity by EMSA (E). Whole cell lysates of NK92 κB-GFB cells pretreated for 30 min with media or piceatannol and stimulated for 30 min with IgG or NKp30 were evaluated by Western blot for phospho-Syk (F, top). The membrane was stripped and reprobed for total Syk (F, bottom). The numbers below the phospho-Syk blot represent the ratio of the phospho-Syk band intensity to the IgG phospho-Syk band. (G) A20 transcripts were quantified using real-time PCR from NK92-κB GFP cells that were pretreated for 30 min with media, or MG132 and then stimulated for 4 h with immobilized IgG or anti-NKp30.
Discussion
We have previously defined a defect in NK cell cytotoxicity in patients having a hypomorphic mutation in the IKBKG gene encoding NEMO (12). These disease-associated mutations preserve NEMO protein expression but impair the ability of TNF receptor family molecules to induce NF-κB activation. Because NK cell cytotoxicity from these patients could still be induced after IL-2 stimulation, it was unlikely that the deficit in function was due to a developmental abnormality of the NK cell lineage. For this reason we hypothesized that the normal process of inducing cytolytic function in NK cells requires NEMO-dependent NF-κB activation. Using the YTS and NK92 cell lines, as well as normal ex vivo NK cells we have found that the ligation of NK cell activation receptors, most notably NKp30, induces rapid IκB degradation and NF-κB nuclear translocation. Others have found that soluble stimuli such as the TLR3 agonist poly I:C (24), the TLR2 agonist lipophosphoglycan (8), IL-2 (9, 25), IL-18 and TNF (26) can induce NF-κB activation in NK cells. The present work, however, is to our knowledge the first demonstration of NF-κB activation by an NK cell lysis receptor. We have also identified that this induction was functional as it was capable of causing NF-κB dependent transcription and resultant protein synthesis within a relatively short time (Fig. 9).
Following our hypothesis that activation receptor-induced NF-κB activation is a NEMO directed process, it was predicted that specific NF-κB heterodimers reliant upon NEMO function would be translocated into the nucleus. The ability of NK cell activation receptors to induce particular NF-κB dimers has also not been previously investigated in human NK cells and represents a means by which functional diversity can be generated within the NF-κB family (13). We found using supershift analysis that NKp30 ligation and the more typical soluble NF-κB inducing factor TNF resulted in nuclear translocation of the classical heterodimer consisting of p65/p50. This provides a connection to the to NEMO-containing IKK complex since this is the most common heterodimer induced by the canonical pathway of NF-κB activation. Furthermore, evidence of the NEMO-independent non-canonical pathway of NF-κB activation was not found at the early timepoints studied here, as there was no p100 degradation identified and no increase in nuclear content of p52 or RelB. Studies of NK cells from mice deficient in c-Rel have demonstrated a requirement of this particular family member in optimal NK cell proliferation and IFN-γ production (11), but this may be due to developmental requirements for this factor in NK cells. A developmental requirement for appropriate NF-κB activation in NK cells has also been raised using studies of mice deficient in IκBα and IκBε (27). Although lysis receptor-induced activation and perhaps function of NK cells involves the canonical pathway of NF-κB activation and p65/p50 heterodimers, our focus has been on rapid events occurring after NK cell activation that could unfold within the timeframe of perforin-mediated cytotoxicity. There may still be a role for the non-canonical pathway of NF-κB activation and alternative NF-κB dimers, however, as longer timeframes are required for induction of the non-canonical pathway of NF-κB in other cell types (18).
