Abstract
Human milk is a complete source of nourishment for the infant. Exclusive breastfeeding not only sustains the infant’s development but also guides the proliferation of a protective intestinal microbiota. Among the many components of milk that modulate the infant gut microbiota, the milk glycans, which comprise free oligosaccharides, glycoproteins, and glycolipids, are increasingly recognized as drivers of microbiota development and overall gut health. These glycans may display pleiotropic functions, conferring protection against infectious diseases and also acting as prebiotics, selecting for the growth of beneficial intestinal bacteria. The prebiotic effect of milk glycans has direct application to prevention of diseases such as necrotizing enterocolitis, a common and devastating disease of preterm infants. In this article, we review the impact of the human (and bovine) milk glycome on gut health through establishment of a milk-oriented microbiota in the neonate.
Keywords: glycobiome, milk bioactives, milk oligosaccharides, bifidobacteria, neonatal microbiota
INTRODUCTION
Breastfeeding and human milk are considered the normative standard for infant feeding, according to the American Academy of Pediatrics, and are associated with both short-term and long-term effects on neonatal health (1). Exclusive breast milk feeding is recommended for the first six months of life and (in combination with solid foods) advised to extend for two years of life (1–3). Breast milk is the sole source of nourishment for infants, through which they acquire all elements necessary for their development. Milk is a complete natural formulation containing hormones, antibodies, glycans, glycoconjugates, antimicrobials, and nourishment for the infant’s microbiota. Indeed, milk is a complex and complete source of bioactive molecules that help protect the newborn against infectious diseases and promote development while selectively enriching a protective and beneficial gut microbiota. Therefore, milk combines infant nutrition and microbial nutrition, which in concert promote infant health. Several studies correlate breastfeeding to reduced infant mortality, lower incidence of infectious diseases, and enhanced immune response. It is well accepted that exclusive breastfeeding protects infants against diarrhea (4, 5). Breastfeeding is also associated with reduced risk for postneonatal death (6), reduced incidence of necrotizing enterocolotis (NEC) (1), better cognitive development (7), and better growth and overall health outcomes (3).
The mother infected with HIV presents a striking example for the array of benefits of breast-feeding. In developed countries, the risk of HIV infection through breastfeeding is such that HIV-positive women are encouraged to feed their infants formula only. However, in developing countries, where infant mortality related to malnutrition is high and formula is often not readily available, breast milk offers a significant advantage for babies of HIV-infected mothers. In a study composed of nearly 300 mother-infant pairs in India, the effects of breastfeeding were associated with higher survival rates of infants born to HIV-positive mothers compared with formula-fed infants (8). The mortality rate of breastfed infants was nearly 10 times lower than that of formula-fed infants (0.68% versus 9.6%, respectively) at 12 months of age (8). A similar trend was also observed in a clinical trial in Botswana, Africa. In a pioneer investigation on the effects of breastfeeding during extended antiretroviral therapy on postnatal transmission of HIV, Thior and collaborators (9) observed that although administration of the drug zidovudine during breast-feeding did not reduce HIV transmission compared with formula-fed infants, mortality rate among breastfed infants was lower. The many bioactive factors in milk may underlie the protection observed in these studies. With growth factors, immunoglobulins, and prebiotics, breastfed infants have a better immune response and development and are likely more resistant to infectious diseases.
BIOACTIVE MOLECULES IN MILK
Milk is a remarkably complex matrix constituted by macronutrients and a plethora of bioactive molecules, including hormones, growth factors, antimicrobials, oligosaccharides, immunoglobulins, mucins, glycolipids, and metabolites. The composition of milk is dynamic and displays variations during lactation stages, feedings, and within mothers, but it is mostly conserved across populations (2). The highest differences in compositions are observed comparing mature milk to colostrum, the milk secreted a few days after birth, which contains the highest concentrations of bioactive milk proteins, such as immunoglobulins and antimicrobials (10). The remarkable fluidity of milk composition during lactation reflects an adjustment to the infant’s needs, as well as the evolutionary role of milk as the sole source of nutrients and immunity to support development of the newborn (10).
The most abundant component of human milk is lactose (70 g/L), followed by lipids (40 g/L), human milk oligosaccharides (HMOs) (5–15 g/L), and proteins (8 g/L) (11). Lactose and lipids represent a major nutrient source for the infant. HMOs are free glycans that display several biological functions; together with many glycoproteins and glycolipids, they represent the total glycoconjugates in milk. It has been estimated that more than 70% of human milk proteins are glycosylated, harboring both N-linked and O-linked glycan moieties (12). The majority of milk glycoproteins are found in skim milk (whey and casein), but the milk fat globule membrane (MFGM) contains a representative amount of total glycoproteins (reviewed in 13). The most abundant human milk glycoproteins are α-lactalbumin (17% of total protein), lactoferrin (Lf) (17%), and secretory IgA (sIgA) (11%), belonging to the whey fraction, and κ-casein, from casein fraction (9%) (13).
Human Milk Oligosaccharides
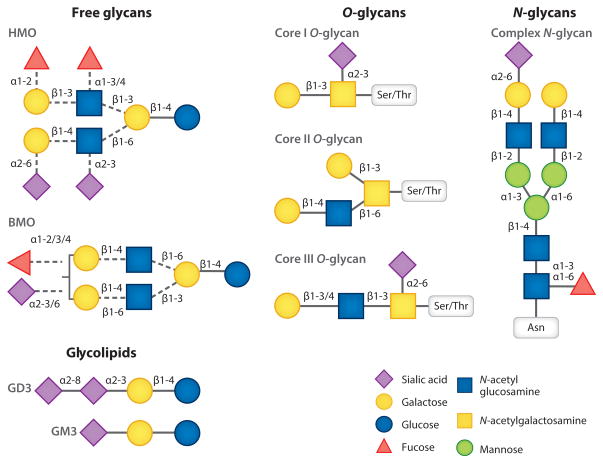
HMOs reportedly comprise the third most abundant component of human milk and encompass a collection of structurally complex sugars displaying an array of linkages. HMOs can be linear or branched, comprising 3–14 monosaccharides. Five monosaccharides are the building blocks of HMOs: D-glucose (Glc), D-galactose (Gal), N-acetylglucosamine (GlcNAc), L-fucose (Fuc), and N-acetylneuraminic acid (NeuAc). HMOs are built on a basic lactose core [Gal(β1–4)Glc] that is extended with N-acetyllactosamine repeating units and can be divided into type I [Gal(β1–3) GlcNAc] or type II [Gal(β1–4)GlcNAc] depending on the specific linkage between Gal and GlcNAc (14). Moreover, HMOs can be decorated with fucose or N-acetylneuraminic acid, mostly in terminal positions. Fucosylation invariably yields Fuc(α1–3) on the reducing end and mostly Fuc(α1–2) on the nonreducing end. Sialylation is also observed in the nonreducing end via α2,3 or α2,6 linkages (14, 15). The basic structure of HMO and the linkages involved are depicted in Figure 1.
Figure 1.
Structural diversity of the milk glycobiome. Abbreviations: BMO, bovine milk oligosaccharide; HMO, human milk oligosaccharide.
Biological synthesis of human milk glycans
HMOs are synthesized in the mammary gland by the action of several glycosyltransferases. Nearly all HMOs contain lactose at the reducing end, so it is postulated that HMO occurs as an extension of lactose biosynthesis, although the processes involved in the generation of the extremely diverse HMO repertoire remain mostly elusive (16). Biosynthesis of lactose takes place in the Golgi, the organelle where protein glycosylation occurs, and requires α-lactalbumin. In the Golgi, the enzyme β-1,4-galactosyltransferase1 (β4gal-T1) binds to UDP-galactose, promoting interaction with α-lactalbumin and modifying specificity of β4gal-T1, which in turn allows galactose transfer to glucose, forming lactose (17). A genetic basis underlies the production of α1–2, α1–3, and α1–4 fucosylated core structures of oligosaccharides, dependent on genes of blood group H, Lewis antigens, and secretor status (18). The α1,2 fucosyltransferase encoded by FUT2 defines the secretor status; a secretor individual produces ABH antigens in body fluids, including milk. The Lewis blood group gene FUT3 encodes α1,3/4 fucosyltransferase that forms α1,4 fucosylation, resulting in Lewis B antigen in secretors and Lewis A antigen in nonsecretors. The FUT2 and FUT3 genes are independent, yielding a combination of phenotypes; 70% of the population expresses both FUT2 and FUT3 (19). Moreover, sialylation at α2,3 and α2,6 contributes to increases in the variability of HMO structures in milk. HMOs are considered resistant to digestion in the gastrointestinal (GI) tract based on in vitro (20) and in vivo observations (18). HMOs have been detected in the stool (21) and urine (22, 23) and circulate in the blood of breastfed infants (19), indicating a potential systemic effect of HMO that extends beyond intestinal modes of action.
One of the biggest challenges in HMO research has been the characterization of individual structures, particularly owing to the formation of multiple isomeric forms for each structure, resulting from the variety of possible linkages. Detection and analysis methods have been optimized (24); with the employment of sophisticated mass spectrometry (MS)-based approaches, some 200 HMOs have been revealed (25). More recently, libraries of neutral (15) and sialylated (26) HMOs have been annotated employing HPLC-chip/time-of-flight (TOF) MS, facilitating study of structure-function relationships.
Bovine Milk Glycans
Several bioactive components present in human milk are also found in bovine milk, albeit in lower amounts. Infant formulas are typically based in part on bovine milk components; however, they may lack the specific bioactive glycans that are presumably important for infant development. Thus, identification of bioactive glycans in bovine milk is of major importance for manipulation of infant formulas to achieve the health-promoting benefits of breast milk, because human milk is not a viable source for commercialization.
Considering the multitude of health-promoting effects of HMOs—which function as both a prebiotic and a glycan decoy to prevent intestinal binding of infectious agents—there is great commercial interest in discovering analogs that would mimic the structural and functional complexity of HMOs (27). An obvious source of such glycans would be from other animal milks. Bovine milk contains several simple and complex oligosaccharides that are structurally analogous to HMOs (28). Despite their structural similarity, oligosaccharides in mature bovine milk are present in significantly lower concentrations than in mature human milk, decreasing progressively along the course of lactation (29).
Both human and bovine milk contain higher concentrations of sialylated oligosaccharides in the early stages of lactation. Before the development and application of high-performance MS for analysis of milk glycans, only a handful of BMO structures were known. A systematic analysis of glycans in bovine colostrum during the first week of lactation, using HPLC-chip/TOF MS, revealed that the decline of sialylated bovine milk oligosaccharides (BMOs) (especially of N-glycoylneuraminic acid-BMO) is accompanied by an increased diversity of neutral oligosaccharides (30). Glycomics methods employing advanced separation techniques, including nanoflow liquid chromatography and mass spectrometry, in combination with selective enzymatic digestion, resulted in the creation of a complete BMO library that includes over 60 structures, of which several are novel and fucosylated (31). The feasibility of isolating BMO from dairy byproducts using membrane filtration has been shown (32). Recent work using HPLC-chip/TOF MS also revealed the presence of nearly 50 BMOs in whey permeate, a by-product of whey protein manufacturing. Given the extremely large and growing production of cheese whey worldwide, it would seem obvious that whey may provide a significant source from which to obtain HMO mimics. This line of research would add a new dimension to profitable use of cheese whey in the dairy industry.
Glycoproteins
Like human milk, bovine milk contains several glycoconjugates, such as α-lactalbumin, glycomacropeptide (GMP), Lf, immunoglobulins, and milk fat globule (MFG) proteins, which, in addition to HMOs, have been the subject of study. The concentration of these glycoconjugates is different between human and bovine milk; however, their biological functions seem to be conserved. Lf is well recognized as a major multifunctional protein from human milk, displaying antimicrobial, iron-chelating, anti-inflammatory, and prebiotic functions (10, 33). Human Lf (hLF) is an 80-KDa glycoprotein very abundant in human milk, present at concentrations of ~7 g/L in colostrum and ~2 g/L in mature milk. Lf is also found in bovine milk, with 68% sequence similarity to hLF (34), although it is present at significantly lower concentrations (50–300 mg/L). Pepsin digestion of hLf and bLf releases a potent bactericidal peptide, lactoferricin (Lfc) (35), that binds lipopolysaccharide from Gram-negative bacteria such as Escherichia coli and Salmonella (36). HLf is a highly glycosylated protein displaying three glycosylation sites (Asn137, Asn478, and Asn623) and harboring fucosylated and sialylated glycans (37). Bovine Lf (bLf) is also glycosylated, containing five potential glycosylation sites (Asn233, Asn281, Asn368, Asn476, and Asn545) (34). Lf glycosylation protects the protein against proteolytic digestion. Indeed, intact Lf has been detected in the stools of breastfed infants, indicating the resistance of this protein to GI tract enzymatic activities (38).
Casein GMP is the C-terminal portion of κ-casein (amino acids 106–109) that is present in both human and bovine milk (39). Two genetic variants are known for GMP, variant A and variant B, which differ in only two amino acids in the protein sequence. The peptide is relatively small, with a molecular weight of 8,000 Da; however, owing to glycosylation and the formation of dimers and trimers, its actual size can be up to 30,000 Da. The carbohydrate moiety of bovine GMP may contain up to five different O-linked oligosaccharide chains, which are typically heavily sialylated. GMP has been reported to have a prebiotic effect on bifidobacteria and lactic acid bacteria, potentially owing to its N-acetylneuraminic acid content (40). GMP could also be an important bioregulator of GI functions, and it was reported to stimulate the release of cholecystokinin in animals and humans (41).
