Abstract
Retinoic acid (RA) is a terpenoid that is synthesized from Vitamin A/retinol (ROL) and binds to the nuclear receptors retinoic acid receptor (RAR)/retinoid X receptor (RXR) to control multiple developmental processes in vertebrates. The available clinic and experimental data provide uncontested evidence for the pleiotropic roles of RA signalling in development of multiple embryonic structures and organs such eyes, central nervous system, gonads, lungs and heart. The development of any of these above-mentioned embryonic organ systems can be effectively utilized to showcase the many strategies utilized by RA signalling. However, it is very likely that the strategies employed to transfer RA signals during cardiac development comprise the majority of the relevant and sophisticated ways through which retinoid signals can be conveyed in a complex biological system. Here, we provide the reader with arguments indicating that RA signalling is exquisitely regulated according to specific phases of cardiac development and that RA signalling itself is one of the major regulators of the timing of cardiac morphogenesis and differentiation. We will focus on the role of signalling by RA receptors (RARs) in early phases of heart development.
Keywords: Retinoic acid, heart development, RAR, RXR, ALDH1A2
1. INTRODUCTION
In vertebrates, the signalling pathways that convey intracellular signals arising from changes in the extracellular RA status always operate with a considerable degree of redundancy. One of the reasons behind this redundancy is the fact that the two nuclear receptor transcription factors that bind retinoids with high affinities, i.e. retinoic acid receptor (RAR) and retinoid X receptor (RXR), are encoded each by very similar gene paralogs (i.e. the paralogs RARα, RARβ, RARγ and RXRα, RXRβ, RXRγ), which arose from sequential duplication from ancestral RAR and RXR genes [1, 2]. Because expression of each of these paralogs displays appreciable overlap in embryonic tissues of mice, chicken and zebrafish, there is hardly, if at all, a cell in the vertebrate embryo that does not possess one, two, or more of these nuclear receptors [1, 3-7]. This ensures that fundamentally all vertebrate embryonic cells can respond to RA, a notion easily derived from the inspection of RA reporter embryos treated with pharmacological doses of retinoids [8, 9].
The redundancy of RAR and RXR paralogs was systematically established by a series of gene targeting and breeding approaches, which provided a highly detailed and much more realistic view of the roles played by RARs in development than achieved with RA teratogenesis alone (reviewed in [10]). For heart development, but not necessarily for the development of other organ systems and tissues (e.g. the developing eye), the complexity of RAR signalling can be reasonably approached using a few simplifying assumptions, which will be discussed below. However, it is important to state that there is one fundamental generalization worth approaching this early, namely: if nearly all embryonic cells can and do respond to RA, then the focus of regulation in this system must reside in other elements of the signalling pathway.
After 15 years of intense research, it is clear that local RA signalling is fundamentally controlled through enzymes that produce, or that inactivate retinoids [11]. Therefore, in this review, we will focus on the various sequential roles that are globally played by all RA receptors during early cardiac development, taking care to mention the cases in which specific roles have been characterized for individual RAR and RXR paralogs.
Why cardiac development is an attractive scenario for nuclear receptor biologists?
Perhaps the best reason to use the cardiovascular system as a model for vertebrate retinoid biology is the terminality of the phenotypes associated with its early developmental disturbances, which betrays the crucial ontological importance of this system and provides unequivocal opportunities for sensitive assays and rescue experiments. This is because, in contrast to other chordates such as tunicates and cephalochordates, vertebrates evolved a peculiar model of circulatory system in which circulatory work is centralized in the chambered heart, which is a highly efficient pump [12]. This chambered configuration is so special that apparently only one other group of animals, the mollusks, adopted a similar design for its circulatory pumps [13, 14]. Moreover, it is only fair to say that research on vertebrate cardiovascular development has progressed to a point in which the major players have been identified and the major interactions between them have at least been sketched, a state of affairs, perhaps, second only to research on the developing spinal cord [15, 16].
2. AN EVO-DEVO PRIMER OF RA SIGNALLING
2.1 The origins of RA signalling
Theodosius Dobzhansky's quip that nothing in biology makes sense except in light of evolution is widely known. In fact, often, the chronological sequence of appearance of important biological characters offers clues about the internal workings of modern regulatory mechanisms. With this in mind, it is safe to say that signalling by RA preceded, by far, the evolution of advanced chambered pumps of vertebrates. How do we know that? One critical clue comes from the well-known roles of aldehyde dehydrogenases (ALDHs) in the detoxification of biogenic or xenobiotic aldehydes. These aldehydes constitute an obvious menace to biological systems due to their capacity to create protein adducts and to thus modify protein function [17]. It just so happens that ALDHs also play important roles in the biosynthesis of signalling compounds [18]. The classic example is the retinoid synthetic pathway, in which the progressive oxidation of retinol (ROL) is completed with formation of RA from retinaldehyde, a reaction catalyzed in vertebrates by retinaldehyde dehydrogenases (ALDH1s). Therefore, based on the classic roles of ALDHs in detoxification, Yoshida et al. (1988) [17] proposed that the signalling roles of ALDH1s evolved from an ancestral enzyme primarily equipped for protection against toxic aldehydes. Many years after this thoughtful suggestion, Sobreira et al. (2011) [19] performed 3D reconstructions of the eukaryote ALDH1/2 ancestor and showed that this putative enzyme was indeed similar to the present-day vertebrate ALDH2, a liver enzyme crucial for acetaldehyde degradation and a target for therapeutic drug development against ischemia [20]. Thus, the current evidence suggests that the first metazoan ALDH enzyme with the capacity to perform RA synthesis arose shortly after the split of fungal, plant and animal lineages, roughly 1.5 billions of years ago [21].
How about RARs? Very recently, the dogma that RA signalling was a chordate invention was definitively superseded by demonstrations that RAR genes are present in non-chordate deuterostomes such as echinoderms, and in protostomes such as mollusks and annelids [22-25]. Therefore, there is solid evidence that RARs were already part of the regulatory complement of the last common ancestor of animals with bilateral symmetry (a.k.a. Urbilateria). Reconstruction of the ancestral chordate RAR reveals that this putative gene had, among other characteristics, an expression pattern similar to the expression profile of present-day RARβ [4, 26]. This led Escriva et al. (2006) to propose that, upon duplication and divergence from an ancestral vertebrate, RARβ-like, RAR ancestor, RARα evolved widespread embryonic expansion, while RARγ underwent an important restriction of expression domains in mammals [1]. The evolutionary analysis of Escriva et al. (2006) is consistent with the major roles in cardiac development established for RARα and the less prominent roles of RARβ and RARγ ([27, 28] and see below).
Why are these evolutionary considerations important? We believe that due consideration of the role of evolution in shaping and sophisticating the vertebrate body plan will reveal that there is no justification for sweeping conclusions about the role of RA signalling in the development of the heart or, by that matter, of any vertebrate organ. As such, there is no meaningful answer to the question of what is “the” function of RA in vertebrate heart development.
Most extant vertebrates possess genomes that underwent one or two, sometimes three, rounds of duplication [29] and massive proliferation of repetitive sequences [30, 31]. These are processes known to be conducive to evolution of new gene functions (neofunctionalization), distribution of ancestral functions among duplicates (subfunctionalization) and emergence of novel gene regulatory networks [31, 32]. Thus, it should not be too difficult to accept that, at least in principle, RA signalling may have as different roles in cardiac development as signalling pathways such as Wingless-related MMTV Integration site (Wnt), Fibroblast Growth Factors (FGF), Bone Morphogenetic Proteins (BMP) and others.
If we fast forward evolution from the metazoan ancestor to the emergence of chordates and vertebrates, we will have no difficulty in realizing that RA signalling seems to be first deeply involved with the demarcation of the posterior embryonic quadrant in early chordate development [33-36] and then with eye function and morphogenesis in vertebrates [37]. Therefore, the comparative study of chordate embryonic RA signalling suggests that the multiple roles of RA signalling in vertebrate development were derived from the above-mentioned ancestral functions.
Even a casual inspection of the published patterns of expression of RA synthesis enzymes will indicate that, in addition to the above-mentioned, presumed, ancestral functions, RA signalling has been called into action many times more. This is true even if we restrict our analysis to the earliest and more relevant RA synthesis enzymes during development [34, 38]. Thus, we can only conclude that it is very likely that RA signalling was recruited multiple times during embryonic ontogenesis to serve distinct functions, not only in the development of different individual organs, but also to execute different roles in the development of a single organ, for which, perhaps, the heart is one of the best characterized examples [39-42].
In summary, what we learned from the last 15 years of studying development is that RA signalling is a multi-faceted and dynamic system characterized by impressive timing and display of multiple windows of opportunity, the latter also a hallmark of embryonic development itself.
3. BASIC FACTS ABOUT RA SIGNALLING
The pathways of retinoid synthesis and degradation have been the subject of some excellent recent reviews [43-45]. Here our objective is to provide the essentials to understand the role of RA signalling in early heart development.
3.1 Signals
The major signals in the RA signalling pathway are conveyed by all-trans-RA (at-RA) and 9-cis-RA (9C) isomers. While at-RA binds both RARs and RXRs, 9C-RA can only bind RXRs [46]. Clearly, at-RA is the relevant player in vertebrate development, while 9C-RA has been reported to be detected in the adult mouse pancreas [47]. Despite that, 9C-RA has not been detected thus far in embryonic tissues and its physiological role in embryonic development is unclear (reviewed in [22]).
3.2 Synthesis
The first reaction in the final oxidative pathway leading to RA is the conversion of retinol into retinaldehyde [48], while the second and final synthetic reaction is the production of RA from retinaldehyde. Both alcohol dehydrogenases (i.e. ADH enzymes) and/or short reductases dehydrogenases (i.e. SDR enzymes) can perform the first step. In general, ADH enzymes are broadly distributed in the early embryo and their knockout alleles display only a few minor developmental defects. SDRs, on the other hand, seemed to be expressed too late in ontogenesis to display any key developmental roles [49-52] (but see below).
Because ADHs had been demonstrated to be critically involved in protection against retinol overload, rather than in the synthesis of developmentally-required RA, and SDRs appeared to be more relevant to the storage of retinoids in the form of retinyl esters [43], we and others have reasoned that the focal point of RA synthesis must be the conversion of retinaldehyde into retinoic acid by ALDH1As (a.k.a. RALDHs). In fact, ALDH1A1, 2 and 3 are expressed in domains that nearly match those of RA signalling [22, 34, 41, 53, 54], with ALDH1A2 clearly displaying a broader tissue distribution ([22] and references therein; [55]). Consistent with this, Aldh1a2-null mouse embryos die very early in development, in contrast with Aldh1a3 knockouts, which die at the peripartum, and Aldh1a1 knockout animals, which are viable [56-58]. When examined from this angle, the fact that the teleost fish Oryzias latipes (Medaka) completes its entire morphogenesis, grows, survives and reproduces with ALDH1A2 as its only retinaldehyde dehydrogenase is entirely consistent with the central role played by ALDH1A2 in vertebrate development [59].
However, once more demonstrating that scientific truths are only provisional, the above-mentioned scenario of RA regulation was destined to be changed by work on RDH10, an SDR enzyme. Taking advantage of an ethylnitrosourea (ENU) mutagenesis screen, Sandell et al. (2007) demonstrated that RDH10 was absolutely required for embryonic development, proving that the conversion of retinol into retinaldehyde was an essential step in embryonic RA biosynthesis. The first targeted alleles of Rdh10, however, were hypomorphs that produced partial phenotypes, which did not include cardiac defects [55]. Thus, these initial results did not authorize the inclusion of RDH10 in the list of relevant players in cardiac RA signalling. Further investigation with a new set of alleles has shown that Rdh10 is indeed essential for heart development. The novel phenotypes are reminiscent of those reported for Aldh1a2 knockout embryos, which include smaller atria, consistent with the role of RA signalling in atrial development [60-63].
However, it must be stressed that efficient targeting of the Rdh10 locus does not eliminate RA synthesis in the mouse embryo, which suggests that there are still additional and relevant ROL-oxidizing activities to be discovered. Consistent with this, DHRS3, a second SDR enzyme related to RDH10, which catalyzes the reduction of retinaldehyde, was recently shown to play a critical role in the prevention of RA excess. As a result, Dhrs3−/− mice display defects in ventricular and atrial septation and outflow tract formation [64] and are not viable.
As it is the case with RDH10, disruption of the expression of DHRS3 does not eliminate retinaldehyde reductase activity in mouse embryonic fibroblasts (MEF) derived from Dhrs3−/− embryos [65]. Therefore, there are other enzymes that could contribute to the interconversion of retinol and retinaldehyde in vivo during embryogenesis but, ultimately, their activity does not compensate for the lack of RDH10, or DHRS3, respectively [66]. Importantly, RDH10 and DHRS3 have been shown to form a functional heterodimer and augment each other's catalytic activity resulting in further fine tuning of the interconversion of retinol and retinaldehyde [65].
Work on RDH10 and DHRS3 has indeed demonstrated that the interconversion of retinol and retinaldehyde is regulated and critical for cardiac development, in particular, and for embryonic development, in general. These studies have also demonstrated that during embryogenesis RDH10 and DHRS3 are the primary enzymes responsible for oxidation of retinol, or reduction of retinaldehyde, respectively. However it is important to note that the most severe expression of RA signalling impairment remains the genetic abrogation of Aldh1a2, which reaffirms the value of using the Aldh1a2 gene as a handle to understand the role of RA signalling in development and evolution [61, 67].
3.3 RA degradation
RA is degraded to inactive, or partially active, metabolites by P450 enzymes, mainly by CYP26A1, B1 and C1, although CYP3As and CY2Cs also metabolize at-RA (reviewed in [68, 69]). CYP26A1 and B1 have high catalytic activity towards at-RA, but lower activity toward 9C-RA, while CYP26C1 processes at-RA and 9C-RA equally well. The major metabolic products of at-RA oxidation are 4-OH-at-RA, 4-oxo-at-RA, 18-OH-at-RA and 5,6-epoxy-RA, while the major oxidative metabolites of 9C-RA by CYP26C1 are the corresponding 4-OH-9C-RA and 4-oxo- 9C–RA [70-75].
RA oxidation products, 4-oxo-, 4-OH-, 18-OH- and 5,6-epoxy-RA, can bind RAR with relevant affinities, retain the potency to transactivate RA reporters in cell culture and may be potent teratogens [76, 77]. However, convincing genetic evidence suggests that the teratogenic phenotypes produced by excess activation of RA signalling reflect abundance of RA per se, rather than increased availability of its catabolic products [78]. Now, it is generally accepted that the abundance of RA during development is dominantly determined by the balance between its synthesis by retinaldehyde dehydrogenases (ALDH1As) and by its degradation through CYP26s.
3.4 Emerging players in RA signalling
3.4.1 Synthesis
Other enzymes involved in RA biosynthesis are ALDH8A1 (a.k.a RALDH4), which is inactive, or performs poorly with all-trans retinal (at-RAL), but does process 9C retinal to 9C-RA with high activity and converts 13-cis retinal to13-cis RA with low activity [79, 80]. Some cytochrome P450 enzymes also catalyze the oxidation of retinal into RA. CYP1A2 and CYP3A6 are capable of oxidizing at-RAL into at-RA [81], while CYP2J4 is active toward at-RAL and 9C-retinal, producing the corresponding RAs [82]. Chambers et al. (2007) described CYP1B1, an enzyme that can catalyze the two oxidation steps of RA biosynthesis, the conversion of at-ROL into at-RAL and the oxidation of at-RAL into at-RA [83].
3.4.2 Transport of RA precursors
In spite of RA's lipophilicity and small, but significant, water solubility [84], recently, it became clear that in some tissues, and at some developmental times, RA signalling utilizes a dedicated carrier system for facilitated, bidirectional, flow of retinoids across cell membranes. This carrier was identified as STRA6, first reported as a RA-stimulated cell-surface protein in cancer cells [85]. STRA6 is a widely expressed membrane receptor with 9 transmembrane domains [86], which does not bind RA directly, but, instead, is a receptor for both ROL-bound (holo), or ROL free (apo) retinol binding protein (RBP) [87]. STRA6 is expressed during embryonic development. In adults STRA6 is vigorously expressed in brain, spleen, kidney, female genital tract and testis, but much less so in heart, or lungs. Early, S35 radioactive in situ hybridization experiments did not suggest significant expression in the embryonic heart [85], which is somewhat puzzling, given the abundance of cardiac defects in humans suffering from the Matthew-Wood/PDAC (Pulmonary hypoplasia/agenesis, Diaphragmatic hernia/eventration, Anophthalmia/microphthalmia, and Cardiac defect) Syndrome, which has been associated with Stra6 mutations [88]. Recent work with Stra6 knockout mice indicate that ocular, not cardiac phenotypes, constitute the major manifestations of STRA6 deficiency [89, 90], which is consistent with the lack of significant STRA6 expression in the embryonic mouse heart [85]. In contrast to mice, morpholino-induced STRA6 deficiency in zebrafish is associated with cardiac defects such as atrial chamber dilatation and lack of looping. However, it remains possible that these changes in cardiac morphology are secondary to the edema displayed by STRA6-deficient zebrafish embryos, which, by itself, can result from changes in cardiac muscle function, rather than from direct interference with morphogenetic processes.