A drawback of the biochemical studies of NF-κB activation is that they largely preclude inducing function in the NK cell by a target cell. This is because a target cell will contribute its nuclear content to the analyses. A prior attempt to address this had utilized K562 target cells mixed with 50-fold more ex vivo NK cells and found that the addition of the NK cells resulted in greater NF-κB content in the combined nuclear fraction (6). Although this is a relevant finding, it does not account for target cells that are activated by NK cells having increased nuclear NF-κB content, which we have observed (Pandey and Orange unpublished). In addition, the nucleus of a K562 target cells is generally more voluminous than that of ex vivo NK cells and thus will contribute a larger proportion of the nuclear proteins. This presents an additional challenge to biochemical approaches using physiologic target cell activation especially if target cells increase their nuclear NF-κB content after exposure to NK cells. Specifically, a majority of the nuclear NF-κB could be derivative from a minority of target cells. Finally, as the data derivative from biochemical techniques are averages of protein content or localization over multiple cells they may still reflect inconsistencies within single NK cells being activated by a target cell, as well as cells that have never conjugated. To address these issues we quantitatively evaluated the localization of NF-κB proteins in individual NK cells that had encountered physiologic target cells microscopically. Using this approach we found that NF-κB p65 accumulates within the NK cell nucleus within 30 min of conjugation with a target cell. Surprisingly this was the only NF-κB protein that was found to shift significantly and uniformly into the nucleus. A lack of reproducible detection of a p50 shift was a likely feature of the high baseline presence of p50 in the NK cell nucleus as demonstrated in supershift assays (Fig. 5) and by Western blot (Fig. 7). Thus, the relative amount of p50 shifting with p65 may be difficult to discern as p50 may be both exiting and entering the nucleus after activation. We propose that the abundant p50 present in the nucleus of unstimulated NK cells represents the baseline presence of p50 homodimers which have been identified in other cell types to serve an inhibitory function (21, 22). These might serve the purpose in NK cells of specifically preventing the typical activation-induced transcriptional programs until lysis receptors are actually engaged. The data do, however, confirm the rapid physiologic induction of canonical NF-κB activation in NK cells.
We also found that antibodies against several NK cell receptors that have been reported to trigger cytotoxicity could all induce rapid NF-κB activation. These included NKp30 in all NK cells evaluated and the YTS cell lysis receptors CD28 and CD11a. In YTS cells, anti-CD56 did not induce function, demonstrating that NF-κB activation was not a nonspecific feature of receptor cross-linking. Furthermore, in experiments using ex vivo NK cells and immobilized IgG as a control for anti-NKp30 (Figure 4), we did not see substantial activation of the NF-κB pathway. This suggests that signaling through the NK cell IgG receptor, CD16 may not induce rapid NF-κB activation. CD16 signaling has been previously demonstrated to be capable of inducing function in an NF-κB independent manner (28). Although not the focus of this work, this conclusion is consistent with the observation that cells from patients with a hypomorphic NEMO can mediate ADCC indistinguishably from controls directly ex vivo (12). This prior study also found that IL-2 could induce cytotoxicity in the presence of a mutant NEMO molecule. IL-2 has been shown to activate NF-κB in NK cells (9, 25) and to drive perforin expression (25). It is quite likely, however, that the ability of IL-2 to acutely induce cytotoxicity in NK cells is independent of NF-κB and NF-κB-mediated increases in perforin expression since most NK cells typically express substantial levels of perforin at rest.
In this current study we have shown that both soluble and polarized signals can induce NF-κB activation and function in NK cells. The polarized signals provided here were induced by immobilized antibodies directed against lysis receptors as well as physiologic target cells themselves. There were some suggestions that the different signals may lead to differing degrees of signal strength. For example, in some experiments TNF stimulation appeared to more completely induce IκB degradation than other stimuli. Similarly, there may be some differences in the time to peak IκB degradation among different stimuli (Fig. 3). Further investigation specifically directed toward understanding these differences will be important in comprehending the relevance of signal strength variation. Although there may be differences in the strength of signal derivative from the soluble as compared to polarized signals, it is likely that there will be a special role for a polarized signal in inducing transcriptional activation through the NF-κB pathway in NK cells. In particular one purpose of activating polarized contacts formed by NK cells (often referred to as immunological synapses) may be in rapidly promoting NF-κB function. In this light the IKK proteins have been shown to polarize to the T cell immunological synapse (29, 30). Another purpose of generating NF-κB activation in a polarized manner, may be to create the intracellular network to traffic components from the immunological synapse into the nucleus and directionally back to the synapse. It will be important to focus future efforts upon the coordination of the NK cell immunological synapse and transcriptional activation and what critical links exist between the two processes. Our data suggest that NKp30-induced Syk, which is polarized to the NK cell immunological synapse (31), is at least an early requirement in this process. The comparison of activating and inhibitory immunological synapses will also be important as it is possible that each may be associated with a distinct effect on the NF-κB system, such as the further induction or maintenance of inhibitory NF-κB homodimers after immunological synapse-directed ligation of inhibitory receptors.
