Abstract
The Octamer-binding proteins (Oct) are a group of highly conserved transcription factors that specifically bind to the octamer motif (ATGCAAAT) and closely related sequences that are found in promoters and enhancers of a wide variety of both ubiquitously expressed and cell type-specific genes. Oct factors belong to the larger family of POU domain factors that are characterized by the presence of a highly conserved bipartite DNA binding domain, consisting of an amino-terminal specific subdomain (POUS) and a carboxyl-terminal homeo-subdomain (POUH). Eleven Oct proteins have been named (Oct1-11), and currently, eight genes encoding Oct proteins (Oct1, Oct2, Oct3/4, Oct6, Oct7, Oct8, Oct9, and Oct11) have been cloned and characterized. Oct1 and Oct2 are widely expressed in adult tissues, while other Oct proteins are much more restricted in their expression patterns. Oct proteins are implicated in crucial and versatile biological events, such as embryogenesis, neurogenesis, immunity, and body glucose and amino acid metabolism. The aberrant expression and null function of Oct proteins have also been linked to various diseases, including deafness, diabetes and cancer. In this review, I will report both the genomic structure and major functions of individual Oct proteins in physiological and pathological processes.
Keywords: Genomic organization, Oct1, Oct2, Oct4, Review, Transcription factors
2. INTRODUCTION
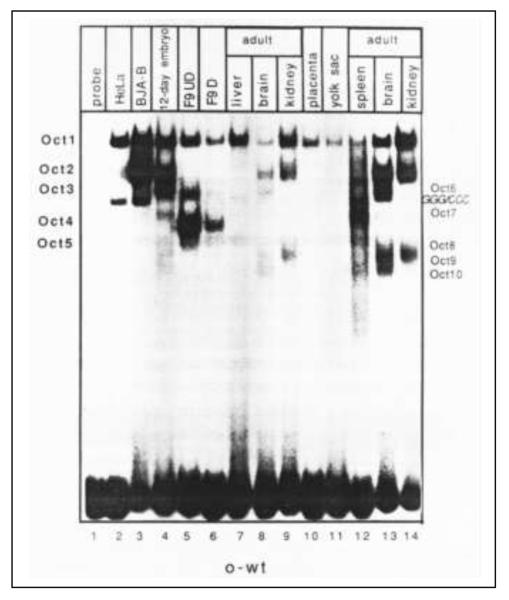
Transcriptional regulation of gene expression is dependent on the interactions of transacting proteins with cis-acting sequence-specific DNA elements located in gene promoter or enhancer regions (1). The octamer motif, ATTTGCAT, and its closely related sequences are a group of cis-acting transcriptional regulatory elements found in the promoters and enhancers of a wide variety of both ubiquitously expressed and cell type-specific genes (2, 3). Nuclear extracts prepared from various adult mouse organs and different developmental stages of mouse embryos form at least 11 distinct binding complexes when incubated with the radiolabeled octamer motif sequence in electrophoretic mobility shift assays (EMSA) (figure 1) (3-5). These binding proteins were named octamer-binding transcription factors, or Oct, and were assigned Oct1 to Oct11 designations. To date, eight genes that encode these Oct proteins, Oct1 (6), Oct2 (7), Oct3/4 (8), Oct6 (9), Oct7 (10, 11), Oct8 (12), Oct9 (13), and Oct11 (5), have been cloned and characterized (table 1 and figure 2A for human sequences). At least some of the remaining band-shift activities may represent degraded products or the splice variants of the above genes. Except for the apparent ubiquitous expression patterns of Oct1 (2, 6) and possibly Oct2 (14), all other members of the Oct proteins exhibit a developmental and tissue-specific expression pattern (15, 16).
Figure 1.
Electrophoretic mobility shift assay of octamer-binding transcription factors (Octs) in adult mouse tissues, embryos and cell lines. HeLa: human cervical carcinoma cell line; BJA-B: human lymphoblastoid cell line; undifferentiated F9 cells (F9 UD): embryonic carcinoma cell line; differentiated F9 cells (F9 D). Adapted from (3).
Table 1.
Genomic location and organization of human Octamer-binding Transcription Factors (Octs)
| Protein | Gene symbol |
Chromosome location1 |
Genomic organization1 | Putative isoforms: aa, (kDa)2 |
|---|---|---|---|---|
| Oct1 | POU2F1 | 1q24.2 |

|
Oct1A: 766 (79) Oct1B: 743 (76) Oct1F 703 (72) Oct1L: 755 (78) |
| Oct2 | POU2F2 | 19q13.2 |

|
Oct2.1 463 (49) Oct2.2 479 (51) Oct2.7 467 (50) |
| Oct3 | 8Q24.21 | Intronless mRNA: NM_001159542; protein: NP_001153014 |
Oct3 359 (39) |
|
| Oct4 | POU5F1 | 6P21.31 |
|
Oct4A 360 (39) Oct4B 265 (30) 190 (21) 164 (18) |
| Oct6 | POU3F1 | 1p34.1 | Intronless mRNA: NM_002699; protein: NP_002690 |
Oct6 451 (45) |
| Oct7 (Brn2) |
POU3F2 | 6q16 | Intronless mRNA: NM_005604; protein: NP_005595 |
Oct7 443 (47) 263 (29) 244 (27) |
| Oct8 (Brn1) |
POU3F3 | 2q12.1 | Intronless mRNA: NM_006236; protein: NP_006227 |
Oct8 500 (50) |
| Oct9 (Brn4) |
POU3F4 | Xq21.1 | Intronless mRNA: NM_000307; protein: NP_000298 |
Oct9 361 (39) |
| Oct11 (Skn- 1a) |
POU2F3 | 11q23.3 |
|
Oct11 436 (47) |
Adapted from the NCBI Gene website (http://www.ncbi.nlm.nih.gov/pubmed?Db=gene&Cmd=retrieve&dopt=full_report&list_uids=“gene id”), version: 12-Jan-2012. In the genomic organization column, each vertical bar represents an exon, and arrows indicate the orientation of the gene. The GenBank Accession Number of each reference sequence is labeled.
aa = amino acids, kDa = deduced molecular mass in kilodaltons.
Figure 2.
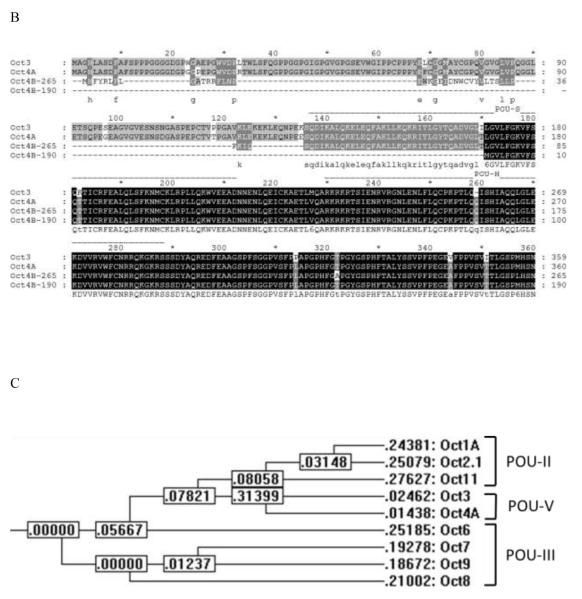
A, B. Alignments of all human Octamer-binding factors (Octs) (A) and Oct3 and Oct4 isoforms (B). The GenBank protein identification numbers of these sequences are NP_002688 (Oct1A), NP_002689.1 (Oct2.1), NP_001153014.1 (Oct3), NP_002692.2 (Oct4A), NP_002690.3 (Oct6), NP_005595 (Oct7), NP_006227.1 (Oct8), NP_000298.2 (Oct9), NP_055167.2 (Oct11), CAA77952 (Oct4B-265) and NP_001167002.1 (Oct4B-190). The alignment was performed with the CLUSTAL-W program with an open gap cost = 10 and a gap extension cost = 0.2. Residues that are highlighted with black shading represent conserved amino acids, and the gray shading indicates 5 or more (A) or 2 or more (B) conserved residues at that position. Positions of the POU-specific domain (POU-S) and the POU homeodomain (POU-H) are given by dashed lines at the top of the sequence alignment. In addition, the conserved amino acids are shown on the bottom of the sequence alignment. C. Phylogenetic tree of Oct factors drawn from the multiple sequence alignment using CLUSTAL-W. The numbers represent tree weights.
Sequence analysis of Oct factors reveals that all of these proteins are members of the POU factor family (17), a family of proteins (>150 family members) characterized by the presence of a POU-specific domain (POUS) and a POU-homeodomain (POUH) (figure 3). Both of these two POU domains are required for high-affinity, site-specific binding to the octamer motif and are involved in protein-protein interactions with other transcription factors or cofactors. Different Oct factors appear to differ in their transcription-activating domains and in their capacity to transactivate target genes. These proteins originate from different highly conserved Oct genes; however, multiple differentially spliced isoforms of the same Oct gene also exist (table 1). This review will report the genomic structure and expression of individual Oct genes and will summarize their known major functions in physiological and pathophysiological processes.
Figure 3.
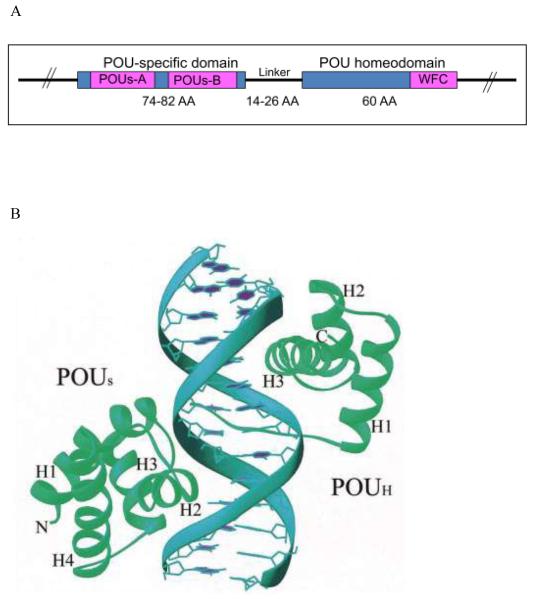
A. Common structural domains of POU transcription factors, including Octamer-binding factors (Octs). AA, amino acids; POUs, POU-specific domain; WFC, WFC motif. B. Crystal structure of DNA binding of the POU-specific domain and the POU-homeodomain (POUH). The helices (H) of each POU domain and the N- and C-termini are labeled. B is adapted from (37).
3. STRUCTURAL CHARACTERISTICS OF OCT PROTEINS
All Oct proteins, like their POU domain-containing family members, share a highly conserved bipartite DNA binding domain, consisting of an approximately 75 (74-82) amino acid amino-terminal POUS subdomain and a 60 amino acid carboxy-terminal POUH subdomain. These domains are tethered by a linker of variable length (14-26 amino acids) (figure 3A).
The POUH is distantly related to the classic homeodomain, which is encoded by homeobox. The homeobox genes encode a large family of transcription factors (>2000), which act as master regulators of anatomical development in animals, fungi and plants (18). The homeodomain forms a triple α-helix that binds to DNA through a helix-turn-helix (HTH) structure (19). The 17 amino acid helix III, referred to as the WFC region (figure 3A) (20), is highly conserved and may function to recognize DNA. Evidence suggests that the homeodomain proteins bind DNA as multimers (19). Although the POUH is related to the classical homeodomain proteins, POUH alone cannot efficiently bind to DNA (21, 22). The mechanism of POUH binding is distinct from that of the classical homeodomain proteins. Efficient and sequence-specific DNA binding requires POUH to cooperate with POUS (21, 22); this cooperation will be discussed in Section 4.
The POUS is only present in POU factors. POUS is more conserved than POUH, with two highly conserved regions: POUS-A and POUS-B (figures 2A and 3A). The solution structure of POUS, determined by nuclear magnetic resonance (NMR), reveals that it forms two long and two short α-helices (23). The POUS binds to the left half of the octamer motif through HTH (figure 3B), and this binding may be mediated by helix III (23, 24).
In addition to DNA binding, the POU domains also mediate specific protein-protein interactions between Oct factors or between Oct factors and other transcription factors or cofactors. These interactions result in cooperation or preclusion of DNA-binding and recruitment of critical factors to activate or inhibit gene expression. For example, Oct1 and Pit1 form heterodimers via their POU domains on the prolactin promoter; this interaction induces prolactin expression (25). The cyclin-dependent kinase-activating kinase (CAK) assembly factor, MAT1, can interact with the POU domain of Oct1 and Oct2 to enhance CAK activity (26). Transcriptional synergism between glucocorticoid receptor (GR) and Oct1 and Oct2 in mouse mammary tumor virus (MMTV) expression is mediated by direct binding between the GR DNA-binding domain and the POU domain of the Oct protein (27). Finally, the herpesvirus trans-activator VP16 specifically interacts with the human Oct1 homeodomain, and a single amino acid exchange transfers this interaction from Oct1 to the Oct2 homeodomain (28).
There are no structural characteristics common to all Oct factors outside of the POU domain. These regions bear the transactivation and inhibitory domains of the proteins, and their activities are isoform-specific. The Oct1 protein contains two 5′ glutamine-rich domains, a serine-threonine-rich domain and a hydrophobic C-terminus region (6, 29). Truncated Oct1 variants, missing either a C-terminal or an N-terminal domain, exhibit different capacities to cause developmental defects in Xenopus embryos (30). Paradoxically, the Oct1 C-terminus represses basal level promoter activity but enhances the hormonal induction of the β-casein gene (31). Two transcriptional activation domains have been identified in Oct2: one in the N-terminal glutamine-rich region and another in the C-terminal serine-, threonine-, and proline-rich region (32). An inhibition domain was also identified in the N-terminus of Oct2 (33). The C-terminal domain of Oct2 is required for its in vivo functions (34). Similarly, transactivation domains have also been identified in both the N- and C-termini of Oct4 (35, 36).
The phylogenetic analyses of all known Oct proteins are consistent with the classifications of the POU domain factors (figure 2C). It appears that only the proteins in POU-II, III and V subclasses exhibit high-affinity binding to the octamer sequences.
