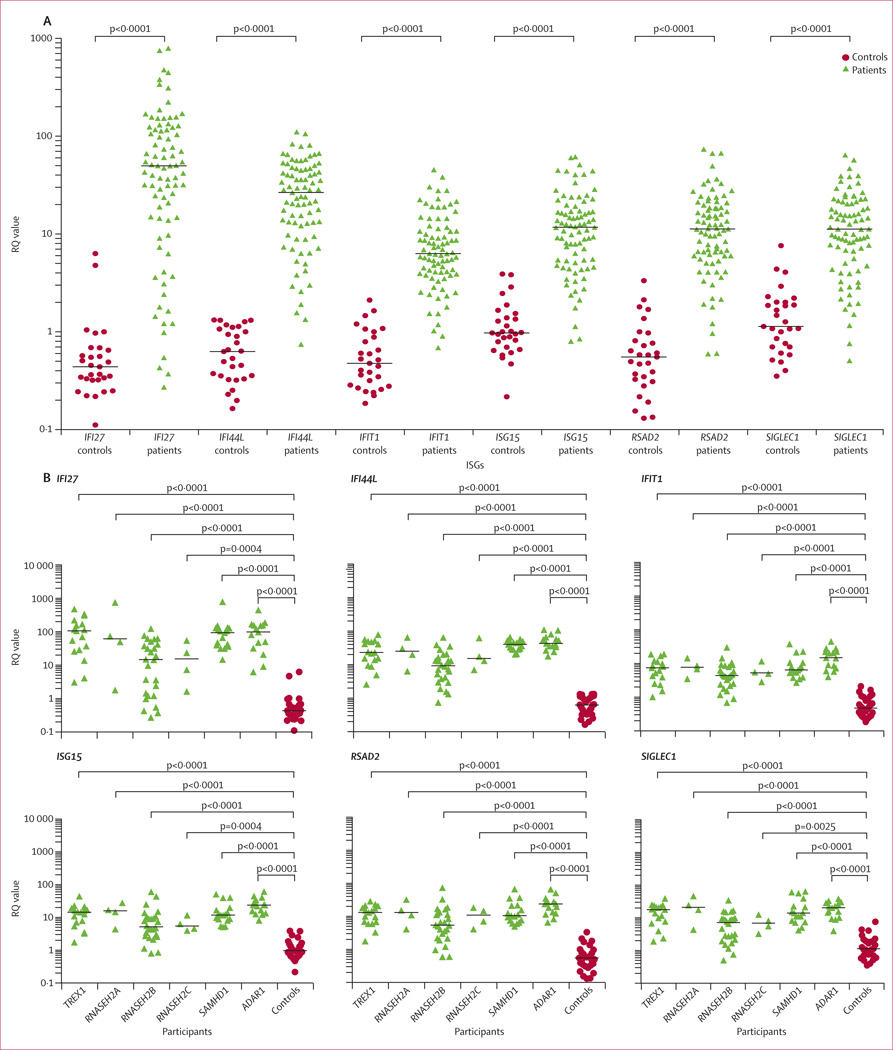
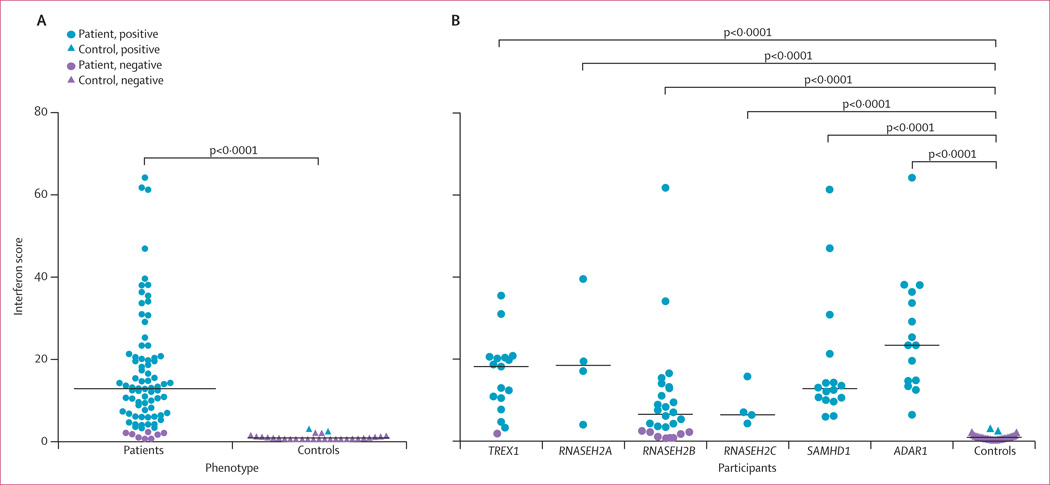
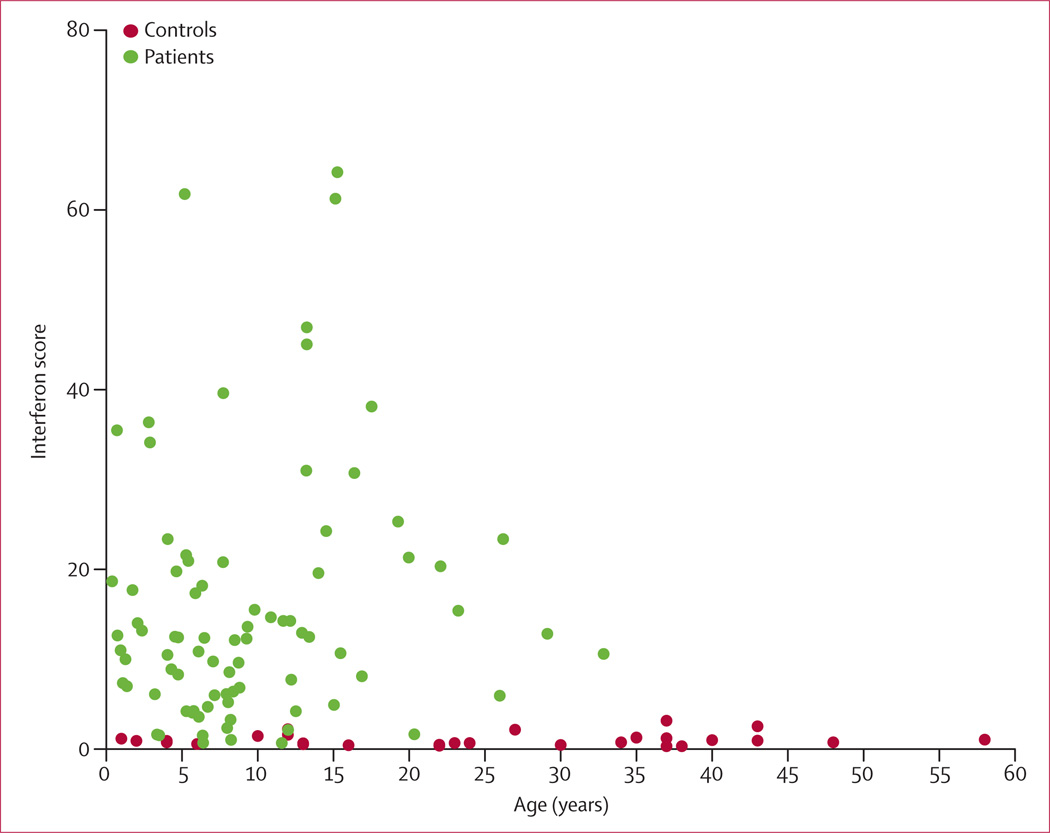
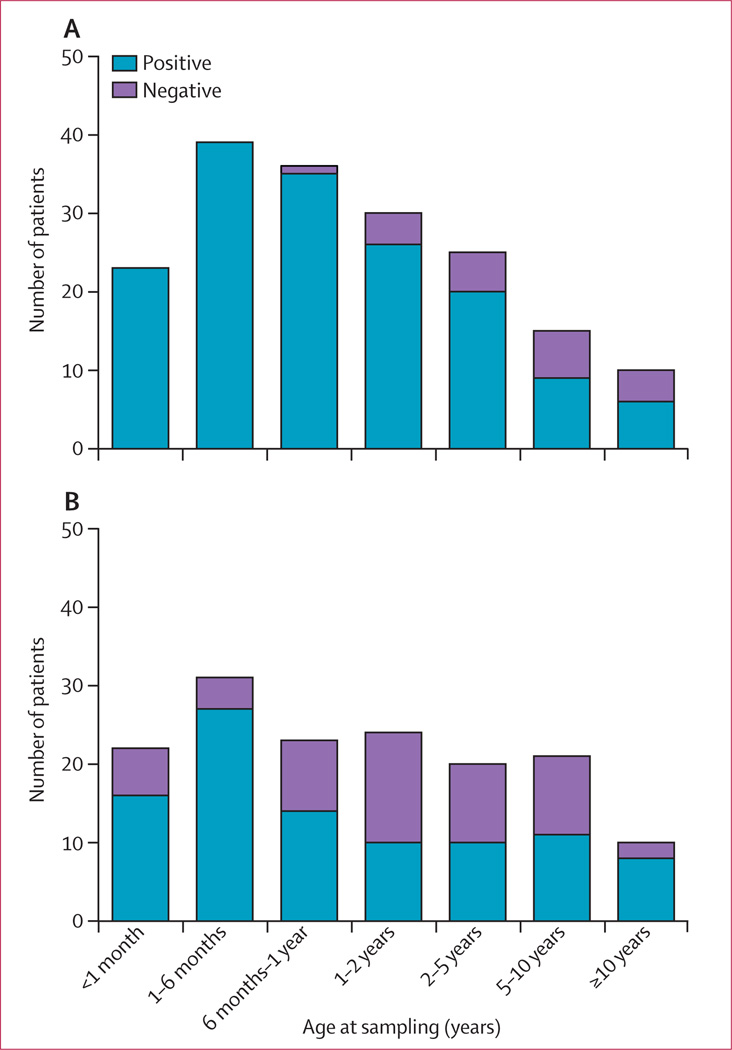
Gillian I Rice
Gillian I Rice
1Manchester Centre for Genomic Medicine, Institute of Human Development, Faculty of Medical and Human Sciences, University of Manchester, Manchester Academic Health Sciences Centre (MAHSC), Manchester, UK (G I Rice PhD, G M A Forte MPhil, M Szynkiewicz MSc, D S Chase PhD, Y J Crow PhD); Erasme University Hospital, Universite Libre de Bruxelles (ULB), Brussels, Belgium, (A Aeby MD, C N Vilain MD, V Wermenbol MD); Medical Molecular Genetics Department, (M S Abdel-Hamid MSc, L Effat PhD), and Clinical Genetics Department, (S A Temtamy PhD, M S Aglan PhD, M S Zaki PhD, G M H Abdel-Salam PhD), Human Genetics and Genome Research Division, National Research Centre, Cairo, Egypt; Bradford Teaching Hospitals NHS Foundation Trust, Bradford, UK (S Ackroyd MB ChB); Department of Clinical Biochemistry, Lancashire Teaching Hospitals NHS Foundation Trust, Preston, UK (R Allcock FRCPath); University Hospitals of Coventry and Warwickshire, Coventry, UK (K M Bailey MRCPCH); Department of Brain and Behavioral Sciences, Unit of Child Neurology and Psychiatry, University of Pavia, Pavia, Italy, (U Balottin MD); Child Neurology and Psychiatry Unit, (U Balottin, I Olivieri MD, S Orcesi MD), and Laboratory of Experimental Neurobiology (C Cereda PhD), C. Mondino National Neurological Institute, Pavia, Italy; Praticien Hospitalier, Unité de Neurologie Pédiatrique et Centre de Référence des Maladies Neuromusculaires, Hôpital Necker-Enfants Malades, Paris, France (C Barnerias MD); Department of Pediatrics, and Department of Neurology and Neurosurgery, Montreal Children’s Hospital, McGill University Health Center, Montreal, QC, Canada (G Bernard MD); Department of Dermatology, Reference Centre for Cutaneous Rare Diseases (MAGEC), Necker-Enfants Malades APHP, Sorbonne Paris Cité, Université Paris Descartes, Institut Imagine, Paris, France (C Bodemer PhD); Department of Pediatrics, Hospital Universitario Araba, Vitoria-Gasteiz, Spain (M P Botella MD); Manchester Centre for Genomic Medicine, Central Manchester University Hospitals NHS Foundation Trust, MAHSC, Manchester, UK (K E Chandler MD, Y J Crow); Department of Paediatric Neurology, University Hospitals of Leicester NHS Trust, Leicester, UK (L Dabydeen MRCPCH); TY Nelson Department of Neurology, and Neuroimmunology Group, the Children’s Hospital at Westmead, University of Sydney, Sydney, NSW, Australia (R C Dale PhD); Nutrition and Metabolism Unit, Hôpital Universitaire des Enfants Reine Fabiola (ULB), Brussels, Belgium (C De Laet MD); Department of Paediatric Neurology, Royal Preston Hospital, Preston, UK (C G E L De Goede FRCPCH); Pediatric Neurology Unit, Hospital Vall d’Hebron, Barcelona, Spain (M del Toro MD); Paediatric Neurology Unit, University Hospital 12 Octubre, Madrid, Spain (N Nunez Enamorado MD, R Simon MD); Department of Clinical and Experimental Sciences, Child Neurology and Psychiatry Unit, University of Brescia, Brescia, Italy (E Fazzi MD); Department of Genetics, BioCruces Health Research Institute, Hospital, Universitario Cruces, Bizkaia, Spain (B Gener MD); Institute of Biomedicine and Translational Medicine, University of Tartu, Tartu, Estonia (M Haldre BSc, P Peterson PhD, K Kisand PhD); General Neurology and Complex Motor Disorders Service, Evelina Children’s Hospital, Guy’s and St Thomas’ NHS Foundation Trust, London, UK (J-P S-M Lin PhD); Department of Paediatric Neurology, Leeds General Infirmary, Leeds, UK (J H Livingston MB ChB); Department of Neurosciences and Behavior Sciences, School of Medicine of Ribeirão Preto, University of São Paulo, São Paulo, Brazil, (C Marques Lourenco MD, W Marques Jr MD); Royal Devon and Exeter Foundation NHS Trust, Exeter, UK (P Oades FRCPCH); Section for Child Neurology, Women and Children’s Division, Oslo University Hospital, Oslo, Norway (M Rasmussen MD); Neuropediatrie, Hopital Gui de Chauliac, and INSERM U1051, INM, Montpellier, France (A Roubertie MD); Center for Genetic Medicine Research and Department of Neurology, Children’s National Medical Center, George Washington University School of Medicine, Washington, DC, USA, (J Loewenstein Schmidt CGC, A Vanderver MD); The Genetic Institute, Ha’Emek Medical Center, Afula and the Rappaport Faculty of Medicine, Technion, Haifa, Israel (S A Shalev MD, R Spiegel MD); Department of Neurology, University of Utah School of Medicine, Salt Lake City, UT, USA (K J Swoboda FACMG); Neurology Department, Royal Manchester Children’s Hospital, Manchester, UK (G Vassallo MD); Centre for Rare Diseases and Personalised Medicine and Department of Medical and Molecular Genetics, School of Clinical and Experimental Medicine, University of Birmingham, UK, and West Midlands Regional Genetics Service, Clinical Genetics Unit, Birmingham Women’s Hospital, Birmingham, UK (J Vogt MRCP); Nottingham Children’s Hospital, Nottingham University Hospitals NHS Trust, Nottingham, UK (W P Whitehouse FRCPCH); Neuropaediatrics, Department of Paediatrics, Mater Dei Hospital, Tal-Qroqq, Malta (D Soler MD); and Université Paris Descartes, EA 1833 and Hôpital Cochin, Service de Virologie, Paris, France, (F Rozenberg MD, P Lebon MD)
1,
Gabriella M A Forte
Gabriella M A Forte
1Manchester Centre for Genomic Medicine, Institute of Human Development, Faculty of Medical and Human Sciences, University of Manchester, Manchester Academic Health Sciences Centre (MAHSC), Manchester, UK (G I Rice PhD, G M A Forte MPhil, M Szynkiewicz MSc, D S Chase PhD, Y J Crow PhD); Erasme University Hospital, Universite Libre de Bruxelles (ULB), Brussels, Belgium, (A Aeby MD, C N Vilain MD, V Wermenbol MD); Medical Molecular Genetics Department, (M S Abdel-Hamid MSc, L Effat PhD), and Clinical Genetics Department, (S A Temtamy PhD, M S Aglan PhD, M S Zaki PhD, G M H Abdel-Salam PhD), Human Genetics and Genome Research Division, National Research Centre, Cairo, Egypt; Bradford Teaching Hospitals NHS Foundation Trust, Bradford, UK (S Ackroyd MB ChB); Department of Clinical Biochemistry, Lancashire Teaching Hospitals NHS Foundation Trust, Preston, UK (R Allcock FRCPath); University Hospitals of Coventry and Warwickshire, Coventry, UK (K M Bailey MRCPCH); Department of Brain and Behavioral Sciences, Unit of Child Neurology and Psychiatry, University of Pavia, Pavia, Italy, (U Balottin MD); Child Neurology and Psychiatry Unit, (U Balottin, I Olivieri MD, S Orcesi MD), and Laboratory of Experimental Neurobiology (C Cereda PhD), C. Mondino National Neurological Institute, Pavia, Italy; Praticien Hospitalier, Unité de Neurologie Pédiatrique et Centre de Référence des Maladies Neuromusculaires, Hôpital Necker-Enfants Malades, Paris, France (C Barnerias MD); Department of Pediatrics, and Department of Neurology and Neurosurgery, Montreal Children’s Hospital, McGill University Health Center, Montreal, QC, Canada (G Bernard MD); Department of Dermatology, Reference Centre for Cutaneous Rare Diseases (MAGEC), Necker-Enfants Malades APHP, Sorbonne Paris Cité, Université Paris Descartes, Institut Imagine, Paris, France (C Bodemer PhD); Department of Pediatrics, Hospital Universitario Araba, Vitoria-Gasteiz, Spain (M P Botella MD); Manchester Centre for Genomic Medicine, Central Manchester University Hospitals NHS Foundation Trust, MAHSC, Manchester, UK (K E Chandler MD, Y J Crow); Department of Paediatric Neurology, University Hospitals of Leicester NHS Trust, Leicester, UK (L Dabydeen MRCPCH); TY Nelson Department of Neurology, and Neuroimmunology Group, the Children’s Hospital at Westmead, University of Sydney, Sydney, NSW, Australia (R C Dale PhD); Nutrition and Metabolism Unit, Hôpital Universitaire des Enfants Reine Fabiola (ULB), Brussels, Belgium (C De Laet MD); Department of Paediatric Neurology, Royal Preston Hospital, Preston, UK (C G E L De Goede FRCPCH); Pediatric Neurology Unit, Hospital Vall d’Hebron, Barcelona, Spain (M del Toro MD); Paediatric Neurology Unit, University Hospital 12 Octubre, Madrid, Spain (N Nunez Enamorado MD, R Simon MD); Department of Clinical and Experimental Sciences, Child Neurology and Psychiatry Unit, University of Brescia, Brescia, Italy (E Fazzi MD); Department of Genetics, BioCruces Health Research Institute, Hospital, Universitario Cruces, Bizkaia, Spain (B Gener MD); Institute of Biomedicine and Translational Medicine, University of Tartu, Tartu, Estonia (M Haldre BSc, P Peterson PhD, K Kisand PhD); General Neurology and Complex Motor Disorders Service, Evelina Children’s Hospital, Guy’s and St Thomas’ NHS Foundation Trust, London, UK (J-P S-M Lin PhD); Department of Paediatric Neurology, Leeds General Infirmary, Leeds, UK (J H Livingston MB ChB); Department of Neurosciences and Behavior Sciences, School of Medicine of Ribeirão Preto, University of São Paulo, São Paulo, Brazil, (C Marques Lourenco MD, W Marques Jr MD); Royal Devon and Exeter Foundation NHS Trust, Exeter, UK (P Oades FRCPCH); Section for Child Neurology, Women and Children’s Division, Oslo University Hospital, Oslo, Norway (M Rasmussen MD); Neuropediatrie, Hopital Gui de Chauliac, and INSERM U1051, INM, Montpellier, France (A Roubertie MD); Center for Genetic Medicine Research and Department of Neurology, Children’s National Medical Center, George Washington University School of Medicine, Washington, DC, USA, (J Loewenstein Schmidt CGC, A Vanderver MD); The Genetic Institute, Ha’Emek Medical Center, Afula and the Rappaport Faculty of Medicine, Technion, Haifa, Israel (S A Shalev MD, R Spiegel MD); Department of Neurology, University of Utah School of Medicine, Salt Lake City, UT, USA (K J Swoboda FACMG); Neurology Department, Royal Manchester Children’s Hospital, Manchester, UK (G Vassallo MD); Centre for Rare Diseases and Personalised Medicine and Department of Medical and Molecular Genetics, School of Clinical and Experimental Medicine, University of Birmingham, UK, and West Midlands Regional Genetics Service, Clinical Genetics Unit, Birmingham Women’s Hospital, Birmingham, UK (J Vogt MRCP); Nottingham Children’s Hospital, Nottingham University Hospitals NHS Trust, Nottingham, UK (W P Whitehouse FRCPCH); Neuropaediatrics, Department of Paediatrics, Mater Dei Hospital, Tal-Qroqq, Malta (D Soler MD); and Université Paris Descartes, EA 1833 and Hôpital Cochin, Service de Virologie, Paris, France, (F Rozenberg MD, P Lebon MD)
1,
Marcin Szynkiewicz
Marcin Szynkiewicz
1Manchester Centre for Genomic Medicine, Institute of Human Development, Faculty of Medical and Human Sciences, University of Manchester, Manchester Academic Health Sciences Centre (MAHSC), Manchester, UK (G I Rice PhD, G M A Forte MPhil, M Szynkiewicz MSc, D S Chase PhD, Y J Crow PhD); Erasme University Hospital, Universite Libre de Bruxelles (ULB), Brussels, Belgium, (A Aeby MD, C N Vilain MD, V Wermenbol MD); Medical Molecular Genetics Department, (M S Abdel-Hamid MSc, L Effat PhD), and Clinical Genetics Department, (S A Temtamy PhD, M S Aglan PhD, M S Zaki PhD, G M H Abdel-Salam PhD), Human Genetics and Genome Research Division, National Research Centre, Cairo, Egypt; Bradford Teaching Hospitals NHS Foundation Trust, Bradford, UK (S Ackroyd MB ChB); Department of Clinical Biochemistry, Lancashire Teaching Hospitals NHS Foundation Trust, Preston, UK (R Allcock FRCPath); University Hospitals of Coventry and Warwickshire, Coventry, UK (K M Bailey MRCPCH); Department of Brain and Behavioral Sciences, Unit of Child Neurology and Psychiatry, University of Pavia, Pavia, Italy, (U Balottin MD); Child Neurology and Psychiatry Unit, (U Balottin, I Olivieri MD, S Orcesi MD), and Laboratory of Experimental Neurobiology (C Cereda PhD), C. Mondino National Neurological Institute, Pavia, Italy; Praticien Hospitalier, Unité de Neurologie Pédiatrique et Centre de Référence des Maladies Neuromusculaires, Hôpital Necker-Enfants Malades, Paris, France (C Barnerias MD); Department of Pediatrics, and Department of Neurology and Neurosurgery, Montreal Children’s Hospital, McGill University Health Center, Montreal, QC, Canada (G Bernard MD); Department of Dermatology, Reference Centre for Cutaneous Rare Diseases (MAGEC), Necker-Enfants Malades APHP, Sorbonne Paris Cité, Université Paris Descartes, Institut Imagine, Paris, France (C Bodemer PhD); Department of Pediatrics, Hospital Universitario Araba, Vitoria-Gasteiz, Spain (M P Botella MD); Manchester Centre for Genomic Medicine, Central Manchester University Hospitals NHS Foundation Trust, MAHSC, Manchester, UK (K E Chandler MD, Y J Crow); Department of Paediatric Neurology, University Hospitals of Leicester NHS Trust, Leicester, UK (L Dabydeen MRCPCH); TY Nelson Department of Neurology, and Neuroimmunology Group, the Children’s Hospital at Westmead, University of Sydney, Sydney, NSW, Australia (R C Dale PhD); Nutrition and Metabolism Unit, Hôpital Universitaire des Enfants Reine Fabiola (ULB), Brussels, Belgium (C De Laet MD); Department of Paediatric Neurology, Royal Preston Hospital, Preston, UK (C G E L De Goede FRCPCH); Pediatric Neurology Unit, Hospital Vall d’Hebron, Barcelona, Spain (M del Toro MD); Paediatric Neurology Unit, University Hospital 12 Octubre, Madrid, Spain (N Nunez Enamorado MD, R Simon MD); Department of Clinical and Experimental Sciences, Child Neurology and Psychiatry Unit, University of Brescia, Brescia, Italy (E Fazzi MD); Department of Genetics, BioCruces Health Research Institute, Hospital, Universitario Cruces, Bizkaia, Spain (B Gener MD); Institute of Biomedicine and Translational Medicine, University of Tartu, Tartu, Estonia (M Haldre BSc, P Peterson PhD, K Kisand PhD); General Neurology and Complex Motor Disorders Service, Evelina Children’s Hospital, Guy’s and St Thomas’ NHS Foundation Trust, London, UK (J-P S-M Lin PhD); Department of Paediatric Neurology, Leeds General Infirmary, Leeds, UK (J H Livingston MB ChB); Department of Neurosciences and Behavior Sciences, School of Medicine of Ribeirão Preto, University of São Paulo, São Paulo, Brazil, (C Marques Lourenco MD, W Marques Jr MD); Royal Devon and Exeter Foundation NHS Trust, Exeter, UK (P Oades FRCPCH); Section for Child Neurology, Women and Children’s Division, Oslo University Hospital, Oslo, Norway (M Rasmussen MD); Neuropediatrie, Hopital Gui de Chauliac, and INSERM U1051, INM, Montpellier, France (A Roubertie MD); Center for Genetic Medicine Research and Department of Neurology, Children’s National Medical Center, George Washington University School of Medicine, Washington, DC, USA, (J Loewenstein Schmidt CGC, A Vanderver MD); The Genetic Institute, Ha’Emek Medical Center, Afula and the Rappaport Faculty of Medicine, Technion, Haifa, Israel (S A Shalev MD, R Spiegel MD); Department of Neurology, University of Utah School of Medicine, Salt Lake City, UT, USA (K J Swoboda FACMG); Neurology Department, Royal Manchester Children’s Hospital, Manchester, UK (G Vassallo MD); Centre for Rare Diseases and Personalised Medicine and Department of Medical and Molecular Genetics, School of Clinical and Experimental Medicine, University of Birmingham, UK, and West Midlands Regional Genetics Service, Clinical Genetics Unit, Birmingham Women’s Hospital, Birmingham, UK (J Vogt MRCP); Nottingham Children’s Hospital, Nottingham University Hospitals NHS Trust, Nottingham, UK (W P Whitehouse FRCPCH); Neuropaediatrics, Department of Paediatrics, Mater Dei Hospital, Tal-Qroqq, Malta (D Soler MD); and Université Paris Descartes, EA 1833 and Hôpital Cochin, Service de Virologie, Paris, France, (F Rozenberg MD, P Lebon MD)
1,
Diana S Chase
Diana S Chase
1Manchester Centre for Genomic Medicine, Institute of Human Development, Faculty of Medical and Human Sciences, University of Manchester, Manchester Academic Health Sciences Centre (MAHSC), Manchester, UK (G I Rice PhD, G M A Forte MPhil, M Szynkiewicz MSc, D S Chase PhD, Y J Crow PhD); Erasme University Hospital, Universite Libre de Bruxelles (ULB), Brussels, Belgium, (A Aeby MD, C N Vilain MD, V Wermenbol MD); Medical Molecular Genetics Department, (M S Abdel-Hamid MSc, L Effat PhD), and Clinical Genetics Department, (S A Temtamy PhD, M S Aglan PhD, M S Zaki PhD, G M H Abdel-Salam PhD), Human Genetics and Genome Research Division, National Research Centre, Cairo, Egypt; Bradford Teaching Hospitals NHS Foundation Trust, Bradford, UK (S Ackroyd MB ChB); Department of Clinical Biochemistry, Lancashire Teaching Hospitals NHS Foundation Trust, Preston, UK (R Allcock FRCPath); University Hospitals of Coventry and Warwickshire, Coventry, UK (K M Bailey MRCPCH); Department of Brain and Behavioral Sciences, Unit of Child Neurology and Psychiatry, University of Pavia, Pavia, Italy, (U Balottin MD); Child Neurology and Psychiatry Unit, (U Balottin, I Olivieri MD, S Orcesi MD), and Laboratory of Experimental Neurobiology (C Cereda PhD), C. Mondino National Neurological Institute, Pavia, Italy; Praticien Hospitalier, Unité de Neurologie Pédiatrique et Centre de Référence des Maladies Neuromusculaires, Hôpital Necker-Enfants Malades, Paris, France (C Barnerias MD); Department of Pediatrics, and Department of Neurology and Neurosurgery, Montreal Children’s Hospital, McGill University Health Center, Montreal, QC, Canada (G Bernard MD); Department of Dermatology, Reference Centre for Cutaneous Rare Diseases (MAGEC), Necker-Enfants Malades APHP, Sorbonne Paris Cité, Université Paris Descartes, Institut Imagine, Paris, France (C Bodemer PhD); Department of Pediatrics, Hospital Universitario Araba, Vitoria-Gasteiz, Spain (M P Botella MD); Manchester Centre for Genomic Medicine, Central Manchester University Hospitals NHS Foundation Trust, MAHSC, Manchester, UK (K E Chandler MD, Y J Crow); Department of Paediatric Neurology, University Hospitals of Leicester NHS Trust, Leicester, UK (L Dabydeen MRCPCH); TY Nelson Department of Neurology, and Neuroimmunology Group, the Children’s Hospital at Westmead, University of Sydney, Sydney, NSW, Australia (R C Dale PhD); Nutrition and Metabolism Unit, Hôpital Universitaire des Enfants Reine Fabiola (ULB), Brussels, Belgium (C De Laet MD); Department of Paediatric Neurology, Royal Preston Hospital, Preston, UK (C G E L De Goede FRCPCH); Pediatric Neurology Unit, Hospital Vall d’Hebron, Barcelona, Spain (M del Toro MD); Paediatric Neurology Unit, University Hospital 12 Octubre, Madrid, Spain (N Nunez Enamorado MD, R Simon MD); Department of Clinical and Experimental Sciences, Child Neurology and Psychiatry Unit, University of Brescia, Brescia, Italy (E Fazzi MD); Department of Genetics, BioCruces Health Research Institute, Hospital, Universitario Cruces, Bizkaia, Spain (B Gener MD); Institute of Biomedicine and Translational Medicine, University of Tartu, Tartu, Estonia (M Haldre BSc, P Peterson PhD, K Kisand PhD); General Neurology and Complex Motor Disorders Service, Evelina Children’s Hospital, Guy’s and St Thomas’ NHS Foundation Trust, London, UK (J-P S-M Lin PhD); Department of Paediatric Neurology, Leeds General Infirmary, Leeds, UK (J H Livingston MB ChB); Department of Neurosciences and Behavior Sciences, School of Medicine of Ribeirão Preto, University of São Paulo, São Paulo, Brazil, (C Marques Lourenco MD, W Marques Jr MD); Royal Devon and Exeter Foundation NHS Trust, Exeter, UK (P Oades FRCPCH); Section for Child Neurology, Women and Children’s Division, Oslo University Hospital, Oslo, Norway (M Rasmussen MD); Neuropediatrie, Hopital Gui de Chauliac, and INSERM U1051, INM, Montpellier, France (A Roubertie MD); Center for Genetic Medicine Research and Department of Neurology, Children’s National Medical Center, George Washington University School of Medicine, Washington, DC, USA, (J Loewenstein Schmidt CGC, A Vanderver MD); The Genetic Institute, Ha’Emek Medical Center, Afula and the Rappaport Faculty of Medicine, Technion, Haifa, Israel (S A Shalev MD, R Spiegel MD); Department of Neurology, University of Utah School of Medicine, Salt Lake City, UT, USA (K J Swoboda FACMG); Neurology Department, Royal Manchester Children’s Hospital, Manchester, UK (G Vassallo MD); Centre for Rare Diseases and Personalised Medicine and Department of Medical and Molecular Genetics, School of Clinical and Experimental Medicine, University of Birmingham, UK, and West Midlands Regional Genetics Service, Clinical Genetics Unit, Birmingham Women’s Hospital, Birmingham, UK (J Vogt MRCP); Nottingham Children’s Hospital, Nottingham University Hospitals NHS Trust, Nottingham, UK (W P Whitehouse FRCPCH); Neuropaediatrics, Department of Paediatrics, Mater Dei Hospital, Tal-Qroqq, Malta (D Soler MD); and Université Paris Descartes, EA 1833 and Hôpital Cochin, Service de Virologie, Paris, France, (F Rozenberg MD, P Lebon MD)
