Abstract
The acquisition of cell motility is an early step in melanoma metastasis. Here we use intravital imaging of signalling reporter cell-lines combined with genome-wide transcriptional analysis to define signalling pathways and genes associated with melanoma metastasis. Intravital imaging revealed heterogeneous cell behaviour in vivo: less than 10% of cells were motile and both singly moving cells and streams of cells were observed. Motile melanoma cells had increased Notch- and SRF-dependent transcription. Subsequent genome-wide analysis identified an overlapping set of genes associated with high Notch and SRF activity. We identified EZH2, a histone methyltransferase in the Polycomb Repressor Complex 2, as a regulator of these genes. Heterogeneity of EZH2 levels is observed in melanoma models and co-ordinated up-regulation of genes positively regulated by EZH2 is associated with melanoma metastasis. EZH2 was also identified as regulating the amelanotic phenotype of motile cells in vivo by suppressing expression of the P-glycoprotein Oca2. Analysis of patient samples confirmed an inverse relationship between EZH2 levels and pigment. EZH2 targeting with siRNA and chemical inhibition reduced invasion in mouse and human melanoma cell lines. The EZH2 regulated SRF target genes KIF2C and KIF22 are required for melanoma cell invasion and important for lung colonisation. We propose that heterogeneity in EZH2 levels leads to heterogeneous expression of a cohort of genes associated with motile behaviour including KIF2C and KIF22. EZH2 dependent increased expression of these genes promotes melanoma cell motility and early steps in metastasis.
Keywords: Intravital microscopy, melanoma, metastasis, cancer cell motility, SRF, Notch, EZH2, Polycomb
Introduction
Melanoma is a skin cancer arising from the uncontrolled proliferation and invasion of melanocytes. Melanoma metastasis starts with the acquisition of motility in the primary tumour. Previous intravital imaging of B16 melanoma showed that motile cells had lower pigment levels and higher Brn2 promoter activity compared to non-motile cells,1 indicating that motile cells may represent a less differentiated subset of cells. This is consistent with studies suggesting that melanoma cells revert to a “neural crest-like” state during metastasis.2,3 It has also been proposed that melanoma cells switch between mutually exclusive invasive and proliferative states.4,5 The proliferative state is characterised by high expression of the melanocyte transcription factor MITF and low Brn2 expression, whereas the invasive state is characterised as having Brn2 expression but low MITF expression.6,7 However, little is known about the signalling pathways that might drive the changes in differentiation and promote a motile state.
We chose to use intravital imaging of transcriptional reporters to monitor signalling of B16 melanoma cells during the early steps of the metastatic process. We focussed on two pathways thought to be important in melanoma metastasis, the Notch, and SRF pathways. Notch signalling is usually activated through membrane bound ligand-receptor interactions leading to Notch receptor activation, cleavage and the release of the active Notch intracellular domain (NICD). NICD binds to the DNA-binding factor RBPSUK (RBPjκ) and leads to increased transcription of target genes.8 Melanoma cells with constitutively active Notch signalling show increased motility in vitro and increased lung colonisation, through Notch mediated regulation of N-cadherin.9-11 Furthermore, the Notch pathway has an important role in maintaining melanocyte stem cells12 and Notch activation in mature melanocytes is sufficient to induce re-programming and de-differentiation to neural crest stem cell–like cells.13
The SRF pathway links changes in the actin cytoskeleton to gene transcription. Actin polymerisation leads to a decrease in the myocardin related transcription factor MRTF-A binding to G-actin. MRTF-A (also called MKL) is a co-activator of SRF and subsequently accumulates in the nucleus leading to increased SRF dependent transcription.14 Loss of MRTF-A in B16 melanoma cells leads to decreased experimental lung metastasis.15 Depletion of the SRF targets MYL9 (myosin light chain) and MYH10 (myosin heavy chain IIb) also reduced invasion and metastasis. Another actomyosin regulator, RhoC, is up-regulated in metastatic melanoma cells16 and can increase SRF signalling.17 Although both SRF and Notch are implicated in metastasis, it is not clear whether their activity is simply permissive for metastasis or whether they are dynamically regulated during different stages of the metastatic process.
In addition to transcription factor activity, the expression of many genes is influenced by post-translational modification of histones. EZH2 is part of the Polycomb repressor complex 2 (PRC2) and catalyses the tri-methylation of histoneH3 at lysine 27 (H3K27me3), a mark usually associated with gene repression.18 During development EZH2 silences differentiation associated genes to maintain cells in a progenitor state.19 EZH2 is up-regulated in many tumour types including melanoma and increases in expression from benign nevi to metastatic melanoma.20,21 Further EZH2 expression levels are coupled to BRAF, a common melanoma oncogene.22 EZH2 has also been implicated in cancer cell EMT and invasion.20,21,23
In this study, we use intravital imaging to show heterogeneous transcriptional activation of the Notch, and SRF pathways in melanoma cells in vivo and activation of these pathways in motile cells. Genome-wide analysis reveals that MRTF/SRF targets and genes up-regulated in the Notch reporter high population show significant overlap. Further, these overlapping genes are regulated by EZH2. We demonstrate key roles for EZH2 in suppressing pigment production in invasive melanoma cells by repressing Oca2 levels. Further, we show that EZH2 enables invasion and metastasis by positively controlling the expression KIF2C and KIF22.
Results
B16 melanoma shows heterogeneous motile behaviour in vivo
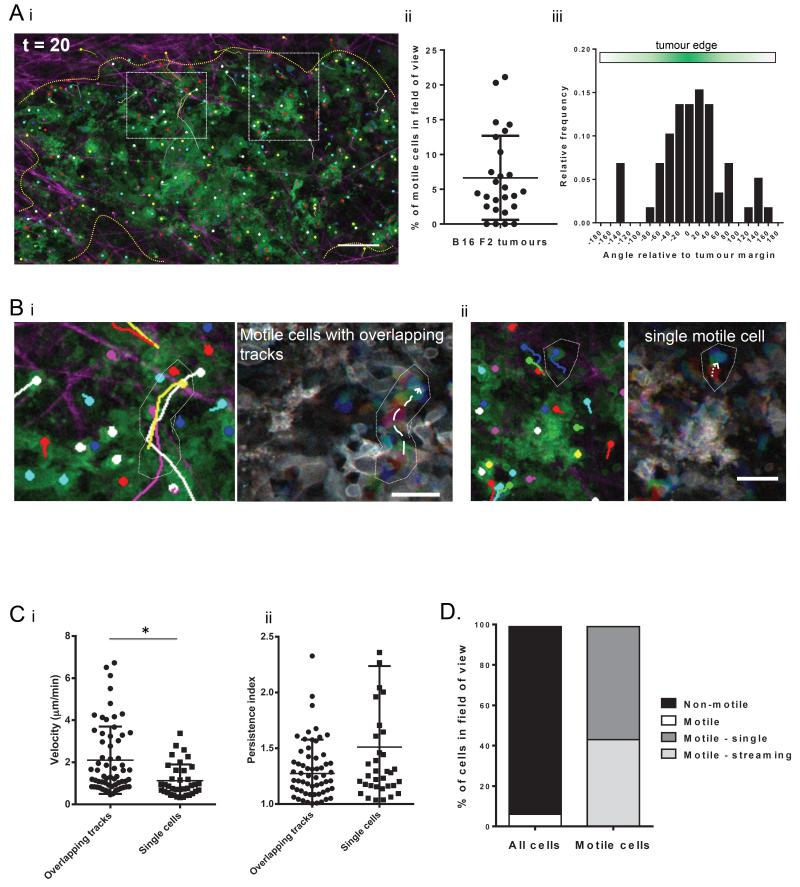
Intravital studies of cancer motility have revealed heterogeneous cancer cell behaviour. 1,24,25 We examined the behaviour of metastatic melanoma cells using intravital imaging of B16 F2 tumours followed by cell tracking. The majority of B16 F2 melanoma cells were non-motile. On average 6.6% of cells were motile, with a range of 0-22% per field of view. (Fig.1Ai,ii and Supp. mov.1). Motile melanoma cells showed speeds between 0.4μm/min to 6.7μm/min. Further, the distribution of directions was significantly non-uniform, with melanoma cells predominantly moving towards the tumour margin (Fig.1Aiii). Cell tracking also revealed that some melanoma cells moved as single cells whereas other cells followed the same path (Fig.1B and Supp. mov.2). Cells with tracks that overlapped in a 15 minute time window (see methods for a more detailed classification) moved significantly faster than cells moving in isolation, however there was no significant difference in persistence (Fig.1C). We propose that cells following the same paths are using multicellular streaming26 as a mode of motility, whereas cells in isolation are using single cell amoeboid motility. Quantification of motile cells showed that, on average, 44% of motile cells exhibited multicellular streaming and 56% display single cell motility (Fig.1D)
Figure 1. B16 F2 melanoma cell behaviour is heterogeneous in vivo.
A. i) Still image of intravital movie of B16 F2 cells with green membrane marker. Individual cell tracks shown as overlay. Collagen fibres in magenta. Dashed boxes depict areas shown in part B. Dotted yellow line indicates the tumour margin. Scale bar indicates 50 μm. ii) Number of motile cells per field of view in a 20 minute timelapse movie of B16 F2 melanoma cells in vivo. Each data point represents a single movie. Bar indicates mean and error bars show standard deviation of 9 mice analysed. iii) Histogram of angle of cell migration relative to tumour margin. Angle 0 is directly to tumour edge. B. Images from intravital movie of B16 F2 tumours showing cells moving (i) with overlapping tracks and (ii) singly. B16 cells have green membrane marker. Left image taken from movie endpoint with individual cell tracks shown as overlay. Right image is motion analysis of motile cells. Images from three different time points of the time-series are shown overlaid in red, green and blue. Scale bars indicate 30 μm. C. i) Speed and ii) Persistence of motile B16 melanoma cells in vivo measured by manual tracking. Each data point represents an individual cell. Data from 3 movies of 3 mice. * indicates p<0.05 in student t-test. D. Quantification of mode of motility of B16 F2 melanoma cells in vivo. Data average from 18 movies of 9 mice.