4. BINDING SPECIFICITY OF OCT PROTEINS
Both POUH and POUS can individually bind to DNA with different sequence specificity. The binding consensuses of the POUH and POUS subdomains are RTAATNA (R = purine) and gAATAT(G/T)CA, respectively (21). These two consensus sequences overlap the right and left halves of the POU domain recognition sequence [a(a/t)TATGC(A/T)AAT(t/a)t], respectively; the core sequence is the octamer motif. However, individual POU subdomains bind to DNA with low affinity and less specificity (21). For the intact POU domain, the two subdomains are folded independently and bind to the opposite faces of the DNA in two adjacent major grooves through HTH (figure 3B) (37). The POUS subdomain enhances the POU domain DNA-binding affinity up to 1000-fold and also enhances the sequence recognition specificity (21).
The length (minimum of 10 to 14 amino acids) and sequence of the hypervariable POU linker also influences the DNA-binding specificity (38, 39). In addition, the bases flanking the core recognition sequence make a modest but significant contribution to the DNA-binding affinity of the POU domain (21). These influences should contribute to the selective binding of a specific Oct factor to a particular promoter.
Interestingly, not all natural Oct-binding elements in gene promoters are optimized for high-affinity binding. The Oct1-binding site of the mouse β-casein gene proximal promoter varies by one base pair from the complement of the classic octamer motif. Mutation of this base to yield the exact octamer motif sequence increases the binding affinity for Oct1 in an in vitro assay, but significantly decreases the basal activity level and hormonal induction of the promoter (31). In addition, the orientation of the Oct-binding site within gene promoters is also critical. Reversing the orientation of the Oct1-binding site of the mouse β-casein gene proximal promoter to the orientation of the classic octamer motif also dramatically reduces the basal activity level and hormonal induction of the promoter (31), possibly due to the disruption of Oct1 interactions with other factors on the promoter.
While the octamer motif can be bound by all Oct factors, individual Oct proteins prefer some sequences more than others. A recent chromatin immunoprecipitation-sequencing (ChIP-seq) study in glioblastoma cancer cells identified a consensus Oct4-binding sequence, TTTkswTw (k=T or G, s=C or G, w=A or T), which is AT-rich like the classic octamer motif (preferably bound by Oct 1 and 2) but is distinguishable from the classical sequence (40).
The DNA-binding activity of Oct can be modulated by phosphorylation, glycosylation, oxidation, ubiquitinylation, and sumoylation of the POU domain and other regions. Mitosis-specific phosphorylation of a serine residue in the POUH subdomain of Oct1 by protein kinase A is associated with inhibition of Oct1 DNA binding in vitro and in vivo (41, 42). Phosphorylation of Oct1 and Oct2 by protein kinase A, protein kinase C and casein kinase 2 in vitro regulates their binding specificity (43). Alternative phosphorylation and glycosylation of several residues in the POU domain, the linker region between the two POU subdomains and the N-terminus of Oct2 are involved in the differential binding behavior of Oct2 to the octamer motif (44). Cysteine oxidation of Oct7 reduces its binding to octamer sequences (45). Sumoylation of the lysine residue (Lys 118) next to the Oct4 POU domain enhances Oct4 stability and DNA binding (46).
5. OCT MEMBERS AND THEIR TISSUE EXPRESSION, GENOMIC ORGANIZATION AND GENERAL FUNCTIONS
5.1. Oct1
[Gene symbol: POU2F1; GenBank Gene ID = 5451 (human) and 18986 (mouse)]
Oct1 was one of the first identified members of the POU factor family (6) and is the most studied member of the Oct transcription factors. It is ubiquitously expressed in many tissues and cells and either positively or negatively regulates the expression of a variety of genes. These genes include the RNA polymerase II-transcribed and ubiquitously expressed histone H2B gene (47, 48), tissue-specific immunoglobulin genes (49, 50), β-casein gene (31, 51), and RNA polymerase II- or III-transcribed snRNA genes (29). Oct1 is also involved in basal transcription from virus promoters (52, 53). Oct1 has been shown to interact with a variety of tissue-specific co-activating factors and viral proteins. The herpesvirus V16 protein and Bob are two such proteins that specifically bind to the POU domain of Oct1 (54-56). Therefore, the ubiquitously expressed Oct1 is able to contribute to tissue-specific expression by requiring tissue-specific co-activators.
A multitude of Oct1 isoforms has been identified in human and mouse tissues and cells; at least four have been found in humans (table 1) and at least seven in mice (2). These multiple isoforms are derived from a single Oct1 gene, located on chromosome 1 in both humans and mice. Oct1 gene, which contains 18 exons, spans over 207 kb in humans and 128 kb in mice (2, 57). The Oct1 human transcript variant 1 (GenBank accession number: NM_002697) encodes the longest isoform, Oct1A, which contains 766 amino acids (aa) and has a predicted molecular weight of 79 kilodaltons (kDa) (table 1) (57). The variant 2 transcript (NM_001198786) lacks an in-frame coding exon and encodes 703 aa Oct1F (NP_001185715), which lacks an internal segment of Oct1A on the N-terminal side of the POU domain. The variant 3 transcript (NM_001198783) has an alternate promoter and first exon, resulting in the 755 aa Oct1L (NP_001185712) with a shorter and distinct N-terminus. However, the major Oct1 isoform is probably Oct1B (P14859), which contains 743 aa (6) and has a mass of 76 kDa. Oct1B lacks the first 23 aa of Oct1A and results from a transcript (not shown in table 1) that does not contain the upstream AUG codon of the Oct1A transcript. Human Oct1B is the most popular human Oct1 sequence deposited in GenBank and also corresponds to the major Oct1 isoform detected in mouse tissues (2). In addition, a human Oct1 variant transcript (NR_017361) has an additional exon in the 5′ region (table 1), resulting in an internal stop codon and a nonfunctional protein. In mouse tissue, the majority of known Oct1 isoforms have different C-termini (2); the counterparts of most of these isoforms likely exist in humans.
All known Oct1 isoforms retain intact POU domains and the linker sequence but differ in their N- or C- terminal segments. Because the N- and C-terminal sequences contain activation and inhibition domains, different isoforms may very likely have different physiological functions. The occurrence of multiple Oct1 isoforms raises the possibility that these isoforms are expressed in a tissue-specific manner and play different roles in different tissues. This hypothesis is supported by the finding that two of the mouse isoforms, Oct-1L and Oct-1R, are only expressed in lymphocytes (58).
5.2. Oct2
[Gene symbol: POU2F2; Gene ID = 5452 (human) and 18987 (mouse)]
Oct2 was originally cloned from B cells (7). It is encoded by a single gene spanning approximately 43 kb and containing 15 exons on human chromosome 19 (table 1) and mouse chromosome 7. Like Oct1, multiple alternatively spliced isoforms have been identified in both humans (table 1) (59) and mice (14, 60). In human tissue, the variant 1 transcript (NM_001207025) encodes Oct2.2 (NP_001193954), a 479 aa protein with a mass of 51 kDa. Variant 2 (NM_002698) uses an alternate in-frame splice junction at the 5′ end of exon 8 and encodes the major isoform Oct2.1 (NP_002689), a 463 aa protein with a mass of 49 kDa. Variant 3 (NM_001207026) uses an alternate in-frame splice junction at the 3′ end of exon 14 and encodes isoform Oct2.7, which has a shorter C-terminus. The counterparts of these isoforms have also been observed in mouse tissues, in addition to at least 4 other Oct 2 isoforms (14).
Oct2 is generally considered to be expressed only in B lymphocytes and neuronal cells (61). However, Oct2 is widely expressed in many tissues at the transcription level (14). B lymphocytes and the mammary gland predominantly express Oct2.1, which includes a C-terminal activation domain that overcomes the effect of an N-terminal inhibitory domain and stimulates transcription of its target genes (14, 33, 61, 62). In contrast, neuronal cells predominantly express Oct2.4 and Oct2.5, which lack the C-terminal activation domain but contain an intact N-terminal inhibitory domain, resulting in a generally repressive effect on transcription (61, 62). Originally, Oct2 was thought to play a critical role in determining the B cell-specific expression of immunoglobulin (Ig) genes, but it was later shown not to be essential for Ig gene expression in Oct2-deficient mice (63, 64) (see Section 6.4).
5.3. Oct3/4
[Gene symbol: POU5F1; Gene ID = 5460 (human) and 18999 (mouse)]
Oct4 (also known as Oct3) is only expressed in totipotent mouse and human embryonic stem (ES) and germ cells. In these cells, it plays a pivotal role in the regulation and maintenance of pluripotency (it is a widely used marker for pluripotency) and self-renewal of ES cells (65). Expression of Oct4 is down-regulated when embryonic stem cells are triggered to differentiate, and expression is lost in normal somatic cells of differentiated tissues (66).
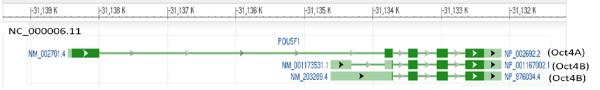
The POU5F1 gene refers to a less than 17 kb sequence located on chromosome 6 (6P21.31), which consists of 6 exons and generates multiple transcripts by utilizing different transcription initiation sites or different splicing mechanisms (table 1 only shows the last 5 exons). The main transcripts (NM_002701 and NM_203289; table 1) were originally cloned from human pancreatic islet cells and encode Oct4A and Oct4B, which consist of 360 and 265 aa (figure 2B), respectively (originally named Oct3A and Oct3B) (67). These two isoforms differ in their cell-specific expression (68), DNA binding, transactivation and capacity to confer self-renewal (69). Recently, our laboratory has cloned multiple transcripts of this gene (DQ486514-DQ486517) from MCF10 cells, a normal human mammary epithelial cell line. These transcripts lack an upstream AUG codon (AGG), which is found in the Celera human genome assembly but not in the GRCh37 primary assembly in GenBank. This polymorphism (reference SNP 3130932) may have functional significance and is thus worth further investigation. In vitro transcription and translation of these cDNAs resulted in no protein products (Zhao et al., manuscript in preparation). However, these transcripts may use a downstream non-AUG (CUG) start codon to produce a 190 aa product (Oct4B-190) (70). Additionally, they may also be able to use a downstream AUG start codon to produce a 164 aa protein (Oct4B-164). Nevertheless, even though these short products are produced in cells, whether they retain Oct4-like biological functions is unclear because these proteins lack half or a majority of the POUS subdomain. Therefore, they may not efficiently bind to octamer sequences of target genes (figure 2B).
Interestingly, from cells of the human breast cancer cell line MCF7, our lab has cloned different sets of transcripts from an intronless Oct4-like gene on human chromosome 8 [DQ486513; Oct3C in (67)] that was thought to be a pseudogene. However, the full cDNA can be transcribed and translated into a protein product in vitro, and its deduced amino acid sequence has 96% sequence identify to the Oct4A described above (figure 2B) (Zhao et al., manuscript in preparation). Based on their differences in deduced protein products and their different chromosome locations and genomic structures, I propose to describe these two sequences as two individual genes by designating the gene on chromosome 6 Oct4 and the gene on chromosome 8 Oct3. Describing these two sequences as two individual genes is further supported by emerging evidence that some cancer cells gain expression of Oct3 but not Oct4 and that Oct3 plays a role in carcinogenesis [Zhao et al., manuscript in preparation; (71)].
In addition, multiple Oct4 pseudogene sequences have been identified on chromosomes 1, 3, 10, and 12 (72-74). Expression of these pseudogenes has been observed in different tumor cells (72, 74, 75); thus, they may have caused some misunderstanding of the roles of Oct4 in cancer.
5.4. Oct6
[Gene symbol: POU3F1; Gene ID = 5453 (human) and 18991 (mouse)]
Oct6 (also known as SCIP and Tst-1), a POU-III sub-class member, is an intronless gene located on chromosome 1 (1p34.1). It encodes a 451 aa protein with a mass of 45 kDa (table 1). The protein contains a glycine/alanine-rich N-terminal region, a short proline/histidine-rich C-terminal region and a histidine-rich sequence that is homologous to a region of kininogen, a precursor for kinin. Oct6 expression is confined to embryonic stem cells, the developing brain, Schwann cells, oligodendrocyte precursors, skin and testes (76-78). Its function has been mainly studied in Schwann cells, where it is required for proper myelination of peripheral nerves (76, 79-81). In addition, Oct6 is also plays roles in the regulation of epidermal keratinocyte differentiation during normal development and wound healing (82) and in the survival and self-renewal of mouse spermatogonial stem cells induced by glial cell line-derived neurotrophic factor (GDNF) (83).
5.5. Oct7
[Gene symbol: POU3F2; Gene ID = 5454 (human) and 18992 (mouse)]
Oct7 [N-Oct3 and Brain-2 (Brn2)], the second member of the POU-III class of transcription factors, is a neural factor involved in neuronal differentiation. The Oct7 gene is also intronless like Oct6 and is located on chromosome 6 (6q16) (10). The predicted full-length open reading frame (ORF) encodes a 443 aa protein with a mass of 47 kDa (table 1); however, two additional ORFs are nested in the same frame and may encode 29 and 27 kDa proteins, respectively (11). Interestingly, the Oct7 cDNA contains an unusually high density of CpG dinucleotides in both the 5′ untranslated region and the coding region; this CpG rich region resembles a CpG island and may have methylation implications (11). In addition, the N-terminus of the protein contains homopolymeric regions of 21 glycines and 21 glutamines (figure 2A). The homopolymeric glutamines are encoded mainly by CAG clusters, which are known to be a hot spot for triplet repeat mutations (11).
Oct7 mRNA is abundantly expressed during all stages of neurogenesis and is expressed at a lower level in distinct neural subsets within the adult central nervous system (CNS), including the paraventricular nuclei (PVN) and the supraoptic nuclei (SON) of the hypothalamus (12). Oct7 plays an essential role in neurogenesis (see Section 6.7). In addition, Oct7 is expressed in most cell derivatives of the neuroectoderm, including neuroblastoma and melanoma cells (84). In melanocytic cells, growth arrest and DNA damage-inducible protein (GADD45) and microphthalmia-associated transcription factor (MITF) are two known targets of Oct7. Expression of GADD45 is activated by Oct7 in response to UVB radiation (85), and Oct7 represses expression of MITF, which controls cell survival, differentiation, proliferation, and migration/metastasis (86). Emerging evidence demonstrates that modulation of MITF levels by Oct7 regulates melanocytic growth and tumorigenesis (see Section 7).