1,
Alec Aeby
Alec Aeby
1Manchester Centre for Genomic Medicine, Institute of Human Development, Faculty of Medical and Human Sciences, University of Manchester, Manchester Academic Health Sciences Centre (MAHSC), Manchester, UK (G I Rice PhD, G M A Forte MPhil, M Szynkiewicz MSc, D S Chase PhD, Y J Crow PhD); Erasme University Hospital, Universite Libre de Bruxelles (ULB), Brussels, Belgium, (A Aeby MD, C N Vilain MD, V Wermenbol MD); Medical Molecular Genetics Department, (M S Abdel-Hamid MSc, L Effat PhD), and Clinical Genetics Department, (S A Temtamy PhD, M S Aglan PhD, M S Zaki PhD, G M H Abdel-Salam PhD), Human Genetics and Genome Research Division, National Research Centre, Cairo, Egypt; Bradford Teaching Hospitals NHS Foundation Trust, Bradford, UK (S Ackroyd MB ChB); Department of Clinical Biochemistry, Lancashire Teaching Hospitals NHS Foundation Trust, Preston, UK (R Allcock FRCPath); University Hospitals of Coventry and Warwickshire, Coventry, UK (K M Bailey MRCPCH); Department of Brain and Behavioral Sciences, Unit of Child Neurology and Psychiatry, University of Pavia, Pavia, Italy, (U Balottin MD); Child Neurology and Psychiatry Unit, (U Balottin, I Olivieri MD, S Orcesi MD), and Laboratory of Experimental Neurobiology (C Cereda PhD), C. Mondino National Neurological Institute, Pavia, Italy; Praticien Hospitalier, Unité de Neurologie Pédiatrique et Centre de Référence des Maladies Neuromusculaires, Hôpital Necker-Enfants Malades, Paris, France (C Barnerias MD); Department of Pediatrics, and Department of Neurology and Neurosurgery, Montreal Children’s Hospital, McGill University Health Center, Montreal, QC, Canada (G Bernard MD); Department of Dermatology, Reference Centre for Cutaneous Rare Diseases (MAGEC), Necker-Enfants Malades APHP, Sorbonne Paris Cité, Université Paris Descartes, Institut Imagine, Paris, France (C Bodemer PhD); Department of Pediatrics, Hospital Universitario Araba, Vitoria-Gasteiz, Spain (M P Botella MD); Manchester Centre for Genomic Medicine, Central Manchester University Hospitals NHS Foundation Trust, MAHSC, Manchester, UK (K E Chandler MD, Y J Crow); Department of Paediatric Neurology, University Hospitals of Leicester NHS Trust, Leicester, UK (L Dabydeen MRCPCH); TY Nelson Department of Neurology, and Neuroimmunology Group, the Children’s Hospital at Westmead, University of Sydney, Sydney, NSW, Australia (R C Dale PhD); Nutrition and Metabolism Unit, Hôpital Universitaire des Enfants Reine Fabiola (ULB), Brussels, Belgium (C De Laet MD); Department of Paediatric Neurology, Royal Preston Hospital, Preston, UK (C G E L De Goede FRCPCH); Pediatric Neurology Unit, Hospital Vall d’Hebron, Barcelona, Spain (M del Toro MD); Paediatric Neurology Unit, University Hospital 12 Octubre, Madrid, Spain (N Nunez Enamorado MD, R Simon MD); Department of Clinical and Experimental Sciences, Child Neurology and Psychiatry Unit, University of Brescia, Brescia, Italy (E Fazzi MD); Department of Genetics, BioCruces Health Research Institute, Hospital, Universitario Cruces, Bizkaia, Spain (B Gener MD); Institute of Biomedicine and Translational Medicine, University of Tartu, Tartu, Estonia (M Haldre BSc, P Peterson PhD, K Kisand PhD); General Neurology and Complex Motor Disorders Service, Evelina Children’s Hospital, Guy’s and St Thomas’ NHS Foundation Trust, London, UK (J-P S-M Lin PhD); Department of Paediatric Neurology, Leeds General Infirmary, Leeds, UK (J H Livingston MB ChB); Department of Neurosciences and Behavior Sciences, School of Medicine of Ribeirão Preto, University of São Paulo, São Paulo, Brazil, (C Marques Lourenco MD, W Marques Jr MD); Royal Devon and Exeter Foundation NHS Trust, Exeter, UK (P Oades FRCPCH); Section for Child Neurology, Women and Children’s Division, Oslo University Hospital, Oslo, Norway (M Rasmussen MD); Neuropediatrie, Hopital Gui de Chauliac, and INSERM U1051, INM, Montpellier, France (A Roubertie MD); Center for Genetic Medicine Research and Department of Neurology, Children’s National Medical Center, George Washington University School of Medicine, Washington, DC, USA, (J Loewenstein Schmidt CGC, A Vanderver MD); The Genetic Institute, Ha’Emek Medical Center, Afula and the Rappaport Faculty of Medicine, Technion, Haifa, Israel (S A Shalev MD, R Spiegel MD); Department of Neurology, University of Utah School of Medicine, Salt Lake City, UT, USA (K J Swoboda FACMG); Neurology Department, Royal Manchester Children’s Hospital, Manchester, UK (G Vassallo MD); Centre for Rare Diseases and Personalised Medicine and Department of Medical and Molecular Genetics, School of Clinical and Experimental Medicine, University of Birmingham, UK, and West Midlands Regional Genetics Service, Clinical Genetics Unit, Birmingham Women’s Hospital, Birmingham, UK (J Vogt MRCP); Nottingham Children’s Hospital, Nottingham University Hospitals NHS Trust, Nottingham, UK (W P Whitehouse FRCPCH); Neuropaediatrics, Department of Paediatrics, Mater Dei Hospital, Tal-Qroqq, Malta (D Soler MD); and Université Paris Descartes, EA 1833 and Hôpital Cochin, Service de Virologie, Paris, France, (F Rozenberg MD, P Lebon MD)
1,
Mohamed S Abdel-Hamid
Mohamed S Abdel-Hamid
1Manchester Centre for Genomic Medicine, Institute of Human Development, Faculty of Medical and Human Sciences, University of Manchester, Manchester Academic Health Sciences Centre (MAHSC), Manchester, UK (G I Rice PhD, G M A Forte MPhil, M Szynkiewicz MSc, D S Chase PhD, Y J Crow PhD); Erasme University Hospital, Universite Libre de Bruxelles (ULB), Brussels, Belgium, (A Aeby MD, C N Vilain MD, V Wermenbol MD); Medical Molecular Genetics Department, (M S Abdel-Hamid MSc, L Effat PhD), and Clinical Genetics Department, (S A Temtamy PhD, M S Aglan PhD, M S Zaki PhD, G M H Abdel-Salam PhD), Human Genetics and Genome Research Division, National Research Centre, Cairo, Egypt; Bradford Teaching Hospitals NHS Foundation Trust, Bradford, UK (S Ackroyd MB ChB); Department of Clinical Biochemistry, Lancashire Teaching Hospitals NHS Foundation Trust, Preston, UK (R Allcock FRCPath); University Hospitals of Coventry and Warwickshire, Coventry, UK (K M Bailey MRCPCH); Department of Brain and Behavioral Sciences, Unit of Child Neurology and Psychiatry, University of Pavia, Pavia, Italy, (U Balottin MD); Child Neurology and Psychiatry Unit, (U Balottin, I Olivieri MD, S Orcesi MD), and Laboratory of Experimental Neurobiology (C Cereda PhD), C. Mondino National Neurological Institute, Pavia, Italy; Praticien Hospitalier, Unité de Neurologie Pédiatrique et Centre de Référence des Maladies Neuromusculaires, Hôpital Necker-Enfants Malades, Paris, France (C Barnerias MD); Department of Pediatrics, and Department of Neurology and Neurosurgery, Montreal Children’s Hospital, McGill University Health Center, Montreal, QC, Canada (G Bernard MD); Department of Dermatology, Reference Centre for Cutaneous Rare Diseases (MAGEC), Necker-Enfants Malades APHP, Sorbonne Paris Cité, Université Paris Descartes, Institut Imagine, Paris, France (C Bodemer PhD); Department of Pediatrics, Hospital Universitario Araba, Vitoria-Gasteiz, Spain (M P Botella MD); Manchester Centre for Genomic Medicine, Central Manchester University Hospitals NHS Foundation Trust, MAHSC, Manchester, UK (K E Chandler MD, Y J Crow); Department of Paediatric Neurology, University Hospitals of Leicester NHS Trust, Leicester, UK (L Dabydeen MRCPCH); TY Nelson Department of Neurology, and Neuroimmunology Group, the Children’s Hospital at Westmead, University of Sydney, Sydney, NSW, Australia (R C Dale PhD); Nutrition and Metabolism Unit, Hôpital Universitaire des Enfants Reine Fabiola (ULB), Brussels, Belgium (C De Laet MD); Department of Paediatric Neurology, Royal Preston Hospital, Preston, UK (C G E L De Goede FRCPCH); Pediatric Neurology Unit, Hospital Vall d’Hebron, Barcelona, Spain (M del Toro MD); Paediatric Neurology Unit, University Hospital 12 Octubre, Madrid, Spain (N Nunez Enamorado MD, R Simon MD); Department of Clinical and Experimental Sciences, Child Neurology and Psychiatry Unit, University of Brescia, Brescia, Italy (E Fazzi MD); Department of Genetics, BioCruces Health Research Institute, Hospital, Universitario Cruces, Bizkaia, Spain (B Gener MD); Institute of Biomedicine and Translational Medicine, University of Tartu, Tartu, Estonia (M Haldre BSc, P Peterson PhD, K Kisand PhD); General Neurology and Complex Motor Disorders Service, Evelina Children’s Hospital, Guy’s and St Thomas’ NHS Foundation Trust, London, UK (J-P S-M Lin PhD); Department of Paediatric Neurology, Leeds General Infirmary, Leeds, UK (J H Livingston MB ChB); Department of Neurosciences and Behavior Sciences, School of Medicine of Ribeirão Preto, University of São Paulo, São Paulo, Brazil, (C Marques Lourenco MD, W Marques Jr MD); Royal Devon and Exeter Foundation NHS Trust, Exeter, UK (P Oades FRCPCH); Section for Child Neurology, Women and Children’s Division, Oslo University Hospital, Oslo, Norway (M Rasmussen MD); Neuropediatrie, Hopital Gui de Chauliac, and INSERM U1051, INM, Montpellier, France (A Roubertie MD); Center for Genetic Medicine Research and Department of Neurology, Children’s National Medical Center, George Washington University School of Medicine, Washington, DC, USA, (J Loewenstein Schmidt CGC, A Vanderver MD); The Genetic Institute, Ha’Emek Medical Center, Afula and the Rappaport Faculty of Medicine, Technion, Haifa, Israel (S A Shalev MD, R Spiegel MD); Department of Neurology, University of Utah School of Medicine, Salt Lake City, UT, USA (K J Swoboda FACMG); Neurology Department, Royal Manchester Children’s Hospital, Manchester, UK (G Vassallo MD); Centre for Rare Diseases and Personalised Medicine and Department of Medical and Molecular Genetics, School of Clinical and Experimental Medicine, University of Birmingham, UK, and West Midlands Regional Genetics Service, Clinical Genetics Unit, Birmingham Women’s Hospital, Birmingham, UK (J Vogt MRCP); Nottingham Children’s Hospital, Nottingham University Hospitals NHS Trust, Nottingham, UK (W P Whitehouse FRCPCH); Neuropaediatrics, Department of Paediatrics, Mater Dei Hospital, Tal-Qroqq, Malta (D Soler MD); and Université Paris Descartes, EA 1833 and Hôpital Cochin, Service de Virologie, Paris, France, (F Rozenberg MD, P Lebon MD)
1,
Sam Ackroyd
Sam Ackroyd
1Manchester Centre for Genomic Medicine, Institute of Human Development, Faculty of Medical and Human Sciences, University of Manchester, Manchester Academic Health Sciences Centre (MAHSC), Manchester, UK (G I Rice PhD, G M A Forte MPhil, M Szynkiewicz MSc, D S Chase PhD, Y J Crow PhD); Erasme University Hospital, Universite Libre de Bruxelles (ULB), Brussels, Belgium, (A Aeby MD, C N Vilain MD, V Wermenbol MD); Medical Molecular Genetics Department, (M S Abdel-Hamid MSc, L Effat PhD), and Clinical Genetics Department, (S A Temtamy PhD, M S Aglan PhD, M S Zaki PhD, G M H Abdel-Salam PhD), Human Genetics and Genome Research Division, National Research Centre, Cairo, Egypt; Bradford Teaching Hospitals NHS Foundation Trust, Bradford, UK (S Ackroyd MB ChB); Department of Clinical Biochemistry, Lancashire Teaching Hospitals NHS Foundation Trust, Preston, UK (R Allcock FRCPath); University Hospitals of Coventry and Warwickshire, Coventry, UK (K M Bailey MRCPCH); Department of Brain and Behavioral Sciences, Unit of Child Neurology and Psychiatry, University of Pavia, Pavia, Italy, (U Balottin MD); Child Neurology and Psychiatry Unit, (U Balottin, I Olivieri MD, S Orcesi MD), and Laboratory of Experimental Neurobiology (C Cereda PhD), C. Mondino National Neurological Institute, Pavia, Italy; Praticien Hospitalier, Unité de Neurologie Pédiatrique et Centre de Référence des Maladies Neuromusculaires, Hôpital Necker-Enfants Malades, Paris, France (C Barnerias MD); Department of Pediatrics, and Department of Neurology and Neurosurgery, Montreal Children’s Hospital, McGill University Health Center, Montreal, QC, Canada (G Bernard MD); Department of Dermatology, Reference Centre for Cutaneous Rare Diseases (MAGEC), Necker-Enfants Malades APHP, Sorbonne Paris Cité, Université Paris Descartes, Institut Imagine, Paris, France (C Bodemer PhD); Department of Pediatrics, Hospital Universitario Araba, Vitoria-Gasteiz, Spain (M P Botella MD); Manchester Centre for Genomic Medicine, Central Manchester University Hospitals NHS Foundation Trust, MAHSC, Manchester, UK (K E Chandler MD, Y J Crow); Department of Paediatric Neurology, University Hospitals of Leicester NHS Trust, Leicester, UK (L Dabydeen MRCPCH); TY Nelson Department of Neurology, and Neuroimmunology Group, the Children’s Hospital at Westmead, University of Sydney, Sydney, NSW, Australia (R C Dale PhD); Nutrition and Metabolism Unit, Hôpital Universitaire des Enfants Reine Fabiola (ULB), Brussels, Belgium (C De Laet MD); Department of Paediatric Neurology, Royal Preston Hospital, Preston, UK (C G E L De Goede FRCPCH); Pediatric Neurology Unit, Hospital Vall d’Hebron, Barcelona, Spain (M del Toro MD); Paediatric Neurology Unit, University Hospital 12 Octubre, Madrid, Spain (N Nunez Enamorado MD, R Simon MD); Department of Clinical and Experimental Sciences, Child Neurology and Psychiatry Unit, University of Brescia, Brescia, Italy (E Fazzi MD); Department of Genetics, BioCruces Health Research Institute, Hospital, Universitario Cruces, Bizkaia, Spain (B Gener MD); Institute of Biomedicine and Translational Medicine, University of Tartu, Tartu, Estonia (M Haldre BSc, P Peterson PhD, K Kisand PhD); General Neurology and Complex Motor Disorders Service, Evelina Children’s Hospital, Guy’s and St Thomas’ NHS Foundation Trust, London, UK (J-P S-M Lin PhD); Department of Paediatric Neurology, Leeds General Infirmary, Leeds, UK (J H Livingston MB ChB); Department of Neurosciences and Behavior Sciences, School of Medicine of Ribeirão Preto, University of São Paulo, São Paulo, Brazil, (C Marques Lourenco MD, W Marques Jr MD); Royal Devon and Exeter Foundation NHS Trust, Exeter, UK (P Oades FRCPCH); Section for Child Neurology, Women and Children’s Division, Oslo University Hospital, Oslo, Norway (M Rasmussen MD); Neuropediatrie, Hopital Gui de Chauliac, and INSERM U1051, INM, Montpellier, France (A Roubertie MD); Center for Genetic Medicine Research and Department of Neurology, Children’s National Medical Center, George Washington University School of Medicine, Washington, DC, USA, (J Loewenstein Schmidt CGC, A Vanderver MD); The Genetic Institute, Ha’Emek Medical Center, Afula and the Rappaport Faculty of Medicine, Technion, Haifa, Israel (S A Shalev MD, R Spiegel MD); Department of Neurology, University of Utah School of Medicine, Salt Lake City, UT, USA (K J Swoboda FACMG); Neurology Department, Royal Manchester Children’s Hospital, Manchester, UK (G Vassallo MD); Centre for Rare Diseases and Personalised Medicine and Department of Medical and Molecular Genetics, School of Clinical and Experimental Medicine, University of Birmingham, UK, and West Midlands Regional Genetics Service, Clinical Genetics Unit, Birmingham Women’s Hospital, Birmingham, UK (J Vogt MRCP); Nottingham Children’s Hospital, Nottingham University Hospitals NHS Trust, Nottingham, UK (W P Whitehouse FRCPCH); Neuropaediatrics, Department of Paediatrics, Mater Dei Hospital, Tal-Qroqq, Malta (D Soler MD); and Université Paris Descartes, EA 1833 and Hôpital Cochin, Service de Virologie, Paris, France, (F Rozenberg MD, P Lebon MD)
1,
Rebecca Allcock
Rebecca Allcock