Generation of Notch and SRF reporter B16 F2 lines
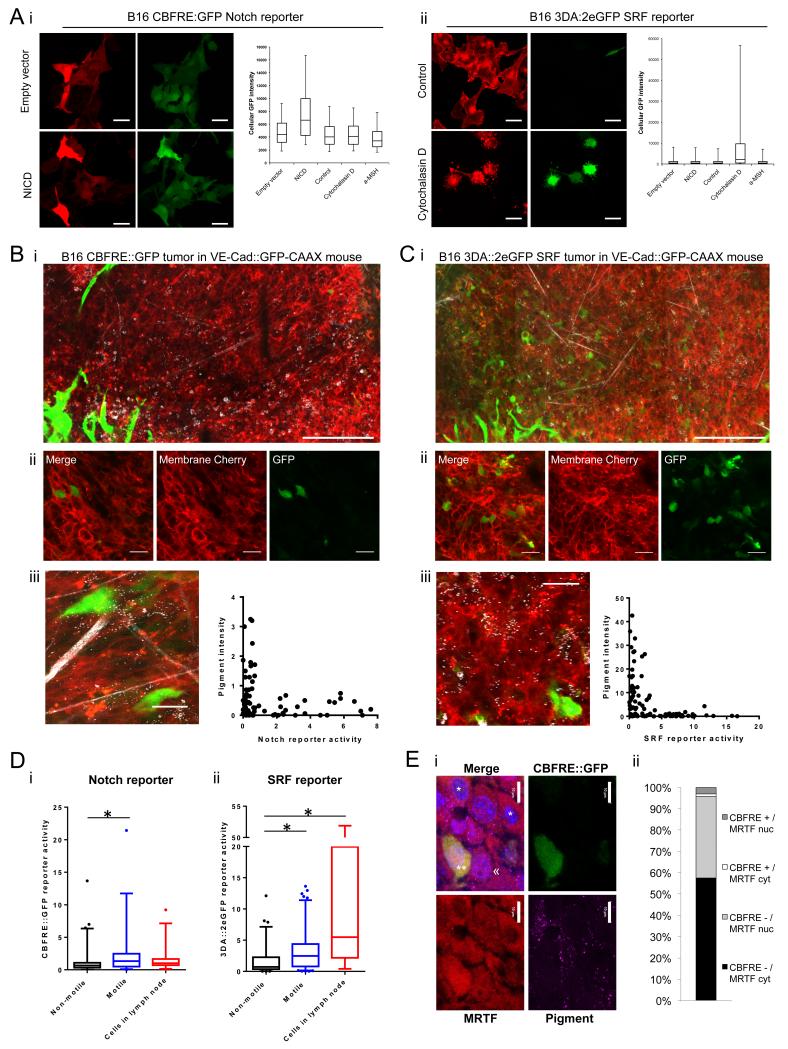
Heterogeneity in signalling within the tumour could account for the different behaviours of non-motile and motile cells. We hypothesized that two signalling pathways implicated in melanoma metastasis, SRF and Notch, might be differentially activated in migratory and non-migratory cells. To test this we generated B16 F2 reporter cell-lines to visualise the signalling status of Notch (using CBFRE::GFP reporter) and SRF signalling (using 3DA::2eGFP reporter -see Supp Fig.1 for more details of reporter constructs). Stable monoclonal reporter cell-lines were made in B16 F2 cells containing a constitutive mRFP membrane label (Supp. Fig.1) to aid visualisation and to ensure that any heterogeneity in signalling observed was not the result of heterogeneity within the starting B16 F2 cell line. Multiple SRF reporter clones were chosen, 1 3DA::2eGFP and 1 3DA::2eGFP Fos3’UTR reporter clone (Fos 3’UTR helps destabilise eGFP mRNA), but only 1 responsive Notch CBFRE::GFP reporter clone was generated. The veracity of the Notch reporter was confirmed by transfection of NICD together with mCherry to label transfected cells. NICD transfection increased the expression of eGFP, whereas empty vector alone had no effect (Fig.2Ai). The 3DA::2eGFP SRF reporter cell-line did not show changes in eGFP expression following NICD transfection. In contrast, 5μM Cytochalasin D, which activates the SRF co-factor MRTF/MAL, increased eGFP expression in nearly all of the 3DA:2eGFP SRF reporter cells (Fig.2Aii). Therefore the reporter cell-lines were responsive to the relevant signal in vitro (Fig.2A).
Figure 2. Notch and SRF signalling is increased in the singly motile cell population in vivo.
A. Confocal images and cellular fluorescence quantification of B16 F2 i) CBFRE::GFP Notch reporter and ii) 3DA::2eGFP SRF reporter cell-lines with membrane targeted mRFP in vitro. CBFRE::GFP Notch reporter co-transfected with empty-vector plus mCherry or NICD plus mCherry. 3DA::2eGFP SRF reporter cells serum starved for 8 hours before treatment with control or 5 μM Cytochalasin D for 16 hours. Scale bars indicate 30 μm. Plots of reporter fluorescence intensity after co-transfection of empty-vector and mCherry, NICD and mCherry, or mCherry and treatment with vehicle control, 5 μM Cytochalasin D, or 10 μM α-MSH. Reporter fluorescence intensity of mCherry transfected cells measured per cell by flow cytometry. Boxes show median and inter-quartile range and whiskers show 95th and 5th percentiles. Intravital confocal tile images of B) B16 F2 CBFRE::GFP Notch reporter and C) 3DA::2eGFP SRF reporter tumours with membrane targeted mRFP in VECad::GFPCAAX mice. Endothelial cells have green membranes, Collagen fibres and pigment in white. Scale bar indicates 200 μm. ii) Reporter tumours in C57Bl6J mice. Scale bar indicates 30 μm. iii) Quantification of cellular pigment and reporter activity in vivo on a cell-by-cell basis. Each data point represents a single cell, at least 60 cells quantified. D. Cellular reporter fluorescence intensity of non-motile and motile cells in primary tumour and cells in lymph node micrometastases from i) B16 F2 CBFRE::GFP Notch reporter tumours and ii) 3DA::2eGFP SRF reporter tumours. For primary tumour analysis, greater than 25 cells analysed from at least 6 movies of 3 mice. For lymph-node analysis at least 18 cells analysed from more than 4 mice. Boxes show median and inter-quartile range and whiskers show 95th and 5th percentiles. Outlying data points shown by dots. Star indicates p-value <0.05 in ANOVA test. E. i) Panels show MRTF/MKL staining (red), pigment (magenta), and CBFRE::GFP (green) staining of B16 CBFRE::GFP tumour. ** indicates a GFP positive cell with nuclear MRTF, << indicates a GFP negative cell with nuclear MRTF, * indicates GFP negative cells with cytoplasmic MRTF. Scale bar is 10μm. ii) Chart shows the proportion of cells in indicated categories. > 400 cells analysed from three tumours.
Notch and SRF transcriptional activity is heterogeneous in B16 cells in vivo
We next imaged the B16 reporter cell-lines in vivo. To obtain more general information about the tissue structure, we imaged collagen second harmonic signal and used C57/BL6 mice with GFP-CAAX labelled endothelial cells.27 Notch reporter tumours showed heterogeneity in Notch signalling with few GFP positive, Notch active cells (Fig. 2Bi,ii and Supp. Fig.1C). However, we did note an exclusive relationship between CBFRE::GFP positive cells and the presence of melanin pigment within cells measured using near infrared stimulation of visible emission (Fig.2Biii).1
The B16 F2 3DA::2eGFP SRF (and 3DA::2eGFP Fos3’UTR) reporter tumours also showed that SRF transcriptional activity was heterogeneous in vivo (Fig.2Ci,ii and Supp. Fig.1C). 3DA::2eGFP positive cells were scattered throughout the tumour, both in small clusters and individual positive cells. Similar to the Notch reporter, we observed that SRF reporter positive cells displayed an exclusive relationship with pigment (Fig.2Ciii and Supp. Fig.2A). Cells with high SRF-dependent transcriptional activity had low pigment levels and vice-versa. This suggests an overlap between the motile low pigment cells reported in Pinner et al1 and cells with both active SRF signalling and Notch signalling.
Motile behaviour of B16 cells in vivo is associated with increased Notch and SRF transcriptional activity
Given the heterogeneity in Notch and SRF signalling and their links to melanoma metastasis we hypothesised that the cells with active Notch, and SRF signalling were more motile. Quantification of Notch reporter tumours showed that motile cells had increased Notch dependent transcription compared to non-motile cells (Fig.2Di and Supp.Mov.3). Similarly, motile cells had increased SRF reporter activity compared to non-motile cells (Fig.2Dii, Supp.Mov.4 and Supp. Fig.2A). This indicates that active Notch and SRF signalling is associated with cell motility in melanoma. However, both Notch and SRF reporter activity was occasionally observed in non-motile cells, suggesting that each pathway alone was not sufficient for motility.
We next investigated subsequent steps in the metastatic process. Metastatic cells in the lymph node of Notch reporter tumours showed little GFP intensity, similar to non-motile cells in the primary tumour, indicating a lack of Notch signalling (Fig.2Di and Supp. Fig.2B). In contrast, 3DA::2eGFP SRF reporter cells in the lymph node were GFP positive, similar to motile cells in the primary tumour, suggesting that SRF signalling remained active in cells that arrived at the metastatic site (Fig. 2Dii and Supp. Fig.2C). Together these data suggest a transient activation of Notch signalling during melanoma metastasis, but a more prolonged activation of SRF signalling.
The data above show that both Notch and SRF signalling are increased in invasive melanoma cells. This observation predicts that ‘double positive’ cells with high levels of both signalling pathways should be observed. We tested this hypothesis by staining CBFRE::GFP Notch reporter tumours for the SRF co-factor MRTF/MKL. Activation of SRF signalling is associated with nuclear translocation of MRTF/MKL, therefore the activity of SRF can be inferred from the localisation of MRTF/MKL. Quantification revealed that cells with both active Notch and SRF signalling formed 5% of the tumour; this number corresponds well with the overall proportion of motile cells in the tumour (6.6% - shown in Fig. 1Aii). Further, Fisher’s exact statistical test revealed a significantly greater fraction of double positive cells than would be expected if SRF and Notch signalling were independently regulated (p=0.0023).
