Abstract
Purpose
To determine whether pretreatment CT texture features can improve patient risk stratification beyond conventional prognostic factors (CPFs) in stage III non-small cell lung cancer (NSCLC).
Methods and Materials
We retrospectively reviewed 91 patients with stage III NSCLC treated with definitive chemoradiation. All patients underwent a pretreatment diagnostic contrast enhanced CT (CE-CT) followed by a 4D-CT for treatment simulation. We used the average (average-CT) and expiratory (T50-CT) images from the 4D-CT along with the CE-CT for texture extraction. Histogram, gradient, co-occurrence, gray-tone difference, and filtration-based techniques were used for texture feature extraction. Penalized Cox regression implementing cross-validation was used for covariate selection and modeling. Models incorporating texture features from the 3 image types and CPFs were compared to models incorporating CPFs alone for overall survival (OS), local-regional control (LRC), and freedom from distant metastases (FFDM). Predictive Kaplan-Meier curves were generated using leave-one-out cross-validation. Patients were stratified based on their predicted outcome being above/below the median. Reproducibility of texture features was evaluated using test-retest scans from independent patients and quantified using concordance correlation coefficients (CCC). We compared models incorporating the reproducibility seen on test-retest scans to our original models and determined the classification reproducibility.
Results
Models incorporating both texture features and CPFs demonstrated a significant improvement in risk stratification compared to models using CPFs alone for OS (p=0.046), LRC (p=0.01), and FFDM (p=0.005). The average CCC was 0.89, 0.91, and 0.67 for texture features extracted from the average-CT, T50-CT, and CE-CT, respectively. Incorporating reproducibility within our models yielded 80.4 (SD=3.7), 78.3 (SD=4.0), and 78.8 (SD=3.9) percent classification reproducibility in terms of OS, LRC, and FFDM, respectively.
Conclusions
Pretreatment tumor texture may provide prognostic information beyond what is obtained from CPFs. Models incorporating feature reproducibility achieved classification rates of ~80%. External validation would be required to establish texture as a prognostic factor.
Introduction
Lung cancer is currently the most common cause of death from cancer in the United States.1 Frequently, patients present with Stage III disease and are not amenable to surgical resection. For these patients, standard of care consists of definitive chemoradiotherapy. Even when treated aggressively, patient 3-year survival is approximately 27%.2 Inoperable non-small-cell lung cancer (NSCLC) patients are a very heterogeneous population with varying degrees tumor extent, comorbidity, etc. This presents a significant challenge to clinicians when attempting to provide optimal treatment. Traditional TNM staging is not ideal for stratifying patients and there is a tremendous need to develop better tools for assessing prognosis.
Efforts have been made to address this issue by identifying prognostic genetic expression signatures and using functional imaging techniques such as FDG-PET.3–5 Recently, tumor heterogeneity as assessed by computed tomography (CT) has yielded promising preliminary results in a variety of cancers.6–8 These techniques assess the spatial variation of tumor density within a patient’s tumor. Since CT is routinely obtained for all patients undergoing radiation therapy, prognostic markers generated in this manner would be less costly and less time consuming than genetic or functional imaging based techniques.
In this study we examine the impact of CT texture features to enhance patient risk stratification beyond conventional prognostic factors (CPFs) for patients with Stage III NSCLC.
Methods and Materials
Patients
We retrospectively reviewed the medical records of patients with stage III NSCLC treated with definitive radiation therapy between July 2004 and January 2012. These dates were chosen in order to include patients receiving 4DCT, which our institution implemented in early 2004, and provide adequate follow-up time. We excluded all patients receiving induction chemotherapy, proton based radiation therapy, <5 years post treatment for solid tumor, multiple primary lesions, non-platin based concurrent chemotherapy, and those not receiving a diagnostic contrast enhanced scan prior to 4DCT treatment planning. Additional patients were excluded for the following reasons: non-identifiable or small primary tumor (16), image restoration error (7), uncertainty in tumor extent (8), having a break from treatment longer than one week (2), and image artifacts (8). This yielded 91 out of the 132 patients for analysis. The median follow-up for all living patients at time of analysis was 59 months (range, 17 – 97 months). The primary endpoints of our analysis were overall survival (OS), local-regional control (LRC), and freedom from distant metastasis (FFDM). The endpoints were defined from first day of definitive radiation treatment until death, local-regional failure, or presentation of metastatic disease for OS, LRC, and FFDM, respectively. Patients not experiencing an event were censored for the corresponding endpoint at the date of death or last follow-up. The institutional review board approved this retrospective chart review study and waived the need for informed consent. We also complied with all Health Insurance Portability and Accountability Act (HIPAA) regulations.