Glycolipids
The majority of glycosphingolipids in both human and bovine milk are gangliosides that contain sialic acid residues in their glycan chains. The chemical structure of gangliosides is characterized by a ceramide lipid chain linked to acidic oligosaccharide chains containing one or more sialic acid residues (42); the ceramide portion is embedded into the membrane of MFGs, whereas the glycan moieties are exposed at the surface, similarly to the conformation of gangliosides that are part of epithelial cell membranes. Gangliosides in human milk are almost exclusively associated with the MFGM (43). The major ganglioside in human colostrum is GD3 (Sia α2–8 Sia α2–3 Gal β1–4 Glc β1–1 ceramide), which totals ~65% of glycolipid content, whereas in mature milk GM3 [Sia α2–3 galactose (Gal) β1–4 glucose (Glc) β1–1 ceramide] is predominant, representing ~70% of all glycolipids. GD3 accounts for ~25% of glycolipid content, whereas GM2 and GM1 are minor components of mature milk but are nonetheless important against enteric pathogens (13). Several gangliosides are also present in mature bovine milk, where the main ganglioside is GD3 (44). The chemical nature of gangliosides renders them difficult to purify and analyze. One study employed matrix-assisted laser desorption/ionization Fourier transform ion cyclotron resonance MS coupled with collision-induced dissociation and infrared multiphoton dissociation MS/MS for ganglioside quantification and characterization. This approach unveiled 15 and 16 structures in bovine and human milk, respectively (43). It detected 10 GM3 and 6 GD3 variants in human milk and 7 GM3 and 6 GD3 variants in bovine milk (43).
A more recent investigation of gangliosides in bovine milk using ultra-high performance liquid chromatography-tandem MS focused on quantifying the specific amounts of gangliosides in bovine milk over lactation. This work revealed considerable quantities of gangliosides at day two (GM3, 0.98 mg/L; GD3, 15.2 mg/L) and showed a rapid decrease at two weeks (GM3, 0.15 mg/L; GD3, 3.3 mg/L) and three months (GM3, 0.15 mg/L; GD3, 2.4 mg/L) (45). The same work suggested that gangliosides were preferentially concentrated during the dairy processing operations in a side stream called buttermilk.
MILK GLYCOME–BASED ENRICHMENT OF A PROTECTIVE INFANT MICROBIOTA: THE CONCEPT OF MILK-ORIENTED MICROBIOTA
The Human Intestinal Microbiota
The human intestinal microbiota is composed by a large and diverse collection of microbes that reside in the gut in a symbiotic relationship with its host. Bacterial numbers increase markedly from proximal to distal throughout the GI tract, with the large intestine estimated to contain 500–1,000 species (46). In children and adults, the phyla Bacteroidetes and Firmicutes are the most abundant, totaling 90% of the phylotypes in the gut (47). Remarkably, the intestinal microbial population exceeds the number of mammalian cells by one order of magnitude (47). The genetic pool of intestinal microbiota contains 10 times more genes than the human genome, and the microbiome encodes functions that complement mammalian physiology, such as the ability to digest otherwise indigestible dietary polysaccharides (48). The gut microbiota contributes to human health in several ways, by supplying vitamins and short chain fatty acids (49), stimulating the immune system, and protecting against enteric infections by a process known as colonization resistance (50). The human microbiome varies from individuals, ages, and diet (51, 52). Alterations in the microbiota composition and dysbiosis are associated with an array of immunological, infectious, and metabolic diseases (reviewed in 53), such as obesity (54, 55), inflammatory bowel disease (56), necrotizing enterocolitis (57, 58), and colorectal cancer (59).
Neonatal Intestine and Development of the Infant Microbiota
A major factor that shapes the human gut microbiota is the early development of the intestinal microbial communities that occurs right after birth. Early colonization of the intestine has major short-term and long-term implications for human physiology. Therefore, it is critical to investigate the changes that intestinal microbiota undergo during neonatal development. Manipulation of the neonatal gut microbiota is particularly relevant for development of preventive strategies directed to preterm infants.
The neonatal intestine at birth is immature, and the complex composition of breast milk provides elements for a microenvironment that ensures gut maturation and protection (60). The neonatal intestine, and particularly the premature intestine, is characterized by a highly immunoresponsive mucosa, and consequently, it is more susceptible to perturbations originated in the luminal compartment (61). Immature enterocytes are more sensitive to inflammatory stimuli, such as lipopolysaccharide and flagellin, expressing lower levels of IκB and releasing more IL-8, causing aberrant inflammatory responses that compromise the integrity of the intestinal mucosa (62). The neonatal ecosystem, composed of the intestinal mucosa, luminal nutrients, and microbiota, is very complex and tightly controlled to ensure homeostasis in the developing gut; hence, the establishment of a health-promoting microbiota in early life is crucial for intestinal health in infants. In this section, we explore the singular aspects of the neonatal intestine and its microbiota, both of which act in concert toward the establishment of a healthy symbiotic relationship to promote development of the newborn.
In the womb, the fetal intestine is a semi-sterile environment that differs entirely from the neonatal intestine. Colonization of the human gut essentially begins at birth, when the newborn is exposed to maternal vaginal and GI microbiota as well as microbes from the external environment (61). Soon after birth, the neonatal intestine is colonized by symbiotic bacteria in a stepwise fashion, which can be divided into four main subsequent steps: (a) acquisition of maternal vaginal, colonic, and skin flora at birth; (b) introduction of oral feedings (breastfeeding or formula feeding); (c) weaning; and (d) acquisition of complete adult colonization (63). Several factors influence early colonization of the infant gut, including mode of delivery (vaginal or cesarean) (64), gestational age at the time of birth (preterm or full-term), early exposure to antibiotics, and feeding mode (breast milk or formula) (65). The composition of the infant’s gut microbiota undergoes successive changes soon after delivery. The maternal gut microbes appear to be the main source of gut microbes for the infant. First colonizers are acquired through the mother, and therefore, the delivery mode has an impact on the bacteria genera that are passed to the newborn. Vaginal delivery accounts for higher levels of bifidobacteria (66) and lactobacilli. Infants born via cesarean delivery have low levels of bifidobacteria and high levels of clostridia. Nonetheless, cesarean-born infants who are breastfed retain a high level of bifidobacteria, highlighting the importance of breastfeeding to select for a protective gut microbiota in the infant.
In the first few days after birth, the intestine is colonized by a heterogeneous microbial population, independent of nutrient sources, that becomes more stable in the first week of life. At that time, facultative anaerobes that belong to Enterobacteriaceae, Streptococcus, Enterococcus, and Staphylococcus are already present, mainly owing to initial oxygen availability in the newborn gut (67). E. coli, Enterococcus faecalis, and Enterococcus faecium are the most represented species among the first colonizers. The gradual oxygen consumption by facultative anaerobic bacteria creates a reduced environment that allows expansion of obligate anaerobes belonging to the genera Bifidobacterium, Bacteroides, and Clostridium, followed by Veillonella, Ruminococcus, and Eubacterium (68). Intestinal colonization undergoes additional changes upon introduction of solid food. A recent study, in which the infant microbiota was analyzed monthly for two years after birth, shows that the genus Bifidobacterium is detected in the first few months of life, predominates in the first year, then declines along the second year. By the end of the second year of life, the infant microbiota is more diverse (69).
Bacterial colonization after birth is essential for intestinal development and maturation of the immune system (70). Early colonization is a determinant of mucosal homeostasis and is mostly dependent on the diet (63). Nutrition plays a major role in selecting for a signature microbiome that is protective for the infant (71). Considering the critical effects of the early colonizers on the baby’s overall health, it is of major importance to develop approaches to ensure the development of a protective gut microbiome early in life.
A Milk-Oriented Microbiota in Breastfed Infants
As the sole nutrient source for newborns, milk stimulates intestinal defenses and concomitantly provides enough caloric intake to sustain normal development and GI homeostasis. A predominance of Bifidobacterium species in the intestine of infants has been documented in several studies (72–74), and the use of next-generation approaches has contributed immensely to a better understanding of the dynamics of microbial communities in the developing intestine. Analyses of the infant microbiome indicate a trend that is consistent with breastfeeding, irrespective of geographic region or age: Breast milk selects for a highly adapted intestinal microbiota, dominated by bifidobacteria or, as suggested recently by our group, a milk-oriented microbiota (MOM). In particular, the species Bifidobacterium longum subsp. longum, B. longum subsp. infantis, and Bifidobacterium breve are most commonly found inhabiting the GI tract of nursing infants.
The Microbiota of Breastfed Versus Formula-Fed Infants
It is well established now that bifidobacteria dominate the intestinal microbial population of infants (72, 74, 75), reaching ~75% of the gut microbial population (51). In fact, Bifidobacterium is the predominant genus in both breastfed and formula-fed infants, with species variations according to the feeding mode. The species B. longum, B. infantis, B. breve, and B. bifidum are commonly detected in breastfed babies (72, 76), and in formula-fed babies, Bifidobacterium adolescentis and Bifidobacterium pseudocatenulatum, which are often found in the intestinal adult microbiota (77). Yatsunenko et al. (51) reported an abundance of the genera Bifidobacterium, Erwinia, Actinomyces, and Haemophilus in breastfed babies, whereas less bifidobacteria and more Firmicutes and Bacteroidetes were observed in formula-fed babies (51). A recent investigation of the gut microbiota in healthy children revealed 24 bacterial taxa that are associated with age, in which the top five are Faecalibacterium prausnitzii, Ruminococcus sp., Lactobacillus ruminis, Dorea longicatena, and B. longum. This study also supports the high abundance of Bifidobacterium sp. in the first few months of age in healthy babies, with a decrease toward the first two years of life (69).
Preterm Infants Lack a Milk-Oriented Microbiota
Babies born prior to 32 weeks gestation are essentially an evolutionarily new population. Human milk evolved to nourish and protect the term infant but in many ways is unable to provide for the needs of the very premature infant. Comparisons between term and very preterm infants provide the most compelling evidence of the importance of the interactions between three central components: human milk, the developing intestinal tract, and the intestinal microbiota. First, milk from mothers who deliver preterm differs in significant ways from that of mothers who deliver at term. Preterm milk is higher in protein content but much more variable in the composition of HMOs than milk from mothers delivering at term (78). This immature production of HMOs likely has relevance in altering the composition of the intestinal microbiota. Second, the intestinal tract of preterm infants is immature in essentially every functional aspect, including motility, absorption, digestion, secretion of mucus and antimicrobial peptides, barrier function, and vascularization (79). One of the striking features of the immature intestine is the excessive inflammatory response to gut luminal microbes (62, 80). Third, the intestinal microbiota differs markedly between the term and premature infant. In contrast to the bifidobacteria-rich microbiota of many breastfed full-term infants, the preterm gut microbiota is characterized by high levels of facultative anaerobes; low levels of Bifidobacterium and Bacteroides; and an abundance of Proteobacteria, especially Enterobacteriaceae (81, 82).
NEC is a common and devastating GI disease that primarily affects premature infants. The incidence of NEC varies; from 2–10% of premature infants with birth weight less than 1,500 g are affected, with annual costs in the United States estimated at $0.5–1 billion (83). Mortality rates vary with the severity of the disease, with as many as 25% of neonates with severe NEC succumbing to the disease. Long-term morbidity, including malabsorption, poor growth, short gut syndrome, and neurodevelopmental delays, is common among survivors.
Predisposing factors include immaturity (the more premature an infant is at birth, the higher the risk of NEC), enteral feeding, and dysbiosis (79). Limited data suggest additional risk factors, including genetic predisposition (84) and maternal smoking (85), though these have not yet been confirmed. Multiple case reports and case series have linked a variety of pathogenic bacteria, viruses, and fungi with NEC, suggesting that NEC may be a common disease pathway following a variety of insults. Recent observations of changes in the intestinal microbiota prior to the onset of NEC are compelling (58, 86–89). Dysbiosis in this population is extremely common and multifactorial. Evidence supporting a direct association between dysbiosis and NEC includes the following: First, antibiotic administration is associated with alterations in the intestinal microbiota and with increased risk of NEC (88, 90); second, acid suppression is associated with alterations in the intestinal microbiota and with increased risk of NEC (91, 92); and third, probiotic administration is associated with a decrease in dysbiosis and a decrease in NEC (93, 94).
Of the many strategies for prevention of NEC, the most promising is the provision of human milk. Multiple studies have demonstrated that premature infants receiving their mother’s own milk have lower rates of NEC (95) as well as fewer episodes of sepsis, lower risk of retinopathy of prematurity, fewer rehospitalizations in the first year of life, and improved neurodevelopmental outcomes (96). As a result, the American Academy of Pediatrics recently recommended that premature infants receive human milk whenever possible and pasteurized donor human milk if the supply from their own mothers is not sufficient (1). Two challenges of providing human milk to very premature infants are worth noting. First, human milk is ideal for the term infant but does not contain adequate protein, calcium, phosphorus, or vitamin D for the very premature infant. To achieve adequate growth and development, most premature infants require fortification of human milk. Fortification has historically meant addition of bovine-based fortifiers with the potential for contamination with bacteria associated with NEC (97) and intolerance of bovine proteins. A more recent approach to fortifying human milk with concentrated, pooled, pasteurized donor milk has been demonstrated to decrease the risk of NEC; however, the trials to date are underpowered (98). Second, pasteurization is essential to decrease bacterial and viral contamination of human milk; however, the standard process of pasteurization decreases the content of a variety of bioactive molecules, including secretory IgA, Lf, lysozyme, growth factors, enzymes, and vitamins. Further research into improved methods of fortification and pasteurization is clearly needed to improve the outcomes of the most premature infants.
MECHANISM OF GLYCAN CONSUMPTION BY A MILK-ORIENTED MICROBIOTA
Catabolism of HMOs
HMO use is a major adaptation that drives the development of the MOM in nursing infants. The phenomenon of HMO consumption by bifidobacteria was observed in the early 1950s by Gyorgy etal.(99), who documented the requirement of a bifidus factor to support growth of an infant fecal isolate then named Lactobacillus bifidus var. pennsylvannicus (today reclassified as B. bifidum by ATCC). The bifidus factor (99) was determined, at the time (100), to include N-acetyl glucosamine (a component of HMO). HMOs are highly diverse in their glycosidic bonds, and their degradation requires a sophisticated repertoire of glycosyl hydrolases that are not produced in the human intestine (101, 102). Consequently, HMOs delivered to the gut during breastfeeding arrive intact in the colon with the sole purpose of nourishing the infant’s microbiota (102).