5.6. Oct8
[Gene symbol: POU3F3; Gene ID = 5455 (human) and 18993 (mouse)]
Oct8 (Brn1) is another member of the class III POU family of transcription factors. Oct8 is an intronless gene like other members of this class and is located on chromosome 2 (2q12.1). The predicted ORF of Oct8 encodes a 500 aa protein with a mass of 50 kDa (table 1); this protein contains alanine, glycine and proline repeats (figure 2A). The presence of homopolymeric amino acid repeats is a major structural feature of most mammalian class III POU genes; these repeats are absent in the non-mammalian homologues. These repeats may be generated via GC pressure in mammals (87). Oct8 is essential for the development and functions of nephrons in the kidney. Oct8-deficient mice have a severely retarded Henle’s loop, distal convoluted tubule and macula densa (88). Oct8 is also able to fully replace Oct6 during Schwann cell development and myelination (89, 90).
5.7. Oct9
[Gene symbol: POU3F4; Gene ID = 5456 (human) and 18994 (mouse)]
The Oct9 (Brn4) gene encodes another member of the POU-III class of neural transcriptional factors. It consists of a single exon and is located on the X chromosome (Xq21.1) (13). The ORF encodes a 39 kDa protein with a length of 361 aa (table 1 and figure 2A). In contrast to other members of the POU-III class and like its non-mammalian homologue, the mammalian Oct9 has no amino acid repeats. Oct9 expression is restricted to only a few regions of the adult forebrain, including the SON and PVN of the hypothalamus (91). Differentiation of neural stem cells requires Oct9 expression (92). Additionally, Oct9 plays a prominent role in inner ear development (93). Evidence strongly suggests that nonfunctional Oct9, resulting from various mutations, results in X chromosome-linked nonsyndromic mixed deafness (DFN3) (94-96) (see section 7).
5.8. Oct11
[Gene symbol: POU2F3; Gene ID = 25833 (human) and 18988 (mouse)]
Oct11 (Skn-1a, Pla-1, and Epoc-1) is a POU domain factor primarily expressed in the epidermis that plays a major role in keratinocyte proliferation and differentiation (82). The Oct11 gene is located on chromosome 11 (11q23.2) and contains 13 exons, which span over 80 kb (table 1). Oct11 generates multiple alternatively spliced transcript variants encoding multiple isoforms with various N-termini (97, 98). The major transcript variant (NM_014352) encodes a 436 aa protein with a mass of 47 kDa (NP_055167). This variant contains two functional domains: a primary C-terminal transactivation domain and a combined N-terminal inhibitory domain and transactivation domain (99). Because of the combined effects of the N-termini, it is not surprising that Oct11 isoforms have different functions in keratinocyte proliferation and differentiation (97, 100). Oct11 has also recently been shown to play a critical role in specifying taste receptor cell lineage (101) and is a candidate cervical cancer suppressor protein (see Section 7).
6. FUNCTIONAL ROLES OF OCT IN SPECIFIC PHYSIOLOGICAL PROCESSES
Oct proteins have been known to function as both positive and negative regulators of gene expression. Genes known to be regulated by Oct include a wide variety of ubiquitously expressed genes and tissue-specific genes. Thus, Oct factors are involved in the regulation of many physiological functions. In this section, some of these functions that have been relatively well studied will be discussed.
6.1. Essential functions of Oct proteins in embryogenesis
The POU factors are known to play critical roles in establishing and maintaining cell fate and cell identity throughout embryonic development; they accomplish these functions by establishing correct spatio-temporal expression patterns of target genes. Currently, we know that at least Oct1-, Oct2- and Oct4- gene-deficient (Oct−/−) mice are embryonic lethal, indicating essential roles of these Oct proteins in embryogenesis. Oct1−/− mice die between embryonic day (E) 12.5 and E18.5 (102-104). Oct1-null embryos have reduced extra-embryonic ectoderm formation, lack the ectoplacental cone, and fail to develop beyond the early streak stage (104). Oct1 appears to be primarily required for the maintenance and differentiation of the trophoblast stem cell compartment during early post-implantation. In addition, the Oct1-deficient embryos lack erythroid precursor cells and often appear anemic (102). Oct2−/− mice survive to birth but die within a few hours of birth with no apparent defects (63). Strikingly, although Oct1+/− or Oct2+/− heterozygotes mice exhibit no apparent differences from wild-type animals, Oct1+/−; Oct2+/− transheterozygotes rarely survive to adulthood (102), indicating a gene dosage effect and interdependent roles of Oct1 and Oct2 in mouse development.
Oct4−/− embryos die at the time of implantation (105). Loss of Oct4 changes the fate of the inner cells of the blastocyst of early mouse embryos; these cells cannot form the inner cell mass (ICM) and differentiate into a trophectoderm lineage. Thus, Oct4 is considered to be a gatekeeper in the early steps of mammalian embryogenesis (106).
Oct7 is essential for the differentiation of specific neuronal lineages in mouse hypothalamus development. The migratory precursor cells for neurons of the PVN and SON of the hypothalamus in transgenic mice carrying a loss-of-function mutation in Oct7 die at approximately E12.5 (107). These animals die within 10 days of birth with a complete deficiency of these neurons.
6.2. Pivotal role of Oct4 in the maintenance of cell pluripotency
Oct4 is only expressed in totipotent mouse and human ES and germ cells. The expression level of Oct4 is vital for the maintenance of pluripotency and early cell differentiation. Knockdown of Oct4 in ES cells promotes cell differentiation (108). Expression of Oct4 is down-regulated when embryonic stem cells are triggered to differentiate, and expression is lost in normal somatic cells of differentiated tissues (66). Oct4-deficient mouse embryos lose pluripotency and differentiate into trophectoderm (105). The pivotal role of Oct4 in cell pluripotency was further demonstrated in a recent study by Takahashi and Yamanaka; they found that forced expression of Oct4 and 3 other genes (Sox 2, c-Myc and KLF4) in an adult mouse fibroblast population induced the characteristics of ES cells (109). When these cells were injected into blastocysts, they contributed to mouse embryonic development and were thus shown to be pluripotent. These cells were termed induced pluripotent (iPS) cells. More recently, iPS cells were produced from human fibroblasts following forced expression of the same set of factors used in mice (110) or a combination of Oct4, Sox3, Nanog and lin28 (111). These procedures have now been confirmed and extended in other somatic cells, including stomach and liver cells (112), and in other species, such as monkey (113) and rat (114). Oct4 has been shown to be the most important factor in making iPS cells. iPS cells can be obtained even if Myc is omitted. Sox-1 can replace sox-2, and Klf 2 or Klf 5 can replace Klf 4. However, Oct4 cannot be replaced by other Oct members (e.g., Oct1 and Oct6) (115). In fact, iPS cells have been generated from adult mouse neural stem cells using Oct4 alone (116).
Because Oct4 is essential for totipotency, it has been postulated that the variations in Oct4 expression levels alone account for the majority of current failures related to somatic cell cloning (117).
6.3. Interactions of Oct proteins with basal transcription machinery in the regulation of gene expression
In eukaryotes, transcription is performed by three different RNA polymerases (I, II and III), which synthesize different classes of RNA. Among them, RNA polymerase II (Pol II) transcribes all protein-coding genes and is composed of 12 subunits (Rbp1 to Rbp12). The function of Pol II requires assistance from a large number of proteins, called general or basal transcription factors (GTF); these proteins include TFIIA, TFIIB, TFIID, TFIIE, TFIIF and TFIIH. Together with Pol II, these GTFs form a preinitiation complex (PIC) that binds to promoter regions of DNA upstream of the gene they regulate (118). Among them, TFIID is one of the primary factors and recruits and helps assemble the other GTFs and Pol II to form the PIC on the promoter. The TATA-binding protein (TBP), a component of TFIID, recognizes and binds to the TATA box, an A/T rich sequence present approximately 25-30 nucleotides upstream of the transcription initiation sites of many protein-coding genes. Another GTF that contacts the promoter is TFIIB, which binds to two TFIIB recognition sequences (BRE) present upstream and downstream of the TATA box.
In the promoters of many genes, such as β-casein, MMTV, Ig, and lipoprotein lipase, an octamer-binding site is closely present upstream of the TATA box. Evidence suggests that Oct proteins may activate transcription of these genes by directly interacting with TBP. Both Oct1 and Oct2 can efficiently associate with TBP in vitro and in vivo through POU domains, and these associations do not depend on the presence of other proteins (119). Oct1 can also interacts with TFIIB at the lipoprotein lipase proximal promoter (120) and with MAT-1, a component of TFIIH (26). Through interactions with TFIIB and TFIIH, Oct factors may be able to functionally replace the role of TBP at some promoters that lack a TATA box. Taken together, Oct proteins likely recruit GTFs to form the PIC or to help stabilize the PIC on promoters to stimulate gene transcription.
6.4. Regulation of immune functions by Oct1/2
The octamer sequence was first identified as a conserved sequence in virtually all Ig variable region promoters and in both the heavy- and k light-chain Ig enhancers (121, 122). The sequence is required for B cell-specific expression of Ig genes (123); a point mutation in the octamer region reduces the expression of an Ig transgene to 5% of its normal level (124). Because of its predominant expression in the B cell lineage, Oct2 was originally thought to play a critical role in determining the B cell-specific expression of Ig genes. However, Oct2 is not essential for Ig gene expression because Oct2-deficient mice express Ig and other B cell-specific genes tested at the pre-B-cell stage of development (63, 64). Thus, it was postulated that the role of Oct2 in the regulation of Ig gene expression could be compensated by ubiquitously expressed Oct1. Oct1 and Oct2 have nearly identical DNA binding specificity, and the Ig promoters are equally responsive to both Oct1 and Oct2 (125). Furthermore, Oct1 may activate Ig heavy chain promoters before Oct2 to enhance transcription from Ig light-chain promoters during B cell differentiation (125). Nevertheless, Oct2-null mutants have fewer mature B cells and reduced secretion of antibodies following stimulation (63), indicating that Oct2 is required for later B-cell maturation. Thus, the function of Oct2 in antibody secretion cannot be adequately complemented or completely replaced by Oct1. Additionally, Oct1 is also dispensable for B cell development and Ig transcription (103). The specific roles of these two factors in B cell development and their functions require further investigation.
Binding of Oct1 or Oct2 to the Ig promoters recruits the B cell-specific coactivator Bob-1/OCA-B/OBF-1 (55, 126). Bob-1 enhances Oct1-, and to a lesser extent, Oct2-mediated promoter activity. Interaction with tissue-specific cofactors enables the ubiquitously expressed Oct1 to contribute to tissue-specific gene expression. The partnership between Oct2 and Bob-1 has been shown to play a critical role in sustaining Ig-secretin cell functions (127).
The expression of Ig genes has also been confirmed in many non-B cancer cells and some normal cells (128, 129). This expression is also dependent on the presence of the octamer element in the Ig promoter, as in B cells. However, Oct1 but not Oct2 regulates the Ig promoter activity and induces Ig gene expression in epithelial cancer cells (130), suggesting distinct regulatory mechanisms for Ig gene expression in B cells and non-B cancer cells.
6.5. Oct1, a sensor of metabolic and stress signals
Recently, Oct1 has been found to function as a sensor of both metabolic and stress/survival signals (131-134). In pancreatic islet cells, Oct1 senses intracellular cAMP levels (132). Elevation of cAMP levels enhances Oct1 phosphorylation and shuttles Oct1 from the nucleus to the cytoplasm. Reduced nuclear Oct1 leads to increased expression of Cdx-2, which, in turn, regulates proglucagon and proinsulin expression (131, 132). Consistently, Oct1-deficient cells result in a coordinated metabolic shift with reduced glucose metabolism coupled with increased mitochondrial activity and amino acid oxidation (133). Although these cells and Oct1−/− embryos use less glucose, they are more metabolically active and survive better in the absence of glucose than their wild-type counterparts. Oct1−/− cells have elevated cellular levels of amino acids of glutamate, threonine, isoleucine, proline and glycine coupled with increased amino acid catabolism. Remarkably, glutamate oxidation is robustly increased (5 fold) in Oct1−/− cells (133). In addition, a recent metabolic footprinting (MFP) study suggested that Oct1 may be involved in cell uptake of all essential amino acids except valine and promotes protein synthesis (135). Thus, Oct1 is emerging as an important regulator of body glucose and amino acid metabolism. Because the metabolic changes of Oct1-deficient cells oppose tumorigenicity, Oct1 could be a target for cancer therapy.
Oct1−/− cells are hypersensitive to stress-induced agents, such as ionizing radiation (IR), doxorubicin and hydrogen peroxide (134). Treatment of cancer cells with these agents induces Oct1 levels in these cells (136). When DNA damage occurs via IR, the Oct1 protein is phosphorylated at Ser/Thr sites within its N-terminus by DNA-dependent protein kinase (DNAPK), leading to increased stability (137). Increased Oct1 levels in the cell down-regulate the expression of Histone H2B and U2 RNA and promote cell survival (138). Replacement of Ser/Thr residues with Ala prevents Oct1 from rescuing Oct1−/− embryonic fibroblasts following IR treatment, indicating the importance of Oct1 and its phosphorylation in the stress response. Stress exposure induces associations of Oct1 with a distinct group of targets, and Oct1 is essential for the post-stress transcriptional response (139).
6.6. Regulation of milk protein gene expression by Oct factors
An octamer-binding site is highly conserved in the proximal promoter regions of major milk protein casein genes (51, 140, 141). Mutating this binding site, reversing its orientation or changing it to the consensus octamer motif severely reduces the induction of the promoter activity for the lactogenic hormones insulin, hydrocortisone and prolactin (31). Both Oct1 and Oct2 can activate the hormonal induction of these genes, and this induction is strongly reduced by addition of either Oct1 or Oct2 siRNA in mammary epithelial cells. The activation is mediated by the physical interactions of Oct1 with prolactin downstream signaling molecule STAT5 and glucocorticoid receptor (GR) (X. Qian and F.Q. Zhao, unpublished observations). The lactogenic hormones, progesterone or a combination of these hormones with estradiol induces octamer binding in the mammary gland (51), indicating that Oct factors may also play a role in mammary development in addition to activating mammary-specific milk gene expression induced by lactogenic hormones.