Notch and SRF signalling is associated with a common transcriptional programme
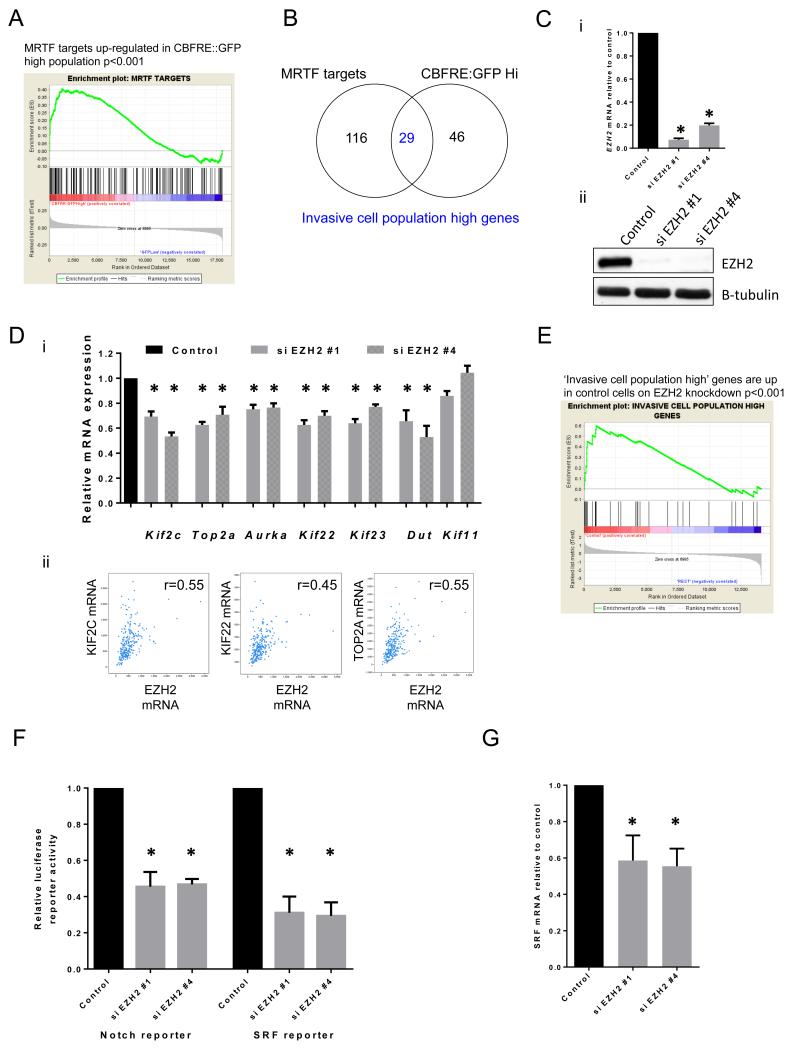
Given that SRF and Notch signalling were increased in motile cell populations, we sought to identify any genes associated with both elevated Notch and SRF signalling, and if these genes might modulate cell migration and metastasis. CBFRE::GFP Notch reporter high and low populations were purified by fluorescent-activated cell sorting (FACS) and subjected to microarray analysis (Supp. File 3). Target genes of the SRF pathway in B16 F2 melanoma cells were obtained from Medjkane et al.15 (Supp.File.3). Gene Set Enrichment Analysis (GSEA, Broad Institute) showed significant overlap (p-value<0.001) between MRTF/MKL target genes and genes up-regulated in the Notch reporter high population (Fig.3A). Out of 145 MRTF target genes and 75 genes up-regulated in the Notch high population, 29 showed overlap between the two populations. These genes will be referred to as ‘invasive cell population high’ genes (Fig.3B, Table 1). Interestingly, many of these 29 genes were not up-regulated when Notch signalling was ectopically activated using NICD over-expression (Supp.Fig.3Ai). This suggests that Notch signalling might be coincidently activated with the ‘invasive cell population high’ genes, but not directly activating them. As a control we confirmed that CBFRE::GFP Notch high genes did show significant overlap with NICD Notch target genes (Supp.Fig.3Aii). We therefore propose that the ‘invasive cell population high’ genes are not necessarily Notch target genes, but that a critical regulator of invasive potential promotes both Notch and SRF pathway activity.
Figure 3. EZH2 regulates ‘invasive cell population high’ genes.
A. Gene Set Enrichment Analysis of MRTF targets (defined in B16 cells) enriched in the CBFRE::GFP Notch reporter high population. B. Diagram showing overlap between MRTF targets and genes up-regulated in CBFRE::GFP high population. C. EZH2 (i) mRNA levels and (ii) protein levels in B16 cells after control and si EZH2 knockdown. D.i) Relative mRNA levels of ‘invasive cell population high’ genes after si EZH2 knockdown in B16 cells. ii) TCGA analysis of KIF2C, KIF22 and TOP2A mRNA expression vs EZH2 mRNA expression as determined by RNASeq. E. Gene Set Enrichment Analysis of ‘invasive cell population high’ genes enriched in control B16 cells compared to siEZH2 treated cells. F. Relative luciferase reporter activity of B16 cells relative to control levels 72 hrs after control and si EZH2 knockdown. G. Srf mRNA levels in B16 cells relative to control levels after control and si EZH2 knockdown. Error bars show s.e.m of 3 experiments. Star indicates p-value <0.05 in ANOVA test.
Table 1. ‘Invasive population high’ genes.
29 MRTF target genes up-regulated in CBFRE::GFP Notch reporter high population
| CCNB1 |
| TOP2A |
| KIF22 |
| KIF11 |
| AURKA |
| CDCA5 |
| KIF2C |
| KIF23 |
| CCNF |
| MSN |
| MXD3 |
| CDCA8 |
| EPB4.1L2 |
| PAK3 |
| DUT |
| MCM3 |
| SH3BGRL |
| PLK1 |
| AXL |
| BUB1 |
| PRKG2 |
| BIRC5 |
| CASP7 |
| SPAG5 |
| CCK |
| PBK |
| CDC20 |
| BICD1 |
| CSPRS |
To gain further evidence that the ‘invasive cell population high’ genes were associated with migratory cells in vivo we investigated their expression in melanoma cells with high Brn2 activity. The activity of a Brn2::GFP reporter is elevated in migratory melanoma cells in vivo1 and both MRTF target genes and CBFRE::GFP high genes are enriched in the Brn2 promoter high population (Supp.Fig.3B). Further the ‘invasive cell population high’ genes were enriched in the Brn2 promoter high cells (Supp.Fig.3C). These data strengthen the link between ‘invasive cell population high’ genes and motile melanoma cells.
EZH2 regulates the transcriptional programme common to cells with high Notch and SRF activity
We next explored what factors may regulate the ‘invasive cell population high’ genes as we hypothesised that a critical regulator of invasive potential promotes expression of these genes. The Molecular Signatures Database (MSigDB, Broad Institute) was used to search for published gene sets with the most overlaps with the ‘invasive cell population high’ genes (Supp. File 1). Interestingly, genes positively regulated by EZH2 in prostate cancer cells (identified by Nuytten et al.28) showed significant overlap (FDR=1.22 ×10−14) with ‘invasive cell population high’ genes (Supp. File 1). Consistent with reports linking RhoA to SRF activity and invasion, RhoA-regulated genes also showed significant overlap with ‘invasive cell population high’ genes (Supp. File 1). We decided to pursue the role of EZH2 in melanoma motility due to its global role in gene transcription regulation and previous reports of a functional role in cancer metastasis23,29. Microarray analysis of cells transfected with two independent EZH2 siRNA identified genes positively regulated by EZH2 in B16 cells (knockdown efficiency shown in Figure 3C and genes listed in Supp. File 1). EZH2 depletion did not affect cell viability nor induced senescence (Supp. Fig. 3D). GO analysis showed that genes positively regulated by EZH2 were enriched for cytoskeletal components (Supp. File 1). We confirmed that EZH2 positively regulates the expression of multiple kinesins and other genes identified in the microarray analysis using QRT-PCR analysis (Figure 3Di). Analysis of TGCA data revealed strong positive correlations between EZH2 mRNA and KIF2C, KIF22, and TOP2A mRNA (Figure 3Dii). Finally, GSEA confirmed a significant overlap of genes positively regulated by EZH2 in B16 cells with the ‘invasive cell population high’ genes (Fig. 3E). These analyses confirm EZH2 as a positive regulator of the ‘invasive cell population high’ genes.
The data above show that EZH2 positively regulates the ‘invasive cell population high’ genes, including multiple kinesins. Although EZH2 is predominantly thought of as a repressor of transcription it can positively regulate genes (Fig.3D).18,30 We hypothesise that EZH2 levels may control activity through the Notch and SRF pathway. In support of this idea, EZH2 depletion reduced Notch and SRF luciferase reporter activity (Fig.3F). EZH2 depletion also reduced SRF transcript levels (Fig.3G), this confirms that EZH2 regulation of the SRF pathway.
EZH2 levels are heterogeneous in mouse and human melanoma
Intravital imaging showed that cell behaviour is heterogeneous in vivo with ~6% of cells being motile. If EZH2 levels regulate the expression of genes up-regulated in motile cells, then EZH2 levels should be heterogeneous in vivo. Immunohistochemistry of two different mouse melanoma models, including the B16 model, revealed heterogeneity in EZH2 levels in vivo (Supp. Fig.4Ai). In both models, the invasive tumour margin had higher levels of EZH2 (Supp. Fig.4Aii). The distribution of EZH2 levels in sections of human melanoma metastases was similar to that found in the mouse models (Supp. Fig.4E). Immunofluorescence staining also revealed that the heterogeneity in EZH2 levels correlated with variations in H3K27Me3 levels (Supp. Fig.4B&C). Depletion of EZH2 in B16 cells significantly decreased H3K27me3 levels (Supp. Fig.4D). These data establish that intratumour heterogeneity in EZH2 expression is a feature of melanoma and correlates with changes in K27Me3 modification of its substrate Histone H3.
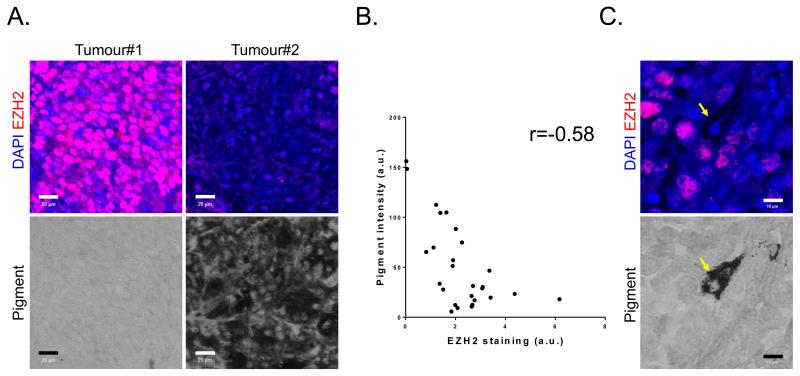
We next investigated variation of EZH2 expression in a panel of 27 human melanomas. This revealed both inter- and intra-tumour heterogeneity in the levels of EZH2. We also observed that melanoma with the highest levels of EZH2 had the lowest levels of pigmentation (Figure 4A&B). This negative correlative was interesting because we had previously found that the invasive sub-population of melanoma cells in the B16 model lack pigment. Even within the same tumour, highly pigmented cells had lower levels of EZH2 expression than their neighbours (Figure 4C). These data demonstrate inverse relationship between EZH2 and pigmentation in human melanoma.
Figure 4. EZH2 levels are heterogeneous in mouse and human melanoma.