Although the first indications of the bifidus factor were over 50 years ago, a thorough assessment and mechanisms involved in HMO consumption were mostly accumulated in the past decade, and it remains a subject of intense investigation. Ward et al. (103) were the first to demonstrate the consumption of HMOs as the sole carbon source by infant-borne bacteria through application of mass-spectrometry analysis of carbohydrate consumption during bacterial growth. In general, some species, such as B. infantis and B. bifidum, are able to efficiently consume HMOs as the sole carbon source and reach high cell densities in vitro (104, 105), being able to metabolize a wide spectrum of HMO structures, with various degrees of polymerization (short or long), but displaying a preference for short-chain HMOs. The capacity to ferment HMOs is well conserved within B. infantis strains, but other Bifidobacterium species present strain variation (106, 107). Others have also shown how individual HMO components are consumed by different bifidobacteria (108).
The extent of HMO use by other intestinal bacteria is variable and not a widespread phenotype. Bacteroides fragilis and Bacteroides vulgatus are also avid HMO consumers, whereas Lactobacillus acidophilus, Clostridium perfringens, E. coli, Eubacterium rectale, Streptococcus thermophilus, E. faecalis, and Veillonella parvula consume HMOs poorly or not at all (104). Little is known of the direct effects of HMO use in vivo, other than correlations between breastfeeding and bifidobacteria predominance. In an elegant study, Marcobal et al. (109) demonstrated how bifidobacterial adaptations to HMO consumption contribute to niche partition in the intestine in the presence of another HMO consumer bacterium, Bacteroides. Along with Bifidobacterium, Bacteroides is a prominent commensal bacterium known to efficiently consume HMOs (104). However, dual colonization of germ-free mice with B. infantis and Bacteroides thetaiotaomicron shows that B. infantis can outcompete Bacteroides in the mouse intestine when a specific HMO, lacto-N-neotetraose (LNnT), is supplied in the diet (109). The ability of B. infantis to successfully compete for colonization of the breastfed infant gut was also shown in a recent clinical trial (93). In that trial, human milk–fed preterm infants were given exogenous B. infantis or Bifidobacterium lactis. These two strains were chosen because B. infantis is capable of consuming HMOs, whereas B. lactis lacks this ability. The results clearly showed an increase in B. infantis in the feces (and lower levels of γ-proteobacteria) after administration of B. infantis in combination with human milk. In contrast, B. lactis failed to successfully colonize human milk–fed preterm infants, supporting the concept that the ability to consume human milk glycans enables successful colonization of the infant’s gut by bifidobacteria and that a synbiotic combination of B. infantis and milk has a greater potential to fight the Proteobacteria overgrowth typically observed in NEC cases.
A molecular dissection of HMO consumption has advanced greatly in the past few years. Genome sequencing of B. infantis revealed a 43-kb cluster composed of genes encoding proteins dedicated to the binding, transport, and degradation of complex HMOs. Additional sequencing and comparative genomic hybridization (110) analyses suggested this HMO cluster performs a central role in HMO metabolism and preceded biochemical characterization on the enzymes involved in HMO uptake and metabolism. Subsequent whole-cell proteomics revealed unique induction of several transport components (surface-bound solute-binding proteins) and a particular array of glycosyl hydrolases involved in HMO consumption that differ from the pattern observed during growth on other prebiotic sugars (111).
B. infantis appears to import intact HMO and degrade these structures intracellularly (Figure 2). B. infantis encodes two α-sialidases, five α-fucosidases, five β-galactosidases, and three β-N-acetylglucosaminidases (101). During B. infantis growth on HMOs in vitro, the expression of several glycosidases of the HMO cluster is activated. Specific genes encoding fucosidases (112, 113), sialidases (114), hexosaminidases (115), galactosidases (116, 117), and lacto-N-biosidases (118) are all induced during growth on pooled and specific HMOs. B. infantis employs the sialidase NanH2, which cleaves α2–6 and α2–3 linkages, to consume sialylated lacto-N-tetraose (114). Notable from this body of work is the functional redundancy within B. infantis for HMO catabolism with multiple homologs of glycosyl hydrolases (enumerated above), although not every enzyme is active on HMO. Although lacking an HMO cluster similar to B. infantis, initial examination of B. breve and B. longum subsp. longum (B. longum) suggests a similar intracellular degradation of HMO components (119, 120). Recently, different isolates of B. breve that possess both GH29 and GH95 family fucosidases were shown to grow to a higher level on HMO and consume more fucosylated HMOs (and, surprisingly, more sialyated HMOs as well) (107).
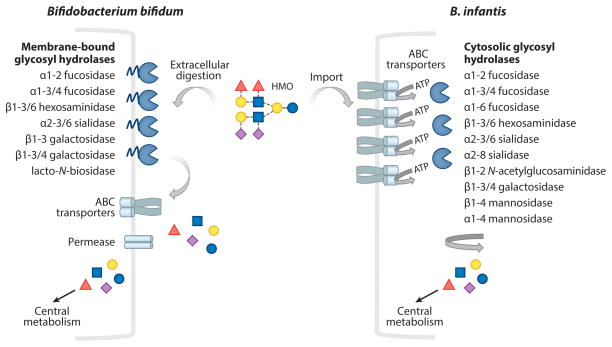
Figure 2.
Consumption of milk glycans by bifidobacteria. This figure illustrates the transport and glycolytic degradation of human milk oligosaccharide (HMO) (as a representative milk glycan), highlighting the differences between the HMO degradation in Bifidobacterium infantis and Bifidobacterium bifidum. HMO consumption in B. infantis involves intracellular import by ABC transporters, then degradation by intracellular glycosyl hydrolases, releasing free monosaccharides that enter the central metabolism. In contrast, HMO is degraded extracellularly in B. bifidum by the action of membrane-anchored extracellular glycosyl hydrolases, followed by intracellular transport of mono- or disaccharides (GNB/LNB) via sugar permeases or ABC transporters that enter central metabolism. Modified with permission from Reference 188.
The mechanisms of HMO consumption by B. bifidum present some differences compared with B. infantis. B. bifidum degrades HMOs extracellularly using extracellular membrane–anchored glycosyl hydrolases (Figure 2). Similarly to B. infantis, B. bifidum encodes α-fucosidases AfcA (113, 121) and AfcB, α-sialidase SiabB2 (122), and β-galactosidases and β-N-acetylglucosaminidases (117). B. bifidum harbors a lacto-N-biosidase that cleaves lacto-N-tetraose (LNT), yielding lactose and lacto-N-biose (LNB), which is imported by the bacteria and degraded intracellularly (118). The distinct mechanism of HMO has major implications for sugar availability in the infant intestine. HMO import and intracellular breakdown by B. infantis sequesters these complex oligosaccharides without resulting in monosaccharide release into the gut lumen. In contrast, metabolism of HMOs by B. bifidum likely results in increased monosaccharide availability in the gut, making them accessible to transient pathogenic bacteria. The monosaccharide release in vivo has major relevance when choosing a probiotic strain, particularly for the prevention of NEC in preterm infants. Most enteric pathogenic bacteria do not harbor a glycolyticpotential to catabolize complex sugars and rely on free monosaccharides that are made available by the microbiota (123, 124).
Catabolism of Milk Glycoconjugates
Along with free milk glycans, glycoproteins and glycolipids from milk have prebiotic effects. Like HMOs, milk glycoconjugates are multifunctional molecules capable of performing antimicrobial, prebiotic, and immunostimulatory functions. GMP and Lf are examples of pleiotropic milk glycojugates. When used as dietary supplements, Lf and GMP can select for growth of commensal bacteria in the gut, which cleaves sugar residues to use as an energy source (125, 126).
Most studies of prebiotic activity of milk glycans report the use of bovine glycoconjugates, owing to the problems involved in conducting large animal studies using human-derived glycans. Bruck et al. (127) observed that GMP can support a bifidobacterial enrichment similar to breast milk in two-stage continuous culture, suggesting that the bifidogenic effect of GMP makes it a suitable supplement to infant formulas. GMP supplementation led to a population dominated by bifidobacteria relative to Bacteroides, Clostridium, and E. coli, a trend observed similarly in nursing infants. Similarly, Chen et al. (128) showed that GMP-fed mice exhibited an increased population of bifidobacteria relative to Enterobacteriaceae.
Lf has also been shown to be bifidogenic (129–131). A study using a pig model and transgenic milk containing hLf indicated that hLf has a bifidogenic effect in vivo (126). Lf can support growth of beneficial bifidobacteria in vitro. Liepke and collaborators (132) identified a bifidogenic peptide derived from Lf, named prebiotic Lf-derived peptide-I, which results from the digestion of Lf with pepsin. The peptide stimulated the growth of B. bifidum DSM 20082, B. bifidum DSM 20215, B. infantis, and B. breve, but not the growth of other gut commensals Clostridium difficile, E. coli, E. faecalis, and Candida albicans, suggesting a potential use of Lf bifidogenic peptide on selecting a healthy microbiota. Lf may also function as a modulator of the gut microbiota owing to prebiotic activity in vivo. Supplementation of diet with recombinant lactoferrampin-Lfc resulted in increased levels of bifidobacteria and lactobacilli in the GI tract of piglets (125).
An initial mechanistic path for catabolism of milk glycoproteins has recently been uncovered. Garrido et al. (133) demonstrated that select bifidobacteria produce cell wall–associated endo-β-N-acetylglucosaminidases that cleave N-glycans from Lf from both bovine and human milk sources. The endoglycosidase activity correlated with the presence of the genes encoding glycosyl hydrolases belonging to families GH18a, GH18b, and GH85 (133), and the purified GH18 family enzymes cleaved a wide array of glycan types (high mannose, complex, and hybrid) from glycoproteins. Kiyohara et al. (134) recently described a novel α-N-acetylgalactosaminidase from infant-borne bifidobacteria belonging to a new glycoside hydrolase family (family 129) that is able to cleave O-linked glycoproteins, such as mucins. These findings suggest that endoglycosidase encoded by bifidobacteria has the potential to release and consume both N- and O-linked glycans present in milk glycoproteins, shedding light into the importance of other milk-glycosylated structures as prebiotics for bifidobacteria.
Milk glycolipids may also act as prebiotics. Studies on bioactivity of milk glycolipids are relatively scarce compared with those for other glycoconjugates. Few studies suggest a prebiotic effect for milk gangliosides. Supplementation of milk formula with gangliosides isolated from porcine brain caused an increase in bifidobacteria populations and a decrease in E. coli levels in the fecal microbiota of preterm infants (135). More studies are necessary to dissect the mechanisms of why a ganglioside-rich diet reduces the E. coli population in infants, which has application for prevention of NEC, because higher E. coli levels are observed in some cases. Lee et al. (136) recently demonstrated that B. infantis, B. bifidum, and B. breve can consume bovine milk ganglioside as a sole carbon source in vitro. Analysis of the spent media by nano-HPLC Chip QTOF showed that B. infantis and B. bifidum can catabolize significant amounts of input GM3 and GD3; B. infantis consumed 63% of GD3 and B. bifidum consumed 100%. In contrast, B. longum, B. adolescentis, and B. lactis degraded ganglioside to a lower extent (30%, 28%, and 48%, respectively).
MILK GLYCOBIOME MEDIATES PROTECTION AGAINST INFECTIOUS DISEASES
The immunological immaturity of the neonatal intestine renders the infant more susceptible to GI and systemic infections. Because it harbors so many protective molecules, human breast milk has been referred to as the “gold standard for protective nutrients” (60, p. 1).
Breastfeeding is associated with reduced incidence of several infectious diseases, such as diarrhea (137), respiratory infection (138), otitis media (139), and infection by protozoa (140). The immune system of the neonate is not fully developed at birth, so the elements, such as antibodies, present in breast milk represent a major source of protection for the neonate against infectious diseases (13). Increasing evidence indicates that additional glycoconjugates in milk are also important players in infant protection via breastfeeding, displaying several functions, such as acting as decoys for pathogens, reducing virulence gene expression, preventing binding to host receptors, and impairing colonization by pathogens in animal models (summarized in Figure 3) (13). Milk contains free glycans and glycoconjugates represented by glycoproteins and glycolipids; collectively, they are referred to as the milk glycobiome. Observations dated from the early 1980s already pointed to a protective role of the milk glycobiome in infectious disease prevention owing to its ability to block binding of pathogenic bacteria to respective receptors expressed by mammalian tissues (141). The antivirulence effects of milk glycoconjugates are discussed in the following section.
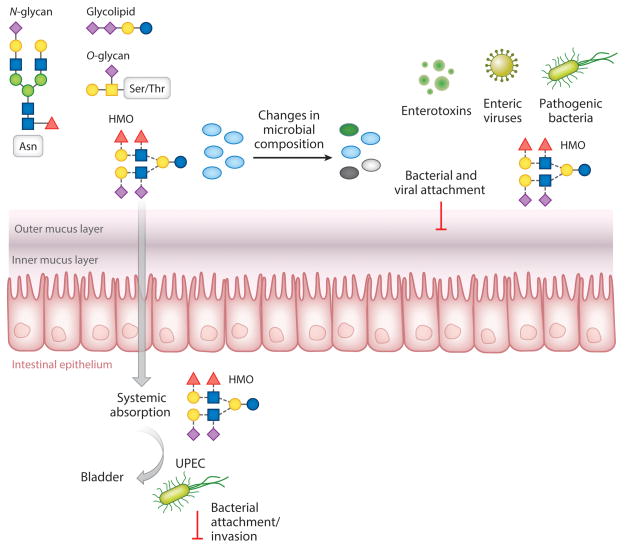
Figure 3.