6.7. Oct proteins in neural development
The class III POU Oct factors, including Oct6 through Oct9, play critical roles in the development and function of the nervous system. Oct6 is required for proper myelination of peripheral nerves (76, 79-81). During the development of the peripheral nervous system, axonal and other extracellular signals stimulate Oct6 expression in Schwann cells via the Oct6 Schwann cell enhancer (SCE) (142). Oct6 initiates the transition from ensheathing, promyelinating Schwann cells to myelinating cells. Schwann cell differentiation is transiently arrested at the promyelination stage in Oct6−/− mice (81). This function of Oct6 may overlap with the function of Oct6’s close relative, Oct7, which is also expressed in the Schwann cell lineage and has an expression pattern similar to that of Oct6. Overexpression of Oct7 in Oct6-deficient Schwann cells partially rescues the developmental delay phenotype (143). Oct6/Oct7 cooperates with Sox10 to up-regulate the major myelin-related zinc finger transcription factor Krox20, which is involved in the regulation process (144-146).
Oct7, 8 and 9 are also called brain factors (Brn): Oct7 (Brn2), Oct8 (Brn1) and Oct9 (Brn4). Together with Brn3, these factors are expressed predominantly in the central nervous system and are widely distributed in the neural tube early in development but are subsequently restricted to different regions of the brain. Oct7 plays an essential role in the determination and development of specific neuronal lineages in the hypothalamus (107). Migratory precursor cells for neurons of the PVN and SON of the hypothalamus die at ~E12.5 in Oct7−/− mice. The importance of Oct7 in neurogenesis was further demonstrated in a recent study in which forced expression of Oct7 together with Asc1, Myt1l and NeuroD1 efficiently converted fetal and postnatal fibroblasts into functional neural cells (147). Both Oct7 and Oct8 are involved in brain hippocampus development and complementarily regulates neocortical development (148). Oct8 is also able to fully replace Oct6 during Schwann cell development and myelination (89, 90). However, replacement of Oct6 with Oct8 in mice leads to severe defects in forebrain development (90), indicating that although overlapping functions exist for Oct6, 7 and 8, individual factors may have specific functions. Finally, Oct9 has been implicated in the regulation of striatal neuron precursor differentiation (149, 150).
Neural tissues also express Oct2, predominantly the Oct2.4 and Oct2.5 isoforms. These isoforms generally have an inhibitory effect on gene expression (62). However, the specific roles of these isoforms in the nervous system are not yet known.
6.8. Oct11 and Oct6 in keratinocyte and taste cell proliferation and differentiation
Keratinocytes are the major epithelial cells of the epidermis. The formation of a stratified epidermis requires a balance between keratinocyte proliferation and differentiation, which is tightly regulated by several families of transcription factors, including POU factors. Three POU factors, Oct1, Oct6 and Oct11, are expressed in keratinocytes. Oct11 and Oct6 are predominantly expressed in suprabasal cells, while Oct1 is present in both undifferentiated and differentiated keratinocytes (82). Oct11 is primarily expressed in the epidermis (151) and is also termed skin factor, or Skn-1a. Oct11 can promote epidermal keratinocyte differentiation in vitro (82, 152) and inhibits keratinocyte expression of K14, a keratin marker of undifferentiated epidermal epithelia (153). Although the skins of both Oct11- and Oct6-deficient mice appear normal, epidermis from mice lacking both Oct11 and Oct6 is hyperplastic (82). These results indicate that Oct11 and Oct6 both function to regulate epidermal differentiation and wound healing.
Recently, Oct11 was also shown to play a critical role in generating and balancing taste receptor cell lineage (101). Oct11-deficient mice completely lack sweet, umami and bitter cells, and they fail to expand sour cells. Thus, these animals do not respond to these tastes.
7. PATHOPHYSIOLOGICAL ROLES OF OCT PROTEINS
Oct1 plays important roles in viral DNA replication and gene expression. It is involved in adenovirus DNA replication. In this process, Oct1 recruits the precursor terminal protein/DNA polymerase heterodimer to the adenovirus origin of replication to initiate viral DNA replication (154). The MMTV proximal promoter contains an octamer-binding site and a glucocorticoid response element. Oct1 stimulates basal transcription of this promoter (52), but its activity can be strongly enhanced by glucocorticoid via direct interaction with the glucocorticoid receptor (GR) (27). Binding of Oct1 and nuclear factor 1 (NF1) to the MMTV promoter induces a partial nucleosome positioning that facilitates the GR-DNA interaction (155). In the herpes simplex virus (HSV) immediate-early (IE) enhancer core, Oct1 interacts with the viral transactivator protein VP16 and binds to the 5′ portion of the core element to initiate the enhancer assembly (156). Oct1 is required for HSV IE gene expression at low multiplicities of infection; infection by HSV is arrested in Oct1-null cells (157). Oct1 also interacts with the Epstein-Barr Virus (EBV) IE protein BRLF1 and enhances BRLF1-mediated disruption of EBV latency independent of DNA binding (158). Furthermore, Oct1 represses virus-induced interferon A (IFN-A11) gene expression (159). Thus, Oct1 plays critical roles in viral infection and may be a useful therapeutic target for controlling virus infection.
Several studies have demonstrated that Oct1 levels are increased in various cancer cells (160-163), and overexpression of Oct1 is pro-tumorigenic (164). Oct1 deficiency opposes oncogenic transformation and tumorigenicity (133, 162, 163). Because Oct1 is a sensor of metabolic stress, it is not surprising to observe an association between multiple variants of Oct1 and type 2 diabetes (165).
Down-regulation of Oct2 and its coactivator Bob-1 has been linked to abnormal Ig expression in Hodgkin and Reed-Sternberg (HRS) cells in Hodgkin disease (166, 167) and in primary effusion lymphoma (PEL) (168). Oct2 and Bob-1 are useful markers for the differential diagnosis of classic Hodgkin lymphoma (169). In PEL tumor cells, Oct2 also inhibits the expression of ORF50, the key regulator of the switch from latency to lytic reactivation of human herpesvirus-8 (HHV-8). Oct2 competes with Oct1, which enhances ORF50 transactivation. Thus, down-regulation of Oct2 may promote viral reactivation of HHV-8 (168).
There is mounting evidence that links Oct3/4 to carcinogenesis. Increased expression of Oct4 (to 150% of wild type expression) in embryonic ES cells shifts the potential of these cells to form tumors in syngeneic hosts from 4% to greater than 80%, while inactivation induces regression of the malignant phenotype (170). Oct4 has been defined as a diagnostic marker for human testicular germ cell tumor precursor carcinoma in situ/intratubular germ cell neoplasia undifferentiated, seminoma, and embryonal carcinoma (171). In addition to expression in germ-cell tumors, Oct3/4 has also been detected in several somatic tumors, including breast, bladder, gastric, pancreatic, lung, and rectal cancers (66, 161, 171-179). In breast cancer cells, Oct3/4 expression is repressed by all-trans-retinoic acid and is coupled with decreased cell proliferation (180). Expression of Oct3/4 in these cells up-regulates expression of fibroblast growth factor-4 (FGF-4) (180), a gene that stimulates MCF-7 cells to become more tumorigenic and metastatic in ovariectomized and tamoxifen-treated nude mice (181).
A transgenic study demonstrated that activation of Oct3/4 in somatic tissues of adult mice in vivo inhibits cell differentiation in a manner similar to that observed in embryonic cells, which results in epithelial tissue dysplastic growth in the small intestine and epidermis coupled with an expansion of progenitor cells (182). The animals die shortly after Oct3/4 expression is activated. Furthermore, chimeric mice, in which somatic tissues are composed of a mixture of wild-type and Oct3/4-inducible cells, develop visible skin tumors three weeks following induction of Oct4 expression, and induced tumors disappear completely after the induction of Oct3/4 is withdrawn (182).
However, expression of Oct4 in somatic tumor cells has been challenged by several studies (183-185). These investigators argue that the observed expression of Oct4 in some somatic tumor cells may be due to false-positive results or could have resulted from amplification of Oct4 pseudogenes. In addition, our lab has noted that breast cancer tissue and MCF7 cells express an Oct4-like gene (Oct3), but not Oct4 (see Section 5.3). Thus, it is essential to verify the transcripts of Oct3/4 genes when investigating their expression in tumors.
In melanoma cells, Oct7 expression is 10-fold higher than in normal human melanocytes, and overexpression of Oct7 is associated with increased cell proliferation and invasiveness (84, 186, 187). Oct7 controls melanocyte migration and proliferation via its regulation of MITF, by the overall Oct7 level and through regulation of Pax3 transcription by Oct7 phosphorylation at T361 and S362 residues (188, 189). Oct7 can counter-regulate NOTCH pathway of melanoma cells with MITF to suppress the differentiated melanocytic phenotype and enhance tumor metastasis (189).
Strong evidence has linked mutations of Oct9 to DFN3, the most prevalent and hereditary X chromosome-linked hearing loss, which is characterized by conductive hearing loss and progressive sensorineural deafness (94-96, 190). These mutations include substitutions (e.g., Gly216Glu, Arg329Pro, Arg330Ser, and Arg323Gly), deletions (e.g., Ser310del) or truncations (e.g., Ala116fs). Most of these mutations are within the regions that encode the DNA-binding domains of Oct9, and thus, these mutations can severely affect Oct9 DNA binding and its capacity to transactivate target genes (95, 96) such as Epha4 in optic mesenchyme cells (191). The hearing loss associated with nonfunctional Oct9 in humans is consistent with observations in Oct9-deficient mice, which reveal defective otic fibrocytes and stria vascularis in the cochlear lateral wall (192, 193).
Oct11 was identified as a candidate cervical cancer suppressor (194). The Oct11 gene lies in a critical loss of heterozygosity (LOH) region on chromosome 11 (11q23.3) in cervical cancer cells. Oct11 expression is lost in more than 50% of cervical tumors and cell lines. Silencing of the Oct11 gene by aberrant DNA methylation of its promoter has been observed in 39% of cervical tumors but has not been noted in normal epithelium (195).
8. Conclusions and perspectives
Multiple Oct factors are expressed in mammalian cells and have been shown to play critical functions in various physiological processes, such as embryogenesis, cell differentiation, immune function and neurogenesis. Because of their similar structural and DNA-binding properties, some Oct proteins share functional properties. For example, Oct1 and Oct2 both regulate immunoglobulin gene expression, and Oct6-9 regulate neurogenesis. However, individual Oct factors may have specific roles that cannot be completely replaced by other Oct proteins.
It is important to note that the specific functions of individual Oct proteins are complicated by the presence of multiple isoforms. These isoforms may exhibit tissue-specific expression and may play different and even opposing roles. In addition, pseudogenes or pseudotranscripts exist for some Oct factors. Thus, it is necessary to verify the specific transcripts and isoforms expressed when studying the roles of Oct factors in a particular tissue or disease.
Oct factors have been linked to various diseases; these include Oct1 in viral infections and diabetes, Oct-2 in Hodgkin diseases, Oct-7 in melanoma, Oct9 in DFN3, and Oct1, Oct4 and Oct11 in various cancers. These Oct proteins offer potential therapeutic targets for the diagnosis and treatment of these diseases.
Finally, novel functions of Oct proteins are still being identified. In addition to the discussed Oct factors above, additional POU domain factors may also bind to octamer sequences and could be new Oct factor candidates.
ACKNOWLEDGEMENTS
FQZ’s work in this review was supported by the USDA Experimental Station Hatch Grant (VT-H01508), the Department of Defense Breast Cancer Research Program Concept Award (#W81XWH-05-1-0432), and the Vermont Cancer Center VCC/LCCRO Pilot Award (#022820).