A. Human melanoma biopsies were stained for EZH2 (red), DAPI (blue), and pigment (transmitted light – grayscale). One highly pigmented and one amelanotic tumour are shown. Scale bar is 20μm. B. Scatter plot shows quantification of average EZH2 intensity and average pigment intensity in 27 human melanoma samples. C. An example intra-tumour heterogeneity in EZH2 expression (red) and pigment (transmitted light – grayscale). Arrow shows cell with low EZH2 and high pigment. Scale bar is 10μm.
EZH2 regulates pigment levels through Oca2
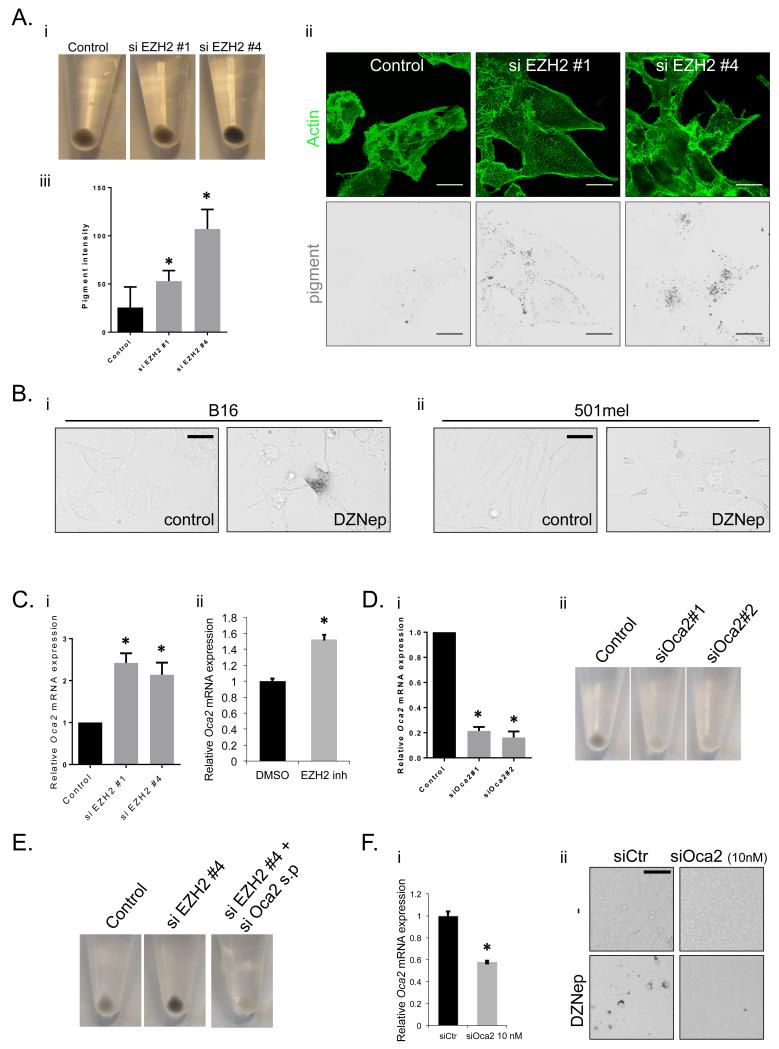
The negative correlation in human tumours between EZH2 levels and pigmentation and previous work showing invasive cells have lower pigment1 prompted a functional analysis of EZH2 on pigment levels. Depletion of EZH2 resulted in a 2-4 fold increase in pigment as determined by cell pellets and two-photon microscopy (Fig.5A). Depletion of another PRC2 component, Suz12, also increased pigment levels (Supp. Fig.5A) suggesting that EZH2 may regulate pigment levels via PRC2-mediated gene repression. We tested the importance of EZH2 catalytic function by using DZNep, which inhibits EZH2 methyl transferase functions.31 Treatment of both B16 and human 501mel cells with DZNep for 48 hours led to a marked increase in melanosomes (Fig. 5B). We did not observe any consistent changes in mRNA of the pigmentation genes Slc45a2, Tyr, Tyrp1 or Dct following EZH2 depletion (Supp. Fig.6A). However, both siRNA depletion and pharmacological targeting of EZH2 caused an increase Oca2 mRNA (Fig.5C). Further, Oca2 and EZH2 are expressed in a mutually exclusive manner across a large number of melanoma samples: tumours with high levels of EZH2 have low Oca2 mRNA and vice-versa (Supp. Fig.6B). Oca2 encodes the P-protein, a transmembrane protein localised to melanosomes32,33 and loss of function mutation leads to a form of albinism.34 Depletion of Oca2 with two independent siRNA dramatically decreased pigment levels in B16 cells (Fig.5D – cell viability was unaffected Supp. Fig.6C). The effect of EZH2 on pigment levels was ablated on combined knockdown of EZH2 and Oca2 (Fig.5E), suggesting that EZH2 affects pigment levels through regulation of the Oca2 gene. Further, pigment production in B16 cells was highly sensitive to the levels of Oca2, because 50% depletion of Oca2 mRNA using a low concentration of siRNA was sufficient to prevent the increase in melanosomes following inhibition of EZH2 (Figure 5F).
Figure 5. EZH2 regulates pigment levels through control of Oca2.
A.i) Cell pellets of B16 cells after control and si EZH2 knockdown showing pigment production. ii) Confocal images of B16 cells after control and si EZH2 knockdown. Actin in green and pigment as determined by 2-photon microscopy shown in black. Scale bar indicates 25 μm. iii) Quantification of pigment levels on a cell by cell basis as determined by 2-photon microscopy after control and si EZH2 knockdown. Error bars show s.e.m of at least 50 cells from 2 experiments. B. Transmitted light images of control and 2μM DZNep treated i) B16 and ii) 501mel cells are shown. Scale bar is 20μm. C. Oca2 mRNA levels in B16 cells relative to control levels after i) EZH2 knockdown and ii) 2μM DZNep treatment. n=3 mean and s.e.m shown. D. Oca2 mRNA levels in B16 cells relative to control levels after transfection of 75nM control and Oca2 siRNA. ii) Cell pellets showing pigment production of B16 cells after transfection of 75nM control and Oca2 siRNA. E. Cell pellets showing pigment production of B16 cells after control, EZH2, and double Oca2 and EZH2 knockdown. F.i) Oca2 mRNA levels in B16 cells relative to control levels after transfection of 10nM control and Oca2 siRNA. n=3 mean and s.d. shown. ii) Transmitted light images of control and 2μM DZNep treated B16 cells with and without transfection of 10nM Oca2 siRNA are shown. Scale bar is 20μm. Error bars show s.e.m of 3 experiments. Star indicates p-value <0.05 in ANOVA test.
EZH2 regulates melanoma invasion and metastasis
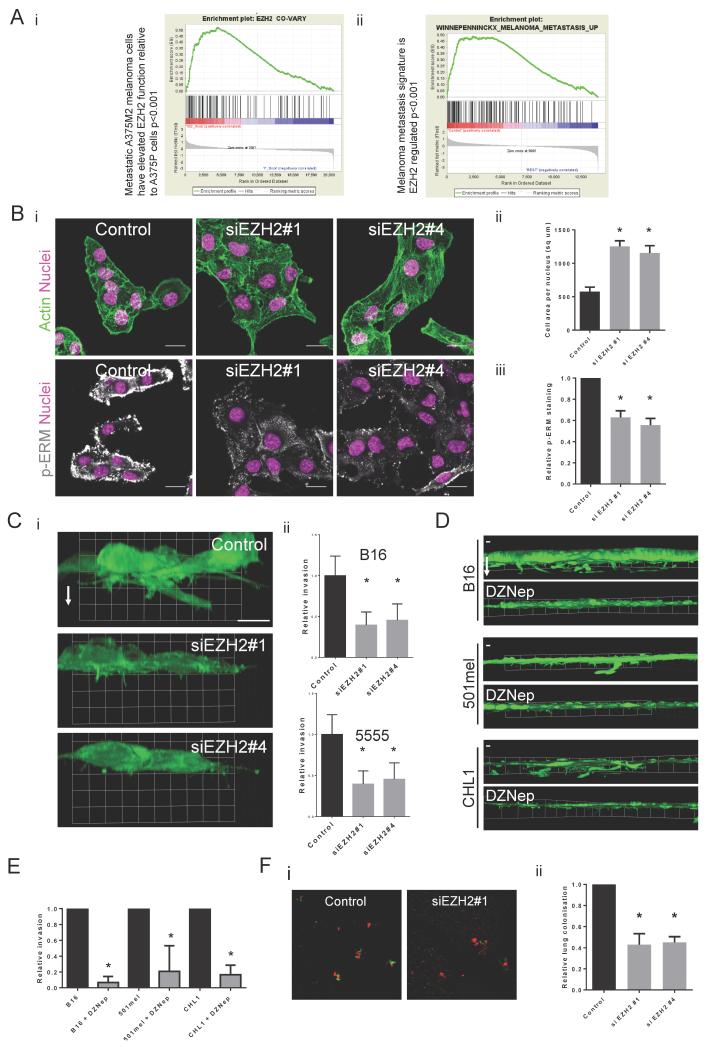
Given that EZH2 can regulate the genes associated with motile behaviour in a mouse model of melanoma, we explored possible links between EZH2 and metastasis in human melanoma. Analysis of microarray data35 comparing A375P (non-metastatic) and A375M2 (metastatic) melanoma cells showed that genes positively regulated by EZH2 are up-regulated in the A375M2 metastatic sub-line compared to the A375P parental population (Fig.6Ai). More strikingly, genes up-regulated in human melanoma patients who developed metastasis within 4 years compared to patients who did not, showed significant overlap with genes positively regulated by EZH2 in B16 melanoma (Fig.6Aii). These data are consistent with a role for EZH2 in regulating melanoma metastasis.
Figure 6. EZH2 promotes invasion and metastasis.
A.i). Genes that positively co-vary with EZH2 in a panel of melanoma cell-lines are enriched in the more metastatic A375 M2 cells. ii) Geneset enrichment analysis showing enrichment of the geneset “Genes up-regulated in melanoma patients who developed metastasis in 4 years” in control B16 cells compared to EZH2 siRNA treated cells. B.i) Confocal images of B16 cells after control and si EZH2 knockdown. Actin in green, DAPI in pink and phospho-ERM staining in white. Scale bar indicates 20 μm. ii) Quantification of B16 cell area after control and si EZH2 knockdown. Error bars show s.e.m of 3 experiments with 5 images each experiment. iii) Quantification of phospho-ERM immunostaining in B16 cells after control and si EZH2 knockdown. Error bars show s.e.m of 2 experiments with 4 images each experiment. C.i) ‘Side-on’ 3D re-constructions of F-actin images of B16 cells transfected with control or EZH2 siRNA plated on collagen/matrigel matrices. White arrow indicates direction of invasion. Grid units are 13μm. ii) Graphs showing relative invasion into collagen/matrigel matrices after control and si EZH2 smartpool knockdown in mouse B16 and 5555 melanoma cells. Error bars show standard deviation. n=3 mean and s.d. shown. D. ‘Side-on’ 3D reconstructions of B16, 501mel and CHL1 cells treated with either DMSO or 2μM DZNep plated on collagen/matrigel matrices. White arrow indicates direction of invasion. Grid units are 50μm. E. Graphs showing relative invasion into collagen/matrigel matrices of B16, 501mel and CHL1 cells after DMSO and 2μM DZNep treatment. n=4 mean and s.d. shown. F.i) Confocal images of lungs after I.V injection of a 50:50 mix of red B16 F10 control cells and green B16 F10 control or EZH2 knockdown cells. Scale bar indicates 200 μm. ii) Graph showing relative lung colonisation of B16 F10 cells after control and si EZH2 knockdown. Error bars show s.e.m of at least 6 mice. Star indicates p-value <0.05 in ANOVA test.