Milk bioactives protect against infectious diseases. Human milk oligosaccharides (HMOs) are ingested during consumption of breast milk and reach the intestine minimally digested. HMOs have local effects in the gastrointestinal tract, altering microbial composition (bifidogenic effect); promoting barrier function; and blocking adhesion sites of pathogenic viruses, bacteria, and bacterial toxins. HMO is also systemically absorbed, traveling through the bloodstream and reaching other organs, such as the bladder. Abbreviation: UPEC, uropathogenic Escherichia coli.
Protection Mediated by HMOs
HMOs are known to protect against several infections by several infection agents, including bacteria, viruses, and parasites. Most studies on HMO-mediated protection against infectious diseases have relied on pathogen deflection and blocking of bacteria-host interactions that mediate initial steps of pathogen colonization. Attachment to host cell surfaces is a common strategy employed by bacterial, viral, and protozoan pathogens. Bacterial adhesion to mammalian cells is one of the first steps during infection and is essential for bacterial dissemination; therefore, anti-adhesion strategies are much coveted to prevent disease (142). One of the classical ways in which milk glycans have been associated with protection against infectious diseases is by their intrinsic ability to mimic cell surface receptors and, therefore, preventing bacteria from interacting with their receptors in the host cells and consequently, inhibiting host colonization by bacteria and viruses (141).
HMOs represent an important protection against viral infection by preventing viral attachment to host cells. Norovirus (NV), a member of the Caliciviridae family, is a major causative agent of outbreaks of acute gastroenteritis (143). Norovirus infection causes ~71,000 deaths in children younger than 5 years old (144). Noroviruses attach to host intestinal epithelial cells by binding to cell surface glycans, including the ABO and Lewis histo blood group antigens (HBGAs). For this reason, ABO and Lewis blood type and host secretor status play a major role in susceptibility to infection (145). HBGA motifs are also found in HMOs, and HMOs with specific Lewis and secretor antigens were shown to block norovirus attachment to HBGA antigens present in saliva (143), and high levels of lacto-N-difucohexaose in maternal milk were associated with lower incidence of diarrhea caused by calicivirus in breastfed infants (146).
In addition to their role as soluble receptor mimics, free milk glycans can protect mammalian cells against cytotoxicity caused by enteric bacteria and protozoans. Enteropathogenic E. coli (EPEC) is a causative agent of infant diarrhea in developing countries (147). Recent work has demonstrated that preincubation with pooled HMO decreased EPEC attachment to epithelial cell lines HeLa and T84 in vitro and decreased intestinal colonization by EPEC in a newborn suckling mouse model of infection (148), indicating the potential of HMOs as inhibitors of bacterial infection at earlier stages of infection. Interestingly, the non-milk-based prebiotic galactooligosaccharides (GOS) did not reduce EPEC adhesion or colonization in vivo. However, more studies are necessary to determine the mechanism through which HMO impacts EPEC intestinal colonization, and also to determine whether this animal model is suitable to evaluate important aspects of EPEC pathogenesis, such as biofilm formation and attaching and effacing lesion formation. The mechanism of action by which HMO confers protection to EPEC is unknown, as is whether HMO can block bacterial attachment to cell receptors or trigger changes in gene expression in the bacteria that end up downregulating expression of fimbriae or other adhesion molecules.
Other studies have assessed the use of individual HMOs, such as fucosyl lactose (FL), on inhibition of bacterial attachment, and these studies are important to test the addition of HMOs into infant formulas to prevent enteric as well as nonenteric bacterial infections. Synthetic 2′FL and 3′FL decreased adhesion of Campylobacter jejuni, Pseudomonas aeruginosa, EPEC, and Salmonella enterica serovar Fyris to Caco-2 cells in vitro (149). However, the observed minimal impact on adhesion (~20%) might not be protective in vivo, particularly for bacteria with a high infectious dose. Additional studies are necessary to evaluate the relevance of purified individual HMOs on bacterial adhesion in vivo, as well as to determine the dose required for infection prevention.
HMOs can also be effective against extraintestinal bacterial infection. The presence of sialylated MOs in the urine of breastfed but not bottle-fed infants raised the possibility that these sugars could act systemically to prevent infectious diseases. Martin-Sosa et al. (150) demonstrated that incubation of extraintestinal E. coli pathovars enterotoxigenic (ETEC) and uropathogenic (UPEC) with humansialylated MOs causes inhibition of fimbriae-mediated erythrocyte agglutination in vitro.
UPEC is a leading cause of urinary tract infection (UTI) worldwide, and recurrent UTI in infants increases the risk of kidney failure (151). UPEC pathogenesis involves attachment and invasion of the bladder epithelium (152). A recent study shows that the rate of UTI in neonates is rising (153). Pretreatment of human bladder epithelial HTB-9 cells with pooled HMO caused a significant decrease in invasion by UPEC (from 10% to 1.7%) and reduced cytotoxicity in a dose-dependent manner, which was associated with HMO internalization by the bladder cells. Interestingly, HMO prevented cell detachment during infection, a common feature of UPEC pathogenesis that contributes to bacterial dissemination. 3-SL sialylated but not neutral HMO was able to mediate protection. A role of sialylated milk glycans in UPEC protection has been reported previously, although the effects were attributed to the role of MOs as soluble receptor mimics (150). The prebiotic GOS was able to reduce bacterial invasion but did not prevent cytotoxicity to the same extent as HMO. This study sheds light on novel mechanisms of HMO action, and it correlates the intracellular metabolism of HMO by the host and protection against deleterious effects that occur upon bacterial infection. Another important aspect of this study is that it provides direct evidence that HMO acquired through breastfeeding can be spread systemically and therefore has potential to prevent nonenteric infectious diseases as well.
HMOs were able to protect intestinal epithelial cell line HT29 against Entamoeba histolytica cytotoxicity, an effect that was mimicked by LNT and prebiotic GOS (154). An exposed galactose residue is required to prevent cytotoxicity, consistent with the fact that E. histolytica recognizes host cell Gal residues via a surface-expressed lectin (154). The fact that GOS, a common additive to infant formula, had protective effects comparable to those of HMOs contrasts with the finding that breastfed infants are more protected against E. histolytica. Breastfed infants are less susceptible to infection with E. histolytica than formula-fed infants; whether this difference is attenuated by the addition of GOS to formula has not yet been studied. In vivo studies are necessary to evaluate the antiparasite activity of HMO. In summary, several reports indicate that HMOs act as inhibitors of adhesion and of the cytotoxicity caused by pathogens. Although the in vitro studies provide a foundation for future investigations of the role of HMOs in infectious disease prevention, animal studies are necessary to confirm such findings and to evaluate the protective effects in vivo.
Protection Mediated by Milk Glycoconjugates
In addition to HMOs, milk glycoproteins have also been investigated for protective strategies against bacterial infection (127).
Lactoferrin (Lf)
Several studies have demonstrated that Lf can inhibit adhesion of enteric bacteria (EPEC, Shigella flexneri, Salmonella typhimurium, Listeria monocytogenes) to eukaryotic cell lines. Lf is a known antimicrobial by its iron-chelating properties. The antipathogenic activity of hLF has been attributed to its glycan moieties. hLf is a heavily glycosylated protein harboring fucosylated and sialylated glycans. Interestingly, hLF glycosylation is dynamic and changes during lactation with diminished overall glycosylation during transition from colostrum to transitional milk, followed by an increase on day 30 of lactation, which coincides with production of mature milk and an increase in Lf fucosylation. Interestingly, a recent study indicates that glycosylation of Lf is involved in the antiadhesion mechanisms (155).
Lf also has antiviral activity. Mechanisms mediating antiviral effects of bovine Lf and its proteolytic product Lfc toward HSV are mostly unknown, but there are reports demonstrating inhibition of HSV-1 intracellular trafficking to the host cell nuclei in vitro (156) and protection against HSV-2 in vivo (157). Marr et al. (156) showed that Lf and Lfc interfere with the HSV-1 infection cycle by drastically reducing viral nuclear trafficking from 47% to 1% in Vero cells. Viral capsids could be seen associated with microtubules, and viral trafficking along the microtubules was not abolished but delayed. Another study showed that hLf presented the same effects of interfering with intercellular spread of both HSV-1 and HSV-2. Glycosylation of these proteins may underlie their antiviral mechanisms (158). In summary, the effects of Lf on the HSV-1 infection cycle observed in these studies shed light on the antiviral properties of bLf and bLfc and their potential against HSV infection.
Secretory IgA
Acquired during breastfeeding, sIgA is the predominant antibody in human milk and represents a major defense mechanism for the newborn (60). During the first 30 days after birth, maternal IgA from breast milk is the sole source of IgA for the neonate. SIgA is a major component of the mucosal defense system in the GI tract, capable of neutralizing infectious agents. Secreted IgA is an important player in gut homeostasis between symbiotic gut bacteria and the epithelium, which limits bacterial association and translocation through the intestinal epithelium (159). Recently, an elegant study using a mouse model demonstrated that neonatal exposure to IgA from breast milk has long-term effects; early exposure to maternal IgA changes microbiota composition in weaned mice through adulthood, alters transcriptional response in the colonic epithelium, and protects against dextran sodium sulphate (DSS)-induced colonic damage (160). Exposure to IgA reduced translocation of aerobic bacteria to mesenteric lymph nodes.
SIgA is a highly glycosylated protein that displays both N-linked (161) and O-linked glycans, containing fucose, galactose, sialic acid, and mannose residues (162). The glycan moieties of IgA present antipathogenic properties. The essential role of sIgA glycans on the interaction with Gram-positive but not Gram-negative commensal gut bacteria has been demonstrated (163). Different glycans from sIgA have been associated with protection against different pathogens, likely owing to distinct residues that mediate bacteria-host interactions for each pathogen. Fucosylated glycans inhibit EPEC adhesion to HEp-2 cells (164) and Helicobacter pylori attachment to human gastric tissue (165). The H. pylori adhesin BabA binds to the fucosylated Lewis histo-blood group antigen expressed by the gastric epithelial lining, a site colonized by H. pylori (166). Mannose-rich glycans can inhibit binding of E. coli to a colonic HT29 cell line mediated by type I fimbriae (167), which is a widespread virulence factor among E. coli pathotypes (147) that interact with host cell surface molecules via mannose residues (168).
Glycomacropeptide (GMP)
In addition to its prebiotic role, κ-casein has antimicrobial activity. During milk digestion, which occurs in the gut, κ-casein undergoes proteolytic cleavage that releases GMP, which is the mediator of antimicrobial effects of this glycoprotein. κ-Casein inhibited attachment of Streptococcus pneumoniae and Haemophilus influenza to human respiratory epithelial cells (169). An antiadhesion effect was also observed for H. pylori attachment to gastric cells; treatment with α-L-fucosidase reduced the inhibitory effect, indicating the importance of fucose residues of κ-casein in mediating antiadhesion effects during H. pylori infection (170).
Gangliosides
The first observations that milk gangliosides can protect against pathogenic bacteria, their secreted toxins, and infection by eukaryotic parasites (171) date from the early 1980s. The protective effects of milk gangliosides have been attributed to their glycan moieties, which function as decoys for pathogen receptors in the host cells. GM1 and GM3 inhibited ETEC adhesion to Caco-2 cells (172). GM1 from milk prevented binding of heat labile toxin (LT) from E. coli in vitro and abolished the fluid accumulation induced by cholera toxin (CT) from Vibrio cholerae in a rabbit intestinal loop model (173). GM1 is the host cell receptor for these toxins. E. coli LT from human isolates and V. cholerae CT are both AB5 enterotoxins with ~80% amino acid identity, which bind cell-surface GM1 and Gd1b gangliosides (147). Preventive effects from bovine milk gangliosides against LT and CT require higher concentrations (174). GD3 was reported to minimally affect adhesion of ETEC and EPEC to intestinal cell lines (172). Globotriaosylceramide (Gb3), which is a neutral glycolipid in milk present in nanomolar concentrations, interacts with Shiga toxins (Stx) produced by Shigella dysenteriae and EHEC in vitro (175). Stx (also known as Verotoxin), an AB5 toxin, is a major virulence factor of EHEC; Stx binds to Gb3 expressed by kidney epithelial cells, causing tissue destruction that leads to hemolytic uremic syndrome as a result of EHEC infection. Stx also destroys intestinal epithelial cells, causing hemorrhagic colitis, which is a hallmark of EHEC infection (176). In vivo studies are necessary to assess the protective effects of milk gangliosides. Considering that antibiotic treatment is not recommended for EHEC infections because it enhances Stx production, leading to hemolytic uremic syndrome (HUS) (176), milk gangliosides could be used to block Stx access to its host cell receptor, preventing HUS in infected individuals.
Milk fat globule (MFG)
MUC1 is an abundant mucin in human milk and a major component of the MFG. Glycosylation of MUC1 was shown to interfere with HIV transmission from mucosal dendritic cells (DC) to CD4+ T cells in vitro (177). DCs play a pivotal role in the immune system as antigen-presenting cells, and HIV exploits DC for increased viral dissemination. Some DC subtypes in the GI mucosa express the lectin DC-SIGN (178). The HIV envelope glycoprotein gp120 binds to DC-SIGN (179), an interaction mediated by multiple N-glycosylation sites and high-mannose sugars (180, 181), enhancing viral transmission to CD4+ T cells, the main target of HIV. MUC1 binds to DC-SIGN receptor, preventing interaction with HIV gp120 (177). Natural antibodies in milk can also block HIV transmission from DC cells to CD4+ T cells via DC-SIGN (182). Milk-derived IgA and IgG that recognize the carbohydrate recognition domain of DC-SIGN significantly reduced HIV attachment to DC-SIGN-expressing HeLa cells (182). Taken together, both studies suggest that milk glycoproteins have major implications for prevention of postnatal HIV transmission during breastfeeding.