3.5 RA receptors
3.5.1 Anatomy of a receptor
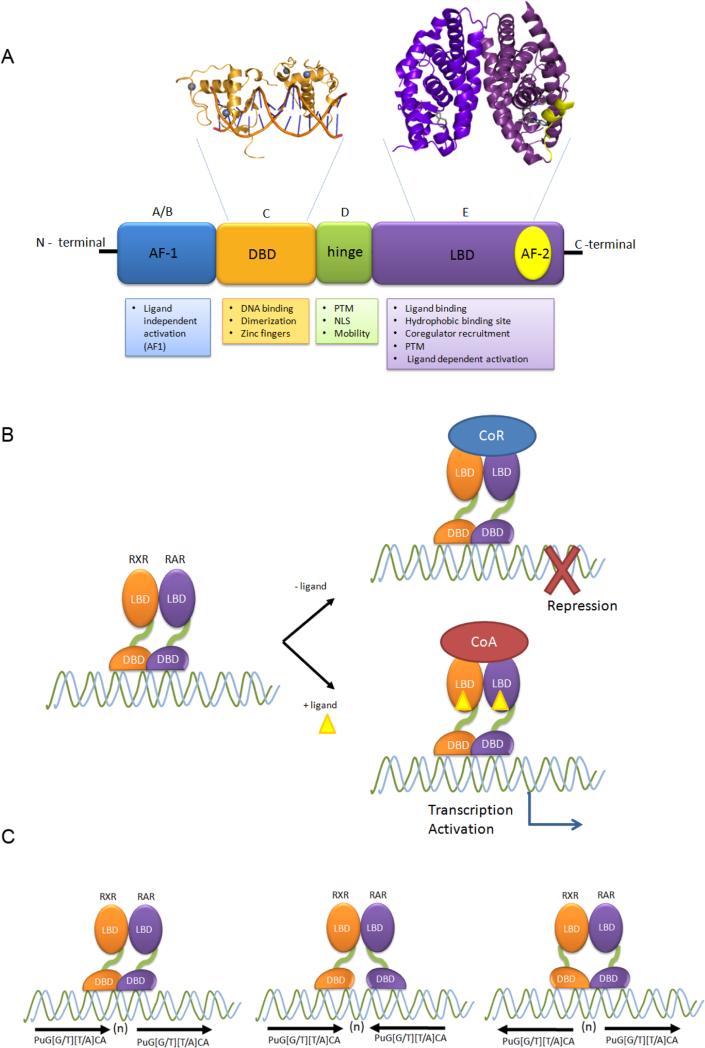
As other nuclear receptors, RAR and RXR are modular proteins that share a multi domain structure (Figure 1A). The N-terminal domain (A/B domain) is the least conserved domain and contains the ligand-independent activation function-1 (AF-1). This is the only domain for which there is no structural information, consistent with biophysical data that suggest that the N-terminal domain is a disordered structure [91].
Figure 1. Schematic representation of RAR structure and RXR/RAR heterodimers binding sites.
A) Multi-domain representation of RAR architecture with details of DBD and LBD tridimensional structures. As the majority of the nuclear receptors, RAR is composed by N-terminal domain (A/B domain, blue), which comprises the ligand independent AF-1; followed by the DNA binding domain (C domain, or DBD, light orange), which displays two zinc finger motifs, is responsible for DNA recognition, and has some dimerization properties. The hinge (D domain, green) is highly variable among NRs and contains nuclear localization signal (NLS). The hinge domain is a target for post-translational modifications (PTM). In RAR the Hinge domain is unusually long, which provides higher degrees of freedom for positioning of DBD and LBD. The last domain (E domain, or LBD, purple) displays a hydrophobic binding site, which is responsible for ligand binding and for conformational modifications that setup the ligand-dependent AF-2 domain (yellow), which acts by recruiting coactivators (PDB Ids: DBD - 1DSZ, LBD -1DKF). B). The RXR/RAR heterodimer binds to DNA in absence of ligands. In this configuration it recruits corepressors and inhibit expression of target genes. In the presence all-trans retinoic acid, 9-cis-retinoic acid, or of both, the RAR-RXR heterodimer underwent conformational changes that lead to corepressor dissociation and to coactivator recruitment and recruits coactivators, which ultimately increases target gene expression. C) Generally, RAR binds DNA as heterodimers with RXR. RA receptor DNA binding sites display numerous different orientations of repetitions of the consensus sequences PuG[G/T][T/A]CA organized in direct repetition [(→(n)→], palindromes [→(n) ←] and inverted repeats [←(n)→], including widely spaced (up to 150 nucleotides) direct repeats.
The DNA binding domain (DBD or C domain) is the most conserved among all the NRs, being well characterized in terms of structure and function. The DBD displays two zinc fingers coordinated by 8 cysteines, which bind and recognize stereotyped DNA sequences and, sometimes, are important for dimerization. [92-96].
The ligand binding domain (LBD, or E domain) is a multifunctional domain primarily responsible for ligand binding. LBDs present only modest sequence similarity, but display similar folding across all NRs. The RAR LBD is composed of 12 α-helices in an antiparallel sandwich [91, 93, 94, 96-98]. RA binding sites in RARs are highly hydrophobic and can accommodate multiple RAR agonists and antagonists. The LBD features a key activation function (AF-2), which is ligand-dependent and responsible for agonist-induced conformational changes. In RAR, as in most NRs, the position and conformation of helix 12 (H12) is key to the processes of corepressor dissociation and coactivator binding, which in turn control the shift from inactive to active receptor states. [91-98]. The LBD also contains the main dimerization interface, which contains H7 and H11, as well as sites for post-translational modifications, which may modulate recruitment of coregulators. Between C and E domains there is a hinge (D domain), which is variable among all the NRs. The hinge domain confers mobility between DBD and LBD, displays nuclear localization signals and sites for post-translational modifications [92, 93, 95]. Interestingly, RAR possesses a very long and unstructured hinge region, which may underlie its versatility to bind a host of different and widely-spaced DNA sites (see below).
3.5.2 RAR at work
Generally, RA receptors are bound to DNA in the nucleus, even in the absence of ligand. For these and, as a matter of fact, for most NRs, the key seems to be the switch between unliganded states (receptor not bound to ligand), which attract complexes of transcriptional co-repressors, and liganded states (receptor bound to ligand), which associate with co-activators of transcription (Figure 1B). It is worth noting that unliganded RARs have also been suggested to be actively involved in normal development through complexing with co-repressors [99, 100]. This is particularly evident, for instance, during Xenopus neural patterning [101], when recruitment of Nuclear Co-repressors (NCor1 or NCor2) is required to protect anterior structures from the posteriorizing effects of low levels of RA. In fact, there is nothing exotic about the role of unliganded receptors, if one considers that all gene transcription is controlled through the integration of multiple inputs. In other words, transcriptional regulation does not have to be (and rarely is) just on and off, it can also be much less than basal, which is a relevant issue for the well known inverse agonist function of some RAR antagonists [102-105]. Therefore, RA deficiency does not equate with a lack of RAR signaling, but reflects a shift in the nature of RAR signaling towards reduced activation and increased repression of target genes.
Our knowledge about the specific DNA sequences that bind nuclear receptors derived from initial work on steroid hormone receptors [106]. Mutational, gel shift, footprinting, as well as selection approaches, showed that it was possible to narrow down steroid response to repetitions of 5′ – PuG[G/T][T/A]CA – 3′ sequences, the core binding site for many nuclear receptors. This is the magic six-nucleotide sequence, often represented in the literature as variations of AGGTCA. Subsequent research showed that, more often than not, nuclear receptors work in pairs, as homodimers or heterodimers, with one monomer binding to a 5’ hexad, and another monomer binding to a 3’ hexad. However, hexads can in fact be arrayed in direct repetitions (DRs), palindromes, inverted (IRs) or everted (ERs) repeats (Figure 1C) [106, 107].
In early models, the binding specificity associated to non-steroid nuclear receptors was thought to be conveyed by the number of intervening nucleotides between 5’ and 3’ AGGTCA hexads. It was at this time that some generalizations were attempted, which were destined to have a profound effect on the perception of non-steroid nuclear receptor signal transduction in general, and on RAR signalling in particular. The initial evidence supported a scheme, the 3, 4, 5 rule, in which three intervening nucleotides between direct repeats of the AGGTCA were interpreted as a typical arrangement for binding of vitamin D receptors, the so-called DR-3 sites, or vitamin D responsive elements (VDREs). Four spacer nucleotides, DR-4, were associated with binding sites for thyroid hormone receptors (TREs) and five interceding nucleotides, DR-5, were linked to RARs (retinoic acid responsive elements (RAREs)) [106, 108-113], reviewed in [114] .
Things started to get more complicated when it came to RA receptors, because it was soon apparent that RXR:RAR heterodimers were able to bind direct repeats spaced by two (DR-2) and one (DR-1) interceding nucleotides, besides the “classic” DR-5s of the 3, 4, 5 rule. Apparently, this was only the tip of the iceberg of a far less well-behaved system than originally envisaged, because it became clear that RXR:RAR dimers could also bind to direct hexad repeats spaced by 0, 3, 8, 10, 13, 14, 15, and even to widely-spaced elements displaying more than fifty, sometimes, hundreds of intervening nucleotides [115]. Inverted repeats of 0, 5, 8 and 9 were also recognized as RAREs, as were complex elements combining direct with inverted repeats (DR2+IR1) [116, 117].
In contrast to classical RAR-RXR targets whose transcription is dependent on RNA Pol-II, a vast majority of potential DR2-type RARE (90%) in the human genome are associated with the short interspersed elements (SINEs) of the Alu-type [118]. Found primarily in primates, Alu are retrotransposons derived from the ribosomal 7SL RNA which are transcribed by RNA Pol-III. It was shown that RAR-dependent transcription of Alu repeats leads to the generation of a small RNA species, termed repeat-induced RNAs (riRNAs), which induce the degradation of specific RNAs that maintain the stem cell identity; thereby, riRNAs allow stem cells to differentiate. This interesting variation of an otherwise conventional mode of gene regulation by RAR requires processing of RAR-induced riRNAs by DICER and recruitment of riRNA by Argonaute 3 (AGO3) to silence stem cell mRNAs [119]. Though these findings have important repercussions on our understanding of the role of RAR and the differentiation of stem cells in primates, it is not yet clear whether the riRNA-mediated silencing mechanism is evolutionarily conserved in rodent animal models which harbor B1 instead of the Alu-type repeats.
To make matters more complicated, the specificity of RAREs is further corroded by the fact that other NRs also utilize some of the same spacer models used by RARs. For instance, orphan receptors such as COUPTF-II and COUPTF-I often bind to DR-0, DR-1, DR-2, DR-4, DR-6, DR8 and DR11 sites, in addition to other models [106]. Recently, a study with human breast cancer cell lines suggested that RAR binds to sequences that are coincident with palindromic binding sites for estrogen receptor and FOX1 [120]. More disconcertingly, some results appear contradictory. For instance, while chromatin immunoprecipitation (ChIP) assays in mouse embryonic stem cells showed that few of the RAR binding sites contain DRs [121], another study on RA induced-differentiation of embryonic stem cells into spinal motor neurons displayed a clear enrichment in classic DR2s and DR5s [122]. Collectively, these studies leave open the daunting possibility that RARs can bind different sites depending on cell type, context, and differentiation stage [123].
The puzzling variability of RAREs finds a clear counterpart in the structural organization of RAR. In contrast to other receptors such as VDR and PPAR, which display much less diverse panoply of responsive elements, RAR is well known to feature a long and unstructured hinge region, which may underlie its capacity to bind to extended and complex modes of responsive elements [117]. Further possible factors that underlie the versatility of RAR include additional contacts among flanking sequences of each hexad and other NR domains, such as the N-terminal domain and hinge, which increases the possibilities of interactions between protein and DNA sequences.
All, however, is not lost. Meticulous studies have shown that, notwithstanding the great potential for confusion, there appears to be some reasonable predictors for RAR binding and/or transactivation potential for a given RARE candidate. It is clear that the magic AGGTCA is a strong indicator of high affinity but, conversely, of very poor specificity. This is because the small sequence departures from the consensus AGGTCA do carry important information as to the binding preference for RAR, or, for instance, thyroid hormone receptors (TREs) [124]. For DNA binding itself, spacing seems to be almost unimportant, at least in the context of comparisons between RAR and TR. However, not all DNA sites that bind RARs display transcriptional activity and this is when an additional lease on life was given to the 3, 4, 5 rule, because efficient RA signalling requires DR-5, while effective TRE signalling is associated with DR-4, at least in the somewhat narrow context of cultured CV-1 cells [124].
From an analytical point, it is perhaps more satisfactory to think about the strength of a given candidate RARE as the result of several layers of regulation in which organism, cell type, cell state, hexad sequence and orientation, spacing, flanking nucleotides, chromatin and co-regulator milieu all potentially have a say in its regulatory potential. Therefore, before attributing RARE status to a candidate sequence, one ideally should first produce a robust set of observations with the aid of molecular, biological and biophysical methods (e.g. gel shifts, chromatin immunoprecipitation, mutations, fluorescence anisotropy) and functional approaches (e.g. expression analyses, reporter gene assays, transgenic animals). Unfortunately, the truth is that it has been proven far more difficult to characterize a bona fide RARE than a simple inspection of DNA sequences.
4. MULTIPLE RA SIGNALLING MECHANISMS
The goal of this section is to highlight that the signalling modes operated by RA in development are not limited to the traditional French flag scheme of concentration gradients [125, 126]. For that we will indicate when and where in development other modes of RA signalling have been suggested.
4.1 RA signalling gradients
Developmental biologists are all justly fascinated with morphogen concentration gradients as transmitters of information. Indeed, morphogen gradients offer elegant solutions for the specification of cell fates across a morphogenetic field. In activin signalling, for instance, it has been shown that cells turn on different sets of genetic programs according to the absolute number of occupied receptors, which is a direct function of local morphogen concentration [127].
Until very recently, the molecular mechanisms involved in information transfer from RA-dominated fields have been comparatively less understood due to the lack of sensitivity of assays for retinoids. Therefore, developmental biologists have turned to artificial reporter assays that measure activation of simple transcription units (e.g. basal promoters fused to multiple RAREs), or to the assessment of expression from endogenous target genes (e.g. homeobox genes) as surrogates for direct RA detection [8, 128]. Even with these relatively limited approaches it was already quite evident that RA could, directly, or indirectly, transmit information through large embryonic domains. Eichelle and Taller estimated that the radioactive-labeled retinoid agonist TTNPB could diffuse about 500 μm of embryonic tissue in 3.5 hours [129], while White et al. demonstrated that RA can activate transcription of a reporter gene in cells 70-300 μm away from a loaded bead [9].
RA signalling gradients are typically established at the borders of ALDH1A expression domains, which act as intense sources of RA. This RA is thought to diffuse into ALDH1A negative tissue and give rise to concentration profiles. Passive diffusion mechanisms are often compounded by addition of an active sink for RA in the form of neighboring expression domains for CYP26s and this is thought to generate steep concentration profiles in tissues located between ALDH1A and CYP26 domains. The first indication that these source and sink arrangements for RA exist came when Swindell et al. (1999) demonstrated that the tissue fated to give rise to posterior chicken hindbrain is sandwiched between a posterior domain of ALDH1A2 in the mesoderm and an anterior domain of CYP26A1 in the ectoderm [72]. Subsequently, Reijntjes et al. showed that the mesodermal layer expresses its own RA-degrading enzyme, CYP26C1, which, likely, constitutes a more important sink for RA produced in posterior mesoderm than the ectodermal CYP26A1 domain. [130]. A more sophisticated and dynamic version of these source and sink RA signalling gradients has been suggested by studies showing that normal and abnormal gene expression profiles and morphological patterns are established in the hindbrain and in pharyngeal arches when the domains of ALDH1A2 or CYP26s move in relation to their original limits [131, 132].
After years suffering “gradient snobbery” from its sister pathways such as Sonic hedgehog (Shh), Wnts, BMPs, FGF and others, the RA signalling pathway was, again, rescued from backstage. Using an elegant approach based on FRET (Förster resonance energy transfer/Fluorescence resonance energy transfer) signals from engineered mouse RARs expressed in transgenic zebrafish to measure free intracellular RA, Shimozono et al. (2013), showed that there are two major RA gradient zones in the embryo, one anterior and other posterior [133]. The two gradient zones lie between the extremes of an ALDH1A2-expressing source region represented by somitic mesoderm and CYP26-expressing sink regions at cranial and caudal embryonic ends. It is possible to see a cranial zone of approximately 200 μm in which the FRET signal gradually increases from very low levels at the cephalic end, as well as a caudal zone of approximately 250 μm, in which the FRET signal gradually decreases between the unsegmented paraxial mesoderm and the tail bud [133]. In summary, these new developments leave little doubt as to the capacity of RA signalling to transmit information through extended embryonic domains via concentration gradients.
4.2 Beyond signalling gradients
As hinted before, the fact that RA signalling gradients do exist and play fundamental roles in embryonic patterning does not, however, exhaust all modes of signalling by this versatile system. In fact, it is hard to believe that the vast domains of ALDH1A expression and RA synthesis exist only to give rise to signalling gradients at their borders (e.g. [8, 9, 133]). It seems likely that when RA synthesis is activated to high levels in tissues such as somites, or in unsegmented paraxial mesoderm, RA signalling is also playing major roles within these tissues. Clear examples of that were provided in the context of neural development, in which a RA rich region, anterior to the neural stem cell zone in the tail bud, effectively pushes neural progenitors out of the cell cycle and directs them towards the neural differentiation pathway [134]. In summary, today it is clear that RA conveys information by way of multiple strategies.
4.3 Field signalling
An important example of this signalling mode occurs in the paraxial mesoderm, where RA works to keep FGF signalling in check [135], to synchronize somite formation [136] and to prevent apoptosis [137]. Another example is the forming eye at the optic cup stage, when ALDH1A2 is expressed in a large domain that generates a RA signal necessary for the late, restricted expression, of ALDH1A1 and ALDH1A3 in the dorsal and ventral retina, respectively [138]. RA signalling fields can also be utilized to establish morphogenetic limits that restrict the development of structures along embryonic axes. Morphogenesis of the pharynx in amphioxus [139, 140] and cardiac specification in vertebrates (see below) are interesting examples of how expression domains of RA receptors, or of synthesizing enzymes, can set up fields of RA signalling that restrict gills slits, or heart to anterior embryonic regions, respectively.
4.4 Drive-through signalling
A peculiar signalling strategy is utilized in spinal cord development, where the lateral subset of neurons of the lateral motor column (LLMC) is specified by RA when they migrate through a field of intense RA signalling established by medial lateral motor column neurons (MLMC), which intensely express ALDH1A2. It has also been proposed that the very early steps of cardiac anterior-posterior (AP) patterning by RA involve a drive-through type of signalling, when ingression of epiblast through the posterior heart-forming region of the primitive streak, at full streak stage, produces heart precursors that migrate through a field of RA synthesized via ALDH1A2, which is thought to specify sino-atrial fates in these cells [141]. Thus, in this unusual strategy, cells that do not express ALDH1A2 may be transiently exposed to intense RA signalling and acquire their definitive fates while on route to their definitive embryonic quarters [142].