Abbreviations
- BRN
brian factor
- DFN
hereditary X chromosome-linked hearing loss
- EBV
Epstein-Barr Virus
- ES
embryonic stem cell
- HSV
herpes simplex virus
- IE
immediate-early
- Ig
immunoglobulin
- kDa
kilodaltons
- MMTV
mouse mammary tumor virus
- Oct
octamer-binding transcription factor
- PEL
primary effusion lymphoma
- POU
POU domain
- POUH
POU homeodomain
- POUS
POU specific subdomain
- PVN
paraventricular nuclei
- SON
supraoptic nuclei
- TBP
TATA-binding protein
10. REFERENCES
- 1.Kadonaga JT. Regulation of RNA polymerase II transcription by sequence-specific DNA binding factors. Cell. 2004;116:247–57. doi: 10.1016/s0092-8674(03)01078-x. [DOI] [PubMed] [Google Scholar]
- 2.Zhao FQ, Zheng Y, Dong B, Oka T. Cloning, genomic organization, expression, and effect on beta-casein promoter activity of a novel isoform of the mouse Oct-1 transcription factor. Gene. 2004;326:175–87. doi: 10.1016/j.gene.2003.10.023. [DOI] [PubMed] [Google Scholar]
- 3.Scholer HR, Hatzopoulos AK, Balling R, Suzuki N, Gruss P. A family of octamer-specific proteins present during mouse embryogenesis: evidence for germline-specific expression of an Oct factor. EMBO J. 1989;8:2543–50. doi: 10.1002/j.1460-2075.1989.tb08392.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scholer HR. Octamania: the POU factors in murine development. Trends Genet. 1991;7:323–9. doi: 10.1016/0168-9525(91)90422-m. [DOI] [PubMed] [Google Scholar]
- 5.Goldsborough AS, Healy LE, Copeland NG, Gilbert DJ, Jenkins NA, Willison KR, Ashworth A. Cloning, chromosomal localization and expression pattern of the POU domain gene Oct-11. Nucleic Acids Res. 1993;21:127–34. doi: 10.1093/nar/21.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sturm RA, Das G, Herr W. The ubiquitous octamer-binding protein Oct-1 contains a POU domain with a homeo box subdomain. Genes Dev. 1988;2:1582–99. doi: 10.1101/gad.2.12a.1582. [DOI] [PubMed] [Google Scholar]
- 7.Clerc RG, Corcoran LM, LeBowitz JH, Baltimore D, Sharp PA. The B-cell-specific Oct-2 protein contains POU box- and homeo box-type domains. Genes Dev. 1988;2:1570–81. doi: 10.1101/gad.2.12a.1570. [DOI] [PubMed] [Google Scholar]
- 8.Rosner MH, Vigano MA, Ozato K, Timmons PM, Poirier F, Rigby PW, Staudt LM. A POU-domain transcription factor in early stem cells and germ cells of the mammalian embryo. Nature. 1990;345:686–92. doi: 10.1038/345686a0. [DOI] [PubMed] [Google Scholar]
- 9.Meijer D, Graus A, Kraay R, Langeveld A, Mulder MP, Grosveld G. The octamer binding factor Oct6: cDNA cloning and expression in early embryonic cells. Nucleic Acids Res. 1990;18:7357–65. doi: 10.1093/nar/18.24.7357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Atanasoski S, Toldo SS, Malipiero U, Schreiber E, Fries R, Fontana A. Isolation of the human genomic brain-2/N-Oct 3 gene (POUF3) and assignment to chromosome 6q16. Genomics. 1995;26:272–80. doi: 10.1016/0888-7543(95)80211-4. [DOI] [PubMed] [Google Scholar]
- 11.Schreiber E, Tobler A, Malipiero U, Schaffner W, Fontana A. cDNA cloning of human N-Oct3, a nervous-system specific POU domain transcription factor binding to the octamer DNA motif. Nucleic Acids Res. 1993;21:253–8. doi: 10.1093/nar/21.2.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He X, Treacy MN, Simmons DM, Ingraham HA, Swanson LW, Rosenfeld MG. Expression of a large family of POU-domain regulatory genes in mammalian brain development. Nature. 1989;340:35–41. doi: 10.1038/340035a0. [DOI] [PubMed] [Google Scholar]
- 13.Douville PJ, Atanasoski S, Tobler A, Fontana A, Schwab ME. The brain-specific POU-box gene Brn4 is a sex-linked transcription factor located on the human and mouse X chromosomes. Mamm Genome. 1994;5:180–2. doi: 10.1007/BF00352353. [DOI] [PubMed] [Google Scholar]
- 14.Dong B, Zhao FQ. Expression of the Oct-2 transcription factor in mouse mammary gland and cloning and characterization of a novel Oct-2 isoform. Cell Tissue Res. 2007;328:595–606. doi: 10.1007/s00441-006-0368-0. [DOI] [PubMed] [Google Scholar]
- 15.Veenstra GJ, van der Vliet PC, Destree OH. POU domain transcription factors in embryonic development. Mol Biol Rep. 1997;24:139–55. doi: 10.1023/a:1006855632268. [DOI] [PubMed] [Google Scholar]
- 16.Latchman DS. POU family transcription factors in the nervous system. J Cell Physiol. 1999;179:126–33. doi: 10.1002/(SICI)1097-4652(199905)179:2<126::AID-JCP2>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 17.Verrijzer CP, Van der Vliet PC. POU domain transcription factors. Biochim Biophys Acta. 1993;1173:1–21. doi: 10.1016/0167-4781(93)90237-8. [DOI] [PubMed] [Google Scholar]
- 18.Christensen KL, Patrick AN, McCoy EL, Ford HL. The six family of homeobox genes in development and cancer. Adv Cancer Res. 2008;101:93–126. doi: 10.1016/S0065-230X(08)00405-3. [DOI] [PubMed] [Google Scholar]
- 19.Wolberger C. Homeodomain interactions. Curr Opin Struct Biol. 1996;6:62–8. doi: 10.1016/s0959-440x(96)80096-0. [DOI] [PubMed] [Google Scholar]
- 20.Herr W, Sturm RA, Clerc RG, Corcoran LM, Baltimore D, Sharp PA, Ingraham HA, Rosenfeld MG, Finney M, Ruvkun G, et al. The POU domain: a large conserved region in the mammalian pit-1, oct-1, oct-2, and Caenorhabditis elegans unc-86 gene products. Genes Dev. 1988;2:1513–6. doi: 10.1101/gad.2.12a.1513. [DOI] [PubMed] [Google Scholar]
- 21.Verrijzer CP, Alkema MJ, van Weperen WW, Van Leeuwen HC, Strating MJ, van der Vliet PC. The DNA binding specificity of the bipartite POU domain and its subdomains. EMBO J. 1992;11:4993–5003. doi: 10.1002/j.1460-2075.1992.tb05606.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schonemann MD, Ryan AK, Erkman L, McEvilly RJ, Bermingham J, Rosenfeld MG. POU domain factors in neural development. Adv Exp Med Biol. 1998;449:39–53. doi: 10.1007/978-1-4615-4871-3_4. [DOI] [PubMed] [Google Scholar]
- 23.Dekker N, Cox M, Boelens R, Verrijzer CP, van der Vliet PC, Kaptein R. Solution structure of the POU-specific DNA-binding domain of Oct-1. Nature. 1993;362:852–5. doi: 10.1038/362852a0. [DOI] [PubMed] [Google Scholar]
- 24.Botfield MC, Jancso A, Weiss MA. Biochemical characterization of the Oct-2 POU domain with implications for bipartite DNA recognition. Biochemistry. 1992;31:5841–8. doi: 10.1021/bi00140a020. [DOI] [PubMed] [Google Scholar]
- 25.Voss JW, Wilson L, Rosenfeld MG. POU-domain proteins Pit-1 and Oct-1 interact to form a heteromeric complex and can cooperate to induce expression of the prolactin promoter. Genes Dev. 1991;5:1309–20. doi: 10.1101/gad.5.7.1309. [DOI] [PubMed] [Google Scholar]
- 26.Inamoto S, Segil N, Pan ZQ, Kimura M, Roeder RG. The cyclin-dependent kinase-activating kinase (CAK) assembly factor, MAT1, targets and enhances CAK activity on the POU domains of octamer transcription factors. J Biol Chem. 1997;272:29852–8. doi: 10.1074/jbc.272.47.29852. [DOI] [PubMed] [Google Scholar]
- 27.Prefontaine GG, Walther R, Giffin W, Lemieux ME, Pope L, Hache RJ. Selective binding of steroid hormone receptors to octamer transcription factors determines transcriptional synergism at the mouse mammary tumor virus promoter. J Biol Chem. 1999;274:26713–9. doi: 10.1074/jbc.274.38.26713. [DOI] [PubMed] [Google Scholar]
- 28.Lai JS, Cleary MA, Herr W. A single amino acid exchange transfers VP16- induced positive control from the Oct-1 to the Oct-2 homeo domain. Genes Dev. 1992;6:2058–65. doi: 10.1101/gad.6.11.2058. [DOI] [PubMed] [Google Scholar]
- 29.Tanaka M, Lai JS, Herr W. Promoter-selective activation domains in Oct-1 and Oct-2 direct differential activation of an snRNA and mRNA promoter. Cell. 1992;68:755–67. doi: 10.1016/0092-8674(92)90150-b. [DOI] [PubMed] [Google Scholar]
- 30.Veenstra GJ, Peterson-Maduro J, Mathu MT, van der Vliet PC, Destree OH. Non-cell autonomous induction of apoptosis and loss of posterior structures by activation domain-specific interactions of Oct-1 in the Xenopus embryo. Cell Death Differ. 1998;5:774–84. doi: 10.1038/sj.cdd.4400416. [DOI] [PubMed] [Google Scholar]
- 31.Dong B, Zhao FQ. Involvement of the ubiquitous Oct-1 transcription factor in hormonal induction of beta-casein gene expression. Biochem J. 2007;401:57–64. doi: 10.1042/BJ20060570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tanaka M, Herr W. Differential transcriptional activation by Oct-1 and Oct-2: interdependent activation domains induce Oct-2 phosphorylation. Cell. 1990;60:375–86. doi: 10.1016/0092-8674(90)90589-7. [DOI] [PubMed] [Google Scholar]
- 33.Lillycrop KA, Dawson SJ, Estridge JK, Gerster T, Matthias P, Latchman DS. Repression of a herpes simplex virus immediate-early promoter by the Oct-2 transcription factor is dependent on an inhibitory region at the N terminus of the protein. Mol Cell Biol. 1994;14:7633–42. doi: 10.1128/mcb.14.11.7633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Corcoran LM, Koentgen F, Dietrich W, Veale M, Humbert PO. All known in vivo functions of the Oct-2 transcription factor require the C-terminal protein domain. J Immunol. 2004;172:2962–2969. doi: 10.4049/jimmunol.172.5.2962. [DOI] [PubMed] [Google Scholar]
- 35.Brehm A, Ohbo K, Scholer H. The carboxy-terminal transactivation domain of Oct-4 acquires cell specificity through the POU domain. Mol Cell Biol. 1997;17:154–62. doi: 10.1128/mcb.17.1.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Imagawa M, Miyamoto A, Shirakawa M, Hamada H, Muramatsu M. Stringent integrity requirements for both trans-activation and DNA-binding in a trans-activator, Oct3. Nucleic Acids Res. 1991;19:4503–8. doi: 10.1093/nar/19.16.4503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Phillips K, Luisi B. The virtuoso of versatility: POU proteins that flex to fit. J Mol Biol. 2000;302:1023–39. doi: 10.1006/jmbi.2000.4107. [DOI] [PubMed] [Google Scholar]
- 38.Aurora R, Herr W. Segments of the POU domain influence one another’s DNA- binding specificity. Mol Cell Biol. 1992;12:455–67. doi: 10.1128/mcb.12.2.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Leeuwen HC, Strating MJ, Rensen M, de Laat W, van der Vliet PC. Linker length and composition influence the flexibility of Oct-1 DNA binding. EMBO J. 1997;16:2043–53. doi: 10.1093/emboj/16.8.2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fang XF, Zhang WY, Zhao N, Yu W, Ding D, Hong X, Li LS, Zhang HR, Zheng S, Lin BY. Genome-wide analysis of OCT4 binding sites in glioblastoma cancer cells. J Zhejiang Univ Sci B. 2011;12:812–9. doi: 10.1631/jzus.B1100059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Segil N, Roberts SB, Heintz N. Mitotic phosphorylation of the Oct-1 homeodomain and regulation of Oct-1 DNA binding activity. Science. 1991;254:1814–6. doi: 10.1126/science.1684878. [DOI] [PubMed] [Google Scholar]
- 42.Segil N, Roberts SB, Heintz N. Cell-cycle-regulated phosphorylation of the transcription factor Oct-1. Cold Spring Harb Symp Quant Biol. 1991;56:285–92. doi: 10.1101/sqb.1991.056.01.035. [DOI] [PubMed] [Google Scholar]
- 43.Grenfell SJ, Latchman DS, Thomas NS. Oct-1 [corrected] and Oct-2 DNA- binding site specificity is regulated in vitro by different kinases. Biochem J. 1996;315(Pt 3):889–93. doi: 10.1042/bj3150889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ahmad I, Hoessli DC, Walker-Nasir E, Rafik SM, Shakoori AR, Nasirud D. Oct-2 DNA binding transcription factor: functional consequences of phosphorylation and glycosylation. Nucl. Acids Res. 2006;34:175–184. doi: 10.1093/nar/gkj401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smith AG, Brightwell G, Smit SE, Parsons PG, Sturm RA. Redox regulation of Brn-2/N-Oct-3 POU domain DNA binding activity and proteolytic formation of N-Oct-5 during melanoma cell nuclear extraction. Melanoma Res. 1998;8:2–10. doi: 10.1097/00008390-199802000-00002. [DOI] [PubMed] [Google Scholar]
- 46.Wei F, Scholer HR, Atchison ML. Sumoylation of Oct4 enhances its stability, DNA binding, and transactivation. J Biol Chem. 2007;282:21551–60. doi: 10.1074/jbc.M611041200. [DOI] [PubMed] [Google Scholar]
- 47.La Bella F, Gallinari P, McKinney J, Heintz N. Histone H1 subtype-specific consensus elements mediate cell cycle-regulated transcription in vitro. Genes Dev. 1989;3:1982–90. doi: 10.1101/gad.3.12a.1982. [DOI] [PubMed] [Google Scholar]
- 48.LaBella F, Sive HL, Roeder RG, Heintz N. Cell-cycle regulation of a human histone H2b gene is mediated by the H2b subtype-specific consensus element. Genes Dev. 1988;2:32–9. doi: 10.1101/gad.2.1.32. [DOI] [PubMed] [Google Scholar]
- 49.Dreyfus M, Doyen N, Rougeon F. The conserved decanucleotide from the immunoglobulin heavy chain promoter induces a very high transcriptional activity in B-cells when introduced into an heterologous promoter. EMBO J. 1987;6:1685–90. doi: 10.1002/j.1460-2075.1987.tb02418.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wirth T, Staudt L, Baltimore D. An octamer oligonucleotide upstream of a TATA motif is sufficient for lymphoid-specific promoter activity. Nature. 1987;329:174–8. doi: 10.1038/329174a0. [DOI] [PubMed] [Google Scholar]
