Abstract
Cranial neural crest (CNC) cells are patterned and coalesce to facial prominences that undergo convergence and extension to generate the craniofacial form. We applied a chemical genetics approach to identify pathways that regulate craniofacial development during embryogenesis. Treatment with the nitric oxide synthase inhibitor TRIM abrogated first pharyngeal arch structures and induced ectopic ceratobranchial formation. TRIM promoted a progenitor CNC fate and inhibited chondrogenic differentiation, which were mediated through impaired nitric oxide (NO) production without appreciable effect on global protein S-nitrosylation. Instead, TRIM perturbed hox gene patterning and caused histone hypoacetylation. Rescue of TRIM phenotype was achieved with over-expression of histone acetyltransferase kat6a, inhibition of histone deacetylase, and complimentary NO. These studies demonstrate that NO signaling and histone acetylation are coordinated mechanisms that regulate CNC patterning, differentiation and convergence during craniofacial morphogenesis.
Keywords: cranial neural crest cell, nitric oxide, histone acetylation, craniofacial, chemical screen, zebrafish
INTRODUCTION
CNC cells are a group of pluripotent cells that delaminate from the neural tube early in embryogenesis and contribute extensively to the formation of vertebrate facial architecture, including cartilage and bone. Dysregulation of CNC development can lead to dramatic congenital defects such as orofacial clefting. All developmental processes including craniofacial morphogenesis are regulated by interplay between genetic and epigenetic mechanisms. Post-translational modifications of histones by acetylation, phosphorylation, methylation and sumoylation have been demonstrated to regulate craniofacial development. Genetic mutations affecting histone acetylation (MYST4) demethylation (KDM6A, PHF8), and sumoylation (SUMO1) result in orofacial clefts (Alkuraya, et al., 2006; Fischer, et al., 2006; Kraft, et al., 2011; Qi, et al., 2010). Further, epigenetic mechanisms may explain variability in phenotypic penetrance of genetic mutations that affect craniofacial development and mediation of environmental factors.
Histone acetylation is catalyzed by histone acetyltransferases (HATs), which transfer acetyl groups to lysines in the tails of core histone (Marmorstein and Roth, 2001). This process generally results in a more relaxed chromatin conformation, exposing binding sites for other modifiers and promoting gene transcription. In contrast, histone deacetylases (HDACs) remove the acetyl group and is generally associated with transcriptional silencing. Hdac8 has been shown to specifically control patterning of the skull in mice by repressing a number of homeobox transcription factors in the CNC cells, highlighting the importance of epigenetic regulation in CNC development (Haberland, et al., 2009). In human, chromosomal translocation disrupting MYST4 histone acetyltransferase results in a Noonan syndrome-like phenotype that includes a cleft palate (Kraft, et al., 2011). Further, the zebrafish ortholog of the human oncogenic histone acetyltransferase KAT6A regulates hox gene expression in CNC cells, and specifies segmental identity in the pharyngeal arches (PA) 2-4 (Miller, et al., 2004). Loss of kat6a function results in homeotic transformations of the second PA into a mirror-image duplicated jaw (Crump, et al., 2006).
Nitric oxide (NO) is another essential mediator of post-translational chromatin modification. Nitric oxide was initially recognized as an important second messenger signaling molecule generated from metabolism of L-arginine by the nitric oxide synthase (NOS) family of enzymes that includes neuronal (nNOS, NOS1), inducible (iNOS, NOS2), and endothelial (eNOS, NOS3) forms (Moncada and Higgs, 1993). There has been intense interest in NO signaling in a wide range of physiologic and disease states, ranging from vascular dilatation and inflammation to cancer progression. It is also increasingly evident that NO-mediated post-translational modification of protein is a fundamental mechanism regulating protein function, where S-nitrosylation of histones and transcription factors exert broad cellular effects. In fact, NO directly leads to chromatin remodelling by S-nitrosylation of histone acetyltransferases and histone deacetylases, leading to a context-dependent response (Nott, et al., 2008).
Chemical genetic screening in the zebrafish embryo is a powerful approach to interrogate development and disease, and helps to close the gap between molecular basis and pharmaceutical targets (Gut, et al., 2013; North, et al., 2007). The work described here reports the first application of chemical genetics toward the study of CNC cells and craniofacial morphogenesis, and uncovers NO signaling as an important regulatory component in early embryonic development. In complementary chemical screens of ~ 3,000 small molecules, we identified 21 compounds that disrupt craniofacial development with specific ethmoid plate and mandibular phenotypes. Notably, we discovered that the NOS inhibitor TRIM impaired CNC maturation, where insufficient NO signal altered CNC patterning, inhibited CNC migration and the chondrocyte-lineage differentiation. Biochemical and functional analysis demonstrate that TRIM plays a dual role in regulating CNC development via inhibition of NO signaling and histone hypoacetylation. This study describes novel finding that NO signaling and histone acetylation are coordinated to regulate CNC patterning, migration and differentiation during craniofacial morphogenesis.
RESULTS
Chemical screen for modulators of craniofacial development
Two complementary chemical screens were carried out to identify small molecules that regulate embryonic craniofacial development. One screen of 2,500 compounds evaluated neural crest development, using expression of progenitor marker crestin in 24 hours post-fertilization (hpf) embryos as the assay (White, et al., 2011). A second phenotypic screen of a subset of the compounds (488) with known biological functions was performed with Alcian blue staining of embryos at 96 hpf to identify small molecules that affect craniofacial morphogenesis (Fig. 1A). Overall, treatments with 21 compounds (5% of the subset Bioactives library) resulted in profound defects in craniofacial development and were selected as candidates for further analysis (Fig. 1B). A summary of the screen and the distinct classes of observed phenotypes are reported (Table S1). The 21 candidate compounds that perturbed craniofacial development were analyzed with regard to the Octanol-Water partition coefficient (log P). Interestingly, we found that all these biological active compounds possess a positive log P value ranged from +1 to +7 with hydrophobic property (Fig. 1C).
Figure 1. Summary and representative phenotypes from chemical library screen.
(A) Schematic diagram displaying the screening strategy for compounds that affect craniofacial development.
(B) The morphology screen identified 21 compounds that affect the development of craniofacial skeleton.
(C) Octanol-Water partition coefficient (Log P value) of the 21 compounds plotted against respective molecular weights.
(D) Representative Alcian blue stained phenotype of craniofacial structures after chemical treatment. Following each chemical, whole-mount craniofacial skeleton was shown in lateral (left), ventral (middle), and dissected palate in flat-mount (right) views. The zebrafish palate consists of ethmoid plate (ep) and trabecula (tr). Scalebars: 100 μm.
(E) Craniofacial malformation after TRIM treatment compared to DMSO. Upper panel, flatten-mount view of TRIM-induced “ectopic” cartilage. Middle panel, ventral view of the jaw skeleton illustrated by Sox10:gfp reporter line. Lower panel, loss of maxillary prominences after TRIM treatment, which normally contribute to trabecula at 48 hpf (arrow). The lower jaw cartilages are Meckel's (m), palatoquadrate (pq), hyosymplectic (hs), ceratohyal (ch), basihyal (bh), and ceratobranchial 1-5 (cb 1-5). A, anterior; P, posterior.
As expected, compounds that abrogated crestin expression lead to profound neural crest deficiency, and resulted in severe phenotypes of total or significant loss of CNC and its derivatives. For example, leflunomide, an inhibitor of dihydroorotate dehydrogenase (DHODH), abrogated crestin expression in all CNC progenitor cells and inhibited terminal differentiation in early embryogenesis, resulting in total absence of craniofacial structures (Fig. 1D). In contrast, screen of molecules with known biologic function using Alcian blue staining allowed us to identify compounds that lead to specific anomalies in craniofacial skeleton, since this was a morphologic screen. Specifically, the phenotypes observed for these 21 compounds can be classified into three patterns: 7 compounds abrogated development of craniofacial structures in general (TRIM in Fig. 1D, Fig. S1), 5 compounds produced embryos with fused ethmoid plate with small or absent lower jaw (GF109203X in Fig. 1D, Fig. S2), and treatment with 9 compounds resulted in abridged, separated ethmoid plate but intact lower jaw and skull base (Pimozide in Fig. 1D, Fig. S3).