4.5 Dynamic signalling
A dynamic signalling mode in which ALDH1A2 expression is turned on and off in different regions of mesenchyme and epithelia is represented in lung development. At early, pre budding stages, ALDH1A2 expression is high in the mesenchyme of the prospective trachea and lung primordia, while it is absent from respiratory epithelium. From then on, ALDH1A2 expression is downregulated in the distal pulmonary mesenchyme, becoming concentrated at proximal mesenchymal sites. At distal lung sites, ALDH1A2 expression is observed only in the pleural mesothelium in shifting patterns that inversely correlate with sites of FGF-10 expression. These quickly changing expression patterns are thought to control the sites of pulmonary branching by inhibiting FGF-10 expression [143, 144]. More recently, the dynamic patterns of activation/deactivation of RA signalling in the mesenchyme during branching morphogenesis of the lung have been associated to the inhibition of airway smooth muscle differentiation [144]. The effects of RA upon lung airway smooth muscle differentiation constitute an interesting parallel to its effects upon coronary smooth muscle [39].
4.6 The role of CYP26s in RA signalling
As with its function in establishing RA signalling gradients together with ALDH1As, CYP26 enzymes are also key players in other forms of RA signalling. Often, CYP26s are expressed in domains that are complementary to the regions of ALDH1A expression. In these domains the role of CYP26 expression seems to be protection of embryonic regions that do not express ALDH1As from the effects of RA diffusing from neighboring regions [145, 146]. In line with this idea, the reciprocal expression of CYP26 and ALDH1As has also been interpreted as the reason why local deficiencies in RA signalling can sometimes be rescued by artificial procedures such as systemic administration of small doses of retinoids [11, 147]. Moreover, in some tissues such as the zebrafish hindbrain, the redundant action of CYP26A1, CYP26B1 and CYP26C1s, rather than ALDH1A expression, has been proposed to control RA signalling mostly because exogenous treatments with low doses of RA can rescue normal development. More recent findings show that CYP26 enzymes are not only expressed in at-RA-free boundaries, but also in at-RA-responsive tissues. Thus, the pattern of expression of CYP26A1 can create more than just RA-present and RA-absent zones but also shape zones in which RA-forms gradients [9].
Here we argue that, rather than to look for evidence of ALDH1A, or CYP26 dominance in RA signalling, perhaps, it is better to think of the recurrent, but varied modes of RA synthesis and degradation in embryonic development, as gene regulatory programs selected by millions of years of evolution in the multiple vertebrate groups. Some programs are important enough to be conserved in their essence, while others have been extensively modified on the account of the different forms associated and/or required for survival in specific ecological niches.
5. AN EVO-DEVO PRIMER OF HEART DEVELOPMENT
5.1 Heart evolution
There is plenty of evidence suggesting that vertebrate (and mollusk) chambered hearts derived from a very simple ancestor contractile vessel. In this view, the ancestor of the chambered heart was a rather humble layer of coelomic-derived contractile myoepithelial, or myocyte, cells that spread through primitive, endothelial-free, communicating channels in the extracellular matrix (ECM) [13, 148]. Figuratively speaking, this ancestor vessel was nothing more than a jacket of muscle enclosing a cylindrical hole in the ECM. These early origins of the heart are still echoed by the ontological sequence of cardiovascular development in vertebrates. Indeed, consistent with this, it is quite clear that the first contraction of the heart is recorded only after morphogenesis of the whole basic circulatory network is completed.
5.2 A brief synopsis of cardiac development
The anatomical development of the heart is the result of the integration of continuous signalling and morphogenetic events that, for didactic convenience, are presented discretely. Although the conceptual divisions are arbitrary, it is interesting to compartmentalize developmental processes according to early and late events, mostly because these divisions are useful for understanding the types of malformations that arise during the various stages of intrauterine and perinatal life in mammals. Thus, the disturbances of the early events are so severe that they are almost invariably associated with embryonic death and only rarely are observed as a cause of congenital heart disease in humans. On the other hand, disturbances in late events, although severe, often do not translate as an immediate threat to the survival of the embryo or fetus within the protected uterine environment. Therefore, these defects will only manifest during or after the drastic transition to extra-uterine life, thus constituting a group of diseases referred to as congenital heart disease, which will not be covered here.
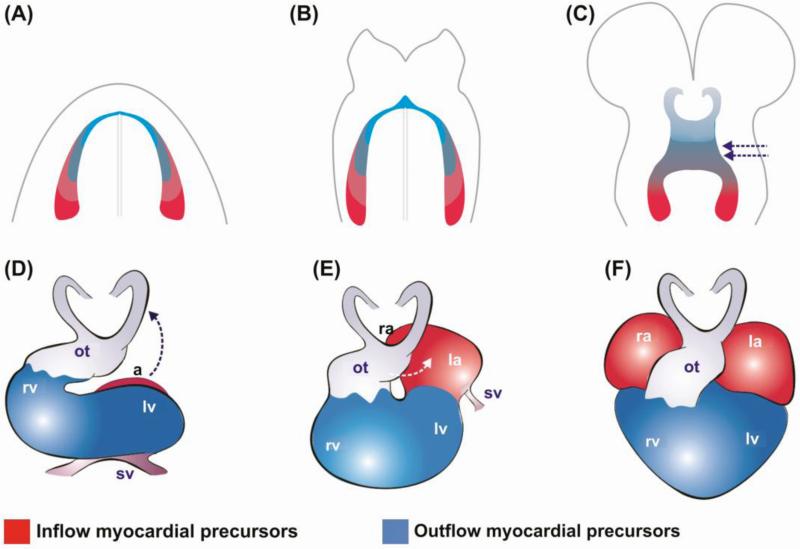
In a nutshell, early heart development requires two major feats of morphogenesis from cardiac progenitor cells that are located in bilateral sheets of post-gastrulation embryonic cells in the cardiogenic lateral mesoderm: 1) the wrapping around of this two-dimensional sheet of cells to form a tridimensional structure, the primordial heart tube (Figure 2A-C); 2) the transformation of this primordial tube into a multi-chambered organ (Figure 2D-F).
Figure 2. Schematic representation of early amniote heart development.
A) During heart development, bilateral cardiac progenitor fields meet above the anterior intestinal portal, forming the so-called cardiac crescent. Inflow (sino-atrial) precursors (red) are localized in the posterior region of the cardiac crescent, while outflow precursors (ventricle and outflow tract) (blue) are located in the anterior region. B) Cranial migration of cardiac precursors is followed by morphogenesis into a midline heart tube. C) Formation of a straight heart tube is followed by the first heartbeat, which initiates the embryonic circulation. All the subsequent morphogenetic events will transform the heart from a peristaltic tube into a new pumping organ, the chambered heart. D) The complex events that lead to cardiac chamber morphogenesis are initiated by a rightward loop of the tube, which places the future right ventricle and the outflow to the embryonic right side (dashed blue arrows), while positioning left ventricular progenitors to the left side. During looping, the heart elongates by addition of myocardial cells at its two poles. After looping is completed, the embryonic heart assumes an “S” shape configuration. At this stage, sino-atrial precursors move dorsally and cranially towards the outflow tract (blue dashed arrows), in the so-called convergence movements E) After convergence, the common outflow tract is further positioned, so that the regions fated to give rise to pulmonary artery and aorta are brought in close contact with the corresponding pulmonary (right) and systemic (left) ventricles. F) Final heart configuration. (ot) outflow tract; (a) atrium; (rv) right ventricle; (lv) left ventricle; (sv) sinus venosus; (ra) right atrium; (la) left atrium. Modified from [149].
A detailed description of cardiac development is not within our scope and the reader is directed to comprehensive reviews [149, 150]. However, it is possible to depict the essence of cardiac development as a succession of dynamic and interconnected events. These include: a) specification of the cell lineages that will form the heart; b) the alignment of cardiac progenitor tissue to anterior-posterior, dorso-ventral and left-right embryonic axes; c) crucial morphogenetic movements and proliferation patterns that together transform the linear, peristaltic, heart tube into a chambered pump; expansion of the muscular ventricular wall; d) growth of valves and septae that will establish near perfect unidirectional blood flow; e) morphogenesis of the heart's own blood supply, the coronary circulation, and f) connection of the heart to the pulmonary and systemic circuits [149, 150].
6. CARDIAC DEVELOPMENTAL PROGRAMS CONTROLLED BY RA SIGNALLING
6.1 Specific roles of RARs and RXRs in cardiac development
As far as cardiac development is concerned, there is solid evidence that the major players are RARα and RXRα [27]. Individual and combined knockout phenotypes for these nuclear receptors produce phenotypes that include all embryonic defects associated with the classic vitamin A deficiency and some more [151]. Many of these effects can be understood as reflections of a critical requirement for RA signalling in the promotion of myocardial growth. For instance, the premature myocardial differentiation associated with insufficient RA signalling is correlated with decreased cardiomyocyte proliferation at the ventricular compact layer, which is directly in contact with the epicardium, an important growth-promoting tissue (see section 6.6). Reduced ventricular proliferation is probably a factor in the failure to grow myocardial trabeculae, which normally grow towards the cardiac lumen. Insufficient trabecular growth may also promote insufficient expansion of the myocardial wall, forming hypoplastic ventricular chambers and lead to the imperfect morphogenesis of the muscular interventricular septum, which, in turn, produces ventricular septal defects [27, 28, 152].
It is important to stress that, judging from knockout phenotypes, RXRα is involved in more critically-relevant cardiac developmental processes than RARα itself. Because RXR also plays a role as a heterodimer partner for other nuclear receptors and can, in cell culture, perform signalling of its own through specific agonists, it was important to establish to what extent RXRα had some direct signalling functions. By specifically targeting AF1 and AF2, or both domains from RXRα, Mascrez and colleagues [153] demonstrated that specific signalling via RXRα is not required for cardiac development. Therefore, it is possible to infer that, at least in most processes of cardiac development, RA signalling is frequently controlled by RXRα and RARα heterodimers and that the major roles demonstrated for RXRα are not due to specific RXR signalling, but are subordinated to its role as a heterodimerization partner for RARα.
6.2 Roles of RA signalling in early cardiac development
RA signalling is intimately involved in cardiac development. The sequential and multifaceted roles of RA signalling in cardiac development were noted when Moss et al. (1998) discovered that RA synthesis enzymes were expressed in the embryonic heart in multiple tissues and in a dynamic fashion. Using an antibody against the then newly-discovered ALDH1A2 enzyme and a mouse RA reporter line [8], the authors mapped its expression from 8.0 to 13.5 dpc mouse hearts and from HH4 to HH23 in chicken and quail embryos and concluded that there were two major developmental programs in the developing amniote heart: 1) an early program (8.0 to 10.5 dpc in mice and HH4-HH23 in chicken) activated in posterior cardiac precursors and linked to cardiac partition into inflow (sino-atrial) and outflow tissues (ventricles and outflow tract); 2) a later program activated in the ventricular epicardium and associated to myocardial growth and coronariogenesis. [13, 141, 154]. Here, instead of offering a chronological description of how concepts on cardiac RA signalling evolved, we will rather describe the ontological sequence of RA programs activated during cardiac development.
6.3 Allocation of fates to cell progenitors in the lateral mesoderm
Perhaps the earliest RA signalling program in heart development is progenitor size control (Figure 3A and Figure 4A). Retrospectively, clues to this role of RA in regulating the size of the cardiac progenitor field were already available from the early pharmacological experiments with exogenous retinoids given to chicken, or to zebrafish embryos [155, 156]. In these experiments, increasing concentrations of RA produced ever decreasing heart sizes, until in concentrations of micromolar order, no hearts could be identified. Consistent with this, treatment of Xenopus embryos with RA prior to cardiomyocyte specification abrogates expression of precardiac mesoderm markers such as nkx2.5 [157, 158]. However, at the time of these studies, it was unclear whether this effect of RA reflected a teratogenic or a physiological mechanism.
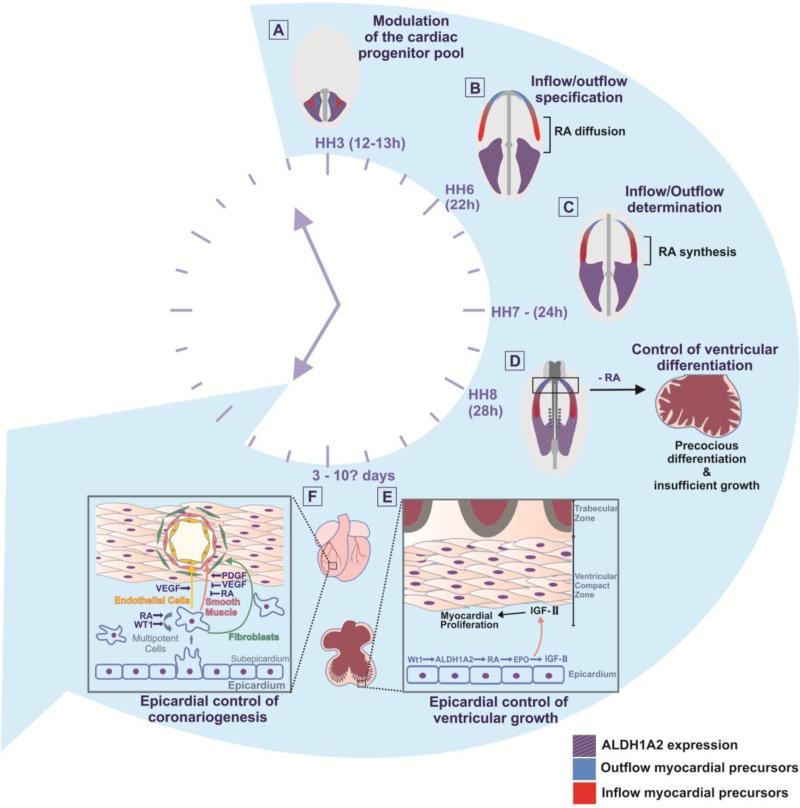
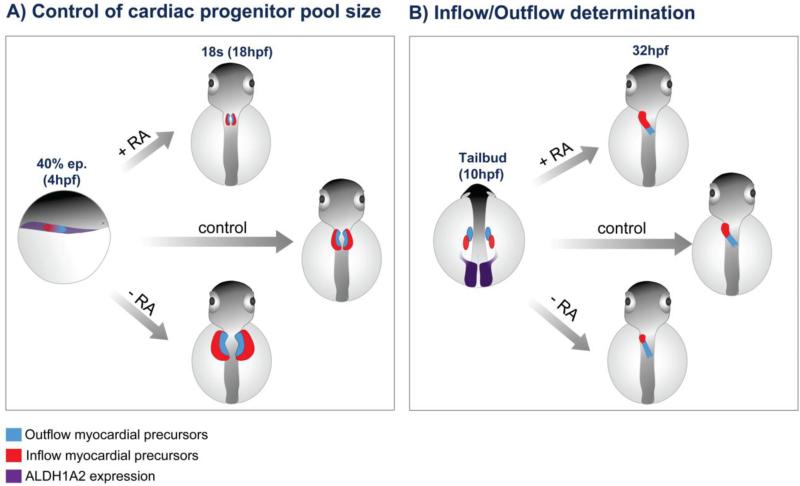
Figure 3. A timeline for the multiple roles of RA signaling during early heart development.
A) Heart progenitor size control. According to data first described in the zebrafish [159] one of the first roles of RA signalling is to control the cardiac progenitor field size at the blastula stage. In chicken embryos, a corresponding phase may be represented in stage HH3, when cardiac, as well as other mesodermal progenitors are still contained within the epiblast, which is in contact with ALDH1A2-expressing tissue. B) Inflow/outflow specification. In HH6 chicken embryos, ALDH1A2 expression (purple) is located in the posterior half of the embryo. RA signals that diffuse from this region form a signaling gradient and contact posterior cardiac precursors (red), specifying them to cardiac inflow fates. C) Inflow/outflow determination. At HH7, a caudorostral wave of ALDH1A2 expression reaches the posterior precursors, endowing them with the ability to synthesize their own RA. High RA concentrations typically associated with ALDH1A2-expressing tissues are thought to irreversibly commit posterior precursors to an inflow, sino-atrial, fate, while ventricular and outflow fates are determined in the absence, or in the presence of low levels of RA signaling. D) Early control of ventricular differentiation. At HH8-HH9 ventricular precursors (blue) are swept by the caudorostral wave of ALDH1A2 at a stage when they are already committed to their fate. At this stage RA signalling is thought to prevent precocious differentiation of ventricular myocardium and to maintain proliferative capacity at the levels required to build a thick ventricular wall and to give rise to a competent muscular septum between the ventricles. In a scenario of insufficient RA signalling (i.e. ALDH1A2 knockout, RXRα knockout, double RXRα/RXRβ knockout mice and vitamin A-deprivation) embryos display precociously differentiated ventricular myocytes and failure to form appropriate trabeculae. E) Epicardial control of ventricular growth. Later in development, at embryonic day 3 (i.e. E3), proepicardial cells start to envelop the myocardium to form the epicardium. Epicardial cells express the transcription factor Wilms tumour 1 (WT1), which in turn activates ALDH1A2 expression and, thus, RA synthesis. RA signalling stimulates the production of Erythropoietin (EPO), and consequent induction of IGF-II, which reaches the myocardium and stimulates myocardium proliferation. F) Epicardial control of coronariogenesis. The epicardium, formed at E3-E4, provides a source of RA signalling that is involved in the maintenance of epicardial-derived cells in an undifferentiated state, within the subepicardium. RA signals from epicardium also act together with Vascular Endothelial Growth Factor (VEGF) to delay smooth muscle differentiation. This delay is crucial to allow building and remodeling of the endothelial network before the final coronary mesh is stabilized by the aggregation of smooth muscle cells, which differentiate by inactivation of RA and VEGF signalling and stimulation of Platelet-Derived Growth Factor (PDGF). Modified from [163].