Human milk mucins were shown to impair rotavirus replication in cell cultures and gastro-enteritis in a mouse model in a manner dependent on sialylated glycans (183). Mucin glycans are also involved in protection against pathogenic bacteria. Sialic-acid glycans from MUC1 prevented attachment of S-fimbriated E. coli to buccal mucosal cells (184). Milk concentrations of MUC1 and MUC4 prevented invasion of Caco-2 by S. typhimurium, although the role of glycans in this phenomenon was not addressed (185).
CONCLUSIONS AND FUTURE STUDIES
The past decade has seen an explosion of research and interest in the human milk glycobiome. The glycan landscape presented in human milk clearly affords significant protection to the neonate, either through direct protection by blocking pathogen binding or indirectly through enrichment of a protective MOM. However, much detail on the specific nature of these interactions between glycans and their target bacterial partners remains unclear. Which specific glycans mediate which functions? Which bacteria are the keystone species, and which consume the saccharide crumbs left behind? In addition, it is hard to assess the relative influence of the different functions of the milk glycome on the ensuing infant microbiota, particularly given the many other microbiota-modulating functions delivered in milk, such as lysozyme, Lf, Lfc, sIgAs, or a range of newly discovered antimicrobial agents in milk (186, 187). Luckily, new analytical tools to comprehensively examine the free glycans and glycoconjugates in feces, blood, and urine provide new vistas upon which to understand the specific glycan drivers of microbial growth in situ. These new tools, combined with metagenomic analyses, enable researchers to link specific bacterial taxa changes with a shifting landscape of glycan substrates traveling through the intestine. The wealth of knowledge generated from this will not only expand the basic understanding of infant development but also create a constellation of diagnostic targets and new tools to enhance clinical options within the nursery and neonatal intensive care unit.
One clear opportunity lies in the coevolved nature of infant-borne bifidobacteria and the cognate milk glycans as a potential synbiotic therapy for premature infants. Moreover, the ability to more readily garner HMO and human glycoconjugate mimics from animal milk sets the stage for more protective and longer lasting establishment of a MOM within the most fragile humans. This conceptual application, however, could readily translate to other therapeutic realms where humans experience dysbiosis and are in need of protection from a MOM.
Acknowledgments
We acknowledge all of the researchers in the UC Davis Foods for Health Institute and the Milk Bioactives Program for their enthusiasm, imagination, and collective contribution to this subject matter. Work by the Milk Bioactives Program has been supported by the UC Davis Research Investments in the Sciences and Engineering Program; the UC Discovery Grant Program; the California Dairy Research Foundation; the Dairy Research Institute; the Bill & Melinda Gates Foundation; and the National Institutes of Health awards R01HD059127, R01HD065122, R01HD061923, R21AT006180, and R01AT007079. D.A.M. acknowledges support as the Peter J. Shields Endowed Chair in Dairy Food Science.
Footnotes
DISCLOSURE STATEMENT
D.A.M. and D.B. own shares in Evolve Biosystems Inc., a company focused on probiotics for infants.
Contributor Information
Alline R. Pacheco, Email: arpacheco@ucdavis.edu.
Daniela Barile, Email: dbarile@ucdavis.edu.
Mark A. Underwood, Email: mark.underwood@ucdmc.ucdavis.edu.
David A. Mills, Email: damills@ucdavis.edu.
LITERATURE CITED
- 1.Am. Acad. Pediatr. Breastfeeding and the use of human milk. Pediatrics. 2012;129:e827–41. doi: 10.1542/peds.2011-3552. [DOI] [PubMed] [Google Scholar]
- 2.Ballard O, Morrow AL. Human milk composition: nutrients and bioactive factors. Pediatr Clin North Am. 2013;60:49–74. doi: 10.1016/j.pcl.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haschke F, Haiden N, Detzel P, Yarnoff B, Allaire B, Haschke-Becher E. Feeding patterns during the first 2 years and health outcome. Ann Nutr Metab. 2013;62(Suppl 3):16–25. doi: 10.1159/000351575. [DOI] [PubMed] [Google Scholar]
- 4.Ruiz-Palacios GM, Calva JJ, Pickering LK, Lopez-Vidal Y, Volkow P, et al. Protection of breast-fed infants against Campylobacter diarrhea by antibodies in human milk. J Pediatr. 1990;116:707–13. doi: 10.1016/s0022-3476(05)82652-6. [DOI] [PubMed] [Google Scholar]
- 5.Clavano NR. Mode of feeding and its effect on infant mortality and morbidity. J Trop Pediatr. 1982;28:287–93. doi: 10.1093/tropej/28.6.287. [DOI] [PubMed] [Google Scholar]
- 6.Chen A, Rogan WJ. Breastfeeding and the risk of postneonatal death in the United States. Pediatrics. 2004;113:e435–39. doi: 10.1542/peds.113.5.e435. [DOI] [PubMed] [Google Scholar]
- 7.Andres A, Cleves MA, Bellando JB, Pivik RT, Casey PH, Badger TM. Developmental status of 1-year-old infants fed breast milk, cow’s milk formula, or soy formula. Pediatrics. 2012;129:1134–40. doi: 10.1542/peds.2011-3121. [DOI] [PubMed] [Google Scholar]
- 8.Alvarez-Uria G, Midde M, Pakam R, Bachu L, Naik PK. Effect of formula feeding and breast-feeding on child growth, infant mortality, and HIV transmission in children born to HIV-infected pregnant women who received triple antiretroviral therapy in a resource-limited setting: data from an HIV cohort study in India. ISRN Pediatr. 2012;2012:763591. doi: 10.5402/2012/763591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thior I, Lockman S, Smeaton LM, Shapiro RL, Wester C, et al. Breastfeeding plus infant zido-vudine prophylaxis for 6 months vs formula feeding plus infant zidovudine for 1 month to reduce mother-to-child HIV transmission in Botswana: a randomized trial: the Mashi Study. JAMA. 2006;296:794–805. doi: 10.1001/jama.296.7.794. [DOI] [PubMed] [Google Scholar]
- 10.Chatterton DE, Nguyen DN, Bering SB, Sangild PT. Anti-inflammatory mechanisms of bioactive milk proteins in the intestine of newborns. Int J Biochem Cell Biol. 2013;45:1730–47. doi: 10.1016/j.biocel.2013.04.028. [DOI] [PubMed] [Google Scholar]
- 11.Zivkovic AM, German JB, Lebrilla CB, Mills DA. Human milk glycobiome and its impact on the infant gastrointestinal microbiota. PNAS. 2011;108(Suppl 1):4653–58. doi: 10.1073/pnas.1000083107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Apweiler R, Hermjakob H, Sharon N. On the frequency of protein glycosylation, as deduced from analysis of the SWISS-PROT database. Biochim Biophys Acta. 1999;1473:4–8. doi: 10.1016/s0304-4165(99)00165-8. [DOI] [PubMed] [Google Scholar]
- 13.Peterson R, Cheah WY, Grinyer J, Packer N. Glycoconjugates in human milk: protecting infants from disease. Glycobiology. 2013;23:1425–38. doi: 10.1093/glycob/cwt072. [DOI] [PubMed] [Google Scholar]
- 14.Ruhaak LR, Lebrilla CB. Analysis and role of oligosaccharides in milk. BMB Rep. 2012;45:442–51. doi: 10.5483/BMBRep.2012.45.8.161. [DOI] [PubMed] [Google Scholar]
- 15.Wu S, Tao N, German JB, Grimm R, Lebrilla CB. Development of an annotated library of neutral human milk oligosaccharides. J Proteome Res. 2010;9:4138–51. doi: 10.1021/pr100362f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bode L. Human milk oligosaccharides: Every baby needs a sugar mama. Glycobiology. 2012;22:1147–62. doi: 10.1093/glycob/cws074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oftedal OT. The evolution of milk secretion and its ancient origins. Animal. 2012;6:355–68. doi: 10.1017/S1751731111001935. [DOI] [PubMed] [Google Scholar]
- 18.Albrecht S, Schols HA, van den Heuvel EG, Voragen AG, Gruppen H. Occurrence of oligosaccharides in feces of breast-fed babies in their first six months of life and the corresponding breast milk. Carbohydr Res. 2011;346:2540–50. doi: 10.1016/j.carres.2011.08.009. [DOI] [PubMed] [Google Scholar]
- 19.Rudloff S, Kunz C. Milk oligosaccharides and metabolism in infants. Adv Nutr. 2012;3:398S–405S. doi: 10.3945/an.111.001594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gnoth MJ, Kunz C, Kinne-Saffran E, Rudloff S. Human milk oligosaccharides are minimally digested in vitro. J Nutr. 2000;130:3014–20. doi: 10.1093/jn/130.12.3014. [DOI] [PubMed] [Google Scholar]
- 21.Albrecht S, Schols HA, van den Heuvel EGHM, Voragen AGJ, Gruppen H. CE-LIF-MSn profiling of oligosaccharides in human milk and feces of breast-fed babies. Electrophoresis. 2010;31:1264–73. doi: 10.1002/elps.200900646. [DOI] [PubMed] [Google Scholar]
- 22.Rudloff S, Pohlentz G, Diekmann L, Egge H, Kunz C. Urinary excretion of lactose and oligosaccharides in preterm infants fed human milk or infant formula. Acta Paediatr. 1996;85:598–603. doi: 10.1111/j.1651-2227.1996.tb14095.x. [DOI] [PubMed] [Google Scholar]
- 23.Rudloff S, Pohlentz G, Borsch C, Lentze MJ, Kunz C. Urinary excretion of in vivo 13C-labelled milk oligosaccharides in breastfed infants. Br J Nutr. 2012;107:957–63. doi: 10.1017/S0007114511004016. [DOI] [PubMed] [Google Scholar]
- 24.Ruhaak LR, Lebrilla CB. Advances in analysis of human milk oligosaccharides. Adv Nutr. 2012;3:406S–14S. doi: 10.3945/an.112.001883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ninonuevo MR, Park Y, Yin H, Zhang J, Ward RE, et al. A strategy for annotating the human milk glycome. J Agric Food Chem. 2006;54:7471–80. doi: 10.1021/jf0615810. [DOI] [PubMed] [Google Scholar]
- 26.Wu S, Grimm R, German JB, Lebrilla CB. Annotation and structural analysis of sialylated human milk oligosaccharides. J Proteome Res. 2011;10:856–68. doi: 10.1021/pr101006u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zivkovic AM, Barile D. Bovine milk as a source of functional oligosaccharides for improving human health. Adv Nutr. 2011;2:284–89. doi: 10.3945/an.111.000455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tao N, DePeters EJ, Freeman S, German JB, Grimm R, Lebrilla CB. Bovine milk glycome. J Dairy Sci. 2008;91:3768–78. doi: 10.3168/jds.2008-1305. [DOI] [PubMed] [Google Scholar]
- 29.Tao N, DePeters EJ, German JB, Grimm R, Lebrilla CB. Variations in bovine milk oligosaccharides during early and middle lactation stages analyzed by high-performance liquid chromatography-chip/mass spectrometry. J Dairy Sci. 2009;92:2991–3001. doi: 10.3168/jds.2008-1642. [DOI] [PubMed] [Google Scholar]
- 30.Barile D, Marotta M, Chu C, Mehra R, Grimm R, et al. Neutral and acidic oligosaccharides in Holstein-Friesian colostrum during the first 3 days of lactation measured by high performance liquid chromatography on a microfluidic chip and time-of-flight mass spectrometry. J Dairy Sci. 2010;93:3940–49. doi: 10.3168/jds.2010-3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aldredge DL, Geronimo MR, Hua S, Nwosu CC, Lebrilla CB, Barile D. Annotation and structural elucidation of bovine milk oligosaccharides and determination of novel fucosylated structures. Glycobiology. 2013;23:664–76. doi: 10.1093/glycob/cwt007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barile D, Tao N, Lebrilla CB, Coisson JD, Arlorio M, German JB. Permeate from cheese whey ultrafiltration is a source of milk oligosaccharides. Int Dairy J. 2009;19:524–30. doi: 10.1016/j.idairyj.2009.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lonnerdal B. Nutritional and physiologic significance of human milk proteins. Am J Clin Nutr. 2003;77:1537S–43S. doi: 10.1093/ajcn/77.6.1537S. [DOI] [PubMed] [Google Scholar]
- 34.Le Parc A, Dallas DC, Duaut S, Leonil J, Martin P, Barile D. Characterization of goat milk lactoferrin N-glycans and comparison with the N-glycomes of human and bovine milk. Electrophoresis. 2014;35:1560–70. doi: 10.1002/elps.201300619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tomita M, Bellamy W, Takase M, Yamauchi K, Wakabayashi H, Kawase K. Potent antibacterial peptides generated by pepsin digestion of bovine lactoferrin. J Dairy Sci. 1991;74:4137–42. doi: 10.3168/jds.S0022-0302(91)78608-6. [DOI] [PubMed] [Google Scholar]
- 36.Yamauchi K, Tomita M, Giehl TJ, Ellison RT., 3rd Antibacterial activity of lactoferrin and a pepsin-derived lactoferrin peptide fragment. Infect Immun. 1993;61:719–28. doi: 10.1128/iai.61.2.719-728.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.O’Riordan N, Kane M, Joshi L, Hickey RM. Structural and functional characteristics of bovine milk protein glycosylation. Glycobiology. 2014;24:220–36. doi: 10.1093/glycob/cwt162. [DOI] [PubMed] [Google Scholar]
- 38.Davidson LA, Lonnerdal B. Persistence of human milk proteins in the breast-fed infant. Acta Paediatr Scand. 1987;76:733–40. doi: 10.1111/j.1651-2227.1987.tb10557.x. [DOI] [PubMed] [Google Scholar]