In the group of compounds that lead to general absence of craniofacial structures, TRIM was unique in generating “ectopic” cartilaginous structures. We first tested the effect of TRIM treatment on CNC and its derived jaw skeletons during the full-time developmental window, staring from 12 hpf, when CNC cells initiate migration, to 96 hpf when the morphogenetic processes occur to shape the craniofacial skeleton. Compared to intact jaw skeletons in DMSO control, the upper and lower jaw structures failed to form in TRIM treated embryos; however, the skeletal elements that do form were paired and ectopically clustered in the dorsolateral region anterior to the otic vesicles, as evidenced by Alcian blue stained cartilages and in sox10:gfp reporter line (Fig. 1E, upper and middle panel). Furthermore, TRIM treatment at 12-48 hpf resulted in failure of the maxillary prominences to develop fully, which normally contribute to trabecula and lateral ethmoid plate in upper jaw (Fig. 1E, lower panel). In summary, TRIM appeared to inhibit the formation of discrete skeletal structures such as the palate and lower jaw cartilages. Meanwhile, the more posterior PA structures are formed, but are lateralized to an ectopic dorsolateral domain, where chondrocytes do not normally migrate. Further, with TRIM treatments at discrete stages and graded durations, we observed that TRIM's effect on CNC development is temporal and regional specific, with most profound inhibition during periods of CNC migration, patterning, and convergence morphogenesis before 48 hpf (Fig. S4).
TRIM treatment abrogates midline convergence of posterior CNC cells
To better understand how the “ectopic” cartilage structure was formed and whether TRIM treatment led to aberrant cell migration (Fig. 1E), cell lineage tracing was performed employing sox10:Kaede (Dougherty, et al., 2012). Using the 10-somite stage as the reference for CNC fate map, specific regions were photoconverted in the context of DMSO or TRIM treatment. When the most anterior CNC cells that normally contribute to the anterior first PA (pa 1a) were labeled (Fig. 2A), the cells migrated anteriorly and formed the ethmoid plate at 4 days post-fertilization (dpf) (Fig. 2B). When the same anterior CNC population was followed in the TRIM-treated embryos, the cells were found scattered in a lateral region around the eyes, failing to condense into the ethmoid plate in the midline (Fig. 2C, C’). Therefore, TRIM treatment did not prevent the anterior CNC cells from migrating to the anterior domain; however, once there, the CNC cells did not organize into the paired trabeculae, and did not converge to the midline to form the ethmoid plate.
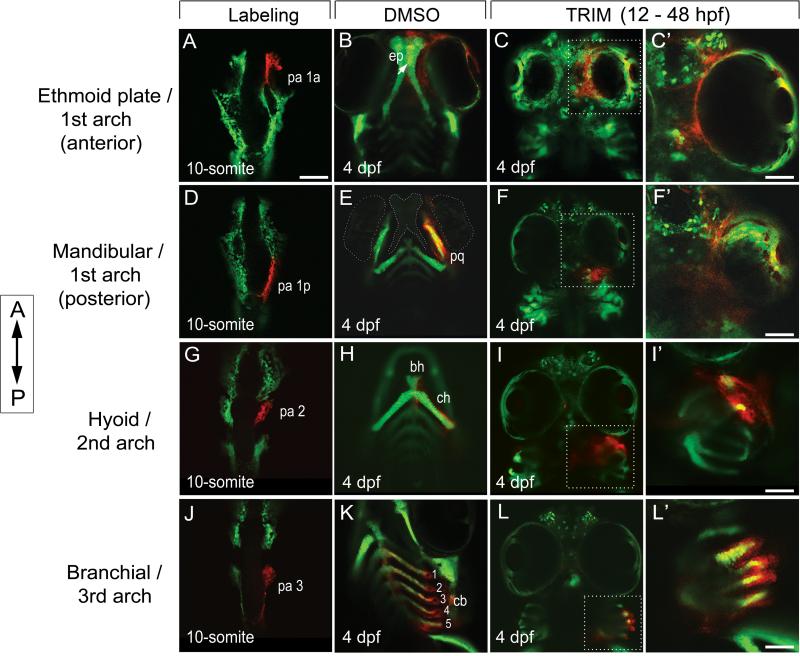
Figure 2. Ectopic cartilages were derived from malformed ceratobranchial as a result of the failure in midline convergence of CNC cells.
Photoconversion labeling of CNC cells in sox10:kaede followed from 10-somite stage (A,D,G,J) to 4 dpf, after treatment with DMSO (B,E,H,K, ventral view) and TRIM (C,F,I,L, ventral view). Anterior is to the top.
(A-C) CNC cells anterior of the eye were fated to the first PA (A, pa1a) and contributed to ep (B, arrow). In TRIM-exposed embryos, the anterior CNC cells failed to converge and condense to the midline (C). C’, enlarged view of the dotted area in C in dorsal focus.
(D-F) The posterior population of the first PA (D, pa 1p) normally populates the lower jaw structures (m, pq) (E). After TRIM treatment, the cells were sequestered in a lateralized domain posterior to the eyes (F). F’, enlarged view of the dotted area in F in dorsal.
(G-I) CNC cells in PA 2 (G) normally populate the bh, ch (H). After TRIM treatment, the cells in PA2 failed to converge in the midline and were stuck in the lateral position posterior to the eyes (I). I’, enlarged view of the dotted area in I in lateral.
(J-L) CNC cells that give rise to the third and posterior PA (J, pa3) formed paired segmented ceratobranchials at 4 dpf (K, cb 1-5). When these cells were followed in TRIM-treated embryos, they remained lateralized without midline convergence, thus failed to form the ceratobranchials (L). L’, enlarged view of the dotted area in L in lateral.
bh, basihyal; ch, ceratohyal; cb, ceratobranchial; ep, ethmoid plate; pq, palatoquadrate. Scalebars: A-L, 50 μm; C’, F’, I’, L’, 20 μm.
When the CNC population that contributes to the posterior first PA (pa 1p) was labeled in (Fig. 2D), lineage tracing confirmed that these cells normally contributed to the Meckel's cartilage and palatoquadrate in mandibular skeleton (Fig. 2E). However, after TRIM exposure, the posterior CNC group did target to the ventral domain but failed to converge to the midline. Instead, the cells were lateralized and the lower jaw structures such as Meckel's cartilage and palatoquadrate failed to form (Fig. 2F, F’).
Similarly, the CNC cells in the second and third PA were followed and found to contribute to the basihyal, ceratohyal, and ceratobranchial structures (Fig. 2G, H, J, K). However, TRIM treatment abrogated midline convergence of CNC cells to form any of the ventral jaw structures, as the cells remained sequestered in the lateral positions corresponding to these pharyngeal domains (Fig. 2I, I’, L, L’). In fact, we found that the lateralized cells that would normally form the ceratobranchials contributed to the ectopic posterior structures that was previously delineated by Alcian blue stain (Fig. 2L, L’). Taken together, our results show that the CNC populations were able to migrate to their respective PA segments. However, once localized to the proper anterior-posterior segment, the CNC cells failed to converge and condense in the midline, thus they were unable to organize into discrete skeletal structures.
TRIM treatment alters antero-posterior patterning of CNC cells
Given the different responses of TRIM treatment in anterior versus posterior CNC as to cartilaginous structures, we next examined whether this difference was due to perturbed antero-posterior patterning of the CNC. In sox10:mCherry 28 hpf embryos, CNC cells migrate and condense into segmentally organized PA structures. Compared with the clearly distinguishable arches in DMSO control, TRIM exposure resulted in tangled arches with blurring of segmental boundaries (Fig. 3A). This observation was supported by the expression profile of the endothelin type-A receptor ednra1 in the migrating CNC cells and ectomesenchymal cells (Nair, et al., 2007), demonstrating TRIM-induced defect in discrete PA patterning (Fig. 3B).
Figure 3. TRIM treatment altered CNC antero-posterior patterning.
(A-B) TRIM-induced defect in patterning of discrete PA, as demonstrated in sox10:mCherry embryos with blurring of segmental boundaries and by endra1 expression.
(C) Expressions of dlx2a (red) and hoxa2b (purple) were significantly reduced in all PA after TRIM exposure.
(D) hoxb2a expression (purple) in PA2 was decreased after TRIM treatment (black dashed line), with more dispersed and enhanced expression pattern in posterior arches (red dashed line). Early CNC marker dlx2a was used as a landmark for PA identification (C-D).
(E-F) Both of the posterior PA markers hoxb1b and hoxb3a were upregulated after TRIM treatment. Notably, the hoxb1b/b3a+ cells ectopically expanded into anterior PA regions, as normally their expression should be restricted in posterior PA 3-7.
(G) A summary of TRIM's effect on hox gene patterning in anterior versus posterior arches.