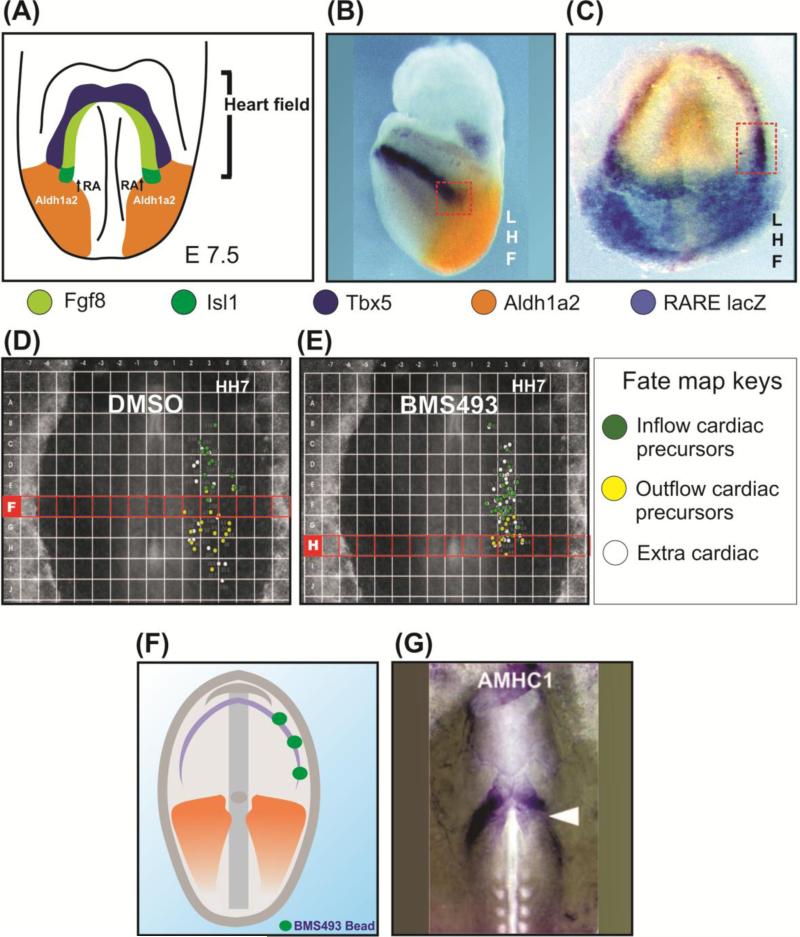
Figure 4. Multiple functions of retinoic acid (RA) signalling in posterior, sino-atrial, cardiac precursors.
A) The RA produced by ALDH1A2 was suggested to establish the posterior limit of a population of cardiac progenitors that expresses Isl-1 and/or FGF-8 (a.k.a. the Second Heart Field, or SHF), but not Tbx-5 (a.k.a. First Heart Field, or FHF). Primary data from [171, 172]. Diagram adapted from [173]. B) RA produced by ALDH1A2 was previously suggested to provide clues for the specification of inflow (sino-atrial) fates to posterior cells of the Tbx-5-expressing progenitor field (FHF). The red rectangle highlights the overlap between Tbx-5 and ALDH1A2 expression [40]. C) Double Tbx-5 in situ hybridization/beta galactosidase stain in a 7.5 d.p.c. (days post coitum) mouse embryo. The red rectangle highlights the overlap between Tbx-5 expression and beta galactosidase activity. This result indicates that cells in the caudal domain of the Tbx-5-expressing cardiac progenitor population (or FHF) activate RA signalling in response to local concentrations of the retinoid produced by ALDH1A2 [40]. D) Chicken cardiac fate map performed in the presence of DMSO (control). Cardiac outflow (ventricular and outflow tract) progenitors (green) and inflow (sino-atrial) progenitors (in yellow) are well separated in the anterior-posterior axis. Note that no outflow progenitors were mapped below line “F” [40]. E) Chicken cardiac fate map performed in the presence of the RAR pan-antagonist BMS493. The limits between outflow and inflow progenitors are blurred. Note that after RA signalling inhibition cardiac outflow progenitors are now mapped well below their usual domain, in line “H” [40]. F) Diagram representing unilateral (right side only) treatment of chicken embryos with agar cylinders containing BMS493 at HH6. G) Results of the experiment represented in F. Local inhibition of RA signalling in the embryonic domain occupied by the ALDH1A2 caudal to rostral wave prevents expression of the inflow marker AMHC1 in the treated side. This results indicates that the RA produced in response to the ALDH1A2 caudal to rostral wave is instructive to posterior cardiac progenitors [40]
These earlier results were placed in the context of zebrafish heart field size regulation when Keegan et al. (2005) showed that RA inhibition by a pan-RAR antagonist during gastrulation produced oversized cardiac precursor domains, while RA administration produced minute cardiac progenitor areas. [159] (Figure 4A). Subsequently, Waxman et al. (2008) suggested that RA signalling normally operates to keep cardiac differentiation in check in the lateral mesoderm, allowing lateral mesoderm progenitors to take on other embryonic fates [160]. Consistent with this, RA inhibition decreases expression of gene markers for competing cell fates in the lateral mesoderm such as pectoral fin [160], pancreatic and pharyngeal pouch [161, 162].
It is important to realize that RA signalling effects on cardiac progenitor size were only investigated in detail in the zebrafish and, according to Keegan et al. (2005) changes in RA status can only produce effects on cardiac size up to the tailbud phase. In summary, while the available evidence strongly suggests that this effect may represent an ancestral function of RA in cardiac development, more work is needed in other species to confirm it, but the prospects seem good.
While the evidence for a role of RA in the regulation of cardiac field size is clear, the conclusion that the former is the only, or even the dominant retinoid function in cardiac development, is clearly unwarranted. It is worth noting that in the experiments of Keegan et al. (2005) and Waxman et al. (2008), RAR antagonists were administered at very early stages of development and maintained for long periods of time, or for the duration of the experiment. It is easy to see that in these approaches, RAR antagonists stay with the embryo for multiple phases of cardiac development. Therefore, the outcomes of these protracted experiments must be interpreted as the net sum of all the effects produced by RA signal manipulation on cardiac development.
Here we argue that much of the zebrafish data, including the classic experiments, which suggest the role of RA in inflow/outflow patterning will remain contradictory and unexplained by current thinking schemes unless due consideration is given to the dynamic, shifting, roles of RA signalling in cardiac development (see below).
6.4 The inflow/outflow division of the cardiogenic mesoderm: static and dynamic signals
The role of RA signalling in the partition of cardiac tissues into inflow and outflow fates (roughly sinus venosa and atria as opposed to ventricles and outflow tract) was the first to be more precisely recognized (Figure 3B,C). The timelines and context have been described in detail [141, 154]. Briefly, there is compelling evidence that, after gastrulation, RA signalling patterns the vertebrate cardiac field along the AP axis to mark broad domains containing precursors of inflow or outflow cardiac tissues, respectively [40, 154]. Integrating experiments from multiple model species and experimental approaches that included detailed fate maps, we concluded that, in amniotes, RA induces the sino-atrial phenotype in posterior cardiac precursors, while anterior precursors must be protected from RA, albeit transiently (see section 6.4), to differentiate into ventricular cells [40, 154].
Highlighting the exquisite sensitivity to developmental timing and the dynamic organization of embryonic RA signalling, we identified two sequential mechanisms that convey RA signals to amniote cardiac precursors [40]. We proposed that, at the time of commitment to cardiac AP fates, RA signals reaches heart progenitors in the lateral mesoderm by two ways, both depending on ALDH1A2.
At specification phases, when sino-atrial fates are still labile [164], posterior cardiac precursors (not coincidentally the cells fated to give rise to sinus venosa and atria) receive RA signals that diffuse from the ALDH1A2-expressing mesoderm caudal to the cardiac crescent (somitic and lateral mesoderm) (Figure 3B). Although not yet established by modern techniques, it is very likely that this scenario constitutes a classical signalling gradient, in which sino-atrial precursors in the posterior mesoderm are exposed to stronger RA signals than ventricular and outflow tract precursors in the anterior mesoderm. Thus, RA diffusing from posterior mesoderm seems to be in the right place and at the right time for the specification of cardiac inflow fates.
Interestingly, amniotes seem to require a subsequent phase of RA signalling before inflow/outflow fates are irreversibly communicated to cardiac progenitors. In fact, at determination stages, a dynamic caudal to rostral wave of ALDH1A2 sweeps the lateral mesoderm and gradually engulfs posterior, sino-atrial, progenitors, endowing them with the ability to synthesize their own RA (Figure 3C). We have proposed that the high RA concentrations typically associated with ALDH1A2-expressing tissues irreversibly commit posterior precursors to an inflow, sino-atrial, fate, while ventricular and outflow fates are consolidated in the absence or in the presence of low levels of RA signalling.
An often asked question about the roles of RA in cardiac inflow/outflow patterning is how one can reconcile the data discussed above with the second heart field (SHF) paradigm proposed by Kelly, Buckingham and colleagues [165, 166]. The crucial points of divergence between the inflow/outflow and SHF interpretations about the mechanisms of cardiac chamber development were reviewed in detail recently [167].
Here it is sufficient to say that the roots of the controversy between those in favor of the SHF concept and those that do not lie in the value attributed to the heterogeneity displayed in the early cardiac field. Buckingham and colleagues think it is justified to treat the cardiac field as two distinct progenitor fields (FHF and SHF), while we believe there is sufficient overlap to treat the whole cardiac field as one unit, at least for the purpose of early inflow/outflow patterning [168-170].
Concerning the SHF AP organization, it was proposed that RA is involved in the establishment of SHF (Isl-1-expressing) posterior boundaries [171-173] through Hoxb1 expression in cells that will later give rise to atria and sub-pulmonary myocardium [174].
We do not disagree with the idea that RA signalling plays important roles in atrial progenitors. In fact, we were the first to show that ALDH1A2 expression provides instructive clues to atrial precursors [40]. Figure 4 taken from the work of Hochgreb and colleagues shows that in the mouse embryo a progenitor population of Tbx-5-expressing atrial progenitors is swept by a caudal rostral wave of RA signalling [40]. This wave of RA signalling includes not only ALDH1A2 expression, but also contains clear indication, in the form of RA reporter activation, that this Tbx-5-expressing atrial progenitor population is in fact responding to RA in the same caudo-rostral pattern demonstrated for ALDH1A2 expression.
The importance of RA signalling through the caudal to rostral wave of ALDH1A2 for cardiac inflow/outflow patterning was further demonstrated in the chicken embryo. Figure 4 shows that the inflow/outflow fate of cardiac progenitors in the lateral mesoderm is directly dependent on RA signalling. If fate maps are produced in the presence of a RAR inhibitor, outflow fates start to appear in positions normally only occupied by atrial progenitors. Moreover, if we locally block RA signalling in the domain occupied by the right caudal to rostral wave it is possible to show that atrial marker expression by AMHC1 is completely abrogated, while the left contralateral expression AMHC1 expression is preserved.
We understand that the SHF concept generated intense interest in the field. However, now it is time to accommodate the SHF data into a clear synthesis that can be presented to the new generation of investigators as a starting point for novel and exciting discoveries. We can only do that if we utilize all the evidence at hand, not only the pieces that fall into this, or that paradigm. The role of RA signaling in posterior cardiac progenitors is a case in point. The fact that there is evidence for a role of RA signaling in establishing posterior boundaries for caudal Isl-1-expressing cardiac progenitors (posterior SHF) does not negate the fact that RA provides instructive clues to specify caudal, Tbx-5-expressing, cardiac precursors (posterior FHF) to an atrial fate (Hochgreb et al., 2003). To us it seems that the effects of RA upon posterior cardiac precursors are indeed in line with the idea that RA roles are dynamic, diverse and directed to different subpopulations in the heart field[167].
6.5 Early, non-epicardial, control of ventricular differentiation
ALDH1A2 knockout embryos display precociously differentiated ventricular myocytes at 8.5 dpc. These cells are flattened and already show organized sarcomeres. Throughout the ventricles, myocytes are interspersed by large intercellular gaps, which are characterized by failure to form appropriate trabeculae [27, 28]. This developmental defect is also observed in other situations of insufficient RA signalling such as vitamin A-deprivation, RXRα knockout and double RXRα/RXRβ knockout mice.
Defects in at least two different pathways have been suggested to produce the altered ventricular myocytes of RXRα and RXRα/RXRβ knockout mice: one pathway controlling differentiation and proliferation and another controlling myocyte shape and intercellular connectivity. Interference with the differentiation and proliferation pathway leads to a phenotype of early differentiation shared by RXRα, RXRβ, RXRα/RXRβ, RARα, RARβ knockout and VAD mouse embryos, while perturbation of the myocyte shape and intercellular connectivity, which specifically requires RXRα, leads to the development of spongy ventricles and ventricular septal defects in RXRα knockout and VAD embryos. Tampering with the myocyte differentiation/proliferation pathway can be tolerated probably because ventricular proliferation deficits of RXRβ, RARα and RARβ knockout embryos can be compensated by other mechanisms such that no spongy ventricles are formed in these embryos. In contrast, the problems of proliferation, cell shape and cell to cell connectivity of RXRα−/− embryos are not tolerated, possibly because of the pleiotropic effects of RXRα in cushion tissue, epicardium and in the placenta [152].
The ventricular phenotypes discussed in the preceding section are curious, because neither ALDH1A2 nor RA reporters are expressed in the 8.5 dpc mouse ventricle [41]. It is thus possible that these ventricular defects are the result of a much earlier requirement for RA in ventricular precursors. Clues to this interpretation came, surprisingly, from work in chicken, rather than mice. Taking advantage of the embryonic resistance to manipulation and comparative larger size of avian than mammal embryos, Hochgreb et al. (2003) showed by fate map analyses that ventricular and truncal precursors express ALDH1A2 through stages HH8-10, which include the cardiac developmental phases that precede the formation of the primordial heart tube [40] (Figure 3D).
Since AP fates in the heart are already determined at stage HH8 (Figure 3C), exposure of anterior cardiac precursors to RA signalling at stage HH8 and HH9 must serve a developmental program distinct from AP patterning. We have suggested that an early window of ALDH1A2 expression in chicken bilateral ventricular precursors when they are still in the lateral mesoderm activates RA signalling in the compact zone of the chicken ventricular myocardium to inhibit precocious differentiation and maintain the high proliferative capacity necessary to septate the ventricles and build a competent heart (Figure 3D). As such, these data suggest that ALDH1A2 expression in mouse embryos at equivalent embryonic stages may constitute the long sought non-epicardial source of RA that inhibits differentiation and keeps proliferation high in 8.5 to 9.5 dpc ventricular myocytes [175].
6.6 Epicardial control of ventricular growth
The discovery that the epicardium was a site of intense RA production was consistent with earlier data that suggested the importance of the epicardium as a signalling center in cardiac development and open new research directions. Prodded by the discoveries of the roles of RXRs and RARs in ventricular growth [27, 28, 175], Chen et al. (1998) [176] and Tran and Sucov (1998) [177] demonstrated that early cardiac growth requirements for RXRα are not rescued by transgenic expression of RXRα in the myocardium in RXRα-null embryos and that RXR-null cells are normally integrated into a functional wild type heart (i.e. the effects of RXRα deficiency are not cell-autonomous to the myocardium) [176, 177]. This, of course, suggested that other tissues in the heart could be responsible for the ventricular phenotype. Together with the discovery that the epicardium is a site of intense ALDH1A2 expression, RA synthesis and RA-induced transcription [41, 42], the available data indicated that the epicardium is a critical controller of myocardial growth [178-181]. These discoveries opened a fruitful avenue for research in cardiac development and regeneration (reviewed in [182, 183]).
Epicardial RA signalling through ALDH1A2 is being increasingly understood as a focal point in the control of ventricular myocardium growth. Working with the concept of the epicardium as an RA signalling center Chen et al. (2002) [184] and Stuckmann et al. (2003) [181] suggested that RA produced in the epicardium activates an autocrine RA signalling pathway, as predicted on the basis of RA reporter expression in the epicardium itself [41, 42]. In the following years new members of this pathway, upstream or downstream from ALDH1A2 were discovered.
As an upstream regulator, the Wilms tumour 1 (WT1) zinc finger transcription factor is expressed in the proepicardium as well as in the epicardium (Figure 3E) and was shown to bind three motifs in the 5’ region of the Aldh1a2 gene. Consistent with this, conditional WT1 knockout in the mouse epicardium reduces Aldh1a2 expression and is associated with decreased thickness of the ventricular compact zone [185]. Additional upstream regulators of epicardial Aldh1a2 are also being slowly discovered. Huang and colleagues provide evidence that CCAAT/enhancer binding proteins (C/EBP) cooperate with HOX and MEIS transcription factors at a 160 bp enhancer in the Aldh1a2 intron 1 (CR2 enhancer). This enhancer is part of a previously-described, highly conserved, 840 bp regulatory region of the Aldh1a2 intron 1, the Aldh1a2 1G enhancer, which drives reporter expression in the dorsal-most subset of the ALDH1A2 domain in the posterior embryonic quarter, more precisely in the dorsal neural tube [186], as well as in the epicardium (Castillo et al., unpublished results).
Inspired by the phenotypes of Erythropoietin (EPO) and EPO receptor knockout mice [187], Stuckmann et al. (2003) provided evidence that EPO signalling works in parallel with RA signalling to induce secretion by the epicardium of a diffusible factor, which, in turn, stimulates proliferation of the ventricular compact zone [181] (Figure 3E). This factor has been suggested to be FGF, perhaps FGF9, or FGF16 [188], although there is some evidence that FGF factors may play more important roles at stages earlier than those controlled by the epicardium (reviewed in [182, 183]). More recently, convincing evidence points to IGF-II as the major epicardial factor in charge of ventricular growth regulation [189] (Figure 3E).