Finally there are potential implications for NF-κB-dependent rapidly transcribed genes after lysis receptor ligation or immunological synapse formation. Some activation-induced NF-κB target genes identified in NK cells have been shown to include both perforin and granzyme B (25, 32). They may include other genes characteristic of NK cells that would typically be accessed by the classic NF-κB heterodimer of p65/p50 such as TNF. The NF-κB-dependent induction of the anti-apoptotic gene A20 (Fig. 9) may indicate a specific role for lysis receptor-induced, NF-κB-dependent survival of activated NK cells. Furthermore, since we found new NF-κB-dependent protein expression by 8 hrs after NKp30 signaling, it is possible that other NF-κB target genes might specifically enable other NK cell functions. NK cell lysis receptors, therefore, should be viewed as inducing multi-faceted signaling paradigms that can induce cytotoxicity, but also induce rapid transcriptional regulation potentially to facilitate cytolytic capacity or access distinct functions.
Acknowledgments
The authors thank Drs. Kerry Campbell, and Pinaki Banerjee, as well as Ms. Laura Solt and Kelli Deering for suggestions, help and feedback.
Footnotes
This work was supported by the US immunodeficiency network N01 AI-22070 (to JSO and MJM), NIH AI-067946, The Pennsylvania Department of Health (the Department specifically disclaims responsibility for any analyses, interpretations or conclusions), and a career development award from the American Academy of Allergy, Asthma and Immunology (to JSO).
Disclosures
The authors have no conflicts of interest.
References
- 1.Moretta L, Moretta A. Unravelling natural killer cell function: triggering and inhibitory human NK receptors. Embo J. 2004;23:255–259. doi: 10.1038/sj.emboj.7600019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Orange JS, Ballas ZK. Natural killer cells in human health and disease. Clin Immunol. 2006;118:1–10. doi: 10.1016/j.clim.2005.10.011. [DOI] [PubMed] [Google Scholar]
- 3.Pende D, Parolini S, Pessino A, Sivori S, Augugliaro R, Morelli L, Marcenaro E, Accame L, Malaspina A, Biassoni R, Bottino C, Moretta L, Moretta A. Identification and molecular characterization of NKp30, a novel triggering receptor involved in natural cytotoxicity mediated by human natural killer cells. J Exp Med. 1999;190:1505–1516. doi: 10.1084/jem.190.10.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hayden MS, West AP, Ghosh S. NF-κB and the immune response. Oncogene. 2006;25:6758–6780. doi: 10.1038/sj.onc.1209943. [DOI] [PubMed] [Google Scholar]
- 5.Scheidereit C. IkappaB kinase complexes: gateways to NF-κB activation and transcription. Oncogene. 2006;25:6685–6705. doi: 10.1038/sj.onc.1209934. [DOI] [PubMed] [Google Scholar]
- 6.Valle Blazquez M, Luque I, Collantes E, Aranda E, Solana R, Pena J, Munoz E. Cellular redox status influences both cytotoxic and NF-κB activation in natural killer cells. Immunology. 1997;90:455–460. doi: 10.1111/j.1365-2567.1997.00455.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoshino K, Tsutsui H, Kawai T, Takeda K, Nakanishi K, Takeda Y, Akira S. Cutting edge: generation of IL-18 receptor-deficient mice: evidence for IL-1 receptor-related protein as an essential IL-18 binding receptor. J Immunol. 1999;162:5041–5044. [PubMed] [Google Scholar]