- 51.Zhao FQ, Adachi K, Oka T. Involvement of Oct-1 in transcriptional regulation of beta-casein gene expression in mouse mammary gland. Biochim Biophys Acta. 2002;1577:27–37. doi: 10.1016/s0167-4781(02)00402-5. [DOI] [PubMed] [Google Scholar]
- 52.Kim MH, Peterson DO. Stimulation of basal transcription from the mouse mammary tumor virus promoter by Oct proteins. J. Virol. 1995;69:4717–4726. doi: 10.1128/jvi.69.8.4717-4726.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu YZ, Latchman DS. The octamer-binding proteins Oct-1 and Oct-2 repress the HIV long terminal repeat promoter and its transactivation by Tat. Biochem J. 1997;322(Pt 1):155–8. doi: 10.1042/bj3220155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stern S, Tanaka M, Herr W. The Oct-1 homoeodomain directs formation of a multiprotein-DNA complex with the HSV transactivator VP16. Nature. 1989;341:624–30. doi: 10.1038/341624a0. [DOI] [PubMed] [Google Scholar]
- 55.Gstaiger M, Knoepfel L, Georgiev O, Schaffner W, Hovens CM. A B-cell coactivator of octamer-binding transcription factors. Nature. 1995;373:360–2. doi: 10.1038/373360a0. [DOI] [PubMed] [Google Scholar]
- 56.Lee L, Stollar E, Chang J, Grossmann JG, O’Brien R, Ladbury J, Carpenter B, Roberts S, Luisi B. Expression of the Oct-1 transcription factor and characterization of its interactions with the Bob1 coactivator. Biochemistry. 2001;40:6580–8. doi: 10.1021/bi010095x. [DOI] [PubMed] [Google Scholar]
- 57.Sturm RA, Cassady JL, Das G, Romo A, Evans GA. Chromosomal structure and expression of the human OTF1 locus encoding the Oct-1 protein. Genomics. 1993;16:333–41. doi: 10.1006/geno.1993.1194. [DOI] [PubMed] [Google Scholar]
- 58.Pankratova EV, Deyev IE, Zhenilo SV, Polanovsky OL. Tissue-specific isoforms of the ubiquitous transcription factor Oct-1. Mol Genet Genomics. 2001;266:239–45. doi: 10.1007/s004380100549. [DOI] [PubMed] [Google Scholar]
- 59.Hatzopoulos AK, Stoykova AS, Erselius JR, Goulding M, Neuman T, Gruss P. Structure and expression of the mouse Oct2a and Oct2b, two differentially spliced products of the same gene. Development. 1990;109:349–62. doi: 10.1242/dev.109.2.349. [DOI] [PubMed] [Google Scholar]
- 60.Wirth T, Priess A, Annweiler A, Zwilling S, Oeler B. Multiple Oct2 isoforms are generated by alternative splicing. Nucleic Acids Res. 1991;19:43–51. doi: 10.1093/nar/19.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Latchman DS. The Oct-2 transcription factor. Int J Biochem Cell Biol. 1996;28:1081–3. doi: 10.1016/1357-2725(96)00050-7. [DOI] [PubMed] [Google Scholar]
- 62.Lillycrop KA, Latchman DS. Alternative splicing of the Oct-2 transcription factor RNA is differentially regulated in neuronal cells and B cells and results in protein isoforms with opposite effects on the activity of octamer/TAATGARAT-containing promoters. J Biol Chem. 1992;267:24960–5. [PubMed] [Google Scholar]
- 63.Corcoran LM, Karvelas M, Nossal GJ, Ye ZS, Jacks T, Baltimore D. Oct-2, although not required for early B-cell development, is critical for later B-cell maturation and for postnatal survival. Genes Dev. 1993;7:570–82. doi: 10.1101/gad.7.4.570. [DOI] [PubMed] [Google Scholar]
- 64.Schubart K, Massa S, Schubart D, Corcoran LM, Rolink AG, Matthias P. B cell development and immunoglobulin gene transcription in the absence of Oct-2 and OBF-1. Nat Immunol. 2001;2:69–74. doi: 10.1038/83190. [DOI] [PubMed] [Google Scholar]
- 65.Pesce M, Scholer HR. Oct-4: control of totipotency and germline determination. Mol Reprod Dev. 2000;55:452–7. doi: 10.1002/(SICI)1098-2795(200004)55:4<452::AID-MRD14>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 66.Tai M, Chang C, Olson L, Trosko J. Oct4 expression in adult human stem cells: evidence in support of the stem cell theory of carcinogenesis. Carcinogenesis. 2005;26:495–502. doi: 10.1093/carcin/bgh321. [DOI] [PubMed] [Google Scholar]
- 67.Takeda J, Seino S, Bell GI. Human Oct3 gene family: cDNA sequences, alternative splicing, gene organization, chromosomal location, and expression at low levels in adult tissues. Nucleic Acids Res. 1992;20:4613–20. doi: 10.1093/nar/20.17.4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Atlasi Y, Mowla SJ, Ziaee SA, Gokhale PJ, Andrews PW. OCT4 spliced variants are differentially expressed in human pluripotent and nonpluripotent cells. Stem Cells. 2008;26:3068–74. doi: 10.1634/stemcells.2008-0530. [DOI] [PubMed] [Google Scholar]
- 69.Lee J, Kim HK, Rho JY, Han YM, Kim J. The human OCT-4 isoforms differ in their ability to confer self-renewal. J Biol Chem. 2006;281:33554–65. doi: 10.1074/jbc.M603937200. [DOI] [PubMed] [Google Scholar]
- 70.Wang X, Zhao Y, Xiao Z, Chen B, Wei Z, Wang B, Zhang J, Han J, Gao Y, Li L, Zhao H, Zhao W, Lin H, Dai J. Alternative translation of OCT4 by an internal ribosome entry site and its novel function in stress response. Stem Cells. 2009;27:1265–75. doi: 10.1002/stem.58. [DOI] [PubMed] [Google Scholar]
- 71.Kastler S, Honold L, Luedeke M, Kuefer R, Moller P, Hoegel J, Vogel W, Maier C, Assum G. POU5F1P1, a putative cancer susceptibility gene, is overexpressed in prostatic carcinoma. Prostate. 2010;70:666–74. doi: 10.1002/pros.21100. [DOI] [PubMed] [Google Scholar]
- 72.Zhao S, Yuan Q, Hao H, Guo Y, Liu S, Zhang Y, Wang J, Liu H, Wang F, Liu K, Ling EA, Hao A. Expression of OCT4 pseudogenes in human tumours: lessons from glioma and breast carcinoma. J Pathol. 2011;223:672–82. doi: 10.1002/path.2827. [DOI] [PubMed] [Google Scholar]
- 73.Pain D, Chirn GW, Strassel C, Kemp DM. Multiple retropseudogenes from pluripotent cell-specific gene expression indicates a potential signature for novel gene identification. J Biol Chem. 2005;280:6265–8. doi: 10.1074/jbc.C400587200. [DOI] [PubMed] [Google Scholar]
- 74.Suo G, Han J, Wang X, Zhang J, Zhao Y, Dai J. Oct4 pseudogenes are transcribed in cancers. Biochem Biophys Res Commun. 2005;337:1047–51. doi: 10.1016/j.bbrc.2005.09.157. [DOI] [PubMed] [Google Scholar]
- 75.Guo X, Tang Y. OCT4 pseudogenes present in human leukemia cells. Clin Exp Med. 2011 doi: 10.1007/s10238-011-0163-4. [DOI] [PubMed] [Google Scholar]
- 76.Monuki ES, Kuhn R, Weinmaster G, Trapp BD, Lemke G. Expression and activity of the POU transcription factor SCIP. Science. 1990;249:1300–3. doi: 10.1126/science.1975954. [DOI] [PubMed] [Google Scholar]
- 77.Suzuki N, Rohdewohld H, Neuman T, Gruss P, Scholer HR. Oct-6: a POU transcription factor expressed in embryonal stem cells and in the developing brain. EMBO J. 1990;9:3723–32. doi: 10.1002/j.1460-2075.1990.tb07585.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Faus I, Hsu HJ, Fuchs E. Oct-6: a regulator of keratinocyte gene expression in stratified squamous epithelia. Mol Cell Biol. 1994;14:3263–75. doi: 10.1128/mcb.14.5.3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ryu EJ, Wang JY, Le N, Baloh RH, Gustin JA, Schmidt RE, Milbrandt J. Misexpression of Pou3f1 results in peripheral nerve hypomyelination and axonal loss. J Neurosci. 2007;27:11552–9. doi: 10.1523/JNEUROSCI.5497-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ilia M. Oct-6 transcription factor. Int Rev Neurobiol. 2004;59:471–89. doi: 10.1016/S0074-7742(04)59018-9. [DOI] [PubMed] [Google Scholar]
- 81.Jaegle M, Mandemakers W, Broos L, Zwart R, Karis A, Visser P, Grosveld F, Meijer D. The POU factor Oct-6 and Schwann cell differentiation. Science. 1996;273:507–10. doi: 10.1126/science.273.5274.507. [DOI] [PubMed] [Google Scholar]
- 82.Andersen B, Weinberg WC, Rennekampff O, McEvilly RJ, Bermingham JR, Jr., Hooshmand F, Vasilyev V, Hansbrough JF, Pittelkow MR, Yuspa SH, Rosenfeld MG. Functions of the POU domain genes Skn-1a/i and Tst-1/Oct-6/SCIP in epidermal differentiation. Genes Dev. 1997;11:1873–84. doi: 10.1101/gad.11.14.1873. [DOI] [PubMed] [Google Scholar]
- 83.Wu X, Oatley JM, Oatley MJ, Kaucher AV, Avarbock MR, Brinster RL. The POU domain transcription factor POU3F1 is an important intrinsic regulator of GDNF-induced survival and self-renewal of mouse spermatogonial stem cells. Biol Reprod. 2010;82:1103–11. doi: 10.1095/biolreprod.109.083097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Eisen T, Easty DJ, Bennett DC, Goding CR. The POU domain transcription factor Brn-2: elevated expression in malignant melanoma and regulation of melanocyte-specific gene expression. Oncogene. 1995;11:2157–64. [PubMed] [Google Scholar]
- 85.Lefort K, Rouault JP, Tondereau L, Magaud JP, Dore JF. The specific activation of gadd45 following UVB radiation requires the POU family gene product N-oct3 in human melanoma cells. Oncogene. 2001;20:7375–85. doi: 10.1038/sj.onc.1204923. [DOI] [PubMed] [Google Scholar]
- 86.Goodall J, Carreira S, Denat L, Kobi D, Davidson I, Nuciforo P, Sturm RA, Larue L, Goding CR. Brn-2 represses microphthalmia-associated transcription factor expression and marks a distinct subpopulation of microphthalmia-associated transcription factor- negative melanoma cells. Cancer Res. 2008;68:7788–94. doi: 10.1158/0008-5472.CAN-08-1053. [DOI] [PubMed] [Google Scholar]
- 87.Sumiyama K, Washio-Watanabe K, Saitou N, Hayakawa T, Ueda S. Class III POU genes: generation of homopolymeric amino acid repeats under GC pressure in mammals. J Mol Evol. 1996;43:170–8. doi: 10.1007/BF02338824. [DOI] [PubMed] [Google Scholar]
- 88.Nakai S, Sugitani Y, Sato H, Ito S, Miura Y, Ogawa M, Nishi M, Jishage K, Minowa O, Noda T. Crucial roles of Brn1 in distal tubule formation and function in mouse kidney. Development. 2003;130:4751–9. doi: 10.1242/dev.00666. [DOI] [PubMed] [Google Scholar]
- 89.Friedrich RP, Schlierf B, Tamm ER, Bosl MR, Wegner M. The class III POU domain protein Brn-1 can fully replace the related Oct-6 during schwann cell development and myelination. Mol Cell Biol. 2005;25:1821–9. doi: 10.1128/MCB.25.5.1821-1829.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wolf M, Lommes P, Sock E, Reiprich S, Friedrich RP, Kriesch J, Stolt CC, Bermingham JR, Jr., Wegner M. Replacement of related POU transcription factors leads to severe defects in mouse forebrain development. Dev Biol. 2009;332:418–28. doi: 10.1016/j.ydbio.2009.06.011. [DOI] [PubMed] [Google Scholar]
- 91.Mathis JM, Simmons DM, He X, Swanson LW, Rosenfeld MG. Brain 4: a novel mammalian POU domain transcription factor exhibiting restricted brain-specific expression. EMBO J. 1992;11:2551–61. doi: 10.1002/j.1460-2075.1992.tb05320.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tan XF, Qin JB, Jin GH, Tian ML, Li HM, Zhu HX, Zhang XH, Shi JH, Huang Z. Effects of Brn-4 on the neuronal differentiation of neural stem cells derived from rat midbrain. Cell Biol Int. 2010;34:877–82. doi: 10.1042/CBI20100214. [DOI] [PubMed] [Google Scholar]
- 93.Braunstein EM, Crenshaw EB, 3rd, Morrow BE, Adams JC. Cooperative function of Tbx1 and Brn4 in the periotic mesenchyme is necessary for cochlea formation. J Assoc Res Otolaryngol. 2008;9:33–43. doi: 10.1007/s10162-008-0110-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.de Kok YJ, van der Maarel SM, Bitner-Glindzicz M, Huber I, Monaco AP, Malcolm S, Pembrey ME, Ropers HH, Cremers FP. Association between X-linked mixed deafness and mutations in the POU domain gene POU3F4. Science. 1995;267:685–8. doi: 10.1126/science.7839145. [DOI] [PubMed] [Google Scholar]
- 95.de Kok YJ, Cremers CW, Ropers HH, Cremers FP. The molecular basis of X- linked deafness type 3 (DFN3) in two sporadic cases: identification of a somatic mosaicism for a POU3F4 missense mutation. Hum Mutat. 1997;10:207–11. doi: 10.1002/(SICI)1098-1004(1997)10:3<207::AID-HUMU5>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 96.Li J, Cheng J, Lu Y, Chen A, Sun Y, Kang D, Zhang X, Dai P, Han D, Yuan H. Identification of a novel mutation in POU3F4 for prenatal diagnosis in a Chinese family with X-linked nonsyndromic hearing loss. J Genet Genomics. 2010;37:787–93. doi: 10.1016/S1673-8527(09)60096-5. [DOI] [PubMed] [Google Scholar]
- 97.Cabral A, Fischer DF, Vermeij WP, Backendorf C. Distinct functional interactions of human Skn-1 isoforms with Ese-1 during keratinocyte terminal differentiation. J Biol Chem. 2003;278:17792–9. doi: 10.1074/jbc.M300508200. [DOI] [PubMed] [Google Scholar]
- 98.Nakajima K, Tamai K, Yamazaki T, Toyomaki Y, Nakano H, Uitto J, Sawamura D. Identification of Skn-1n, a splice variant induced by high calcium concentration and specifically expressed in normal human keratinocytes. J Invest Dermatol. 2008;128:1336–9. doi: 10.1038/sj.jid.5701143. [DOI] [PubMed] [Google Scholar]
- 99.Hildesheim J, Foster RA, Chamberlin ME, Vogel JC. Characterization of the regulatory domains of the human skn-1a/Epoc-1/Oct-11 POU transcription factor. J Biol Chem. 1999;274:26399–406. doi: 10.1074/jbc.274.37.26399. [DOI] [PubMed] [Google Scholar]