Challengingly, it has also been suggested that the role played by RA in ventricular growth is independent of the epicardium. Brade et al. (2011) showed that RA signalling in the liver stimulates hepatic production of EPO, which then acts to induce IGF-II secretion by the epicardium [190]. There is clear support for this new connection uncovered between hepatic RA, epicardial EPO signalling and IGF-II secretion. However, hepatic RA signalling does not necessarily deny a role RA signalling by the epicardium. In fact, the argument against the role of epicardial RA signalling in the ventricular myocardium is based on the fact that the hearts of RXRα knockout mice display a severe thinning of the compact zone, but, oddly, retain epicardial RA signalling, as demonstrated by activation of the RARE lacZ reporter. However, it remains formally possible that, in the case of ventricular growth regulation, there is a specific requirement for RXRα that cannot be met by other RXR paralogs. Consistent with this idea, RA-induced proliferation in the ventricle has been demonstrated in isolated, cultured ventricular slices [181]. In any event, it is clear that RA signalling is only one, albeit, an important factor among the myriad of pathways in endocardium, myocardium and epicardium [182, 183, 189, 191] that intersect to produce a vigorous contracting ventricular wall and a leak-proof, muscular, interventricular septum.
6.7 Epicardial control of coronariogenesis
One of the most fascinating processes during cardiac development is represented by the neat ordering of coronary morphogenesis and differentiation. It is well known that vertebrate blood vessels are formed by concentric layers that include the inner endothelium, which is in direct contact with blood. The outer layers are represented, in sequence, by rings of smooth muscle (i.e. the muscular layer) and of connective tissue (the adventitial layer). During coronariogenesis, endothelial cells differentiate first and quickly organize themselves in the form of interconnecting tubes that constitute the primordial mesh of the coronary bed [192, 193]. It is only very much later that smooth muscle precursors are attracted to these endothelial tubes. Only then smooth muscle precursors complete their differentiation programs [193]. The reasons for this exquisitely precise timing of smooth muscle integration into coronary vessels were unknown. Working with smooth muscle cultures and chicken-quail embryonic approaches, Azambuja et al. (2010) showed that, together with vascular endothelial growth factor (VEGF), RA signalling acts in both autocrine and in paracrine modes to inhibit the expression of smooth muscle markers such as CRP2, GATA-6, SRF, and SM22A and to delay smooth muscle differentiation (Figure 3F). Azambuja et al. (2010) suggest that this physiological delay is crucial because it allows for extensive building and remodeling of the endothelial network before the final coronary mesh is stabilized by the aggregation of smooth muscle [39]. This is one of the documented direct actions of RA signalling on timing differentiation processes during cardiac development and it is likely that this timing information will be useful to develop prospective strategies in coronary regeneration research [39].
6.8 The need to integrate the multiple roles of RA signalling during cardiac development: the zebrafish case
In our opinion, further progress in our understanding of the role of RAR signaling in cardiac development will require a fresh synthesis and a critical reevaluation of the evidence and conclusions derived from previous experiments. Initially, RA signalling in zebrafish has been associated with the partitioning of inflow versus outflow cardiac tissues. According to Stainier and Fishman (1992), low doses of exogenous RA produced hearts with atrial predominance and with selective ventricular truncation. In contrast, higher RA doses transformed the heart into a small, completely “atrialized” organ, before treatments with even higher doses left the embryo with no heart cells. These authors interpreted their results as an indication that RA can modify the normal AP pattern of cardiac progenitors in the lateral mesoderm. Admittedly, studies with exogenous, teratogenic, doses of RA do not necessarily inform us about normal physiological development, although, often, they do provide useful clues. Despite that it was clear that these initial results in the zebrafish were completely consistent with previous data in chicken [155] and with other experiments in mice [40, 60-63]. These experiments revealed that the atrial dominance associated with RA excess was just the opposite manifestation of the ventricular dominance obtained with RA loss-of-function experiments with ALDH1A inhibitors and pan-RAR antagonists. In other words, before RA loss-of-function experiments were conducted in the zebrafish, the only possible interpretation was that, as in amniotes, RA signaling in zebrafish also seemed to have a role in the partitioning of hearts between inflow and outflow tissues [42, 154].
Following up on early zebrafish results, Yelon and colleagues investigated the function of RA in heart development, this time with more modern techniques that included the necessary loss-of-function approaches. The first result of their efforts was the discovery of the above-mentioned role of RA in controlling cardiac progenitor size [159] (Figure 5A). Their subsequent interpretations, however, proved progressively more difficult to integrate with previous knowledge. Using high RA doses at very early embryonic times (i.e. 40% epiboly), Waxman et al. (2009) [194] confirmed Stainier and Fishman's results and showed the same pattern of progressive cardiac truncation, in which atrialized phenotypes are produced due to an increased sensitivity of ventricular cells to the deleterious effects of RA. Moreover, with low RA doses administered at the same early stages, Waxman et al. (2009) showed a 70% increase in the atrial domain, which, essentially, confirmed the atrial dominance induced by RA treatment in chicken, mice and in the zebrafish itself [156] (Figure 5B).
Figure 5. Schematic representations of the roles played by retinoic acid (RA) signalling during early cardiac development in the zebrafish.
A) RA signalling controls cardiac progenitor size during the blastula period. At 40% epiboly, cardiac progenitors, including both inflow (red) and outflow (blue) precursors, are positioned at the ventrolateral marginal zone, engulfed by an intense circular domain of expression of Aldh1a2, the major RA synthesis enzyme (purple). Exogenous administration of RA at 40% epiboly leads to diminutive cardiac progenitor areas, while inhibition of RA signalling produces oversized cardiac precursors [159]. B) RA signalling patterns cardiac field into inflow/outflow domains at the onset of somitogenesis. Soon after the tailbud stage (10hpf), Aldh1a2 expression is observed in the somitic and lateral mesoderm (purple) [195]. RA signals produced from these cells are thought to expand anteriorly and reach inflow precursors, endowing them with the ability to synthesize their own RA and determining them to a sino-atrial cell fate. At the tailbud stage (10hpf), treatment with exogenous RA results in hearts with inflow (sino-atrial) dominance, while treatment with the pan-RA receptor antagonist BMS 493 induces hearts with outflow (ventricular) dominance (Figure 13 in [13] and Costa et al., unpublished).
However, and rather surprisingly, the authors argued that their results did not support the roles previously suggested for RA as a promoter of atrial identity through transformation of ventricular identity [40]. Along the same lines, Waxman et al. (2008) [160] concluded, on the basis of rather long (from 40% epiboly to 48 hours of development) treatments with RA synthesis inhibitors and RAR antagonists, that loss of RA signalling increases both atrial and ventricular cell numbers. Finally, these authors concluded from their zebrafish data that RA does not play a role in the formation of atrial myocytes at the expense of ventriculocytes and, thus, that they see no support for the hypothesis that RA signalling may have been involved in the origin of vertebrate chambered hearts as suggested before.
As proponents of the inflow/outflow hypothesis of cardiac chamber evolution, the conclusions of Yelon and colleagues came as a shock for a number of reasons. As discussed in detail before [13], and further stressed here, we believe that there is an important flaw in the reasoning of Waxman et al. (2008) and Waxman et al. (2009). The point is easy enough to understand and can be summarized as follows: if we are led to think that the only role played by RA in early zebrafish heart development is to control the overall size of the cardiac progenitor population (inflow and outflow together), how could the phenotypes of atrial dominance/ ventricular truncation induced by RA be repeatedly observed in the experiments of Stainier and Fishman and of Waxman et al. (2009)? Indeed, none of the experimental approaches utilized by Yelon and colleagues can exclude the alternative hypothesis that their results could reflect an interaction between two sequential, potentially overlapping, RA programs, namely: 1) regulation of heart progenitor size and; 2) promotion of atrial dominance. Indeed, these two programs do overlap. According to Waxman et al. (2008), the first cardiac progenitor size program is active from 40% epiboly up to the tail bud stage (5-10 hours post-fertilization) [160], while the atrial dominance program is active from the tail bud stage to the 9-somite stage (10-13.5 hours post-fertilization [156].
We suggest that the internal contradictions in the work of Waxman et al., 2008 and 2009 and its inconsistency with previous data in the zebrafish and amniotes can be resolved if one postulates that during zebrafish early cardiac development there are at least two major programs, one earlier program that controls progenitor size (Figure 5A) and subsequent one that induces atrial dominance associated with ventricular truncation (Figure 5B).
6.9 How can evolution inform the controversy?
The role of RA signalling as a mechanism of cardiac partition into inflow/outflow domains in amniotes has been already discussed [141, 154]. However, assuming, for the sake of argument, that RA signalling has no role in cardiac inflow/outflow patterning in a teleost such as the zebrafish, is it possible to exclude the possibility that retinoid signalling was involved in the partition of the ancestral vertebrate heart into inflow/outflow domains? In our opinion, this is not reasonable. First of all, teleosts are not basal animals [196] and should not be used as examples of the ancestral vertebrate condition when compared to amniotes [196]. In that regard, the challenge mounted by Waxman et al., (2008) would have been much more effective if performed in cartilaginous fish, or in agnathans, rather than in teleosts. Amniotes and teleosts had a last common ancestor almost 500 millions of years ago [21]). Thus, sufficient time has already elapsed that ancestral mechanisms could now be extensively modified in zebrafish (and in amniotes as well). Another problem with this conclusion can be illustrated with a more mundane, commonplace, example. For instance, it is not justifiable to conclude that because zebrafish do not display the arrangement of stylopod, zeugopod and autopod for their paired appendixes, then none of these structures must really exist in the vertebrate ancestor, or in amniotes. A more sensible conclusion would be to say that, although nearly all vertebrates conserve a seemingly ancestral plan for their paired appendixes, both amniotes and teleosts display clear derivations from this plan. Currently it is thought that these above-mentioned derivations reflect an overall simplification in teleosts and the emergence of the autopod somewhere along the sarcopterygean lineage that includes amniotes [197, 198].
There is good evidence in amniotes to support the notion that increases in the strength of RA signalling during the critical phases of cardiac inflow/outflow patterning are associated with an increased representation of atrial tissues in their embryonic hearts [40]. As we have reviewed before, the mechanisms behind the RA-induced “atrialization” of the embryonic amniote heart remain to be established, but presumably include: switch from ventricular to atrial fates, cell-cycle withdrawal of ventricular precursors, ventricular progenitor apoptosis, or switch of ventricular precursors to a non-cardiac fate.
Again, if we assume, for the sake of argument, that changes in RA signalling status have nothing to do with switches between atrial and ventricular fates, can we effectively eliminate the possibility that RA signalling was involved in the partition of the ancestral heart into inflow/outflow domains? In our opinion this is not a defensible conclusion. By basing their challenge of the inflow/outflow hypothesis only on switches from ventricular to atrial cells, when in reality there are several possible alternative mechanisms for the obvious effects of RA in promoting atrial dominance in zebrafish [156] and amniotes [40, 63], Yelon and colleagues incur at the risk of oversimplification and sidestepping the issue at hand. In fact, it is well possible that the selectable trait conserved throughout vertebrates is a functional balance between atrial and ventricular cardiac domains, which can be established by alternative mechanisms in the various vertebrate groups.
7. CONCLUSION
One of our major objectives in this review was to provide evidence that cardiac RAR signalling is fundamentally complex in embryonic time and space. Another major goal was to place cardiac RAR signalling where it belongs, i.e. as an ancestral signalling pathway whose roles in extant vertebrates reflect a kaleidoscope of species-specific variations against a clearly identified background of functions that presumably date back to the first vertebrate embryo.
As a corollary for the first objective, we conclude that cardiac RA signalling cannot be reduced to a single function. As our understanding of this inherent complex system evolves, so must our experimental approaches. It is, thus, far from satisfactory to use extensive treatments, or early loss-of-function approaches that will influence a sequence of different developmental processes. Rather, it is our opinion that the roles of RAR signalling in cardiac development should be interrogated with short and well-defined manipulations that match the critical time windows at which RA synthetic enzymes are expressed close to, or within the tissue under observation.
As a corollary for the second objective, we believe that a firm grasp on evolution is crucial to integrate results obtained from different vertebrate model systems and to propose conclusions that do not reflect the individual preferences for one model organisms over another. In other words, for highly conserved processes such as vertebrate embryonic heart development, experience tells us that it is not possible to interrogate your preferred vertebrate species and to establish that a given mechanism is right or wrong for the entire suite of vertebrates. A more realistic way to deal with the diversity of developmental mechanisms is to ask first whether there are some common themes that reflect common evolutionary origins and to then identify the distinguishing features in the several evolutionary lineages that evolved from the last common ancestor.
Highlights.
Retinoic acid (RA) signalling is an ancestral feature of bilaterian animals
RA plays time-sensitive, sequential and multifaceted roles in cardiac development
Bona fide RA gradients have been recently objectively demonstrated
RA signalling is regulated by a balance between RA synthesis and degradation
RA roles in early zebrafish cardiac development remain controversial
ACKNOWLEDGEMENTS
We would like to thank Vincent Laudet for the opportunity and two anonymous reviewers for criticisms and suggestions. We are also indebted to Murilo de Carvalho and Wellington Cardoso for thoughtful comments. This work was supported by Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP - grants 2011/15273-7; 2012/14859-0; 2013/12008-6) and Conselho Nacional do Desenvolvimento Científico e Tecnológico (CNPq - grant 481983/2013-9).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Escriva H, Bertrand S, Germain P, Robinson-Rechavi M, Umbhauer M, Cartry J, Duffraisse M, Holland L, Gronemeyer H, Laudet V. Neofunctionalization in vertebrates: the example of retinoic acid receptors. PLoS Genet. 2006;2:e102. doi: 10.1371/journal.pgen.0020102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Philip S, Castro LF, da Fonseca RR, Reis-Henriques MA, Vasconcelos V, Santos MM, Antunes A. Adaptive evolution of the Retinoid X receptor in vertebrates. Genomics. 2012;99:81–89. doi: 10.1016/j.ygeno.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 3.Cui J, Michaille JJ, Jiang W, Zile MH. Retinoid receptors and vitamin A deficiency: differential patterns of transcription during early avian development and the rapid induction of RARs by retinoic acid. Dev Biol. 2003;260:496–511. doi: 10.1016/s0012-1606(03)00257-4. [DOI] [PubMed] [Google Scholar]