(H-K) Expressions of jag1b, hey1, edn1 (24 hpf), and bapx1 (48 hpf) in the anterior CNC are preferentially ablated (black dotted lines), whereas their posterior expressions are largely unaffected (red dotted lines). Embryos were oriented with anterior toward the left. A, anterior; P, posterior. Scalebars: A, B, H-K, 50 μm; C-F, 200 μm.
To investigate the gene regulatory mechanisms underlying the perturbed CNC patterning, we then examined the expression of pharyngeal homeobox (hox) genes, which provide spatial identity for CNC cells to elaborate PA along the antero-posterior axis (Minoux and Rijli, 2010). In particular, Hox paralogue group 2 genes participate in PA patterning, and the two members hoxa2b and hoxb2a were found to function redundantly in patterning the second and more posterior PA along the antero-posterior axis (Pasqualetti, et al., 2000). After TRIM treatment, we observed general depletion of hoxa2b expression in all PA2 and PA3-7 compared to DMSO control (Fig. 3C). In contrast, although the expression of hoxb2a was inhibited anteriorly, its posterior expression appeared to be dispersed and enhanced (Fig. 3D).
Further, we observed that TRIM treatment significantly increased the expression of posterior PA markers hoxb1b and hoxb3a, by both increasing the level of expression as well as ectopically expanding the expression domain (Fig. 3E, F). After TRIM treatment, hoxb1b+ and hoxb3a+ cells were no longer restricted in the posterior PA 3-7, but extended into the more anterior PA 1-2, leading to abnormal CNC segment identity along antero-posterior axis. Taken together, these results demonstrate that TRIM treatment altered the antero-posterior patterning of CNC by inactivating of the arch 2 hox cluster and expanding the posterior pattern to a more anterior segment (Fig. 3G).
Since TRIM-treated embryos exhibited differential defects between the anterior and posterior CNC populations, with respect to hox code genes, we also explored markers of other known pathways regulating CNC patterning (Fig. 3H-K). Among these, jagged-notch and endothelin-1 signals play key roles in dorsoventral patterning of cartilaginous skeleton, through regulation of a complex set of gene expression such as dlx3/5/6, nkx3.2 in facial skeletal precursors (Zuniga, et al., 2010). As the ligand for Notch receptor, jag1b is expressed in the dorsal arches, where the activated Notch signal in CNC-derived mesenchyme induces expression of downstream effector gene hey1. Here we found that both jag1b and hey1 expression were markedly reduced or lost in anterior arches of TRIM-exposed embryos compared with that of DMSO controls (Fig. 3H-I, black dotted line). Strikingly, expression of jag1b and hey1 was largely unaffected in the posterior arches (Fig. 3H-I, red dotted line). This data suggest that insufficient jagged-notch signal is at least partially responsible for the TRIM induced patterning defect in loss of anterior jaw structures. This observation was further supported by analysis of the ectoderm and pharyngeal pouch development. Edn1 is primarily secreted from the PA ectoderm (also from paraxial mesoderm and pharyngeal pouch endoderm), and it participates with homeobox genes in patterning of intermediate and ventral arch domains (Medeiros and Crump, 2012). We observed that edn1 expression is dramatically decreased in the anterior ectoderm and pharyngeal pouch in TRIM treated embryos (Fig. 3 J, black dotted line). Similarly, its downstream effector bapx1, which is primarily required for mandibular arch skeleton and joint development, is entirely missing in the anterior domain, whereas persistent but fused bapx1 expression in the posterior ceratobranchials was detected, partially recapitulating TRIM's Alcian blue phenotype (Fig. 3K).
It is important to note that expression of hoxa2b and hoxb2a were reduced by TRIM treatment, with ectopic expression of hoxb1b and hoxb3a in the anterior domain, and these changes were associated with reduction of jagged-notch and endothelin-1 signal from anterior ectoderm and pharyngeal endoderm. One interpretation of this finding is that ectopic hox expression in the first 2 arches inhibits skeletal development, as evidenced from expanded expression of hoxa1 and hoxa2b from previous study (Alexandre, et al., 1996). Further, since the pharyngeal ectoderm and endoderm provide the major sources of signals (e.g. notch; edn1) to guide cell survival, proliferation, migration, and differentiation within adjacent CNC cells, this lead us to examine if these CNC cell behaviors were consequently affected in TRIM context, thereby resulting in skeletal hypoplasia and jaw defects in TRIM phenotype.
TRIM disrupts chondrocyte-lineage differentiation and promotes CNC progenitor character
We carried out marker gene analysis to better define the effect of TRIM on CNC cell development. Leflunomide, a reported chemical to completely abrogate neural crest development and inhibit melanoma growth, was also identified in this screen and therefore used as a positive control (Fig. 1E). At 24 hpf, TRIM treated embryos show a robust expansion in the number of crestin+ progenitors, as opposed to the inhibitory effect of leflunomide (Fig. 4A). In CNC development, crestin is normally downregulated after the terminal differentiation of CNC progenitors. This finding therefore suggests that TRIM treatment promotes the maintenance or quiescence of CNC progenitors, which in turn leads to onset latency of genes required for CNC migration and downstream lineage differentiation.
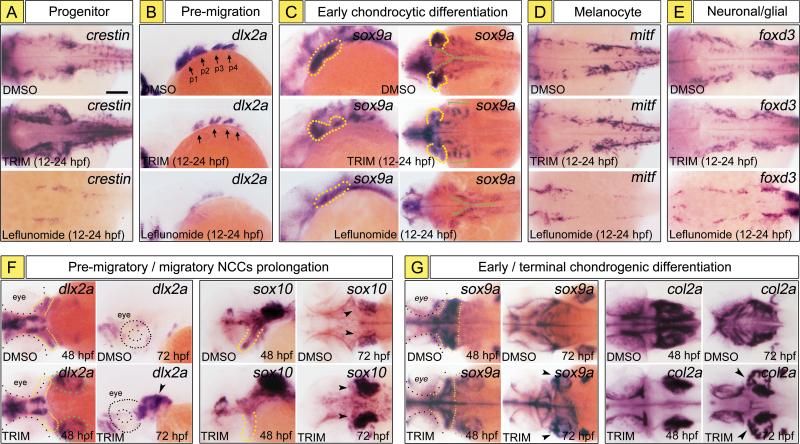
Figure 4. TRIM promotes a CNC progenitor cell fate and disrupts chondrogenic differentiation.
(A-E) CNC marker analysis at 24 hpf demonstrate that TRIM treatment expanded the number of crestin+ CNC progenitors (A), inhibited dlx2a+ migratory cells (B, arrows) and the sox9a+ pre-chondrogenic cells (C, yellow dotted) in all PA. However, the mitf + melanocytes (D) and foxd3+ neuronal/glial cells (E) were not seriously affected by TRIM. Notably, sox9a expression was localized to an aberrant lateralized location (C, green brackets) compared to the ventral-lateral distribution pattern in DMSO control. Conversely, leflunomide exerted a more general and profound depletion effect on all CNC cells and their derivatives (A-E, bottom panel).
(F) At 48 and 72 hpf, TRIM treatment caused loss of dlx2a+ or sox10+ migratory cells in the anterior PA (yellow dotted), whereas aberrant dlx2a+ or sox10+ CNC cells were sequestered in the posterior domain and retained in a progenitor state (green dotted, arrowheads).
(G) At 48 and 72 hpf, chondrogenic differentiation of sox9a+ CNC cells or terminal col2a+ chondrocytes in anterior PA was significantly inhibited by TRIM (yellow dotted). Instead, ectopic posterior expression of sox9a and col2a was trapped in the lateralized ceratobranchial (green dotted, arrowheads).
Scalebar: 100 μm.
Indeed, this explanation was evidenced by our observations in gene expression of the pre-migratory marker dlx2a, as well as the early chondrocyte-lineage differentiation marker sox9a (Fig. 4 B, C). CNC gene dlx2a is normally expressed in 4-paired PA along the dorsoventral axis. However, with TRIM treatment, dlx2a expression was significantly reduced (Fig. 4B, arrows), indicating TRIM induced migratory defect in CNC cells. Moreover, TRIM-treated embryos showed significant reduction of sox9a expression in anterior arches, especially in the first 2 anterior migratory streams corresponding to the first arch (Fig. 4C, dotted area), demonstrating dramatically decreased chondrogenic differentiation of CNC progenitors. Unexpectedly, we found that TRIM treatment also resulted in ectopic expression of sox9a, where 4-paired cell clusters emerged dorsolaterally (Fig. 4C, brackets), compared with the normal ventral-lateral migration pattern of sox9a+ cells in DMSO control (Fig. 4C, green dotted line). We speculated that these sox9a+ cell clusters were resulted from aberrant migration of the CNC cells that exit the posterior PA to form the ceratobranchials.
