Abstract
Centromere sequences are not conserved between species, and there is compelling evidence for epigenetic regulation of centromere identity, with location being dictated by the presence of chromatin containing the histone H3 variant CENP-A. Paradoxically, in most organisms CENP-A chromatin generally occurs on particular sequences. To investigate the contribution of primary DNA sequence to establishment of CENP-A chromatin in vivo, we utilised the fission yeast Schizosaccharomyces pombe. CENP-ACnp1 chromatin is normally assembled on ∼10 kb of central domain DNA within these regional centromeres. We demonstrate that overproduction of S. pombe CENP-ACnp1 bypasses the usual requirement for adjacent heterochromatin in establishing CENP-ACnp1 chromatin, and show that central domain DNA is a preferred substrate for de novo establishment of CENP-ACnp1 chromatin. When multimerised, a 2 kb sub-region can establish CENP-ACnp1 chromatin and form functional centromeres. Randomization of the 2 kb sequence to generate a sequence that maintains AT content and predicted nucleosome positioning is unable to establish CENP-ACnp1 chromatin. These analyses indicate that central domain DNA from fission yeast centromeres contains specific information that promotes CENP-ACnp1 incorporation into chromatin. Numerous transcriptional start sites were detected on the forward and reverse strands within the functional 2 kb sub-region and active promoters were identified. RNAPII is enriched on central domain DNA in wild-type cells, but only low levels of transcripts are detected, consistent with RNAPII stalling during transcription of centromeric DNA. Cells lacking factors involved in restarting transcription—TFIIS and Ubp3—assemble CENP-ACnp1 on central domain DNA when CENP-ACnp1 is at wild-type levels, suggesting that persistent stalling of RNAPII on centromere DNA triggers chromatin remodelling events that deposit CENP-ACnp1. Thus, sequence-encoded features of centromeric DNA create an environment of pervasive low quality RNAPII transcription that is an important determinant of CENP-ACnp1 assembly. These observations emphasise roles for both genetic and epigenetic processes in centromere establishment.
Author Summary
The kinetochore directs the separation of chromosomes and is assembled at a special region of the chromosome—the centromere. DNA is wrapped around particles called nucleosomes, which contain histone proteins. The nucleosomes at centromeres are specialized, and contain the centromere-specific histone CENP-A. CENP-A nucleosomes form the platform upon which the kinetochore is built. Thus, CENP-A and centromere function go hand-in-hand. How the cell ensures that CENP-A is deposited at centromeres and not elsewhere is not well understood. We investigated the role that DNA sequence plays in defining centromere function in fission yeast. Our observations suggest that it is not the DNA sequence per se that is important for attracting CENP-A, but rather, the particular environment that the sequence creates. During transcription of centromeric DNA, RNA polymerase (RNAPII) appears to get stuck or stalled. Particular proteins—such as TFIIS and Ubp3—are known to help restart RNAPII so it can continue transcribing. We found that when cells lack Ubp3 or TFIIS, CENP-A becomes deposited on centromere sequences. We propose that persistent stalling of RNAPII on centromere DNA attracts factors that help deposit CENP-A. This study highlights the influence of DNA sequence in creating an attractive environment for CENP-A assembly.
Introduction
Centromeres are the chromosomal sites upon which kinetochores are assembled to ensure accurate segregation of sister chromatids into daughter cells. Most kinetochores are built upon a specialized type of chromatin in which canonical histone H3 is replaced by the histone variant CENP-A. Although the centromere-kinetochore complex performs conserved essential functions, and kinetochore proteins are generally conserved [1], centromeric DNA is not conserved, even between related species, and a huge variety of centromere sequences and structures exist [2–5]. The point centromeres of budding yeast consist of 125 bp of DNA and utilize an essential centromere-specific DNA binding protein [6]. At the other extreme, the nematode, Caenorhabditis elegans, has holocentric centromeres, in which kinetochore proteins assemble at multiple loci along each chromosome arm [7,8]. The majority of centromeres studied to date are regional. Centromeres in various plant and animal species are composed of arrays of different types of satellite, repetitive sequences and transposable elements, for instance, human centromeres encompass several megabases of tandem repetitive arrays of alpha-satellite sequence [9–11]. Fission yeast centromeres represent another type of regional centromere, in which a unique central core of 4–7 kb is flanked by inverted repeat elements and blocks of relatively large repeat units, resulting in centromeres of 40–120 kb [12]. Even the centromeres of different chromosomes in individual species are not necessarily homologous; each Candida albicans centromere has a unique central core, whilst chicken and potato each utilize both repeat-rich and unique sequence centromeres [13–15]. Thus, functional centromeres are assembled on diverse types of sequences in different organisms and it remains unknown if there is a universal fundamental property that defines centromeric sequences.
Abundant evidence indicates that centromeres are epigenetically regulated [16]. Although rare, neocentromeres have been observed in many species, forming on DNA sequences that do not normally possess centromere function and share no sequence homology with normal centromeres [17]. The best-characterized example in human is the neocentromere in 10q25 on the long arm of chromosome 10 that arose upon deletion of the centromere and loss of the entire alpha satellite array [18]. In S. pombe, neocentromeres form in close proximity to telomeres following the engineered deletion of a centromere [19]. Conversely, centromeres can be inactivated on dicentric human chromosomes despite the continued presence of alpha-satellite sequence at both centromeric loci [20]. In S. pombe one centromere on a dicentric chromosome can be inactivated by mechanisms such as heterochromatinisation or formation of a domain of histone hypoacetylation [21]. These and numerous other examples demonstrate that centromeric sequences are neither necessary nor sufficient for kinetochore assembly.
The histone H3 variant, CENP-A acts as the epigenetic mark that specifies centromere identity [22–24]. CENP-A is found only at active centromeres, including neocentromeres, and is absent at inactivated centromeres. The forced recruitment of CENP-A either by directly tethering CENP-A or its chaperone (HJURP) to a non-centromeric locus leads to the accumulation of CENP-A and kinetochore proteins at that location [24,25]. It is thought that continued deposition of CENP-A at centromere regions through cell and organism generations involves a self-propagation mechanism in which CENP-A chromatin, or features of the kinetochore which is assembled upon it, are recognized and attract additional CENP-A [26,27].
In most organisms there is no obligate coupling of sequence and CENP-A assembly, yet kinetochores are normally assembled upon particular centromeric sequences in any given species [4]. This suggests that centromeric sequences possess underlying properties that promote CENP-A incorporation. Alternatively, the preponderance of particular sequences at centromeres could be driven by properties of CENP-A chromatin or kinetochores themselves [28]. However, centromeric DNA allows the de novo assembly of functional centromeres following its introduction into cells in many organisms. Alpha satellite arrays are able to direct the de novo assembly of centromeres when introduced into certain cell lines as naked DNA [29,30]. De novo assembly of centromeres also occurs when centromeric DNA from S. pombe is introduced into cells. However, de novo establishment does not seem to be a universal property: despite promiscuous neocentromere formation in C. albicans, transformation with bone fide centromeric sequences does not result in kinetochore assembly[13]. At the other extreme, many sequences introduced into the holocentric organism C. elegans appear able to assemble CENP-A chromatin [31,32]. Thus, the relationship between centromeric sequence and the establishment and maintenance of CENP-A chromatin is enigmatic.
Transcription has received a lot of attention as a possible contributor to assembly of CENP-A chromatin. Transcripts emanating from centromeric regions have been detected in many organisms, including maize, human, rice, budding yeast, fission yeast and tammar wallaby [33–38]. Interfering with the chromatin status or transcriptional properties of centromeric repeats affects maintenance of CENP-A chromatin and segregation function on human artificial chromosomes (HACs) [39,40]. RNA Polymerase II (RNAPII) has been detected at mitotic mammalian centromeres where it may influence centromere function [38]. In fission yeast, transient H2B ubiquitylation may loosen centromeric chromatin to promote transcription and CENP-ACnp1 incorporation and defective reassembly of H3 chromatin behind elongating RNAPII aids CENP-ACnp1 incorporation [41,42]. However, although there are numerous tantalising hints that transcriptional activity contributes to centromere function or identity, much remains to be understood [33,38,43,44].
Here we investigate the contribution of DNA sequence to the establishment of CENP-A chromatin in fission yeast, an organism in which epigenetic mechanisms clearly influence centromere identity. Normally proximal heterochromatin is required to facilitate establishment of CENP-ACnp1 chromatin on centromere central domain sequences [45,46]. We show that this requirement can be bypassed by overexpression of CENP-ACnp1 and that central domain DNA is a preferred substrate for establishment of CENP-ACnp1 chromatin. We find that there is functional redundancy within the central domain but that sub-regions are non-equivalent in their ability to establish CENP-ACnp1 chromatin. Analysis of a 2 kb region capable of directing CENP-ACnp1 assembly indicates that it contains numerous transcriptional start sites, along with promoter elements, and that relatively high levels of RNAPII are recruited, despite low levels of transcripts produced, consistent with the presence of stalled RNAPII. Our observations suggest that redundant sequence features in the centromere central domain create a unique transcriptional environment that is permissive for CENP-ACnp1 establishment. Consistent with this, defective transcriptional elongation where stalled RNAPII is increased promotes the establishment of CENP-ACnp1 chromatin.
Results
Elevated CENP-ACnp1 levels bypass the requirement for heterochromatin in establishing CENP-ACnp1 chromatin
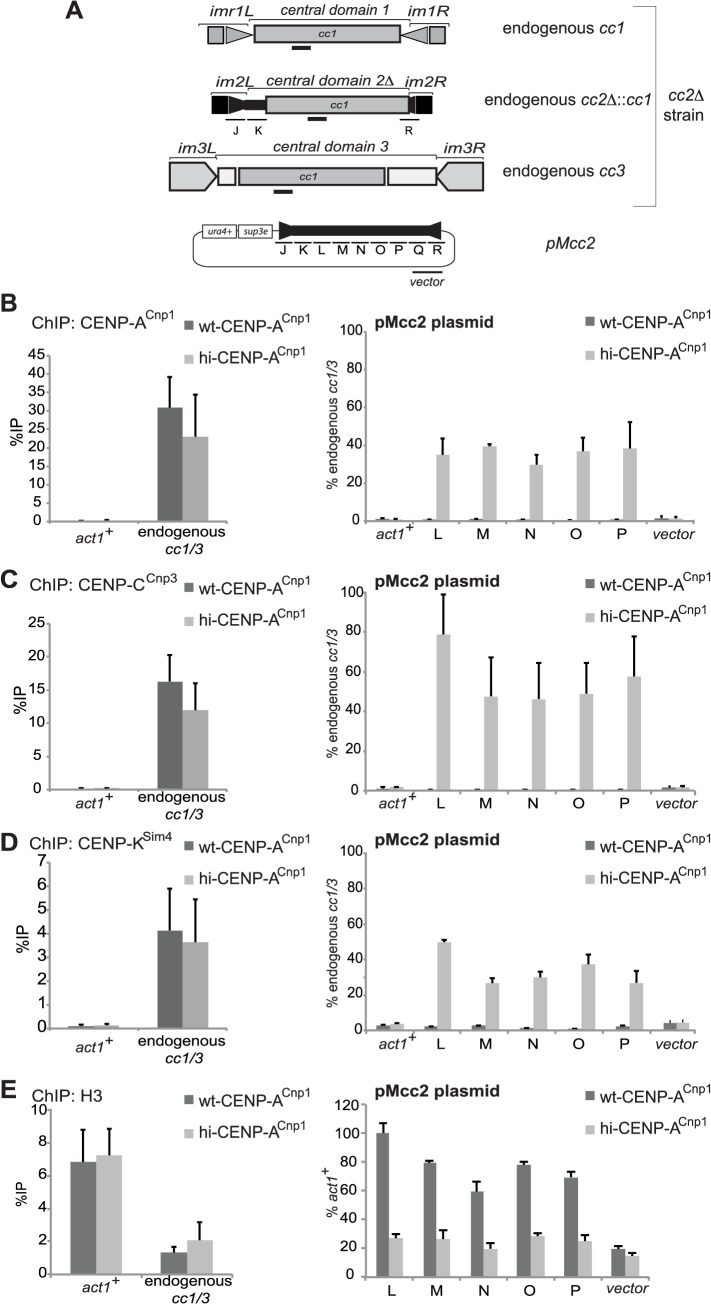
In wild-type fission yeast cells, de novo CENP-ACnp1 chromatin establishment on circular plasmid-based minichromosomes requires an outer repeat or tethered Clr4 histone H3K9 methyltransferase to form a block of heterochromatin in close proximity to central domain DNA from centromeres [45,46]. CENP-ACnp1 can also be deposited at other non-centromeric locations in the genome when it is overexpressed, however the level incorporated at these ectopic sites is much lower than that detected at natural centromeres [41,47]. To determine whether central domain DNA is a preferential substrate for the establishment of CENP-ACnp1 chromatin, plasmid pMcc2 bearing 8.5 kb of central domain from cen2 (imr2-cc2-imr2) sequence, but no heterochromatic outer repeat sequences, was transformed into cells expressing additional GFP-CENP-ACnp1 (Fig. 1A).
Fig 1. Elevated levels of CENP-ACnp1 are sufficient to establish centromeric chromatin in the absence of heterochromatin.
(A) Schematic representation of the three endogenous centromeres in cc2Δ strains and the plasmid (pMcc2) used in the study. In cc2Δ strains, part of the central core 2 (regions L to Q, each 1 kb) is replaced by 5.5 kb of DNA from central core 1, allowing the analysis of the L-Q sequence within pMcc2. Primer pairs used for quantification at endogenous centromeres recognise the region of homology shared between cc1 and cc3. The product amplified by qPCR for the endogenous centromeres is represented as a black bar. (B) ChIP analysis of CENP-ACnp1 levels at endogenous centromere (cc1/3), act1 +, plasmid backbone (vector) and at cc2 on the plasmid (pMcc2: L-P) in wild-type (wt-CENP-ACnp1) or in the presence of high levels of CENP-ACnp1 (hi-CENP-ACnp1). (C) ChIP analysis for the kinetochore protein CENP-CCnp3. (D) ChIP analysis for the kinetochore protein CENP-KSim4. (E) ChIP analysis of histone H3 levels. For the kinetochore proteins analysed, ChIPs are reported as %IP for endogenous centromeres or as relative to cc1/3 for the pMcc2 plasmid. For the levels of H3, the ChIP analyses are reported as %IP for cc1/3 and as relative to act1 + for the pMcc2 plasmid (n = 3).