- 100.Takemoto H, Tamai K, Akasaka E, Rokunohe D, Takiyoshi N, Umegaki N, Nakajima K, Aizu T, Kaneko T, Nakano H, Sawamura D. Relation between the expression levels of the POU transcription factors Skn-1a and Skn-1n and keratinocyte differentiation. J Dermatol Sci. 2010;60:203–5. doi: 10.1016/j.jdermsci.2010.10.006. [DOI] [PubMed] [Google Scholar]
- 101.Matsumoto I, Ohmoto M, Narukawa M, Yoshihara Y, Abe K. Skn-1a (Pou2f3) specifies taste receptor cell lineage. Nat Neurosci. 2011;14:685–7. doi: 10.1038/nn.2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wang VE, Schmidt T, Chen J, Sharp PA, Tantin D. Embryonic lethality, decreased erythropoiesis, and defective octamer-dependent promoter activation in Oct-1- deficient mice. Mol Cell Biol. 2004;24:1022–32. doi: 10.1128/MCB.24.3.1022-1032.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wang VE, Tantin D, Chen J, Sharp PA. B cell development and immunoglobulin transcription in Oct-1-deficient mice. Proc Natl Acad Sci U S A. 2004;101:2005–10. doi: 10.1073/pnas.0307304101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sebastiano V, Dalvai M, Gentile L, Schubart K, Sutter J, Wu GM, Tapia N, Esch D, Ju JY, Hubner K, Bravo MJ, Scholer HR, Cavaleri F, Matthias P. Oct1 regulates trophoblast development during early mouse embryogenesis. Development. 2010;137:3551–60. doi: 10.1242/dev.047027. [DOI] [PubMed] [Google Scholar]
- 105.Nichols J, Zevnik B, Anastassiadis K, Niwa H, Klewe-Nebenius D, Chambers I, Scholer H, Smith A. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell. 1998;95:379–91. doi: 10.1016/s0092-8674(00)81769-9. [DOI] [PubMed] [Google Scholar]
- 106.Pesce M, Scholer HR. Oct-4: Gatekeeper in the Beginnings of Mammalian Development. Stem Cells. 2001;19:271–278. doi: 10.1634/stemcells.19-4-271. [DOI] [PubMed] [Google Scholar]
- 107.Nakai S, Kawano H, Yudate T, Nishi M, Kuno J, Nagata A, Jishage K, Hamada H, Fujii H, Kawamura K, et al. The POU domain transcription factor Brn-2 is required for the determination of specific neuronal lineages in the hypothalamus of the mouse. Genes Dev. 1995;9:3109–21. doi: 10.1101/gad.9.24.3109. [DOI] [PubMed] [Google Scholar]
- 108.Zaehres H, Lensch MW, Daheron L, Stewart SA, Itskovitz-Eldor J, Daley GQ. High-efficiency RNA interference in human embryonic stem cells. Stem Cells. 2005;23:299–305. doi: 10.1634/stemcells.2004-0252. [DOI] [PubMed] [Google Scholar]
- 109.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–76. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 110.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–72. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 111.Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, Slukvin II, Thomson JA. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–20. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 112.Aoi T, Yae K, Nakagawa M, Ichisaka T, Okita K, Takahashi K, Chiba T, Yamanaka S. Generation of pluripotent stem cells from adult mouse liver and stomach cells. Science. 2008;321:699–702. doi: 10.1126/science.1154884. [DOI] [PubMed] [Google Scholar]
- 113.Liu H, Zhu F, Yong J, Zhang P, Hou P, Li H, Jiang W, Cai J, Liu M, Cui K, Qu X, Xiang T, Lu D, Chi X, Gao G, Ji W, Ding M, Deng H. Generation of induced pluripotent stem cells from adult rhesus monkey fibroblasts. Cell Stem Cell. 2008;3:587–90. doi: 10.1016/j.stem.2008.10.014. [DOI] [PubMed] [Google Scholar]
- 114.Liao J, Cui C, Chen S, Ren J, Chen J, Gao Y, Li H, Jia N, Cheng L, Xiao H, Xiao L. Generation of induced pluripotent stem cell lines from adult rat cells. Cell Stem Cell. 2009;4:11–5. doi: 10.1016/j.stem.2008.11.013. [DOI] [PubMed] [Google Scholar]
- 115.Nakagawa M, Koyanagi M, Tanabe K, Takahashi K, Ichisaka T, Aoi T, Okita K, Mochiduki Y, Takizawa N, Yamanaka S. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat Biotechnol. 2008;26:101–6. doi: 10.1038/nbt1374. [DOI] [PubMed] [Google Scholar]
- 116.Kim JB, Sebastiano V, Wu G, Arauzo-Bravo MJ, Sasse P, Gentile L, Ko K, Ruau D, Ehrich M, van den Boom D, Meyer J, Hubner K, Bernemann C, Ortmeier C, Zenke M, Fleischmann BK, Zaehres H, Scholer HR. Oct4-induced pluripotency in adult neural stem cells. Cell. 2009;136:411–9. doi: 10.1016/j.cell.2009.01.023. [DOI] [PubMed] [Google Scholar]
- 117.Boiani M, Eckardt S, Scholer HR, McLaughlin KJ. Oct4 distribution and level in mouse clones: consequences for pluripotency. Genes Dev. 2002;16:1209–1219. doi: 10.1101/gad.966002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Thomas MC, Chiang CM. The general transcription machinery and general cofactors. Crit Rev Biochem Mol Biol. 2006;41:105–78. doi: 10.1080/10409230600648736. [DOI] [PubMed] [Google Scholar]
- 119.Zwilling S, Annweiler A, Wirth T. The POU domains of the Oct1 and Oct2 transcription factors mediate specific interaction with TBP. Nucleic Acids Res. 1994;22:1655–62. doi: 10.1093/nar/22.9.1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Nakshatri H, Nakshatri P, Currie RA. Interaction of Oct-1 with TFIIB. Implications for a novel response elicited through the proximal octamer site of the lipoprotein lipase promoter. J Biol Chem. 1995;270:19613–23. doi: 10.1074/jbc.270.33.19613. [DOI] [PubMed] [Google Scholar]
- 121.Parslow TG, Blair DL, Murphy WJ, Granner DK. Structure of the 5′ ends of immunoglobulin genes: a novel conserved sequence. Proc Natl Acad Sci U S A. 1984;81:2650–4. doi: 10.1073/pnas.81.9.2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Nelms K, Van Ness B. Identification of an octamer-binding site in the human kappa light-chain enhancer. Mol Cell Biol. 1990;10:3843–6. doi: 10.1128/mcb.10.7.3843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Staudt LM, Lenardo MJ. Immunoglobulin gene transcription. Annu Rev Immunol. 1991;9:373–98. doi: 10.1146/annurev.iy.09.040191.002105. [DOI] [PubMed] [Google Scholar]
- 124.Poellinger L, Yoza BK, Roeder RG. Functional cooperativity between protein molecules bound at two distinct sequence elements of the immunoglobulin heavy-chain promoter. Nature. 1989;337:573–6. doi: 10.1038/337573a0. [DOI] [PubMed] [Google Scholar]
- 125.Kemler I, Bucher E, Seipel K, Muller-Immergluck MM, Schaffner W. Promoters with the octamer DNA motif (ATGCAAAT) can be ubiquitous or cell type-specific depending on binding affinity of the octamer site and Oct-factor concentration. Nucleic Acids Res. 1991;19:237–42. doi: 10.1093/nar/19.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Strubin M, Newell JW, Matthias P. OBF-1, a novel B cell-specific coactivator that stimulates immunoglobulin promoter activity through association with octamer-binding proteins. Cell. 1995;80:497–506. doi: 10.1016/0092-8674(95)90500-6. [DOI] [PubMed] [Google Scholar]
- 127.Salas M, Eckhardt LA. Critical role for the Oct-2/OCA-B partnership in Ig-secreting cells. J Immunol. 2003;171:6589–98. doi: 10.4049/jimmunol.171.12.6589. [DOI] [PubMed] [Google Scholar]
- 128.Zheng J, Huang J, Mao Y, Liu S, Sun X, Zhu X, Ma T, Zhang L, Ji J, Zhang Y, Yin CC, Qiu X. Immunoglobulin gene transcripts have distinct VHDJH recombination characteristics in human epithelial cancer cells. J Biol Chem. 2009;284:13610–9. doi: 10.1074/jbc.M809524200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Huang J, Zhang L, Ma T, Zhang P, Qiu X. Expression of immunoglobulin gene with classical V-(D)-J rearrangement in mouse testis and epididymis. J Histochem Cytochem. 2009;57:339–49. doi: 10.1369/jhc.2008.951434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Zhu X, Wu L, Zhang L, Hao P, Zhang S, Huang J, Zheng J, Liu Y, Li W, Zhang Y, Zhou C, Yin CC, Qiu X. Distinct regulatory mechanism of immunoglobulin gene transcription in epithelial cancer cells. Cell Mol Immunol. 2010;7:279–86. doi: 10.1038/cmi.2010.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Wang P, Jin T. Oct-1 functions as a sensor for metabolic and stress signals. Islets. 2010;2:46–8. doi: 10.4161/isl.2.1.10017. [DOI] [PubMed] [Google Scholar]
- 132.Wang P, Wang Q, Sun J, Wu J, Li H, Zhang N, Huang Y, Su B, Li RK, Liu L, Zhang Y, Elsholtz HP, Hu J, Gaisano HY, Jin T. POU homeodomain protein Oct-1 functions as a sensor for cyclic AMP. J Biol Chem. 2009;284:26456–65. doi: 10.1074/jbc.M109.030668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Shakya A, Cooksey R, Cox JE, Wang V, McClain DA, Tantin D. Oct1 loss of function induces a coordinate metabolic shift that opposes tumorigenicity. Nat Cell Biol. 2009;11:320–7. doi: 10.1038/ncb1840. [DOI] [PubMed] [Google Scholar]
- 134.Tantin D, Schild-Poulter C, Wang V, Hache RJ, Sharp PA. The octamer binding transcription factor Oct-1 is a stress sensor. Cancer Res. 2005;65:10750–8. doi: 10.1158/0008-5472.CAN-05-2399. [DOI] [PubMed] [Google Scholar]
- 135.Bellance N, Pabst L, Allen G, Rossignol R, Nagrath D. Oncosecretomics coupled to bioenergetics identifies alpha-amino adipic acid, isoleucine and GABA as potential biomarkers of cancer: Differential expression of c-Myc, Oct1 and KLF4 coordinates metabolic changes. Biochim Biophys Acta. 2012 doi: 10.1016/j.bbabio.2012.07.004. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 136.Zhao H, Jin S, Fan F, Fan W, Tong T, Zhan Q. Activation of the Transcription Factor Oct-1 in Response to DNA Damage. Cancer Res. 2000;60:6276–6280. [PubMed] [Google Scholar]
- 137.Schild-Poulter C, Shih A, Tantin D, Yarymowich NC, Soubeyrand S, Sharp PA, Hache RJ. DNA-PK phosphorylation sites on Oct-1 promote cell survival following DNA damage. Oncogene. 2007;26:3980–8. doi: 10.1038/sj.onc.1210165. [DOI] [PubMed] [Google Scholar]
- 138.Schild-Poulter C, Shih A, Yarymowich NC, Hache RJ. Down-regulation of histone H2B by DNA-dependent protein kinase in response to DNA damage through modulation of octamer transcription factor 1. Cancer Res. 2003;63:7197–205. [PubMed] [Google Scholar]
- 139.Kang J, Gemberling M, Nakamura M, Whitby FG, Handa H, Fairbrother WG, Tantin D. A general mechanism for transcription regulation by Oct1 and Oct4 in response to genotoxic and oxidative stress. Genes Dev. 2009;23:208–22. doi: 10.1101/gad.1750709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Winklehner-Jennewein P, Geymayer S, Lechner J, Welte T, Hansson L, Geley S, Doppler W. A distal enhancer region in the human beta-casein gene mediates the response to prolactin and glucocorticoid hormones. Gene. 1998;217:127–39. doi: 10.1016/s0378-1119(98)00380-1. [DOI] [PubMed] [Google Scholar]
- 141.Groenen MA, Dijkhof RJ, van der Poel JJ, van Diggelen R, Verstege E. Multiple octamer binding sites in the promoter region of the bovine alpha s2-casein gene. Nucleic Acids Res. 1992;20:4311–8. doi: 10.1093/nar/20.16.4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Mandemakers W, Zwart R, Jaegle M, Walbeehm E, Visser P, Grosveld F, Meijer D. A distal Schwann cell-specific enhancer mediates axonal regulation of the Oct-6 transcription factor during peripheral nerve development and regeneration. EMBO J. 2000;19:2992–3003. doi: 10.1093/emboj/19.12.2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Jaegle M, Ghazvini M, Mandemakers W, Piirsoo M, Driegen S, Levavasseur F, Raghoenath S, Grosveld F, Meijer D. The POU proteins Brn-2 and Oct-6 share important functions in Schwann cell development. Genes Dev. 2003;17:1380–91. doi: 10.1101/gad.258203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Jagalur NB, Ghazvini M, Mandemakers W, Driegen S, Maas A, Jones EA, Jaegle M, Grosveld F, Svaren J, Meijer D. Functional dissection of the Oct6 Schwann cell enhancer reveals an essential role for dimeric Sox10 binding. J Neurosci. 2011;31:8585–94. doi: 10.1523/JNEUROSCI.0659-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Ghislain J, Charnay P. Control of myelination in Schwann cells: a Krox20 cis- regulatory element integrates Oct6, Brn2 and Sox10 activities. EMBO Rep. 2006;7:52–8. doi: 10.1038/sj.embor.7400573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Svaren J, Meijer D. The molecular machinery of myelin gene transcription in Schwann cells. Glia. 2008;56:1541–51. doi: 10.1002/glia.20767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Pang ZP, Yang N, Vierbuchen T, Ostermeier A, Fuentes DR, Yang TQ, Citri A, Sebastiano V, Marro S, Sudhof TC, Wernig M. Induction of human neuronal cells by defined transcription factors. Nature. 2011;476:220–3. doi: 10.1038/nature10202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Sugitani Y, Nakai S, Minowa O, Nishi M, Jishage K, Kawano H, Mori K, Ogawa M, Noda T. Brn-1 and Brn-2 share crucial roles in the production and positioning of mouse neocortical neurons. Genes Dev. 2002;16:1760–5. doi: 10.1101/gad.978002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Shimazaki T, Arsenijevic Y, Ryan AK, Rosenfeld MG, Weiss S. A role for the POU-III transcription factor Brn-4 in the regulation of striatal neuron precursor differentiation. EMBO J. 1999;18:444–56. doi: 10.1093/emboj/18.2.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Shi J, Jin G, Zhu H, Tian M, Zhang X, Qin J, Tan X. The role of Brn-4 in the regulation of neural stem cell differentiation into neurons. Neurosci Res. 2010;67:8–17. doi: 10.1016/j.neures.2010.01.007. [DOI] [PubMed] [Google Scholar]