Imaging
All patients received a diagnostic contrast enhanced CT (CE-CT) and a non-contrasted 4DCT scan prior to treatment. For contrast enhanced scans, patients were scanned using 120 kVp, 400–1160mA, and an exposure time of 265–570ms. All images were reconstructed using the standard reconstruction kernel. Axial images were 512 × 512 pixels with voxel dimensions of 0.059–0.090cm × 0.059–0.090cm × 0.25cm.
For the 4D-CT scans, the average intensity projection (AVG-CT) and expiratory phase (T50-CT) images were used in this study. Patients were scanned using 120 kVp, 100–200mA, and an exposure time of 500–800ms. All images were reconstructed using the standard reconstruction kernel. Axial images were 512 × 512 pixels with voxel dimensions of 0.096cm × 0.096cm × 0.25–0.30cm.
Conventional Prognostic Factors
The CPFs included in our analysis were T stage (T1/2 vs T3/4), N (N0/1 vs N2/3 Stage, Overall Stage (IIIa vs. IIIb), age (≥65 vs. <65), gender, histology (SCC vs. Other), Charlson Comorbidity Index (CCI) (>0 vs. 0), ECOG performance status(>0 vs 0), Karnofsky Performance Status (KPS) (≥80 vs. <80), smoking status (current vs. former/never), estimated pack years (continuous), and gross tumor volume (GTV) (continuous). These factors were included as they have all been suggested to be prognostic in stage III NSCLC.9 All TNM staging was performed according to the 7th edition American Joint Committee on Cancer staging manual.10 GTV was measured by the volume of the initial tumor contoured for patient treatment which included both the primary and nodal disease.
Texture Analysis
Tumor texture analysis was conducted using the gross tumor volume contour delineated by each patient’s treating physician. The nodal tumor volumes were excluded from texture analysis. In some cases further contour modification was performed. The reason for further modification is due to the goal of analyzing tissue with an extremely high likelihood of representing tumor whereas clinically physicians routinely include any and all tissue with a reasonable likelihood of representing tumor. Therefore, overly generous portions of the contour such as invasion into bone and other normal tissue structures such as the aorta were modified (DF). Contours were extracted and analyzed directly from our treatment planning system using in-house software built using a commercial software package (Matlab version 8.1.0. Natick, Massachusetts: The MathWorks Inc., 2013). For the AVG-CT and T50-CT, a lower and upper threshold of −100 to 200 Hounsfield units (HU) was implemented to exclude lung tissue, air, and/or bone in order to determine our final region of interest (ROI). A lower threshold of −100 HU was used for the contrast enhanced images with no upper threshold. Only voxels within the defined threshold bounds were included in the texture analysis. We extracted 66 texture features from the windowed tumor contours for each of the three image types (AVG-CT, T50-CT, and CE-CT) and these are shown below in Table 1 organized by method. The methods used were histogram (IHIST), absolute gradient (GRAD), nearest gray tone difference matrix (NGTDM)11, co-occurrence matrix (COM)12, Laplacian of Gaussian filtration (LoG)13. When reporting texture metrics, the notation convention of method_feature will be used. For instance, the NGTDM coarseness feature would be identified by NGTDM_coarseness.
Table 1.
Extracted Texture Features
| Intensity Histogram (IHIST) |
Absolute Gradient (Grad) |
Nearest Gray Tone Difference Matrix (NGTDM) |
Co-Occurrence Matrix (COM) |
Laplacian of Gaussian Filtration Metrics (LoG)* |
|---|---|---|---|---|
| Mean Variance Skewness Kurtosis Entropy Uniformity |
Mean Variance Skewness Kurtosis % non-zero |
Coarseness Contrast Busyness |
Angular 2nd Moment Contrast Correlation Sum of Squares Inv. Diff. Moment Sum Average Sum Variance Sum Entropy Entropy Diff. Entropy Infomc1 Infomc2 |
Mean Uniformity Standard Deviation Entropy |
These features were calculated for sigma values used for the Laplacian of Gaussian Filter of: 1.0, 1.5, 1.8, 2.0, and 2.5 for the largest axial (LA) slice and for the entire tumor
The ROIs were scaled into 8-bit images and then filtered using a Wiener filter, a two dimensional adaptive noise-removal filter, in an attempt to reduce any Gaussian noise present within the ROI. This was done for all texture feature methods except the LoG based method as this employs its own blurring step within the filtration process. For metrics using a histogram for calculation (IHIST, LoG_Uniformity, and LoG_Entropy) 256 bins were utilized (i.e. 4096/256 = 16 HU/bin). Calculation of the NGTDM and COM features were performed for three dimensions (i.e. each reference pixel has 26 neighbors [NGTDM] and 13 unique directions [COM]). The process is the same as described by Haralick and Amadasum, however averages over all neighbors (NGTDM) and directions (COM) were performed.11,12 This allowed for COM features to be non-directional and NGTDM features to use all adjacent pixel values in calculations and not only those within the same axial slice.