1Manchester Centre for Genomic Medicine, Institute of Human Development, Faculty of Medical and Human Sciences, University of Manchester, Manchester Academic Health Sciences Centre (MAHSC), Manchester, UK (G I Rice PhD, G M A Forte MPhil, M Szynkiewicz MSc, D S Chase PhD, Y J Crow PhD); Erasme University Hospital, Universite Libre de Bruxelles (ULB), Brussels, Belgium, (A Aeby MD, C N Vilain MD, V Wermenbol MD); Medical Molecular Genetics Department, (M S Abdel-Hamid MSc, L Effat PhD), and Clinical Genetics Department, (S A Temtamy PhD, M S Aglan PhD, M S Zaki PhD, G M H Abdel-Salam PhD), Human Genetics and Genome Research Division, National Research Centre, Cairo, Egypt; Bradford Teaching Hospitals NHS Foundation Trust, Bradford, UK (S Ackroyd MB ChB); Department of Clinical Biochemistry, Lancashire Teaching Hospitals NHS Foundation Trust, Preston, UK (R Allcock FRCPath); University Hospitals of Coventry and Warwickshire, Coventry, UK (K M Bailey MRCPCH); Department of Brain and Behavioral Sciences, Unit of Child Neurology and Psychiatry, University of Pavia, Pavia, Italy, (U Balottin MD); Child Neurology and Psychiatry Unit, (U Balottin, I Olivieri MD, S Orcesi MD), and Laboratory of Experimental Neurobiology (C Cereda PhD), C. Mondino National Neurological Institute, Pavia, Italy; Praticien Hospitalier, Unité de Neurologie Pédiatrique et Centre de Référence des Maladies Neuromusculaires, Hôpital Necker-Enfants Malades, Paris, France (C Barnerias MD); Department of Pediatrics, and Department of Neurology and Neurosurgery, Montreal Children’s Hospital, McGill University Health Center, Montreal, QC, Canada (G Bernard MD); Department of Dermatology, Reference Centre for Cutaneous Rare Diseases (MAGEC), Necker-Enfants Malades APHP, Sorbonne Paris Cité, Université Paris Descartes, Institut Imagine, Paris, France (C Bodemer PhD); Department of Pediatrics, Hospital Universitario Araba, Vitoria-Gasteiz, Spain (M P Botella MD); Manchester Centre for Genomic Medicine, Central Manchester University Hospitals NHS Foundation Trust, MAHSC, Manchester, UK (K E Chandler MD, Y J Crow); Department of Paediatric Neurology, University Hospitals of Leicester NHS Trust, Leicester, UK (L Dabydeen MRCPCH); TY Nelson Department of Neurology, and Neuroimmunology Group, the Children’s Hospital at Westmead, University of Sydney, Sydney, NSW, Australia (R C Dale PhD); Nutrition and Metabolism Unit, Hôpital Universitaire des Enfants Reine Fabiola (ULB), Brussels, Belgium (C De Laet MD); Department of Paediatric Neurology, Royal Preston Hospital, Preston, UK (C G E L De Goede FRCPCH); Pediatric Neurology Unit, Hospital Vall d’Hebron, Barcelona, Spain (M del Toro MD); Paediatric Neurology Unit, University Hospital 12 Octubre, Madrid, Spain (N Nunez Enamorado MD, R Simon MD); Department of Clinical and Experimental Sciences, Child Neurology and Psychiatry Unit, University of Brescia, Brescia, Italy (E Fazzi MD); Department of Genetics, BioCruces Health Research Institute, Hospital, Universitario Cruces, Bizkaia, Spain (B Gener MD); Institute of Biomedicine and Translational Medicine, University of Tartu, Tartu, Estonia (M Haldre BSc, P Peterson PhD, K Kisand PhD); General Neurology and Complex Motor Disorders Service, Evelina Children’s Hospital, Guy’s and St Thomas’ NHS Foundation Trust, London, UK (J-P S-M Lin PhD); Department of Paediatric Neurology, Leeds General Infirmary, Leeds, UK (J H Livingston MB ChB); Department of Neurosciences and Behavior Sciences, School of Medicine of Ribeirão Preto, University of São Paulo, São Paulo, Brazil, (C Marques Lourenco MD, W Marques Jr MD); Royal Devon and Exeter Foundation NHS Trust, Exeter, UK (P Oades FRCPCH); Section for Child Neurology, Women and Children’s Division, Oslo University Hospital, Oslo, Norway (M Rasmussen MD); Neuropediatrie, Hopital Gui de Chauliac, and INSERM U1051, INM, Montpellier, France (A Roubertie MD); Center for Genetic Medicine Research and Department of Neurology, Children’s National Medical Center, George Washington University School of Medicine, Washington, DC, USA, (J Loewenstein Schmidt CGC, A Vanderver MD); The Genetic Institute, Ha’Emek Medical Center, Afula and the Rappaport Faculty of Medicine, Technion, Haifa, Israel (S A Shalev MD, R Spiegel MD); Department of Neurology, University of Utah School of Medicine, Salt Lake City, UT, USA (K J Swoboda FACMG); Neurology Department, Royal Manchester Children’s Hospital, Manchester, UK (G Vassallo MD); Centre for Rare Diseases and Personalised Medicine and Department of Medical and Molecular Genetics, School of Clinical and Experimental Medicine, University of Birmingham, UK, and West Midlands Regional Genetics Service, Clinical Genetics Unit, Birmingham Women’s Hospital, Birmingham, UK (J Vogt MRCP); Nottingham Children’s Hospital, Nottingham University Hospitals NHS Trust, Nottingham, UK (W P Whitehouse FRCPCH); Neuropaediatrics, Department of Paediatrics, Mater Dei Hospital, Tal-Qroqq, Malta (D Soler MD); and Université Paris Descartes, EA 1833 and Hôpital Cochin, Service de Virologie, Paris, France, (F Rozenberg MD, P Lebon MD)
1,
Kathryn M Bailey
Kathryn M Bailey
1Manchester Centre for Genomic Medicine, Institute of Human Development, Faculty of Medical and Human Sciences, University of Manchester, Manchester Academic Health Sciences Centre (MAHSC), Manchester, UK (G I Rice PhD, G M A Forte MPhil, M Szynkiewicz MSc, D S Chase PhD, Y J Crow PhD); Erasme University Hospital, Universite Libre de Bruxelles (ULB), Brussels, Belgium, (A Aeby MD, C N Vilain MD, V Wermenbol MD); Medical Molecular Genetics Department, (M S Abdel-Hamid MSc, L Effat PhD), and Clinical Genetics Department, (S A Temtamy PhD, M S Aglan PhD, M S Zaki PhD, G M H Abdel-Salam PhD), Human Genetics and Genome Research Division, National Research Centre, Cairo, Egypt; Bradford Teaching Hospitals NHS Foundation Trust, Bradford, UK (S Ackroyd MB ChB); Department of Clinical Biochemistry, Lancashire Teaching Hospitals NHS Foundation Trust, Preston, UK (R Allcock FRCPath); University Hospitals of Coventry and Warwickshire, Coventry, UK (K M Bailey MRCPCH); Department of Brain and Behavioral Sciences, Unit of Child Neurology and Psychiatry, University of Pavia, Pavia, Italy, (U Balottin MD); Child Neurology and Psychiatry Unit, (U Balottin, I Olivieri MD, S Orcesi MD), and Laboratory of Experimental Neurobiology (C Cereda PhD), C. Mondino National Neurological Institute, Pavia, Italy; Praticien Hospitalier, Unité de Neurologie Pédiatrique et Centre de Référence des Maladies Neuromusculaires, Hôpital Necker-Enfants Malades, Paris, France (C Barnerias MD); Department of Pediatrics, and Department of Neurology and Neurosurgery, Montreal Children’s Hospital, McGill University Health Center, Montreal, QC, Canada (G Bernard MD); Department of Dermatology, Reference Centre for Cutaneous Rare Diseases (MAGEC), Necker-Enfants Malades APHP, Sorbonne Paris Cité, Université Paris Descartes, Institut Imagine, Paris, France (C Bodemer PhD); Department of Pediatrics, Hospital Universitario Araba, Vitoria-Gasteiz, Spain (M P Botella MD); Manchester Centre for Genomic Medicine, Central Manchester University Hospitals NHS Foundation Trust, MAHSC, Manchester, UK (K E Chandler MD, Y J Crow); Department of Paediatric Neurology, University Hospitals of Leicester NHS Trust, Leicester, UK (L Dabydeen MRCPCH); TY Nelson Department of Neurology, and Neuroimmunology Group, the Children’s Hospital at Westmead, University of Sydney, Sydney, NSW, Australia (R C Dale PhD); Nutrition and Metabolism Unit, Hôpital Universitaire des Enfants Reine Fabiola (ULB), Brussels, Belgium (C De Laet MD); Department of Paediatric Neurology, Royal Preston Hospital, Preston, UK (C G E L De Goede FRCPCH); Pediatric Neurology Unit, Hospital Vall d’Hebron, Barcelona, Spain (M del Toro MD); Paediatric Neurology Unit, University Hospital 12 Octubre, Madrid, Spain (N Nunez Enamorado MD, R Simon MD); Department of Clinical and Experimental Sciences, Child Neurology and Psychiatry Unit, University of Brescia, Brescia, Italy (E Fazzi MD); Department of Genetics, BioCruces Health Research Institute, Hospital, Universitario Cruces, Bizkaia, Spain (B Gener MD); Institute of Biomedicine and Translational Medicine, University of Tartu, Tartu, Estonia (M Haldre BSc, P Peterson PhD, K Kisand PhD); General Neurology and Complex Motor Disorders Service, Evelina Children’s Hospital, Guy’s and St Thomas’ NHS Foundation Trust, London, UK (J-P S-M Lin PhD); Department of Paediatric Neurology, Leeds General Infirmary, Leeds, UK (J H Livingston MB ChB); Department of Neurosciences and Behavior Sciences, School of Medicine of Ribeirão Preto, University of São Paulo, São Paulo, Brazil, (C Marques Lourenco MD, W Marques Jr MD); Royal Devon and Exeter Foundation NHS Trust, Exeter, UK (P Oades FRCPCH); Section for Child Neurology, Women and Children’s Division, Oslo University Hospital, Oslo, Norway (M Rasmussen MD); Neuropediatrie, Hopital Gui de Chauliac, and INSERM U1051, INM, Montpellier, France (A Roubertie MD); Center for Genetic Medicine Research and Department of Neurology, Children’s National Medical Center, George Washington University School of Medicine, Washington, DC, USA, (J Loewenstein Schmidt CGC, A Vanderver MD); The Genetic Institute, Ha’Emek Medical Center, Afula and the Rappaport Faculty of Medicine, Technion, Haifa, Israel (S A Shalev MD, R Spiegel MD); Department of Neurology, University of Utah School of Medicine, Salt Lake City, UT, USA (K J Swoboda FACMG); Neurology Department, Royal Manchester Children’s Hospital, Manchester, UK (G Vassallo MD); Centre for Rare Diseases and Personalised Medicine and Department of Medical and Molecular Genetics, School of Clinical and Experimental Medicine, University of Birmingham, UK, and West Midlands Regional Genetics Service, Clinical Genetics Unit, Birmingham Women’s Hospital, Birmingham, UK (J Vogt MRCP); Nottingham Children’s Hospital, Nottingham University Hospitals NHS Trust, Nottingham, UK (W P Whitehouse FRCPCH); Neuropaediatrics, Department of Paediatrics, Mater Dei Hospital, Tal-Qroqq, Malta (D Soler MD); and Université Paris Descartes, EA 1833 and Hôpital Cochin, Service de Virologie, Paris, France, (F Rozenberg MD, P Lebon MD)
1,
Umberto Balottin
Umberto Balottin
1Manchester Centre for Genomic Medicine, Institute of Human Development, Faculty of Medical and Human Sciences, University of Manchester, Manchester Academic Health Sciences Centre (MAHSC), Manchester, UK (G I Rice PhD, G M A Forte MPhil, M Szynkiewicz MSc, D S Chase PhD, Y J Crow PhD); Erasme University Hospital, Universite Libre de Bruxelles (ULB), Brussels, Belgium, (A Aeby MD, C N Vilain MD, V Wermenbol MD); Medical Molecular Genetics Department, (M S Abdel-Hamid MSc, L Effat PhD), and Clinical Genetics Department, (S A Temtamy PhD, M S Aglan PhD, M S Zaki PhD, G M H Abdel-Salam PhD), Human Genetics and Genome Research Division, National Research Centre, Cairo, Egypt; Bradford Teaching Hospitals NHS Foundation Trust, Bradford, UK (S Ackroyd MB ChB); Department of Clinical Biochemistry, Lancashire Teaching Hospitals NHS Foundation Trust, Preston, UK (R Allcock FRCPath); University Hospitals of Coventry and Warwickshire, Coventry, UK (K M Bailey MRCPCH); Department of Brain and Behavioral Sciences, Unit of Child Neurology and Psychiatry, University of Pavia, Pavia, Italy, (U Balottin MD); Child Neurology and Psychiatry Unit, (U Balottin, I Olivieri MD, S Orcesi MD), and Laboratory of Experimental Neurobiology (C Cereda PhD), C. Mondino National Neurological Institute, Pavia, Italy; Praticien Hospitalier, Unité de Neurologie Pédiatrique et Centre de Référence des Maladies Neuromusculaires, Hôpital Necker-Enfants Malades, Paris, France (C Barnerias MD); Department of Pediatrics, and Department of Neurology and Neurosurgery, Montreal Children’s Hospital, McGill University Health Center, Montreal, QC, Canada (G Bernard MD); Department of Dermatology, Reference Centre for Cutaneous Rare Diseases (MAGEC), Necker-Enfants Malades APHP, Sorbonne Paris Cité, Université Paris Descartes, Institut Imagine, Paris, France (C Bodemer PhD); Department of Pediatrics, Hospital Universitario Araba, Vitoria-Gasteiz, Spain (M P Botella MD); Manchester Centre for Genomic Medicine, Central Manchester University Hospitals NHS Foundation Trust, MAHSC, Manchester, UK (K E Chandler MD, Y J Crow); Department of Paediatric Neurology, University Hospitals of Leicester NHS Trust, Leicester, UK (L Dabydeen MRCPCH); TY Nelson Department of Neurology, and Neuroimmunology Group, the Children’s Hospital at Westmead, University of Sydney, Sydney, NSW, Australia (R C Dale PhD); Nutrition and Metabolism Unit, Hôpital Universitaire des Enfants Reine Fabiola (ULB), Brussels, Belgium (C De Laet MD); Department of Paediatric Neurology, Royal Preston Hospital, Preston, UK (C G E L De Goede FRCPCH); Pediatric Neurology Unit, Hospital Vall d’Hebron, Barcelona, Spain (M del Toro MD); Paediatric Neurology Unit, University Hospital 12 Octubre, Madrid, Spain (N Nunez Enamorado MD, R Simon MD); Department of Clinical and Experimental Sciences, Child Neurology and Psychiatry Unit, University of Brescia, Brescia, Italy (E Fazzi MD); Department of Genetics, BioCruces Health Research Institute, Hospital, Universitario Cruces, Bizkaia, Spain (B Gener MD); Institute of Biomedicine and Translational Medicine, University of Tartu, Tartu, Estonia (M Haldre BSc, P Peterson PhD, K Kisand PhD); General Neurology and Complex Motor Disorders Service, Evelina Children’s Hospital, Guy’s and St Thomas’ NHS Foundation Trust, London, UK (J-P S-M Lin PhD); Department of Paediatric Neurology, Leeds General Infirmary, Leeds, UK (J H Livingston MB ChB); Department of Neurosciences and Behavior Sciences, School of Medicine of Ribeirão Preto, University of São Paulo, São Paulo, Brazil, (C Marques Lourenco MD, W Marques Jr MD); Royal Devon and Exeter Foundation NHS Trust, Exeter, UK (P Oades FRCPCH); Section for Child Neurology, Women and Children’s Division, Oslo University Hospital, Oslo, Norway (M Rasmussen MD); Neuropediatrie, Hopital Gui de Chauliac, and INSERM U1051, INM, Montpellier, France (A Roubertie MD); Center for Genetic Medicine Research and Department of Neurology, Children’s National Medical Center, George Washington University School of Medicine, Washington, DC, USA, (J Loewenstein Schmidt CGC, A Vanderver MD); The Genetic Institute, Ha’Emek Medical Center, Afula and the Rappaport Faculty of Medicine, Technion, Haifa, Israel (S A Shalev MD, R Spiegel MD); Department of Neurology, University of Utah School of Medicine, Salt Lake City, UT, USA (K J Swoboda FACMG); Neurology Department, Royal Manchester Children’s Hospital, Manchester, UK (G Vassallo MD); Centre for Rare Diseases and Personalised Medicine and Department of Medical and Molecular Genetics, School of Clinical and Experimental Medicine, University of Birmingham, UK, and West Midlands Regional Genetics Service, Clinical Genetics Unit, Birmingham Women’s Hospital, Birmingham, UK (J Vogt MRCP); Nottingham Children’s Hospital, Nottingham University Hospitals NHS Trust, Nottingham, UK (W P Whitehouse FRCPCH); Neuropaediatrics, Department of Paediatrics, Mater Dei Hospital, Tal-Qroqq, Malta (D Soler MD); and Université Paris Descartes, EA 1833 and Hôpital Cochin, Service de Virologie, Paris, France, (F Rozenberg MD, P Lebon MD)
1,
Christine Barnerias
Christine Barnerias
1Manchester Centre for Genomic Medicine, Institute of Human Development, Faculty of Medical and Human Sciences, University of Manchester, Manchester Academic Health Sciences Centre (MAHSC), Manchester, UK (G I Rice PhD, G M A Forte MPhil, M Szynkiewicz MSc, D S Chase PhD, Y J Crow PhD); Erasme University Hospital, Universite Libre de Bruxelles (ULB), Brussels, Belgium, (A Aeby MD, C N Vilain MD, V Wermenbol MD); Medical Molecular Genetics Department, (M S Abdel-Hamid MSc, L Effat PhD), and Clinical Genetics Department, (S A Temtamy PhD, M S Aglan PhD, M S Zaki PhD, G M H Abdel-Salam PhD), Human Genetics and Genome Research Division, National Research Centre, Cairo, Egypt; Bradford Teaching Hospitals NHS Foundation Trust, Bradford, UK (S Ackroyd MB ChB); Department of Clinical Biochemistry, Lancashire Teaching Hospitals NHS Foundation Trust, Preston, UK (R Allcock FRCPath); University Hospitals of Coventry and Warwickshire, Coventry, UK (K M Bailey MRCPCH); Department of Brain and Behavioral Sciences, Unit of Child Neurology and Psychiatry, University of Pavia, Pavia, Italy, (U Balottin MD); Child Neurology and Psychiatry Unit, (U Balottin, I Olivieri MD, S Orcesi MD), and Laboratory of Experimental Neurobiology (C Cereda PhD), C. Mondino National Neurological Institute, Pavia, Italy; Praticien Hospitalier, Unité de Neurologie Pédiatrique et Centre de Référence des Maladies Neuromusculaires, Hôpital Necker-Enfants Malades, Paris, France (C Barnerias MD); Department of Pediatrics, and Department of Neurology and Neurosurgery, Montreal Children’s Hospital, McGill University Health Center, Montreal, QC, Canada (G Bernard MD); Department of Dermatology, Reference Centre for Cutaneous Rare Diseases (MAGEC), Necker-Enfants Malades APHP, Sorbonne Paris Cité, Université Paris Descartes, Institut Imagine, Paris, France (C Bodemer PhD); Department of Pediatrics, Hospital Universitario Araba, Vitoria-Gasteiz, Spain (M P Botella MD); Manchester Centre for Genomic Medicine, Central Manchester University Hospitals NHS Foundation Trust, MAHSC, Manchester, UK (K E Chandler MD, Y J Crow); Department of Paediatric Neurology, University Hospitals of Leicester NHS Trust, Leicester, UK (L Dabydeen MRCPCH); TY Nelson Department of Neurology, and Neuroimmunology Group, the Children’s Hospital at Westmead, University of Sydney, Sydney, NSW, Australia (R C Dale PhD); Nutrition and Metabolism Unit, Hôpital Universitaire des Enfants Reine Fabiola (ULB), Brussels, Belgium (C De Laet MD); Department of Paediatric Neurology, Royal Preston Hospital, Preston, UK (C G E L De Goede FRCPCH); Pediatric Neurology Unit, Hospital Vall d’Hebron, Barcelona, Spain (M del Toro MD); Paediatric Neurology Unit, University Hospital 12 Octubre, Madrid, Spain (N Nunez Enamorado MD, R Simon MD); Department of Clinical and Experimental Sciences, Child Neurology and Psychiatry Unit, University of Brescia, Brescia, Italy (E Fazzi MD); Department of Genetics, BioCruces Health Research Institute, Hospital, Universitario Cruces, Bizkaia, Spain (B Gener MD); Institute of Biomedicine and Translational Medicine, University of Tartu, Tartu, Estonia (M Haldre BSc, P Peterson PhD, K Kisand PhD); General Neurology and Complex Motor Disorders Service, Evelina Children’s Hospital, Guy’s and St Thomas’ NHS Foundation Trust, London, UK (J-P S-M Lin PhD); Department of Paediatric Neurology, Leeds General Infirmary, Leeds, UK (J H Livingston MB ChB); Department of Neurosciences and Behavior Sciences, School of Medicine of Ribeirão Preto, University of São Paulo, São Paulo, Brazil, (C Marques Lourenco MD, W Marques Jr MD); Royal Devon and Exeter Foundation NHS Trust, Exeter, UK (P Oades FRCPCH); Section for Child Neurology, Women and Children’s Division, Oslo University Hospital, Oslo, Norway (M Rasmussen MD); Neuropediatrie, Hopital Gui de Chauliac, and INSERM U1051, INM, Montpellier, France (A Roubertie MD); Center for Genetic Medicine Research and Department of Neurology, Children’s National Medical Center, George Washington University School of Medicine, Washington, DC, USA, (J Loewenstein Schmidt CGC, A Vanderver MD); The Genetic Institute, Ha’Emek Medical Center, Afula and the Rappaport Faculty of Medicine, Technion, Haifa, Israel (S A Shalev MD, R Spiegel MD); Department of Neurology, University of Utah School of Medicine, Salt Lake City, UT, USA (K J Swoboda FACMG); Neurology Department, Royal Manchester Children’s Hospital, Manchester, UK (G Vassallo MD); Centre for Rare Diseases and Personalised Medicine and Department of Medical and Molecular Genetics, School of Clinical and Experimental Medicine, University of Birmingham, UK, and West Midlands Regional Genetics Service, Clinical Genetics Unit, Birmingham Women’s Hospital, Birmingham, UK (J Vogt MRCP); Nottingham Children’s Hospital, Nottingham University Hospitals NHS Trust, Nottingham, UK (W P Whitehouse FRCPCH); Neuropaediatrics, Department of Paediatrics, Mater Dei Hospital, Tal-Qroqq, Malta (D Soler MD); and Université Paris Descartes, EA 1833 and Hôpital Cochin, Service de Virologie, Paris, France, (F Rozenberg MD, P Lebon MD)