- 39.Wang B, Brand-Miller J, McVeagh P, Petocz P. Concentration and distribution of sialic acid in human milk and infant formulas. Am J Clin Nutr. 2001;74:510–15. doi: 10.1093/ajcn/74.4.510. [DOI] [PubMed] [Google Scholar]
- 40.Idota T, Kawakami H, Nakajima I. Growth-promoting effects of N-acetylneuraminic acid-containing substances on Bifidobacteria. Biosci Biotechnol Biochem. 1994;58:1720–22. [Google Scholar]
- 41.Yvon M, Beucher S, Guilloteau P, Le Huerou-Luron I, Corring T. Effects of caseinomacropeptide (CMP) on digestion regulation. Reprod Nutr Dev. 1994;34:527–37. doi: 10.1051/rnd:19940602. [DOI] [PubMed] [Google Scholar]
- 42.Lloyd KO, Furukawa K. Biosynthesis and functions of gangliosides: recent advances. Glycoconj J. 1998;15:627–36. doi: 10.1023/a:1006924128550. [DOI] [PubMed] [Google Scholar]
- 43.Lee H, An HJ, Lerno LA, Jr, German JB, Lebrilla CB. Rapid profiling of bovine and human milk gangliosides by matrix-assisted laser desorption/ionization Fourier transform ion cyclotron resonance mass spectrometry. Int J Mass Spectrom. 2011;305:138–50. doi: 10.1016/j.ijms.2010.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pan XL, Izumi T. Variation of the ganglioside compositions of human milk, cow’s milk and infant formulas. Early Hum Dev. 2000;57:25–31. doi: 10.1016/s0378-3782(99)00051-1. [DOI] [PubMed] [Google Scholar]
- 45.Lee H, German JB, Kjelden R, Lebrilla CB, Barile D. Quantitative analysis of gangliosides in bovine milk and colostrum-based dairy products by ultrahigh performance liquid chromatography-tandem mass spectrometry. J Agric Food Chem. 2013;61:9689–96. doi: 10.1021/jf402255g. [DOI] [PubMed] [Google Scholar]
- 46.Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006;124:837–48. doi: 10.1016/j.cell.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 47.Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–38. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Backhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science. 2005;307:1915–20. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- 49.Nicholson JK, Holmes E, Kinross J, Burcelin R, Gibson G, et al. Host-gut microbiota metabolic interactions. Science. 2012;336:1262–67. doi: 10.1126/science.1223813. [DOI] [PubMed] [Google Scholar]
- 50.Spees AM, Lopez CA, Kingsbury DD, Winter SE, Bäumler AJ. Colonization resistance: Battle of the bugs or ménage à trois with the host? PLOS Pathog. 2013;9:e1003730. doi: 10.1371/journal.ppat.1003730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–27. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–63. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cho I, Blaser MJ. The human microbiome: at the interface of health and disease. Nat Rev Genet. 2012;13:260–70. doi: 10.1038/nrg3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ley RE, Backhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. PNAS. 2005;102:11070–75. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–31. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 56.Manichanh C, Rigottier-Gois L, Bonnaud E, Gloux K, Pelletier E, et al. Reduced diversity of faecal microbiota in Crohn’s disease revealed by a metagenomic approach. Gut. 2006;55:205–11. doi: 10.1136/gut.2005.073817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Normann E, Fahlen A, Engstrand L, Lilja HE. Intestinal microbial profiles in extremely preterm infants with and without necrotizing enterocolitis. Acta Paediatr. 2013;102:129–36. doi: 10.1111/apa.12059. [DOI] [PubMed] [Google Scholar]
- 58.Mai V, Young CM, Ukhanova M, Wang X, Sun Y, et al. Fecal microbiota in premature infants prior to necrotizing enterocolitis. PLOS ONE. 2011;6:e20647. doi: 10.1371/journal.pone.0020647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wu N, Yang X, Zhang R, Li J, Xiao X, et al. Dysbiosis signature of fecal microbiota in colorectal cancer patients. Microb Ecol. 2013;66:462–70. doi: 10.1007/s00248-013-0245-9. [DOI] [PubMed] [Google Scholar]
- 60.Walker A. Breast milk as the gold standard for protective nutrients. J Pediatr. 2010;156:S3–7. doi: 10.1016/j.jpeds.2009.11.021. [DOI] [PubMed] [Google Scholar]
- 61.Caicedo RA, Schanler RJ, Li N, Neu J. The developing intestinal ecosystem: implications for the neonate. Pediatr Res. 2005;58:625–28. doi: 10.1203/01.PDR.0000180533.09295.84. [DOI] [PubMed] [Google Scholar]
- 62.Claud EC, Lu L, Anton PM, Savidge T, Walker WA, Cherayil BJ. Developmentally regulated IκB expression in intestinal epithelium and susceptibility to flagellin-induced inflammation. PNAS. 2004;101:7404–8. doi: 10.1073/pnas.0401710101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Walker WA. Initial intestinal colonization in the human infant and immune homeostasis. Ann Nutr Metab. 2013;63(Suppl 2):8–15. doi: 10.1159/000354907. [DOI] [PubMed] [Google Scholar]
- 64.Biasucci G, Rubini M, Riboni S, Morelli L, Bessi E, Retetangos C. Mode of delivery affects the bacterial community in the newborn gut. Early Hum Dev. 2010;86(Suppl 1):13–15. doi: 10.1016/j.earlhumdev.2010.01.004. [DOI] [PubMed] [Google Scholar]
- 65.Penders J, Thijs C, Vink C, Stelma FF, Snijders B, et al. Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics. 2006;118:511–21. doi: 10.1542/peds.2005-2824. [DOI] [PubMed] [Google Scholar]
- 66.Huurre A, Kalliomaki M, Rautava S, Rinne M, Salminen S, Isolauri E. Mode of delivery—effects on gut microbiota and humoral immunity. Neonatology. 2008;93:236–40. doi: 10.1159/000111102. [DOI] [PubMed] [Google Scholar]
- 67.Fanaro S, Chierici R, Guerrini P, Vigi V. Intestinal microflora in early infancy: composition and development. Acta Paediatr Suppl. 2003;91:48–55. doi: 10.1111/j.1651-2227.2003.tb00646.x. [DOI] [PubMed] [Google Scholar]
- 68.Mackie RI, Sghir A, Gaskins HR. Developmental microbial ecology of the neonatal gastrointestinal tract. Am J Clin Nutr. 1999;69:1035S–45S. doi: 10.1093/ajcn/69.5.1035s. [DOI] [PubMed] [Google Scholar]
- 69.Subramanian S, Huq S, Yatsunenko T, Haque R, Mahfuz M, et al. Persistent gut microbiota immaturity in malnourished Bangladeshi children. Nature. 2014;510:417–21. doi: 10.1038/nature13421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Scholtens PA, Oozeer R, Martin R, Amor KB, Knol J. The early settlers: intestinal microbiology in early life. Annu Rev Food Sci Technol. 2012;3:425–47. doi: 10.1146/annurev-food-022811-101120. [DOI] [PubMed] [Google Scholar]
- 71.Berni Canani R, Passariello A, Buccigrossi V, Terrin G, Guarino A. The nutritional modulation of the evolving intestine. J Clin Gastroenterol. 2008;42(Suppl 3 Pt 2):S197–200. doi: 10.1097/MCG.0b013e31817da155. [DOI] [PubMed] [Google Scholar]
- 72.Turroni F, Peano C, Pass DA, Foroni E, Severgnini M, et al. Diversity of bifidobacteria within the infant gut microbiota. PLOS ONE. 2012;7:e36957. doi: 10.1371/journal.pone.0036957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Avershina E, Storro O, Oien T, Johnsen R, Wilson R, et al. Bifidobacterial succession and correlation networks in a large unselected cohort of mothers and their children. Appl Environ Microbiol. 2013;79:497–507. doi: 10.1128/AEM.02359-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Harmsen HJ, Wildeboer-Veloo AC, Raangs GC, Wagendorp AA, Klijn N, et al. Analysis of intestinal flora development in breast-fed and formula-fed infants by using molecular identification and detection methods. J Pediatr Gastroenterol Nutr. 2000;30:61–67. doi: 10.1097/00005176-200001000-00019. [DOI] [PubMed] [Google Scholar]
- 75.Jost T, Lacroix C, Braegger CP, Chassard C. New insights in gut microbiota establishment in healthy breast fed neonates. PLOS ONE. 2012;7:e44595. doi: 10.1371/journal.pone.0044595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Roger LC, McCartney AL. Longitudinal investigation of the faecal microbiota of healthy full-term infants using fluorescence in situ hybridization and denaturing gradient gel electrophoresis. Microbiology. 2010;156:3317–28. doi: 10.1099/mic.0.041913-0. [DOI] [PubMed] [Google Scholar]
- 77.Turroni F, Foroni E, Pizzetti P, Giubellini V, Ribbera A, et al. Exploring the diversity of the bifidobacterial population in the human intestinal tract. Appl Environ Microbiol. 2009;75:1534–45. doi: 10.1128/AEM.02216-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.De Leoz MLA, Gaerlan SC, Strum JS, Dimapasoc LM, Mirmiran M, et al. Lacto-N-tetraose, fucosylation, and secretor status are highly variable in human milk oligosaccharides from women delivering preterm. J Proteome Res. 2012;11:4662–72. doi: 10.1021/pr3004979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Neu J, Walker WA. Necrotizing enterocolitis. N Engl J Med. 2011;364:255–64. doi: 10.1056/NEJMra1005408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fusunyan RD, Nanthakumar NN, Baldeon ME, Walker WA. Evidence for an innate immune response in the immature human intestine: toll-like receptors on fetal enterocytes. Pediatr Res. 2001;49:589–93. doi: 10.1203/00006450-200104000-00023. [DOI] [PubMed] [Google Scholar]
- 81.Arboleya S, Binetti A, Salazar N, Fernandez N, Solis G, et al. Establishment and development of intestinal microbiota in preterm neonates. FEMS Microbiol Ecol. 2012;79:763–72. doi: 10.1111/j.1574-6941.2011.01261.x. [DOI] [PubMed] [Google Scholar]
- 82.Butel MJ, Suau A, Campeotto F, Magne F, Aires J, et al. Conditions of bifidobacterial colonization in preterm infants: a prospective analysis. J Pediatr Gastroenterol Nutr. 2007;44:577–82. doi: 10.1097/MPG.0b013e3180406b20. [DOI] [PubMed] [Google Scholar]
- 83.Johnson TJ, Patel AL, Jegier BJ, Engstrom JL, Meier PP. Cost of morbidities in very low birth weight infants. J Pediatr. 2013;162:243–49.e1. doi: 10.1016/j.jpeds.2012.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Morrow AL, Meinzen-Derr J, Huang P, Schibler KR, Cahill T, et al. Fucosyltransferase 2 non-secretor and low secretor status predicts severe outcomes in premature infants. J Pediatr. 2011;158:745–51. doi: 10.1016/j.jpeds.2010.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Downard CD, Grant SN, Maki AC, Krupski MC, Matheson PJ, et al. Maternal cigarette smoking and the development of necrotizing enterocolitis. Pediatrics. 2012;130:78–82. doi: 10.1542/peds.2011-3808. [DOI] [PubMed] [Google Scholar]
- 86.Torrazza RM, Ukhanova M, Wang X, Sharma R, Hudak ML, et al. Intestinal microbial ecology and environmental factors affecting necrotizing enterocolitis. PLOS ONE. 2013;8:e83304. doi: 10.1371/journal.pone.0083304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Morrow AL, Lagomarcino AJ, Schibler KR, Taft DH, Yu Z, et al. Early microbial and metabolomic signatures predict later onset of necrotizing enterocolitis in preterm infants. Microbiome. 2013;1:13. doi: 10.1186/2049-2618-1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang Y, Hoenig JD, Malin KJ, Qamar S, Petrof EO, et al. 16S rRNA gene-based analysis of fecal microbiota from preterm infants with and without necrotizing enterocolitis. ISME J. 2009;3:944–54. doi: 10.1038/ismej.2009.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Jenke AC, Postberg J, Mariel B, Hensel K, Foell D, et al. S100A12 and hBD2 correlate with the composition of the fecal microflora in ELBW infants and expansion of E. coli is associated with NEC. BioMed Res Int. 2013;2013:150372. doi: 10.1155/2013/150372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cotten CM, Taylor S, Stoll B, Goldberg RN, Hansen NI, et al. Prolonged duration of initial empirical antibiotic treatment is associated with increased rates of necrotizing enterocolitis and death for extremely low birth weight infants. Pediatrics. 2009;123:58–66. doi: 10.1542/peds.2007-3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gupta RW, Tran L, Norori J, Ferris MJ, Eren AM, et al. Histamine-2 receptor blockers alter the fecal microbiota in premature infants. J Pediatr Gastroenterol Nutr. 2013;56:397–400. doi: 10.1097/MPG.0b013e318282a8c2. [DOI] [PubMed] [Google Scholar]
- 92.Terrin G, Passariello A, De Curtis M, Manguso F, Salvia G, et al. Ranitidine is associated with infections, necrotizing enterocolitis, and fatal outcome in newborns. Pediatrics. 2012;129:e40–45. doi: 10.1542/peds.2011-0796. [DOI] [PubMed] [Google Scholar]
- 93.Underwood MA, Kalanetra KM, Bokulich NA, Lewis ZT, Mirmiran M, et al. A comparison of two probiotic strains of bifidobacteria in premature infants. J Pediatr. 2013;163:1585–91.e9. doi: 10.1016/j.jpeds.2013.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.AlFaleh K, Anabrees J, Bassler D, Al-Kharfi T. Probiotics for prevention of necrotizing enterocolitis in preterm infants. Cochrane Database Syst Rev. 2014;4:CD005496. doi: 10.1002/14651858.CD005496.pub3. [DOI] [PubMed] [Google Scholar]