All strains used have 6 kb of cen2 central domain DNA replaced with 5.5 kb of cen1 central domain DNA (cc2Δ::cc1—Fig. 1A, S1 Fig.) so that only 2.5 kb of normal cen2 central domain DNA remains at this modified cen2 (imr2L, regions J, K, R; Fig. 1A). The resulting deletion of fragments L-Q from the cen2 central domain allows detailed and specific analysis of 6 kb of central domain DNA when borne by plasmid-based minichromosomes. Quantitative chromatin immunoprecipitation assays (qChIP) shows that CENP-ACnp1 chromatin does not assemble on regions L, M N, O or P when a plasmid (pMcc2) containing the 8.5 kb cc2 sequence, but lacking heterochromatin, was transformed into wild-type cells [45]. However, when pMcc2 was transformed into cells over-expressing CENP-ACnp1 (hi-CENP-ACnp1; ∼15 fold more than wild-type cells [41]), CENP-ACnp1 and the kinetochore proteins CENP-CCnp3 and CENP-KSim4 were easily detected over the central domain of pMcc2 by qChIP (Fig. 1B-E). Importantly, these centromeric proteins were enriched on centromeric DNA but not on the plasmid backbone, indicating that CENP-ACnp1 chromatin assembles specifically on central domain DNA from centromeres (Fig. 1). The relative level of enrichment of CENP-ACnp1 and the other kinetochore proteins on different parts of pMcc2 suggests all proteins are distributed uniformly across this plasmid-borne central domain (Fig. 1B-D). Furthermore, the levels of histone H3 associated with the L-P regions of pMcc2 were reduced in cells expressing additional CENP-ACnp1 compared to control cells (Fig. 1E). We conclude that H3 chromatin is normally assembled on central domain DNA on pMcc2 in wild-type cells but CENP-ACnp1 chromatin assembles instead when pMcc2 is placed in hi-CENP-ACnp1 cells.
CENP-ACnp1 can assemble on pre-chromatinised substrates and is trans-generationally inherited
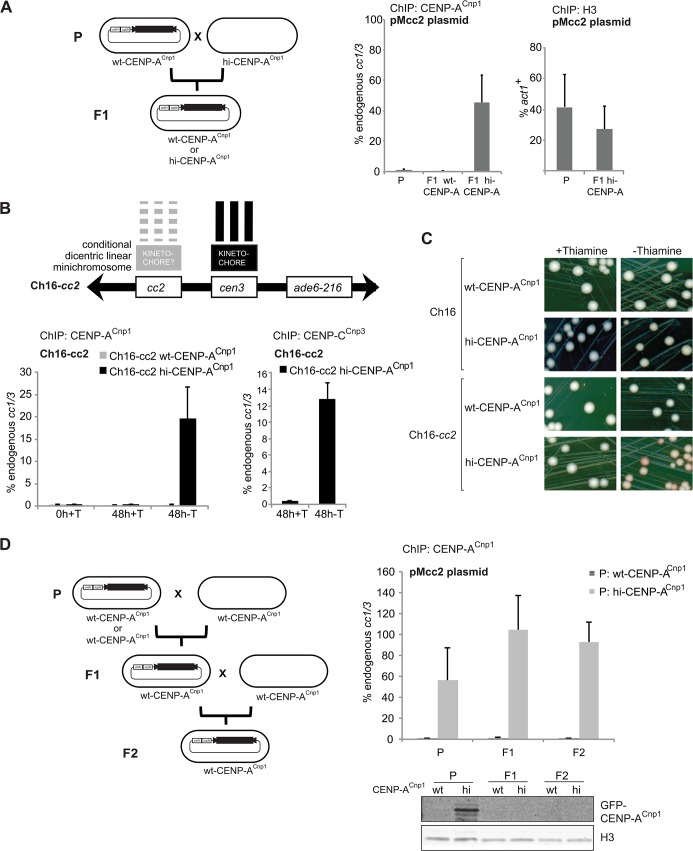
To determine whether CENP-ACnp1 can become established on plasmids that are already assembled in chromatin, the pMcc2 plasmid was transformed into cells expressing wt-CENP-ACnp1 levels and subsequently crossed with hi-CENP-ACnp1 cells. qChIP analyses indicate that CENP-ACnp1 is initially absent from pMcc2 in the wt-CENP-ACnp1 parental strain and then becomes assembled in CENP-ACnp1 chromatin when transferred into the hi-CENP-ACnp1 environment, indicating that plasmid-borne cc2 initially assembled in normal (H3) chromatin can be converted to CENP-ACnp1 chromatin (Fig. 2A).
Fig 2. CENP-ACnp1 chromatin behaves as an epigenetic mark.
(A) A wild-type strain (parental: P; wt-CENP-ACnp1) was transformed with the pMcc2 plasmid and then crossed to a strain overexpressing CENP-ACnp1 (hi-CENP-ACnp1). From the first generation (F1) cells containing pMcc2 and expressing wild-type level or overexpressing CENP-ACnp1 were selected and analysed by ChIP for CENP-ACnp1 and H3 levels on the pMcc2 plasmid (P: n = 2, F1: n = 4) (B) cc2 was integrated on the arm of Ch16, that contains cen3, creating Ch16-cc2. Ch16-cc2 was crossed into a strain containing nmt41-GFP-CENP-ACnp1. In presence of thiamine (0h+T) GFP-CENP-ACnp1 is repressed. The same cells were diluted in medium containing thiamine (48+T; GFP-CENP-ACnp1 repressed) or without thiamine (48h-T; GFP-CENP-ACnp1 expressed) for 48h. ChIP was performed for CENP-ACnp1 and CENP-CCnp3 levels. (C) Colony colour sectoring assay for cells containing minichromosome Ch16 or Ch16-cc2. Red sectors indicate loss of minichromosomes. (D) A wild-type strain (wt-CENP-ACnp1) or a strain overexpressing CENP-ACnp1 (hi-CENP-ACnp1) were transformed with the pMcc2 plasmid (parental: P) and then crossed to a wt-CENP-ACnp1 strain. From the first generation (F1), wt-CENP-ACnp1 strains containing pMcc2 were selected and subsequently crossed to wt-CENP-ACnp1 strain. Progeny resulting from this second cross are referred to as second generation (F2). ChIP analysis for CENP-ACnp1 levels on the pMcc2 plasmid in the different generations relative to cc1/3; region M analysed by qPCR (P: n = 2, F1: n = 4, F2: n = 8).
In addition, a copy of cc2 (8.5 kb) was inserted on the arm of the 530 kb Ch16 linear minichromosome which carries a complete cen3 [48] (Ch16-cc2; Fig. 2B). When the expression of additional GFP-CENP-ACnp1 was repressed (0h+T), no CENP-ACnp1 was detected on cc2. However, when GFP-CENP-ACnp1 was induced (48h-T) both CENP-ACnp1 and CENP-CCnp3 were detected on cc2 (Fig. 2B). Thus, cc2 borne on a linear minichromosome can be converted from a pre-chromatinised state to a CENP-ACnp1 state. Moreover, colony colour assays indicate that hi-CENP-ACnp1 expression induces increased loss of Ch16-cc2, which is consistent with a second functional kinetochore being formed at cc2 on Ch16 (Fig. 2C). Thus, Ch16-cc2 behaves as an inducible dicentric chromosome controlled by CENP-ACnp1 levels.
It is possible that high levels of CENP-ACnp1 are continuously required to maintain CENP-ACnp1 on pMcc2, or alternatively, once established, CENP-ACnp1 and kinetochore proteins may persist even when CENP-ACnp1 is returned to wild-type levels (wt-CENP-ACnp1). To investigate the maintenance of CENP-ACnp1 chromatin, pMcc2 was first transformed into hi-CENP-ACnp1 cells to allow the assembly of centromeric chromatin and subsequently these pMcc2-containing cells were crossed with wt-CENP-ACnp1 cells to transfer the pMcc2 plasmid into cells expressing wild-type CENP-ACnp1 levels. ChIP analyses show that CENP-ACnp1 persisted on the pMcc2 in this wild-type background (Fig. 2D). Western analysis of extracts from Parental, F1 and F2 cells confirmed that GFP-CENP-ACnp1 was lost in F1 and F2 cells (Fig. 2D). Thus, CENP-ACnp1 chromatin behaves as a true epigenetic entity in that once established it carries its own efficient propagation mechanism, persisting even though the original stimulus has been removed. More remarkably, this CENP-ACnp1 chromatin is maintained through 2 rounds of meiosis and at least 50 mitotic divisions. Thus central domain sequences are particularly receptive to the establishment and maintenance of CENP-ACnp1 chromatin.
Central domain sequence and length affect de novo CENP-ACnp1 deposition
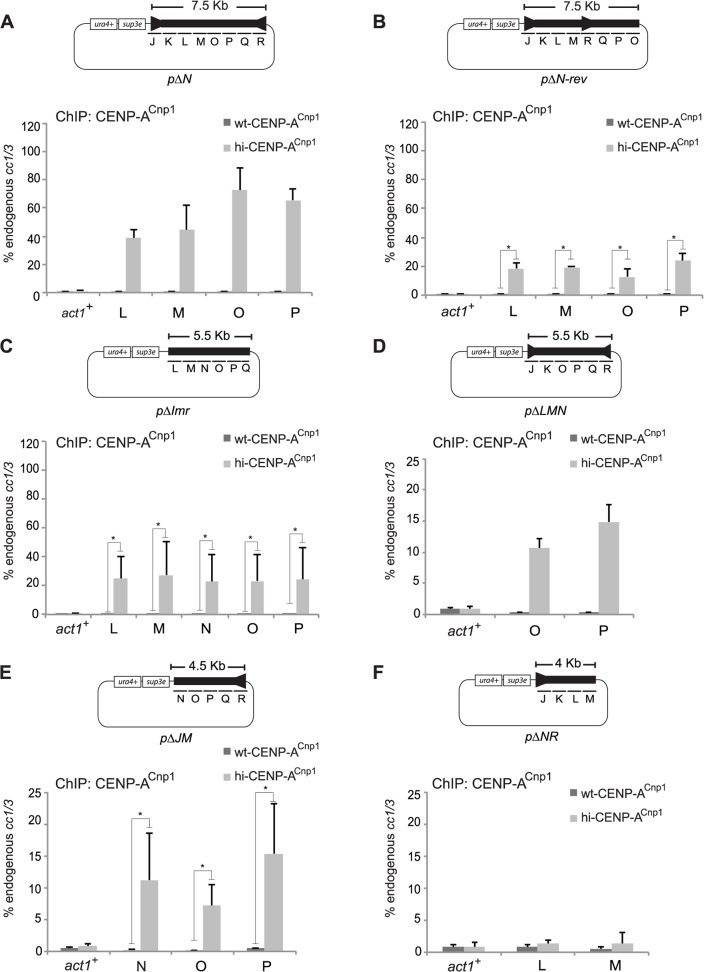
To determine if specific regions from the central domain of cen2 are required to establish CENP-ACnp1 chromatin, plasmids bearing different sub-fragments from cc2 were transformed into wt-CENP-ACnp1 or hi-CENP-ACnp1 cells (Fig. 3). We used an unbiased approach to divide the 8.5 kb cc2 into 1 kb regions (J-R). Deletion of 1 kb from the centre of cc2 (N) does not affect CENP-ACnp1 establishment (pΔN; Fig. 3A, compare with pMcc2, Fig. 1B). Notably, CENP-ACnp1 incorporation on a plasmid carrying identical centromeric DNA as pΔN but with the right half (O-R) inverted relative to the native sequence, was less efficient (pΔN-rev; Fig. 3B). Thus, the relative orientation of central domain sequences within cc2 influences the degree of CENP-ACnp1 deposition; directionality or the juxtaposition of certain sequences may be important for promoting CENP-ACnp1 incorporation. The central CENP-ACnp1 domain at endogenous fission yeast centromeres is composed of inverted imr repeats that flank the central core. ChIP analyses demonstrated that, in a plasmid-based establishment assay, the imr repeats are dispensable for de novo CENP-ACnp1 incorporation on the remaining central domain sequences (pΔimr; Fig. 3C).
Fig 3. Centromeric DNA sequence affects CENP-ACnp1 chromatin establishment.
Plasmids containing fragments of cc2 DNA were transformed into wild-type (wt-CENP-ACnp1) or cells expressing high levels of CENP-ACnp1 (hi-CENP-ACnp1). Plasmids contained: (A) Deletion of central 1 kb (N) from cc2, (B) Left half of cc2, (J to M: 4 kb) plus region O to R (3.5 kb) in inverted orientation (region N absent), (C) imr2 regions (J and R) deleted along with 1 kb of cc2 (K), leaving LMNOPQ, (D) Region L to N deleted, leaving JKOPQR. (E) Deletion of J to M, leaving N to R, (F) Deletion from N to R, leaving J to M. The enrichment of CENP-ACnp1 on the plasmids was analysed by ChIP and calculated relative to the endogenous cc1/3 (n = 3). In (B), (C) and (E) p-value was calculated. Asterisks indicate p <0.05.