It's important to note that, unlike leflunomide, TRIM treatment affected the developmental process that directs CNC progenitors to chondrocyte lineage, as we didn't observe significant decrease of mitf and foxd3, which was selectively expressed in CNC cells and required to differentiate melanocyte and neuron/glial lineages (Pavan and Raible, 2012), respectively (Fig. 4D, E).
To further explore our findings, which suggested that TRIM treatment at 24 hpf caused a reduction in chondrogenesis in anterior CNC and aberrant CNC marker expression in posterior segments, we examined CNC migration and skeletal marker expression at later timepoints of craniofacial form. At 48 hpf, dlx2a expression was reduced in anterior CNC in TRIM-treated embryos compared with controls (Fig. 4F, yellow dotted). However, robust dlx2a expression was detected in the posterior CNC population anterior to the otic vesicle (Fig. 4F, green dotted). Normally at 72 hpf there is little CNC cell migration and dlx2a expression in CNC is almost turned off, whereas under TRIM treatment, even high level of dlx2a expression persisted aberrantly in the pharyngeal domain (Fig. 4F, arrowhead).
Consistent with prolonged dlx2a expression in the posterior CNC population with TRIM treatment, we observed almost same expression pattern of CNC progenitor and migratory marker sox10. In 48 hpf control embryos, sox10+ cells had migrated below the eyes into the pharynx to further contribute to the jaw cartilage. However in TRIM-exposed embryo, the pharyngeal sox10 expression did not occur (Fig. 4F, yellow dotted). By 72 hpf, TRIM treatment resulted in high and sustained sox10 expression in the posterior dorsolateral cell population, similar to that of dlx2a pattern (Fig. 4F, arrowheads). Taken together, the ectopic and persistent dlx2a/sox10 expression suggests that the CNC cells that were sequestered in the posterior domain retained in a progenitor state.
To examine chondrogenic differentiation of the aberrant dlx2a+/sox10+ CNC cells, we analyzed the expression of early and terminal chondrogenic markers sox9a and col2a, respectively. We found that TRIM treatment reduced the number of anterior sox9a+ cells, which normally reside in PA and contribute to jaw skeleton at 48 hpf (Fig. 4G, yellow dotted). This finding is consistent with the loss of anterior skeletal structures such as the ethmoid plate and Meckel's cartilage in TRIM phenotype. However, we also found there was aberrant sox9a expression in the ectopic dorsolateral CNC at both 48 hpf and 72 hpf (Fig. 4G, green dotted, arrowhead). Further, these sox9a+ cells can undergo terminal chondrogenic differentiation, as evidenced by col2a expression in the paired ceratobranchial structures that we previously characterized by Alcian blue and lineage tracing analysis (Fig. 4G, arrowheads). Collectively, these results demonstrate that ectopic chondrogenesis in TRIM-exposed embryos was prefigured by aberrant dlx2a/sox10 expression in posterior CNC, and that loss of craniofacial skeleton in TRIM phenotype was caused, at least partially, by severe inhibition of cell migration and chondrogenic differentiation in anterior CNC.
CNC cell survival or proliferation does not account for TRIM phenotype
Changes in gene expression and loss of craniofacial structures following TRIM treatment could also be caused by generalized decrease in CNC cell survival and/or cell proliferation (Kamel, et al., 2013). We assayed cell apoptosis despite that the increased number of crestin+ cells provides strong evidence to help exclude the possibility of TRIM-induced cell death due to toxicity (Fig. 4A). At 28 hpf, when TRIM-induced decreases in dlx2a/sox9a expression in PA1-2 became evident, there was no significant increase in cell death (Fig. S5, A-L). We examined whether the TRIM-mediated phenotype is caused by impairment in CNC cell proliferation and found no discernible difference in mitotic rate in CNC cells (Fig. S5, M-T). Together, these results exclude generalized apoptosis and decreased proliferation to account for the failure of the CNC cells to contribute to the craniofacial skeleton.
Potentiating histone acetylation counteracts TRIM-induced craniofacial defect
In order to further understand the molecular basis of TRIM-induced reduction in CNC marker expression (e.g. hoxa2b, Fig. 3C; dlx2a, Fig. 4B), we asked whether there are the epigenetics mechanisms that are involved in TRIM and normal craniofacial developmental context. Epigenetic CNC gene is mediated at least in part by histone acetyltransferase KAT6A and histone binding proteins. Since decrease in hox2 gene expression was both observed with TRIM treatment and in KAT6A mutant, we first examined whether TRIM defect was partially mediated by changes in histone acetylation in CNC.
By analyzing the nuclei lysates from TRIM-treated embryos with DMSO control, we detected a significant decrease in the level of acetylated histone H4 by 50% (Fig. 5A). We next measured the direct effect of TRIM exposure on HAT activity, which catalyzes the transfer of acetyl groups to core histone. Compared to DMSO control, a dose-dependent inhibition effect was detected in TRIM-treat embryos (Fig. 5B, left). These data together indicates that, in TRIM treated embryos, attenuated HAT action may at least partially account for the unbalanced status of histone hypoacetylation, which consequently leads to transcriptional inactivation of genes required in early CNC development. To test this hypothesis, we took the gain-of-function approach by genetic interference of HAT expression or chemical interference of HDAC activity, to investigate if targeted acetylation could compromise TRIM-induced defect in CNC gene expression and the resulting skeletal form (Fig. 5B, middle).
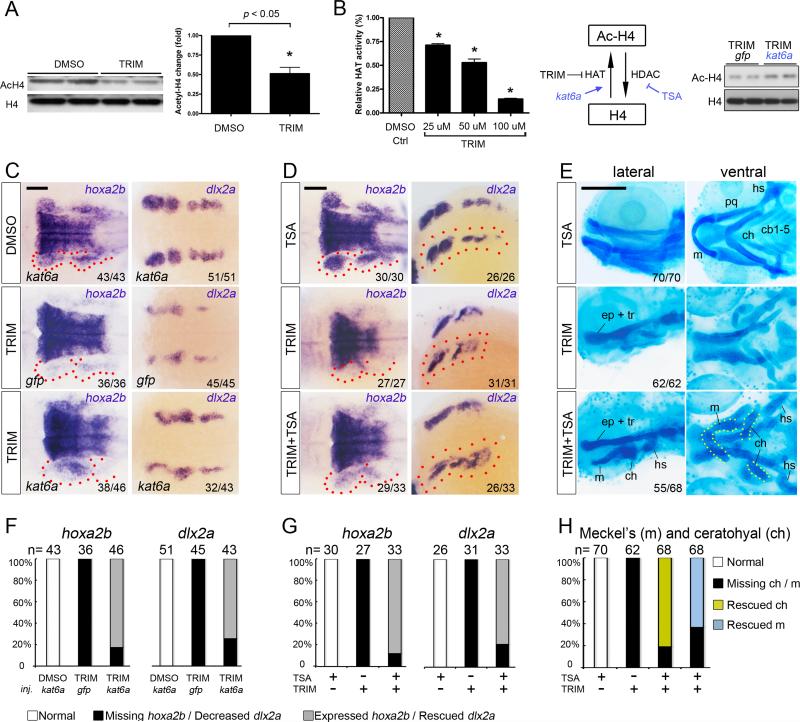
Figure 5. Potentiating histone acetylation counteracts TRIM-induced craniofacial defect.
(A) Impaired histone H4 acetylation was detected in TRIM-treated embryos compared to DMSO control. Quantification data is presented as mean ± s.e.m. *P < 0.05.
(B) Treatment with TRIM inhibited the activity of histone acetyltransferase in a dose-dependent manner. Injection of kat6a mRNA partially counteracts TRIM-induced hypoacetylation.
(C) Overexpression of kat6a mRNA antagonizes TRIM-induced genetic defect by partially rescuing expression of CNC marker hoxa2b and dlx2a in PA (24 hpf; middle versus bottom panels). Scalebar 100 μm.
(D) Chemical potentiation of histone acetylation by TSA partially restored TRIM-induced hoxa2b and dlx2a depletion in PA (24 hpf; middle versus bottom panels). Scalebar 100 μm.