We next investigated the functional role of EZH2 in melanoma motility and metastasis. Depletion of EZH2 led to changes in actin cytoskeleton regulation (Fig.6B). EZH2 depleted cells had an increase in cell area (Fig.6Bii) and a decrease in phosphorylation of the plasma membrane-cortex linker proteins, ERM (Fig.6Biii). We also observed a similar change in cell morphology on depletion of Suz12 (Supp.Fig.5C) suggesting a more general PRC2 regulation of the cytoskeleton. EZH2 depletion decreased invasive protrusions into collagen-matrigel matrices and subsequently decreased invasion in both B16 and 5555 mouse melanoma cell-lines (Fig.6C). DZNep inhibition of EZH2 catalytic function also reduced invasion of B16 and 5555 cells. Importantly, we confirmed this finding in two human melanoma cell lines (501mel and CHL-1 – Figure 6D&E). Many regulators of cell migration and invasion are also required for extra-vasation and lung colonisation during metastasis. EZH2-depleted cells were significantly under-represented in the lungs 48hrs after co-injection intra-venously with control cells (Fig. 6F). These data establish that EZH2 is required for efficient lung colonisation by metastatic melanoma.
EZH2-regulated kinesins are required for invasion and metastasis
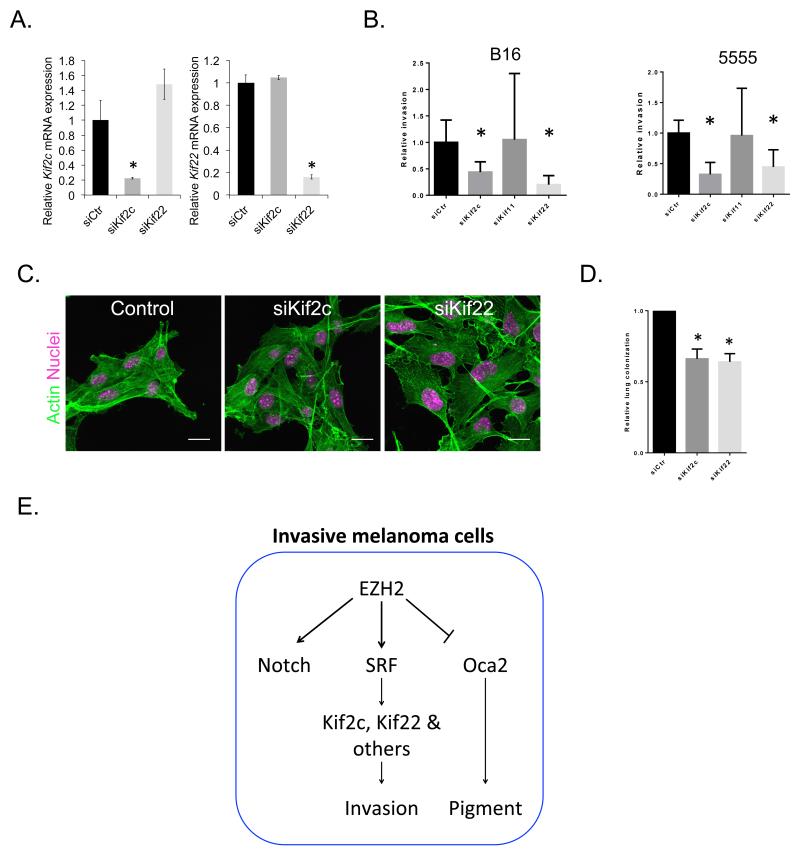
We next tested whether any of the ‘invasive cell population high’ genes might be involved in EZH2-mediated melanoma invasion and metastasis. We focused on two cytoskeletal genes whose expression is regulated by EZH2 (additionally confirmed using DZNep– Supp. Fig. 7A) and correlates with EZH2 mRNA levels across a large cohort of melanoma patients (Fig. 3Dii). Depletion of the kinesins KIF2C and KIF22 significantly reduced invasion into collagen-matrigel matrices in both B16 and 5555 mouse melanoma cell-lines (Fig.7A). Neither kinesin was required for cell viability (Supp. Fig. 8B). In contrast, depletion of the KIF11, which is not EZH2 regulated (Fig. 3Di), had no effect. Further, KIF22 depletion also led to an increase in the area of B16 cells (Fig. 7C). Consistent with the identification of KIF2C and KIF22 as MRTF/SRF-regulated genes, siRNA-mediated depletion of SRF reduced the invasion of melanoma cells (Supp Fig.8B). Depletion of both KIF2C and KIF22 significantly reduced lung colonisation in a short-term metastasis assay (Fig. 7D). Therefore we propose that EZH2 regulation of KIF2C and KIF22 promotes the invasive phenotype of mouse melanoma cells.
Figure 7. KIF2c and KIF22 are required for invasion and metastasis.
A. Graphs show relative mRNA levels of KIF2C and KIF22 in B16 cells following transfection of either 75nM control, KIF2C or KIF22 siRNA. n=3 mean and s.d. shown. B. Graphs showing relative invasion into collagen/matrigel matrices after control and si KIF2C, KIF11, KIF22 smartpool knockdown in mouse B16 and 5555 melanoma cells. Error bars show standard deviation. C. Confocal images of B16 cells after control and si KIF2C and KIF22 smartpool knockdown. Actin in green, DAPI in magenta. D. Graph showing relative lung colonisation of B16 F10 cells after control and si KIF2C and KIF22 knockdown. Error bars show s.e.m of at least 6 mice. Star indicates p-value <0.05 in ANOVA test. E. Both motile behaviour and EZH2 levels are heterogeneous in vivo. Invasive cells in vivo have increased EZH2 levels. High EZH2 levels promote Notch and SRF pathway activity and activation of the downstream SRF target genes KIF2C and KIF22 which are required for optimal invasion.
Discussion
Relatively little is known about the initial steps of melanoma metastasis in vivo. We show that even in an aggressive mouse melanoma cell-line, 6-7% of cells are motile in vivo. This is similar to murine breast cancer models.24 Most motile cells use an amoeboid-like single cell mode of motility, however roughly 40% exhibit a streaming mode of motility whereby motile cells share the same tracks. Both the splitting and merging of groups of cells moving along common tracks was observed. This suggests that multicellular streaming is not associated with stable cell-cell junctions that persist for long periods (>15 minutes). Streaming cells are also faster than singly motile cells and may be following pre-generated tracks in the ECM, potentially made by fibroblasts, as seen in vitro.36
We show that Notch- and SRF-dependent transcriptional activation is heterogeneous in vivo and increased in motile cells. More detailed analysis reveals that SRF signalling is increased in both singly moving and streaming cells (Supp.Fig.2B). Notch signalling is clearly elevated in singly moving cells, but the data is more ambiguous in streaming cells (Supp.Fig.2B). This may reflect the lower range of Notch reporter cells or some mechanistic difference between singly moving and streaming cells. The cell-lines are clonal in origin, therefore heterogeneity in SRF and Notch activity is unlikely to result from genetic differences. SRF signalling is responsive to both soluble factors, such as LPA and SDF-1, and external physical tension.37 Notch signalling can be activated through cell-cell interactions and hypoxia.38,39 Thus many cell extrinsic factors can modulate SRF and Notch signalling potentially leading to signalling heterogeneity. We demonstrate that EZH2 function influences activity of SRF and Notch reporters. The increased levels of EZH2 observed at the tumour margin (Fig.4) suggest that stromal cells enriched in this region may be responsible for the heterogeneous activation of SRF- and Notch-dependent transcription.
Increased Notch- and SRF- dependent transcriptional activation in motile melanoma cell population is consistent with previous work showing the importance of these pathways for melanoma metastasis.9,15 However, we noted that activation of SRF was not sufficient to drive cell invasion when EZH2 function was blocked (Supp. Fig.8C) indicating that there are likely to be additional mechanisms by which EZH2 controls invasive potential. Although Notch activity is associated with melanoma metastasis it does not appear to be directly involved in regulation of cell migration. Depletion of RBPjK had no effect on invasion (Supp.Fig.8B) and NICD over-expressing cells did not show elevated motility in vivo (data not shown). Notch signalling may promote metastasis by mechanisms unrelated to cell migration.
Combining the transcriptional reporters with genome-wide analysis enabled identification of genes associated with melanoma cell motility. Many of these ‘invasive cell population high’ genes were associated with the cell cycle and the microtubule cytoskeleton. Our observations are similar to previous reports in breast cancer where more than half of differentially regulated genes in the motile cell population in vivo were associated with the cell-cycle and further the Polycomb proteins EZH1 and EED were also up-regulated.40 Multiple kinesins were up-regulated in the motile population of both B16 melanoma and breast cancer40 such as KIF2C and KIF23. Thus, the ‘invasive cell population high’ genes are likely to be relevant in many tumour types. We show that EZH2 can positively regulate the expression of KIF2C and KIF22. How EZH2 positively regulates these genes is unknown as EZH2 and PRC2 are classically considered to silence gene expression.18 EZH2 may indirectly regulate these genes through silencing a transcriptional repressor, or it may directly methylate and activate proteins that positively regulate SRF and Notch signalling. EZH2 has a modest effect on the basal activity of SRF and Notch reporter constructs (Fig.3) and may make melanoma cells more responsive to cues that activate SRF and Notch signalling. The heterogeneity in EZH2 levels in both human and mouse melanoma is consistent with a role for EZH2 in regulating the switch to the relatively rare motile state. The epigenetic mechanism of action of EZH2 could imply that melanoma cells would remain in this more motile state for a pro-longed period.