1,
Genevieve Bernard
Genevieve Bernard
1Manchester Centre for Genomic Medicine, Institute of Human Development, Faculty of Medical and Human Sciences, University of Manchester, Manchester Academic Health Sciences Centre (MAHSC), Manchester, UK (G I Rice PhD, G M A Forte MPhil, M Szynkiewicz MSc, D S Chase PhD, Y J Crow PhD); Erasme University Hospital, Universite Libre de Bruxelles (ULB), Brussels, Belgium, (A Aeby MD, C N Vilain MD, V Wermenbol MD); Medical Molecular Genetics Department, (M S Abdel-Hamid MSc, L Effat PhD), and Clinical Genetics Department, (S A Temtamy PhD, M S Aglan PhD, M S Zaki PhD, G M H Abdel-Salam PhD), Human Genetics and Genome Research Division, National Research Centre, Cairo, Egypt; Bradford Teaching Hospitals NHS Foundation Trust, Bradford, UK (S Ackroyd MB ChB); Department of Clinical Biochemistry, Lancashire Teaching Hospitals NHS Foundation Trust, Preston, UK (R Allcock FRCPath); University Hospitals of Coventry and Warwickshire, Coventry, UK (K M Bailey MRCPCH); Department of Brain and Behavioral Sciences, Unit of Child Neurology and Psychiatry, University of Pavia, Pavia, Italy, (U Balottin MD); Child Neurology and Psychiatry Unit, (U Balottin, I Olivieri MD, S Orcesi MD), and Laboratory of Experimental Neurobiology (C Cereda PhD), C. Mondino National Neurological Institute, Pavia, Italy; Praticien Hospitalier, Unité de Neurologie Pédiatrique et Centre de Référence des Maladies Neuromusculaires, Hôpital Necker-Enfants Malades, Paris, France (C Barnerias MD); Department of Pediatrics, and Department of Neurology and Neurosurgery, Montreal Children’s Hospital, McGill University Health Center, Montreal, QC, Canada (G Bernard MD); Department of Dermatology, Reference Centre for Cutaneous Rare Diseases (MAGEC), Necker-Enfants Malades APHP, Sorbonne Paris Cité, Université Paris Descartes, Institut Imagine, Paris, France (C Bodemer PhD); Department of Pediatrics, Hospital Universitario Araba, Vitoria-Gasteiz, Spain (M P Botella MD); Manchester Centre for Genomic Medicine, Central Manchester University Hospitals NHS Foundation Trust, MAHSC, Manchester, UK (K E Chandler MD, Y J Crow); Department of Paediatric Neurology, University Hospitals of Leicester NHS Trust, Leicester, UK (L Dabydeen MRCPCH); TY Nelson Department of Neurology, and Neuroimmunology Group, the Children’s Hospital at Westmead, University of Sydney, Sydney, NSW, Australia (R C Dale PhD); Nutrition and Metabolism Unit, Hôpital Universitaire des Enfants Reine Fabiola (ULB), Brussels, Belgium (C De Laet MD); Department of Paediatric Neurology, Royal Preston Hospital, Preston, UK (C G E L De Goede FRCPCH); Pediatric Neurology Unit, Hospital Vall d’Hebron, Barcelona, Spain (M del Toro MD); Paediatric Neurology Unit, University Hospital 12 Octubre, Madrid, Spain (N Nunez Enamorado MD, R Simon MD); Department of Clinical and Experimental Sciences, Child Neurology and Psychiatry Unit, University of Brescia, Brescia, Italy (E Fazzi MD); Department of Genetics, BioCruces Health Research Institute, Hospital, Universitario Cruces, Bizkaia, Spain (B Gener MD); Institute of Biomedicine and Translational Medicine, University of Tartu, Tartu, Estonia (M Haldre BSc, P Peterson PhD, K Kisand PhD); General Neurology and Complex Motor Disorders Service, Evelina Children’s Hospital, Guy’s and St Thomas’ NHS Foundation Trust, London, UK (J-P S-M Lin PhD); Department of Paediatric Neurology, Leeds General Infirmary, Leeds, UK (J H Livingston MB ChB); Department of Neurosciences and Behavior Sciences, School of Medicine of Ribeirão Preto, University of São Paulo, São Paulo, Brazil, (C Marques Lourenco MD, W Marques Jr MD); Royal Devon and Exeter Foundation NHS Trust, Exeter, UK (P Oades FRCPCH); Section for Child Neurology, Women and Children’s Division, Oslo University Hospital, Oslo, Norway (M Rasmussen MD); Neuropediatrie, Hopital Gui de Chauliac, and INSERM U1051, INM, Montpellier, France (A Roubertie MD); Center for Genetic Medicine Research and Department of Neurology, Children’s National Medical Center, George Washington University School of Medicine, Washington, DC, USA, (J Loewenstein Schmidt CGC, A Vanderver MD); The Genetic Institute, Ha’Emek Medical Center, Afula and the Rappaport Faculty of Medicine, Technion, Haifa, Israel (S A Shalev MD, R Spiegel MD); Department of Neurology, University of Utah School of Medicine, Salt Lake City, UT, USA (K J Swoboda FACMG); Neurology Department, Royal Manchester Children’s Hospital, Manchester, UK (G Vassallo MD); Centre for Rare Diseases and Personalised Medicine and Department of Medical and Molecular Genetics, School of Clinical and Experimental Medicine, University of Birmingham, UK, and West Midlands Regional Genetics Service, Clinical Genetics Unit, Birmingham Women’s Hospital, Birmingham, UK (J Vogt MRCP); Nottingham Children’s Hospital, Nottingham University Hospitals NHS Trust, Nottingham, UK (W P Whitehouse FRCPCH); Neuropaediatrics, Department of Paediatrics, Mater Dei Hospital, Tal-Qroqq, Malta (D Soler MD); and Université Paris Descartes, EA 1833 and Hôpital Cochin, Service de Virologie, Paris, France, (F Rozenberg MD, P Lebon MD)
1,
Christine Bodemer
Christine Bodemer
1Manchester Centre for Genomic Medicine, Institute of Human Development, Faculty of Medical and Human Sciences, University of Manchester, Manchester Academic Health Sciences Centre (MAHSC), Manchester, UK (G I Rice PhD, G M A Forte MPhil, M Szynkiewicz MSc, D S Chase PhD, Y J Crow PhD); Erasme University Hospital, Universite Libre de Bruxelles (ULB), Brussels, Belgium, (A Aeby MD, C N Vilain MD, V Wermenbol MD); Medical Molecular Genetics Department, (M S Abdel-Hamid MSc, L Effat PhD), and Clinical Genetics Department, (S A Temtamy PhD, M S Aglan PhD, M S Zaki PhD, G M H Abdel-Salam PhD), Human Genetics and Genome Research Division, National Research Centre, Cairo, Egypt; Bradford Teaching Hospitals NHS Foundation Trust, Bradford, UK (S Ackroyd MB ChB); Department of Clinical Biochemistry, Lancashire Teaching Hospitals NHS Foundation Trust, Preston, UK (R Allcock FRCPath); University Hospitals of Coventry and Warwickshire, Coventry, UK (K M Bailey MRCPCH); Department of Brain and Behavioral Sciences, Unit of Child Neurology and Psychiatry, University of Pavia, Pavia, Italy, (U Balottin MD); Child Neurology and Psychiatry Unit, (U Balottin, I Olivieri MD, S Orcesi MD), and Laboratory of Experimental Neurobiology (C Cereda PhD), C. Mondino National Neurological Institute, Pavia, Italy; Praticien Hospitalier, Unité de Neurologie Pédiatrique et Centre de Référence des Maladies Neuromusculaires, Hôpital Necker-Enfants Malades, Paris, France (C Barnerias MD); Department of Pediatrics, and Department of Neurology and Neurosurgery, Montreal Children’s Hospital, McGill University Health Center, Montreal, QC, Canada (G Bernard MD); Department of Dermatology, Reference Centre for Cutaneous Rare Diseases (MAGEC), Necker-Enfants Malades APHP, Sorbonne Paris Cité, Université Paris Descartes, Institut Imagine, Paris, France (C Bodemer PhD); Department of Pediatrics, Hospital Universitario Araba, Vitoria-Gasteiz, Spain (M P Botella MD); Manchester Centre for Genomic Medicine, Central Manchester University Hospitals NHS Foundation Trust, MAHSC, Manchester, UK (K E Chandler MD, Y J Crow); Department of Paediatric Neurology, University Hospitals of Leicester NHS Trust, Leicester, UK (L Dabydeen MRCPCH); TY Nelson Department of Neurology, and Neuroimmunology Group, the Children’s Hospital at Westmead, University of Sydney, Sydney, NSW, Australia (R C Dale PhD); Nutrition and Metabolism Unit, Hôpital Universitaire des Enfants Reine Fabiola (ULB), Brussels, Belgium (C De Laet MD); Department of Paediatric Neurology, Royal Preston Hospital, Preston, UK (C G E L De Goede FRCPCH); Pediatric Neurology Unit, Hospital Vall d’Hebron, Barcelona, Spain (M del Toro MD); Paediatric Neurology Unit, University Hospital 12 Octubre, Madrid, Spain (N Nunez Enamorado MD, R Simon MD); Department of Clinical and Experimental Sciences, Child Neurology and Psychiatry Unit, University of Brescia, Brescia, Italy (E Fazzi MD); Department of Genetics, BioCruces Health Research Institute, Hospital, Universitario Cruces, Bizkaia, Spain (B Gener MD); Institute of Biomedicine and Translational Medicine, University of Tartu, Tartu, Estonia (M Haldre BSc, P Peterson PhD, K Kisand PhD); General Neurology and Complex Motor Disorders Service, Evelina Children’s Hospital, Guy’s and St Thomas’ NHS Foundation Trust, London, UK (J-P S-M Lin PhD); Department of Paediatric Neurology, Leeds General Infirmary, Leeds, UK (J H Livingston MB ChB); Department of Neurosciences and Behavior Sciences, School of Medicine of Ribeirão Preto, University of São Paulo, São Paulo, Brazil, (C Marques Lourenco MD, W Marques Jr MD); Royal Devon and Exeter Foundation NHS Trust, Exeter, UK (P Oades FRCPCH); Section for Child Neurology, Women and Children’s Division, Oslo University Hospital, Oslo, Norway (M Rasmussen MD); Neuropediatrie, Hopital Gui de Chauliac, and INSERM U1051, INM, Montpellier, France (A Roubertie MD); Center for Genetic Medicine Research and Department of Neurology, Children’s National Medical Center, George Washington University School of Medicine, Washington, DC, USA, (J Loewenstein Schmidt CGC, A Vanderver MD); The Genetic Institute, Ha’Emek Medical Center, Afula and the Rappaport Faculty of Medicine, Technion, Haifa, Israel (S A Shalev MD, R Spiegel MD); Department of Neurology, University of Utah School of Medicine, Salt Lake City, UT, USA (K J Swoboda FACMG); Neurology Department, Royal Manchester Children’s Hospital, Manchester, UK (G Vassallo MD); Centre for Rare Diseases and Personalised Medicine and Department of Medical and Molecular Genetics, School of Clinical and Experimental Medicine, University of Birmingham, UK, and West Midlands Regional Genetics Service, Clinical Genetics Unit, Birmingham Women’s Hospital, Birmingham, UK (J Vogt MRCP); Nottingham Children’s Hospital, Nottingham University Hospitals NHS Trust, Nottingham, UK (W P Whitehouse FRCPCH); Neuropaediatrics, Department of Paediatrics, Mater Dei Hospital, Tal-Qroqq, Malta (D Soler MD); and Université Paris Descartes, EA 1833 and Hôpital Cochin, Service de Virologie, Paris, France, (F Rozenberg MD, P Lebon MD)
1,
Maria P Botella
Maria P Botella
1Manchester Centre for Genomic Medicine, Institute of Human Development, Faculty of Medical and Human Sciences, University of Manchester, Manchester Academic Health Sciences Centre (MAHSC), Manchester, UK (G I Rice PhD, G M A Forte MPhil, M Szynkiewicz MSc, D S Chase PhD, Y J Crow PhD); Erasme University Hospital, Universite Libre de Bruxelles (ULB), Brussels, Belgium, (A Aeby MD, C N Vilain MD, V Wermenbol MD); Medical Molecular Genetics Department, (M S Abdel-Hamid MSc, L Effat PhD), and Clinical Genetics Department, (S A Temtamy PhD, M S Aglan PhD, M S Zaki PhD, G M H Abdel-Salam PhD), Human Genetics and Genome Research Division, National Research Centre, Cairo, Egypt; Bradford Teaching Hospitals NHS Foundation Trust, Bradford, UK (S Ackroyd MB ChB); Department of Clinical Biochemistry, Lancashire Teaching Hospitals NHS Foundation Trust, Preston, UK (R Allcock FRCPath); University Hospitals of Coventry and Warwickshire, Coventry, UK (K M Bailey MRCPCH); Department of Brain and Behavioral Sciences, Unit of Child Neurology and Psychiatry, University of Pavia, Pavia, Italy, (U Balottin MD); Child Neurology and Psychiatry Unit, (U Balottin, I Olivieri MD, S Orcesi MD), and Laboratory of Experimental Neurobiology (C Cereda PhD), C. Mondino National Neurological Institute, Pavia, Italy; Praticien Hospitalier, Unité de Neurologie Pédiatrique et Centre de Référence des Maladies Neuromusculaires, Hôpital Necker-Enfants Malades, Paris, France (C Barnerias MD); Department of Pediatrics, and Department of Neurology and Neurosurgery, Montreal Children’s Hospital, McGill University Health Center, Montreal, QC, Canada (G Bernard MD); Department of Dermatology, Reference Centre for Cutaneous Rare Diseases (MAGEC), Necker-Enfants Malades APHP, Sorbonne Paris Cité, Université Paris Descartes, Institut Imagine, Paris, France (C Bodemer PhD); Department of Pediatrics, Hospital Universitario Araba, Vitoria-Gasteiz, Spain (M P Botella MD); Manchester Centre for Genomic Medicine, Central Manchester University Hospitals NHS Foundation Trust, MAHSC, Manchester, UK (K E Chandler MD, Y J Crow); Department of Paediatric Neurology, University Hospitals of Leicester NHS Trust, Leicester, UK (L Dabydeen MRCPCH); TY Nelson Department of Neurology, and Neuroimmunology Group, the Children’s Hospital at Westmead, University of Sydney, Sydney, NSW, Australia (R C Dale PhD); Nutrition and Metabolism Unit, Hôpital Universitaire des Enfants Reine Fabiola (ULB), Brussels, Belgium (C De Laet MD); Department of Paediatric Neurology, Royal Preston Hospital, Preston, UK (C G E L De Goede FRCPCH); Pediatric Neurology Unit, Hospital Vall d’Hebron, Barcelona, Spain (M del Toro MD); Paediatric Neurology Unit, University Hospital 12 Octubre, Madrid, Spain (N Nunez Enamorado MD, R Simon MD); Department of Clinical and Experimental Sciences, Child Neurology and Psychiatry Unit, University of Brescia, Brescia, Italy (E Fazzi MD); Department of Genetics, BioCruces Health Research Institute, Hospital, Universitario Cruces, Bizkaia, Spain (B Gener MD); Institute of Biomedicine and Translational Medicine, University of Tartu, Tartu, Estonia (M Haldre BSc, P Peterson PhD, K Kisand PhD); General Neurology and Complex Motor Disorders Service, Evelina Children’s Hospital, Guy’s and St Thomas’ NHS Foundation Trust, London, UK (J-P S-M Lin PhD); Department of Paediatric Neurology, Leeds General Infirmary, Leeds, UK (J H Livingston MB ChB); Department of Neurosciences and Behavior Sciences, School of Medicine of Ribeirão Preto, University of São Paulo, São Paulo, Brazil, (C Marques Lourenco MD, W Marques Jr MD); Royal Devon and Exeter Foundation NHS Trust, Exeter, UK (P Oades FRCPCH); Section for Child Neurology, Women and Children’s Division, Oslo University Hospital, Oslo, Norway (M Rasmussen MD); Neuropediatrie, Hopital Gui de Chauliac, and INSERM U1051, INM, Montpellier, France (A Roubertie MD); Center for Genetic Medicine Research and Department of Neurology, Children’s National Medical Center, George Washington University School of Medicine, Washington, DC, USA, (J Loewenstein Schmidt CGC, A Vanderver MD); The Genetic Institute, Ha’Emek Medical Center, Afula and the Rappaport Faculty of Medicine, Technion, Haifa, Israel (S A Shalev MD, R Spiegel MD); Department of Neurology, University of Utah School of Medicine, Salt Lake City, UT, USA (K J Swoboda FACMG); Neurology Department, Royal Manchester Children’s Hospital, Manchester, UK (G Vassallo MD); Centre for Rare Diseases and Personalised Medicine and Department of Medical and Molecular Genetics, School of Clinical and Experimental Medicine, University of Birmingham, UK, and West Midlands Regional Genetics Service, Clinical Genetics Unit, Birmingham Women’s Hospital, Birmingham, UK (J Vogt MRCP); Nottingham Children’s Hospital, Nottingham University Hospitals NHS Trust, Nottingham, UK (W P Whitehouse FRCPCH); Neuropaediatrics, Department of Paediatrics, Mater Dei Hospital, Tal-Qroqq, Malta (D Soler MD); and Université Paris Descartes, EA 1833 and Hôpital Cochin, Service de Virologie, Paris, France, (F Rozenberg MD, P Lebon MD)
1,
Cristina Cereda
Cristina Cereda