Reproducibility Analysis
We obtained test-retest scans from 10, 10, and 13 independent patients for the AVG-CT, T50-CT, and CE-CT, respectively. The test-retest scans of the AVG-CT and T50-CT images were taken at our institution and on average separated by 27 min (range: 16–47). We were unable to acquire CE-CT test-retest images from patients within a close time period. We therefore used contrast enhanced scans taken outside our institution prior to treatment and compared them to the diagnostic CE-CT taken within our institution. The average separation between these scans was 38 days (range: 17–72). The contours for the test-retest scans were performed by a single observer (DF) on separate occasions for the test and retest scans in order to incorporate intra-observer contour variability. We calculated the classification reproducibility of our models incorporating the reproducibility seen via the test-retest scans. The classification reproducibility is defined as what percent of patients were categorized into the same group as our original models when incorporating the test-retest variation into our texture parameters. This was done for the models incorporating texture and CPFs.
Statistical Analysis
All statistical analyses were conducted in R 3.0.2 with the following R packages: survival v(2.37–4), penalized (0.9–42), survcomp v(1.10.0), and risksetROC v(1.0.4). The 13 clinical factors and 66 texture features from each image type (198 total texture features) were entered into a penalized multivariate Cox proportional hazards model which simultaneously performs covariate selection in addition to model development. This process is governed by the L1 penalty parameter which governs the balance between model fit and model complexity. The L1 parameter was chosen since it results in many covariate coefficients being penalized down to zero yielding few covariates with non-zero coefficients. Covariates with non-zero coefficients can be seen as the only ones “included” in the model as covariates with coefficients of zero will not contribute. This penalty parameter is estimated based on the cross-validated likelihood to optimize the model performance from a predictive perspective for future observations. All covariates were standardized prior to penalization in order to disregard the different scales present between texture features. The resulting covariate coefficients are rescaled to their original values when reporting the final model.14
When determining the out-of-sample performance of our generated models, we predominantly used methodologies suggested by Simon et al.15 We used leave-one-out cross-validation when generating Kaplan-Meier curves rather than data refitting. Cross-validated Kaplan-Meier curves use a model prediction for a particular patient when they are not used during the model training. The patient which is left out is changed and the process repeated such that each observation in the sample has a prediction from when it was not involved in model development. These predictions are used to stratify patients into risk groups which removes the bias associated with testing a model on the same dataset from which it was generated. High risk patients were defined as having predicted outcomes less than the median. Low risk patients were defined as having predicted outcomes greater than or equal to the median. When no covariates were selected during model development, a random split was used for illustrative purposes to stratify into high and low risk.
To determine if models generated using both texture and CPFs out performed models using CPFs alone in terms of risk stratification, we took the difference of the log-rank statistic associated with their respective Kaplan-Meier curves to calculate the associated p-value. In order to calculate p-values of our cross-validated stratification, we randomly permuted the outcomes with respect to the texture features and CPFs and re-ran our original analysis. This process was repeated 200 times in order to determine what proportion of randomly permuted data achieved a log-rank score greater than our original models (i.e. the p-value). P-values less than 0.05 were considered to indicate statistical significance.
Reproducibility of texture features was evaluated using concordance correlation coefficients (CCC).16 To analyze the influence of texture feature reproducibility on our model predictions, we determined the mean and standard deviation of the differences between each texture feature acquired from the test-retest images. We randomly sampled a normal distribution with the aformentioned mean and standard deviation for each texture feature and added this uncertainty to our features and re-predicted outcomes.
Results
CPFs and treatment characteristics of all patients are listed in Table 2.
Table 2.