(E) TRIM-induced skeletal abrogation was partially rescued by TSA co-treatment, as demonstrated by formation of the 1st and 2nd PA cartilage structures, including the Meckel's cartilage (m), ceratohyal (ch), and hyosymplectic (hs) (96 hpf, middle versus bottom panels). Scalebar 250 μm.
(F, G, H) Quantification of the rescuing phenotype observed in C, D, E, respectively. The statistical data are presented as percentage of embryos with corresponding phenotype.
First, we augmented HAT activity by coinjection of kat6a mRNA into TRIM-exposed embryos, which correspondingly enhanced total histone acetylation (Fig. 5B, right). As expected, overexpression of kat6a mRNA compromised TRIM-induced hoxa2b depletion in PA flanking the rhombomeres, compared to embryos injected with gfp mRNA control (Fig. 5C, F, red dotted). Similarly, kat6a overexpression also achieved partial rescue of dlx2a expression in TRIM treated embryos (Fig. 5C, F). This data suggests that sufficient HAT activity is required for recovered expression of key CNC genes.
Correspondingly, since HDAC-mediated histone deacetylation generally represses transcription, we tested whether co-treatment of Trichostatin A (TSA), an inhibitor of classes I and II HDAC, could antagonize the deacetylation process and consequently rescue the TRIM-induced craniofacial defect. Consistent with HAT overexpression result, TSA and TRIM co-treatment partially restored the expression of hoxa2b and dlx2a in the pharyngeal CNC cells (Fig. 5D, G, red dotted). Intriguingly, we observed that TSA co-treatment with TRIM mitigated the loss of chondrogenic development observed with TRIM alone, as evidenced by partial rescue of the 1st and 2nd pharyngeal cartilage structures, including the Meckel's cartilage, ceratohyal, and hyosymplectic (Fig. 5E, H). Taken together, these data suggest that either chemical or genetic means to augment histone acetylation can mitigate the inhibitory effect of TRIM on CNC gene expression and craniofacial morphogenesis, providing evidence to support a role for histone acetylation to regulate in CNC and skeletal development.
Nitric oxide signal promotes histone acetylation and is critical for CNC and craniofacial development
Previous in vitro physiological and toxicological studies have indicated that TRIM is a potent chemical inhibitor of both nNOS and iNOS (Handy and Moore, 1997). To elucidate the relationship between TRIM and histone acetylation, we next examined whether TRIM-induced NOS inhibition coordinates with impaired acetylation to play a role in the disrupted CNC form. Compared to DMSO control, TRIM treatment resulted in significant decrease of NO production in the pharyngeal region (Fig. 6A). To further validate this data, we next introduced the NO donor S-nitroso-N-acetyl-DL-penicillamine (SNAP) for rescue, and indeed, SNAP co-treatment with TRIM robustly restored endogenous NO production to the normal level as in control embryos (Fig. 6A). Quantification of NO labeling was reflected by the percentage change compared to DMSO control embryos (Fig. 6B).
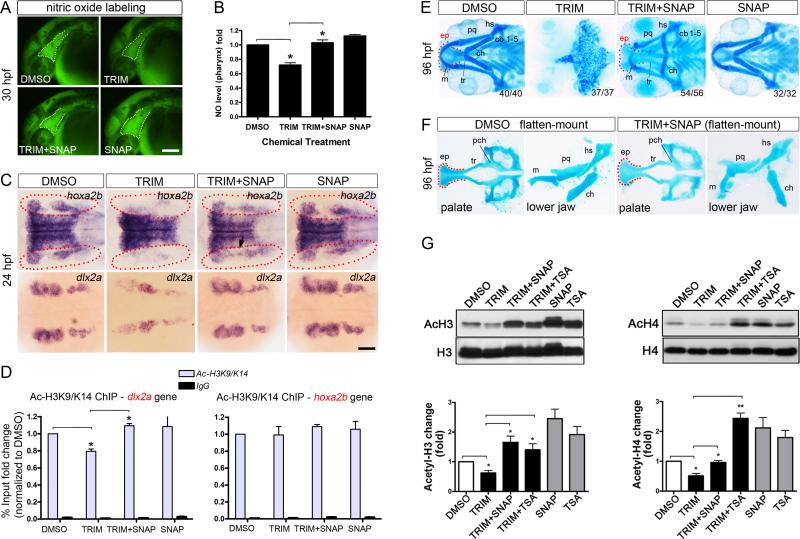
Figure 6. Nitric oxide signal plays a critical role in CNC development via coordinating with histone acetylation.
(A-B) TRIM treatment decreased endogenous NO production in pharynx compared to DMSO control (30 hpf; dotted area). Co-treatment with the SNAP (NO donor) rescued TRIM-induced NO insufficiency. SNAP treatment alone has an enhanced effect on NO production.
(C) Potentiating NO production by SNAP robustly recovered TRIM-induced hoxa2b depletion and dlx2a downregulation in PA at 24 hpf (TRIM versus TRIM+SNAP).
(D) ChIP assay using acetylated H3K9/K14 were performed as the conditions in (C). Co-treatment with SNAP robustly recovered TRIM-induced decrease in histone H3K9/14 acetylation of the CNC gene dlx2a.
(E) SNAP successfully rescued TRIM-induced craniofacial abrogation, as demonstrated by fully restored structures in both upper (ep, tr) and lower jaw (m, pq, hs, ch, cb) at 96 hpf.
(F) SNAP-mediated rescue of TRIM phenotype had fully accomplished palate (ep, tr) as well as the mesodermally derived parachordal (pch). Lower jaw elements were fully developed except that the apex of the ceratohyal (ch) points caudal rather than cephalic.
(G) Western blot of acetylated H3/H4 and quantification (normalized to total histone). TRIM-induced histone hypoacetylation (DMSO versus TRIM) was rescued by SNAP co-treatment (TRIM versus TRIM+SNAP). Data are expressed as mean ± s.e.m. *P < 0.05, **P < 0.01.
Scalebar 100 μm (A, C).
We next evaluated if changes in NO level is associated with CNC gene expression. By 24 hpf, SNAP co-treatment significantly rescued the TRIM-induced hoxa2b depletion in CNC cells almost to the wildtype level, albeit with blurring of the anterior and posterior PA boundary (Fig. 6C, red dotted). Meanwhile, large numbers of dlx2a+ cells were also restored and shaped back to normal pattern (Fig. 6C, TRIM versus TRIM+SNAP). This data demonstrated that potentiating NO production successfully recovered the expression of patterning and migratory genes in CNC, which are indispensable for craniofacial development. Employing chromatin immunoprecipitation (ChIP) assays, we found that TRIM-treated embryos exhibit decreased H3K9K14 acetylation in the promoter of dlx2a gene, which was subsequently recovered by SNAP cotreatment (Fig. 6D), indicating NO level is associated with CNC gene expression and histone acetylation. However, we did not detect such changes in the hoxa2b gene promoter, likely due to the predominant hoxa2b expression in the rhombomeres masking the down-regulation in the CNC we were able to detect by RNA in situ. Most strikingly, SNAP-mediated rescue restored all the cartilaginous structures of both upper (ep, tr) and lower jaw (m, pq, hs, ch, cb) at 96 hpf (Fig. 6E), whereas TRIM-treated embryo only formed scattered chondrocyte clusters. In a flatten-mount view, embryos co-treated with SNAP and TRIM had fully developed PA1-derived palate as well as the mesodermally derived parachordal (pch), to the same extent as DMSO control (Fig. 6F). Further, all the lower jaw elements (m, pq, hs, ch) were intact except that 2nd arch derived ceratohyal (ch) was malpositioned caudally rather than cephalically (Fig. 6F). In addition to SNAP, we tested S-nitrosothiol type of NO donor such as S-Nitrosoglutathione (GSNO) and observed similar potent effect in rescue of TRIM-induced craniofacial defects, but diazeniumdiolates (NONOates) type of NO donors (e.g. SPER/NO, DETA-NO), failed to rescue TRIM phenotype (data not shown). Together, these results demonstrate that TRIM phenotype resulted from impaired NO production in vivo, suggesting sufficient NO signal is critical in maintenance of CNC gene expression and craniofacial morphogenesis.