- 8.Becker I, Salaiza N, Aguirre M, Delgado J, Carrillo-Carrasco N, Kobeh LG, Ruiz A, Cervantes R, Torres AP, Cabrera N, Gonzalez A, Maldonado C, Isibasi A. Leishmania lipophosphoglycan (LPG) activates NK cells through toll-like receptor-2. Mol Biochem Parasitol. 2003;130:65–74. doi: 10.1016/s0166-6851(03)00160-9. [DOI] [PubMed] [Google Scholar]
- 9.Jyothi MD, Khar A. Interleukin-2-induced nitric oxide synthase and nuclear factor-kappaB activity in activated natural killer cells and the production of interferon-gamma. Scand J Immunol. 2000;52:148–155. doi: 10.1046/j.1365-3083.2000.00762.x. [DOI] [PubMed] [Google Scholar]
- 10.Tato CM, Villarino A, Caamano JH, Boothby M, Hunter CA. Inhibition of NF-kappa B activity in T and NK cells results in defective effector cell expansion and production of IFN-γ required for resistance to Toxoplasma gondii. J Immunol. 2003;170:3139–3146. doi: 10.4049/jimmunol.170.6.3139. [DOI] [PubMed] [Google Scholar]
- 11.Tato CM, Mason N, Artis D, Shapira S, Caamano JC, Bream JH, Liou HC, Hunter CA. Opposing roles of NF-κB family members in the regulation of NK cell proliferation and production of IFN-gamma. Int Immunol. 2006;18:505–513. doi: 10.1093/intimm/dxh391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Orange JS, Brodeur SR, Jain A, Bonilla FA, Schneider LC, Kretschmer R, Nurko S, Rasmussen WL, Kohler JR, Gellis SE, Ferguson BM, Strominger JL, Zonana J, Ramesh N, Ballas ZK, Geha RS. Deficient natural killer cell cytotoxicity in patients with IKK-γ/NEMO mutations. J Clin Invest. 2002;109:1501–1509. doi: 10.1172/JCI14858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoffmann A, Leung TH, Baltimore D. Genetic analysis of NF-κB/Rel transcription factors defines functional specificities. Embo J. 2003;22:5530–5539. doi: 10.1093/emboj/cdg534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Drexler HG, Matsuo Y. Malignant hematopoietic cell lines: in vitro models for the study of natural killer cell leukemia-lymphoma. Leukemia. 2000;14:777–782. doi: 10.1038/sj.leu.2401778. [DOI] [PubMed] [Google Scholar]
- 15.Lyss G, Knorre A, Schmidt TJ, Pahl HL, Merfort I. The anti-inflammatory sesquiterpene lactone helenalin inhibits the transcription factor NF-κB by directly targeting p65. J Biol Chem. 1998;273:33508–33516. doi: 10.1074/jbc.273.50.33508. [DOI] [PubMed] [Google Scholar]
- 16.Jiang K, Zhong B, Gilvary DL, Corliss BC, Vivier E, Hong-Geller E, Wei S, Djeu JY. Syk regulation of phosphoinositide 3-kinase-dependent NK cell function. J Immunol. 2002;168:3155–3164. doi: 10.4049/jimmunol.168.7.3155. [DOI] [PubMed] [Google Scholar]
- 17.Hoffmann A, Levchenko A, Scott ML, Baltimore D. The IκB-NF-κB signaling module: temporal control and selective gene activation. Science. 2002;298:1241–1245. doi: 10.1126/science.1071914. [DOI] [PubMed] [Google Scholar]
- 18.Dejardin E, Droin NM, Delhase M, Haas E, Cao Y, Makris C, Li ZW, Karin M, Ware CF, Green DR. The lymphotoxin-beta receptor induces different patterns of gene expression via two NF-κB pathways. Immunity. 2002;17:525–535. doi: 10.1016/s1074-7613(02)00423-5. [DOI] [PubMed] [Google Scholar]
- 19.Chen X, Allan DS, Krzewski K, Ge B, Kopcow H, Strominger JL. CD28-stimulated ERK2 phosphorylation is required for polarization of the microtubule organizing center and granules in YTS NK cells. Proc Natl Acad Sci U S A. 2006;103:10346–10351. doi: 10.1073/pnas.0604236103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barber DF, Faure M, Long EO. LFA-1 contributes an early signal for NK cell cytotoxicity. J Immunol. 2004;173:3653–3659. doi: 10.4049/jimmunol.173.6.3653. [DOI] [PubMed] [Google Scholar]