Previous work has shown that the motile cells in B16 melanoma are less pigmented than non-motile cells.1 We show that EZH2 can control this phenotype change associated with motility, through regulation of Oca2/P-protein expression. We hypothesise that this is via direct PRC2 dependent regulation of the Oca2 locus as high H3K27me3 levels could be detected in multiple lymphoma cell-lines that do not express Oca2.31 Depletion of EZH2 leads to changes in the actin cytoskeleton and a reduction in invasion and metastasis of melanoma cells. We identified two kinesins, KIF2C and KIF22, as playing a role in EZH2-dependent melanoma invasion. Previous reports show that KIF2C is associated with motility and metastasis in multiple cancer types.41,42
Taken together, these data place EZH2 as a central regulator of both the amelanotic and migratory phenotype of a subset of melanoma cells associated with metastasis. In conclusion, we propose that heterogeneity in EZH2 levels leads to heterogeneous expression of a cohort of genes associated with motile behaviour in vivo. These genes include many kinesins, and a subset are regulated by SRF. Elevated expression of these genes is important for melanoma cells to respond to pro-migratory cues in the primary tumour micro-environment, and favours their extravasation into the lung parenchyma.
Materials and Methods
Cells and tissue
B16, CHL-1, 5555 and 501mel were kind gifts from Richard Treisman, Michael Way, Richard Marais and Chris Marshall respectively. Brn2::GFP reporter cells as described in Pinner et al.1 Cells were grown in DMEM (Invitrogen) supplemented with 10% fetal bovine serum (PAA Laboratories) and penicillin-streptomycin (Gibco). Human melanoma tissue array obtained from US Biomax (#BCC38218). 2 Human melanoma metastasis samples obtained from Royal Marsden Hospital, London in accordance with UK ethical procedures.
Reagents, plasmids, antibodies and transfections
Cytochalasin D (Tocris), α-MSH (Tocris), DZNep (MERCK Millipore). pCBFRE::GFP and CBFRE::Luciferase were gifts from Julian Lewis. p3DA::2eGFP, p3DA::2eGFP Fos 3’UTR, p3DA::luciferase and pSRF-VP16 were a gift from Richard Treisman. pNICD was a gift from David Ish-Horowicz. Anti-EZH2, Cell Signalling #5246. Anti-Phospho-ERM (Ezrin-Thr567, Radixin Thr564, Moesin Thr558) Cell Signalling #3141. Anti-Beta Tubulin, Sigma (T7816). Anti-Suz12 Abcam ab12073. Anti-MRTF, Santa Cruz sc21588DNA. Transfections were performed with Fugene6 (Promega) and siRNA performed with Lipofectamine 2000(Invitrogen) and Dharmafect 1 (Dharmacon) for B16 and 5555 cells respectively. Unless stated, cells were assayed 48 hours post transfection. siRNA sequences (Dharmacon) in Supp. File 2.
Mice and Imaging
Tumour imaging was performed as described previously.43 Mice were injected subcutaneously on the flank with 1×106 melanoma cells. On day of imaging, mice were anaesthetised and tumour exposed as described previously43. All images acquired with LSM780 (Carl Zeiss). VE-Cadherin:GFPCAAX27 and C57BL6 mice used for intravital imaging. C57BL6 mice used for lymph node endpoint studies. Two-photon Ti-Sapphire laser (Coherent) used to image pigment both in vitro and in vivo with pigment emission between 500 and 550 nm.
Image Analysis and Quantification
Quantification of confocal images of melanoma cell motility, reporter and pigment intensity in LSM Zen 2009 (Carl Zeiss) and pigment/EZH2 intensity in human tissue array performed in ImageJ. Cellular pigment or reporter intensity was corrected for background levels and then normalised to the average intensity for the whole image. Cell area measured in Volocity (Perkin Elmer). Cell tracking was performed with ImageJ Manual Tracking Plug-in. Overlapping tracks of motile cells were defined as parallel tracks within 5μm of each other laterally, with overlap of longer than 12μm and cells present within 15 minutes of each other. Persistence was calculated by actual distance travelled by the cell divided by the Euclidean distance.
Invasion Assays
Cells were plated on collagen/matrigel matrices (approx. 4.6 mg/ml collagen and 2.2 mg/ml matrigel) and left to invade down in to the gel for 48 hours. The number of cells invaded into the gel was normalised to non-invading cells.
Lung Colonisation assays
1×106 B16 F10 cells containing 50:50 mix of mCherry and GFP cells transfected with control or EZH2 targeted siRNA were injected into mouse tail-vein. After 48 hours mice were sacrificed, lungs dissected and imaged using 10x lens (Zeiss). Images were analysed for area of mCherry vs GFP cells using Volocity (Perkin Elmer). The ratio of control transfected B16 F10 to EZH2 siRNA transfected B16 F10 cells was normalised to the ratio of mCherry:GFP cells injected and from control mice.
Microarray, GSEA and TCGA analysis
Microarray analysis was performed together with The Genome Centre at The Barts London using Illumina chips (REF6V3). Fold changes were calculated as average for replicates. 3 replicates of CBFRE:GFP Notch reporter high versus low, 2 replicates of Brn2::GFP reporter high versus low and 2 replicates of B16 control and EZH2 siRNA treatment were performed. GSEA was performed using free software from The Broad Institute of MIT following the program guidelines. The “MRTF targets” geneset was defined as genes down-regulated by greater than 1.5 fold in B16 F2 cells on MRTF depletion performed in 15. The ‘invasive cell population high’ genes were defined as genes both down-regulated greater than 1.5 fold on MRTF depletion and up-regulated greater than 1.2 fold in the CBFRE::GFP High population. TCGA data was used from cBioportal website.
Supplementary Material
Supplementary Figure Legends Supplementary Figure 1 A. Schematic of the retroviral constitutive mRFP-CAAX construct. B.i) Schematic of the Notch reporter construct and CBF responsive element. CBF-1 binding site shown in red. SV40 M.P denotes the simian virus 40 minimal promoter. Graph showing percentage of responsive cells in a transient transfection assay to test the Notch reporter (CBFRE::GFP). CBFRE::GFP was co-transfected with mCherry and either NICD or the empty vector in a ratio of 2:1. The ratio of GFP positive responsive cells to mCherry positive cells was calculated and converted to a percentage. Error bars show standard error. Stars indicate a p-value <0.05 in an ANOVA test. ii) Schematic showing SRF reporter construct used (3DA::2eGFP). Red indicates SRF binding sequences. xActin M.P denotes Xenopus type 5 Actin minimal promoter containing the TATA box and transcription start site. Graph showing SRF reporter activity after transient transfection assay to test the SRF reporter. The SRF reporter construct was co-transfected with a constitutive CFP plasmid. The cells were serum starved and then stimulated with either 15% FCS or 5 μM cytochalasin D for 5 or 10 hours. Boxes show median and inter-quartile range and whiskers show 95th and 5th percentiles. C. Histograms of cellular Notch and SRF reporter fluorescence in vivo.
Supplementary Figure 2 A.i) Cellular fluorescence quantification of B16 F2 3DA::2eGFP Fos3’UTR SRF reporter cells serum starved for 8 hours before treatment with control or 5 μM Cytochalasin D for 16 hours. ii) Quantification of cellular pigment and reporter activity of 3DA::2eGFP Fos3’UTR SRF reporter tumours with membrane targeted mRFP. Each data point represents a single cell. iii) Cellular reporter fluorescence intensity of non-motile and motile cells in primary tumour 3DA::2eGFP Fos 3’UTR SRF reporter tumours. Greater than 25 cells analysed from at least 9 movies of 3 mice. B. Cellular reporter fluorescence intensity of non-motile, singly motile and streaming cells in B16 CBFRE::GFP and B16 3DA::2eGFP primary tumours. C. Intravital confocal images of B16 F2 CBFRE::GFP Notch reporter and 3DA:2eGFP SRF reporter tumours with membrane targeted mRFP in lymph node micrometastases. Scale bar indicates 20 μm.
Supplementary Figure 3 A.i) Geneset enrichment analysis showing lack of enrichment of ‘invasive cell population high’ genes in NICD Notch target genes. ii) Geneset enrichment analysis showing enrichment of NICD Notch targets in CBFRE::GFP Notch reporter high population. B.i) Geneset enrichment analysis showing enrichment of CBFRE::GFP High genes in Brn2 promoter high population. ii) Geneset enrichment analysis showing enrichment of MRTF target genes in Brn2 promoter high population. C. Geneset enrichment analysis showing enrichment of ‘invasive cell population high’ genes in B16 Brn2::GFP reporter high population. D.i) Histogram shows the proportion of acid β-gal positive cells in control and EZH2 depleted B16 cells n=3. ii) Histogram shows the proportion of viable cells (assessed by Toluedene blue exclusion) in control and EZH2 depleted cultures n=3.
Supplementary Figure 4 A.i) Immunohistochemistry of paraffin fixed B16 and 5555 mouse melanoma for EZH2. Arrows indicate cells with high EZH2 levels. M indicates tumour margin and T indicates tumour mass. Scale bars in left panels indicate 50 μm and in right panels indicate 150 μm. ii) Histograms of EZH2 staining intensity in the whole tumour or tumour edge in B16 and 5555 melanoma. B. Immunofluorecence staining of frozen sections of human melanoma metastasis 2 for DAPI (white) and EZH2 (blue) and tri-methylated lysine 27 on histone H3 (H3K27me3) (red). Scale bar indicates 50 μm. C. Graph showing correlation between relative nuclear EZH2 and H3K27me3 intensities. p-value indicates statistically significant correlation using Spearmans rank test. D. Quantification of H3K27me3 immunostaining in B16 cells on control and si EZH2 depletion. Data from at least 5 images of 3 independent experiments. Error bars show the standard error and stars indicate p-value<0.05 in ANOVA statistical test. E. EZH2 immunostaining and histograms of cellular EZH2 levels of frozen human metastatic melanoma tumours. EZH2 in white, actin in green, DAPI in purple. Scale bar indicates 50 μm.
Supplementary Figure 5 A.i) Western blot showing Suz12 protein levels in B16 cells after transfection of control of Suz12 siRNA. ii) Quantification of cellular pigment levels after control and si Suz12 depletion in B16 cells. B. Confocal images of B16 cells after control and si Suz12 knockdown. Actin in white. Scale bar indicates 25 μm.
Supplementary Figure 6 A. Table shows the relative levels of Slc45a2, Tyr, Tyrp1, Oca2 and Dct mRNA in B16 cells after mock transfection and transfection with control and EZH2 siRNA. B. TCGA analysis of Oca2 mRNA levels vs EZH2 mRNA levels as determined by RNASeq. C. Graph shows proportion of viable cells (assessed by Toluedene blue exclusion) in control and Oca2 siRNA depleted B16 cells. n=3.