Having established the involvement of NO in TRIM-mediated craniofacial malformation and phenotypic rescue, we then examined the coordinated effect between NO and histone acetylation. Compared to DMSO control, we found that NO donor SNAP significantly increased the levels of acetylated histones H3 and H4 by at lease 2 folds, which was functionally equivalent to TSA (Fig. 6G). As a result, SNAP co-treatment with TRIM achieved greatly elevated levels of histone acetylation compared to TRIM treatment alone (Fig. 6G, TRIM versus TRIM+SNAP), thus providing one possible epigenetic explanation as to the restored transcriptional activation such as hoxa2b and dlx2a in SNAP-rescued embryos. Consistent with our data in manipulation of HAT/HDAC activity for TRIM rescue (Fig. 5), these results together suggest a model where NO signal and histone acetylation are coordinated to regulate CNC development.
In addition to histone acetylation, NO also mediates dynamic post-translational modification of proteins through S-nitrosylation (Schonhoff and Benhar, 2011). We next examined whether TRIM and NO donor (SNAP, GSNO) treatment altered total protein S-nitrosylation, to determine the extent of CNC development attributable to alternative NO-mediated mechanisms. Using biotin-switch assay, we did not observe any difference in the degree of total protein S-nitrosylation between DMSO and TRIM treatment groups, or between TRIM and SNAP/GSNO rescued groups (Fig. S7). This result indicates that NO-mediated nitrosylation is not the primary mechanism compared to alterations of histone acetylation in generating the TRIM phenotype.
To further evaluate the significance of NO signal on CNC and craniofacial development, we took the genetic loss-of-function approach by performing morpholino oligonucleotides (MO)-mediated gene knockdown of both nos1 (nNOS) and nos2 (iNOS) to attenuate NO production and histone acetylation (Fig. 7 B, C, Fig. S6). At 48 hpf, we observed dramatic development defect in both cranial and spinal skeleton in nos1+nos2 morphant (Fig. 7A, upper panel). Further, nos1+nos2 knockdown led to loss of facial soft tissues and shortened upper jaw at 96 hpf (Fig. 7A, arrow in lower panel). Compared to control, all the jaw elements in nos1+nos2 deficient embryos appear to be hypoplastic, especially with a truncated ethmoid plate that failed to fully develop (Fig. 7D, MM versus MO). Consistently, expression of migratory gene dlx2a and pattern gene hoxa2b in PA was markedly decreased, which was partially recapitulated in TRIM phenotype (Fig. 7E, MM versus MO). Moreover, the nos1+nos2 knockdown defects in craniofacial phenotype and expression of CNC genes were partially rescued by co-treatment with NO donor SNAP (Fig. 7 D, E). Taken together, these results suggest a critical role of NO signal in maintenance of CNC gene activity for craniofacial development.
Figure 7. Genetic validation of nitric oxide signal in CNC development.
(A-E) Loss-of-function study of nos1/nos2 reveals the critical role of NO signal in CNC development. Compared to mismatch (MM) control, morpholino (MO) knockdown of nos1/nos2 resulted in dramatic developmental defect in cranial and spinal skeleton (A), NO deficiency (B), histone hypoacetylation (C), hypoplastic jaw structure (D), and decreased or even loss of gene expression in PA (E). The nos1/2 morphant partially recapitulates the TRIM phenotype, and can be rescued by SNAP cotreatment (D, E).
(F) Schematic diagram summarizes the coordinated regulation of NO signal and histone acetylation in CNC development. In response to TRIM-induced NO deficiency, HAT activity is insufficient, resulting in histone hypoacetylation. Red color stands for TRIM's inhibitory effect on histone acetylation. Genetic enhancement of HAT (kat6a overexpression), chemical interference of HDAC (TSA), or complementary NO (SNAP) (labeled in green) can reverse this defect and promote acetylation, which presumably favors the transcriptional activation of key genes (hoxa2b, dlx2a) required in craniofacial development, and ultimately achieves partial or almost complete rescue of TRIM phenotype.
ch, ceratohyal; cb, ceratobranchial; ep, ethmoid plate; hs, hyosymplectic; m, Mechel's; pq, palatoquadrate. Scalebar: A (upper, 500 μm; lower, 100 μm); B, E, 100 μm.
DISCUSSION
Application of chemical screen in a developmental context provides complementary approach to gain molecular insight into developmental mechanisms. We employed assays of crestin expression in CNC progenitors and morphologic analysis, where the latter screen focused on a subset of compounds with annotated bioactivity. TRIM became the ideal candidate as it caused a graded craniofacial disruption, resulting in loss of anterior PA derivatives (ethmoid plate, trabecula, and mandible) and formation of ectopic posterior structures (Fig. 1E). Identification of the targeted biological pathway is a core challenge in chemical genetics. In this study, we thoroughly characterized the TRIM-induced craniofacial defect, combined with chemical and genetic manipulations uncovering histone acetylation and nitric oxide synthase as the relevant pathways.
Mechanistically, loss of craniofacial skeleton and gain of ectopic cartilage following TRIM treatment could be reasoned by several developmental events, such as aberrant migration of CNC cells to proper segments along the antero-posterior axis, failure in identity specification within arch segments, changes in cell proliferation and survival, and/or defects in chondrogenic differentiation. In our stepwise approach to identify the biologic target, two basic questions need to be answered first - where is the ectopic cartilage come from and why the anterior skeleton fail to form. From lineage tracing we determined that malformed ceratobranchial (PA 3-7 derived) contributed to the ectopic cartilage, whereas failed midline convergence of PA 1-2 populations accounted for the missing jaw elements. More importantly, these different responses in anterior versus posterior to TRIM treatment indicated a patterning defect in antero-posterior axis, which was subsequently illustrated by gene expression analysis of two groups of patterning pathway: hox codes and notch/endothelin genes. Our study suggested that ectopic hox expression in the first 2 arches inhibits pharyngeal arch development, as further evidenced by missing key signals including jagged-notch and edn1-bapx1 in the anterior pharyngeal ectoderm, endoderm, and CNC-derived mesenchyme (Fig. 3H-K). TRIM-induced deficiency of signals could be disastrous to the adjacent CNC cells, where normally the process of cell migration, multi-lineage differentiation in elaborating the craniofacial form was profoundly influenced by wealth of signals in these cranial microenvironment (Ragland and Raible, 2004).
Interestingly, CNC marker analysis indeed demonstrated that chondrogenic differentiation of CNC cells is much vulnerable to TRIM treatment. The expansion of crestin+ progenitors excludes the generalized cell death or TRIM toxicity, meanwhile it suggests another possible mechanism as to quiescence or onset latency of gene expressions (e.g. hoxa2b, dlx2a, sox9a) that are required to direct CNC progenitor to cranial chondrocytic lineage. This explanation was supported by our observation of aberrant dlx2a, sox10, and sox9a expressions in later developmental stages (48 and 72 hpf, Fig. 4F, G). Further, gene inactivation could be a subsequent effect of TRIM-induced impaired histone acetylation. Our gain-of-function approach by potentiating histone acetylation achieved partial rescue of CNC genes (kat6a, TSA in Fig. 5), and established the critical role of histone acetylation in regulating craniofacial development. The strongest rescue of TRIM phenotype was achieved by manipulation of the NO production using SNAP or GSNO. Complementary NO potently rescued TRIM-induced gene and phenotypic defect, which was shown to be coordinated with gain-of-function effect in histone acetylation (Fig. 6G). However, we noticed that TRIM+SNAP rescue compares favorably with TRIM+TSA rescue, indicating that NO might function upstream of histone acetylation and/or through non-acetylation pathways (e.g. through S-nitrosylation, or NO may directly target on the expression of chondrogenic genes). These data collectively suggest an important role for NO signal in CNC development. The importance of NO signal was further evidenced by loss-of-function study, where combined knockdown of nos1+nos2 lead to severe craniofacial anomaly in zebrafish. Taken together, these studies proposed a novel mechanism that NO signal and histone acetylation are coordinated mechanisms regulating CNC patterning and craniofacial morphogenesis.
Epigenetic regulation of craniofacial formation and malformation
The developing CNC cells must be patterned, undergo specific migratory paths, and coalesce to facial prominences that undergo convergence and extension to generate the craniofacial form. The important role of epigenetic regulation in these processes is underscored by several lines of evidence. First, several enzymes that catalyze in histone modification regulate craniofacial development, such as histone deacetylase 4 (hdac4) and acetyltransferases kat6a (Crump, et al., 2006; Delaurier, et al., 2012;). Next, human mutations disrupting histone acetylation (MYST4) demethylation (KDM6A, PHF8), and sumoylation (SUMO1) result in orofacial clefts (Alkuraya, et al., 2006; Fischer, et al., 2006; Kraft, et al., 2011; Qi, et al., 2010). Further, orofacial clefts such as the common cleft lip and palate are usually non-syndromic and exhibit non-Mendelian patterns of inheritance, where epigenetic effect on the penetrance of a genetic predisposition presents a possible explanation (Spritz, 2001). Lastly, population studies demonstrate environmental contribution to the occurrence of orofacial clefts, where epigenetic modifications may serve as the mediating factor. Therefore, application of chemical genetics to uncover the role of NO signaling and histone acetylation in CNC development highlights a central developmental process that may have broader implications in organogenesis at large.