1Manchester Centre for Genomic Medicine, Institute of Human Development, Faculty of Medical and Human Sciences, University of Manchester, Manchester Academic Health Sciences Centre (MAHSC), Manchester, UK (G I Rice PhD, G M A Forte MPhil, M Szynkiewicz MSc, D S Chase PhD, Y J Crow PhD); Erasme University Hospital, Universite Libre de Bruxelles (ULB), Brussels, Belgium, (A Aeby MD, C N Vilain MD, V Wermenbol MD); Medical Molecular Genetics Department, (M S Abdel-Hamid MSc, L Effat PhD), and Clinical Genetics Department, (S A Temtamy PhD, M S Aglan PhD, M S Zaki PhD, G M H Abdel-Salam PhD), Human Genetics and Genome Research Division, National Research Centre, Cairo, Egypt; Bradford Teaching Hospitals NHS Foundation Trust, Bradford, UK (S Ackroyd MB ChB); Department of Clinical Biochemistry, Lancashire Teaching Hospitals NHS Foundation Trust, Preston, UK (R Allcock FRCPath); University Hospitals of Coventry and Warwickshire, Coventry, UK (K M Bailey MRCPCH); Department of Brain and Behavioral Sciences, Unit of Child Neurology and Psychiatry, University of Pavia, Pavia, Italy, (U Balottin MD); Child Neurology and Psychiatry Unit, (U Balottin, I Olivieri MD, S Orcesi MD), and Laboratory of Experimental Neurobiology (C Cereda PhD), C. Mondino National Neurological Institute, Pavia, Italy; Praticien Hospitalier, Unité de Neurologie Pédiatrique et Centre de Référence des Maladies Neuromusculaires, Hôpital Necker-Enfants Malades, Paris, France (C Barnerias MD); Department of Pediatrics, and Department of Neurology and Neurosurgery, Montreal Children’s Hospital, McGill University Health Center, Montreal, QC, Canada (G Bernard MD); Department of Dermatology, Reference Centre for Cutaneous Rare Diseases (MAGEC), Necker-Enfants Malades APHP, Sorbonne Paris Cité, Université Paris Descartes, Institut Imagine, Paris, France (C Bodemer PhD); Department of Pediatrics, Hospital Universitario Araba, Vitoria-Gasteiz, Spain (M P Botella MD); Manchester Centre for Genomic Medicine, Central Manchester University Hospitals NHS Foundation Trust, MAHSC, Manchester, UK (K E Chandler MD, Y J Crow); Department of Paediatric Neurology, University Hospitals of Leicester NHS Trust, Leicester, UK (L Dabydeen MRCPCH); TY Nelson Department of Neurology, and Neuroimmunology Group, the Children’s Hospital at Westmead, University of Sydney, Sydney, NSW, Australia (R C Dale PhD); Nutrition and Metabolism Unit, Hôpital Universitaire des Enfants Reine Fabiola (ULB), Brussels, Belgium (C De Laet MD); Department of Paediatric Neurology, Royal Preston Hospital, Preston, UK (C G E L De Goede FRCPCH); Pediatric Neurology Unit, Hospital Vall d’Hebron, Barcelona, Spain (M del Toro MD); Paediatric Neurology Unit, University Hospital 12 Octubre, Madrid, Spain (N Nunez Enamorado MD, R Simon MD); Department of Clinical and Experimental Sciences, Child Neurology and Psychiatry Unit, University of Brescia, Brescia, Italy (E Fazzi MD); Department of Genetics, BioCruces Health Research Institute, Hospital, Universitario Cruces, Bizkaia, Spain (B Gener MD); Institute of Biomedicine and Translational Medicine, University of Tartu, Tartu, Estonia (M Haldre BSc, P Peterson PhD, K Kisand PhD); General Neurology and Complex Motor Disorders Service, Evelina Children’s Hospital, Guy’s and St Thomas’ NHS Foundation Trust, London, UK (J-P S-M Lin PhD); Department of Paediatric Neurology, Leeds General Infirmary, Leeds, UK (J H Livingston MB ChB); Department of Neurosciences and Behavior Sciences, School of Medicine of Ribeirão Preto, University of São Paulo, São Paulo, Brazil, (C Marques Lourenco MD, W Marques Jr MD); Royal Devon and Exeter Foundation NHS Trust, Exeter, UK (P Oades FRCPCH); Section for Child Neurology, Women and Children’s Division, Oslo University Hospital, Oslo, Norway (M Rasmussen MD); Neuropediatrie, Hopital Gui de Chauliac, and INSERM U1051, INM, Montpellier, France (A Roubertie MD); Center for Genetic Medicine Research and Department of Neurology, Children’s National Medical Center, George Washington University School of Medicine, Washington, DC, USA, (J Loewenstein Schmidt CGC, A Vanderver MD); The Genetic Institute, Ha’Emek Medical Center, Afula and the Rappaport Faculty of Medicine, Technion, Haifa, Israel (S A Shalev MD, R Spiegel MD); Department of Neurology, University of Utah School of Medicine, Salt Lake City, UT, USA (K J Swoboda FACMG); Neurology Department, Royal Manchester Children’s Hospital, Manchester, UK (G Vassallo MD); Centre for Rare Diseases and Personalised Medicine and Department of Medical and Molecular Genetics, School of Clinical and Experimental Medicine, University of Birmingham, UK, and West Midlands Regional Genetics Service, Clinical Genetics Unit, Birmingham Women’s Hospital, Birmingham, UK (J Vogt MRCP); Nottingham Children’s Hospital, Nottingham University Hospitals NHS Trust, Nottingham, UK (W P Whitehouse FRCPCH); Neuropaediatrics, Department of Paediatrics, Mater Dei Hospital, Tal-Qroqq, Malta (D Soler MD); and Université Paris Descartes, EA 1833 and Hôpital Cochin, Service de Virologie, Paris, France, (F Rozenberg MD, P Lebon MD)
1,
Kate E Chandler
Kate E Chandler
1Manchester Centre for Genomic Medicine, Institute of Human Development, Faculty of Medical and Human Sciences, University of Manchester, Manchester Academic Health Sciences Centre (MAHSC), Manchester, UK (G I Rice PhD, G M A Forte MPhil, M Szynkiewicz MSc, D S Chase PhD, Y J Crow PhD); Erasme University Hospital, Universite Libre de Bruxelles (ULB), Brussels, Belgium, (A Aeby MD, C N Vilain MD, V Wermenbol MD); Medical Molecular Genetics Department, (M S Abdel-Hamid MSc, L Effat PhD), and Clinical Genetics Department, (S A Temtamy PhD, M S Aglan PhD, M S Zaki PhD, G M H Abdel-Salam PhD), Human Genetics and Genome Research Division, National Research Centre, Cairo, Egypt; Bradford Teaching Hospitals NHS Foundation Trust, Bradford, UK (S Ackroyd MB ChB); Department of Clinical Biochemistry, Lancashire Teaching Hospitals NHS Foundation Trust, Preston, UK (R Allcock FRCPath); University Hospitals of Coventry and Warwickshire, Coventry, UK (K M Bailey MRCPCH); Department of Brain and Behavioral Sciences, Unit of Child Neurology and Psychiatry, University of Pavia, Pavia, Italy, (U Balottin MD); Child Neurology and Psychiatry Unit, (U Balottin, I Olivieri MD, S Orcesi MD), and Laboratory of Experimental Neurobiology (C Cereda PhD), C. Mondino National Neurological Institute, Pavia, Italy; Praticien Hospitalier, Unité de Neurologie Pédiatrique et Centre de Référence des Maladies Neuromusculaires, Hôpital Necker-Enfants Malades, Paris, France (C Barnerias MD); Department of Pediatrics, and Department of Neurology and Neurosurgery, Montreal Children’s Hospital, McGill University Health Center, Montreal, QC, Canada (G Bernard MD); Department of Dermatology, Reference Centre for Cutaneous Rare Diseases (MAGEC), Necker-Enfants Malades APHP, Sorbonne Paris Cité, Université Paris Descartes, Institut Imagine, Paris, France (C Bodemer PhD); Department of Pediatrics, Hospital Universitario Araba, Vitoria-Gasteiz, Spain (M P Botella MD); Manchester Centre for Genomic Medicine, Central Manchester University Hospitals NHS Foundation Trust, MAHSC, Manchester, UK (K E Chandler MD, Y J Crow); Department of Paediatric Neurology, University Hospitals of Leicester NHS Trust, Leicester, UK (L Dabydeen MRCPCH); TY Nelson Department of Neurology, and Neuroimmunology Group, the Children’s Hospital at Westmead, University of Sydney, Sydney, NSW, Australia (R C Dale PhD); Nutrition and Metabolism Unit, Hôpital Universitaire des Enfants Reine Fabiola (ULB), Brussels, Belgium (C De Laet MD); Department of Paediatric Neurology, Royal Preston Hospital, Preston, UK (C G E L De Goede FRCPCH); Pediatric Neurology Unit, Hospital Vall d’Hebron, Barcelona, Spain (M del Toro MD); Paediatric Neurology Unit, University Hospital 12 Octubre, Madrid, Spain (N Nunez Enamorado MD, R Simon MD); Department of Clinical and Experimental Sciences, Child Neurology and Psychiatry Unit, University of Brescia, Brescia, Italy (E Fazzi MD); Department of Genetics, BioCruces Health Research Institute, Hospital, Universitario Cruces, Bizkaia, Spain (B Gener MD); Institute of Biomedicine and Translational Medicine, University of Tartu, Tartu, Estonia (M Haldre BSc, P Peterson PhD, K Kisand PhD); General Neurology and Complex Motor Disorders Service, Evelina Children’s Hospital, Guy’s and St Thomas’ NHS Foundation Trust, London, UK (J-P S-M Lin PhD); Department of Paediatric Neurology, Leeds General Infirmary, Leeds, UK (J H Livingston MB ChB); Department of Neurosciences and Behavior Sciences, School of Medicine of Ribeirão Preto, University of São Paulo, São Paulo, Brazil, (C Marques Lourenco MD, W Marques Jr MD); Royal Devon and Exeter Foundation NHS Trust, Exeter, UK (P Oades FRCPCH); Section for Child Neurology, Women and Children’s Division, Oslo University Hospital, Oslo, Norway (M Rasmussen MD); Neuropediatrie, Hopital Gui de Chauliac, and INSERM U1051, INM, Montpellier, France (A Roubertie MD); Center for Genetic Medicine Research and Department of Neurology, Children’s National Medical Center, George Washington University School of Medicine, Washington, DC, USA, (J Loewenstein Schmidt CGC, A Vanderver MD); The Genetic Institute, Ha’Emek Medical Center, Afula and the Rappaport Faculty of Medicine, Technion, Haifa, Israel (S A Shalev MD, R Spiegel MD); Department of Neurology, University of Utah School of Medicine, Salt Lake City, UT, USA (K J Swoboda FACMG); Neurology Department, Royal Manchester Children’s Hospital, Manchester, UK (G Vassallo MD); Centre for Rare Diseases and Personalised Medicine and Department of Medical and Molecular Genetics, School of Clinical and Experimental Medicine, University of Birmingham, UK, and West Midlands Regional Genetics Service, Clinical Genetics Unit, Birmingham Women’s Hospital, Birmingham, UK (J Vogt MRCP); Nottingham Children’s Hospital, Nottingham University Hospitals NHS Trust, Nottingham, UK (W P Whitehouse FRCPCH); Neuropaediatrics, Department of Paediatrics, Mater Dei Hospital, Tal-Qroqq, Malta (D Soler MD); and Université Paris Descartes, EA 1833 and Hôpital Cochin, Service de Virologie, Paris, France, (F Rozenberg MD, P Lebon MD)
1,
Lyvia Dabydeen
Lyvia Dabydeen
1Manchester Centre for Genomic Medicine, Institute of Human Development, Faculty of Medical and Human Sciences, University of Manchester, Manchester Academic Health Sciences Centre (MAHSC), Manchester, UK (G I Rice PhD, G M A Forte MPhil, M Szynkiewicz MSc, D S Chase PhD, Y J Crow PhD); Erasme University Hospital, Universite Libre de Bruxelles (ULB), Brussels, Belgium, (A Aeby MD, C N Vilain MD, V Wermenbol MD); Medical Molecular Genetics Department, (M S Abdel-Hamid MSc, L Effat PhD), and Clinical Genetics Department, (S A Temtamy PhD, M S Aglan PhD, M S Zaki PhD, G M H Abdel-Salam PhD), Human Genetics and Genome Research Division, National Research Centre, Cairo, Egypt; Bradford Teaching Hospitals NHS Foundation Trust, Bradford, UK (S Ackroyd MB ChB); Department of Clinical Biochemistry, Lancashire Teaching Hospitals NHS Foundation Trust, Preston, UK (R Allcock FRCPath); University Hospitals of Coventry and Warwickshire, Coventry, UK (K M Bailey MRCPCH); Department of Brain and Behavioral Sciences, Unit of Child Neurology and Psychiatry, University of Pavia, Pavia, Italy, (U Balottin MD); Child Neurology and Psychiatry Unit, (U Balottin, I Olivieri MD, S Orcesi MD), and Laboratory of Experimental Neurobiology (C Cereda PhD), C. Mondino National Neurological Institute, Pavia, Italy; Praticien Hospitalier, Unité de Neurologie Pédiatrique et Centre de Référence des Maladies Neuromusculaires, Hôpital Necker-Enfants Malades, Paris, France (C Barnerias MD); Department of Pediatrics, and Department of Neurology and Neurosurgery, Montreal Children’s Hospital, McGill University Health Center, Montreal, QC, Canada (G Bernard MD); Department of Dermatology, Reference Centre for Cutaneous Rare Diseases (MAGEC), Necker-Enfants Malades APHP, Sorbonne Paris Cité, Université Paris Descartes, Institut Imagine, Paris, France (C Bodemer PhD); Department of Pediatrics, Hospital Universitario Araba, Vitoria-Gasteiz, Spain (M P Botella MD); Manchester Centre for Genomic Medicine, Central Manchester University Hospitals NHS Foundation Trust, MAHSC, Manchester, UK (K E Chandler MD, Y J Crow); Department of Paediatric Neurology, University Hospitals of Leicester NHS Trust, Leicester, UK (L Dabydeen MRCPCH); TY Nelson Department of Neurology, and Neuroimmunology Group, the Children’s Hospital at Westmead, University of Sydney, Sydney, NSW, Australia (R C Dale PhD); Nutrition and Metabolism Unit, Hôpital Universitaire des Enfants Reine Fabiola (ULB), Brussels, Belgium (C De Laet MD); Department of Paediatric Neurology, Royal Preston Hospital, Preston, UK (C G E L De Goede FRCPCH); Pediatric Neurology Unit, Hospital Vall d’Hebron, Barcelona, Spain (M del Toro MD); Paediatric Neurology Unit, University Hospital 12 Octubre, Madrid, Spain (N Nunez Enamorado MD, R Simon MD); Department of Clinical and Experimental Sciences, Child Neurology and Psychiatry Unit, University of Brescia, Brescia, Italy (E Fazzi MD); Department of Genetics, BioCruces Health Research Institute, Hospital, Universitario Cruces, Bizkaia, Spain (B Gener MD); Institute of Biomedicine and Translational Medicine, University of Tartu, Tartu, Estonia (M Haldre BSc, P Peterson PhD, K Kisand PhD); General Neurology and Complex Motor Disorders Service, Evelina Children’s Hospital, Guy’s and St Thomas’ NHS Foundation Trust, London, UK (J-P S-M Lin PhD); Department of Paediatric Neurology, Leeds General Infirmary, Leeds, UK (J H Livingston MB ChB); Department of Neurosciences and Behavior Sciences, School of Medicine of Ribeirão Preto, University of São Paulo, São Paulo, Brazil, (C Marques Lourenco MD, W Marques Jr MD); Royal Devon and Exeter Foundation NHS Trust, Exeter, UK (P Oades FRCPCH); Section for Child Neurology, Women and Children’s Division, Oslo University Hospital, Oslo, Norway (M Rasmussen MD); Neuropediatrie, Hopital Gui de Chauliac, and INSERM U1051, INM, Montpellier, France (A Roubertie MD); Center for Genetic Medicine Research and Department of Neurology, Children’s National Medical Center, George Washington University School of Medicine, Washington, DC, USA, (J Loewenstein Schmidt CGC, A Vanderver MD); The Genetic Institute, Ha’Emek Medical Center, Afula and the Rappaport Faculty of Medicine, Technion, Haifa, Israel (S A Shalev MD, R Spiegel MD); Department of Neurology, University of Utah School of Medicine, Salt Lake City, UT, USA (K J Swoboda FACMG); Neurology Department, Royal Manchester Children’s Hospital, Manchester, UK (G Vassallo MD); Centre for Rare Diseases and Personalised Medicine and Department of Medical and Molecular Genetics, School of Clinical and Experimental Medicine, University of Birmingham, UK, and West Midlands Regional Genetics Service, Clinical Genetics Unit, Birmingham Women’s Hospital, Birmingham, UK (J Vogt MRCP); Nottingham Children’s Hospital, Nottingham University Hospitals NHS Trust, Nottingham, UK (W P Whitehouse FRCPCH); Neuropaediatrics, Department of Paediatrics, Mater Dei Hospital, Tal-Qroqq, Malta (D Soler MD); and Université Paris Descartes, EA 1833 and Hôpital Cochin, Service de Virologie, Paris, France, (F Rozenberg MD, P Lebon MD)
1,
Russell C Dale
Russell C Dale
1Manchester Centre for Genomic Medicine, Institute of Human Development, Faculty of Medical and Human Sciences, University of Manchester, Manchester Academic Health Sciences Centre (MAHSC), Manchester, UK (G I Rice PhD, G M A Forte MPhil, M Szynkiewicz MSc, D S Chase PhD, Y J Crow PhD); Erasme University Hospital, Universite Libre de Bruxelles (ULB), Brussels, Belgium, (A Aeby MD, C N Vilain MD, V Wermenbol MD); Medical Molecular Genetics Department, (M S Abdel-Hamid MSc, L Effat PhD), and Clinical Genetics Department, (S A Temtamy PhD, M S Aglan PhD, M S Zaki PhD, G M H Abdel-Salam PhD), Human Genetics and Genome Research Division, National Research Centre, Cairo, Egypt; Bradford Teaching Hospitals NHS Foundation Trust, Bradford, UK (S Ackroyd MB ChB); Department of Clinical Biochemistry, Lancashire Teaching Hospitals NHS Foundation Trust, Preston, UK (R Allcock FRCPath); University Hospitals of Coventry and Warwickshire, Coventry, UK (K M Bailey MRCPCH); Department of Brain and Behavioral Sciences, Unit of Child Neurology and Psychiatry, University of Pavia, Pavia, Italy, (U Balottin MD); Child Neurology and Psychiatry Unit, (U Balottin, I Olivieri MD, S Orcesi MD), and Laboratory of Experimental Neurobiology (C Cereda PhD), C. Mondino National Neurological Institute, Pavia, Italy; Praticien Hospitalier, Unité de Neurologie Pédiatrique et Centre de Référence des Maladies Neuromusculaires, Hôpital Necker-Enfants Malades, Paris, France (C Barnerias MD); Department of Pediatrics, and Department of Neurology and Neurosurgery, Montreal Children’s Hospital, McGill University Health Center, Montreal, QC, Canada (G Bernard MD); Department of Dermatology, Reference Centre for Cutaneous Rare Diseases (MAGEC), Necker-Enfants Malades APHP, Sorbonne Paris Cité, Université Paris Descartes, Institut Imagine, Paris, France (C Bodemer PhD); Department of Pediatrics, Hospital Universitario Araba, Vitoria-Gasteiz, Spain (M P Botella MD); Manchester Centre for Genomic Medicine, Central Manchester University Hospitals NHS Foundation Trust, MAHSC, Manchester, UK (K E Chandler MD, Y J Crow); Department of Paediatric Neurology, University Hospitals of Leicester NHS Trust, Leicester, UK (L Dabydeen MRCPCH); TY Nelson Department of Neurology, and Neuroimmunology Group, the Children’s Hospital at Westmead, University of Sydney, Sydney, NSW, Australia (R C Dale PhD); Nutrition and Metabolism Unit, Hôpital Universitaire des Enfants Reine Fabiola (ULB), Brussels, Belgium (C De Laet MD); Department of Paediatric Neurology, Royal Preston Hospital, Preston, UK (C G E L De Goede FRCPCH); Pediatric Neurology Unit, Hospital Vall d’Hebron, Barcelona, Spain (M del Toro MD); Paediatric Neurology Unit, University Hospital 12 Octubre, Madrid, Spain (N Nunez Enamorado MD, R Simon MD); Department of Clinical and Experimental Sciences, Child Neurology and Psychiatry Unit, University of Brescia, Brescia, Italy (E Fazzi MD); Department of Genetics, BioCruces Health Research Institute, Hospital, Universitario Cruces, Bizkaia, Spain (B Gener MD); Institute of Biomedicine and Translational Medicine, University of Tartu, Tartu, Estonia (M Haldre BSc, P Peterson PhD, K Kisand PhD); General Neurology and Complex Motor Disorders Service, Evelina Children’s Hospital, Guy’s and St Thomas’ NHS Foundation Trust, London, UK (J-P S-M Lin PhD); Department of Paediatric Neurology, Leeds General Infirmary, Leeds, UK (J H Livingston MB ChB); Department of Neurosciences and Behavior Sciences, School of Medicine of Ribeirão Preto, University of São Paulo, São Paulo, Brazil, (C Marques Lourenco MD, W Marques Jr MD); Royal Devon and Exeter Foundation NHS Trust, Exeter, UK (P Oades FRCPCH); Section for Child Neurology, Women and Children’s Division, Oslo University Hospital, Oslo, Norway (M Rasmussen MD); Neuropediatrie, Hopital Gui de Chauliac, and INSERM U1051, INM, Montpellier, France (A Roubertie MD); Center for Genetic Medicine Research and Department of Neurology, Children’s National Medical Center, George Washington University School of Medicine, Washington, DC, USA, (J Loewenstein Schmidt CGC, A Vanderver MD); The Genetic Institute, Ha’Emek Medical Center, Afula and the Rappaport Faculty of Medicine, Technion, Haifa, Israel (S A Shalev MD, R Spiegel MD); Department of Neurology, University of Utah School of Medicine, Salt Lake City, UT, USA (K J Swoboda FACMG); Neurology Department, Royal Manchester Children’s Hospital, Manchester, UK (G Vassallo MD); Centre for Rare Diseases and Personalised Medicine and Department of Medical and Molecular Genetics, School of Clinical and Experimental Medicine, University of Birmingham, UK, and West Midlands Regional Genetics Service, Clinical Genetics Unit, Birmingham Women’s Hospital, Birmingham, UK (J Vogt MRCP); Nottingham Children’s Hospital, Nottingham University Hospitals NHS Trust, Nottingham, UK (W P Whitehouse FRCPCH); Neuropaediatrics, Department of Paediatrics, Mater Dei Hospital, Tal-Qroqq, Malta (D Soler MD); and Université Paris Descartes, EA 1833 and Hôpital Cochin, Service de Virologie, Paris, France, (F Rozenberg MD, P Lebon MD)