Clinical and Treatment Characteristics
| Conventional Prognostic Factors |
N | % | Treatment Characteristics |
N | % | ||
|---|---|---|---|---|---|---|---|
| No. Patients | 91 | NA | Radiation Dose | ||||
| Median Age (years) | 65 | NA | 60 Gy @ 2 Gy/fx | 6 | 7 | ||
| Mean GTV (cc) | 132.2 | NA | 63 Gy @ 1.8 Gy/fx | 42 | 46 | ||
| Gender | 66 Gy @ 2 Gy/fx | 7 | 8 | ||||
| Male | 55 | 60 | 70Gy @ 2 Gy/fx | 20 | 22 | ||
| Female | 36 | 40 | 74 Gy @ 2 Gy/fx | 4 | 4 | ||
| T Stage | Other | 12 | 13 | ||||
| T1 | 10 | 11 | Radiation Type | ||||
| T2 | 33 | 36 | 3DCRT | 5 | 6 | ||
| T3 | 23 | 25 | IMRT | 86 | 94 | ||
| T4 | 25 | 28 | Concurrent Chemotherapy | ||||
| N Stage | Carboplatin Based | 78 | 86 | ||||
| N0/N1 | 11 | 12 | Cisplatin Based | 13 | 14 | ||
| N2/N3 | 80 | 88 | Adjuvant Chemotherapy | ||||
| Overall Stage | Yes | 37 | 41 | ||||
| IIIa | 45 | 50 | No | 54 | 59 | ||
| IIIb | 46 | 50 | |||||
| Histology | |||||||
| Squamous Cell Carcinoma | 46 | 50 | |||||
| Adenocarcinoma/Other | 45 | 50 | |||||
| Smoking Status | |||||||
| Never | 5 | 6 | |||||
| Former | 65 | 71 | |||||
| Current | 21 | 23 | |||||
| Pack Years | |||||||
| 0–24 | 13 | 14 | |||||
| 25–49 | 37 | 41 | |||||
| 50–74 | 22 | 24 | |||||
| 75+ | 19 | 21 | |||||
| CCI | |||||||
| 0–1 | 80 | 88 | |||||
| 2–3 | 9 | 10 | |||||
| 4 | 2 | 2 | |||||
| Performance Status (ECOG) | |||||||
| 0–1 | 89 | 98 | |||||
| 2 | 2 | 2 | |||||
| Performance Status (KPS) | |||||||
| 100-90 | 37 | 41 | |||||
| 80-70 | 53 | 58 | |||||
| <70 | 1 | 1 | |||||
Model Development and Analysis
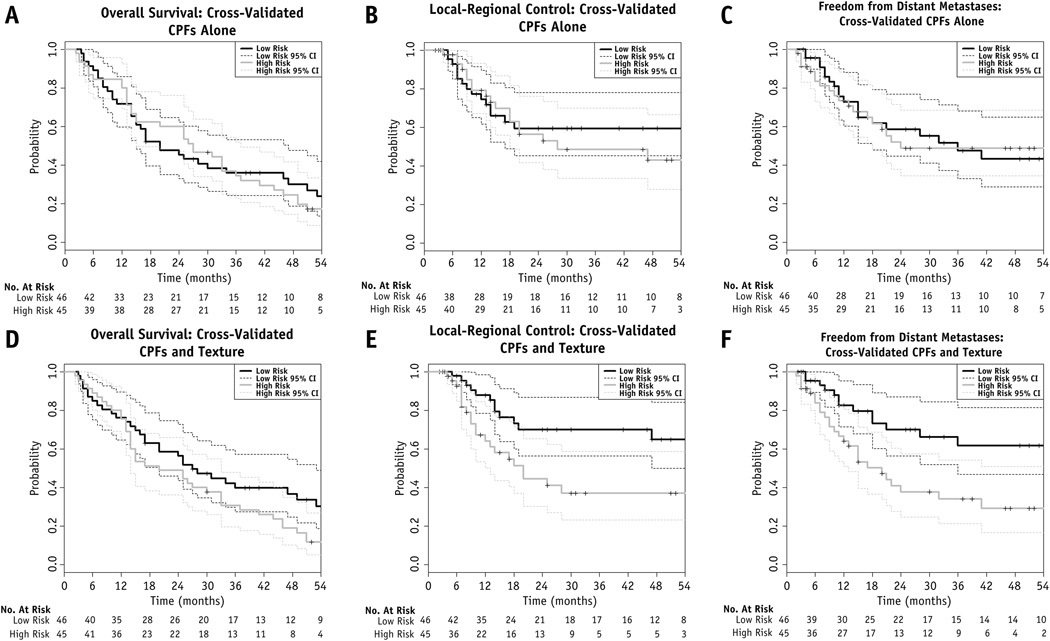
Using L1 penalized Cox proportional hazards regression; models for OS, LRC, and FFDM were generated. Covariates with non-zero coefficients are shown in Table 3. To illustrate patient stratification using these models, cross-validated Kaplan-Meier curves for OS, LRC, and FFDM were developed along with their associated 95% confidence intervals (95% CI). Models with and without texture features are shown in Figure 1. Models using both texture features and CPFs demonstrated a statistically significant improvement in stratification compared to models using CPFs alone for OS (p=0.046), LRC (p=0.01), and FFDM (p=0.005).
Table 3.