It is noteworthy that disruption of general epigenetic mechanisms can result in specific phenotypes, rather than global failures in organogenesis or embryonic lethality. Mutations in histone demethylases (PHF8 and KDM6A), sumoylation (SUMO1) result in cleft palate. Mutations in zebrafish kat6a resulted in homeotic transformation of PA2 identity (Crump, et al., 2006). Knockdown of histone deacetylase 4 (hdac4) produced discrete defects in the fusion line between the median ethmoid plate and the trabeculae, analogous to a cleft between the frontonasal and maxillary processes of amniotes (Delaurier, et al., 2012). Therefore, examples in clinical presentation and in animal model phenotypes corroborate that disruption of general epigenetic mechanisms may result in specific orofacial anomalies.
Nitric oxide as second messenger regulating CNC development
NO influences gene expression in a very general and profound manner, thus NO is implicated in many physiologic and pathologic processes, ranging from vascular homeostasis and hematopoiesis, to atherosclerosis and carcinogenesis (Foster, et al., 2009; North, et al., 2009). Here we show that treatment of developing embryos with TRIM, a competitive inhibitor of NOS, altered hox gene patterning, promoted CNC progenitor fate but did not affect the general migratory trajectory of CNC cells from early somite stages to their pharyngeal segment. However, the uncoupling of normal CNC cell differentiation and migration resulted in loss of ethmoid plate and lower jaw structures, and led to ectopic formation of posterior elements that normally contribute to the ceratobranchials. During hematopoiesis, it was recently demonstrated that nos1 is required in a cell-autonomous manner for hematopoietic stem cell (HSC) development, and that NO donors regulated HSC number independent of blood flow (North, et al., 2009). Therefore in CNC development, as in hematopoiesis, NO levels appear to fine tune cell fate.
In regard to orofacial cleft pathogenesis, identification of NO to regulate CNC behavior links placental circulation, fetal stress, maternal and environmental effects to CNC development, not only in providing a pathophysiological basis, but also in identifying potential pharmacologic strategies to prevent orofacial clefts.
Further, we show that in together with NOS inhibition and NO deficiency, TRIM exerts its effect on hox gene patterning and craniofacial morphogenesis through histone acetylation. Therefore, it raises the question as to whether NO-mediated S-nitrosylation of histones is also affected by TRIM, implicating S-nitrosylation as another mechanism regulating CNC development. Using the biotin switch assay, we did not detect a defect in S-nitrosylation of total protein with TRIM treatment. Future work aims to parse out whether S-nitrosylation of specific histone modifying enzymes or transcription factors regulating CNC development may be affected.
MATERIALS AND METHODS
Zebrafish husbandry and genetic strains
Zebrafish were raised and maintained under established protocols as per Subcommittee on Research Animal Care, Massachusetts General Hospital. Sox10:egfp, sox10:mCherry and sox10: Kaede lines were generated using a 7.2 kb sox10 promoter.
Wholemount RNA in situ hybridization and cartilage staining
Wholemount RNA in situ hybridization was performed as described (Kamel, et al., 2013). Imaging of zebrafish cartilage was achieved by Alcian blue staining, captured using Nikon SMZ1000 and Nikon Eclipse 80i microscopes.
Small molecule screen
Over 2,980 compounds were screened from the ICCB Known Bioactives Library (Enzo), including additional compounds identified from a chemical screen (BIOMOL 480, Sigma LOPAC1280, and the Children's Hospital Boston Chemical Screening facility) based on crestin expression (White, et al., 2011). In phenotypic screen, embryos were exposed to chemicals from 5-somite to 48 hpf, with average concentration of 10-100 μM. After treatment, embryos were rinsed and incubated in E3 to 4 dpf for Alcian blue stain.
Nitric oxide donors SNAP (100 μM), GSNO (100 μM), SPER/NO (25 μM), and DETANO (180 μM) were used in the chemical rescue experiments and S-nitrosylation assay. TRIM treatment at 30 μM generates consistent phenotype.
Lineage tracing analysis
CNC cells in regions of interest were photoconverted at 10-somite stage in sox10:kaede embryo using a Nikon Eclipse Ti AIR Confocal microscope with a 404 nm laser. Green and red fluorescence signals were captured simultaneously by using the 488 nm and 562 nm laser wavelengths, respectively (Dougherty, et al., 2012).
mRNA, morpholinos, and microinjection
Kat6a mRNA was synthesized using mMESSAGE mMACHINE T7 Ultra Kit (Ambion), and purified using the MEGAclear Kit (Ambion). nos1 (5’ACGCTGGGCTCTGATTCCTGCATTG) and nos2 (5’AGTGGTTTGTGCTTGTCTTCCCATC) morpholinos were synthesized by GeneTools. About 2 nl mRNA or morpholino solution was injected into 1-cell stage embryo.
In vivo nitric oxide labeling and S-nitrosylation by biotin-switch assay
NO labeling was performed as described (Lepiller, et al., 2007). Fluorescent images of DAF-FM-DA labeled embryos were taken using Nikon smz1000 stereomicroscopes with appropriate filter (EX 470/40). Fluorescence intensities of pharynx labeling were measured using ImageJ (NIH) software.
The biotin-switch assay was used to detect S-nitrosylated proteins from the embryos extract, via specifically labelling nitrosylated cysteines with a biotin moiety as previous described (Schonhoff and Benhar, 2011). The covalently labeled biotin in targeted proteins was further detected by western blot.
Histone protein extraction, western blotting, and HAT activity assay
Total Histone protein extraction was performed according to Abcam protocol online. Western blotting was performed using antibodies specific for histone H3 (cat. 9715), histone H4 (cat. 2592), acetyl-histone H3 (Lys9/Lys14) (cat. 9677), acetyl-histone H4 (Lys5) (cat. 9672) (Cell Signaling). The effect of TRIM-exposure on HAT activity was detected in vitro using the Histone Acetyltransferase Activity Assay Kit (Abcam, ab65352).
Chromatin immunoprecipitation (ChIP) assay
ChIP assay was performed using whole embryos and indicated antibodies. Embryos were treated from 12 to 24 hpf for sample preparation. Real-time PCR analysis was performed using primers for the promoter region of dlx2a (5’TTCATGATTGACCACGCATT3’; 5’ TGTGTGGCGATGGTAAACTG3’); and hoxa2b (5’ATTTGTCTACGCGCAATGTG3’; 5’TCCCATTAATCCCGAGTCTG3’).
Supplementary Material
SIGNIFICANCE.
The work described here reports the first application of chemical genetics toward the study of CNC cells and craniofacial morphogenesis, and uncovers NO signaling as an important regulatory component in early embryonic development. Elucidating the mechanistic action of TRIM demonstrates that NO signaling and histone acetylation are coordinated mechanisms that regulate CNC patterning, differentiation and convergence during craniofacial morphogenesis. This study also demonstrates the utility of zebrafish model to discover compounds that can be developed for pharmacologic manipulation of craniofacial development.
ACKNOWLEDGEMENTS
We are grateful to Jenna Galloway (Massachusetts General Hospital) for detailed review of the manuscript, and Renee Ethier for excellent management of our aquatics facility. We thank Lei Zhong, Jinzhong Qin and Chiachi Sun for their helpful technical assistance. This work was funded by grants to E.C.L. from the March of Dimes Basil O'Connor Award, Plastic Surgery Foundation, American Surgical Association and Shriners Hospitals for Children. Y.K. was funded by Shriners Hospitals for Children Research Fellowship Award. L.I.Z. was supported by HHMI and NIH/NCI R01 CA103846. L.I.Z. is a founder and stockholder of Fate, Inc., a founder and stockholder of Scholar Rock, and a scientific advisor for Stemgent.