1,
Corinne De Laet
Corinne De Laet
1Manchester Centre for Genomic Medicine, Institute of Human Development, Faculty of Medical and Human Sciences, University of Manchester, Manchester Academic Health Sciences Centre (MAHSC), Manchester, UK (G I Rice PhD, G M A Forte MPhil, M Szynkiewicz MSc, D S Chase PhD, Y J Crow PhD); Erasme University Hospital, Universite Libre de Bruxelles (ULB), Brussels, Belgium, (A Aeby MD, C N Vilain MD, V Wermenbol MD); Medical Molecular Genetics Department, (M S Abdel-Hamid MSc, L Effat PhD), and Clinical Genetics Department, (S A Temtamy PhD, M S Aglan PhD, M S Zaki PhD, G M H Abdel-Salam PhD), Human Genetics and Genome Research Division, National Research Centre, Cairo, Egypt; Bradford Teaching Hospitals NHS Foundation Trust, Bradford, UK (S Ackroyd MB ChB); Department of Clinical Biochemistry, Lancashire Teaching Hospitals NHS Foundation Trust, Preston, UK (R Allcock FRCPath); University Hospitals of Coventry and Warwickshire, Coventry, UK (K M Bailey MRCPCH); Department of Brain and Behavioral Sciences, Unit of Child Neurology and Psychiatry, University of Pavia, Pavia, Italy, (U Balottin MD); Child Neurology and Psychiatry Unit, (U Balottin, I Olivieri MD, S Orcesi MD), and Laboratory of Experimental Neurobiology (C Cereda PhD), C. Mondino National Neurological Institute, Pavia, Italy; Praticien Hospitalier, Unité de Neurologie Pédiatrique et Centre de Référence des Maladies Neuromusculaires, Hôpital Necker-Enfants Malades, Paris, France (C Barnerias MD); Department of Pediatrics, and Department of Neurology and Neurosurgery, Montreal Children’s Hospital, McGill University Health Center, Montreal, QC, Canada (G Bernard MD); Department of Dermatology, Reference Centre for Cutaneous Rare Diseases (MAGEC), Necker-Enfants Malades APHP, Sorbonne Paris Cité, Université Paris Descartes, Institut Imagine, Paris, France (C Bodemer PhD); Department of Pediatrics, Hospital Universitario Araba, Vitoria-Gasteiz, Spain (M P Botella MD); Manchester Centre for Genomic Medicine, Central Manchester University Hospitals NHS Foundation Trust, MAHSC, Manchester, UK (K E Chandler MD, Y J Crow); Department of Paediatric Neurology, University Hospitals of Leicester NHS Trust, Leicester, UK (L Dabydeen MRCPCH); TY Nelson Department of Neurology, and Neuroimmunology Group, the Children’s Hospital at Westmead, University of Sydney, Sydney, NSW, Australia (R C Dale PhD); Nutrition and Metabolism Unit, Hôpital Universitaire des Enfants Reine Fabiola (ULB), Brussels, Belgium (C De Laet MD); Department of Paediatric Neurology, Royal Preston Hospital, Preston, UK (C G E L De Goede FRCPCH); Pediatric Neurology Unit, Hospital Vall d’Hebron, Barcelona, Spain (M del Toro MD); Paediatric Neurology Unit, University Hospital 12 Octubre, Madrid, Spain (N Nunez Enamorado MD, R Simon MD); Department of Clinical and Experimental Sciences, Child Neurology and Psychiatry Unit, University of Brescia, Brescia, Italy (E Fazzi MD); Department of Genetics, BioCruces Health Research Institute, Hospital, Universitario Cruces, Bizkaia, Spain (B Gener MD); Institute of Biomedicine and Translational Medicine, University of Tartu, Tartu, Estonia (M Haldre BSc, P Peterson PhD, K Kisand PhD); General Neurology and Complex Motor Disorders Service, Evelina Children’s Hospital, Guy’s and St Thomas’ NHS Foundation Trust, London, UK (J-P S-M Lin PhD); Department of Paediatric Neurology, Leeds General Infirmary, Leeds, UK (J H Livingston MB ChB); Department of Neurosciences and Behavior Sciences, School of Medicine of Ribeirão Preto, University of São Paulo, São Paulo, Brazil, (C Marques Lourenco MD, W Marques Jr MD); Royal Devon and Exeter Foundation NHS Trust, Exeter, UK (P Oades FRCPCH); Section for Child Neurology, Women and Children’s Division, Oslo University Hospital, Oslo, Norway (M Rasmussen MD); Neuropediatrie, Hopital Gui de Chauliac, and INSERM U1051, INM, Montpellier, France (A Roubertie MD); Center for Genetic Medicine Research and Department of Neurology, Children’s National Medical Center, George Washington University School of Medicine, Washington, DC, USA, (J Loewenstein Schmidt CGC, A Vanderver MD); The Genetic Institute, Ha’Emek Medical Center, Afula and the Rappaport Faculty of Medicine, Technion, Haifa, Israel (S A Shalev MD, R Spiegel MD); Department of Neurology, University of Utah School of Medicine, Salt Lake City, UT, USA (K J Swoboda FACMG); Neurology Department, Royal Manchester Children’s Hospital, Manchester, UK (G Vassallo MD); Centre for Rare Diseases and Personalised Medicine and Department of Medical and Molecular Genetics, School of Clinical and Experimental Medicine, University of Birmingham, UK, and West Midlands Regional Genetics Service, Clinical Genetics Unit, Birmingham Women’s Hospital, Birmingham, UK (J Vogt MRCP); Nottingham Children’s Hospital, Nottingham University Hospitals NHS Trust, Nottingham, UK (W P Whitehouse FRCPCH); Neuropaediatrics, Department of Paediatrics, Mater Dei Hospital, Tal-Qroqq, Malta (D Soler MD); and Université Paris Descartes, EA 1833 and Hôpital Cochin, Service de Virologie, Paris, France, (F Rozenberg MD, P Lebon MD)
1,
Christian G E L De Goede
Christian G E L De Goede
1Manchester Centre for Genomic Medicine, Institute of Human Development, Faculty of Medical and Human Sciences, University of Manchester, Manchester Academic Health Sciences Centre (MAHSC), Manchester, UK (G I Rice PhD, G M A Forte MPhil, M Szynkiewicz MSc, D S Chase PhD, Y J Crow PhD); Erasme University Hospital, Universite Libre de Bruxelles (ULB), Brussels, Belgium, (A Aeby MD, C N Vilain MD, V Wermenbol MD); Medical Molecular Genetics Department, (M S Abdel-Hamid MSc, L Effat PhD), and Clinical Genetics Department, (S A Temtamy PhD, M S Aglan PhD, M S Zaki PhD, G M H Abdel-Salam PhD), Human Genetics and Genome Research Division, National Research Centre, Cairo, Egypt; Bradford Teaching Hospitals NHS Foundation Trust, Bradford, UK (S Ackroyd MB ChB); Department of Clinical Biochemistry, Lancashire Teaching Hospitals NHS Foundation Trust, Preston, UK (R Allcock FRCPath); University Hospitals of Coventry and Warwickshire, Coventry, UK (K M Bailey MRCPCH); Department of Brain and Behavioral Sciences, Unit of Child Neurology and Psychiatry, University of Pavia, Pavia, Italy, (U Balottin MD); Child Neurology and Psychiatry Unit, (U Balottin, I Olivieri MD, S Orcesi MD), and Laboratory of Experimental Neurobiology (C Cereda PhD), C. Mondino National Neurological Institute, Pavia, Italy; Praticien Hospitalier, Unité de Neurologie Pédiatrique et Centre de Référence des Maladies Neuromusculaires, Hôpital Necker-Enfants Malades, Paris, France (C Barnerias MD); Department of Pediatrics, and Department of Neurology and Neurosurgery, Montreal Children’s Hospital, McGill University Health Center, Montreal, QC, Canada (G Bernard MD); Department of Dermatology, Reference Centre for Cutaneous Rare Diseases (MAGEC), Necker-Enfants Malades APHP, Sorbonne Paris Cité, Université Paris Descartes, Institut Imagine, Paris, France (C Bodemer PhD); Department of Pediatrics, Hospital Universitario Araba, Vitoria-Gasteiz, Spain (M P Botella MD); Manchester Centre for Genomic Medicine, Central Manchester University Hospitals NHS Foundation Trust, MAHSC, Manchester, UK (K E Chandler MD, Y J Crow); Department of Paediatric Neurology, University Hospitals of Leicester NHS Trust, Leicester, UK (L Dabydeen MRCPCH); TY Nelson Department of Neurology, and Neuroimmunology Group, the Children’s Hospital at Westmead, University of Sydney, Sydney, NSW, Australia (R C Dale PhD); Nutrition and Metabolism Unit, Hôpital Universitaire des Enfants Reine Fabiola (ULB), Brussels, Belgium (C De Laet MD); Department of Paediatric Neurology, Royal Preston Hospital, Preston, UK (C G E L De Goede FRCPCH); Pediatric Neurology Unit, Hospital Vall d’Hebron, Barcelona, Spain (M del Toro MD); Paediatric Neurology Unit, University Hospital 12 Octubre, Madrid, Spain (N Nunez Enamorado MD, R Simon MD); Department of Clinical and Experimental Sciences, Child Neurology and Psychiatry Unit, University of Brescia, Brescia, Italy (E Fazzi MD); Department of Genetics, BioCruces Health Research Institute, Hospital, Universitario Cruces, Bizkaia, Spain (B Gener MD); Institute of Biomedicine and Translational Medicine, University of Tartu, Tartu, Estonia (M Haldre BSc, P Peterson PhD, K Kisand PhD); General Neurology and Complex Motor Disorders Service, Evelina Children’s Hospital, Guy’s and St Thomas’ NHS Foundation Trust, London, UK (J-P S-M Lin PhD); Department of Paediatric Neurology, Leeds General Infirmary, Leeds, UK (J H Livingston MB ChB); Department of Neurosciences and Behavior Sciences, School of Medicine of Ribeirão Preto, University of São Paulo, São Paulo, Brazil, (C Marques Lourenco MD, W Marques Jr MD); Royal Devon and Exeter Foundation NHS Trust, Exeter, UK (P Oades FRCPCH); Section for Child Neurology, Women and Children’s Division, Oslo University Hospital, Oslo, Norway (M Rasmussen MD); Neuropediatrie, Hopital Gui de Chauliac, and INSERM U1051, INM, Montpellier, France (A Roubertie MD); Center for Genetic Medicine Research and Department of Neurology, Children’s National Medical Center, George Washington University School of Medicine, Washington, DC, USA, (J Loewenstein Schmidt CGC, A Vanderver MD); The Genetic Institute, Ha’Emek Medical Center, Afula and the Rappaport Faculty of Medicine, Technion, Haifa, Israel (S A Shalev MD, R Spiegel MD); Department of Neurology, University of Utah School of Medicine, Salt Lake City, UT, USA (K J Swoboda FACMG); Neurology Department, Royal Manchester Children’s Hospital, Manchester, UK (G Vassallo MD); Centre for Rare Diseases and Personalised Medicine and Department of Medical and Molecular Genetics, School of Clinical and Experimental Medicine, University of Birmingham, UK, and West Midlands Regional Genetics Service, Clinical Genetics Unit, Birmingham Women’s Hospital, Birmingham, UK (J Vogt MRCP); Nottingham Children’s Hospital, Nottingham University Hospitals NHS Trust, Nottingham, UK (W P Whitehouse FRCPCH); Neuropaediatrics, Department of Paediatrics, Mater Dei Hospital, Tal-Qroqq, Malta (D Soler MD); and Université Paris Descartes, EA 1833 and Hôpital Cochin, Service de Virologie, Paris, France, (F Rozenberg MD, P Lebon MD)
1,
Mireia del Toro
Mireia del Toro
1Manchester Centre for Genomic Medicine, Institute of Human Development, Faculty of Medical and Human Sciences, University of Manchester, Manchester Academic Health Sciences Centre (MAHSC), Manchester, UK (G I Rice PhD, G M A Forte MPhil, M Szynkiewicz MSc, D S Chase PhD, Y J Crow PhD); Erasme University Hospital, Universite Libre de Bruxelles (ULB), Brussels, Belgium, (A Aeby MD, C N Vilain MD, V Wermenbol MD); Medical Molecular Genetics Department, (M S Abdel-Hamid MSc, L Effat PhD), and Clinical Genetics Department, (S A Temtamy PhD, M S Aglan PhD, M S Zaki PhD, G M H Abdel-Salam PhD), Human Genetics and Genome Research Division, National Research Centre, Cairo, Egypt; Bradford Teaching Hospitals NHS Foundation Trust, Bradford, UK (S Ackroyd MB ChB); Department of Clinical Biochemistry, Lancashire Teaching Hospitals NHS Foundation Trust, Preston, UK (R Allcock FRCPath); University Hospitals of Coventry and Warwickshire, Coventry, UK (K M Bailey MRCPCH); Department of Brain and Behavioral Sciences, Unit of Child Neurology and Psychiatry, University of Pavia, Pavia, Italy, (U Balottin MD); Child Neurology and Psychiatry Unit, (U Balottin, I Olivieri MD, S Orcesi MD), and Laboratory of Experimental Neurobiology (C Cereda PhD), C. Mondino National Neurological Institute, Pavia, Italy; Praticien Hospitalier, Unité de Neurologie Pédiatrique et Centre de Référence des Maladies Neuromusculaires, Hôpital Necker-Enfants Malades, Paris, France (C Barnerias MD); Department of Pediatrics, and Department of Neurology and Neurosurgery, Montreal Children’s Hospital, McGill University Health Center, Montreal, QC, Canada (G Bernard MD); Department of Dermatology, Reference Centre for Cutaneous Rare Diseases (MAGEC), Necker-Enfants Malades APHP, Sorbonne Paris Cité, Université Paris Descartes, Institut Imagine, Paris, France (C Bodemer PhD); Department of Pediatrics, Hospital Universitario Araba, Vitoria-Gasteiz, Spain (M P Botella MD); Manchester Centre for Genomic Medicine, Central Manchester University Hospitals NHS Foundation Trust, MAHSC, Manchester, UK (K E Chandler MD, Y J Crow); Department of Paediatric Neurology, University Hospitals of Leicester NHS Trust, Leicester, UK (L Dabydeen MRCPCH); TY Nelson Department of Neurology, and Neuroimmunology Group, the Children’s Hospital at Westmead, University of Sydney, Sydney, NSW, Australia (R C Dale PhD); Nutrition and Metabolism Unit, Hôpital Universitaire des Enfants Reine Fabiola (ULB), Brussels, Belgium (C De Laet MD); Department of Paediatric Neurology, Royal Preston Hospital, Preston, UK (C G E L De Goede FRCPCH); Pediatric Neurology Unit, Hospital Vall d’Hebron, Barcelona, Spain (M del Toro MD); Paediatric Neurology Unit, University Hospital 12 Octubre, Madrid, Spain (N Nunez Enamorado MD, R Simon MD); Department of Clinical and Experimental Sciences, Child Neurology and Psychiatry Unit, University of Brescia, Brescia, Italy (E Fazzi MD); Department of Genetics, BioCruces Health Research Institute, Hospital, Universitario Cruces, Bizkaia, Spain (B Gener MD); Institute of Biomedicine and Translational Medicine, University of Tartu, Tartu, Estonia (M Haldre BSc, P Peterson PhD, K Kisand PhD); General Neurology and Complex Motor Disorders Service, Evelina Children’s Hospital, Guy’s and St Thomas’ NHS Foundation Trust, London, UK (J-P S-M Lin PhD); Department of Paediatric Neurology, Leeds General Infirmary, Leeds, UK (J H Livingston MB ChB); Department of Neurosciences and Behavior Sciences, School of Medicine of Ribeirão Preto, University of São Paulo, São Paulo, Brazil, (C Marques Lourenco MD, W Marques Jr MD); Royal Devon and Exeter Foundation NHS Trust, Exeter, UK (P Oades FRCPCH); Section for Child Neurology, Women and Children’s Division, Oslo University Hospital, Oslo, Norway (M Rasmussen MD); Neuropediatrie, Hopital Gui de Chauliac, and INSERM U1051, INM, Montpellier, France (A Roubertie MD); Center for Genetic Medicine Research and Department of Neurology, Children’s National Medical Center, George Washington University School of Medicine, Washington, DC, USA, (J Loewenstein Schmidt CGC, A Vanderver MD); The Genetic Institute, Ha’Emek Medical Center, Afula and the Rappaport Faculty of Medicine, Technion, Haifa, Israel (S A Shalev MD, R Spiegel MD); Department of Neurology, University of Utah School of Medicine, Salt Lake City, UT, USA (K J Swoboda FACMG); Neurology Department, Royal Manchester Children’s Hospital, Manchester, UK (G Vassallo MD); Centre for Rare Diseases and Personalised Medicine and Department of Medical and Molecular Genetics, School of Clinical and Experimental Medicine, University of Birmingham, UK, and West Midlands Regional Genetics Service, Clinical Genetics Unit, Birmingham Women’s Hospital, Birmingham, UK (J Vogt MRCP); Nottingham Children’s Hospital, Nottingham University Hospitals NHS Trust, Nottingham, UK (W P Whitehouse FRCPCH); Neuropaediatrics, Department of Paediatrics, Mater Dei Hospital, Tal-Qroqq, Malta (D Soler MD); and Université Paris Descartes, EA 1833 and Hôpital Cochin, Service de Virologie, Paris, France, (F Rozenberg MD, P Lebon MD)
1,
Laila Effat
Laila Effat