Supplementary Figure 7 A. KIF2c and KIF22 mRNA levels in B16 cells relative to control levels after 2μM DZNep treatment. n=3 mean and s.d. shown. B. Histogram shows the proportion of viable cells (assessed by Toluedene blue exclusion) in control, KIF2c, and KIF22 depleted cultures - n=3.
Supplementary Figure 8 A. Confocal images of B16 F2 cells after control and siRBPSUH and siSRF smart pool knockdown – F-actin in green. B. Graph showing relative invasion of B16 cells into collagen/matrigel matrices after control and siRBPSUH and siSRF smart pool knockdown. n=3 mean and s.d. shown C. Graph showing relative invasion of B16 cells into collagen/matrigel matrices treated with either DMSO or 2μM DZNep and transfected with control, SRF-VP16, or NICD-GFP. n=3 mean and s.d. shown
Acknowledgements
We would like to thank members of the Tumour Cell Biology laboratory for their discussion and comments. We thank Richard Treisman’s laboratory for reagents. We are indebted to the Experimental Histopathology laboratory, FACS laboratory, and Biological Resources Unit at the London Research Institute and Rosamond Nuamah and Charles Mein at Barts and The London Genome Centre. We are extremely grateful for PhD sponsorship of C.S.M from The McGrath Charitable Trust via Cancer Research UK.
Funding This study was supported by Cancer Research UK.
Abbreviations
- SRF
Serum Response Factor
- TGF-β
Transforming Growth Factor Beta
- ERM
Ezrin, Radixin, Moesin proteins
Footnotes
Disclosure of potential conflict of interests There are no potential conflicts of interest to disclose.
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc)
References
- 1.Pinner S, Jordan P, Sharrock K, Bazley L, Collinson L, Marais R, et al. Intravital imaging reveals transient changes in pigment production and Brn2 expression during metastatic melanoma dissemination. Cancer research. 2009;69(20):7969–77. doi: 10.1158/0008-5472.CAN-09-0781. Epub 2009/10/15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gupta PB, Kuperwasser C, Brunet JP, Ramaswamy S, Kuo WL, Gray JW, et al. The melanocyte differentiation program predisposes to metastasis after neoplastic transformation. Nature genetics. 2005;37(10):1047–54. doi: 10.1038/ng1634. Epub 2005/09/06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bailey CM, Morrison JA, Kulesa PM. Melanoma revives an embryonic migration program to promote plasticity and invasion. Pigment cell & melanoma research. 2012;25(5):573–83. doi: 10.1111/j.1755-148X.2012.01025.x. Epub 2012/06/12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoek KS, Eichhoff OM, Schlegel NC, Dobbeling U, Kobert N, Schaerer L, et al. In vivo switching of human melanoma cells between proliferative and invasive states. Cancer research. 2008;68(3):650–6. doi: 10.1158/0008-5472.CAN-07-2491. Epub 2008/02/05. [DOI] [PubMed] [Google Scholar]
- 5.Hoek KS, Goding CR. Cancer stem cells versus phenotype-switching in melanoma. Pigment cell & melanoma research. 2010;23(6):746–59. doi: 10.1111/j.1755-148X.2010.00757.x. Epub 2010/08/24. [DOI] [PubMed] [Google Scholar]
- 6.Carreira S, Goodall J, Denat L, Rodriguez M, Nuciforo P, Hoek KS, et al. Mitf regulation of Dia1 controls melanoma proliferation and invasiveness. Genes & development. 2006;20(24):3426–39. doi: 10.1101/gad.406406. Epub 2006/12/22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goodall J, Carreira S, Denat L, Kobi D, Davidson I, Nuciforo P, et al. Brn-2 represses microphthalmia-associated transcription factor expression and marks a distinct subpopulation of microphthalmia-associated transcription factor-negative melanoma cells. Cancer research. 2008;68(19):7788–94. doi: 10.1158/0008-5472.CAN-08-1053. Epub 2008/10/03. [DOI] [PubMed] [Google Scholar]
- 8.Jarriault S, Brou C, Logeat F, Schroeter EH, Kopan R, Israel A. Signalling downstream of activated mammalian Notch. Nature. 1995;377(6547):355–8. doi: 10.1038/377355a0. Epub 1995/09/28. [DOI] [PubMed] [Google Scholar]
- 9.Balint K, Xiao M, Pinnix CC, Soma A, Veres I, Juhasz I, et al. Activation of Notch1 signaling is required for beta-catenin-mediated human primary melanoma progression. The Journal of clinical investigation. 2005;115(11):3166–76. doi: 10.1172/JCI25001. Epub 2005/10/22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu ZJ, Xiao M, Balint K, Smalley KS, Brafford P, Qiu R, et al. Notch1 signaling promotes primary melanoma progression by activating mitogen-activated protein kinase/phosphatidylinositol 3-kinase-Akt pathways and up-regulating N-cadherin expression. Cancer research. 2006;66(8):4182–90. doi: 10.1158/0008-5472.CAN-05-3589. Epub 2006/04/19. [DOI] [PubMed] [Google Scholar]
- 11.Pinnix CC, Lee JT, Liu ZJ, McDaid R, Balint K, Beverly LJ, et al. Active Notch1 confers a transformed phenotype to primary human melanocytes. Cancer research. 2009;69(13):5312–20. doi: 10.1158/0008-5472.CAN-08-3767. Epub 2009/06/25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moriyama M, Osawa M, Mak SS, Ohtsuka T, Yamamoto N, Han H, et al. Notch signaling via Hes1 transcription factor maintains survival of melanoblasts and melanocyte stem cells. The Journal of cell biology. 2006;173(3):333–9. doi: 10.1083/jcb.200509084. Epub 2006/05/03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zabierowski SE, Baubet V, Himes B, Li L, Fukunaga-Kalabis M, Patel S, et al. Direct reprogramming of melanocytes to neural crest stem-like cells by one defined factor. Stem Cells. 2011;29(11):1752–62. doi: 10.1002/stem.740. Epub 2011/09/29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Olson EN, Nordheim A. Linking actin dynamics and gene transcription to drive cellular motile functions. Nature reviews Molecular cell biology. 2010;11(5):353–65. doi: 10.1038/nrm2890. Epub 2010/04/24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Medjkane S, Perez-Sanchez C, Gaggioli C, Sahai E, Treisman R. Myocardin-related transcription factors and SRF are required for cytoskeletal dynamics and experimental metastasis. Nature cell biology. 2009;11(3):257–68. doi: 10.1038/ncb1833. Epub 2009/02/10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clark EA, Golub TR, Lander ES, Hynes RO. Genomic analysis of metastasis reveals an essential role for RhoC. Nature. 2000;406(6795):532–5. doi: 10.1038/35020106. Epub 2000/08/22. [DOI] [PubMed] [Google Scholar]
- 17.Kitzing TM, Wang Y, Pertz O, Copeland JW, Grosse R. Formin-like 2 drives amoeboid invasive cell motility downstream of RhoC. Oncogene. 2010;29(16):2441–8. doi: 10.1038/onc.2009.515. Epub 2010/01/27. [DOI] [PubMed] [Google Scholar]
- 18.Morey L, Helin K. Polycomb group protein-mediated repression of transcription. Trends in biochemical sciences. 2010;35(6):323–32. doi: 10.1016/j.tibs.2010.02.009. Epub 2010/03/30. [DOI] [PubMed] [Google Scholar]
- 19.Ezhkova E, Pasolli HA, Parker JS, Stokes N, Su IH, Hannon G, et al. Ezh2 orchestrates gene expression for the stepwise differentiation of tissue-specific stem cells. Cell. 2009;136(6):1122–35. doi: 10.1016/j.cell.2008.12.043. Epub 2009/03/24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McHugh JB, Fullen DR, Ma L, Kleer CG, Su LD. Expression of polycomb group protein EZH2 in nevi and melanoma. Journal of cutaneous pathology. 2007;34(8):597–600. doi: 10.1111/j.1600-0560.2006.00678.x. Epub 2007/07/21. [DOI] [PubMed] [Google Scholar]
- 21.Asangani IA, Harms PW, Dodson L, Pandhi M, Kunju LP, Maher CA, et al. Genetic and epigenetic loss of microRNA-31 leads to feed-forward expression of EZH2 in melanoma. Oncotarget. 2012;3(9):1011–25. doi: 10.18632/oncotarget.622. Epub 2012/09/06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hou P, Liu D, Dong J, Xing M. The BRAF(V600E) causes widespread alterations in gene methylation in the genome of melanoma cells. Cell Cycle. 2012;11(2):286–95. doi: 10.4161/cc.11.2.18707. Epub 2011/12/23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tiwari N, Tiwari VK, Waldmeier L, Balwierz PJ, Arnold P, Pachkov M, et al. Sox4 is a master regulator of epithelial-mesenchymal transition by controlling ezh2 expression and epigenetic reprogramming. Cancer cell. 2013;23(6):768–83. doi: 10.1016/j.ccr.2013.04.020. Epub 2013/06/15. [DOI] [PubMed] [Google Scholar]
- 24.Giampieri S, Manning C, Hooper S, Jones L, Hill CS, Sahai E. Localized and reversible TGFbeta signalling switches breast cancer cells from cohesive to single cell motility. Nature cell biology. 2009;11(11):1287–96. doi: 10.1038/ncb1973. Epub 2009/10/20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sanz-Moreno V, Gadea G, Ahn J, Paterson H, Marra P, Pinner S, et al. Rac activation and inactivation control plasticity of tumor cell movement. Cell. 2008;135(3):510–23. doi: 10.1016/j.cell.2008.09.043. Epub 2008/11/06. [DOI] [PubMed] [Google Scholar]
- 26.Friedl P, Locker J, Sahai E, Segall JE. Classifying collective cancer cell invasion. Nature cell biology. 2012;14(8):777–83. doi: 10.1038/ncb2548. Epub 2012/08/03. [DOI] [PubMed] [Google Scholar]
- 27.Manning CS, Jenkins R, Hooper S, Gerhardt H, Marais R, Adams S, et al. Intravital imaging reveals conversion between distinct tumor vascular morphologies and localized vascular response to Sunitinib. Intravital. 2013;2(1):33–44. [Google Scholar]
- 28.Nuytten M, Beke L, Van Eynde A, Ceulemans H, Beullens M, Van Hummelen P, et al. The transcriptional repressor NIPP1 is an essential player in EZH2-mediated gene silencing. Oncogene. 2008;27(10):1449–60. doi: 10.1038/sj.onc.1210774. Epub 2007/08/29. [DOI] [PubMed] [Google Scholar]
- 29.Ren G, Baritaki S, Marathe H, Feng J, Park S, Beach S, et al. Polycomb protein EZH2 regulates tumor invasion via the transcriptional repression of the metastasis suppressor RKIP in breast and prostate cancer. Cancer research. 2012;72(12):3091–104. doi: 10.1158/0008-5472.CAN-11-3546. Epub 2012/04/17. [DOI] [PubMed] [Google Scholar]