Full Models for OS, LRC, and FFDM
| Coefficient in OS Model |
Coefficient in LRC Model |
Coefficient in FFDM Model |
|
|---|---|---|---|
| Conventional Prognostic Factors: | |||
| Age (65> vs ≤65) | 0.089 | NI | NI |
| ECOG (0/1 vs 2) | 0.34 | NI | NI |
| Histology (SCC vs Other) | 6.5×10−4 | NI | NI |
| Gender (Male vs Female) | −0.05 | NI | −0.44 |
| GTV | 0.0024 | NI | 0.0011 |
| Texture Features: | |||
| CE-CT | |||
| LoG_LA_Averageσ=1 | 0.14 | NI | 0.14 |
| LoG_Averageσ=1 | NI | 0.29 | NI |
| IHIST_kurtosis | −0.022 | NI | −0.019 |
| NGTDM_busyness | NI | NI | 26.0 |
| COM_infomc1 | 2.5 | NI | |
| AVG-CT | |||
| LoG_SDσ=1 | 0.024 | NI | 0.065 |
| LoG_LA_Uniformityσ=1 | 0.43 | NI | 6.2×10−4 |
| LoG_LA_Uniformityσ=2.5 | 0.55 | NI | NI |
| T50-CT | |||
| GRAD_kurtosis | NI | NI | 0.038 |
| LoG_LA_Averageσ=1.5 | −0.080 | NI | NI |
| COM_sosvariance | 0.0011 | NI | NI |
| LoG_LA_Uniformityσ=1.5 | NI | −0.48 | NI |
Abbreviations: NI-not included in model, SCC-squamous cell carcinoma, GTV-gross tumor volume, SD-standard deviation, LA-largest axial slice
Figure 1.
Cross-validated Kaplan-Meier Curves for Models Using Texture Features and CPFs (A–C) versus Models using CPFs alone (D–F)
To confirm our results were not due to overfitting and establish p-values for the model stratification, we randomly permuted the outcomes with respect to the CPFs and texture features and performed the same cross-validation/model fitting used during our original model development. This was done 200 times to test how often random data would yield a prognostic model with a log-rank statistic greater than that generated using the true outcomes. The log-rank statistic from the permuted outcomes was greater than the original log-rank statistic in 11/200 (p = 0.055), 0/200 (p < 0.005), and 1/200 (p = 0.005) for OS, LRC, and FFDM, respectively.
Reproducibility Analysis
Data from test-retest scans from 10, 10, and 13 independent patients for the AVG-CT, T50-CT, and CE-CT, respectively were used for our assessment of reproducibility. We found that 85(56/66), 75(50/66), and 23(15/66) percent of texture features had a CCC>0.9 for features generated from T50-CT, Average-CT, and CE-CT, respectively.
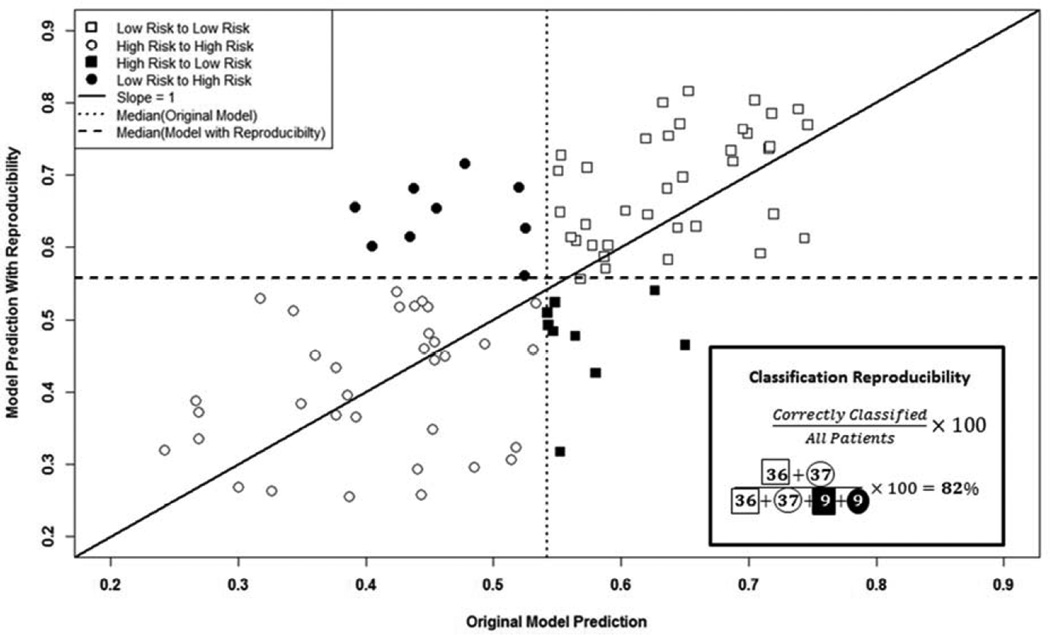
Incorporating reproducibility within our models yielded 80.4 (SD=3.7), 78.3 (SD=4.0), and 78.8 (SD=3.9) percent classification reproducibility in terms of OS, LRC, and FFDM, respectively. Figure 2 illustrates an example iteration where we compared the predicted outcome with reproducibility to the original predicted outcome in terms of FFDM and calculate the classification reproducibility.