- 21.Zhong H, May MJ, Jimi E, Ghosh S. The phosphorylation status of nuclear NF-κB determines its association with CBP/p300 or HDAC-1. Mol Cell. 2002;9:625–636. doi: 10.1016/s1097-2765(02)00477-x. [DOI] [PubMed] [Google Scholar]
- 22.Guan H, Hou S, Ricciardi RP. DNA binding of repressor nuclear factor-κB p50/p50 depends on phosphorylation of Ser337 by the protein kinase A catalytic subunit. J Biol Chem. 2005;280:9957–9962. doi: 10.1074/jbc.M412180200. [DOI] [PubMed] [Google Scholar]
- 23.Krikos A, Laherty CD, Dixit VM. Transcriptional activation of the tumor necrosis factor alpha-inducible zinc finger protein, A20, is mediated by κB elements. J Biol Chem. 1992;267:17971–17976. [PubMed] [Google Scholar]
- 24.Schmidt KN, Leung B, Kwong M, Zarember KA, Satyal S, Navas TA, Wang F, Godowski PJ. APC-independent activation of NK cells by the Toll-like receptor 3 agonist double-stranded RNA. J Immunol. 2004;172:138–143. doi: 10.4049/jimmunol.172.1.138. [DOI] [PubMed] [Google Scholar]
- 25.Zhou J, Zhang J, Lichtenheld MG, Meadows GG. A role for NF-κB activation in perforin expression of NK cells upon IL-2 receptor signaling. J Immunol. 2002;169:1319–1325. doi: 10.4049/jimmunol.169.3.1319. [DOI] [PubMed] [Google Scholar]
- 26.Suzuki N, Chen NJ, Millar DG, Suzuki S, Horacek T, Hara H, Bouchard D, Nakanishi K, Penninger JM, Ohashi PS, Yeh WC. IL-1 receptor-associated kinase 4 is essential for IL-18-mediated NK and Th1 cell responses. J Immunol. 2003;170:4031–4035. doi: 10.4049/jimmunol.170.8.4031. [DOI] [PubMed] [Google Scholar]
- 27.Samson SI, Memet S, Vosshenrich CA, Colucci F, Richard O, Ndiaye D, Israel A, Di Santo JP. Combined deficiency in IκBα and IκBε reveals a critical window of NF-κB activity in natural killer cell differentiation. Blood. 2004;103:4573–4580. doi: 10.1182/blood-2003-08-2975. [DOI] [PubMed] [Google Scholar]
- 28.Jewett A. Activation of c-Jun N-terminal kinase in the absence of NFκB function prior to induction of NK cell death triggered by a combination of anti-class I and anti-CD16 antibodies. Hum Immunol. 2001;62:320–331. doi: 10.1016/s0198-8859(01)00218-x. [DOI] [PubMed] [Google Scholar]
- 29.Weil R, Schwamborn K, Alcover A, Bessia C, Di Bartolo V, Israel A. Induction of the NF-κB cascade by recruitment of the scaffold molecule NEMO to the T cell receptor. Immunity. 2003;18:13–26. doi: 10.1016/s1074-7613(02)00506-x. [DOI] [PubMed] [Google Scholar]
- 30.Wang D, Matsumoto R, You Y, Che T, Lin XY, Gaffen SL, Lin X. CD3/CD28 costimulation-induced NF-κB activation is mediated by recruitment of protein kinase C-theta, Bcl10, and IκB kinase beta to the immunological synapse through CARMA1. Mol Cell Biol. 2004;24:164–171. doi: 10.1128/MCB.24.1.164-171.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vyas YM, Mehta KM, Morgan M, Maniar H, Butros L, Jung S, Burkhardt JK, Dupont B. Spatial organization of signal transduction molecules in the NK cell immune synapses during MHC class I-regulated noncytolytic and cytolytic interactions. J Immunol. 2001;167:4358–4367. doi: 10.4049/jimmunol.167.8.4358. [DOI] [PubMed] [Google Scholar]
- 32.Huang C, Bi E, Hu Y, Deng W, Tian Z, Dong C, Sun B. A novel NF-κB binding site controls human granzyme B gene transcription. J Immunol. 2006;176:4173–4181. doi: 10.4049/jimmunol.176.7.4173. [DOI] [PubMed] [Google Scholar]