- 30.Lee ST, Li Z, Wu Z, Aau M, Guan P, Karuturi RK, et al. Context-specific regulation of NF-kappaB target gene expression by EZH2 in breast cancers. Molecular cell. 2011;43(5):798–810. doi: 10.1016/j.molcel.2011.08.011. Epub 2011/09/03. [DOI] [PubMed] [Google Scholar]
- 31.McCabe MT, Ott HM, Ganji G, Korenchuk S, Thompson C, Van Aller GS, et al. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature. 2012;492(7427):108–12. doi: 10.1038/nature11606. Epub 2012/10/12. [DOI] [PubMed] [Google Scholar]
- 32.Rosemblat S, Durham-Pierre D, Gardner JM, Nakatsu Y, Brilliant MH, Orlow SJ. Identification of a melanosomal membrane protein encoded by the pink-eyed dilution (type II oculocutaneous albinism) gene. Proceedings of the National Academy of Sciences of the United States of America. 1994;91(25):12071–75. doi: 10.1073/pnas.91.25.12071. Epub 1994/12/06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen K, Manga P, Orlow SJ. Pink-eyed dilution protein controls the processing of tyrosinase. Molecular biology of the cell. 2002;13(6):1953–64. doi: 10.1091/mbc.02-02-0022.. Epub 2002/06/12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Donnelly MP, Paschou P, Grigorenko E, Gurwitz D, Barta C, Lu RB, et al. A global view of the OCA2-HERC2 region and pigmentation. Human genetics. 2012;131(5):683–96. doi: 10.1007/s00439-011-1110-x. Epub 2011/11/09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sanz-Moreno V, Gaggioli C, Yeo M, Albrengues J, Wallberg F, Viros A, et al. ROCK and JAK1 signaling cooperate to control actomyosin contractility in tumor cells and stroma. Cancer cell. 2011;20(2):229–45. doi: 10.1016/j.ccr.2011.06.018. Epub 2011/08/16. [DOI] [PubMed] [Google Scholar]
- 36.Gaggioli C, Hooper S, Hidalgo-Carcedo C, Grosse R, Marshall JF, Harrington K, et al. Fibroblast-led collective invasion of carcinoma cells with differing roles for RhoGTPases in leading and following cells. Nature cell biology. 2007;9(12):1392–400. doi: 10.1038/ncb1658. Epub 2007/11/27. [DOI] [PubMed] [Google Scholar]
- 37.Zhao XH, Laschinger C, Arora P, Szaszi K, Kapus A, McCulloch CA. Force activates smooth muscle alpha-actin promoter activity through the Rho signaling pathway. Journal of cell science. 2007;120(Pt 10):1801–9. doi: 10.1242/jcs.001586. Epub 2007/04/26. [DOI] [PubMed] [Google Scholar]
- 38.Andersson ER, Sandberg R, Lendahl U. Notch signaling: simplicity in design, versatility in function. Development. 2011;138(17):3593–612. doi: 10.1242/dev.063610. Epub 2011/08/11. [DOI] [PubMed] [Google Scholar]
- 39.Sahlgren C, Gustafsson MV, Jin S, Poellinger L, Lendahl U. Notch signaling mediates hypoxia-induced tumor cell migration and invasion. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(17):6392–7. doi: 10.1073/pnas.0802047105. Epub 2008/04/23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Goswami S, Wang W, Wyckoff JB, Condeelis JS. Breast cancer cells isolated by chemotaxis from primary tumors show increased survival and resistance to chemotherapy. Cancer research. 2004;64(21):7664–7. doi: 10.1158/0008-5472.CAN-04-2027. Epub 2004/11/03. [DOI] [PubMed] [Google Scholar]
- 41.Ishikawa K, Kamohara Y, Tanaka F, Haraguchi N, Mimori K, Inoue H, et al. Mitotic centromere-associated kinesin is a novel marker for prognosis and lymph node metastasis in colorectal cancer. British journal of cancer. 2008;98(11):1824–9. doi: 10.1038/sj.bjc.6604379. Epub 2008/05/29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nakamura Y, Tanaka F, Haraguchi N, Mimori K, Matsumoto T, Inoue H, et al. Clinicopathological and biological significance of mitotic centromere-associated kinesin overexpression in human gastric cancer. British journal of cancer. 2007;97(4):543–9. doi: 10.1038/sj.bjc.6603905. Epub 2007/07/27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sahai E, Wyckoff J, Philippar U, Segall JE, Gertler F, Condeelis J. Simultaneous imaging of GFP, CFP and collagen in tumors in vivo using multiphoton microscopy. BMC biotechnology. 2005;5:14. doi: 10.1186/1472-6750-5-14. Epub 2005/05/25. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure Legends Supplementary Figure 1 A. Schematic of the retroviral constitutive mRFP-CAAX construct. B.i) Schematic of the Notch reporter construct and CBF responsive element. CBF-1 binding site shown in red. SV40 M.P denotes the simian virus 40 minimal promoter. Graph showing percentage of responsive cells in a transient transfection assay to test the Notch reporter (CBFRE::GFP). CBFRE::GFP was co-transfected with mCherry and either NICD or the empty vector in a ratio of 2:1. The ratio of GFP positive responsive cells to mCherry positive cells was calculated and converted to a percentage. Error bars show standard error. Stars indicate a p-value <0.05 in an ANOVA test. ii) Schematic showing SRF reporter construct used (3DA::2eGFP). Red indicates SRF binding sequences. xActin M.P denotes Xenopus type 5 Actin minimal promoter containing the TATA box and transcription start site. Graph showing SRF reporter activity after transient transfection assay to test the SRF reporter. The SRF reporter construct was co-transfected with a constitutive CFP plasmid. The cells were serum starved and then stimulated with either 15% FCS or 5 μM cytochalasin D for 5 or 10 hours. Boxes show median and inter-quartile range and whiskers show 95th and 5th percentiles. C. Histograms of cellular Notch and SRF reporter fluorescence in vivo.
Supplementary Figure 2 A.i) Cellular fluorescence quantification of B16 F2 3DA::2eGFP Fos3’UTR SRF reporter cells serum starved for 8 hours before treatment with control or 5 μM Cytochalasin D for 16 hours. ii) Quantification of cellular pigment and reporter activity of 3DA::2eGFP Fos3’UTR SRF reporter tumours with membrane targeted mRFP. Each data point represents a single cell. iii) Cellular reporter fluorescence intensity of non-motile and motile cells in primary tumour 3DA::2eGFP Fos 3’UTR SRF reporter tumours. Greater than 25 cells analysed from at least 9 movies of 3 mice. B. Cellular reporter fluorescence intensity of non-motile, singly motile and streaming cells in B16 CBFRE::GFP and B16 3DA::2eGFP primary tumours. C. Intravital confocal images of B16 F2 CBFRE::GFP Notch reporter and 3DA:2eGFP SRF reporter tumours with membrane targeted mRFP in lymph node micrometastases. Scale bar indicates 20 μm.
Supplementary Figure 3 A.i) Geneset enrichment analysis showing lack of enrichment of ‘invasive cell population high’ genes in NICD Notch target genes. ii) Geneset enrichment analysis showing enrichment of NICD Notch targets in CBFRE::GFP Notch reporter high population. B.i) Geneset enrichment analysis showing enrichment of CBFRE::GFP High genes in Brn2 promoter high population. ii) Geneset enrichment analysis showing enrichment of MRTF target genes in Brn2 promoter high population. C. Geneset enrichment analysis showing enrichment of ‘invasive cell population high’ genes in B16 Brn2::GFP reporter high population. D.i) Histogram shows the proportion of acid β-gal positive cells in control and EZH2 depleted B16 cells n=3. ii) Histogram shows the proportion of viable cells (assessed by Toluedene blue exclusion) in control and EZH2 depleted cultures n=3.
Supplementary Figure 4 A.i) Immunohistochemistry of paraffin fixed B16 and 5555 mouse melanoma for EZH2. Arrows indicate cells with high EZH2 levels. M indicates tumour margin and T indicates tumour mass. Scale bars in left panels indicate 50 μm and in right panels indicate 150 μm. ii) Histograms of EZH2 staining intensity in the whole tumour or tumour edge in B16 and 5555 melanoma. B. Immunofluorecence staining of frozen sections of human melanoma metastasis 2 for DAPI (white) and EZH2 (blue) and tri-methylated lysine 27 on histone H3 (H3K27me3) (red). Scale bar indicates 50 μm. C. Graph showing correlation between relative nuclear EZH2 and H3K27me3 intensities. p-value indicates statistically significant correlation using Spearmans rank test. D. Quantification of H3K27me3 immunostaining in B16 cells on control and si EZH2 depletion. Data from at least 5 images of 3 independent experiments. Error bars show the standard error and stars indicate p-value<0.05 in ANOVA statistical test. E. EZH2 immunostaining and histograms of cellular EZH2 levels of frozen human metastatic melanoma tumours. EZH2 in white, actin in green, DAPI in purple. Scale bar indicates 50 μm.
Supplementary Figure 5 A.i) Western blot showing Suz12 protein levels in B16 cells after transfection of control of Suz12 siRNA. ii) Quantification of cellular pigment levels after control and si Suz12 depletion in B16 cells. B. Confocal images of B16 cells after control and si Suz12 knockdown. Actin in white. Scale bar indicates 25 μm.
Supplementary Figure 6 A. Table shows the relative levels of Slc45a2, Tyr, Tyrp1, Oca2 and Dct mRNA in B16 cells after mock transfection and transfection with control and EZH2 siRNA. B. TCGA analysis of Oca2 mRNA levels vs EZH2 mRNA levels as determined by RNASeq. C. Graph shows proportion of viable cells (assessed by Toluedene blue exclusion) in control and Oca2 siRNA depleted B16 cells. n=3.
Supplementary Figure 7 A. KIF2c and KIF22 mRNA levels in B16 cells relative to control levels after 2μM DZNep treatment. n=3 mean and s.d. shown. B. Histogram shows the proportion of viable cells (assessed by Toluedene blue exclusion) in control, KIF2c, and KIF22 depleted cultures - n=3.
Supplementary Figure 8 A. Confocal images of B16 F2 cells after control and siRBPSUH and siSRF smart pool knockdown – F-actin in green. B. Graph showing relative invasion of B16 cells into collagen/matrigel matrices after control and siRBPSUH and siSRF smart pool knockdown. n=3 mean and s.d. shown C. Graph showing relative invasion of B16 cells into collagen/matrigel matrices treated with either DMSO or 2μM DZNep and transfected with control, SRF-VP16, or NICD-GFP. n=3 mean and s.d. shown