Deletion of additional regions (LMN) of cc2 markedly decreased the efficiency of CENP-ACnp1 incorporation relative to pMcc2 and pΔN ( Fig. 3D; compare Fig. 1B, Fig. 3A), suggesting that either the LM region is critical for promoting CENP-ACnp1 incorporation or that the overall reduced centromeric DNA length diminishes CENP-ACnp1 deposition (Fig. 3D). However, further investigation using plasmids bearing smaller cc2 fragments suggests that the specific sequences present have a more significant influence on CENP-ACnp1 deposition than the overall length of cc2 DNA present (Fig. 3E,F). For example, pΔJM and pΔNR differ by only 500 bp, however, pΔJM incorporated substantially more CENP-ACnp1 than pΔNR (Fig. 3F). We conclude that specific sequences from the central domain of fission yeast centromeres, combined with their overall length, promote the efficient de novo assembly of CENP-ACnp1 chromatin.
A 2 kb region of centromeric DNA is sufficient to direct de novo CENP-ACnp1 chromatin assembly
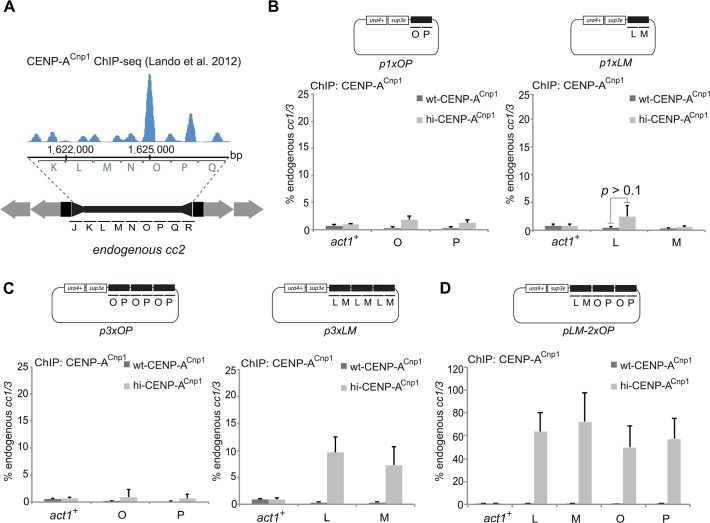
It is possible that shorter fragments of centromere DNA from within the central domain can actively promote CENP-ACnp1 assembly but that because longer total lengths are required to stabilise incorporated CENP-ACnp1 the activity of shorter fragments cannot be detected. To address this possibility we selected two distinct sequences from the central domain of cen2 for analyses. The 2 kb OP region was present on all the pMcc2 derivatives with which we detected significant CENP-ACnp1 incorporation following transformation into hi-CENP-ACnp1 cells (Fig. 3). In addition, ChIP-seq analysis indicates particularly high CENP-ACnp1 nucleosome occupancy within OP at endogenous cen2 [49 and Fig. 4A]. In contrast, the 2 kb LM region appears to be dispensable for de novo CENP-ACnp1 assembly on pMcc2 derived plasmids and exhibits low CENP-ACnp1 nucleosome occupancy (Fig. 3, Fig. 4A).
Fig 4. A 2 kb region of cc2 is sufficient to establish CENP-ACnp1 chromatin.
(A) Profile of ChIP-SEQ for CENP-ACnp1 [49] and corresponding position on endogenous cc2. (B) ChIP analysis of CENP-ACnp1 levels on plasmids containing one copy (p1xLM) or one copy of OP (p1xOP) transformed into wt-CENP-ACnp1 or hi-CENP-ACnp1. (C) CENP-ACnp1 levels on plasmids containing three copies of a 2 kb fragment, either OP or LM, cloned in tandem repeats (p3xOP and p3xLM) and transformed into wild-type (wt-CENP-ACnp1) or into cells expressing high levels of CENP-ACnp1 (hi-CENP-ACnp1). (D) A plasmid containing one copy of the LM fragment adjacent to two copies of OP (direct repeats) was transformed into wt-CENP-ACnp1 and hi-CENP-ACnp1 and CENP-ACnp1 levels analysed by ChIP (n = 3).
Initial tests showed that neither OP (p1xOP) nor LM (p1xLM) sequences alone were capable of inducing significant de novo CENP-ACnp1 incorporation when introduced into hi-CENP-ACnp1 cells (Fig. 4B). This finding is consistent with a minimal length of central domain DNA being required for stable CENP-ACnp1 chromatin assembly and retention. To satisfy this apparent length requirement, the OP and the LM fragments were multimerised as tandem repeats to create 3xOP and 3xLM (p3xOP, p3xLM; Fig. 4C). Remarkably, when transformed into hi-CENP-ACnp1 cells no CENP-ACnp1 was detectable on p3xOP whereas p3xLM allowed a reasonable level of CENP-ACnp1 incorporation (Fig. 4C). This suggests that in isolation the OP region is unable to promote CENP-ACnp1 deposition even though in the context of an entire central domain it normally accepts CENP-ACnp1 and ends up with high CENP-ACnp1 nucleosome occupancy (Fig. 4A). We note that the removal of the LM region from the central domain of pMcc2 derived plasmids greatly reduced the level of CENP-ACnp1 incorporated on OP (compare pΔN Fig. 3A with pΔLMN Fig. 3D). Thus, in contrast to OP, the LM region appears to have the ability to induce CENP-ACnp1 deposition. To directly test this possibility, a single copy of LM was placed adjacent to two tandem copies of OP (pLM-2xOP) and transformed into hi-CENP-ACnp1 cells. High levels of CENP-ACnp1 were detected on both the LM and OP regions of pLM-2xOP (Fig. 4D), thus the LM region has an innate ability to stimulate CENP-ACnp1 deposition on the OP region. A different arrangement of the same sequences (pOPLMOP) also attracted CENP-ACnp1 in hi-CENP-ACnp1 cells (S2 Fig.). These analyses indicate that the 2 kb LM sequence contains all the features that are required to promote and accept CENP-ACnp1 assembly, and thus LM defines a 2 kb region of S. pombe centromeric sequence that allows the de novo assembly of CENP-ACnp1 chromatin.
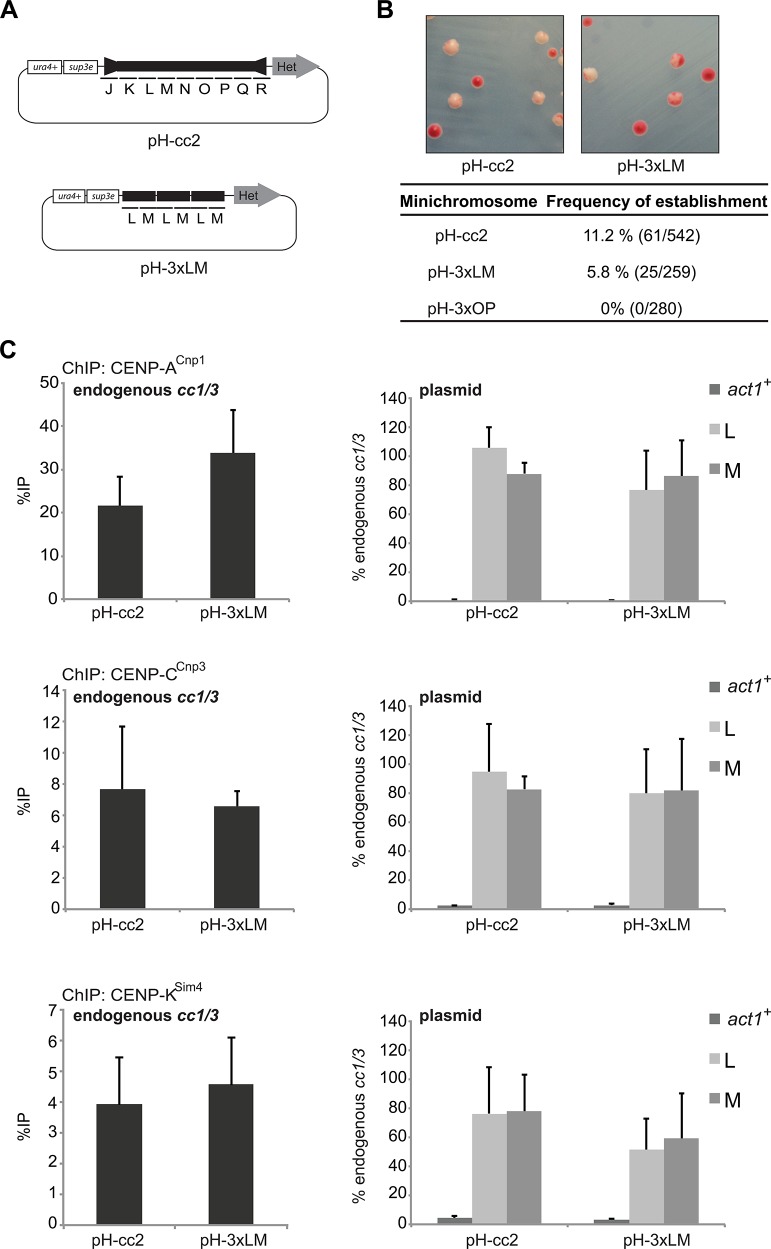
The 2 kb LM element is sufficient to form functional centromeres
Plasmids bearing an entire central core domain flanked by outer heterochromatin repeats assemble functional centromeres when transformed into wild-type cells [45,50]. To determine if the 2 kb LM region imparts centromere function, a plasmid carrying the 3xLM tandem repeat adjacent to a 5 kb outer repeat heterochromatin forming element (pH-3xLM) was transformed into wild-type cells expressing CENP-ACnp1 at normal levels (Fig. 5A). The establishment of functional centromeres in the resulting transformants was monitored by an ade6-based colony colour sectoring assay [51]. Minichromosomes carrying full-length cc2 and 5 kb of outer repeat heterochromatin were able to establish functional centromeres upon transformation (Fig. 5B and S2 Fig.). pH-3xLM and pH-LM-2xOP transformants also established functional centromeres, but at lower frequency than pH-cc2 (Fig. 5B and S2 Fig.). Differences in the ability of various constructs to form functional centromeres may reflect the particular configuration of sequences in individual minichromosomes. In contrast, pH-3xOP (3xOP flanked by heterochromatin) was unable to establish functional centromeres. Thus the LM sequence in a 3x tandem array, flanked by heterochromatin, is sufficient to form functional centromeres. ChIP analyses confirmed that kinetochores were assembled on pH-3xLM since CENP-ACnp1 and the kinetochore proteins CENP-CCnp3 and CENP-KSim4 were enriched over the LM sequences at levels comparable to endogenous centromeres (Fig. 5C). We conclude that the LM sequence within pH-3xLM not only promotes incorporation of CENP-ACnp1 into chromatin but also supports the assembly of a functional centromere.
Fig 5. Tandem copies of the 2 kb LM region adjacent to heterochromatin/outer repeat are sufficient to establish a functional centromere in wild-type cells.
(A) Schematic representation of plasmids containing 5.6 kb of heterochromatin-forming outer repeat element flanking a full length cc2 (pH-cc2) or three tandem repeats of the LM region (pH-3xLM). (B) Establishment assay in wild-type cells. pH-cc2, pH-3xLM and pH-3xOP were transformed in wild-type cells and the percentage of transformants that contained plasmids with centromere function was assessed. White colonies with red sectors indicate formation of functional centromeres (details in M&M). (C) ChIP analysis of CENP-ACnp1, CENP-CCnp3 and CENP-KSim4 levels reported as %IP for endogenous centromeres (cc1/3) and relative to cc1/3 at act1+ and on plasmids (L and M) (n = 3).
Features within centromeric central domain DNA are required to promote CENP-ACnp1 chromatin establishment
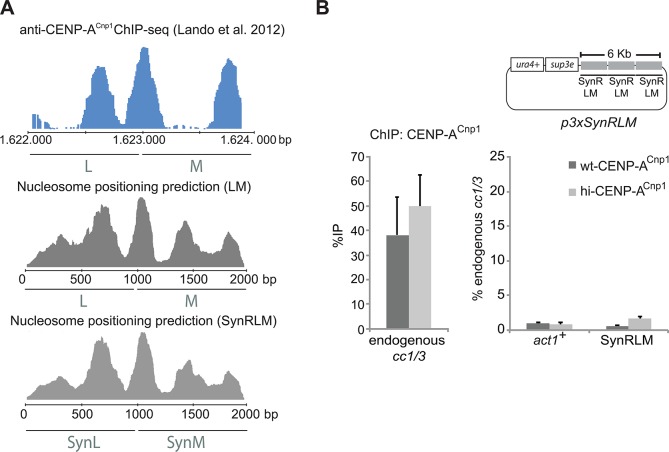
Nucleosome occupancy is known to be influenced by a combination of DNA sequence and the action of chromatin remodelers [52]. Primary DNA sequence itself influences nucleosome occupancy since DNA sequences with a high GC content and periodic dinucleotide patterns, that are devoid of poly(dA:dT) sequences, are strongly favored for nucleosome occupancy because of biophysical constraints that allow such sequences to wrap more easily around nucleosomes. These constraints have led to the development of algorithms that predict the probability of nucleosome occupancy [53,54]. In common with centromeres of many organisms, fission yeast centromeric DNA is AT-rich with a higher frequency of poly(dA:dT) tracts. It is therefore possible that H3 nucleosomes have a lower affinity for such sequences whereas CENP-A nucleosomes may be unperturbed by such AT rich DNA. To examine the underlying sequence specificity within centromeric DNA that favours the deposition of CENP-ACnp1 nucleosomes, the sequence of LM DNA was altered by randomisation using a 5 bp sliding window throughout the entire 2 kb element. This generated a synthetic LM sequence (SynR-LM) that is 62.6% identical to the wild-type LM sequence, retaining the same AT content and dinucleotide periodicity, and thus the same predicted nucleosome occupancy as the wild-type LM element (Fig. 6A) [55].
Fig 6. Genetic information contained within DNA sequence is required for centromeric chromatin establishment.