REFERENCES
- Alexandre D, Clarke JD, Oxtoby E, Yan YL, Jowett T, Holder N. Ectopic expression of Hoxa-1 in the zebrafish alters the fate of the mandibular arch neural crest and phenocopies a retinoic acid-induced phenotype. Development. 1996;122:735–746. doi: 10.1242/dev.122.3.735. [DOI] [PubMed] [Google Scholar]
- Alkuraya FS, Saadi I, Lund JJ, Turbe-Doan A, Morton CC, Maas RL. SUMO1 haploinsufficiency leads to cleft lip and palate. Science. 2006;313:1751. doi: 10.1126/science.1128406. [DOI] [PubMed] [Google Scholar]
- Crump JG, Swartz ME, Eberhart JK, Kimmel CB. Moz-dependent Hox expression controls segment-specific fate maps of skeletal precursors in the face. Development. 2006;133:2661–2669. doi: 10.1242/dev.02435. [DOI] [PubMed] [Google Scholar]
- Delaurier A, Nakamura Y, Braasch I, Khanna V, Kato H, Wakitani S, Postlethwait JH, Kimmel CB. Histone deacetylase-4 is required during early cranial neural crest development for generation of the zebrafish palatal skeleton. BMC Dev Biol. 2012;12:16. doi: 10.1186/1471-213X-12-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougherty M, Kamel G, Shubinets V, Hickey G, Grimaldi M, Liao. EC. Embryonic Fate Map of First Pharyngeal Arch Structures in the sox10:kaede Zebrafish Transgenic Model. Journal of Craniofacial Surgery. 2012 doi: 10.1097/SCS.0b013e318260f20b. [DOI] [PubMed] [Google Scholar]
- Fischer S, Ludecke HJ, Wieczorek D, Bohringer S, Gillessen-Kaesbach G, Horsthemke B. Histone acetylation dependent allelic expression imbalance of BAPX1 in patients with the oculo-auriculo-vertebral spectrum. Hum Mol Genet. 2006;15:581–587. doi: 10.1093/hmg/ddi474. [DOI] [PubMed] [Google Scholar]
- Foster MW, Hess DT, Stamler JS. Protein S-nitrosylation in health and disease: a current perspective. Trends Mol Med. 2009;15:391–404. doi: 10.1016/j.molmed.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gut P, Baeza-Raja B, Andersson O, Hasenkamp L, Hsiao J, Hesselson D, Akassoglou K, Verdin E, Hirschey MD, Stainier DY. Whole-organism screening for gluconeogenesis identifies activators of fasting metabolism. Nat Chem Biol. 2013;9:97–104. doi: 10.1038/nchembio.1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberland M, Mokalled MH, Montgomery RL, Olson EN. Epigenetic control of skull morphogenesis by histone deacetylase 8. Genes Dev. 2009;23:1625–1630. doi: 10.1101/gad.1809209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handy RL, Moore PK. Mechanism of the inhibition of neuronal nitric oxide synthase by 1-(2-trifluoromethylphenyl) imidazole (TRIM). Life Sci. 1997;60:389–394. doi: 10.1016/s0024-3205(97)00295-6. [DOI] [PubMed] [Google Scholar]
- Kamel G, Hoyos T, Rochard L, Dougherty M, Kong Y, Tse W, Shubinets V, Grimaldi M, Liao EC. Requirement for frzb and fzd7a in cranial neural crest convergence and extension mechanisms during zebrafish palate and jaw morphogenesis. Dev Biol. 2013;381:423–433. doi: 10.1016/j.ydbio.2013.06.012. [DOI] [PubMed] [Google Scholar]
- Kraft M, Cirstea IC, Voss AK, Thomas T, Goehring I, Sheikh BN, Gordon L, Scott H, Smyth GK, Ahmadian MR, et al. Disruption of the histone acetyltransferase MYST4 leads to a Noonan syndrome-like phenotype and hyperactivated MAPK signaling in humans and mice. J Clin Invest. 2011;121:3479–3491. doi: 10.1172/JCI43428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepiller S, Laurens V, Bouchot A, Herbomel P, Solary E, Chluba J. Imaging of nitric oxide in a living vertebrate using a diamino-fluorescein probe. Free radical biology & medicine. 2007;43:619–627. doi: 10.1016/j.freeradbiomed.2007.05.025. [DOI] [PubMed] [Google Scholar]
- Marmorstein R, Roth SY. Histone acetyltransferases: function, structure, and catalysis. Curr Opin Genet Dev. 2001;11:155–161. doi: 10.1016/s0959-437x(00)00173-8. [DOI] [PubMed] [Google Scholar]
- Medeiros DM, Crump JG. New perspectives on pharyngeal dorsoventral patterning in development and evolution of the vertebrate jaw. Dev Biol. 371:121–135. doi: 10.1016/j.ydbio.2012.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CT, Maves L, Kimmel CB. moz regulates Hox expression and pharyngeal segmental identity in zebrafish. Development. 2004;131:2443–2461. doi: 10.1242/dev.01134. [DOI] [PubMed] [Google Scholar]
- Minoux M, Rijli FM. Molecular mechanisms of cranial neural crest cell migration and patterning in craniofacial development. Development. 2010;137:2605–2621. doi: 10.1242/dev.040048. [DOI] [PubMed] [Google Scholar]
- Moncada S, Higgs A. The L-arginine-nitric oxide pathway. N Engl J Med. 1993;329:2002–2012. doi: 10.1056/NEJM199312303292706. [DOI] [PubMed] [Google Scholar]
- Nair S, Li W, Cornell R, Schilling TF. Requirements for Endothelin type-A receptors and Endothelin-1 signaling in the facial ectoderm for the patterning of skeletogenic neural crest cells in zebrafish. Development. 2007;134:335–345. doi: 10.1242/dev.02704. [DOI] [PubMed] [Google Scholar]
- North TE, Goessling W, Peeters M, Li P, Ceol C, Lord AM, Weber GJ, Harris J, Cutting CC, Huang P, et al. Hematopoietic stem cell development is dependent on blood flow. Cell. 2009;137:736–748. doi: 10.1016/j.cell.2009.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- North TE, Goessling W, Walkley CR, Lengerke C, Kopani KR, Lord AM, Weber GJ, Bowman TV, Jang IH, Grosser T, et al. Prostaglandin E2 regulates vertebrate haematopoietic stem cell homeostasis. Nature. 2007;447:1007–1011. doi: 10.1038/nature05883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nott A, Watson PM, Robinson JD, Crepaldi L, Riccio A. S-Nitrosylation of histone deacetylase 2 induces chromatin remodelling in neurons. Nature. 2008;455:411–415. doi: 10.1038/nature07238. [DOI] [PubMed] [Google Scholar]
- Pasqualetti M, Ori M, Nardi I, Rijli FM. Ectopic Hoxa2 induction after neural crest migration results in homeosis of jaw elements in Xenopus. Development. 2000;127:5367–5378. doi: 10.1242/dev.127.24.5367. [DOI] [PubMed] [Google Scholar]
- Pavan WJ, Raible DW. Specification of neural crest into sensory neuron and melanocyte lineages. Dev Biol. 2012;366:55–63. doi: 10.1016/j.ydbio.2012.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi HH, Sarkissian M, Hu GQ, Wang Z, Bhattacharjee A, Gordon DB, Gonzales M, Lan F, Ongusaha PP, Huarte M, et al. Histone H4K20/H3K9 demethylase PHF8 regulates zebrafish brain and craniofacial development. Nature. 2010;466:503–507. doi: 10.1038/nature09261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragland JW, Raible DW. Signals derived from the underlying mesoderm are dispensable for zebrafish neural crest induction. Dev Biol. 2004;276:16–30. doi: 10.1016/j.ydbio.2004.08.017. [DOI] [PubMed] [Google Scholar]
- Schonhoff CM, Benhar M. Coligan John E., editor. Analysis of protein S-nitrosylation. Current protocols in protein science / editorial board. 2011 doi: 10.1002/0471140864.ps1406s63. Chapter 14, Unit14 16. [DOI] [PubMed] [Google Scholar]
- Spritz RA. The genetics and epigenetics of orofacial clefts. Curr Opin Pediatr. 2001;13:556–560. doi: 10.1097/00008480-200112000-00011. [DOI] [PubMed] [Google Scholar]
- White RM, Cech J, Ratanasirintrawoot S, Lin CY, Rahl PB, Burke CJ, Langdon E, Tomlinson ML, Mosher J, Kaufman C, et al. DHODH modulates transcriptional elongation in the neural crest and melanoma. Nature. 2011;471:518–522. doi: 10.1038/nature09882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuniga E, Stellabotte F, Crump JG. Jagged-Notch signaling ensures dorsal skeletal identity in the vertebrate face. Development. 2010;137:1843–1852. doi: 10.1242/dev.049056. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.