1Manchester Centre for Genomic Medicine, Institute of Human Development, Faculty of Medical and Human Sciences, University of Manchester, Manchester Academic Health Sciences Centre (MAHSC), Manchester, UK (G I Rice PhD, G M A Forte MPhil, M Szynkiewicz MSc, D S Chase PhD, Y J Crow PhD); Erasme University Hospital, Universite Libre de Bruxelles (ULB), Brussels, Belgium, (A Aeby MD, C N Vilain MD, V Wermenbol MD); Medical Molecular Genetics Department, (M S Abdel-Hamid MSc, L Effat PhD), and Clinical Genetics Department, (S A Temtamy PhD, M S Aglan PhD, M S Zaki PhD, G M H Abdel-Salam PhD), Human Genetics and Genome Research Division, National Research Centre, Cairo, Egypt; Bradford Teaching Hospitals NHS Foundation Trust, Bradford, UK (S Ackroyd MB ChB); Department of Clinical Biochemistry, Lancashire Teaching Hospitals NHS Foundation Trust, Preston, UK (R Allcock FRCPath); University Hospitals of Coventry and Warwickshire, Coventry, UK (K M Bailey MRCPCH); Department of Brain and Behavioral Sciences, Unit of Child Neurology and Psychiatry, University of Pavia, Pavia, Italy, (U Balottin MD); Child Neurology and Psychiatry Unit, (U Balottin, I Olivieri MD, S Orcesi MD), and Laboratory of Experimental Neurobiology (C Cereda PhD), C. Mondino National Neurological Institute, Pavia, Italy; Praticien Hospitalier, Unité de Neurologie Pédiatrique et Centre de Référence des Maladies Neuromusculaires, Hôpital Necker-Enfants Malades, Paris, France (C Barnerias MD); Department of Pediatrics, and Department of Neurology and Neurosurgery, Montreal Children’s Hospital, McGill University Health Center, Montreal, QC, Canada (G Bernard MD); Department of Dermatology, Reference Centre for Cutaneous Rare Diseases (MAGEC), Necker-Enfants Malades APHP, Sorbonne Paris Cité, Université Paris Descartes, Institut Imagine, Paris, France (C Bodemer PhD); Department of Pediatrics, Hospital Universitario Araba, Vitoria-Gasteiz, Spain (M P Botella MD); Manchester Centre for Genomic Medicine, Central Manchester University Hospitals NHS Foundation Trust, MAHSC, Manchester, UK (K E Chandler MD, Y J Crow); Department of Paediatric Neurology, University Hospitals of Leicester NHS Trust, Leicester, UK (L Dabydeen MRCPCH); TY Nelson Department of Neurology, and Neuroimmunology Group, the Children’s Hospital at Westmead, University of Sydney, Sydney, NSW, Australia (R C Dale PhD); Nutrition and Metabolism Unit, Hôpital Universitaire des Enfants Reine Fabiola (ULB), Brussels, Belgium (C De Laet MD); Department of Paediatric Neurology, Royal Preston Hospital, Preston, UK (C G E L De Goede FRCPCH); Pediatric Neurology Unit, Hospital Vall d’Hebron, Barcelona, Spain (M del Toro MD); Paediatric Neurology Unit, University Hospital 12 Octubre, Madrid, Spain (N Nunez Enamorado MD, R Simon MD); Department of Clinical and Experimental Sciences, Child Neurology and Psychiatry Unit, University of Brescia, Brescia, Italy (E Fazzi MD); Department of Genetics, BioCruces Health Research Institute, Hospital, Universitario Cruces, Bizkaia, Spain (B Gener MD); Institute of Biomedicine and Translational Medicine, University of Tartu, Tartu, Estonia (M Haldre BSc, P Peterson PhD, K Kisand PhD); General Neurology and Complex Motor Disorders Service, Evelina Children’s Hospital, Guy’s and St Thomas’ NHS Foundation Trust, London, UK (J-P S-M Lin PhD); Department of Paediatric Neurology, Leeds General Infirmary, Leeds, UK (J H Livingston MB ChB); Department of Neurosciences and Behavior Sciences, School of Medicine of Ribeirão Preto, University of São Paulo, São Paulo, Brazil, (C Marques Lourenco MD, W Marques Jr MD); Royal Devon and Exeter Foundation NHS Trust, Exeter, UK (P Oades FRCPCH); Section for Child Neurology, Women and Children’s Division, Oslo University Hospital, Oslo, Norway (M Rasmussen MD); Neuropediatrie, Hopital Gui de Chauliac, and INSERM U1051, INM, Montpellier, France (A Roubertie MD); Center for Genetic Medicine Research and Department of Neurology, Children’s National Medical Center, George Washington University School of Medicine, Washington, DC, USA, (J Loewenstein Schmidt CGC, A Vanderver MD); The Genetic Institute, Ha’Emek Medical Center, Afula and the Rappaport Faculty of Medicine, Technion, Haifa, Israel (S A Shalev MD, R Spiegel MD); Department of Neurology, University of Utah School of Medicine, Salt Lake City, UT, USA (K J Swoboda FACMG); Neurology Department, Royal Manchester Children’s Hospital, Manchester, UK (G Vassallo MD); Centre for Rare Diseases and Personalised Medicine and Department of Medical and Molecular Genetics, School of Clinical and Experimental Medicine, University of Birmingham, UK, and West Midlands Regional Genetics Service, Clinical Genetics Unit, Birmingham Women’s Hospital, Birmingham, UK (J Vogt MRCP); Nottingham Children’s Hospital, Nottingham University Hospitals NHS Trust, Nottingham, UK (W P Whitehouse FRCPCH); Neuropaediatrics, Department of Paediatrics, Mater Dei Hospital, Tal-Qroqq, Malta (D Soler MD); and Université Paris Descartes, EA 1833 and Hôpital Cochin, Service de Virologie, Paris, France, (F Rozenberg MD, P Lebon MD)
1,
Noemi Nunez Enamorado
Noemi Nunez Enamorado
1Manchester Centre for Genomic Medicine, Institute of Human Development, Faculty of Medical and Human Sciences, University of Manchester, Manchester Academic Health Sciences Centre (MAHSC), Manchester, UK (G I Rice PhD, G M A Forte MPhil, M Szynkiewicz MSc, D S Chase PhD, Y J Crow PhD); Erasme University Hospital, Universite Libre de Bruxelles (ULB), Brussels, Belgium, (A Aeby MD, C N Vilain MD, V Wermenbol MD); Medical Molecular Genetics Department, (M S Abdel-Hamid MSc, L Effat PhD), and Clinical Genetics Department, (S A Temtamy PhD, M S Aglan PhD, M S Zaki PhD, G M H Abdel-Salam PhD), Human Genetics and Genome Research Division, National Research Centre, Cairo, Egypt; Bradford Teaching Hospitals NHS Foundation Trust, Bradford, UK (S Ackroyd MB ChB); Department of Clinical Biochemistry, Lancashire Teaching Hospitals NHS Foundation Trust, Preston, UK (R Allcock FRCPath); University Hospitals of Coventry and Warwickshire, Coventry, UK (K M Bailey MRCPCH); Department of Brain and Behavioral Sciences, Unit of Child Neurology and Psychiatry, University of Pavia, Pavia, Italy, (U Balottin MD); Child Neurology and Psychiatry Unit, (U Balottin, I Olivieri MD, S Orcesi MD), and Laboratory of Experimental Neurobiology (C Cereda PhD), C. Mondino National Neurological Institute, Pavia, Italy; Praticien Hospitalier, Unité de Neurologie Pédiatrique et Centre de Référence des Maladies Neuromusculaires, Hôpital Necker-Enfants Malades, Paris, France (C Barnerias MD); Department of Pediatrics, and Department of Neurology and Neurosurgery, Montreal Children’s Hospital, McGill University Health Center, Montreal, QC, Canada (G Bernard MD); Department of Dermatology, Reference Centre for Cutaneous Rare Diseases (MAGEC), Necker-Enfants Malades APHP, Sorbonne Paris Cité, Université Paris Descartes, Institut Imagine, Paris, France (C Bodemer PhD); Department of Pediatrics, Hospital Universitario Araba, Vitoria-Gasteiz, Spain (M P Botella MD); Manchester Centre for Genomic Medicine, Central Manchester University Hospitals NHS Foundation Trust, MAHSC, Manchester, UK (K E Chandler MD, Y J Crow); Department of Paediatric Neurology, University Hospitals of Leicester NHS Trust, Leicester, UK (L Dabydeen MRCPCH); TY Nelson Department of Neurology, and Neuroimmunology Group, the Children’s Hospital at Westmead, University of Sydney, Sydney, NSW, Australia (R C Dale PhD); Nutrition and Metabolism Unit, Hôpital Universitaire des Enfants Reine Fabiola (ULB), Brussels, Belgium (C De Laet MD); Department of Paediatric Neurology, Royal Preston Hospital, Preston, UK (C G E L De Goede FRCPCH); Pediatric Neurology Unit, Hospital Vall d’Hebron, Barcelona, Spain (M del Toro MD); Paediatric Neurology Unit, University Hospital 12 Octubre, Madrid, Spain (N Nunez Enamorado MD, R Simon MD); Department of Clinical and Experimental Sciences, Child Neurology and Psychiatry Unit, University of Brescia, Brescia, Italy (E Fazzi MD); Department of Genetics, BioCruces Health Research Institute, Hospital, Universitario Cruces, Bizkaia, Spain (B Gener MD); Institute of Biomedicine and Translational Medicine, University of Tartu, Tartu, Estonia (M Haldre BSc, P Peterson PhD, K Kisand PhD); General Neurology and Complex Motor Disorders Service, Evelina Children’s Hospital, Guy’s and St Thomas’ NHS Foundation Trust, London, UK (J-P S-M Lin PhD); Department of Paediatric Neurology, Leeds General Infirmary, Leeds, UK (J H Livingston MB ChB); Department of Neurosciences and Behavior Sciences, School of Medicine of Ribeirão Preto, University of São Paulo, São Paulo, Brazil, (C Marques Lourenco MD, W Marques Jr MD); Royal Devon and Exeter Foundation NHS Trust, Exeter, UK (P Oades FRCPCH); Section for Child Neurology, Women and Children’s Division, Oslo University Hospital, Oslo, Norway (M Rasmussen MD); Neuropediatrie, Hopital Gui de Chauliac, and INSERM U1051, INM, Montpellier, France (A Roubertie MD); Center for Genetic Medicine Research and Department of Neurology, Children’s National Medical Center, George Washington University School of Medicine, Washington, DC, USA, (J Loewenstein Schmidt CGC, A Vanderver MD); The Genetic Institute, Ha’Emek Medical Center, Afula and the Rappaport Faculty of Medicine, Technion, Haifa, Israel (S A Shalev MD, R Spiegel MD); Department of Neurology, University of Utah School of Medicine, Salt Lake City, UT, USA (K J Swoboda FACMG); Neurology Department, Royal Manchester Children’s Hospital, Manchester, UK (G Vassallo MD); Centre for Rare Diseases and Personalised Medicine and Department of Medical and Molecular Genetics, School of Clinical and Experimental Medicine, University of Birmingham, UK, and West Midlands Regional Genetics Service, Clinical Genetics Unit, Birmingham Women’s Hospital, Birmingham, UK (J Vogt MRCP); Nottingham Children’s Hospital, Nottingham University Hospitals NHS Trust, Nottingham, UK (W P Whitehouse FRCPCH); Neuropaediatrics, Department of Paediatrics, Mater Dei Hospital, Tal-Qroqq, Malta (D Soler MD); and Université Paris Descartes, EA 1833 and Hôpital Cochin, Service de Virologie, Paris, France, (F Rozenberg MD, P Lebon MD)
1,
Elisa Fazzi
Elisa Fazzi
1Manchester Centre for Genomic Medicine, Institute of Human Development, Faculty of Medical and Human Sciences, University of Manchester, Manchester Academic Health Sciences Centre (MAHSC), Manchester, UK (G I Rice PhD, G M A Forte MPhil, M Szynkiewicz MSc, D S Chase PhD, Y J Crow PhD); Erasme University Hospital, Universite Libre de Bruxelles (ULB), Brussels, Belgium, (A Aeby MD, C N Vilain MD, V Wermenbol MD); Medical Molecular Genetics Department, (M S Abdel-Hamid MSc, L Effat PhD), and Clinical Genetics Department, (S A Temtamy PhD, M S Aglan PhD, M S Zaki PhD, G M H Abdel-Salam PhD), Human Genetics and Genome Research Division, National Research Centre, Cairo, Egypt; Bradford Teaching Hospitals NHS Foundation Trust, Bradford, UK (S Ackroyd MB ChB); Department of Clinical Biochemistry, Lancashire Teaching Hospitals NHS Foundation Trust, Preston, UK (R Allcock FRCPath); University Hospitals of Coventry and Warwickshire, Coventry, UK (K M Bailey MRCPCH); Department of Brain and Behavioral Sciences, Unit of Child Neurology and Psychiatry, University of Pavia, Pavia, Italy, (U Balottin MD); Child Neurology and Psychiatry Unit, (U Balottin, I Olivieri MD, S Orcesi MD), and Laboratory of Experimental Neurobiology (C Cereda PhD), C. Mondino National Neurological Institute, Pavia, Italy; Praticien Hospitalier, Unité de Neurologie Pédiatrique et Centre de Référence des Maladies Neuromusculaires, Hôpital Necker-Enfants Malades, Paris, France (C Barnerias MD); Department of Pediatrics, and Department of Neurology and Neurosurgery, Montreal Children’s Hospital, McGill University Health Center, Montreal, QC, Canada (G Bernard MD); Department of Dermatology, Reference Centre for Cutaneous Rare Diseases (MAGEC), Necker-Enfants Malades APHP, Sorbonne Paris Cité, Université Paris Descartes, Institut Imagine, Paris, France (C Bodemer PhD); Department of Pediatrics, Hospital Universitario Araba, Vitoria-Gasteiz, Spain (M P Botella MD); Manchester Centre for Genomic Medicine, Central Manchester University Hospitals NHS Foundation Trust, MAHSC, Manchester, UK (K E Chandler MD, Y J Crow); Department of Paediatric Neurology, University Hospitals of Leicester NHS Trust, Leicester, UK (L Dabydeen MRCPCH); TY Nelson Department of Neurology, and Neuroimmunology Group, the Children’s Hospital at Westmead, University of Sydney, Sydney, NSW, Australia (R C Dale PhD); Nutrition and Metabolism Unit, Hôpital Universitaire des Enfants Reine Fabiola (ULB), Brussels, Belgium (C De Laet MD); Department of Paediatric Neurology, Royal Preston Hospital, Preston, UK (C G E L De Goede FRCPCH); Pediatric Neurology Unit, Hospital Vall d’Hebron, Barcelona, Spain (M del Toro MD); Paediatric Neurology Unit, University Hospital 12 Octubre, Madrid, Spain (N Nunez Enamorado MD, R Simon MD); Department of Clinical and Experimental Sciences, Child Neurology and Psychiatry Unit, University of Brescia, Brescia, Italy (E Fazzi MD); Department of Genetics, BioCruces Health Research Institute, Hospital, Universitario Cruces, Bizkaia, Spain (B Gener MD); Institute of Biomedicine and Translational Medicine, University of Tartu, Tartu, Estonia (M Haldre BSc, P Peterson PhD, K Kisand PhD); General Neurology and Complex Motor Disorders Service, Evelina Children’s Hospital, Guy’s and St Thomas’ NHS Foundation Trust, London, UK (J-P S-M Lin PhD); Department of Paediatric Neurology, Leeds General Infirmary, Leeds, UK (J H Livingston MB ChB); Department of Neurosciences and Behavior Sciences, School of Medicine of Ribeirão Preto, University of São Paulo, São Paulo, Brazil, (C Marques Lourenco MD, W Marques Jr MD); Royal Devon and Exeter Foundation NHS Trust, Exeter, UK (P Oades FRCPCH); Section for Child Neurology, Women and Children’s Division, Oslo University Hospital, Oslo, Norway (M Rasmussen MD); Neuropediatrie, Hopital Gui de Chauliac, and INSERM U1051, INM, Montpellier, France (A Roubertie MD); Center for Genetic Medicine Research and Department of Neurology, Children’s National Medical Center, George Washington University School of Medicine, Washington, DC, USA, (J Loewenstein Schmidt CGC, A Vanderver MD); The Genetic Institute, Ha’Emek Medical Center, Afula and the Rappaport Faculty of Medicine, Technion, Haifa, Israel (S A Shalev MD, R Spiegel MD); Department of Neurology, University of Utah School of Medicine, Salt Lake City, UT, USA (K J Swoboda FACMG); Neurology Department, Royal Manchester Children’s Hospital, Manchester, UK (G Vassallo MD); Centre for Rare Diseases and Personalised Medicine and Department of Medical and Molecular Genetics, School of Clinical and Experimental Medicine, University of Birmingham, UK, and West Midlands Regional Genetics Service, Clinical Genetics Unit, Birmingham Women’s Hospital, Birmingham, UK (J Vogt MRCP); Nottingham Children’s Hospital, Nottingham University Hospitals NHS Trust, Nottingham, UK (W P Whitehouse FRCPCH); Neuropaediatrics, Department of Paediatrics, Mater Dei Hospital, Tal-Qroqq, Malta (D Soler MD); and Université Paris Descartes, EA 1833 and Hôpital Cochin, Service de Virologie, Paris, France, (F Rozenberg MD, P Lebon MD)
1,
Blanca Gener
Blanca Gener
1Manchester Centre for Genomic Medicine, Institute of Human Development, Faculty of Medical and Human Sciences, University of Manchester, Manchester Academic Health Sciences Centre (MAHSC), Manchester, UK (G I Rice PhD, G M A Forte MPhil, M Szynkiewicz MSc, D S Chase PhD, Y J Crow PhD); Erasme University Hospital, Universite Libre de Bruxelles (ULB), Brussels, Belgium, (A Aeby MD, C N Vilain MD, V Wermenbol MD); Medical Molecular Genetics Department, (M S Abdel-Hamid MSc, L Effat PhD), and Clinical Genetics Department, (S A Temtamy PhD, M S Aglan PhD, M S Zaki PhD, G M H Abdel-Salam PhD), Human Genetics and Genome Research Division, National Research Centre, Cairo, Egypt; Bradford Teaching Hospitals NHS Foundation Trust, Bradford, UK (S Ackroyd MB ChB); Department of Clinical Biochemistry, Lancashire Teaching Hospitals NHS Foundation Trust, Preston, UK (R Allcock FRCPath); University Hospitals of Coventry and Warwickshire, Coventry, UK (K M Bailey MRCPCH); Department of Brain and Behavioral Sciences, Unit of Child Neurology and Psychiatry, University of Pavia, Pavia, Italy, (U Balottin MD); Child Neurology and Psychiatry Unit, (U Balottin, I Olivieri MD, S Orcesi MD), and Laboratory of Experimental Neurobiology (C Cereda PhD), C. Mondino National Neurological Institute, Pavia, Italy; Praticien Hospitalier, Unité de Neurologie Pédiatrique et Centre de Référence des Maladies Neuromusculaires, Hôpital Necker-Enfants Malades, Paris, France (C Barnerias MD); Department of Pediatrics, and Department of Neurology and Neurosurgery, Montreal Children’s Hospital, McGill University Health Center, Montreal, QC, Canada (G Bernard MD); Department of Dermatology, Reference Centre for Cutaneous Rare Diseases (MAGEC), Necker-Enfants Malades APHP, Sorbonne Paris Cité, Université Paris Descartes, Institut Imagine, Paris, France (C Bodemer PhD); Department of Pediatrics, Hospital Universitario Araba, Vitoria-Gasteiz, Spain (M P Botella MD); Manchester Centre for Genomic Medicine, Central Manchester University Hospitals NHS Foundation Trust, MAHSC, Manchester, UK (K E Chandler MD, Y J Crow); Department of Paediatric Neurology, University Hospitals of Leicester NHS Trust, Leicester, UK (L Dabydeen MRCPCH); TY Nelson Department of Neurology, and Neuroimmunology Group, the Children’s Hospital at Westmead, University of Sydney, Sydney, NSW, Australia (R C Dale PhD); Nutrition and Metabolism Unit, Hôpital Universitaire des Enfants Reine Fabiola (ULB), Brussels, Belgium (C De Laet MD); Department of Paediatric Neurology, Royal Preston Hospital, Preston, UK (C G E L De Goede FRCPCH); Pediatric Neurology Unit, Hospital Vall d’Hebron, Barcelona, Spain (M del Toro MD); Paediatric Neurology Unit, University Hospital 12 Octubre, Madrid, Spain (N Nunez Enamorado MD, R Simon MD); Department of Clinical and Experimental Sciences, Child Neurology and Psychiatry Unit, University of Brescia, Brescia, Italy (E Fazzi MD); Department of Genetics, BioCruces Health Research Institute, Hospital, Universitario Cruces, Bizkaia, Spain (B Gener MD); Institute of Biomedicine and Translational Medicine, University of Tartu, Tartu, Estonia (M Haldre BSc, P Peterson PhD, K Kisand PhD); General Neurology and Complex Motor Disorders Service, Evelina Children’s Hospital, Guy’s and St Thomas’ NHS Foundation Trust, London, UK (J-P S-M Lin PhD); Department of Paediatric Neurology, Leeds General Infirmary, Leeds, UK (J H Livingston MB ChB); Department of Neurosciences and Behavior Sciences, School of Medicine of Ribeirão Preto, University of São Paulo, São Paulo, Brazil, (C Marques Lourenco MD, W Marques Jr MD); Royal Devon and Exeter Foundation NHS Trust, Exeter, UK (P Oades FRCPCH); Section for Child Neurology, Women and Children’s Division, Oslo University Hospital, Oslo, Norway (M Rasmussen MD); Neuropediatrie, Hopital Gui de Chauliac, and INSERM U1051, INM, Montpellier, France (A Roubertie MD); Center for Genetic Medicine Research and Department of Neurology, Children’s National Medical Center, George Washington University School of Medicine, Washington, DC, USA, (J Loewenstein Schmidt CGC, A Vanderver MD); The Genetic Institute, Ha’Emek Medical Center, Afula and the Rappaport Faculty of Medicine, Technion, Haifa, Israel (S A Shalev MD, R Spiegel MD); Department of Neurology, University of Utah School of Medicine, Salt Lake City, UT, USA (K J Swoboda FACMG); Neurology Department, Royal Manchester Children’s Hospital, Manchester, UK (G Vassallo MD); Centre for Rare Diseases and Personalised Medicine and Department of Medical and Molecular Genetics, School of Clinical and Experimental Medicine, University of Birmingham, UK, and West Midlands Regional Genetics Service, Clinical Genetics Unit, Birmingham Women’s Hospital, Birmingham, UK (J Vogt MRCP); Nottingham Children’s Hospital, Nottingham University Hospitals NHS Trust, Nottingham, UK (W P Whitehouse FRCPCH); Neuropaediatrics, Department of Paediatrics, Mater Dei Hospital, Tal-Qroqq, Malta (D Soler MD); and Université Paris Descartes, EA 1833 and Hôpital Cochin, Service de Virologie, Paris, France, (F Rozenberg MD, P Lebon MD)