(A) Comparison between ChIP-seq data for CENP-ACnp1([49]; blue) and nucleosome positioning prediction (algorithm developed by [55]; in grey) for the LM region of cc2 or a mutagenised synthetic version of LM (SynRLM). The algorithm predicts nucleosome peaks within the central domain that match those mapped in vivo by ChIP-seq. SynRLM was designed by randomising the LM sequence in a 5 bp window and taking into consideration both periodicity of nucleosome positions and AT/GC content distribution of the original LM sequence. (B) ChIP analysis of CENP-ACnp1 levels of wild-type (wt-CENP-ACnp1) and cells overexpressing CENP-ACnp1 (hi-CENP-ACnp1) transformed with a plasmid containing three tandem repeats of SynRLM (p3xSynRLM). CENP-ACnp1 levels are shown relative to cc1/3 (n = 3).
Synthesised SynR-LM assembled as a 3xSynR-LM tandem array was placed in the same plasmid backbone as p3xLM to generate p3xSynR-LM. p3xSynR-LM was transformed into wt-CENP-ACnp1 and hi-CENP-ACnp1 cells. In contrast to p3xLM, CENP-ACnp1 was not detectable on the shuffled LM sequence of pSynR-LM (Fig. 6B, compare with Fig. 4B). These analyses demonstrate that preservation of nucleotide composition (AT-content, dinucleotide periodicity) and predicted nucleosome occupancy within an altered centromeric DNA is not sufficient to allow CENP-ACnp1 deposition. The fact that the natural 2 kb LM sequence is active whereas the artificial SynR-LM is inactive reveals that the primary sequence of wild-type centromeric LM DNA encodes properties that somehow allow its recognition in vivo and consequent de novo assembly of CENP-ACnp1 chromatin.
Centromeric DNA produces an unusual transcriptional environment
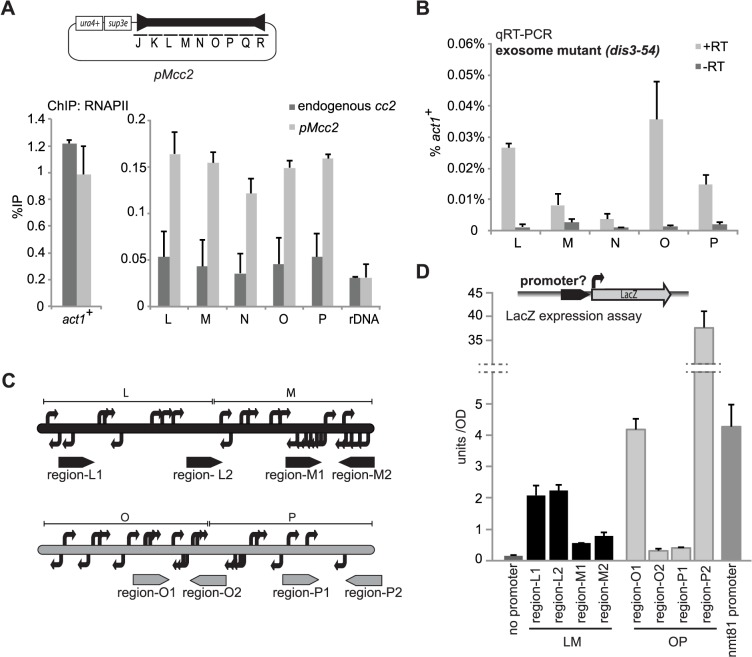
Upon transformation into cells innate features within 3xLM sequence must allow it to be either immediately assembled in CENP-ACnp1 chromatin, or, initially assembled in H3 chromatin with subsequent remodelling that exchanges canonical H3 for CENP-ACnp1. The process of transcription is obviously accompanied by chromatin remodelling and non-coding transcripts synthesised from within the central CENP-ACnp1 domains of fission yeast centromeres have been detected [37,42]. The transcription of central domain DNA might influence the assembly of CENP-ACnp1 chromatin. In cells expressing CENP-ACnp1 at wild-type levels, plasmid-borne central domain sequences are assembled in H3 rather than CENP-ACnp1 chromatin (Fig. 1E). Higher levels of RNAPII are detected on plasmid-borne central domain sequences (pMcc2) introduced into wild-type cells than when cc2 is assembled in CENP-ACnp1 chromatin on pMcc2 or at endogenous centromeres (Fig. 7A, S3 Fig.). Although relatively high levels of RNAPII associate with the pMcc2 central domain when assembled as H3 chromatin in wild-type cells (10–30% of levels at act1 +) (Fig. 1E, Fig. 7A), the level of transcripts emanating from the central domain is very low (<0.1% of act1 +), even when analysed in exosome defective cells (dis3–54; Fig. 7B). Thus, although ample RNAPII is recruited to the central domain of pMcc2 few transcripts are generated, suggesting that transcriptional stalling occurs.
Fig 7. High levels of RNAPII but low levels of transcripts are found at central domain.
(A) ChIP of RNA polymerase II (RNAPII) at endogenous cc2 and at the pMcc2 plasmid in wild-type cells (n = 3). rDNA: negative control. (B) qRT-PCR performed on total RNA extracted from exosome mutant cells (dis3–54) lacking endogenous cc2 and transformed with the pMcc2 plasmid (n = 3). Transcript levels are shown relative to act1 + (n = 3). (C) 5’RACE-PCR was performed on poly(A) purified RNA extracted from exosome mutant (dis3–54) cells lacking endogenous cc2 and transformed with the p3xLM or p3xOP plasmid. In the figure, TSSs are represented as bent arrows. (D) Analysis of promoter activity: ∼200 bp fragments from cc2 (region-L1,-L2,-M1,-M2,-O1,-O2,-P1,-P2) were placed upstream of a LacZ reporter gene. The levels of LacZ expression was assessed by measuring absorbance at 420 nm of cell lysates incubated with 2-Nitrophenyl-β-D-galactopyranoside (ONPG). nmt81: positive control with nmt81 promoter (n = 3).
To map transcriptional start sites (TSSs) within the LM and OP regions, 5’ RACE was performed on RNA extracted from dis3–54 exosome mutant cells harbouring p3xLM or p3xOP (Fig. 7C, S4 Fig.). Many TSSs were identified within LM and OP, suggesting that these regions contain several promoters (Fig. 7C). 200 bp regions from both LM and OP were tested for their ability to drive production of β-galactosidase when placed upstream of a lacZ reporter in fission yeast and as shown in Fig. 7D, the regions displayed promoter activity. Mutated or inverted versions of promoter region M2 did not promote transcription of LacZ (S4 Fig.). Whilst most regions of LM and OP exhibit promoter activity that is lower than that of nmt81 control promoter, it is notable that region-O1 and region-P2 from OP have equivalent and 10-fold higher activity, respectively (Fig. 7D). It is possible that the higher promoter activity possessed by some regions of OP may affect its ability to establish CENP-ACnp1. We surmise that the central domain from cen2 is peppered with promoters that can drive the production of transcripts on both strands. Their relative arrangement along with the strength and pattern of transcription may affect CENP-ACnp1 incorporation.
CENP-ACnp1 establishment is enhanced in mutants that increase RNAPII stalling
The progression of RNAPII is impeded by obstacles such as nucleosomes, DNA damage, bound proteins and by sequences that are intrinsically difficult to transcribe, causing transcriptional pausing, stalling or arrest [56]. RNAPII-associated proteins ease the passage of RNAPII through such impediments, contributing to the processivity of the polymerase [57]. TFIIS facilitates transcriptional elongation of stalled/backtracked RNAPII by stimulating cleavage of nascent transcripts [58–60]. Upon stalling an elongating RNAPII becomes mono- then poly-ubiquitylated on the largest Rpb1 subunit. A rescue pathway involving de-ubiquitylation by the ubiquitin hydrolase Ubp3 is deployed to restart stalled RNAPII [56,61].
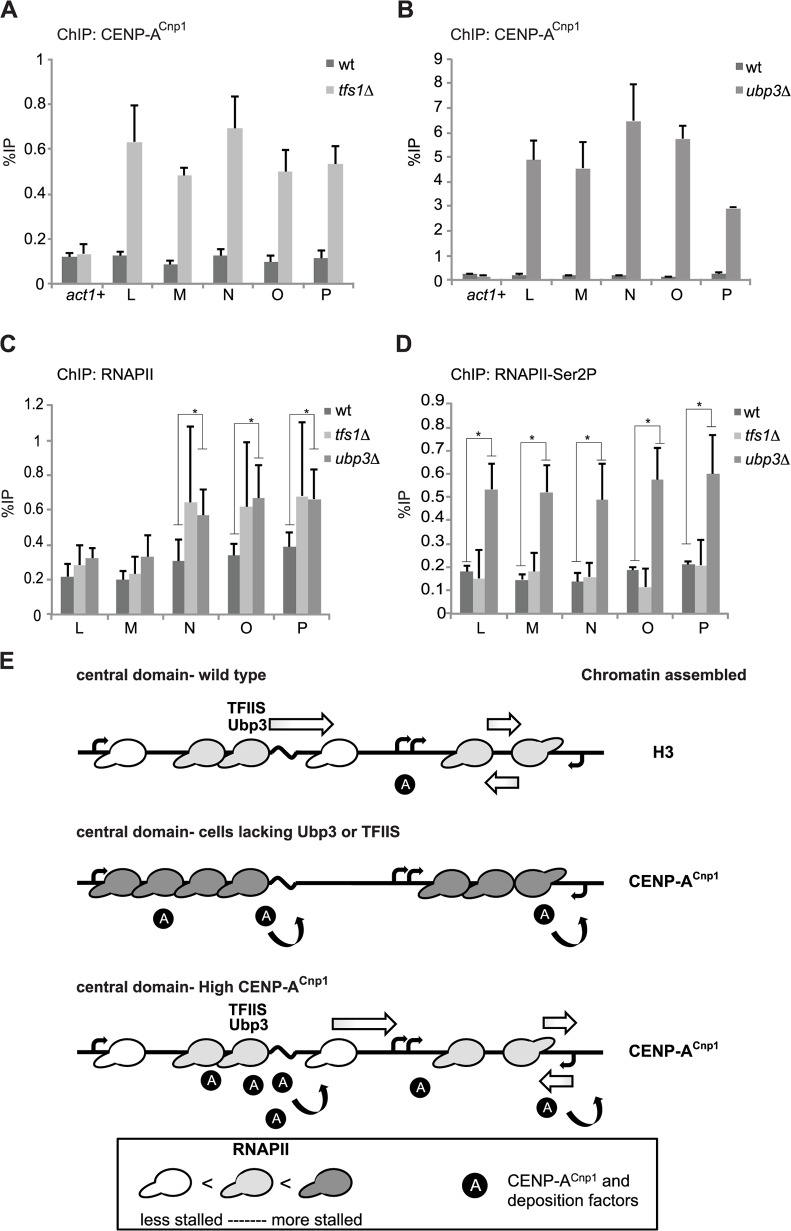
Our analyses suggest that the central domain chromatin landscape contains numerous promoters on both strands and multiple TSSs. In addition, long poly(dA:dT) tracts are likely to be an intrinsically problematic sequence for RNAPII transcription and present a barrier to RNAPII elongation [62,63]. We reasoned that mutants that are defective in the response to transcriptional stalling might influence the ability of the central domain to become assembled in CENP-ACnp1 chromatin. To test this possibility, wild-type and TFIIS (tfs1Δ) mutant cells expressing hi-CENP-ACnp1 were transformed with pMcc2. Surprisingly, slightly increased levels of CENP-ACnp1 were detected on pMcc2 in the tfs1Δ mutant compared to wild-type cells, suggesting that loss of TFSII promotes CENP-ACnp1 deposition (S5 Fig.). Consistent with this, even when pMcc2 was transformed into tfs1Δ cells expressing wt-CENP-ACnp1 levels, CENP-ACnp1 was detected on the pMcc2 central domain (Fig. 8A). In order to determine whether the effect on CENP-ACnp1 establishment was specific to tfs1Δ or a general consequence of increased RNAPII stalling, we also investigated if loss of the ubiquitin hydrolase Ubp3, which normally rescues arrested RNAPII, affects CENP-ACnp1 deposition. Strikingly, CENP-ACnp1 was detected at high levels on central domain sequences in ubp3Δ cells transformed with pMcc2. CENP-ACnp1 was also detected on p3xLM, but not p3xOP in ubp3Δ (Fig. 8B, S6 Fig.). CENP-CCnp3 and CENP-KSim4 centromere proteins were also significantly enriched on pMcc2 in ubp3Δ cells (S8 Fig.). These effects were not due to increased abundance of CENP-ACnp1 in tfs1Δ or ubp3Δ cells as protein levels were similar to wild-type cells (S7 Fig.). In fact, a reduction in CENP-ACnp1 and CENP-CCnp3 levels was detected at endogenous centromeres in ubp3Δ, but not tfs1Δ cells (S8 Fig.). Tfs1 and Ubp3 were previously reported to modulate RNAi-independent heterochromatin assembly [64]. To test whether the effect on CENP-ACnp1 establishment in tfs1Δ or ubp3Δ cells could be due to spurious assembly of heterochromatin on pMcc2, H3K9me2 ChIP was performed. The level of H3K9me2 on pMcc2 in tfs1Δand ubp3Δ was similar to that on a negative control locus, act1 +, and assembly of CENP-ACnp1 on pMcc2 in these mutants was not dependent on the H3K9-methyltransferase Clr4 (S9 Fig.). Thus, CENP-ACnp1 assembly on pMcc2 in the absence of TFIIS or Ubp3 does not result from induction by ectopic heterochromatin.
Fig 8. Mutants that affect RNAPII elongation allow de novo establishment of CENP-ACnp1 chromatin.