- 4.Hale LA, Tallafuss A, Yan YL, Dudley L, Eisen JS, Postlethwait JH. Characterization of the retinoic acid receptor genes raraa, rarab and rarg during zebrafish development. Gene Expr Patterns. 2006;6:546–555. doi: 10.1016/j.modgep.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 5.Mollard R, Viville S, Ward SJ, Decimo D, Chambon P, Dolle P. Tissue-specific expression of retinoic acid receptor isoform transcripts in the mouse embryo. Mech Dev. 2000;94:223–232. doi: 10.1016/s0925-4773(00)00303-8. [DOI] [PubMed] [Google Scholar]
- 6.Romeih M, Cui J, Michaille JJ, Jiang W, Zile MH. Function of RARgamma and RARalpha2 at the initiation of retinoid signaling is essential for avian embryo survival and for distinct events in cardiac morphogenesis. Dev Dyn. 2003;228:697–708. doi: 10.1002/dvdy.10419. [DOI] [PubMed] [Google Scholar]
- 7.Ruberte E, Dolle P, Krust A, Zelent A, Morriss-Kay G, Chambon P. Specific spatial and temporal distribution of retinoic acid receptor gamma transcripts during mouse embryogenesis. Development. 1990;108:213–222. doi: 10.1242/dev.108.2.213. [DOI] [PubMed] [Google Scholar]
- 8.Rossant J, Zirngibl R, Cado D, Shago M, Giguere V. Expression of a retinoic acid response element-hsplacZ transgene defines specific domains of transcriptional activity during mouse embryogenesis. Genes Dev. 1991;5:1333–1344. doi: 10.1101/gad.5.8.1333. [DOI] [PubMed] [Google Scholar]
- 9.White RJ, Nie Q, Lander AD, Schilling TF. Complex regulation of cyp26a1 creates a robust retinoic acid gradient in the zebrafish embryo. PLoS Biol. 2007;5:e304. doi: 10.1371/journal.pbio.0050304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mark M, Ghyselinck NB, Chambon P. Function of retinoid nuclear receptors: lessons from genetic and pharmacological dissections of the retinoic acid signaling pathway during mouse embryogenesis. Annu Rev Pharmacol Toxicol. 2006;46:451–480. doi: 10.1146/annurev.pharmtox.46.120604.141156. [DOI] [PubMed] [Google Scholar]
- 11.White RJ, Schilling TF. How degrading: Cyp26s in hindbrain development. Dev Dyn. 2008;237:2775–2790. doi: 10.1002/dvdy.21695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gans C, Northcutt RG. Neural Crest and the Origin of Vertebrates: A New Head. Science. 1983;220:268–273. doi: 10.1126/science.220.4594.268. [DOI] [PubMed] [Google Scholar]
- 13.Xavier-Neto J, Davidson B, Simoes-Costa MS, Castro RA, Castillo HA, Sampaio AC, Azambuja AP. Evolutionary Origins of Hearts. In: Press A, editor. Heart Development and Regeneration. Elsevier; London: 2010. pp. 3–45. [Google Scholar]
- 14.Xavier-Neto J, Castro RA, Sampaio AC, Azambuja AP, Castillo HA, Cravo RM, Simoes-Costa MS. Parallel avenues in the evolution of hearts and pumping organs. Cell Mol Life Sci. 2007;64:719–734. doi: 10.1007/s00018-007-6524-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Olson EN. Gene regulatory networks in the evolution and development of the heart. Science. 2006;313:1922–1927. doi: 10.1126/science.1132292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alaynick WA, Jessell TM, Pfaff SL. SnapShot: spinal cord development. Cell. 2011;146:178–178. e171. doi: 10.1016/j.cell.2011.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yoshida A, Rzhetsky A, Hsu LC, Chang C. Human aldehyde dehydrogenase gene family. Eur J Biochem. 1998;251:549–557. doi: 10.1046/j.1432-1327.1998.2510549.x. [DOI] [PubMed] [Google Scholar]
- 18.Kikonyogo A, Pietruszko R. Aldehyde dehydrogenase from adult human brain that dehydrogenates gamma-aminobutyraldehyde: purification, characterization, cloning and distribution. Biochem J. 1996;316(Pt 1):317–324. doi: 10.1042/bj3160317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sobreira TJ, Marletaz F, Simoes-Costa M, Schechtman D, Pereira AC, Brunet F, Sweeney S, Pani A, Aronowicz J, Lowe CJ, Davidson B, Laudet V, Bronner M, de Oliveira PS, Schubert M, Xavier-Neto J. Structural shifts of aldehyde dehydrogenase enzymes were instrumental for the early evolution of retinoid-dependent axial patterning in metazoans. Proc Natl Acad Sci U S A. 2011;108:226–231. doi: 10.1073/pnas.1011223108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen CH, Budas GR, Churchill EN, Disatnik MH, Hurley TD, Mochly-Rosen D. Activation of aldehyde dehydrogenase-2 reduces ischemic damage to the heart. Science. 2008;321:1493–1495. doi: 10.1126/science.1158554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hedges SB, Blair JE, Venturi ML, Shoe JL. A molecular timescale of eukaryote evolution and the rise of complex multicellular life. BMC Evol Biol. 2004;4:2. doi: 10.1186/1471-2148-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Simoes-Costa MS, Azambuja AP, Xavier-Neto J. The search for non-chordate retinoic acid signaling: lessons from chordates. J Exp Zoolog B Mol Dev Evol. 2008;310:54–72. doi: 10.1002/jez.b.21139. [DOI] [PubMed] [Google Scholar]
- 23.Campo-Paysaa F, Marletaz F, Laudet V, Schubert M. Retinoic acid signaling in development: tissue-specific functions and evolutionary origins. Genesis. 2008;46:640–656. doi: 10.1002/dvg.20444. [DOI] [PubMed] [Google Scholar]
- 24.Albalat R, Canestro C. Identification of Aldh1a, Cyp26 and RAR orthologs in protostomes pushes back the retinoic acid genetic machinery in evolutionary time to the bilaterian ancestor. Chem Biol Interact. 2009;178:188–196. doi: 10.1016/j.cbi.2008.09.017. [DOI] [PubMed] [Google Scholar]
- 25.Fujiwara S, Kawamura K. Acquisition of retinoic acid signaling pathway and innovation of the chordate body plan. Zoolog Sci. 2003;20:809–818. doi: 10.2108/zsj.20.809. [DOI] [PubMed] [Google Scholar]
- 26.Waxman JS, Yelon D. Comparison of the expression patterns of newly identified zebrafish retinoic acid and retinoid X receptors. Dev Dyn. 2007;236:587–595. doi: 10.1002/dvdy.21049. [DOI] [PubMed] [Google Scholar]
- 27.Kastner P, Grondona JM, Mark M, Gansmuller A, LeMeur M, Decimo D, Vonesch JL, Dolle P, Chambon P. Genetic analysis of RXR alpha developmental function: convergence of RXR and RAR signaling pathways in heart and eye morphogenesis. Cell. 1994;78:987–1003. doi: 10.1016/0092-8674(94)90274-7. [DOI] [PubMed] [Google Scholar]
- 28.Sucov HM, Dyson E, Gumeringer CL, Price J, Chien KR, Evans RM. RXR alpha mutant mice establish a genetic basis for vitamin A signaling in heart morphogenesis. Genes Dev. 1994;8:1007–1018. doi: 10.1101/gad.8.9.1007. [DOI] [PubMed] [Google Scholar]
- 29.Ohno S. Gene duplication and the uniqueness of vertebrate genomes circa 1970-1999. Semin Cell Dev Biol. 1999;10:517–522. doi: 10.1006/scdb.1999.0332. [DOI] [PubMed] [Google Scholar]
- 30.Baertsch R, Diekhans M, Kent WJ, Haussler D, Brosius J. Retrocopy contributions to the evolution of the human genome. BMC Genomics. 2008;9:466. doi: 10.1186/1471-2164-9-466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lowe CB, Bejerano G, Haussler D. Thousands of human mobile element fragments undergo strong purifying selection near developmental genes. Proc Natl Acad Sci U S A. 2007;104:8005–8010. doi: 10.1073/pnas.0611223104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Force A, Lynch M, Pickett FB, Amores A, Yan YL, Postlethwait J. Preservation of duplicate genes by complementary, degenerative mutations. Genetics. 1999;151:1531–1545. doi: 10.1093/genetics/151.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nagatomo K, Fujiwara S. Expression of Raldh2, Cyp26 and Hox-1 in normal and retinoic acid-treated Ciona intestinalis embryos. Gene Expr Patterns. 2003;3:273–277. doi: 10.1016/s1567-133x(03)00051-6. [DOI] [PubMed] [Google Scholar]
- 34.Niederreither K, McCaffery P, Drager UC, Chambon P, Dolle P. Restricted expression and retinoic acid-induced downregulation of the retinaldehyde dehydrogenase type 2 (RALDH-2) gene during mouse development. Mech Dev. 1997;62:67–78. doi: 10.1016/s0925-4773(96)00653-3. [DOI] [PubMed] [Google Scholar]
- 35.Sobreira TJ, Marletaz F, Simoes-Costa M, Schechtman D, Pereira AC, Brunet F, Sweeney S, Pani A, Aronowicz J, Lowe CJ, Davidson B, Laudet V, Bronner M, de Oliveira PS, Schubert M, Xavier-Neto J. Structural shifts of aldehyde dehydrogenase enzymes were instrumental for the early evolution of retinoid-dependent axial patterning in metazoans. Proc Natl Acad Sci U S A. 2010;108:226–231. doi: 10.1073/pnas.1011223108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marletaz F, Holland LZ, Laudet V, Schubert M. Retinoic acid signaling and the evolution of chordates. Int J Biol Sci. 2006;2:38–47. doi: 10.7150/ijbs.2.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Drager UC, Wagner E, McCaffery P. Aldehyde dehydrogenases in the generation of retinoic acid in the developing vertebrate: a central role of the eye. J Nutr. 1998;128:463S–466S. doi: 10.1093/jn/128.2.463S. [DOI] [PubMed] [Google Scholar]
- 38.Ulven SM, Gundersen TE, Weedon MS, Landaas VO, Sakhi AK, Fromm SH, Geronimo BA, Moskaug JO, Blomhoff R. Identification of endogenous retinoids, enzymes, binding proteins, and receptors during early postimplantation development in mouse: important role of retinal dehydrogenase type 2 in synthesis of all-trans-retinoic acid. Dev Biol. 2000;220:379–391. doi: 10.1006/dbio.2000.9634. [DOI] [PubMed] [Google Scholar]
- 39.Azambuja AP, Portillo-Sanchez V, Rodrigues MV, Omae SV, Schechtman D, Strauss BE, Costanzi-Strauss E, Krieger JE, Perez-Pomares JM, Xavier-Neto J. Retinoic acid and VEGF delay smooth muscle relative to endothelial differentiation to coordinate inner and outer coronary vessel wall morphogenesis. Circ Res. 2010;107:204–216. doi: 10.1161/CIRCRESAHA.109.214650. [DOI] [PubMed] [Google Scholar]
- 40.Hochgreb T, Linhares VL, Menezes DC, Sampaio AC, Yan CY, Cardoso WV, Rosenthal N, Xavier-Neto J. A caudorostral wave of RALDH2 conveys anteroposterior information to the cardiac field. Development. 2003;130:5363–5374. doi: 10.1242/dev.00750. [DOI] [PubMed] [Google Scholar]
- 41.Moss JB, Xavier-Neto J, Shapiro MD, Nayeem SM, McCaffery P, Drager UC, Rosenthal N. Dynamic patterns of retinoic acid synthesis and response in the developing mammalian heart. Dev Biol. 1998;199:55–71. doi: 10.1006/dbio.1998.8911. [DOI] [PubMed] [Google Scholar]
- 42.Xavier-Neto J, Shapiro MD, Houghton L, Rosenthal N. Sequential programs of retinoic acid synthesis in the myocardial and epicardial layers of the developing avian heart. Dev Biol. 2000;219:129–141. doi: 10.1006/dbio.1999.9588. [DOI] [PubMed] [Google Scholar]
- 43.Duester G. Retinoic acid synthesis and signaling during early organogenesis. Cell. 2008;134:921–931. doi: 10.1016/j.cell.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Niederreither K, Dolle P. Retinoic acid in development: towards an integrated view. Nature reviews. Genetics. 2008;9:541–553. doi: 10.1038/nrg2340. [DOI] [PubMed] [Google Scholar]
- 45.Rhinn M, Dolle P. Retinoic acid signalling during development. Development. 2012;139:843–858. doi: 10.1242/dev.065938. [DOI] [PubMed] [Google Scholar]
- 46.Mangelsdorf DJ, Evans RM. The RXR heterodimers and orphan receptors. Cell. 1995;83:841–850. doi: 10.1016/0092-8674(95)90200-7. [DOI] [PubMed] [Google Scholar]
- 47.Kane MA, Folias AE, Pingitore A, Perri M, Obrochta KM, Krois CR, Cione E, Ryu JY, Napoli JL. Identification of 9-cis-retinoic acid as a pancreas-specific autacoid that attenuates glucose-stimulated insulin secretion. Proc Natl Acad Sci U S A. 2010;107:21884–21889. doi: 10.1073/pnas.1008859107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Duester G. Involvement of alcohol dehydrogenase, short-chain dehydrogenase/reductase, aldehyde dehydrogenase, and cytochrome P450 in the control of retinoid signaling by activation of retinoic acid synthesis. Biochemistry. 1996;35:12221–12227. doi: 10.1021/bi961176+. [DOI] [PubMed] [Google Scholar]
- 49.Boerman MH, Napoli JL. Characterization of a microsomal retinol dehydrogenase: a short-chain alcohol dehydrogenase with integral and peripheral membrane forms that interacts with holo-CRBP (type I) Biochemistry. 1995;34:7027–7037. doi: 10.1021/bi00021a014. [DOI] [PubMed] [Google Scholar]
- 50.Chai X, Zhai Y, Napoli JL. cDNA cloning and characterization of a cis-retinol/3alpha-hydroxysterol short-chain dehydrogenase. The Journal of biological chemistry. 1997;272:33125–33131. doi: 10.1074/jbc.272.52.33125. [DOI] [PubMed] [Google Scholar]
- 51.Romert A, Tuvendal P, Simon A, Dencker L, Eriksson U. The identification of a 9-cis retinol dehydrogenase in the mouse embryo reveals a pathway for synthesis of 9-cis retinoic acid. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:4404–4409. doi: 10.1073/pnas.95.8.4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang M, Chen W, Smith SM, Napoli JL. Molecular characterization of a mouse short chain dehydrogenase/reductase active with all-trans-retinol in intact cells, mRDH1. The Journal of biological chemistry. 2001;276:44083–44090. doi: 10.1074/jbc.M105748200. [DOI] [PubMed] [Google Scholar]
- 53.Blentic A, Gale E, Maden M. Retinoic acid signalling centres in the avian embryo identified by sites of expression of synthesising and catabolising enzymes. Dev Dyn. 2003;227:114–127. doi: 10.1002/dvdy.10292. [DOI] [PubMed] [Google Scholar]
- 54.Li H, Wagner E, McCaffery P, Smith D, Andreadis A, Drager UC. A retinoic acid synthesizing enzyme in ventral retina and telencephalon of the embryonic mouse. Mech Dev. 2000;95:283–289. doi: 10.1016/s0925-4773(00)00352-x. [DOI] [PubMed] [Google Scholar]
- 55.Sandell LL, Sanderson BW, Moiseyev G, Johnson T, Mushegian A, Young K, Rey JP, Ma JX, Staehling-Hampton K, Trainor PA. RDH10 is essential for synthesis of embryonic retinoic acid and is required for limb, craniofacial, and organ development. Genes Dev. 2007;21:1113–1124. doi: 10.1101/gad.1533407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dupe V, Matt N, Garnier JM, Chambon P, Mark M, Ghyselinck NB. A newborn lethal defect due to inactivation of retinaldehyde dehydrogenase type 3 is prevented by maternal retinoic acid treatment. Proc Natl Acad Sci U S A. 2003;100:14036–14041. doi: 10.1073/pnas.2336223100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fan X, Molotkov A, Manabe S, Donmoyer CM, Deltour L, Foglio MH, Cuenca AE, Blaner WS, Lipton SA, Duester G. Targeted disruption of Aldh1a1 (Raldh1) provides evidence for a complex mechanism of retinoic acid synthesis in the developing retina. Mol Cell Biol. 2003;23:4637–4648. doi: 10.1128/MCB.23.13.4637-4648.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Matt N, Dupe V, Garnier JM, Dennefeld C, Chambon P, Mark M, Ghyselinck NB. Retinoic acid-dependent eye morphogenesis is orchestrated by neural crest cells. Development. 2005;132:4789–4800. doi: 10.1242/dev.02031. [DOI] [PubMed] [Google Scholar]
- 59.Canestro C, Catchen JM, Rodriguez-Mari A, Yokoi H, Postlethwait JH. Consequences of lineage-specific gene loss on functional evolution of surviving paralogs: ALDH1A and retinoic acid signaling in vertebrate genomes. PLoS Genet. 2009;5:e1000496. doi: 10.1371/journal.pgen.1000496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chazaud C, Chambon P, Dolle P. Retinoic acid is required in the mouse embryo for left-right asymmetry determination and heart morphogenesis. Development. 1999;126:2589–2596. doi: 10.1242/dev.126.12.2589. [DOI] [PubMed] [Google Scholar]
- 61.Niederreither K, Subbarayan V, Dolle P, Chambon P. Embryonic retinoic acid synthesis is essential for early mouse post-implantation development. Nat Genet. 1999;21:444–448. doi: 10.1038/7788. [DOI] [PubMed] [Google Scholar]
- 62.Niederreither K, Vermot J, Messaddeq N, Schuhbaur B, Chambon P, Dolle P. Embryonic retinoic acid synthesis is essential for heart morphogenesis in the mouse. Development. 2001;128:1019–1031. doi: 10.1242/dev.128.7.1019. [DOI] [PubMed] [Google Scholar]
- 63.Xavier-Neto J, Neville CM, Shapiro MD, Houghton L, Wang GF, Nikovits W, Jr., Stockdale FE, Rosenthal N. A retinoic acid-inducible transgenic marker of sino-atrial development in the mouse heart. Development. 1999;126:2677–2687. doi: 10.1242/dev.126.12.2677. [DOI] [PubMed] [Google Scholar]
- 64.Billings SE, Pierzchalski K, Butler Tjaden NE, Pang XY, Trainor PA, Kane MA, Moise AR. The retinaldehyde reductase DHRS3 is essential for preventing the formation of excess retinoic acid during embryonic development. FASEB J. 2013;27:4877–4889. doi: 10.1096/fj.13-227967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Adams MK, Belyaeva OV, Wu L, Kedishvili NY. The retinaldehyde reductase activity of DHRS3 is reciprocally activated by Retinol Dehydrogenase 10 to control retinoid homeostasis. J Biol Chem. 2014 doi: 10.1074/jbc.M114.552257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Farjo KM, Moiseyev G, Nikolaeva O, Sandell LL, Trainor PA, Ma JX. RDH10 is the primary enzyme responsible for the first step of embryonic Vitamin A metabolism and retinoic acid synthesis. Dev Biol. 2011;357:347–355. doi: 10.1016/j.ydbio.2011.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rhinn M, Schuhbaur B, Niederreither K, Dolle P. Involvement of retinol dehydrogenase 10 in embryonic patterning and rescue of its loss of function by maternal retinaldehyde treatment. Proc Natl Acad Sci U S A. 2011;108:16687–16692. doi: 10.1073/pnas.1103877108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ross AC, Zolfaghari R. Cytochrome P450s in the regulation of cellular retinoic acid metabolism. Annual review of nutrition. 2011;31:65–87. doi: 10.1146/annurev-nutr-072610-145127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Thatcher JE, Isoherranen N. The role of CYP26 enzymes in retinoic acid clearance. Expert opinion on drug metabolism & toxicology. 2009;5:875–886. doi: 10.1517/17425250903032681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chithalen JV, Luu L, Petkovich M, Jones G. HPLC-MS/MS analysis of the products generated from all-trans-retinoic acid using recombinant human CYP26A. Journal of lipid research. 2002;43:1133–1142. doi: 10.1194/jlr.m100343-jlr200. [DOI] [PubMed] [Google Scholar]