Figure 2.
Impact of Texture Feature Reproducibility on FFDM Estimates
Discussion
We have shown that models incorporating both texture features and CPFs demonstrated a statistically significant improvement in stratification compared to models using CPFs alone in cross-validated Kaplan-Meier curves in terms of OS (p=0.046), LRC (p=0.01), and FFDM (p=0.005). Furthermore, we have shown that even when incorporating the reproducibility of our metrics within our models, classification rates of approximately 80 percent are still achieved.
We utilized cross-validation for both model selection and model validation. Commonly, researchers split their data into test and validation sets in order to validate their findings. This is problematic since both test and validation sets subsequently have reduced sample size and thus reduced statistical power when compared to the initial cohort. To overcome this, we used a cross validation approach which employs repeated data-splitting to prevent over fitting while simultaneously generating accurate estimates of the model coefficients. This process is equivalent to data splitting in terms of producing validated model coefficients, but its use of data is more efficient than a dichotomous split into test and validation sets.17 With the number of features tested, there remains a high risk of a false positive result due to multiplicity of testing. Our methods were designed to minimize this likelihood however this possibility certainly still exists.
Preliminary evidence has suggested that texture analysis has the capability to aid clinicians in cancer diagnosis, staging/prognostication, and response assessment. Table 4 summarizes the literature regarding the use of texture analysis in NSCLC patients.6–8, 18–22 It can be seen that significant heterogeneity exists in the literature in terms of study size, patient stage(s), imaging modality examined, and patient treatment. Prior work has established a relationship between tumor texture and patient outcomes post treatment; however, to the best of our knowledge this is the first study to examine this relationship in NSCLC patients of the same stage receiving chemoradiotherapy.
Table 4.
Applications of Texture Analysis in Lung Cancer Outcomes or Prognostic Factors
| Author | N | Tumor Stage(s) |
Treatment Type |
Imaging Type |
Factor(s) Associated with Tumor Texture |
|---|---|---|---|---|---|
| Al Kadi et al.6 | 15 | I–IV | NA | CE-CT |
|
| Cook et al.18 | 53 | I–IV | CRT | PET |
|
| Ganeshan et al.20 | 54 | I–IV | Unknown | NCE-CT |
|
| Ganeshan et al.7 | 17 | Unknown | NA | NCE-CT |
|
| Ganeshan et al.21 | 18 | I–IV | NA | CE-CT |
|
| Gevaert et al.19 | 26 | I–IV | PORT | NCE-CT |
|
| Mattonen et al.22 | 22 | Stage I & recurrences | NA | NCE-CT |
|
| Win et al.8 | 66 | I–IV | Unknown | NCE-CT |
|
| Current Study | 91 | III | CRT | NCE-CT & CE-CT |
|
Abbreviations: CRT, chemoradiotherapy; PORT, post-operative radiation therapy; CE-CT, contrast enhanced computed tomography; NCE-CT, non-contrast enhanced computed tomography; PET, positron emission tomography
Texture analysis has the potential to develop into a clinically useful tool using medical images that are already obtained during routine patient staging. Therefore, implementation would require little to no added cost and would not require additional time, discomfort, or radiation dose to patients. The ability to stratify patients in ways shown to be superior to current staging methods might allow physicians to deliver more optimized, patient-specific, treatment. Some may argue that for those with a poor prognosis this knowledge is usually not beneficial since patients are already receiving the maximum tolerable treatment. We would argue that treatment optimization does not necessarily equate to treatment escalation. In some instances, perhaps de-escalation of treatment and initiation of early palliative care may provide the best care for the patient. An accurate prognosis would also be beneficial to patients and their caregivers. Those identified as having a poor prognosis may be better equipped to make decisions regarding palliative versus definitive treatment and the role of hospice care.23 A study conducted by Huskamp et al. reported that half of all patients with metastatic lung cancer are not approached about hospice care until 2 months before their death.24 It is important to keep in mind that our analysis was only able to moderately separate patient overall survival. While our models appeared more efficacious in terms of LRC and FFDM, ultimately stratifying patients in terms of OS is would generate the most benefit to physicians and patients alike.