(A) ChIP for CENP-ACnp1 on a pMcc2 plasmid transformed in wild-type (wt) or a strain deleted for TFIISTfs1 (tfs1Δ) (n = 3). (B) ChIP for CENP-ACnp1 on a pMcc2 plasmid transformed in wild-type (wt) or a strain deleted for Ubp3 (ubp3Δ) (n = 3). (C) ChIP for RNAPII on pMcc2 plasmid in wt, tfs1Δ and ubp3Δ. p<0.05 is indicated with an asterisk. (D) Enrichment of RNAPII phosphorylated at Ser2 of its CTD (RNAPII-Ser2P) at the pMcc2 plasmid in wt, tfs1Δ and ubp3Δ. p<0.05 is indicated with an asterisk (n = 3) (E) Model for establishment of CENP-ACnp1 on central domain DNA. Top panel: Central domain DNA contains numerous weak promoters on both strands, leading to collisions between RNAPII complexes and stalling (depicted on right side). In addition, regions that are difficult to transcribe (wavy black line) may cause frequent stalling of RNAPII. RNAPII is depicted as an elongated ellipse and depth of shading indicates degree of stalling. Stalling is transient as factors such as TFIIS and Ubp3 help clear stalling so that elongation of RNAPII can proceed. Shaded-white arrow represents restarting of transcription. Middle panel: Persistent stalling occurs in cells lacking Ubp3 or TFIIS and this RNAPII status attracts deposition factors leading to the incorporation of CENP-ACnp1 in central domain chromatin. Black circles labelled ‘A’ represent CENP-ACnp1 and deposition factors. Curved black arrows represent CENP-ACnp1 deposition. Bottom panel: Even transient RNAPII stalling is sufficient to recruit CENP-ACnp1 and deposition factors when CENP-ACnp1 is present at high levels.
If lack of TFIIS or Ubp3 hinders transcriptional elongation, an increased level of RNAPII would be expected on affected chromatin templates. Elevated levels of Rpb1/RNAPII were detected on the central domain of pMcc2 in tfs1Δ (TFIIS) and ubp3Δ cells (Fig. 8C). In addition, increased levels of the elongation-specific Phospho-Ser2 form of RNAPII were observed on the central domain of pMcc2 in ubp3Δ cells, suggestive of failure to efficiently clear stalled RNAPII (Fig. 8D). Thus, two mutants, which perturb the progress of RNAPII elongation complexes in different ways, lead to deposition of CENP-ACnp1. These observations suggest that altering the transcriptional properties of the central domain chromatin through increased RNAPII stalling creates an environment that is permissive for establishment of CENP-ACnp1 chromatin in place of H3 chromatin.
Discussion
It is thought that once established, CENP-A chromatin has the ability to be ‘self-propagating’, and through the recruitment of factors that are themselves involved in deposition of CENP-A, it ensures its own maintenance [16,17,23,24,26,65]. Epigenetic inheritance can be defined as the propagation of a state in the absence of the initial inducer of that state. In this study, the inducer—overexpression of CENP-ACnp1—causes an event that would not normally occur, the assembly of CENP-ACnp1 chromatin on episomal centromeric DNA (pMcc2). When CENP-ACnp1-assembled pMcc2 is crossed from hi-CENP-ACnp1 cells into wt-CENP-ACnp1 cells, CENP-ACnp1 is propagated in the absence of the initial inducer through many generations and through meiosis. These observations further strengthen the evidence that CENP-A behaves as a bona fide epigenetic entity [24].
It is clear that both epigenetic and genetic factors influence CENP-A assembly. We have investigated the role of DNA sequence in establishment of CENP-A chromatin in fission yeast, an organism where analysis is not confounded by repetitive arrays of short satellite sequences. CENP-ACnp1 is normally restricted to the central domain of centromeres where it forms the basis for the kinetochore. Central domain DNA is a preferred substrate for establishment of CENP-ACnp1 chromatin upon overexpression, whilst other genomic loci do not support accumulation of high levels of CENP-ACnp1 [47], and even vector DNA adjacent to the central domain is not a good substrate. Conditions and mechanisms that influence assembly of CENP-ACnp1 on naïve plasmid DNA are also able to convert pre-chromatinised cc2 present on episomal plasmids or linear minichromosomes. What makes central domain DNA a preferred site for CENP-ACnp1 assembly? The lack of homology between cc2 and cc1/cc3 sequences suggests that it is not a simple case of specific sequence that is critical [66–68]. Our analyses indicate that there is functional redundancy within the central domain and no one particular sequence is either necessary or sufficient for CENP-ACnp1 establishment, consistent with previous findings [50]. Despite this redundancy it appears that there are inherent distinctions between different regions of cc2. The 2 kb sub-regions, LM and OP, are functionally non-equivalent and consistently behaved differently when challenged to assemble CENP-ACnp1 chromatin. LM is competent to establish centromeric chromatin upon CENP-ACnp1 overexpression, contains sufficient information to make a functional centromere when placed next to heterochromatin (pH-3xLM), and assembles CENP-ACnp1 chromatin in cells lacking Ubp3. On the other hand, the OP region fails to become assembled in CENP-ACnp1 chromatin in all these situations, yet can accept CENP-ACnp1 when adjacent to one copy of LM, which apparently acts as an initiator. The ability of LM, but not OP, to substitute for full-length cc2 sequence indicates that not all sequences are equivalent and LM must contain all information necessary to make this region permissive for CENP-ACnp1 establishment. It is possible that the observed higher promoter activity observed in the OP region (Fig. 7D) prevents stabilisation of CENP-ACnp1 nucleosomes on this sequence.
In common with many organisms, the central domain of S. pombe centromeres is AT rich and this property might contribute to the propensity of centromeric DNA to attract CENP-A [5,68]. S. pombe central domain DNA has an AT content of 72% (genome average of 64%), as does the establishment competent LM sequence. However, other regions that alone fail to support CENP-ACnp1 establishment have a similar AT content, such as OP (71% AT) and intergenic regions (72% AT). Moreover, randomisation of the LM sequence resulted in SynR-LM that, even with identical nucleotide composition (72% AT), was incompetent for CENP-ACnp1 establishment. Thus, high AT content alone, even when it mimics natural nucleosome positioning predictions, is not a defining factor in CENP-ACnp1 assembly. Together our observations indicate that rather than there being a specific critical sequence, central domain sequences encode unique properties capable of triggering or promoting the establishment of CENP-ACnp1 chromatin.
Transcription-coupled remodelling is associated with the deposition of histone variants and could potentially contribute to the assembly of CENP-A chromatin [69,70]. However, the simple act of transcription cannot be sufficient to provide specificity to the deposition of CENP-A. Our observations suggest that the transcriptional landscape of the centromeric central domain is unusual: scattered promoters of various strengths resulting in pervasive low quality transcription and numerous TSSs on both strands, in conjunction with poly(dA:dT) tracts that are inherently difficult to transcribe are likely to cause collision between convergently transcribing RNAPIIs and pile-ups at difficult sequences [63,71]. The relatively high density of RNAPII on pMcc2 contrasts with very low levels of transcripts (Fig. 7), consistent with inefficient progress of transcription by RNAPII on cc2, and many stalled elongation complexes. In addition, long tracts of poly(dA:dT) are known to disfavour nucleosome assembly, consistent with the apparently wide spacing of nucleosomes at endogenous centromeres [49,72]. These regions may be de facto nucleosome free regions, similar to those at promoters, allowing cryptic initiation of transcription to occur [72,73]. The randomized synthetic sequence SynR-LM that is a poor substrate for CENP-ACnp1 deposition has similar long A tracts, but transcription-related sequence-sensitive elements—such as promoters and transcription factor binding sites—would be destroyed. Thus, the central domain, due to its sequence-encoded properties, may produce a distinctive chromatin and transcriptional environment.
CENP-ACnp1 chromatin does not assemble de novo on cc2 sequence alone in wild-type cells expressing normal CENP-ACnp1 levels [45]. Instead, we envisage that the unique transcriptional chromatin environment created by the cc2 sequence renders it permissive for CENP-ACnp1 establishment, but that establishment occurs only if other favourable conditions exist. CENP-ACnp1 is preferentially incorporated on these central domain sequences upon overexpression, when adjacent to heterochromatin, and in the absence of factors that usually enhance transcriptional elongation. Any explanation of CENP-ACnp1 chromatin establishment on central domain DNA must also account for how CENP-ACnp1 is incorporated instead of H3. Serine 2 in the CTD heptad repeat of Rpb1 is phosphorylated in elongating RNAPII, and this Ser2P-Rbp1/RNAPII becomes ubiquitylated upon stalling [74–76]. The ubiquitin hydrolase Ubp3 normally acts as a proof-reading activity to prevent degradation of stalled but rescuable RNAPII [56,61]. Absence of Ubp3 compromises the processing of stalled RNAPII, resulting in the accumulation of ubiquitylated Ser2P-Rbp1/RNAPII complexes. We propose that such modifications contribute to the distinctive status of central domain chromatin, leading to recruitment of factors that promote CENP-ACnp1 deposition (Fig. 8E). Alternatively, it may create an environment in which H3 nucleosomes are efficiently turned over/evicted, whereas CENP-ACnp1 nucleosomes are poorly evicted specifically in the context of stalled RNAPII. In cells lacking Ubp3, severe or prolonged stalling, even with normal levels of CENP-ACnp1, would provide extended opportunities for CENP-ACnp1 recruitment, or poor eviction of CENP-ACnp1 during prolonged stalling. TFIIS promotes transcriptional elongation by cleaving nascent transcripts in the context of stalled/backtracked RNAPII [57,58,77]. Although the effects of TFIIS deletion are more subtle than lack of Ubp3, the accumulation of RNAPII correlates with assembly CENP-ACnp1 chromatin, supporting a mechanism where persistent RNAPII stalling within central domain triggers remodelling that results in CENP-ACnp1 deposition.
In this model, when naïve central domain DNA (pMcc2) is introduced into wild-type cells, transient stalling occurs but it is efficiently cleared with the aid of factors such as TFIIS and Ubp3 (Fig. 8E). Because in wild-type cells CENP-ACnp1 levels are extremely low compared to histone H3 there would be little opportunity for CENP-ACnp1 to gain access to cc2, and with efficient clearing of stalled RNAPII, CENP-ACnp1 would fail to accumulate in cc2 [78]. CENP-ACnp1 overexpression would increase the probability of interaction with the transiently stalled RNAPII in central domain chromatin, increasing the likelihood of recruitment. Alternatively, increased access coupled with poor eviction would lead to CENP-ACnp1 accumulation. In addition, CENP-ACnp1 nucleosomes themselves, which have distinct N-terminal tails that lack the conserved lysine residues of H3 whose modification aids transcription, are likely to present a greater barrier to transcription than H3 nucleosomes [79]. Thus, once incorporated, CENP-ACnp1 nucleosomes might exacerbate the poor transcriptional elongation, creating conditions permissive for recruitment of more CENP-ACnp1 in a self-perpetuating system. Longer regions of central domain DNA would have greater probability of triggering stalling events and thus be more likely to initiate the incorporation of CENP-ACnp1. In the context of this model, heterochromatin could promote establishment of CENP-ACnp1 chromatin on adjacent cc2 sequence by drawing plasmids to sites of endogenous heterochromatin such as the spindle pole body where they would encounter a higher concentration of CENP-ACnp1 than non-heterochromatinized plasmids located in the nuclear interior [80]. Alternatively, heterochromatin-associated chromatin modifying activities may influence transcriptional elongation by RNAPII within cc2, causing enhanced stalling and deposition of CENP-ACnp1[41].
Following establishment of CENP-A chromatin and kinetochore assembly, transcription could play a proof-reading role that evicts H3 deposited at centromeres during S phase [81]. Indeed, transcription and RNAPII have been detected at centromeres in mammalian cells and transcription/RNAPII may play a role in centromere integrity [33,34,38]. Transcription of human α-satellite arrays introduced as HACs is known to occur. Although CENP-A assembly is compatible with targeting of mild transcriptional activators, targeting of a strong transcriptional activator is deleterious [30,38,82]. Thus transcription and/or the transcription-coupled histone modifications detected at centromeres may promote CENP-A deposition at mammalian centromeres.
In conclusion, we show that the sequence of fission yeast centromere central domain DNA is important only in so far as it encodes for certain properties that contribute to the region’s unusual chromatin and transcriptional landscape. Establishment of CENP-ACnp1 chromatin is driven by these sequence-encoded properties that when combined with the presence of nearby heterochromatin, overexpressed CENP-ACnp1 or increased RNAPII stalling, tips the balance in favour CENP-ACnp1 chromatin assembly. It seems likely that a similar combination of factors, which together favour CENP-A incorporation, must also contribute to the formation of neocentromeres at novel chromosomal locations.
Materials and Methods
Cell growth and manipulation
Standard genetic and molecular techniques were followed. Fission yeast methods were as described [83]. Fission yeast strains are listed in Table 1. Minichromosomes used in this study were transformed by electroporation. Transformants were selected by growth on PMG—ura—ade at 32°C. As circular minichromosomes lack heterochromatin and therefore centromeric cohesion, plasmids were maintained in cells by selection in medium lacking adenine and uracil. 3 independent colonies from each transformation were analysed for the presence of kinetochore proteins by chromatin immunoprecipitation (ChIP).