- 71.Reijntjes S, Blentic A, Gale E, Maden M. The control of morphogen signalling: regulation of the synthesis and catabolism of retinoic acid in the developing embryo. Dev Biol. 2005;285:224–237. doi: 10.1016/j.ydbio.2005.06.019. [DOI] [PubMed] [Google Scholar]
- 72.Swindell EC, Thaller C, Sockanathan S, Petkovich M, Jessell TM, Eichele G. Complementary domains of retinoic acid production and degradation in the early chick embryo. Dev Biol. 1999;216:282–296. doi: 10.1006/dbio.1999.9487. [DOI] [PubMed] [Google Scholar]
- 73.Taimi M, Helvig C, Wisniewski J, Ramshaw H, White J, Amad M, Korczak B, Petkovich M. A novel human cytochrome P450, CYP26C1, involved in metabolism of 9-cis and alltrans isomers of retinoic acid. The Journal of biological chemistry. 2004;279:77–85. doi: 10.1074/jbc.M308337200. [DOI] [PubMed] [Google Scholar]
- 74.White JA, Guo YD, Baetz K, Beckett-Jones B, Bonasoro J, Hsu KE, Dilworth FJ, Jones G, Petkovich M. Identification of the retinoic acid-inducible all-trans-retinoic acid 4-hydroxylase. The Journal of biological chemistry. 1996;271:29922–29927. doi: 10.1074/jbc.271.47.29922. [DOI] [PubMed] [Google Scholar]
- 75.White JA, Ramshaw H, Taimi M, Stangle W, Zhang A, Everingham S, Creighton S, Tam SP, Jones G, Petkovich M. Identification of the human cytochrome P450, P450RAI-2, which is predominantly expressed in the adult cerebellum and is responsible for all-trans-retinoic acid metabolism. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:6403–6408. doi: 10.1073/pnas.120161397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Idres N, Marill J, Flexor MA, Chabot GG. Activation of retinoic acid receptor-dependent transcription by all-trans-retinoic acid metabolites and isomers. J Biol Chem. 2002;277:31491–31498. doi: 10.1074/jbc.M205016200. [DOI] [PubMed] [Google Scholar]
- 77.Pijnappel WW, Hendriks HF, Folkers GE, van den Brink CE, Dekker EJ, Edelenbosch C, van der Saag PT, Durston AJ. The retinoid ligand 4-oxo-retinoic acid is a highly active modulator of positional specification. Nature. 1993;366:340–344. doi: 10.1038/366340a0. [DOI] [PubMed] [Google Scholar]
- 78.Niederreither K, Abu-Abed S, Schuhbaur B, Petkovich M, Chambon P, Dolle P. Genetic evidence that oxidative derivatives of retinoic acid are not involved in retinoid signaling during mouse development. Nat Genet. 2002;31:84–88. doi: 10.1038/ng876. [DOI] [PubMed] [Google Scholar]
- 79.Lin M, Zhang M, Abraham M, Smith SM, Napoli JL. Mouse retinal dehydrogenase 4 (RALDH4), molecular cloning, cellular expression, and activity in 9-cis-retinoic acid biosynthesis in intact cells. J Biol Chem. 2003;278:9856–9861. doi: 10.1074/jbc.M211417200. [DOI] [PubMed] [Google Scholar]
- 80.Sima A, Parisotto M, Mader S, Bhat PV. Kinetic characterization of recombinant mouse retinal dehydrogenase types 3 and 4 for retinal substrates. Biochim Biophys Acta. 2009;1790:1660–1664. doi: 10.1016/j.bbagen.2009.09.004. [DOI] [PubMed] [Google Scholar]
- 81.Roberts ES, Vaz AD, Coon MJ. Role of isozymes of rabbit microsomal cytochrome P-450 in the metabolism of retinoic acid, retinol, and retinal. Mol Pharmacol. 1992;41:427–433. [PubMed] [Google Scholar]
- 82.Zhang QY, Raner G, Ding X, Dunbar D, Coon MJ, Kaminsky LS. Characterization of the cytochrome P450 CYP2J4: expression in rat small intestine and role in retinoic acid biotransformation from retinal. Arch Biochem Biophys. 1998;353:257–264. doi: 10.1006/abbi.1998.0654. [DOI] [PubMed] [Google Scholar]
- 83.Chambers D, Wilson L, Maden M, Lumsden A. RALDH-independent generation of retinoic acid during vertebrate embryogenesis by CYP1B1. Development. 2007;134:1369–1383. doi: 10.1242/dev.02815. [DOI] [PubMed] [Google Scholar]
- 84.Szuts EZ, Harosi FI. Solubility of retinoids in water. Arch Biochem Biophys. 1991;287:297–304. doi: 10.1016/0003-9861(91)90482-x. [DOI] [PubMed] [Google Scholar]
- 85.Bouillet P, Sapin V, Chazaud C, Messaddeq N, Decimo D, Dolle P, Chambon P. Developmental expression pattern of Stra6, a retinoic acid-responsive gene encoding a new type of membrane protein. Mechanisms of development. 1997;63:173–186. doi: 10.1016/s0925-4773(97)00039-7. [DOI] [PubMed] [Google Scholar]
- 86.Kawaguchi R, Yu J, Wiita P, Ter-Stepanian M, Sun H. Mapping the membrane topology and extracellular ligand binding domains of the retinol binding protein receptor. Biochemistry. 2008;47:5387–5395. doi: 10.1021/bi8002082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kawaguchi R, Yu J, Honda J, Hu J, Whitelegge J, Ping P, Wiita P, Bok D, Sun H. A membrane receptor for retinol binding protein mediates cellular uptake of vitamin A. Science. 2007;315:820–825. doi: 10.1126/science.1136244. [DOI] [PubMed] [Google Scholar]
- 88.Pasutto F, Sticht H, Hammersen G, Gillessen-Kaesbach G, Fitzpatrick DR, Nurnberg G, Brasch F, Schirmer-Zimmermann H, Tolmie JL, Chitayat D, Houge G, Fernandez-Martinez L, Keating S, Mortier G, Hennekam RC, von der Wense A, Slavotinek A, Meinecke P, Bitoun P, Becker C, Nurnberg P, Reis A, Rauch A. Mutations in STRA6 cause a broad spectrum of malformations including anophthalmia, congenital heart defects, diaphragmatic hernia, alveolar capillary dysplasia, lung hypoplasia, and mental retardation. American journal of human genetics. 2007;80:550–560. doi: 10.1086/512203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Berry DC, Jacobs H, Marwarha G, Gely-Pernot A, O'Byrne SM, DeSantis D, Klopfenstein M, Feret B, Dennefeld C, Blaner WS, Croniger CM, Mark M, Noy N, Ghyselinck NB. The STRA6 receptor is essential for retinol-binding protein-induced insulin resistance but not for maintaining vitamin A homeostasis in tissues other than the eye. J Biol Chem. 2013;288:24528–24539. doi: 10.1074/jbc.M113.484014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ruiz A, Mark M, Jacobs H, Klopfenstein M, Hu J, Lloyd M, Habib S, Tosha C, Radu RA, Ghyselinck NB, Nusinowitz S, Bok D. Retinoid content, visual responses, and ocular morphology are compromised in the retinas of mice lacking the retinol-binding protein receptor, STRA6. Invest Ophthalmol Vis Sci. 2012;53:3027–3039. doi: 10.1167/iovs.11-8476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chandra V, Huang P, Hamuro Y, Raghuram S, Wang Y, Burris TP, Rastinejad F. Structure of the intact PPAR-gamma-RXR- nuclear receptor complex on DNA. Nature. 2008;456:350–356. doi: 10.1038/nature07413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Helsen C, Claessens F. Looking at nuclear receptors from a new angle. Molecular and cellular endocrinology. 2014;382:97–106. doi: 10.1016/j.mce.2013.09.009. [DOI] [PubMed] [Google Scholar]
- 93.Rastinejad F, Huang P, Chandra V, Khorasanizadeh S. Understanding nuclear receptor form and function using structural biology. Journal of molecular endocrinology. 2013;51:T1–T21. doi: 10.1530/JME-13-0173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Rastinejad F, Wagner T, Zhao Q, Khorasanizadeh S. Structure of the RXR-RAR DNA-binding complex on the retinoic acid response element DR1. The EMBO journal. 2000;19:1045–1054. doi: 10.1093/emboj/19.5.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wei LN. Retinoid receptors and their coregulators. Annual review of pharmacology and toxicology. 2003;43:47–72. doi: 10.1146/annurev.pharmtox.43.100901.140301. [DOI] [PubMed] [Google Scholar]
- 96.Wurtz JM, Bourguet W, Renaud JP, Vivat V, Chambon P, Moras D, Gronemeyer H. A canonical structure for the ligand-binding domain of nuclear receptors. Nature structural biology. 1996;3:87–94. doi: 10.1038/nsb0196-87. [DOI] [PubMed] [Google Scholar]
- 97.Bourguet W, Ruff M, Chambon P, Gronemeyer H, Moras D. Crystal structure of the ligand-binding domain of the human nuclear receptor RXR-alpha. Nature. 1995;375:377–382. doi: 10.1038/375377a0. [DOI] [PubMed] [Google Scholar]
- 98.Nagy L, Schwabe JW. Mechanism of the nuclear receptor molecular switch. Trends in biochemical sciences. 2004;29:317–324. doi: 10.1016/j.tibs.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 99.Weston AD, Hoffman LM, Underhill TM. Revisiting the role of retinoid signaling in skeletal development. Birth Defects Res C Embryo Today. 2003;69:156–173. doi: 10.1002/bdrc.10010. [DOI] [PubMed] [Google Scholar]
- 100.Weston AD, Blumberg B, Underhill TM. Active repression by unliganded retinoid receptors in development: less is sometimes more. J Cell Biol. 2003;161:223–228. doi: 10.1083/jcb.200211117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Blumberg B, Bolado J, Jr., Moreno TA, Kintner C, Evans RM, Papalopulu N. An essential role for retinoid signaling in anteroposterior neural patterning. Development. 1997;124:373–379. doi: 10.1242/dev.124.2.373. [DOI] [PubMed] [Google Scholar]
- 102.Germain P, Gaudon C, Pogenberg V, Sanglier S, Van Dorsselaer A, Royer CA, Lazar MA, Bourguet W, Gronemeyer H. Differential action on coregulator interaction defines inverse retinoid agonists and neutral antagonists. Chem Biol. 2009;16:479–489. doi: 10.1016/j.chembiol.2009.03.008. [DOI] [PubMed] [Google Scholar]
- 103.Germain P, Staels B, Dacquet C, Spedding M, Laudet V. Overview of nomenclature of nuclear receptors. Pharmacol Rev. 2006;58:685–704. doi: 10.1124/pr.58.4.2. [DOI] [PubMed] [Google Scholar]
- 104.Klein ES, Pino ME, Johnson AT, Davies PJ, Nagpal S, Thacher SM, Krasinski G, Chandraratna RA. Identification and functional separation of retinoic acid receptor neutral antagonists and inverse agonists. J Biol Chem. 1996;271:22692–22696. doi: 10.1074/jbc.271.37.22692. [DOI] [PubMed] [Google Scholar]
- 105.le Maire A, Teyssier C, Erb C, Grimaldi M, Alvarez S, de Lera AR, Balaguer P, Gronemeyer H, Royer CA, Germain P, Bourguet W. A unique secondary-structure switch controls constitutive gene repression by retinoic acid receptor. Nat Struct Mol Biol. 2010;17:801–807. doi: 10.1038/nsmb.1855. [DOI] [PubMed] [Google Scholar]
- 106.Laudet V, Gronemeyer H. The Nuclear Receptor Factsbook. Gulf Professional Publishing; 2002. [Google Scholar]
- 107.Chambon P. A decade of molecular biology of retinoic acid receptors. Faseb J. 1996;10:940–954. [PubMed] [Google Scholar]
- 108.Forman BM, Casanova J, Raaka BM, Ghysdael J, Samuels HH. Half-site spacing and orientation determines whether thyroid hormone and retinoic acid receptors and related factors bind to DNA response elements as monomers, homodimers, or heterodimers. Mol Endocrinol. 1992;6:429–442. doi: 10.1210/mend.6.3.1316541. [DOI] [PubMed] [Google Scholar]
- 109.Kurokawa R, Yu VC, Naar A, Kyakumoto S, Han Z, Silverman S, Rosenfeld MG, Glass CK. Differential orientations of the DNA-binding domain and carboxy-terminal dimerization interface regulate binding site selection by nuclear receptor heterodimers. Genes Dev. 1993;7:1423–1435. doi: 10.1101/gad.7.7b.1423. [DOI] [PubMed] [Google Scholar]
- 110.Naar AM, Boutin JM, Lipkin SM, Yu VC, Holloway JM, Glass CK, Rosenfeld MG. The orientation and spacing of core DNA-binding motifs dictate selective transcriptional responses to three nuclear receptors. Cell. 1991;65:1267–1279. doi: 10.1016/0092-8674(91)90021-p. [DOI] [PubMed] [Google Scholar]
- 111.Perlmann T, Rangarajan PN, Umesono K, Evans RM. Determinants for selective RAR and TR recognition of direct repeat HREs. Genes Dev. 1993;7:1411–1422. doi: 10.1101/gad.7.7b.1411. [DOI] [PubMed] [Google Scholar]
- 112.Umesono K, Murakami KK, Thompson CC, Evans RM. Direct repeats as selective response elements for the thyroid hormone, retinoic acid, and vitamin D3 receptors. Cell. 1991;65:1255–1266. doi: 10.1016/0092-8674(91)90020-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Vivanco Ruiz MM, Bugge TH, Hirschmann P, Stunnenberg HG. Functional characterization of a natural retinoic acid responsive element. EMBO J. 1991;10:3829–3838. doi: 10.1002/j.1460-2075.1991.tb04952.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Glass CK. Differential recognition of target genes by nuclear receptor monomers, dimers, and heterodimers. Endocr Rev. 1994;15:391–407. doi: 10.1210/edrv-15-3-391. [DOI] [PubMed] [Google Scholar]
- 115.Kato S, Sasaki H, Suzawa M, Masushige S, Tora L, Chambon P, Gronemeyer H. Widely spaced, directly repeated PuGGTCA elements act as promiscuous enhancers for different classes of nuclear receptors. Mol Cell Biol. 1995;15:5858–5867. doi: 10.1128/mcb.15.11.5858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Balmer JE, Blomhoff R. A robust characterization of retinoic acid response elements based on a comparison of sites in three species. J Steroid Biochem Mol Biol. 2005;96:347–354. doi: 10.1016/j.jsbmb.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 117.Moutier E, Ye T, Choukrallah MA, Urban S, Osz J, Chatagnon A, Delacroix L, Langer D, Rochel N, Moras D, Benoit G, Davidson I. Retinoic acid receptors recognize the mouse genome through binding elements with diverse spacing and topology. J Biol Chem. 2012;287:26328–26341. doi: 10.1074/jbc.M112.361790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Laperriere D, Wang TT, White JH, Mader S. Widespread Alu repeat-driven expansion of consensus DR2 retinoic acid response elements during primate evolution. BMC Genomics. 2007;8:23. doi: 10.1186/1471-2164-8-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hu Q, Tanasa B, Trabucchi M, Li W, Zhang J, Ohgi KA, Rose DW, Glass CK, Rosenfeld MG. DICER- and AGO3-dependent generation of retinoic acid-induced DR2 Alu RNAs regulates human stem cell proliferation. Nat Struct Mol Biol. 2012;19:1168–1175. doi: 10.1038/nsmb.2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Hua S, Kittler R, White KP. Genomic antagonism between retinoic acid and estrogen signaling in breast cancer. Cell. 2009;137:1259–1271. doi: 10.1016/j.cell.2009.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Delacroix L, Moutier E, Altobelli G, Legras S, Poch O, Choukrallah MA, Bertin I, Jost B, Davidson I. Cell-specific interaction of retinoic acid receptors with target genes in mouse embryonic fibroblasts and embryonic stem cells. Mol Cell Biol. 2010;30:231–244. doi: 10.1128/MCB.00756-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Mahony S, Mazzoni EO, McCuine S, Young RA, Wichterle H, Gifford DK. Ligand-dependent dynamics of retinoic acid receptor binding during early neurogenesis. Genome Biol. 2011;12:R2. doi: 10.1186/gb-2011-12-1-r2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Lalevee S, Anno YN, Chatagnon A, Samarut E, Poch O, Laudet V, Benoit G, Lecompte O, Rochette-Egly C. Genome-wide in silico identification of new conserved and functional retinoic acid receptor response elements (direct repeats separated by 5 bp) J Biol Chem. 2011;286:33322–33334. doi: 10.1074/jbc.M111.263681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Phan TQ, Jow MM, Privalsky ML. DNA recognition by thyroid hormone and retinoic acid receptors: 3,4,5 rule modified. Mol Cell Endocrinol. 2010;319:88–98. doi: 10.1016/j.mce.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Gilbert SF. Developmental Biology. 9th ed. 01375. Sinauer Associates, Inc.; Sunderland, MA: 2010. [Google Scholar]
- 126.Slack JMW. From Egg to Embryo: Regional Specification in Early Development. Cambridge University Press; 1991. [Google Scholar]
- 127.Gurdon JB, Standley H, Dyson S, Butler K, Langon T, Ryan K, Stennard F, Shimizu K, Zorn A. Single cells can sense their position in a morphogen gradient. Development. 1999;126:5309–5317. doi: 10.1242/dev.126.23.5309. [DOI] [PubMed] [Google Scholar]
- 128.Izpisua-Belmonte JC, Tickle C, Dolle P, Wolpert L, Duboule D. Expression of the homeobox Hox-4 genes and the specification of position in chick wing development. Nature. 1991;350:585–589. doi: 10.1038/350585a0. [DOI] [PubMed] [Google Scholar]
- 129.Eichele G, Thaller C. Characterization of concentration gradients of a morphogenetically active retinoid in the chick limb bud. J Cell Biol. 1987;105:1917–1923. doi: 10.1083/jcb.105.4.1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Reijntjes S, Gale E, Maden M. Generating gradients of retinoic acid in the chick embryo: Cyp26C1 expression and a comparative analysis of the Cyp26 enzymes. Dev Dyn. 2004;230:509–517. doi: 10.1002/dvdy.20025. [DOI] [PubMed] [Google Scholar]
- 131.Niederreither K, Vermot J, Le Roux I, Schuhbaur B, Chambon P, Dolle P. The regional pattern of retinoic acid synthesis by RALDH2 is essential for the development of posterior pharyngeal arches and the enteric nervous system. Development. 2003;130:2525–2534. doi: 10.1242/dev.00463. [DOI] [PubMed] [Google Scholar]