In addition to impacting treatment decisions, accurate, validated models may be used in the future to develop more efficient clinical trials. Models could be integrated into the inclusion and exclusion criteria. For instance, trials examining the role of local therapies such as surgery and radiation may benefit from a prior knowledge regarding a patient’s probability of tumor metastasis. This would allow better selection of patients who would be more likely to address the underlying question of the trial. Furthermore, previously conducted trials could be reanalyzed in order to determine if a particular therapy is only beneficial to those with a poor/favorable prognosis.
While the addition of tumor texture analysis into survival models has shown significant potential, various downsides certainly exist. Data is still only available from preliminary studies which require external validation and appropriate assessment of predictive power/accuracy. Careful consideration needs be taken in deciding which texture/analysis methods are appropriate for particular tasks. This includes consideration of values for parameters involved with each methodology as well as whatever preprocessing steps are applied. Differing quantification methods and their associated parameters have the potential to greatly impact study results. Texture analysis is also not applicable to all patients. Those with a small primary tumors or severe imaging artifacts are not appropriate to undergo analysis. Advances in robust, auto segmentation methods would also be exceedingly useful in this field in order to standardize tumor contouring. Using physician generated contours is most commonly used in these types of analyses but is far from perfect. Thresholding is a useful strategy to enhance contour reproducibility, particularly in lung tumors. Other factors that would influence reproducibility which were not included in our analysis are the stability of the CPFs between institutions/physicians such as staging, performance status, tumor volume, etc. Additionally, in this study the imaging protocols were well controlled. The impact of changing image parameters (tube voltage, reconstruction algorithm, pixel size, manufacturer, etc.) should be considered when evaluating data from multiple institutions.25
In the future, we hope to conduct additional studies examining the use of texture analysis in other imaging modalities. Contrasted CT, PET, and MRI could potentially all yield complementary information which would facilitate the creation of more accurate models. A variety of future studies are needed in order to better assess the applicability of texture analysis and its ultimate potential. Prospective trials are needed in order to determine if these techniques could one day be used in the clinic.
Patient outcome modeling has significant implications in many fields of medicine and particularly in oncology. Our study found that the combination of CT texture and CPFs can be used to generate superior outcome models when compared to CPFs alone in terms of OS, LRC, and FFDM. This additional information could be of use to physicians, patients, and caregivers. Further work needs to be done in order to generate widely applicable, accurate risk prediction tools capable of being implemented clinically.
Acknowledgement
Financial Support:
Supported in part by the American Legion Auxiliary, the American Association of Physicists in Medicine Graduate Fellowship, the University of Texas Graduate School of Biomedical Sciences at Houston, and the National Cancer Institute (R03CA178495-01).
The University of Texas MD Anderson Cancer Center is supported by the National Institutes of Health through Core Grant CA 16672.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest: None
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Hanna N, Neubauer M, Yiannoutsos C, et al. Phase iii study of cisplatin, etoposide, and concurrent chest radiation with or without consolidation docetaxel in patients with inoperable stage iii non-small-cell lung cancer: The hoosier oncology group and u.S. Oncology. J Clin Oncol. 2008;26:5755–5760. doi: 10.1200/JCO.2008.17.7840. [DOI] [PubMed] [Google Scholar]
- 3.Chen HY, Yu SL, Chen CH, et al. A five-gene signature and clinical outcome in non-small-cell lung cancer. N Engl J Med. 2007;356:11–20. doi: 10.1056/NEJMoa060096. [DOI] [PubMed] [Google Scholar]
- 4.Machtay M, Duan F, Siegel BA, et al. Prediction of survival by [18f]fluorodeoxyglucose positron emission tomography in patients with locally advanced non-small-cell lung cancer undergoing definitive chemoradiation therapy: Results of the acrin 6668/rtog 0235 trial. J Clin Oncol. 2013;31:3823–3830. doi: 10.1200/JCO.2012.47.5947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Raz DJ, Ray MR, Kim JY, et al. A multigene assay is prognostic of survival in patients with early-stage lung adenocarcinoma. Clin Cancer Res. 2008;14:5565–5570. doi: 10.1158/1078-0432.CCR-08-0544. [DOI] [PubMed] [Google Scholar]