Table 1. Schizosaccharomyces pombe strains.
| A7373 | cc2Δ::cc1 ade6-704-HYGMX6 ura4- |
| A7408 | cc2Δ::cc1 ars1:nmt41-GFP-cnp1-NAT ade6-704-HYGMX6 ura4- |
| A7528 | cc2Δ::cc1 dis3–54 ade6-704-HYGMX6 ura4- |
| A9235 | cc2Δ::cc1 tfs1Δ::leu2 ade6-704-HYGMX6 ura4- |
| A9391 | cc2Δ::cc1 ubp3Δ::KAN ade6-704-HYGMX6 ura4- |
| A9491 | cc2Δ::cc1 tfs1Δ::leu2 clr4Δ::NAT ade6-704-HYGMX6 ura4- |
| A9477 | cc2Δ::cc1 ubp3Δ::KAN clr4Δ::NAT ade6-704-HYGMX6 ura4- |
| A9048 | cc2Δ::cc1 [Ch16 m23:ura4::cc2-KAN ade6-216] ade6-210 leu1-32 ura4- |
| A9054 | cc2Δ::cc1 [Ch16 m23:ura4::cc2-KAN ade6-216] ars1:nmt41-GFP-Cnp1-NAT ade6–210 ura4- |
| A9059 | cc2Δ::cc1 [Ch16-ade6-216] ade6-210 leu1-32 ura4- |
| A9066 | cc2Δ::cc1 [Ch16-ade6-216] ars1:nmt41-GFP-Cnp1-NAT ade6-210 ura4- |
Centromere plasmids and minichromosomes
Plasmids bearing centromere fragments contained a minimal ars1 element to ensure efficient replication in S. pombe, in addition to selectable markers sup3-5 (complements ade6-704), ura4 + and KANR. 8.5 kb of central domain DNA (cc2 plus inner part of imr2L and imr2R) was cloned into the multiple cloning site as a SalI-NcoI fragment to create pMcc2. Various sub-fragments of cc2 (J-Q) were amplified by PCR and cloned into the multiple cloning site as BamHI/BglII fragments. 5.6 kb of heterochromatin-forming outer repeat sequence was inserted adjacent to central domain sequences to test ability to form functional centromeres.
A plasmid, pMC28, bearing cc2, a KAN resistance marker and an inverted ura4 sequence was constructed from pMcc2. Linearisation of the plasmid at NotI within the inverted ura4 sequence allowed integration at ura4 + located on the arm of Ch16-m23:ura4 +. Ch16-m23: ura4 + is a derivative of Ch16, a 530 kb minichromosome, itself derived from Chromosome III [48]. It also bears the ade6-216 allele which complements the ade6-210 allele present on endogenous Chromosome III by interallelic complementation. Integration of linearised pMC28 on Ch16-m23:ura4 allowed selection on the counter-selective drug 5-fluoro-orotic acid and G418 (KAN). Cells that lost the Ch16-m23:ura4::cc2-KAN (abbreviated as Ch16-cc2) became red on limiting adenine and were sensitive to G418. For growth in liquid, cells containing Ch16-cc2 were grown in media lacking adenine.
ChIP
ChIP was performed as previously described [84] using anti-CENP-Acnp1 antibody, anti CENP-CCnp3 antibody, anti-CENP-KSim4 antibody, anti-H3 antibody (ab1791; Abcam,), anti-H3K9me2 antibody (T. Urano) and anti-total RNA polymerase II (4F8; 61081, Active Motif), anti-Rpb1-Ser2P (3E10; 61083, Active Motif) and analysed by qPCR. Primers are listed in Table 2. P-values were calculated by standard t-test on 3 replicates between wild-type and mutant; p<0.05 was considered significant.
Table 2. Primers used in this study.
| qact1 Fw | CCCAAATCCAACCGTGAGAAGATG |
| qact1 rev | CCAGAGTCCAAGACGATACCAGTG |
| qcnt1 fw | CAGACAATCGCATGGTACTATC |
| qcnt1 rev | AGGTGAAGCGTAAGTGAGTG |
| qpDF16-vect2 fw | ATAATACCGCGCCACATAGC |
| qpDF16-vect2 rev | CCAGAAACGCTGGTGAAAGT |
| qcnt2 L fw | GCATCTATTGTACTCTCTC |
| qcnt2 L rev | GAAGGATGGATATGCACGT |
| qcnt2 M fw | GTTAATTGCCATTCTTTGGCG |
| qcnt2 M rev | ATGACATGGCGTGGAAAGTC |
| qcnt2 N fw | CATTAAACAAACAACGGCACAC |
| qcnt2 N rev | TAAGCCAGCAAATTCCTTGAG |
| qcnt2 O fw | GACTATAACTAGACCACTCAG |
| qcnt2 O rev | CTAGATGAATACTCAAGAAAGC |
| qcnt2 P fw | CTGCATATTCGACATCTTGAG |
| qcnt2 P rev | AGCCTGTCCATCGCAAAAGG |
| CC2-F1 (J) | TACTACGGATCCCCCCATGGAATAAT |
| CC2-R1(J) | TACTACGTCGACCTCGAGATCTCAGTGCTCGCTTCTGTGTAA |
| CC2-F2 (K) | TACTACGGATCCGCGAGCACTGTTTACATCTAA |
| CC2-R2(K) | TACTACGTCGACCTCGAGATCTCGTCAAGTTTACAACTCGGT |
| CC2-F3(L) | TACTACGGATCCACCGAGTTGTAAACTTGACG |
| CC2-R3(L) | TACTACGTCGACCTCGAGATCTACAAATAGAATATGCTAACGC |
| CC2-F4(M) | TACTACGGATCCGCGTTAGCATATTCTATTTGTC |
| CC2-R4(M) | TACTACGTCGACCTCGAGATCTTGCGAGTTATGTGTATCTAC |
| CC2-F5(N) | TACTACGGATCCGTAGATACACATAACTCGCA |
| CC2-R5(N) | TACTACGTCGACCTCGAGATCTGGGGTCCTAGATTACGTCTT |
| CC2-F6(O) | TACTACGGATCCAATCTAGGACCCCTACGTTTT |
| CC2-R6(O) | TACTACGTCGACCTCGAGATCTCCCTTGGAAATACCTAGC |
| CC2-F7(P) | TACTACGGATCCGCTAGGTATTTCCAAGGGCTCC |
| CC2-R7(P) | TACTACGTCGACCTCGAGATCTTCCACCACAAATAGTTCAGC |
| CC2-F8(Q) | TACTACGGATCCGCTGAACTATTTGTGGTGGAC |
| CC2-R8(Q) | TACTACGTCGACCTCGAGATCTAAAGGGTATAAACGGCTATC |
| RegionL1-Lacz Fw | TACTACCTGCAGCTTGTCGCAAAACATAAAGC |
| RegionL1-Lacz rev | TACTACCTCGAGGTAAGTTTCTAAACAGGA |
| RegionL2-Lacz Fw | TACTACCTGCAGCGATAAACGTATGATTGCTTTCACC |
| RegionL2-Lacz rev | TACTACCTCGAGGAGAGAGTACAATAGATGC |
| RegionM1-Lacz Fw | TACTACCTGCAGCGTACCTTTTGACCTTTAAG |
| RegionM1-Lacz rev | TACTACGTCGACTCGAG CCGTATTTCACTTAAACGAG |
| RegionM2-Lacz Fw | TACTACCTGCAGTGCGAGTTATGTGTATCTAC |
| RegionM2-Lacz rev | TACTACGTCGACTCGAGCTCGTTTAAGTGAAATACGG |
| RegionO1-Lacz Fw | TACTACCTGCAGCCCTTGCCAGTAATGTGTAT |
| RegionO1-Lacz rev | TACTACCTCGAGCGCATTGAATTTTAGACTG |
| RegionO2-Lacz Fw | TACTACCTGCAGCATTCAAATTGCGTCAAC |
| RegionO2-Lacz rev | TACTACCTCGAGCCAAGTATATGTGTTCTACC |
| RegionP1-Lacz Fw | TACTACCTGCAGAACCAGATTTAATTAACGGCC |
| RegionP1-Lacz rev | TACTACCTCGAGGAAGCACTACTAAGATTAC |
| RegionP2-Lacz Fw | TACTACCTGCAGGTCGATTTTTCGAACTTTCTGC |
| RegionP2-Lacz rev | TACTACCTCGAGTTGGATTCGAAGGTCTTTAC |
| probe cc2 F | CGTGCACATTTGTGAAAAGG |
| probe cc2 rev | TCTCGCGATTAGTTTGTAAAG |
| probe cc1 F | CCATTTGCTAAGTTCGACTC |
| probe cc1 rev | CAGTATTTGTATCGTAGTGG |
Establishment assay
For the establishment assay, cells were transformed with minichromosomes (containing 5.6 kb of outer repeat sequence in addition to cc2 sequences), by electroporation with ∼200 ng of DNA and plated on selective medium. Resultant colonies were replicated onto rich medium containing limiting adenine. The presence of pale pink/white colonies indicates establishment of a functional centromere on the minichromosome. Establishment efficiency is calculated as percentage of these colonies divided by the total number of transformants. Colonies were streaked on limiting adenine plates to confirm the presence of sectoring that is indicative of centromere function.
Real-time PCR (qPCR)
Quantitative PCR reactions were carried out in 10 μl volume, with 5μl Light Cycler 480 SybrGreen Master Mix (Roche), 0.5μl each primer (10 μM) and 3μl ChIP or total template. The data were analysed using Light Cycler 480 Software 1.5 (Roche).
5’RACE-PCR and RT-PCR
5’RACE-PCR was performed as previously described [37]. In brief, RNA was isolated with RNeasy mini/midi kit (Qiagen) according to the manufacturer’s protocol. Poly(A) containing RNA was purified from 500 μg of total RNA by affinity purification with biotinylated oligo-dT using PolyATtract mRNA Isolation Systems (Promega). 5’RACE PCR was performed using SMARTer 5’/3’ RACE (Clontech) according to the manufacturer’s protocol. PCR products were then run on 1% agarose gel, purified and cloned into pGEM-T Easy vector (Promega) and subsequently sequenced. Reverse transcription reaction for 5’RACE and qRT-PCR was performed using Superscript III Reverse Transcriptase (Invitrogen) using RNA extracted from 3 independent colonies. For qRT-PCR, transcript levels were normalized over gDNA to take into account differences in copy number between plasmids and normalized relative to act1 +.
LacZ assay
LacZ assay was performed as described [85]. pREP81X-LacZ was digested with XhoI and PstI and the nmt81 promoter upstream of LacZ replaced with sequences from centromere 2. Plasmids were transformed into wild-type and grown on minimal medium (n = 3).
Southern analysis
DNA was extracted as previously described [83]. The DNA was digested with BglII/SpeI or SphI/SpeI, run on a 1% agarose gel, blotted on nylon membrane (Hybond N, Amersham) and UV-crosslinked. The membrane was hybridized with DNA probes specific for central domain 1 or central domain 2. To make the probes, PCR products were used as template in the labelling reaction using High Prime (Roche). Primers sequences are listed in Table 2.
Western analysis
Western analysis was performed as described previously using anti-GFP antibody (Roche) and anti-H3 antibody (ab1794-abcam) [86]. The intensities of GFP and H3 signals were acquired using LICOR Odyssey Infrared Imaging System software (Li-COR Bioscience).
Supporting Information
(A) Schematic representation of endogenous centromere 1 (cnt1), centromere 2 (cnt2) and central core 2 deletion (cc2Δ). The probes on cc2 (grey) and cc1 (black) used for Southern analysis and the expected size of DNA fragments following restriction enzyme digestion are indicated. (B) Genomic DNA was extracted from wild-type cells and cc2Δ strain. DNA was digested with BglII and SpeI and the Southern hybridised with the 2.8 kb cc1/3 probe. (C) Genomic DNA was extracted from wild-type cells and the cc2Δ strain. The DNA was digested with SphI and SpeI and the Southern hybridised with the 2.8 kb cc2 probe. The absence of the band for cc2 indicates its deletion from the endogenous cen2 locus. (D) Spotting assay of wild-type or clr4Δ cells containing centromere 2 sequence at the endogenous centromere (cc2+) or wild-type cells with cc2 sequence replaced by cc1 (cc2Δ). YES: complete medium. Phloxin B stains dead cells red. Cells with impaired centromere function (e.g. clr4Δ) show sensitivity to the microtubule destabilizing drug thiabendazole (TBZ), while cc2 + and cc2Δ are able to grow in the presence of the drug TBZ suggesting that cc2Δ does not affect centromere function.
(EPS)
(A) ChIP analysis of CENP-ACnp1 levels of wild-type (wt-CENP-ACnp1) and cells overexpressing CENP-ACnp1 (hi-CENP-ACnp1) transformed with a plasmid containing a combination of LM and OP fragment (OP-LM-OP). (B) Establishment assay using plasmids containing 5.6 kb of heterochromatin-forming outer repeat element flanking a full length cc2 (pH-cc2), three tandem repeats of the LM region (pH-3xLM), three tandem repeats of the OP region (pH-3xOP) or one copy of the LM fragment adjacent to two copies of OP (pH-LM-2xOP). pH-cc2, pH-3xLM, pH-3xOP and pH-LM-2xOP were transformed in wild-type cells and the percentage of transformants that contained plasmids with centromere function was assessed by replica plating onto limiting adenine medium. White colonies indicate formation of functional centromeres.
(EPS)
ChIP of RNA polymerase II (RNAPII) on the p3xLM (A) and p3xOP (B) plasmids transformed into wild-type cells. Positions of the PCR products quantified for each plasmid are indicated. (C) ChIP of RNA polymerase II (RNAPII) on the pMcc2 plasmid transformed into wild-type (wt-CENP-ACnp1) and cells overexpressing CENP-ACnp1 (hi-CENP-ACnp1). The enrichment is shown as relative to act1 + (n = 3).
(EPS)
(A) Example of RT-PCR products obtained from the 5’RACE reaction. Generally, multiple products were identified for the centromeric regions, while one single product was identified from the act1 gene. (B) Sequences of TSSs detected by 5’RACE in LM and OP regions. (C) Analysis of promoter activity of the ∼200 bp regions from cc2. Region-M1 and region-M2 were placed upstream of a LacZ reporter gene. The levels of LacZ expression were assessed by measuring absorbance at 420 nm of cell lysates incubated with 2-Nitrophenyl-β-D-galactopyranoside (ONPG). region-M2 inv: region cloned inverted; region-M2 mut: point mutations inserted in the M2sequence. nmt81: positive control with nmt81 promoter (n = 3).
(EPS)
Enrichment levels of CENP-ACnp1 at the endogenous centromeres (cc1/cc3) and at the pMcc2 plasmid transformed in wild-type cells (wt), in cells expressing high levels of CENP-ACnp1 (hi-CENP-ACnp1) and in hi-CENP-ACnp1 in which the transcription factor TFIIS (Tfs1) is deleted (tfs1Δ+hi-CENP-ACnp1) (n = 3).