- 95.Meinzen-Derr J, Poindexter B, Wrage L, Morrow AL, Stoll B, Donovan EF. Role of human milk in extremely low birth weight infants’ risk of necrotizing enterocolitis or death. J Perinatol. 2009;29:57–62. doi: 10.1038/jp.2008.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Underwood MA. Human milk for the premature infant. Pediatr Clin North Am. 2013;60:189–207. doi: 10.1016/j.pcl.2012.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hunter CJ, Bean JF. Cronobacter: an emerging opportunistic pathogen associated with neonatal meningitis, sepsis and necrotizing enterocolitis. J Perinatol. 2013;33:581–85. doi: 10.1038/jp.2013.26. [DOI] [PubMed] [Google Scholar]
- 98.Cristofalo EA, Schanler RJ, Blanco CL, Sullivan S, Trawoeger R, et al. Randomized trial of exclusive human milk versus preterm formula diets in extremely premature infants. J Pediatr. 2013;163:1592–95.e1. doi: 10.1016/j.jpeds.2013.07.011. [DOI] [PubMed] [Google Scholar]
- 99.Gyorgy P, Norris RF, Rose CS. Bifidus factor. I A variant of Lactobacillus bifidus requiring a special growth factor. Arch Biochem Biophys. 1954;48:193–201. doi: 10.1016/0003-9861(54)90323-9. [DOI] [PubMed] [Google Scholar]
- 100.Gauhe A, Gyorgy P, Hoover JRE, Kuhn R, Rose CS, et al. Bifidus factor. 4 Preparations obtained from human milk. Arch Biochem Biophys. 1954;48:214–24. doi: 10.1016/0003-9861(54)90326-4. [DOI] [PubMed] [Google Scholar]
- 101.Garrido D, Dallas DC, Mills DA. Consumption of human milk glycoconjugates by infant-associated bifidobacteria: mechanisms and implications. Microbiology. 2013;159:649–64. doi: 10.1099/mic.0.064113-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Marcobal A, Sonnenburg JL. Human milk oligosaccharide consumption by intestinal microbiota. Clin Microbiol Infect. 2012;18(Suppl 4):12–15. doi: 10.1111/j.1469-0691.2012.03863.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ward RE, Niñonuevo M, Mills DA, Lebrilla CB, German JB. In vitro fermentation of breast milk oligosaccharides by Bifidobacterium infantis and Lactobacillus gasseri. Appl Environ Microbiol. 2006;72:4497–99. doi: 10.1128/AEM.02515-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Marcobal A, Barboza M, Froehlich JW, Block DE, German JB, et al. Consumption of human milk oligosaccharides by gut-related microbes. J Agric Food Chem. 2010;58:5334–40. doi: 10.1021/jf9044205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.LoCascio RG, Niñonuevo MR, Freeman SL, Sela DA, Grimm R, et al. Glycoprofiling of bifidobacterial consumption of human milk oligosaccharides demonstrates strain specific, preferential consumption of small chain glycans secreted in early human lactation. J Agric Food Chem. 2007;55:8914–19. doi: 10.1021/jf0710480. [DOI] [PubMed] [Google Scholar]
- 106.Locascio RG, Niñonuevo MR, Kronewitter SR, Freeman SL, German JB, et al. A versatile and scalable strategy for glycoprofiling bifidobacterial consumption of human milk oligosaccharides. Microb Biotechnol. 2009;2:333–42. doi: 10.1111/j.1751-7915.2008.00072.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ruiz-Moyano S, Totten SM, Garrido D, Smilowitz JT, German JB, et al. Variation in consumption of human milk oligosaccharides by infant-gut associated strains of Bifidobacterium breve. Appl Environ Microbiol. 2013;79:6040–49. doi: 10.1128/AEM.01843-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Yu ZT, Chen C, Newburg DS. Utilization of major fucosylated and sialylated human milk oligosaccharides by isolated human gut microbes. Glycobiology. 2013;23:1281–92. doi: 10.1093/glycob/cwt065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Marcobal A, Barboza M, Sonnenburg ED, Pudlo N, Martens EC, et al. Bacteroides in the infant gut consume milk oligosaccharides via mucus-utilization pathways. Cell Host Microbe. 2011;10:507–14. doi: 10.1016/j.chom.2011.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.LoCascio RG, Desai P, Sela DA, Weimer B, Mills DA. Broad conservation of milk utilization genes in Bifidobacterium longum subsp infantis as revealed by comparative genomic hybridization. Appl Environ Microbiol. 2010;76:7373–81. doi: 10.1128/AEM.00675-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kim JH, An HJ, Garrido D, German JB, Lebrilla CB, Mills DA. Proteomic analysis of Bifidobacterium longum subsp infantis reveals the metabolic insight on consumption of prebiotics and host glycans. PLOS ONE. 2013;8:e57535. doi: 10.1371/journal.pone.0057535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Sela DA, Garrido D, Lerno L, Wu S, Tan K, et al. Bifidobacterium longum subsp infantis ATCC 15697 α-fucosidases are active on fucosylated human milk oligosaccharides. Appl Environ Microbiol. 2012;78:795–803. doi: 10.1128/AEM.06762-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ashida H, Miyake A, Kiyohara M, Wada J, Yoshida E, et al. Two distinct α-L-fucosidases from Bifidobacterium bifidum are essential for the utilization of fucosylated milk oligosaccharides and glycoconjugates. Glycobiology. 2009;19:1010–17. doi: 10.1093/glycob/cwp082. [DOI] [PubMed] [Google Scholar]
- 114.Sela DA, Li Y, Lerno L, Wu S, Marcobal AM, et al. An infant-associated bacterial commensal utilizes breast milk sialyloligosaccharides. J Biol Chem. 2011;286:11909–18. doi: 10.1074/jbc.M110.193359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Garrido D, Ruiz-Moyano S, Mills DA. Release and utilization of N-acetyl-D-glucosamine from human milk oligosaccharides by Bifidobacterium longum subsp infantis. Anaerobe. 2012;18:430–35. doi: 10.1016/j.anaerobe.2012.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Yoshida E, Sakurama H, Kiyohara M, Nakajima M, Kitaoka M, et al. Bifidobacterium longum subsp infantis uses two different β-galactosidases for selectively degrading type-1 and type-2 human milk oligosaccharides. Glycobiology. 2012;22:361–68. doi: 10.1093/glycob/cwr116. [DOI] [PubMed] [Google Scholar]
- 117.Miwa M, Horimoto T, Kiyohara M, Katayama T, Kitaoka M, et al. Cooperation of β-galactosidase and β-N-acetylhexosaminidase from bifidobacteria in assimilation of human milk oligosaccharides with type 2 structure. Glycobiology. 2010;20:1402–9. doi: 10.1093/glycob/cwq101. [DOI] [PubMed] [Google Scholar]
- 118.Wada J, Ando T, Kiyohara M, Ashida H, Kitaoka M, et al. Bifidobacterium bifidum lacto-N-biosidase, a critical enzyme for the degradation of human milk oligosaccharides with a type 1 structure. Appl Environ Microbiol. 2008;74:3996–4004. doi: 10.1128/AEM.00149-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Sela DA, Mills DA. Nursing our microbiota: molecular linkages between bifidobacteria and milk oligosaccharides. Trends Microbiol. 2010;18:298–307. doi: 10.1016/j.tim.2010.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Egan M, O’Connell Motherway M, Ventura M, van Sinderen D. Metabolism of sialic acid by Bifidobacterium breve UCC2003. Appl Environ Microbiol. 2014;80:4414–26. doi: 10.1128/AEM.01114-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Katayama T, Sakuma A, Kimura T, Makimura Y, Hiratake J, et al. Molecular cloning and characterization of Bifidobacterium bifidum 1,2-α-L-fucosidase (AfcA), a novel inverting glycosidase (glycoside hydrolase family 95) J Bacteriol. 2004;186:4885–93. doi: 10.1128/JB.186.15.4885-4893.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kiyohara M, Tanigawa K, Chaiwangsri T, Katayama T, Ashida H, Yamamoto K. An exo-α-sialidase from bifidobacteria involved in the degradation of sialyloligosaccharides in human milk and intestinal glycoconjugates. Glycobiology. 2011;21:437–47. doi: 10.1093/glycob/cwq175. [DOI] [PubMed] [Google Scholar]
- 123.Pacheco AR, Curtis MM, Ritchie JM, Munera D, Waldor MK, et al. Fucose sensing regulates bacterial intestinal colonization. Nature. 2012;492:113–17. doi: 10.1038/nature11623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ng KM, Ferreyra JA, Higginbottom SK, Lynch JB, Kashyap PC, et al. Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature. 2013;502:96–99. doi: 10.1038/nature12503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Tang XS, Shao H, Li TJ, Tang ZR, Huang RL, et al. Dietary supplementation with bovine lactoferrampin-lactoferricin produced by Pichia pastoris fed-batch fermentation affects intestinal microflora in weaned piglets. Appl Biochem Biotechnol. 2012;168:887–98. doi: 10.1007/s12010-012-9827-0. [DOI] [PubMed] [Google Scholar]
- 126.Hu W, Zhao J, Wang J, Yu T, Li N. Transgenic milk containing recombinant human lactoferrin modulates the intestinal flora in piglets. Biochem Cell Biol. 2012;90:485–96. doi: 10.1139/o2012-003. [DOI] [PubMed] [Google Scholar]
- 127.Bruck WM, Graverholt G, Gibson GR. A two-stage continuous culture system to study the effect of supplemental α-lactalbumin and glycomacropeptide on mixed cultures of human gut bacteria challenged with enteropathogenic Escherichia coli and Salmonella serotype Typhimurium. J Appl Microbiol. 2003;95:44–53. doi: 10.1046/j.1365-2672.2003.01959.x. [DOI] [PubMed] [Google Scholar]
- 128.Chen Q, Cao J, Jia Y, Liu X, Yan Y, Pang G. Modulation of mice fecal microbiota by administration of casein glycomacropeptide. Microbiol Res. 2012;3:e3. [Google Scholar]
- 129.Rahman MM, Kim WS, Ito T, Kumura H, Shimazaki K. Growth promotion and cell binding ability of bovine lactoferrin to Bifidobacterium longum. Anaerobe. 2009;15:133–37. doi: 10.1016/j.anaerobe.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 130.Petschow BW, Talbott RD, Batema RP. Ability of lactoferrin to promote the growth of Bifidobacterium spp. in vitro is independent of receptor binding capacity and iron saturation level. J Med Microbiol. 1999;48:541–49. doi: 10.1099/00222615-48-6-541. [DOI] [PubMed] [Google Scholar]
- 131.Oda H, Wakabayashi H, Yamauchi K, Sato T, Xiao JZ, et al. Isolation of a bifidogenic peptide from the pepsin hydrolysate of bovine lactoferrin. Appl Environ Microbiol. 2013;79:1843–49. doi: 10.1128/AEM.03343-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Liepke C, Adermann K, Raida M, Magert HJ, Forssmann WG, Zucht HD. Human milk provides peptides highly stimulating the growth of bifidobacteria. Eur J Biochem. 2002;269:712–18. doi: 10.1046/j.0014-2956.2001.02712.x. [DOI] [PubMed] [Google Scholar]
- 133.Garrido D, Nwosu C, Ruiz-Moyano S, Aldredge D, German JB, et al. Endo-β-N-acetylglucosaminidases from infant gut-associated bifidobacteria release complex N-glycans from human milk glycoproteins. Mol Cell Proteomics. 2012;11:775–85. doi: 10.1074/mcp.M112.018119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Kiyohara M, Nakatomi T, Kurihara S, Fushinobu S, Suzuki H, et al. α-N-acetylgalactosaminidase from infant-associated bifidobacteria belonging to novel glycoside hydrolase family 129 is implicated in alternative mucin degradation pathway. J Biol Chem. 2012;287:693–700. doi: 10.1074/jbc.M111.277384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Rueda R, Sabatel JL, Maldonado J, Molina-Font JA, Gil A. Addition of gangliosides to an adapted milk formula modifies levels of fecal Escherichia coli in preterm newborn infants. J Pediatr. 1998;133:90–94. doi: 10.1016/s0022-3476(98)70184-2. [DOI] [PubMed] [Google Scholar]
- 136.Lee H, Garrido D, Mills DA, Barile D. Hydrolysis of milk gangliosides by infant-gut associated bifidobacteria determined by microfluidic chips and high-resolution mass spectrometry. Electrophoresis. 2014;35:1742–50. doi: 10.1002/elps.201300653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Morrow AL, Rangel JM. Human milk protection against infectious diarrhea: implications for prevention and clinical care. Semin Pediatr Infect Dis. 2004;15:221–28. doi: 10.1053/j.spid.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 138.Lanari M, Prinelli F, Adorni F, Di Santo S, Faldella G, et al. Maternal milk protects infants against bronchiolitis during the first year of life. Results from an Italian cohort of newborns. Early Hum Dev. 2013;89(Suppl 1):S51–57. doi: 10.1016/S0378-3782(13)70016-1. [DOI] [PubMed] [Google Scholar]
- 139.Duffy LC, Faden H, Wasielewski R, Wolf J, Krystofik D. Exclusive breastfeeding protects against bacterial colonization and day care exposure to otitis media. Pediatrics. 1997;100:E7. doi: 10.1542/peds.100.4.e7. [DOI] [PubMed] [Google Scholar]
- 140.Abdel-Hafeez EH, Belal US, Abdellatif MZ, Naoi K, Norose K. Breast-feeding protects infantile diarrhea caused by intestinal protozoan infections. Korean J Parasitol. 2013;51:519–24. doi: 10.3347/kjp.2013.51.5.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Newburg DS, Ruiz-Palacios GM, Morrow AL. Human milk glycans protect infants against enteric pathogens. Annu Rev Nutr. 2005;25:37–58. doi: 10.1146/annurev.nutr.25.050304.092553. [DOI] [PubMed] [Google Scholar]