- 132.Sirbu IO, Gresh L, Barra J, Duester G. Shifting boundaries of retinoic acid activity control hindbrain segmental gene expression. Development. 2005;132:2611–2622. doi: 10.1242/dev.01845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Shimozono S, Iimura T, Kitaguchi T, Higashijima S, Miyawaki A. Visualization of an endogenous retinoic acid gradient across embryonic development. Nature. 2013;496:363–366. doi: 10.1038/nature12037. [DOI] [PubMed] [Google Scholar]
- 134.Diez del Corral R, Olivera-Martinez I, Goriely A, Gale E, Maden M, Storey K. Opposing FGF and retinoid pathways control ventral neural pattern, neuronal differentiation, and segmentation during body axis extension. Neuron. 2003;40:65–79. doi: 10.1016/s0896-6273(03)00565-8. [DOI] [PubMed] [Google Scholar]
- 135.Diez del Corral R, Storey KG. Opposing FGF and retinoid pathways: a signalling switch that controls differentiation and patterning onset in the extending vertebrate body axis. Bioessays. 2004;26:857–869. doi: 10.1002/bies.20080. [DOI] [PubMed] [Google Scholar]
- 136.Vermot J, Pourquie O. Retinoic acid coordinates somitogenesis and left-right patterning in vertebrate embryos. Nature. 2005;435:215–220. doi: 10.1038/nature03488. [DOI] [PubMed] [Google Scholar]
- 137.Maden M, Graham A, Zile M, Gale E. Abnormalities of somite development in the absence of retinoic acid. Int J Dev Biol. 2000;44:151–159. [PubMed] [Google Scholar]
- 138.Mic FA, Molotkov A, Molotkova N, Duester G. Raldh2 expression in optic vesicle generates a retinoic acid signal needed for invagination of retina during optic cup formation. Developmental dynamics : an official publication of the American Association of Anatomists. 2004;231:270–277. doi: 10.1002/dvdy.20128. [DOI] [PubMed] [Google Scholar]
- 139.Escriva H, Holland ND, Gronemeyer H, Laudet V, Holland LZ. The retinoic acid signaling pathway regulates anterior/posterior patterning in the nerve cord and pharynx of amphioxus, a chordate lacking neural crest. Development. 2002;129:2905–2916. doi: 10.1242/dev.129.12.2905. [DOI] [PubMed] [Google Scholar]
- 140.Schubert M, Yu JK, Holland ND, Escriva H, Laudet V, Holland LZ. Retinoic acid signaling acts via Hox1 to establish the posterior limit of the pharynx in the chordate amphioxus. Development. 2005;132:61–73. doi: 10.1242/dev.01554. [DOI] [PubMed] [Google Scholar]
- 141.Rosenthal N, Xavier-Neto J. From the bottom of the heart: anteroposterior decisions in cardiac muscle differentiation. Curr Opin Cell Biol. 2000;12:742–746. doi: 10.1016/s0955-0674(00)00162-9. [DOI] [PubMed] [Google Scholar]
- 142.Sockanathan S, Jessell TM. Motor neuron-derived retinoid signaling specifies the subtype identity of spinal motor neurons. Cell. 1998;94:503–514. doi: 10.1016/s0092-8674(00)81591-3. [DOI] [PubMed] [Google Scholar]
- 143.Malpel S, Mendelsohn C, Cardoso WV. Regulation of retinoic acid signaling during lung morphogenesis. Development. 2000;127:3057–3067. doi: 10.1242/dev.127.14.3057. [DOI] [PubMed] [Google Scholar]
- 144.Chen F, Marquez H, Kim YK, Qian J, Shao F, Fine A, Cruikshank WW, Quadro L, Cardoso WV. Prenatal retinoid deficiency leads to airway hyperresponsiveness in adult mice. J Clin Invest. 124:801–811. doi: 10.1172/JCI70291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Dranse HJ, Sampaio AV, Petkovich M, Underhill TM. Genetic deletion of Cyp26b1 negatively impacts limb skeletogenesis by inhibiting chondrogenesis. J Cell Sci. 2011;124:2723–2734. doi: 10.1242/jcs.084699. [DOI] [PubMed] [Google Scholar]
- 146.Sakai Y, Meno C, Fujii H, Nishino J, Shiratori H, Saijoh Y, Rossant J, Hamada H. The retinoic acid-inactivating enzyme CYP26 is essential for establishing an uneven distribution of retinoic acid along the anterio-posterior axis within the mouse embryo. Genes Dev. 2001;15:213–225. doi: 10.1101/gad.851501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Hernandez RE, Putzke AP, Myers JP, Margaretha L, Moens CB. Cyp26 enzymes generate the retinoic acid response pattern necessary for hindbrain development. Development. 2007;134:177–187. doi: 10.1242/dev.02706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Ruppert EE, Carle KJ. Morphology of metazoan circulatory systems. Zoomorphology. 1983;103:193–208. [Google Scholar]
- 149.Kirby ML, Waldo KL. Neural crest and cardiovascular patterning. Circ Res. 1995;77:211–215. doi: 10.1161/01.res.77.2.211. [DOI] [PubMed] [Google Scholar]
- 150.Christoffels VM, Habets PE, Franco D, Campione M, de Jong F, Lamers WH, Bao ZZ, Palmer S, Biben C, Harvey RP, Moorman AF. Chamber formation and morphogenesis in the developing mammalian heart. Dev Biol. 2000;223:266–278. doi: 10.1006/dbio.2000.9753. [DOI] [PubMed] [Google Scholar]
- 151.Mark M, Ghyselinck NB, Chambon P. Function of retinoic acid receptors during embryonic development. Nucl Recept Signal. 2009;7:e002. doi: 10.1621/nrs.07002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Gruber PJ, Kubalak SW, Pexieder T, Sucov HM, Evans RM, Chien KR. RXR alpha deficiency confers genetic susceptibility for aortic sac, conotruncal, atrioventricular cushion, and ventricular muscle defects in mice. J Clin Invest. 1996;98:1332–1343. doi: 10.1172/JCI118920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Mascrez B, Ghyselinck NB, Chambon P, Mark M. A transcriptionally silent RXRalpha supports early embryonic morphogenesis and heart development. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:4272–4277. doi: 10.1073/pnas.0813143106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Xavier-Neto J, Rosenthal N, Silva FA, Matos TG, Hochgreb T, Linhares VL. Retinoid signaling and cardiac anteroposterior segmentation. Genesis. 2001;31:97–104. doi: 10.1002/gene.10009. [DOI] [PubMed] [Google Scholar]
- 155.Osmond MK, Butler AJ, Voon FC, Bellairs R. The effects of retinoic acid on heart formation in the early chick embryo. Development. 1991;113:1405–1417. doi: 10.1242/dev.113.4.1405. [DOI] [PubMed] [Google Scholar]
- 156.Stainier DY, Fishman MC. Patterning the zebrafish heart tube: acquisition of anteroposterior polarity. Dev Biol. 1992;153:91–101. doi: 10.1016/0012-1606(92)90094-w. [DOI] [PubMed] [Google Scholar]
- 157.Drysdale TA, Patterson KD, Saha M, Krieg PA. Retinoic acid can block differentiation of the myocardium after heart specification. Dev Biol. 1997;188:205–215. doi: 10.1006/dbio.1997.8623. [DOI] [PubMed] [Google Scholar]
- 158.Jiang Y, Drysdale TA, Evans T. A role for GATA-4/5/6 in the regulation of Nkx2.5 expression with implications for patterning of the precardiac field. Dev Biol. 1999;216:57–71. doi: 10.1006/dbio.1999.9469. [DOI] [PubMed] [Google Scholar]
- 159.Keegan BR, Feldman JL, Begemann G, Ingham PW, Yelon D. Retinoic acid signaling restricts the cardiac progenitor pool. Science. 2005;307:247–249. doi: 10.1126/science.1101573. [DOI] [PubMed] [Google Scholar]
- 160.Waxman JS, Keegan BR, Roberts RW, Poss KD, Yelon D. Hoxb5b acts downstream of retinoic acid signaling in the forelimb field to restrict heart field potential in zebrafish. Dev Cell. 2008;15:923–934. doi: 10.1016/j.devcel.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Begemann G, Schilling TF, Rauch GJ, Geisler R, Ingham PW. The zebrafish neckless mutation reveals a requirement for raldh2 in mesodermal signals that pattern the hindbrain. Development. 2001;128:3081–3094. doi: 10.1242/dev.128.16.3081. [DOI] [PubMed] [Google Scholar]
- 162.Stafford D, Prince VE. Retinoic acid signaling is required for a critical early step in zebrafish pancreatic development. Curr Biol. 2002;12:1215–1220. doi: 10.1016/s0960-9822(02)00929-6. [DOI] [PubMed] [Google Scholar]
- 163.Azambuja AP. Departamento de Biologia Celular e do Desenvolvimento. PhD, Universidade de São Paulo; São Paulo: 2009. Mecanismos embrionários de diferenciação de precursores coronários: princípios para aplicação em terapia celular. p. 123. [Google Scholar]
- 164.Yutzey K, Gannon M, Bader D. Diversification of cardiomyogenic cell lineages in vitro. Dev Biol. 1995;170:531–541. doi: 10.1006/dbio.1995.1234. [DOI] [PubMed] [Google Scholar]
- 165.Buckingham M, Meilhac S, Zaffran S. Building the mammalian heart from two sources of myocardial cells. Nat Rev Genet. 2005;6:826–835. doi: 10.1038/nrg1710. [DOI] [PubMed] [Google Scholar]
- 166.Kelly RG, Brown NA, Buckingham ME. The arterial pole of the mouse heart forms from Fgf10-expressing cells in pharyngeal mesoderm. Dev Cell. 2001;1:435–440. doi: 10.1016/s1534-5807(01)00040-5. [DOI] [PubMed] [Google Scholar]
- 167.Xavier-Neto J, Sura-Trueba S, Stolfi A, Souza HM, Sobreira TJ, Schubert M, Castillo HA. An unauthorized biography of the second heart field and a pioneer/scaffold model for cardiac development. Curr Top Dev Biol. 2012;100:67–105. doi: 10.1016/B978-0-12-387786-4.00003-8. [DOI] [PubMed] [Google Scholar]
- 168.Abu-Issa R. Heart fields: spatial polarity and temporal dynamics. Anat Rec (Hoboken) 2014;297:175–182. doi: 10.1002/ar.22831. [DOI] [PubMed] [Google Scholar]
- 169.Cai CL, Liang X, Shi Y, Chu PH, Pfaff SL, Chen J, Evans S. Isl1 identifies a cardiac progenitor population that proliferates prior to differentiation and contributes a majority of cells to the heart. Dev Cell. 2003;5:877–889. doi: 10.1016/s1534-5807(03)00363-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Yuan S, Schoenwolf GC. Islet-1 marks the early heart rudiments and is asymmetrically expressed during early rotation of the foregut in the chick embryo. Anat Rec. 2000;260:204–207. doi: 10.1002/1097-0185(20001001)260:2<204::AID-AR90>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 171.Ryckebusch L, Wang Z, Bertrand N, Lin SC, Chi X, Schwartz R, Zaffran S, Niederreither K. Retinoic acid deficiency alters second heart field formation. Proc Natl Acad Sci U S A. 2008 doi: 10.1073/pnas.0712344105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Sirbu IO, Zhao X, Duester G. Retinoic acid controls heart anteroposterior patterning by down-regulating Isl1 through the Fgf8 pathway. Dev Dyn. 2008;237:1627–1635. doi: 10.1002/dvdy.21570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Zaffran S, el Robrini N, Bertrand N. Retinoids and Cardiac Development. J. Dev. Biol. 2014;2:50–71. [Google Scholar]
- 174.Bertrand N, Roux M, Ryckebusch L, Niederreither K, Dolle P, Moon A, Capecchi M, Zaffran S. Hox genes define distinct progenitor sub-domains within the second heart field. Dev Biol. 2011;353:266–274. doi: 10.1016/j.ydbio.2011.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Kastner P, Messaddeq N, Mark M, Wendling O, Grondona JM, Ward S, Ghyselinck N, Chambon P. Vitamin A deficiency and mutations of RXRalpha, RXRbeta and RARalpha lead to early differentiation of embryonic ventricular cardiomyocytes. Development. 1997;124:4749–4758. doi: 10.1242/dev.124.23.4749. [DOI] [PubMed] [Google Scholar]
- 176.Chen J, Kubalak SW, Chien KR. Ventricular muscle-restricted targeting of the RXRalpha gene reveals a non-cell-autonomous requirement in cardiac chamber morphogenesis. Development. 1998;125:1943–1949. doi: 10.1242/dev.125.10.1943. [DOI] [PubMed] [Google Scholar]
- 177.Tran CM, Sucov HM. The RXRalpha gene functions in a non-cell-autonomous manner during mouse cardiac morphogenesis. Development. 1998;125:1951–1956. doi: 10.1242/dev.125.10.1951. [DOI] [PubMed] [Google Scholar]
- 178.Gittenberger-de Groot AC, Vrancken Peeters MP, Bergwerff M, Mentink MM, Poelmann RE. Epicardial outgrowth inhibition leads to compensatory mesothelial outflow tract collar and abnormal cardiac septation and coronary formation. Circ Res. 2000;87:969–971. doi: 10.1161/01.res.87.11.969. [DOI] [PubMed] [Google Scholar]
- 179.Pennisi DJ, Ballard VL, Mikawa T. Epicardium is required for the full rate of myocyte proliferation and levels of expression of myocyte mitogenic factors FGF2 and its receptor, FGFR-1, but not for transmural myocardial patterning in the embryonic chick heart. Dev Dyn. 2003;228:161–172. doi: 10.1002/dvdy.10360. [DOI] [PubMed] [Google Scholar]
- 180.Perez-Pomares JM, Phelps A, Sedmerova M, Carmona R, Gonzalez-Iriarte M, Munoz-Chapuli R, Wessels A. Experimental studies on the spatiotemporal expression of WT1 and RALDH2 in the embryonic avian heart: a model for the regulation of myocardial and valvuloseptal development by epicardially derived cells (EPDCs) Dev Biol. 2002;247:307–326. doi: 10.1006/dbio.2002.0706. [DOI] [PubMed] [Google Scholar]
- 181.Stuckmann I, Evans S, Lassar AB. Erythropoietin and retinoic acid, secreted from the epicardium, are required for cardiac myocyte proliferation. Dev Biol. 2003;255:334–349. doi: 10.1016/s0012-1606(02)00078-7. [DOI] [PubMed] [Google Scholar]
- 182.Perez-Pomares JM, de la Pompa JL. Signaling during epicardium and coronary vessel development. Circ Res. 2011;109:1429–1442. doi: 10.1161/CIRCRESAHA.111.245589. [DOI] [PubMed] [Google Scholar]
- 183.Sucov HM, Gu Y, Thomas S, Li P, Pashmforoush M. Epicardial control of myocardial proliferation and morphogenesis. Pediatr Cardiol. 2009;30:617–625. doi: 10.1007/s00246-009-9391-8. [DOI] [PubMed] [Google Scholar]
- 184.Chen T, Chang TC, Kang JO, Choudhary B, Makita T, Tran CM, Burch JB, Eid H, Sucov HM. Epicardial induction of fetal cardiomyocyte proliferation via a retinoic acid-inducible trophic factor. Dev Biol. 2002;250:198–207. doi: 10.1006/dbio.2002.0796. [DOI] [PubMed] [Google Scholar]
- 185.Guadix JA, Ruiz-Villalba A, Lettice L, Velecela V, Munoz-Chapuli R, Hastie ND, Perez-Pomares JM, Martinez-Estrada OM. Wt1 controls retinoic acid signalling in embryonic epicardium through transcriptional activation of Raldh2. Development. 2011;138:1093–1097. doi: 10.1242/dev.044594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Castillo HA, Cravo RM, Azambuja AP, Simoes-Costa MS, Sura-Trueba S, Gonzalez J, Slonimsky E, Almeida K, Abreu JG, de Almeida MA, Sobreira TP, de Oliveira SH, de Oliveira PS, Signore IA, Colombo A, Concha ML, Spengler TS, Bronner-Fraser M, Nobrega M, Rosenthal N, Xavier-Neto J. Insights into the organization of dorsal spinal cord pathways from an evolutionarily conserved raldh2 intronic enhancer. Development. 2010;137:507–518. doi: 10.1242/dev.043257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Wu H, Lee SH, Gao J, Liu X, Iruela-Arispe ML. Inactivation of erythropoietin leads to defects in cardiac morphogenesis. Development. 1999;126:3597–3605. doi: 10.1242/dev.126.16.3597. [DOI] [PubMed] [Google Scholar]
- 188.Lavine KJ, Yu K, White AC, Zhang X, Smith C, Partanen J, Ornitz DM. Endocardial and epicardial derived FGF signals regulate myocardial proliferation and differentiation in vivo. Dev Cell. 2005;8:85–95. doi: 10.1016/j.devcel.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 189.Li P, Cavallero S, Gu Y, Chen TH, Hughes J, Hassan AB, Bruning JC, Pashmforoush M, Sucov HM. IGF signaling directs ventricular cardiomyocyte proliferation during embryonic heart development. Development. 2011;138:1795–1805. doi: 10.1242/dev.054338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Brade T, Kumar S, Cunningham TJ, Chatzi C, Zhao X, Cavallero S, Li P, Sucov HM, Ruiz-Lozano P, Duester G. Retinoic acid stimulates myocardial expansion by induction of hepatic erythropoietin which activates epicardial Igf2. Development. 2011;138:139–148. doi: 10.1242/dev.054239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Rossant J. Mouse mutants and cardiac development: new molecular insights into cardiogenesis. Circ Res. 1996;78:349–353. doi: 10.1161/01.res.78.3.349. [DOI] [PubMed] [Google Scholar]
- 192.Lavine KJ, Long F, Choi K, Smith C, Ornitz DM. Hedgehog signaling to distinct cell types differentially regulates coronary artery and vein development. Development. 2008;135:3161–3171. doi: 10.1242/dev.019919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Vrancken Peeters MP, Gittenberger-de Groot AC, Mentink MM, Hungerford JE, Little CD, Poelmann RE. The development of the coronary vessels and their differentiation into arteries and veins in the embryonic quail heart. Dev Dyn. 1997;208:338–348. doi: 10.1002/(SICI)1097-0177(199703)208:3<338::AID-AJA5>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 194.Waxman JS, Yelon D. Increased Hox activity mimics the teratogenic effects of excess retinoic acid signaling. Dev Dyn. 2009;238:1207–1213. doi: 10.1002/dvdy.21951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Grandel H, Lun K, Rauch GJ, Rhinn M, Piotrowski T, Houart C, Sordino P, Kuchler AM, Schulte-Merker S, Geisler R, Holder N, Wilson SW, Brand M. Retinoic acid signalling in the zebrafish embryo is necessary during pre-segmentation stages to pattern the anterior-posterior axis of the CNS and to induce a pectoral fin bud. Development. 2002;129:2851–2865. doi: 10.1242/dev.129.12.2851. [DOI] [PubMed] [Google Scholar]
- 196.Metscher BD, Ahlberg PE. Zebrafish in context: uses of a laboratory model in comparative studies. Dev Biol. 1999;210:1–14. doi: 10.1006/dbio.1999.9230. [DOI] [PubMed] [Google Scholar]
- 197.Freitas R, Zhang G, Cohn MJ. Biphasic Hoxd gene expression in shark paired fins reveals an ancient origin of the distal limb domain. PLoS One. 2007;2:e754. doi: 10.1371/journal.pone.0000754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Schneider I, Shubin NH. The origin of the tetrapod limb: from expeditions to enhancers. Trends Genet. 2013;29:419–426. doi: 10.1016/j.tig.2013.01.012. [DOI] [PubMed] [Google Scholar]