(EPS)
ChIP for CENP-ACnp1 on a p3xLM and p3xOP plasmids transformed into wild-type (wt) or a strain deleted for Ubp3 (ubp3Δ). CENP-ACnp1 levels are shown relative to cc1/3.
(EPS)
Western analysis and protein levels comparison between wild-type (wt), ubp3Δ and tfs1Δ of endogenous N-terminal tagged GFP-CENP-ACnp1 and H3. Ponceau staining as internal loading control.
(EPS)
(A) ChIP for CENP-ACnp1, CENP-CCnp3 and CENP-KSim4 levels at endogenous centromere cc1/3 in wild-type (wt), tfs1Δ and ubp3Δ. p<0.05 is indicated with an asterisk (n = 3). (B) ChIP for CENP-CCnp3 and CENP-KSim4 levels at pMcc2 plasmid transformed into wt, tfs1Δ and ubp3Δ. p<0.05 is indicated with an asterisk (n = 3).
(EPS)
(A) ChIP for H3K9me2 (A) and CENP-ACnp1 (B) at act1 + and at pMcc2 plasmid transformed in wt, clr4Δ, ubp3Δ and ubp3Δclr4Δ. CENP-ACnp1 is deposited on pcc2 in ubp3 regardless of whether Clr4 is present or absent. (C) ChIP for H3K9me2 at act1 + and on the pcc2 plasmid transformed into wt, tfs1Δ and tfs1Δclr4Δ cells. The absence of H3K9me2 on the plasmid suggests that heterochromatinisation is not responsible for CENP-ACnp1 deposition. (D) CENP-ACnp1 levels on act1 + and at the pcc2 plasmid in wt, tfs1Δ and tfs1Δclr4Δ. The absence of the methyltranferase Clr4 does not affect CENP-ACnp1 deposition on pMcc2.
(EPS)
Acknowledgments
We thank all members of the Allshire lab for helpful advice and discussion. In particular, P. Tong for bioinformatics support, and R. Ard, P. Audergon, and M. Shukla for comments on the manuscript. We also thank S. Webb for designing the program for randomization of DNA sequence. We are grateful to S. Forsburg for providing the pRep81X-LacZ plasmid and T. Urano for providing the H3K9me2 antibody (mAb5.1.1).
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by the Wellcome Trust [095021] to RCA and [086574] to SC along with funding from the European Commission Network of Excellence EpiGeneSys- (HEALTH-F4-2010-257082) to RCA. The Wellcome Trust Centre for Cell Biology is supported by core funding from the Wellcome Trust [092076]. www.epigenesys.eu/en/www.wellcome.ac.uk/ The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Westermann S, Schleiffer A (2013) Family matters: structural and functional conservation of centromere-associated proteins from yeast to humans. Trends in Cell Biology 23: 260–269. 10.1016/j.tcb.2013.01.010 [DOI] [PubMed] [Google Scholar]
- 2. Henikoff S, Ahmad K, Malik HS (2001) The centromere paradox: stable inheritance with rapidly evolving DNA. Science 293: 1098–1102. [DOI] [PubMed] [Google Scholar]
- 3. Meraldi P, McAinsh AD, Rheinbay E, Sorger PK (2006) Phylogenetic and structural analysis of centromeric DNA and kinetochore proteins. Genome Biol 7: R23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buscaino A, Allshire R, Pidoux AL (2010) Building centromeres: home sweet home or a nomadic existence? Current Opinion in Genetics & Development: 1–9. [DOI] [PubMed]
- 5. Melters DP, Bradnam KR, Young HA, Telis N, May MR, et al. (2013) Comparative analysis of tandem repeats from hundreds of species reveals unique insights into centromere evolution. Genome Biol 14: R10 10.1186/gb-2013-14-1-r10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Biggins S (2013) The composition, functions, and regulation of the budding yeast kinetochore. Genetics 194: 817–846. 10.1534/genetics.112.145276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Steiner FA, Henikoff S (2014) Holocentromeres are dispersed point centromeres localized at transcription factor hotspots. eLife Sciences 3: e02025 10.7554/eLife.02025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Maddox PS, Oegema K, Desai A, Cheeseman IM (2004) “Holo”er than thou: chromosome segregation and kinetochore function in C. elegans. Chromosome Res 12: 641–653. [DOI] [PubMed] [Google Scholar]
- 9. Willard HF (1985) Chromosome-specific organization of human alpha satellite DNA. Am J Hum Genet 37: 524–532. [PMC free article] [PubMed] [Google Scholar]
- 10. Willard HF (1989) The genomics of long tandem arrays of satellite DNA in the human genome. Genome 31: 737–744. [DOI] [PubMed] [Google Scholar]
- 11. Marçais B, Laurent AM, Charlieu JP, Roizès G (1993) Organization of the variant domains of alpha satellite DNA on human chromosome 21. J Mol Evol 37: 171–178. [DOI] [PubMed] [Google Scholar]
- 12. Pidoux AL, Allshire RC (2004) Kinetochore and heterochromatin domains of the fission yeast centromere. Chromosome Res 12: 521–534. [DOI] [PubMed] [Google Scholar]
- 13. Sanyal K, Baum M, Carbon J (2004) Centromeric DNA sequences in the pathogenic yeast Candida albicans are all different and unique. Proc Natl Acad Sci USA 101: 11374–11379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shang WH, Hori T, Toyoda A, Kato J, Popendorf K, et al. (2010) Chickens possess centromeres with both extended tandem repeats and short non-tandem-repetitive sequences. Genome Research 20: 1219–1228. 10.1101/gr.106245.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gong Z, Wu Y, Koblízková A, Torres GA, Wang K, et al. (2012) Repeatless and repeat-based centromeres in potato: implications for centromere evolution. Plant Cell 24: 3559–3574. 10.1105/tpc.112.100511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Allshire RC, Karpen GH (2008) Epigenetic regulation of centromeric chromatin: old dogs, new tricks? Nat Rev Genet 9: 923–937. 10.1038/nrg2466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Burrack LS, Berman J (2012) Neocentromeres and epigenetically inherited features of centromeres. Chromosome Res 20: 607–619. 10.1007/s10577-012-9296-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Marshall OJ, Chueh AC, Wong LH, Choo KHA (2008) Neocentromeres: new insights into centromere structure, disease development, and karyotype evolution. Am J Hum Genet 82: 261–282. 10.1016/j.ajhg.2007.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ishii K, Ogiyama Y, Chikashige Y, Soejima S, Masuda F, et al. (2008) Heterochromatin Integrity Affects Chromosome Reorganization After Centromere Dysfunction. Science 321: 1088–1091. 10.1126/science.1158699 [DOI] [PubMed] [Google Scholar]
- 20. Stimpson KM, Matheny JE, Sullivan BA (2012) Dicentric chromosomes: unique models to study centromere function and inactivation. Chromosome Res 20: 595–605. 10.1007/s10577-012-9302-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sato H, Masuda F, Takayama Y, Takahashi K, Saitoh S (2012) Epigenetic inactivation and subsequent heterochromatinization of a centromere stabilize dicentric chromosomes. Curr Biol 22: 658–667. 10.1016/j.cub.2012.02.062 [DOI] [PubMed] [Google Scholar]
- 22. Catania S, Allshire RC (2014) Anarchic centromeres: deciphering order from apparent chaos. Current Opinion in Cell Biology 26: 41–50. 10.1016/j.ceb.2013.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Falk SJ, Black BE (2012) Centromeric chromatin and the pathway that drives its propagation. Biochim Biophys Acta 1819: 313–321. 10.1016/j.bbagrm.2011.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mendiburo MJ, Padeken J, Fülöp S, Schepers A, Heun P (2011) Drosophila CENH3 is sufficient for centromere formation. Science 334: 686–690. 10.1126/science.1206880 [DOI] [PubMed] [Google Scholar]
- 25. Barnhart MC, Kuich PHJL, Stellfox ME, Ward JA, Bassett EA, et al. (2011) HJURP is a CENP-A chromatin assembly factor sufficient to form a functional de novo kinetochore. The Journal of Cell Biology 194: 229–243. 10.1083/jcb.201012017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fachinetti D, Diego Folco H, Nechemia-Arbely Y, Valente LP, Nguyen K, et al. (2013) A two-step mechanism for epigenetic specification of centromere identity and function. Nature Cell Biology 15: 1056–1066. 10.1038/ncb2805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Valente LP, Dehé P-M, Klutstein M, Aligianni S, Watt S, et al. (2013) Myb-domain protein Teb1 controls histone levels and centromere assembly in fission yeast. EMBO J. [DOI] [PMC free article] [PubMed]
- 28. Malik HS, Henikoff S (2009) Major evolutionary transitions in centromere complexity. Cell 138: 1067–1082. 10.1016/j.cell.2009.08.036 [DOI] [PubMed] [Google Scholar]
- 29. Nakano M (2003) Epigenetic assembly of centromeric chromatin at ectopic-satellite sites on human chromosomes. Journal of Cell Science 116: 4021–4034. [DOI] [PubMed] [Google Scholar]
- 30. Bergmann JH, Martins NMC, Larionov V, Masumoto H, Earnshaw WC (2012) HACking the centromere chromatin code: insights from human artificial chromosomes. Chromosome Res 20: 505–519. 10.1007/s10577-012-9293-0 [DOI] [PubMed] [Google Scholar]
- 31. Mello CC, Kramer JM, Stinchcomb D, Ambros V (1991) Efficient gene transfer in C.elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J 10: 3959–3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yuen KWY, Nabeshima K, Oegema K, Desai A (2011) Rapid de novo centromere formation occurs independently of heterochromatin protein 1 in C. elegans embryos. Curr Biol 21: 1800–1807. 10.1016/j.cub.2011.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scott KC (2013) Transcription and ncRNAs: at the cent(rome)re of kinetochore assembly and maintenance. Chromosome Res. [DOI] [PubMed]
- 34. Rošić S, Köhler F, Erhardt S (2014) Repetitive centromeric satellite RNA is essential for kinetochore formation and cell division. The Journal of Cell Biology 207: 335–349. 10.1083/jcb.201404097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ohkuni K, Kitagawa K (2011) Endogenous Transcription at the Centromere Facilitates Centromere Activity in Budding Yeast. Curr Biol. [DOI] [PMC free article] [PubMed]
- 36. Topp CN, Zhong CX, Dawe RK (2004) Centromere-encoded RNAs are integral components of the maize kinetochore. Proc Natl Acad Sci USA 101: 15986–15991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Choi ES, Stralfors A, Castillo AG, Durand-Dubief M, Ekwall K, et al. (2011) Identification of noncoding transcripts from within CENP-A chromatin at fission yeast centromeres. J Biol Chem 286: 23600–23607. 10.1074/jbc.M111.228510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chan FL, Marshall OJ, Saffery R, Won Kim B, Earle E, et al. (2012) Active transcription and essential role of RNA polymerase II at the centromere during mitosis. Proceedings of the National Academy of Sciences 109: 1979–1984. 10.1073/pnas.1108705109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bergmann JH, Rodríguez MG, Martins NMC, Kimura H, Kelly DA, et al. (2011) Epigenetic engineering shows H3K4me2 is required for HJURP targeting and CENP-A assembly on a synthetic human kinetochore. EMBO J 30: 328–340. 10.1038/emboj.2010.329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bergmann JH, Jakubsche JN, Martins NM, Kagansky A, Nakano M, et al. (2012) Epigenetic engineering: histone H3K9 acetylation is compatible with kinetochore structure and function. Journal of Cell Science 125: 411–421. 10.1242/jcs.090639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Choi ES, Strålfors A, Catania S, Castillo AG, Svensson JP, et al. (2012) Factors That Promote H3 Chromatin Integrity during Transcription Prevent Promiscuous Deposition of CENP-ACnp1 in Fission Yeast. PLoS Genet 8: e1002985 10.1371/journal.pgen.1002985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sadeghi L, Siggens L, Svensson JP, Ekwall K (2014) Centromeric histone H2B monoubiquitination promotes noncoding transcription and chromatin integrity. Nat Struct Mol Biol: 236–243. [DOI] [PubMed]
- 43. Chueh AC, Northrop EL, Brettingham-Moore KH, Choo KHA, Wong LH (2009) LINE retrotransposon RNA is an essential structural and functional epigenetic component of a core neocentromeric chromatin. PLoS Genet 5: e1000354 10.1371/journal.pgen.1000354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Quénet D, Dalal Y (2014) A long non-coding RNA is required for targeting centromeric protein A to the human centromere. eLife Sciences: e03254. [DOI] [PMC free article] [PubMed]
- 45. Folco HD, Pidoux AL, Urano T, Allshire RC (2008) Heterochromatin and RNAi Are Required to Establish CENP-A Chromatin at Centromeres. Science 319: 94–97. 10.1126/science.1150944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kagansky A, Folco HD, Almeida R, Pidoux AL, Boukaba A, et al. (2009) Synthetic Heterochromatin Bypasses RNAi and Centromeric Repeats to Establish Functional Centromeres. Science 324: 1716–1719. 10.1126/science.1172026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Castillo AG, Pidoux AL, Catania S, Durand-Dubief M, Choi ES, et al. (2013) Telomeric Repeats Facilitate CENP-A(Cnp1) Incorporation via Telomere Binding Proteins. PLoS ONE 8: e69673 10.1371/journal.pone.0069673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Niwa O, Matsumoto T, Chikashige Y, Yanagida M (1989) Characterization of Schizosaccharomyces pombe minichromosome deletion derivatives and a functional allocation of their centromere. EMBO J 8: 3045–3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lando D, Endesfelder U, Berger H, Subramanian L, Dunne PD, et al. (2012) Quantitative single-molecule microscopy reveals that CENP-A(Cnp1) deposition occurs during G2 in fission yeast. Open Biol 2: 120078 10.1098/rsob.120078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Baum M, Ngan VK, Clarke L (1994) The centromeric K-type repeat and the central core are together sufficient to establish a functional Schizosaccharomyces pombe centromere. Mol Biol Cell 5: 747–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Allshire RC, Nimmo ER, Ekwall K, Javerzat JP, Cranston G (1995) Mutations derepressing silent centromeric domains in fission yeast disrupt chromosome segregation. Genes & Development 9: 218–233. [DOI] [PubMed] [Google Scholar]