- 142.Krachler AM, Orth K. Targeting the bacteria-host interface: strategies in anti-adhesion therapy. Virulence. 2013;4:284–94. doi: 10.4161/viru.24606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Jiang X, Huang PW, Zhong WM, Morrow AL, Ruiz-Palacios GM, Pickering LK. Human milk contains elements that block binding of noroviruses to histo-blood group antigens in saliva. Prot Infants Hum Milk. 2004;554:447–50. doi: 10.1007/978-1-4757-4242-8_62. [DOI] [PubMed] [Google Scholar]
- 144.Lanata CF, Fischer-Walker CL, Olascoaga AC, Torres CX, Aryee MJ, Black RE. Global causes of diarrheal disease mortality in children <5 years of age: a systematic review. PLOS ONE. 2013;8:e72788. doi: 10.1371/journal.pone.0072788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Zhang XF, Tan M, Chhabra M, Dai YC, Meller J, Jiang X. Inhibition of histo-blood group antigen binding as a novel strategy to block norovirus infections. PLOS ONE. 2013;8:e69379. doi: 10.1371/journal.pone.0069379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Morrow AL, Ruiz-Palacios GM, Altaye M, Jiang X, Guerrero ML, et al. Human milk oligosaccharides are associated with protection against diarrhea in breast-fed infants. J Pediatr. 2004;145:297–303. doi: 10.1016/j.jpeds.2004.04.054. [DOI] [PubMed] [Google Scholar]
- 147.Kaper JB, Nataro JP, Mobley HL. Pathogenic Escherichia coli. Nat Rev Microbiol. 2004;2:123–40. doi: 10.1038/nrmicro818. [DOI] [PubMed] [Google Scholar]
- 148.Manthey CF, Autran CA, Eckmann L, Bode L. Human milk oligosaccharides protect against enteropathogenic Escherichia coli attachment in vitro and EPEC colonization in suckling mice. J Pediatr Gastroenterol Nutr. 2013;58:167–70. doi: 10.1097/MPG.0000000000000172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Weichert S, Jennewein S, Hufner E, Weiss C, Borkowski J, et al. Bioengineered 2′-fucosyllactose and 3-fucosyllactose inhibit the adhesion of Pseudomonas aeruginosa and enteric pathogens to human intestinal and respiratory cell lines. Nutr Res. 2013;33:831–38. doi: 10.1016/j.nutres.2013.07.009. [DOI] [PubMed] [Google Scholar]
- 150.Martin-Sosa S, Martin MJ, Hueso P. The sialylated fraction of milk oligosaccharides is partially responsible for binding to enterotoxigenic and uropathogenic Escherichia coli human strains. J Nutr. 2002;132:3067–72. doi: 10.1093/jn/131.10.3067. [DOI] [PubMed] [Google Scholar]
- 151.Lin AE, Autran CA, Espanola SD, Bode L, Nizet V. Human milk oligosaccharides protect bladder epithelial cells against uropathogenic Escherichia coli invasion and cytotoxicity. J Infect Dis. 2014;209:389–98. doi: 10.1093/infdis/jit464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Hannan TJ, Totsika M, Mansfield KJ, Moore KH, Schembri MA, Hultgren SJ. Host-pathogen checkpoints and population bottlenecks in persistent and intracellular uropathogenic Escherichia coli bladder infection. FEMS Microbiol Rev. 2012;36:616–48. doi: 10.1111/j.1574-6976.2012.00339.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Greenhow TL, Hung YY, Herz AM, Losada E, Pantell RH. The changing epidemiology of serious bacterial infections in young infants. Pediatr Infect Dis J. 2013;33:595–99. doi: 10.1097/INF.0000000000000225. [DOI] [PubMed] [Google Scholar]
- 154.Jantscher-Krenn E, Lauwaet T, Bliss LA, Reed SL, Gillin FD, Bode L. Human milk oligosaccharides reduce Entamoeba histolytica attachment and cytotoxicity in vitro. Br J Nutr. 2012;108:1839–46. doi: 10.1017/S0007114511007392. [DOI] [PubMed] [Google Scholar]
- 155.Barboza M, Pinzon J, Wickramasinghe S, Froehlich JW, Moeller I, et al. Glycosylation of human milk lactoferrin exhibits dynamic changes during early lactation enhancing its role in pathogenic bacteria-host interactions. Mol Cell Proteomics. 2012;11:M111015248. doi: 10.1074/mcp.M111.015248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Marr AK, Jenssen H, Moniri MR, Hancock RE, Pante N. Bovine lactoferrin and lactoferricin interfere with intracellular trafficking of Herpes simplex virus-1. Biochimie. 2009;91:160–64. doi: 10.1016/j.biochi.2008.05.016. [DOI] [PubMed] [Google Scholar]
- 157.Shestakov A, Jenssen H, Nordstrom I, Eriksson K. Lactoferricin but not lactoferrin inhibit herpes simplex virus type 2 infection in mice. Antiviral Res. 2012;93:340–45. doi: 10.1016/j.antiviral.2012.01.003. [DOI] [PubMed] [Google Scholar]
- 158.Valimaa H, Tenovuo J, Waris M, Hukkanen V. Human lactoferrin but not lysozyme neutralizes HSV-1 and inhibits HSV-1 replication and cell-to-cell spread. Virol J. 2009;6:53. doi: 10.1186/1743-422X-6-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Hooper LV, Macpherson AJ. Immune adaptations that maintain homeostasis with the intestinal microbiota. Nat Rev Immunol. 2010;10:159–69. doi: 10.1038/nri2710. [DOI] [PubMed] [Google Scholar]
- 160.Rogier EW, Frantz AL, Bruno ME, Wedlund L, Cohen DA, et al. Secretory antibodies in breast milk promote long-term intestinal homeostasis by regulating the gut microbiota and host gene expression. PNAS. 2014;111:3074–79. doi: 10.1073/pnas.1315792111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Hughes GJ, Reason AJ, Savoy L, Jaton J, Frutiger-Hughes S. Carbohydrate moieties in human secretory component. Biochim Biophys Acta. 1999;1434:86–93. doi: 10.1016/s0167-4838(99)00168-5. [DOI] [PubMed] [Google Scholar]
- 162.Royle L, Roos A, Harvey DJ, Wormald MR, Van Gijlswijk-Janssen D, et al. Secretory IgA N- and O-glycans provide a link between the innate and adaptive immune systems. J Biol Chem. 2003;278:20140–53. doi: 10.1074/jbc.M301436200. [DOI] [PubMed] [Google Scholar]
- 163.Mathias A, Corthesy B. Recognition of gram-positive intestinal bacteria by hybridoma- and colostrum-derived secretory immunoglobulin A is mediated by carbohydrates. J Biol Chem. 2011;286:17239–47. doi: 10.1074/jbc.M110.209015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Cravioto A, Tello A, Villafán H, Ruiz J, del Vedovo S, Neeser JR. Inhibition of localized adhesion of enteropathogenic Escherichia coli to HEp-2 cells by immunoglobulin and oligosaccharide fractions of human colostrum and breast milk. J Infect Dis. 1991;163:1247–55. doi: 10.1093/infdis/163.6.1247. [DOI] [PubMed] [Google Scholar]
- 165.Falk P, Roth KA, Boren T, Westblom TU, Gordon JI, Normark S. An in vitro adherence assay reveals that Helicobacter pylori exhibits cell lineage-specific tropism in the human gastric epithelium. PNAS. 1993;90:2035–39. doi: 10.1073/pnas.90.5.2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Ilver D, Arnqvist A, Ogren J, Frick IM, Kersulyte D, et al. Helicobacter pylori adhesin binding fucosylated histo-blood group antigens revealed by retagging. Science. 1998;279:373–77. doi: 10.1126/science.279.5349.373. [DOI] [PubMed] [Google Scholar]
- 167.Wold AE, Mestecky J, Tomana M, Kobata A, Ohbayashi H, et al. Secretory immunoglobulin A carries oligosaccharide receptors for Escherichia coli type 1 fimbrial lectin. Infect Immun. 1990;58:3073–77. doi: 10.1128/iai.58.9.3073-3077.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Connell I, Agace W, Klemm P, Schembri M, Marild S, Svanborg C. Type 1 fimbrial expression enhances Escherichia coli virulence for the urinary tract. PNAS. 1996;93:9827–32. doi: 10.1073/pnas.93.18.9827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Aniansson G, Andersson B, Lindstedt R, Svanborg C. Anti-adhesive activity of human casein against Streptococcus pneumoniae and Haemophilus influenzae. Microb Pathog. 1990;8:315–23. doi: 10.1016/0882-4010(90)90090-d. [DOI] [PubMed] [Google Scholar]
- 170.Stromqvist M, Falk P, Bergstrom S, Hansson L, Lonnerdal B, et al. Human milk kappa-casein and inhibition of Helicobacter pylori adhesion to human gastric mucosa. J Pediatr Gastroenterol Nutr. 1995;21:288–96. doi: 10.1097/00005176-199510000-00006. [DOI] [PubMed] [Google Scholar]
- 171.Rueda R. The role of dietary gangliosides on immunity and the prevention of infection. Br J Nutr. 2007;98(Suppl 1):S68–73. doi: 10.1017/S0007114507832946. [DOI] [PubMed] [Google Scholar]
- 172.Idota T, Kawakami H. Inhibitory effects of milk gangliosides on the adhesion of Escherichia coli to human intestinal carcinoma cells. Biosci Biotechnol Biochem. 1995;59:69–72. doi: 10.1271/bbb.59.69. [DOI] [PubMed] [Google Scholar]
- 173.Otnaess AB, Laegreid A, Ertresvag K. Inhibition of enterotoxin from Escherichia coli and Vibrio cholerae by gangliosides from human milk. Infect Immun. 1983;40:563–69. doi: 10.1128/iai.40.2.563-569.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Laegreid A, Otnaess AB, Fuglesang J. Human and bovine milk: comparison of ganglioside composition and enterotoxin-inhibitory activity. Pediatr Res. 1986;20:416–21. doi: 10.1203/00006450-198605000-00008. [DOI] [PubMed] [Google Scholar]
- 175.Newburg DS, Ashkenazi S, Cleary TG. Human milk contains the Shiga toxin and Shiga-like toxin receptor glycolipid Gb3. J Infect Dis. 1992;166:832–36. doi: 10.1093/infdis/166.4.832. [DOI] [PubMed] [Google Scholar]
- 176.Pacheco AR, Sperandio V. Shiga toxin in enterohemorrhagic E. coli: regulation and novel anti-virulence strategies. Front Cell Infect Microbiol. 2012;2:81. doi: 10.3389/fcimb.2012.00081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Saeland E, de Jong MA, Nabatov AA, Kalay H, Geijtenbeek TB, van Kooyk Y. MUC1 in human milk blocks transmission of human immunodeficiency virus from dendritic cells to T cells. Mol Immunol. 2009;46:2309–16. doi: 10.1016/j.molimm.2009.03.025. [DOI] [PubMed] [Google Scholar]
- 178.Wu L, KewalRamani VN. Dendritic-cell interactions with HIV: infection and viral dissemination. Nat Rev Immunol. 2006;6:859–68. doi: 10.1038/nri1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Geijtenbeek TB, Kwon DS, Torensma R, van Vliet SJ, van Duijnhoven GC, et al. DC-SIGN, a dendritic cell-specific HIV-1-binding protein that enhances transinfection of T cells. Cell. 2000;100:587–97. doi: 10.1016/s0092-8674(00)80694-7. [DOI] [PubMed] [Google Scholar]
- 180.Hong PW, Flummerfelt KB, de Parseval A, Gurney K, Elder JH, Lee B. Human immunodeficiency virus envelope (gp120) binding to DC-SIGN and primary dendritic cells is carbohydrate dependent but does not involve 2G12 or cyanovirin binding sites: implications for structural analyses of gp120-DC-SIGN binding. J Virol. 2002;76:12855–65. doi: 10.1128/JVI.76.24.12855-12865.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Hong PW, Nguyen S, Young S, Su SV, Lee B. Identification of the optimal DC-SIGN binding site on human immunodeficiency virus type 1 gp120. J Virol. 2007;81:8325–36. doi: 10.1128/JVI.01765-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Requena M, Bouhlal H, Nasreddine N, Saidi H, Gody JC, et al. Inhibition of HIV-1 transmission in trans from dendritic cells to CD4+ T lymphocytes by natural antibodies to the CRD domain of DC-SIGN purified from breast milk and intravenous immunoglobulins. Immunology. 2008;123:508–18. doi: 10.1111/j.1365-2567.2007.02717.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Yolken RH, Peterson JA, Vonderfecht SL, Fouts ET, Midthun K, Newburg DS. Human milk mucin inhibits rotavirus replication and prevents experimental gastroenteritis. J Clin Investig. 1992;90:1984–91. doi: 10.1172/JCI116078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Schroten H, Hanisch FG, Plogmann R, Hacker J, Uhlenbruck G, et al. Inhibition of adhesion of S-fimbriated Escherichia coli to buccal epithelial cells by human milk fat globule membrane components: a novel aspect of the protective function of mucins in the nonimmunoglobulin fraction. Infect Immun. 1992;60:2893–99. doi: 10.1128/iai.60.7.2893-2899.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Liu B, Yu Z, Chen C, Kling DE, Newburg DS. Human milk mucin 1 and mucin 4 inhibit Salmonella enterica serovar Typhimurium invasion of human intestinal epithelial cells in vitro. J Nutr. 2012;142:1504–9. doi: 10.3945/jn.111.155614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Dallas DC, Guerrero A, Khaldi N, Castillo PA, Martin WF, et al. Extensive in vivo human milk peptidomics reveals specific proteolysis yielding protective antimicrobial peptides. J Proteome Res. 2013;12:2295–304. doi: 10.1021/pr400212z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Hakansson AP, Roche-Hakansson H, Mossberg AK, Svanborg C. Apoptosis-like death in bacteria induced by HAMLET, a human milk lipid-protein complex. PLOS ONE. 2011;6:e17717. doi: 10.1371/journal.pone.0017717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Garrido D, Dallas DC, Mills DA. Consumption of human milk glycoconjugates by infant-associated bifidobacteria: mechanisms and implications. Microbiology. 2013;159:649–64. doi: 10.1099/mic.0.064113-0. [DOI] [PMC free article] [PubMed] [Google Scholar]