- 6.Al-Kadi OS, Watson D. Texture analysis of aggressive and nonaggressive lung tumor ce ct images. IEEE Trans Biomed Eng. 2008;55:1822–1830. doi: 10.1109/TBME.2008.919735. [DOI] [PubMed] [Google Scholar]
- 7.Ganeshan B, Abaleke S, Young RC, et al. Texture analysis of non-small cell lung cancer on unenhanced computed tomography: Initial evidence for a relationship with tumour glucose metabolism and stage. Cancer Imaging. 2010;10:137–143. doi: 10.1102/1470-7330.2010.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Win T, Miles KA, Janes SM, et al. Tumor heterogeneity and permeability as measured on the ct component of pet/ct predict survival in patients with non-small cell lung cancer. Clin Cancer Res. 2013;19:3591–3599. doi: 10.1158/1078-0432.CCR-12-1307. [DOI] [PubMed] [Google Scholar]
- 9.Berghmans T, Paesmans M, Sculier JP. Prognostic factors in stage iii non-small cell lung cancer: A review of conventional, metabolic and new biological variables. Ther Adv Med Oncol. 2011;3:127–138. doi: 10.1177/1758834011401951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hari DM, Leung AM, Lee JH, et al. Ajcc cancer staging manual 7th edition criteria for colon cancer: Do the complex modifications improve prognostic assessment? J Am Coll Surg. 2013;217:181–190. doi: 10.1016/j.jamcollsurg.2013.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Amadasun M, King R. Textural features corresponding to textural properties. Ieee T Syst Man Cyb. 1989;19:1264–1274. [Google Scholar]
- 12.Haralick RM, Shanmuga K, Dinstein I. Textural features for image classification. Ieee T Syst Man Cyb. 1973;Smc3:610–621. [Google Scholar]
- 13.Miles KA, Ganeshan B, Hayball MP. Ct texture analysis using the filtration-histogram method: What do the measurements mean? Cancer Imaging. 2013;13:400–406. doi: 10.1102/1470-7330.2013.9045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goeman JJ. L1 penalized estimation in the cox proportional hazards model. Biometrical journal Biometrische Zeitschrift. 2010;52:70–84. doi: 10.1002/bimj.200900028. [DOI] [PubMed] [Google Scholar]
- 15.Simon RM, Subramanian J, Li MC, et al. Using cross-validation to evaluate predictive accuracy of survival risk classifiers based on high-dimensional data. Briefings in bioinformatics. 2011;12:203–214. doi: 10.1093/bib/bbr001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin LI. A concordance correlation coefficient to evaluate reproducibility. Biometrics. 1989;45:255–268. [PubMed] [Google Scholar]
- 17.Harrell FE, Jr, Lee KL, Mark DB. Multivariable prognostic models: Issues in developing models, evaluating assumptions and adequacy, and measuring and reducing errors. Stat Med. 1996;15:361–387. doi: 10.1002/(SICI)1097-0258(19960229)15:4<361::AID-SIM168>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 18.Cook GJ, Yip C, Siddique M, et al. Are pretreatment 18f-fdg pet tumor textural features in non-small cell lung cancer associated with response and survival after chemoradiotherapy? J Nucl Med. 2013;54:19–26. doi: 10.2967/jnumed.112.107375. [DOI] [PubMed] [Google Scholar]
- 19.Gevaert O, Xu J, Hoang CD, et al. Non-small cell lung cancer: Identifying prognostic imaging biomarkers by leveraging public gene expression microarray data--methods and preliminary results. Radiology. 2012;264:387–396. doi: 10.1148/radiol.12111607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ganeshan B, Panayiotou E, Burnand K, et al. Tumour heterogeneity in non-small cell lung carcinoma assessed by ct texture analysis: A potential marker of survival. Eur Radiol. 2012;22:796–802. doi: 10.1007/s00330-011-2319-8. [DOI] [PubMed] [Google Scholar]
- 21.Ganeshan B, Goh V, Mandeville HC, et al. Non-small cell lung cancer: Histopathologic correlates for texture parameters at ct. Radiology. 2013;266:326–336. doi: 10.1148/radiol.12112428. [DOI] [PubMed] [Google Scholar]
- 22.Mattonen SA, Palma DA, Haasbeek CJ, et al. Distinguishing radiation fibrosis from tumour recurrence after stereotactic ablative radiotherapy (sabr) for lung cancer: A quantitative analysis of ct density changes. Acta Oncol. 2013;52:910–918. doi: 10.3109/0284186X.2012.731525. [DOI] [PubMed] [Google Scholar]
- 23.Mack JW, Smith TJ. Reasons why physicians do not have discussions about poor prognosis, why it matters, and what can be improved. J Clin Oncol. 2012;30:2715–2717. doi: 10.1200/JCO.2012.42.4564. [DOI] [PubMed] [Google Scholar]
- 24.Huskamp HA, Keating NL, Malin JL, et al. Discussions with physicians about hospice among patients with metastatic lung cancer. Arch Intern Med. 2009;169:954–962. doi: 10.1001/archinternmed.2009.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hunter LA, Krafft S, Stingo F, et al. High quality machine-robust image features: Identification in nonsmall cell lung cancer computed tomography images. Med Phys. 2013;40:121916. doi: 10.1118/1.4829514. [DOI] [PMC free article] [PubMed] [Google Scholar]