- 52. Struhl K, Segal E (2013) Determinants of nucleosome positioning. Nature Publishing Group 20: 267–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kanizay L, Dawe RK (2009) Centromeres: long intergenic spaces with adaptive features. Funct Integr Genomics 9: 287–292. 10.1007/s10142-009-0124-0 [DOI] [PubMed] [Google Scholar]
- 54. Segal E, Widom J (2009) What controls nucleosome positions? Trends in Genetics 25: 335–343. 10.1016/j.tig.2009.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kaplan N, Moore IK, Fondufe-Mittendorf Y, Gossett AJ, Tillo D, et al. (2009) The DNA-encoded nucleosome organization of a eukaryotic genome. Nature 458: 362–366. 10.1038/nature07667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wilson MD, Harreman M, Taschner M, Reid J, Walker J, et al. (2013) Proteasome-mediated processing of def1, a critical step in the cellular response to transcription stress. Cell 154: 983–995. 10.1016/j.cell.2013.07.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Selth LA, Sigurdsson S, Svejstrup JQ (2010) Transcript Elongation by RNA Polymerase II. Annu Rev Biochem 79: 271–293. 10.1146/annurev.biochem.78.062807.091425 [DOI] [PubMed] [Google Scholar]
- 58. Kulish D, Struhl K (2001) TFIIS enhances transcriptional elongation through an artificial arrest site in vivo. Molecular and Cellular Biology 21: 4162–4168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Cheung ACM, Cramer P (2011) Structural basis of RNA polymerase II backtracking, arrest and reactivation. Nature 471: 249–253. 10.1038/nature09785 [DOI] [PubMed] [Google Scholar]
- 60. Sigurdsson S, Dirac-Svejstrup AB, Svejstrup JQ (2010) Evidence that Transcript Cleavage Is Essential for RNA Polymerase II Transcription and Cell Viability. Molecular Cell 38: 202–210. 10.1016/j.molcel.2010.02.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kvint K, Uhler JP, Taschner MJ, Sigurdsson S, Erdjument-Bromage H, et al. (2008) Reversal of RNA polymerase II ubiquitylation by the ubiquitin protease Ubp3. Molecular Cell 30: 498–506. 10.1016/j.molcel.2008.04.018 [DOI] [PubMed] [Google Scholar]
- 62. Linton MF, Raabe M, Pierotti V, Young SG (1997) Reading-frame restoration by transcriptional slippage at long stretches of adenine residues in mammalian cells. J Biol Chem 272: 14127–14132. [DOI] [PubMed] [Google Scholar]
- 63. Kiyama R, Oishi M (1996) In Vitro Transcription of a Poly(dA){middle dot}Poly(dT)-Containing Sequence is Inhibited by Interaction between the Template and Its Transcripts. Nucleic Acids Research 24: 4577–4583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Reyes-Turcu FE, Zhang K, Zofall M, Chen E, Grewal SIS (2011) Defects in RNA quality control factors reveal RNAi-independent nucleation of heterochromatin. Nature Structural & Molcular Biology 18: 1132–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Black BE, Cleveland DW (2011) Epigenetic Centromere Propagation and the Nature of CENP-A Nucleosomes. Cell 144: 471–479. 10.1016/j.cell.2011.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Takahashi K, Murakami S, Chikashige Y, Funabiki H, Niwa O, et al. (1992) A low copy number central sequence with strict symmetry and unusual chromatin structure in fission yeast centromere. Mol Biol Cell 3: 819–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Polizzi C, Clarke L (1991) The chromatin structure of centromeres from fission yeast: differentiation of the central core that correlates with function. The Journal of Cell Biology 112: 191–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Wood V, Gwilliam R, Rajandream M- A, Lyne M, Lyne R, et al. (2002) The genome sequence of Schizosaccharomyces pombe. Nature 415: 871–880 [DOI] [PubMed] [Google Scholar]
- 69. Weber CM, Henikoff S (2014) Histone variants: dynamic punctuation in transcription. Genes & Development 28: 672–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Ray-Gallet D, Almouzni G (2010) Nucleosome dynamics and histone variants. Essays Biochem 48: 75–87. 10.1042/bse0480075 [DOI] [PubMed] [Google Scholar]
- 71. Kiyama R, Oishi M (1994) Instability of plasmid DNA maintenance caused by transcription of poly(dT)-containing sequences in Escherichia coli. Gene 150: 57–61. [DOI] [PubMed] [Google Scholar]
- 72. Segal E, Widom J (2009) Poly(dA:dT) tracts: major determinants of nucleosome organization. Curr Opin Struct Biol 19: 65–71. 10.1016/j.sbi.2009.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Raveh-Sadka T, Levo M, Shabi U, Shany B, Keren L, et al. (2012) Manipulating nucleosome disfavoring sequences allows fine-tune regulation of gene expression in yeast. Nat Genet 44: 743–750. 10.1038/ng.2305 [DOI] [PubMed] [Google Scholar]
- 74. Buratowski S (2009) Progression through the RNA Polymerase II CTD Cycle. Molecular Cell 36: 541–546. 10.1016/j.molcel.2009.10.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Heidemann M, Hintermair C, Voß K, Eick D (2013) Dynamic phosphorylation patterns of RNA polymerase II CTD during transcription. Biochim Biophys Acta 1829: 55–62. 10.1016/j.bbagrm.2012.08.013 [DOI] [PubMed] [Google Scholar]
- 76. Somesh BP, Reid J, Liu W-F, Søgaard TMM, Erdjument-Bromage H, et al. (2005) Multiple mechanisms confining RNA polymerase II ubiquitylation to polymerases undergoing transcriptional arrest. Cell 121: 913–923. [DOI] [PubMed] [Google Scholar]
- 77. Martinez-Rucobo FW, Cramer P (2013) Structural basis of transcription elongation. Biochim Biophys Acta 1829: 9–19. 10.1016/j.bbagrm.2012.09.002 [DOI] [PubMed] [Google Scholar]
- 78. Marguerat S, Schmidt A, Codlin S, Chen W, Aebersold R, et al. (2012) Quantitative analysis of fission yeast transcriptomes and proteomes in proliferating and quiescent cells. Cell 151: 671–683. 10.1016/j.cell.2012.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shandilya J, Senapati P, Hans F, Menoni H, Bouvet P, et al. (2014) Centromeric histone variant CENP-A represses acetylation-dependent chromatin transcription that is relieved by histone chaperone NPM1. J Biochem. [DOI] [PubMed]
- 80. Funabiki H, Hagan I, Uzawa S, Yanagida M (1993) Cell cycle-dependent specific positioning and clustering of centromeres and telomeres in fission yeast. The Journal of Cell Biology 121: 961–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Dunleavy EM, Almouzni G, Karpen GH (2011) H3.3 is deposited at centromeres in S phase as a placeholder for newly assembled CENP-A in G₁ phase. Nucleus 2: 146–157. 10.4161/nucl.2.2.15211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Nakano M, Cardinale S, Noskov VN, Gassmann R, Vagnarelli P, et al. (2008) Inactivation of a Human Kinetochore by Specific Targeting of Chromatin Modifiers. Developmental Cell 14: 507–522. 10.1016/j.devcel.2008.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Moreno S, Klar A, Nurse P (1991) Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Meth Enzymol 194: 795–823. [DOI] [PubMed] [Google Scholar]
- 84. Castillo AG, Mellone BG, Partridge JF, Richardson W, Hamilton GL, et al. (2007) Plasticity of Fission Yeast CENP-A Chromatin Driven by Relative Levels of Histone H3 and H4. PLoS Genet 3: e121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Guarente L (1983) Yeast promoters and lacZ fusions designed to study expression of cloned genes in yeast. Meth Enzymol 101: 181–191. [DOI] [PubMed] [Google Scholar]
- 86. Pidoux AL, Choi ES, Abbott JKR, Liu X, Kagansky A, et al. (2009) Fission Yeast Scm3: A CENP-A Receptor Required for Integrity of Subkinetochore Chromatin. Molecular Cell 33: 299–311. 10.1016/j.molcel.2009.01.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Schematic representation of endogenous centromere 1 (cnt1), centromere 2 (cnt2) and central core 2 deletion (cc2Δ). The probes on cc2 (grey) and cc1 (black) used for Southern analysis and the expected size of DNA fragments following restriction enzyme digestion are indicated. (B) Genomic DNA was extracted from wild-type cells and cc2Δ strain. DNA was digested with BglII and SpeI and the Southern hybridised with the 2.8 kb cc1/3 probe. (C) Genomic DNA was extracted from wild-type cells and the cc2Δ strain. The DNA was digested with SphI and SpeI and the Southern hybridised with the 2.8 kb cc2 probe. The absence of the band for cc2 indicates its deletion from the endogenous cen2 locus. (D) Spotting assay of wild-type or clr4Δ cells containing centromere 2 sequence at the endogenous centromere (cc2+) or wild-type cells with cc2 sequence replaced by cc1 (cc2Δ). YES: complete medium. Phloxin B stains dead cells red. Cells with impaired centromere function (e.g. clr4Δ) show sensitivity to the microtubule destabilizing drug thiabendazole (TBZ), while cc2 + and cc2Δ are able to grow in the presence of the drug TBZ suggesting that cc2Δ does not affect centromere function.
(EPS)
(A) ChIP analysis of CENP-ACnp1 levels of wild-type (wt-CENP-ACnp1) and cells overexpressing CENP-ACnp1 (hi-CENP-ACnp1) transformed with a plasmid containing a combination of LM and OP fragment (OP-LM-OP). (B) Establishment assay using plasmids containing 5.6 kb of heterochromatin-forming outer repeat element flanking a full length cc2 (pH-cc2), three tandem repeats of the LM region (pH-3xLM), three tandem repeats of the OP region (pH-3xOP) or one copy of the LM fragment adjacent to two copies of OP (pH-LM-2xOP). pH-cc2, pH-3xLM, pH-3xOP and pH-LM-2xOP were transformed in wild-type cells and the percentage of transformants that contained plasmids with centromere function was assessed by replica plating onto limiting adenine medium. White colonies indicate formation of functional centromeres.
(EPS)
ChIP of RNA polymerase II (RNAPII) on the p3xLM (A) and p3xOP (B) plasmids transformed into wild-type cells. Positions of the PCR products quantified for each plasmid are indicated. (C) ChIP of RNA polymerase II (RNAPII) on the pMcc2 plasmid transformed into wild-type (wt-CENP-ACnp1) and cells overexpressing CENP-ACnp1 (hi-CENP-ACnp1). The enrichment is shown as relative to act1 + (n = 3).
(EPS)
(A) Example of RT-PCR products obtained from the 5’RACE reaction. Generally, multiple products were identified for the centromeric regions, while one single product was identified from the act1 gene. (B) Sequences of TSSs detected by 5’RACE in LM and OP regions. (C) Analysis of promoter activity of the ∼200 bp regions from cc2. Region-M1 and region-M2 were placed upstream of a LacZ reporter gene. The levels of LacZ expression were assessed by measuring absorbance at 420 nm of cell lysates incubated with 2-Nitrophenyl-β-D-galactopyranoside (ONPG). region-M2 inv: region cloned inverted; region-M2 mut: point mutations inserted in the M2sequence. nmt81: positive control with nmt81 promoter (n = 3).
(EPS)
Enrichment levels of CENP-ACnp1 at the endogenous centromeres (cc1/cc3) and at the pMcc2 plasmid transformed in wild-type cells (wt), in cells expressing high levels of CENP-ACnp1 (hi-CENP-ACnp1) and in hi-CENP-ACnp1 in which the transcription factor TFIIS (Tfs1) is deleted (tfs1Δ+hi-CENP-ACnp1) (n = 3).
(EPS)
ChIP for CENP-ACnp1 on a p3xLM and p3xOP plasmids transformed into wild-type (wt) or a strain deleted for Ubp3 (ubp3Δ). CENP-ACnp1 levels are shown relative to cc1/3.
(EPS)
Western analysis and protein levels comparison between wild-type (wt), ubp3Δ and tfs1Δ of endogenous N-terminal tagged GFP-CENP-ACnp1 and H3. Ponceau staining as internal loading control.
(EPS)
(A) ChIP for CENP-ACnp1, CENP-CCnp3 and CENP-KSim4 levels at endogenous centromere cc1/3 in wild-type (wt), tfs1Δ and ubp3Δ. p<0.05 is indicated with an asterisk (n = 3). (B) ChIP for CENP-CCnp3 and CENP-KSim4 levels at pMcc2 plasmid transformed into wt, tfs1Δ and ubp3Δ. p<0.05 is indicated with an asterisk (n = 3).
(EPS)
(A) ChIP for H3K9me2 (A) and CENP-ACnp1 (B) at act1 + and at pMcc2 plasmid transformed in wt, clr4Δ, ubp3Δ and ubp3Δclr4Δ. CENP-ACnp1 is deposited on pcc2 in ubp3 regardless of whether Clr4 is present or absent. (C) ChIP for H3K9me2 at act1 + and on the pcc2 plasmid transformed into wt, tfs1Δ and tfs1Δclr4Δ cells. The absence of H3K9me2 on the plasmid suggests that heterochromatinisation is not responsible for CENP-ACnp1 deposition. (D) CENP-ACnp1 levels on act1 + and at the pcc2 plasmid in wt, tfs1Δ and tfs1Δclr4Δ. The absence of the methyltranferase Clr4 does not affect CENP-ACnp1 deposition on pMcc2.
(EPS)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.