- 151.Andersen B, Schonemann MD, Flynn SE, Pearse RV, 2nd, Singh H, Rosenfeld MG. Skn-1a and Skn-1i: two functionally distinct Oct-2-related factors expressed in epidermis. Science. 1993;260:78–82. doi: 10.1126/science.7682011. [DOI] [PubMed] [Google Scholar]
- 152.Hildesheim J, Kuhn U, Yee CL, Foster RA, Yancey KB, Vogel JC. The hSkn- 1a POU transcription factor enhances epidermal stratification by promoting keratinocyte proliferation. J Cell Sci. 2001;114:1913–23. doi: 10.1242/jcs.114.10.1913. [DOI] [PubMed] [Google Scholar]
- 153.Sugihara TM, Kudryavtseva EI, Kumar V, Horridge JJ, Andersen B. The POU domain factor Skin-1a represses the keratin 14 promoter independent of DNA binding. A possible role for interactions between Skn-1a and CREB-binding protein/p300. J Biol Chem. 2001;276:33036–44. doi: 10.1074/jbc.M103000200. [DOI] [PubMed] [Google Scholar]
- 154.Botting CH, Hay RT. Characterisation of the adenovirus preterminal protein and its interaction with the POU homeodomain of NFIII (Oct-1) Nucleic Acids Res. 1999;27:2799–805. doi: 10.1093/nar/27.13.2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Belikov S, Holmqvist PH, Astrand C, Wrange O. Nuclear factor 1 and octamer transcription factor 1 binding preset the chromatin structure of the mouse mammary tumor virus promoter for hormone induction. J Biol Chem. 2004;279:49857–67. doi: 10.1074/jbc.M409713200. [DOI] [PubMed] [Google Scholar]
- 156.Liu Y, Gong W, Huang CC, Herr W, Cheng X. Crystal structure of the conserved core of the herpes simplex virus transcriptional regulatory protein VP16. Genes Dev. 1999;13:1692–703. doi: 10.1101/gad.13.13.1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Nogueira ML, Wang VE, Tantin D, Sharp PA, Kristie TM. Herpes simplex virus infections are arrested in Oct-1-deficient cells. Proc Natl Acad Sci U S A. 2004;101:1473–8. doi: 10.1073/pnas.0307300101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Robinson AR, Kwek SS, Hagemeier SR, Wille CK, Kenney SC. Cellular transcription factor Oct-1 interacts with the Epstein-Barr virus BRLF1 protein to promote disruption of viral latency. J Virol. 2011;85:8940–53. doi: 10.1128/JVI.00569-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Mesplede T, Island ML, Christeff N, Petek F, Doly J, Navarro S. The POU transcription factor Oct-1 represses virus-induced interferon A gene expression. Mol Cell Biol. 2005;25:8717–31. doi: 10.1128/MCB.25.19.8717-8731.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Almeida R, Almeida J, Shoshkes M, Mendes N, Mesquita P, Silva E, Van Seuningen I, Reis CA, Santos-Silva F, David L. OCT-1 is over-expressed in intestinal metaplasia and intestinal gastric carcinomas and binds to, but does not transactivate, CDX2 in gastric cells. J Pathol. 2005;207:396–401. doi: 10.1002/path.1861. [DOI] [PubMed] [Google Scholar]
- 161.Jin T, Branch DR, Zhang X, Qi S, Youngson B, Goss PE. Examination of POU homeobox gene expression in human breast cancer cells. Int J Cancer. 1999;81:104–112. doi: 10.1002/(sici)1097-0215(19990331)81:1<104::aid-ijc18>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 162.Obinata D, Takayama K, Urano T, Murata T, Kumagai J, Fujimura T, Ikeda K, Horie-Inoue K, Homma Y, Ouchi Y, Takahashi S, Inoue S. Oct1 regulates cell growth of LNCaP cells and is a prognostic factor for prostate cancer. Int J Cancer. 2012;130:1021–8. doi: 10.1002/ijc.26043. [DOI] [PubMed] [Google Scholar]
- 163.Zhou C, Tong Y, Wawrowsky K, Bannykh S, Donangelo I, Melmed S. Oct-1 induces pituitary tumor transforming gene expression in endocrine tumors. Endocr Relat Cancer. 2008;15:817–31. doi: 10.1677/ERC-08-0060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Qin XF, Luo Y, Suh H, Wayne J, Misulovin Z, Roeder RG, Nussenzweig MC. Transformation by homeobox genes can be mediated by selective transcriptional repression. EMBO J. 1994;13:5967–76. doi: 10.1002/j.1460-2075.1994.tb06942.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Ng MC, Lam VK, Tam CH, Chan AW, So WY, Ma RC, Zee BC, Waye MM, Mak WW, Hu C, Wang CR, Tong PC, Jia WP, Chan JC. Association of the POU class 2 homeobox 1 gene (POU2F1) with susceptibility to Type 2 diabetes in Chinese populations. Diabet Med. 2010;27:1443–9. doi: 10.1111/j.1464-5491.2010.03124.x. [DOI] [PubMed] [Google Scholar]
- 166.Theil J, Laumen H, Marafioti T, Hummel M, Lenz G, Wirth T, Stein H. Defective octamer-dependent transcription is responsible for silenced immunoglobulin transcription in Reed-Sternberg cells. Blood. 2001;97:3191–6. doi: 10.1182/blood.v97.10.3191. [DOI] [PubMed] [Google Scholar]
- 167.Re D, Muschen M, Ahmadi T, Wickenhauser C, Staratschek-Jox A, Holtick U, Diehl V, Wolf J. Oct-2 and Bob-1 deficiency in Hodgkin and Reed Sternberg cells. Cancer Res. 2001;61:2080–4. [PubMed] [Google Scholar]
- 168.Di Bartolo DL, Hyjek E, Keller S, Guasparri I, Deng H, Sun R, Chadburn A, Knowles DM, Cesarman E. Role of defective Oct-2 and OCA-B expression in immunoglobulin production and Kaposi’s sarcoma-associated herpesvirus lytic reactivation in primary effusion lymphoma. J Virol. 2009;83:4308–15. doi: 10.1128/JVI.02196-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Browne P, Petrosyan K, Hernandez A, Chan JA. The B-cell transcription factors BSAP, Oct-2, and BOB.1 and the pan-B-cell markers CD20, CD22, and CD79a are useful in the differential diagnosis of classic Hodgkin lymphoma. Am J Clin Pathol. 2003;120:767–77. doi: 10.1309/YCH8-DWUF-FQBK-GPVB. [DOI] [PubMed] [Google Scholar]
- 170.Gidekel S, Pizov G, Bergman Y, Pikarsky E. Oct-3/4 is a dose-dependent oncogenic fate determinant. Cancer Cell. 2003;4:361–370. doi: 10.1016/s1535-6108(03)00270-8. [DOI] [PubMed] [Google Scholar]
- 171.de Jong J, Looijenga LH. Stem Cell Marker OCT3/4 in Tumor Biology and Germ Cell Tumor Diagnostics: History and Future. Crit Rev Oncog. 2006;12:171–203. doi: 10.1615/critrevoncog.v12.i3-4.10. [DOI] [PubMed] [Google Scholar]
- 172.Monk M, Holding C. Human embryonic genes re-expressed in cancer cells. Oncogene. 2001;20:8085–8091. doi: 10.1038/sj.onc.1205088. [DOI] [PubMed] [Google Scholar]
- 173.Atlasi Y, Mowla SJ, Ziaee SA, Bahrami AR. OCT-4, an embryonic stem cell marker, is highly expressed in bladder cancer. Int J Cancer. 2007;120:1598–602. doi: 10.1002/ijc.22508. [DOI] [PubMed] [Google Scholar]
- 174.Chen Z, Xu WR, Qian H, Zhu W, Bu XF, Wang S, Yan YM, Mao F, Gu HB, Cao HL, Xu XJ. Oct4, a novel marker for human gastric cancer. J Surg Oncol. 2009;99:414–9. doi: 10.1002/jso.21270. [DOI] [PubMed] [Google Scholar]
- 175.Iki K, Pour PM. Expression of Oct4, a stem cell marker, in the hamster pancreatic cancer model. Pancreatology. 2006;6:406–13. doi: 10.1159/000094317. [DOI] [PubMed] [Google Scholar]
- 176.Karoubi G, Gugger M, Schmid R, Dutly A. OCT4 expression in human non-small cell lung cancer: implications for therapeutic intervention. Interact Cardiovasc Thorac Surg. 2009;8:393–7. doi: 10.1510/icvts.2008.193995. [DOI] [PubMed] [Google Scholar]
- 177.Saigusa S, Tanaka K, Toiyama Y, Yokoe T, Okugawa Y, Ioue Y, Miki C, Kusunoki M. Correlation of CD133, OCT4, and SOX2 in rectal cancer and their association with distant recurrence after chemoradiotherapy. Ann Surg Oncol. 2009;16:3488–98. doi: 10.1245/s10434-009-0617-z. [DOI] [PubMed] [Google Scholar]
- 178.Xu K, Zhu Z, Zeng F. Expression and significance of Oct4 in bladder cancer. J Huazhong Univ Sci Technolog Med Sci. 2007;27:675–7. doi: 10.1007/s11596-007-0614-z. [DOI] [PubMed] [Google Scholar]
- 179.Zhu Z, Wen J, Zheng X, Wang D, Wang Q, Fan C. Expression of transcription factor Oct4 in bladder cancer cell line T24 and its effects on the biological characteristics of the cells. J Huazhong Univ Sci Technolog Med Sci. 2009;29:73–6. doi: 10.1007/s11596-009-0115-3. [DOI] [PubMed] [Google Scholar]
- 180.Wang P, Branch DR, Bali M, Schultz GA, Goss PE, Jin T. The POU homeodomain protein OCT3 as a potential transcriptional activator for fibroblast growth factor-4 (FGF-4) in human breast cancer cells. Biochem J. 2003;375:199–205. doi: 10.1042/BJ20030579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.McLeskey SW, Kurebayashi J, Honig SF, Zwiebel J, Lippman ME, Dickson RB, Kern FG. Fibroblast growth factor 4 transfection of MCF-7 cells produces cell lines that are tumorigenic and metastatic in ovariectomized or tamoxifen-treated athymic nude mice. Cancer Res. 1993;53:2168–77. [PubMed] [Google Scholar]
- 182.Hochedlinger K, Yamada Y, Beard C, Jaenisch R. Ectopic Expression of Blocks Progenitor-Cell Differentiation and Causes Dysplasia in Epithelial Tissues. Cell. 2005;121:465–477. doi: 10.1016/j.cell.2005.02.018. [DOI] [PubMed] [Google Scholar]
- 183.Mueller T, Luetzkendorf J, Nerger K, Schmoll HJ, Mueller LP. Analysis of OCT4 expression in an extended panel of human tumor cell lines from multiple entities and in human mesenchymal stem cells. Cell Mol Life Sci. 2009;66:495–503. doi: 10.1007/s00018-008-8623-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Cantz T, Key G, Bleidissel M, Gentile L, Han DW, Brenne A, Scholer HR. Absence of OCT4 expression in somatic tumor cell lines. Stem Cells. 2008;26:692–7. doi: 10.1634/stemcells.2007-0657. [DOI] [PubMed] [Google Scholar]
- 185.Suo G, Han J, Wang X, Zhang J, Zhao Y, Zhao Y, Dai J. Oct4 pseudogenes are transcribed in cancers. Biochem. Biophys. Res. Commun. 2005;337:1047. doi: 10.1016/j.bbrc.2005.09.157. [DOI] [PubMed] [Google Scholar]
- 186.Cook AL, Sturm RA. POU domain transcription factors: BRN2 as a regulator of melanocytic growth and tumourigenesis. Pigment Cell Melanoma Res. 2008;21:611–26. doi: 10.1111/j.1755-148X.2008.00510.x. [DOI] [PubMed] [Google Scholar]
- 187.Boyle GM, Woods SL, Bonazzi VF, Stark MS, Hacker E, Aoude LG, Dutton-Regester K, Cook AL, Sturm RA, Hayward NK. Melanoma cell invasiveness is regulated by miR-211 suppression of the BRN2 transcription factor. Pigment Cell Melanoma Res. 2011;24:525–37. doi: 10.1111/j.1755-148X.2011.00849.x. [DOI] [PubMed] [Google Scholar]
- 188.Berlin I, Denat L, Steunou AL, Puig I, Champeval D, Colombo S, Roberts K, Bonvin E, Bourgeois Y, Davidson I, Delmas V, Nieto L, Goding CR, Larue L. Phosphorylation of BRN2 modulates its interaction with the Pax3 promoter to control melanocyte migration and proliferation. Mol Cell Biol. 2012;32:1237–47. doi: 10.1128/MCB.06257-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Thurber AE, Douglas G, Sturm EC, Zabierowski SE, Smit DJ, Ramakrishnan SN, Hacker E, Leonard JH, Herlyn M, Sturm RA. Inverse expression states of the BRN2 and MITF transcription factors in melanoma spheres and tumour xenografts regulate the NOTCH pathway. Oncogene. 2011;30:3036–48. doi: 10.1038/onc.2011.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Lee HK, Song MH, Kang M, Lee JT, Kong KA, Choi SJ, Lee KY, Venselaar H, Vriend G, Lee WS, Park HJ, Kwon TK, Bok J, Kim UK. Clinical and molecular characterizations of novel POU3F4 mutations reveal that DFN3 is due to null function of POU3F4 protein. Physiol Genomics. 2009;39:195–201. doi: 10.1152/physiolgenomics.00100.2009. [DOI] [PubMed] [Google Scholar]
- 191.Coate TM, Raft S, Zhao X, Ryan AK, Crenshaw EB, 3rd, Kelley MW. Otic mesenchyme cells regulate spiral ganglion axon fasciculation through a Pou3f4/EphA4 signaling pathway. Neuron. 2012;73:49–63. doi: 10.1016/j.neuron.2011.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Song MH, Choi SY, Wu L, Oh SK, Lee HK, Lee DJ, Shim DB, Choi JY, Kim UK, Bok J. Pou3f4 deficiency causes defects in otic fibrocytes and stria vascularis by different mechanisms. Biochem Biophys Res Commun. 2011;404:528–33. doi: 10.1016/j.bbrc.2010.12.019. [DOI] [PubMed] [Google Scholar]
- 193.Phippard D, Lu L, Lee D, Saunders JC, Crenshaw EB., 3rd Targeted mutagenesis of the POU-domain gene Brn4/Pou3f4 causes developmental defects in the inner ear. J Neurosci. 1999;19:5980–9. doi: 10.1523/JNEUROSCI.19-14-05980.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Enomoto Y, Enomoto K, Kitamura T, Kanda T. Keratinocyte-specific POU transcription factor hSkn-1a represses the growth of cervical cancer cell lines. Oncogene. 2004;23:5014–22. doi: 10.1038/sj.onc.1207653. [DOI] [PubMed] [Google Scholar]
- 195.Zhang Z, Huettner PC, Nguyen L, Bidder M, Funk MC, Li J, Rader JS. Aberrant promoter methylation and silencing of the POU2F3 gene in cervical cancer. Oncogene. 2006;25:5436–45. doi: 10.1038/sj.onc.1209530. [DOI] [PubMed] [Google Scholar]