1,
Madli Haldre
Madli Haldre
1Manchester Centre for Genomic Medicine, Institute of Human Development, Faculty of Medical and Human Sciences, University of Manchester, Manchester Academic Health Sciences Centre (MAHSC), Manchester, UK (G I Rice PhD, G M A Forte MPhil, M Szynkiewicz MSc, D S Chase PhD, Y J Crow PhD); Erasme University Hospital, Universite Libre de Bruxelles (ULB), Brussels, Belgium, (A Aeby MD, C N Vilain MD, V Wermenbol MD); Medical Molecular Genetics Department, (M S Abdel-Hamid MSc, L Effat PhD), and Clinical Genetics Department, (S A Temtamy PhD, M S Aglan PhD, M S Zaki PhD, G M H Abdel-Salam PhD), Human Genetics and Genome Research Division, National Research Centre, Cairo, Egypt; Bradford Teaching Hospitals NHS Foundation Trust, Bradford, UK (S Ackroyd MB ChB); Department of Clinical Biochemistry, Lancashire Teaching Hospitals NHS Foundation Trust, Preston, UK (R Allcock FRCPath); University Hospitals of Coventry and Warwickshire, Coventry, UK (K M Bailey MRCPCH); Department of Brain and Behavioral Sciences, Unit of Child Neurology and Psychiatry, University of Pavia, Pavia, Italy, (U Balottin MD); Child Neurology and Psychiatry Unit, (U Balottin, I Olivieri MD, S Orcesi MD), and Laboratory of Experimental Neurobiology (C Cereda PhD), C. Mondino National Neurological Institute, Pavia, Italy; Praticien Hospitalier, Unité de Neurologie Pédiatrique et Centre de Référence des Maladies Neuromusculaires, Hôpital Necker-Enfants Malades, Paris, France (C Barnerias MD); Department of Pediatrics, and Department of Neurology and Neurosurgery, Montreal Children’s Hospital, McGill University Health Center, Montreal, QC, Canada (G Bernard MD); Department of Dermatology, Reference Centre for Cutaneous Rare Diseases (MAGEC), Necker-Enfants Malades APHP, Sorbonne Paris Cité, Université Paris Descartes, Institut Imagine, Paris, France (C Bodemer PhD); Department of Pediatrics, Hospital Universitario Araba, Vitoria-Gasteiz, Spain (M P Botella MD); Manchester Centre for Genomic Medicine, Central Manchester University Hospitals NHS Foundation Trust, MAHSC, Manchester, UK (K E Chandler MD, Y J Crow); Department of Paediatric Neurology, University Hospitals of Leicester NHS Trust, Leicester, UK (L Dabydeen MRCPCH); TY Nelson Department of Neurology, and Neuroimmunology Group, the Children’s Hospital at Westmead, University of Sydney, Sydney, NSW, Australia (R C Dale PhD); Nutrition and Metabolism Unit, Hôpital Universitaire des Enfants Reine Fabiola (ULB), Brussels, Belgium (C De Laet MD); Department of Paediatric Neurology, Royal Preston Hospital, Preston, UK (C G E L De Goede FRCPCH); Pediatric Neurology Unit, Hospital Vall d’Hebron, Barcelona, Spain (M del Toro MD); Paediatric Neurology Unit, University Hospital 12 Octubre, Madrid, Spain (N Nunez Enamorado MD, R Simon MD); Department of Clinical and Experimental Sciences, Child Neurology and Psychiatry Unit, University of Brescia, Brescia, Italy (E Fazzi MD); Department of Genetics, BioCruces Health Research Institute, Hospital, Universitario Cruces, Bizkaia, Spain (B Gener MD); Institute of Biomedicine and Translational Medicine, University of Tartu, Tartu, Estonia (M Haldre BSc, P Peterson PhD, K Kisand PhD); General Neurology and Complex Motor Disorders Service, Evelina Children’s Hospital, Guy’s and St Thomas’ NHS Foundation Trust, London, UK (J-P S-M Lin PhD); Department of Paediatric Neurology, Leeds General Infirmary, Leeds, UK (J H Livingston MB ChB); Department of Neurosciences and Behavior Sciences, School of Medicine of Ribeirão Preto, University of São Paulo, São Paulo, Brazil, (C Marques Lourenco MD, W Marques Jr MD); Royal Devon and Exeter Foundation NHS Trust, Exeter, UK (P Oades FRCPCH); Section for Child Neurology, Women and Children’s Division, Oslo University Hospital, Oslo, Norway (M Rasmussen MD); Neuropediatrie, Hopital Gui de Chauliac, and INSERM U1051, INM, Montpellier, France (A Roubertie MD); Center for Genetic Medicine Research and Department of Neurology, Children’s National Medical Center, George Washington University School of Medicine, Washington, DC, USA, (J Loewenstein Schmidt CGC, A Vanderver MD); The Genetic Institute, Ha’Emek Medical Center, Afula and the Rappaport Faculty of Medicine, Technion, Haifa, Israel (S A Shalev MD, R Spiegel MD); Department of Neurology, University of Utah School of Medicine, Salt Lake City, UT, USA (K J Swoboda FACMG); Neurology Department, Royal Manchester Children’s Hospital, Manchester, UK (G Vassallo MD); Centre for Rare Diseases and Personalised Medicine and Department of Medical and Molecular Genetics, School of Clinical and Experimental Medicine, University of Birmingham, UK, and West Midlands Regional Genetics Service, Clinical Genetics Unit, Birmingham Women’s Hospital, Birmingham, UK (J Vogt MRCP); Nottingham Children’s Hospital, Nottingham University Hospitals NHS Trust, Nottingham, UK (W P Whitehouse FRCPCH); Neuropaediatrics, Department of Paediatrics, Mater Dei Hospital, Tal-Qroqq, Malta (D Soler MD); and Université Paris Descartes, EA 1833 and Hôpital Cochin, Service de Virologie, Paris, France, (F Rozenberg MD, P Lebon MD)
1,
Jean-Pierre S-M Lin
Jean-Pierre S-M Lin
1Manchester Centre for Genomic Medicine, Institute of Human Development, Faculty of Medical and Human Sciences, University of Manchester, Manchester Academic Health Sciences Centre (MAHSC), Manchester, UK (G I Rice PhD, G M A Forte MPhil, M Szynkiewicz MSc, D S Chase PhD, Y J Crow PhD); Erasme University Hospital, Universite Libre de Bruxelles (ULB), Brussels, Belgium, (A Aeby MD, C N Vilain MD, V Wermenbol MD); Medical Molecular Genetics Department, (M S Abdel-Hamid MSc, L Effat PhD), and Clinical Genetics Department, (S A Temtamy PhD, M S Aglan PhD, M S Zaki PhD, G M H Abdel-Salam PhD), Human Genetics and Genome Research Division, National Research Centre, Cairo, Egypt; Bradford Teaching Hospitals NHS Foundation Trust, Bradford, UK (S Ackroyd MB ChB); Department of Clinical Biochemistry, Lancashire Teaching Hospitals NHS Foundation Trust, Preston, UK (R Allcock FRCPath); University Hospitals of Coventry and Warwickshire, Coventry, UK (K M Bailey MRCPCH); Department of Brain and Behavioral Sciences, Unit of Child Neurology and Psychiatry, University of Pavia, Pavia, Italy, (U Balottin MD); Child Neurology and Psychiatry Unit, (U Balottin, I Olivieri MD, S Orcesi MD), and Laboratory of Experimental Neurobiology (C Cereda PhD), C. Mondino National Neurological Institute, Pavia, Italy; Praticien Hospitalier, Unité de Neurologie Pédiatrique et Centre de Référence des Maladies Neuromusculaires, Hôpital Necker-Enfants Malades, Paris, France (C Barnerias MD); Department of Pediatrics, and Department of Neurology and Neurosurgery, Montreal Children’s Hospital, McGill University Health Center, Montreal, QC, Canada (G Bernard MD); Department of Dermatology, Reference Centre for Cutaneous Rare Diseases (MAGEC), Necker-Enfants Malades APHP, Sorbonne Paris Cité, Université Paris Descartes, Institut Imagine, Paris, France (C Bodemer PhD); Department of Pediatrics, Hospital Universitario Araba, Vitoria-Gasteiz, Spain (M P Botella MD); Manchester Centre for Genomic Medicine, Central Manchester University Hospitals NHS Foundation Trust, MAHSC, Manchester, UK (K E Chandler MD, Y J Crow); Department of Paediatric Neurology, University Hospitals of Leicester NHS Trust, Leicester, UK (L Dabydeen MRCPCH); TY Nelson Department of Neurology, and Neuroimmunology Group, the Children’s Hospital at Westmead, University of Sydney, Sydney, NSW, Australia (R C Dale PhD); Nutrition and Metabolism Unit, Hôpital Universitaire des Enfants Reine Fabiola (ULB), Brussels, Belgium (C De Laet MD); Department of Paediatric Neurology, Royal Preston Hospital, Preston, UK (C G E L De Goede FRCPCH); Pediatric Neurology Unit, Hospital Vall d’Hebron, Barcelona, Spain (M del Toro MD); Paediatric Neurology Unit, University Hospital 12 Octubre, Madrid, Spain (N Nunez Enamorado MD, R Simon MD); Department of Clinical and Experimental Sciences, Child Neurology and Psychiatry Unit, University of Brescia, Brescia, Italy (E Fazzi MD); Department of Genetics, BioCruces Health Research Institute, Hospital, Universitario Cruces, Bizkaia, Spain (B Gener MD); Institute of Biomedicine and Translational Medicine, University of Tartu, Tartu, Estonia (M Haldre BSc, P Peterson PhD, K Kisand PhD); General Neurology and Complex Motor Disorders Service, Evelina Children’s Hospital, Guy’s and St Thomas’ NHS Foundation Trust, London, UK (J-P S-M Lin PhD); Department of Paediatric Neurology, Leeds General Infirmary, Leeds, UK (J H Livingston MB ChB); Department of Neurosciences and Behavior Sciences, School of Medicine of Ribeirão Preto, University of São Paulo, São Paulo, Brazil, (C Marques Lourenco MD, W Marques Jr MD); Royal Devon and Exeter Foundation NHS Trust, Exeter, UK (P Oades FRCPCH); Section for Child Neurology, Women and Children’s Division, Oslo University Hospital, Oslo, Norway (M Rasmussen MD); Neuropediatrie, Hopital Gui de Chauliac, and INSERM U1051, INM, Montpellier, France (A Roubertie MD); Center for Genetic Medicine Research and Department of Neurology, Children’s National Medical Center, George Washington University School of Medicine, Washington, DC, USA, (J Loewenstein Schmidt CGC, A Vanderver MD); The Genetic Institute, Ha’Emek Medical Center, Afula and the Rappaport Faculty of Medicine, Technion, Haifa, Israel (S A Shalev MD, R Spiegel MD); Department of Neurology, University of Utah School of Medicine, Salt Lake City, UT, USA (K J Swoboda FACMG); Neurology Department, Royal Manchester Children’s Hospital, Manchester, UK (G Vassallo MD); Centre for Rare Diseases and Personalised Medicine and Department of Medical and Molecular Genetics, School of Clinical and Experimental Medicine, University of Birmingham, UK, and West Midlands Regional Genetics Service, Clinical Genetics Unit, Birmingham Women’s Hospital, Birmingham, UK (J Vogt MRCP); Nottingham Children’s Hospital, Nottingham University Hospitals NHS Trust, Nottingham, UK (W P Whitehouse FRCPCH); Neuropaediatrics, Department of Paediatrics, Mater Dei Hospital, Tal-Qroqq, Malta (D Soler MD); and Université Paris Descartes, EA 1833 and Hôpital Cochin, Service de Virologie, Paris, France, (F Rozenberg MD, P Lebon MD)
1,
John H Livingston
John H Livingston
1Manchester Centre for Genomic Medicine, Institute of Human Development, Faculty of Medical and Human Sciences, University of Manchester, Manchester Academic Health Sciences Centre (MAHSC), Manchester, UK (G I Rice PhD, G M A Forte MPhil, M Szynkiewicz MSc, D S Chase PhD, Y J Crow PhD); Erasme University Hospital, Universite Libre de Bruxelles (ULB), Brussels, Belgium, (A Aeby MD, C N Vilain MD, V Wermenbol MD); Medical Molecular Genetics Department, (M S Abdel-Hamid MSc, L Effat PhD), and Clinical Genetics Department, (S A Temtamy PhD, M S Aglan PhD, M S Zaki PhD, G M H Abdel-Salam PhD), Human Genetics and Genome Research Division, National Research Centre, Cairo, Egypt; Bradford Teaching Hospitals NHS Foundation Trust, Bradford, UK (S Ackroyd MB ChB); Department of Clinical Biochemistry, Lancashire Teaching Hospitals NHS Foundation Trust, Preston, UK (R Allcock FRCPath); University Hospitals of Coventry and Warwickshire, Coventry, UK (K M Bailey MRCPCH); Department of Brain and Behavioral Sciences, Unit of Child Neurology and Psychiatry, University of Pavia, Pavia, Italy, (U Balottin MD); Child Neurology and Psychiatry Unit, (U Balottin, I Olivieri MD, S Orcesi MD), and Laboratory of Experimental Neurobiology (C Cereda PhD), C. Mondino National Neurological Institute, Pavia, Italy; Praticien Hospitalier, Unité de Neurologie Pédiatrique et Centre de Référence des Maladies Neuromusculaires, Hôpital Necker-Enfants Malades, Paris, France (C Barnerias MD); Department of Pediatrics, and Department of Neurology and Neurosurgery, Montreal Children’s Hospital, McGill University Health Center, Montreal, QC, Canada (G Bernard MD); Department of Dermatology, Reference Centre for Cutaneous Rare Diseases (MAGEC), Necker-Enfants Malades APHP, Sorbonne Paris Cité, Université Paris Descartes, Institut Imagine, Paris, France (C Bodemer PhD); Department of Pediatrics, Hospital Universitario Araba, Vitoria-Gasteiz, Spain (M P Botella MD); Manchester Centre for Genomic Medicine, Central Manchester University Hospitals NHS Foundation Trust, MAHSC, Manchester, UK (K E Chandler MD, Y J Crow); Department of Paediatric Neurology, University Hospitals of Leicester NHS Trust, Leicester, UK (L Dabydeen MRCPCH); TY Nelson Department of Neurology, and Neuroimmunology Group, the Children’s Hospital at Westmead, University of Sydney, Sydney, NSW, Australia (R C Dale PhD); Nutrition and Metabolism Unit, Hôpital Universitaire des Enfants Reine Fabiola (ULB), Brussels, Belgium (C De Laet MD); Department of Paediatric Neurology, Royal Preston Hospital, Preston, UK (C G E L De Goede FRCPCH); Pediatric Neurology Unit, Hospital Vall d’Hebron, Barcelona, Spain (M del Toro MD); Paediatric Neurology Unit, University Hospital 12 Octubre, Madrid, Spain (N Nunez Enamorado MD, R Simon MD); Department of Clinical and Experimental Sciences, Child Neurology and Psychiatry Unit, University of Brescia, Brescia, Italy (E Fazzi MD); Department of Genetics, BioCruces Health Research Institute, Hospital, Universitario Cruces, Bizkaia, Spain (B Gener MD); Institute of Biomedicine and Translational Medicine, University of Tartu, Tartu, Estonia (M Haldre BSc, P Peterson PhD, K Kisand PhD); General Neurology and Complex Motor Disorders Service, Evelina Children’s Hospital, Guy’s and St Thomas’ NHS Foundation Trust, London, UK (J-P S-M Lin PhD); Department of Paediatric Neurology, Leeds General Infirmary, Leeds, UK (J H Livingston MB ChB); Department of Neurosciences and Behavior Sciences, School of Medicine of Ribeirão Preto, University of São Paulo, São Paulo, Brazil, (C Marques Lourenco MD, W Marques Jr MD); Royal Devon and Exeter Foundation NHS Trust, Exeter, UK (P Oades FRCPCH); Section for Child Neurology, Women and Children’s Division, Oslo University Hospital, Oslo, Norway (M Rasmussen MD); Neuropediatrie, Hopital Gui de Chauliac, and INSERM U1051, INM, Montpellier, France (A Roubertie MD); Center for Genetic Medicine Research and Department of Neurology, Children’s National Medical Center, George Washington University School of Medicine, Washington, DC, USA, (J Loewenstein Schmidt CGC, A Vanderver MD); The Genetic Institute, Ha’Emek Medical Center, Afula and the Rappaport Faculty of Medicine, Technion, Haifa, Israel (S A Shalev MD, R Spiegel MD); Department of Neurology, University of Utah School of Medicine, Salt Lake City, UT, USA (K J Swoboda FACMG); Neurology Department, Royal Manchester Children’s Hospital, Manchester, UK (G Vassallo MD); Centre for Rare Diseases and Personalised Medicine and Department of Medical and Molecular Genetics, School of Clinical and Experimental Medicine, University of Birmingham, UK, and West Midlands Regional Genetics Service, Clinical Genetics Unit, Birmingham Women’s Hospital, Birmingham, UK (J Vogt MRCP); Nottingham Children’s Hospital, Nottingham University Hospitals NHS Trust, Nottingham, UK (W P Whitehouse FRCPCH); Neuropaediatrics, Department of Paediatrics, Mater Dei Hospital, Tal-Qroqq, Malta (D Soler MD); and Université Paris Descartes, EA 1833 and Hôpital Cochin, Service de Virologie, Paris, France, (F Rozenberg MD, P Lebon MD)
1,
Charles Marques Lourenco
Charles Marques Lourenco
1Manchester Centre for Genomic Medicine, Institute of Human Development, Faculty of Medical and Human Sciences, University of Manchester, Manchester Academic Health Sciences Centre (MAHSC), Manchester, UK (G I Rice PhD, G M A Forte MPhil, M Szynkiewicz MSc, D S Chase PhD, Y J Crow PhD); Erasme University Hospital, Universite Libre de Bruxelles (ULB), Brussels, Belgium, (A Aeby MD, C N Vilain MD, V Wermenbol MD); Medical Molecular Genetics Department, (M S Abdel-Hamid MSc, L Effat PhD), and Clinical Genetics Department, (S A Temtamy PhD, M S Aglan PhD, M S Zaki PhD, G M H Abdel-Salam PhD), Human Genetics and Genome Research Division, National Research Centre, Cairo, Egypt; Bradford Teaching Hospitals NHS Foundation Trust, Bradford, UK (S Ackroyd MB ChB); Department of Clinical Biochemistry, Lancashire Teaching Hospitals NHS Foundation Trust, Preston, UK (R Allcock FRCPath); University Hospitals of Coventry and Warwickshire, Coventry, UK (K M Bailey MRCPCH); Department of Brain and Behavioral Sciences, Unit of Child Neurology and Psychiatry, University of Pavia, Pavia, Italy, (U Balottin MD); Child Neurology and Psychiatry Unit, (U Balottin, I Olivieri MD, S Orcesi MD), and Laboratory of Experimental Neurobiology (C Cereda PhD), C. Mondino National Neurological Institute, Pavia, Italy; Praticien Hospitalier, Unité de Neurologie Pédiatrique et Centre de Référence des Maladies Neuromusculaires, Hôpital Necker-Enfants Malades, Paris, France (C Barnerias MD); Department of Pediatrics, and Department of Neurology and Neurosurgery, Montreal Children’s Hospital, McGill University Health Center, Montreal, QC, Canada (G Bernard MD); Department of Dermatology, Reference Centre for Cutaneous Rare Diseases (MAGEC), Necker-Enfants Malades APHP, Sorbonne Paris Cité, Université Paris Descartes, Institut Imagine, Paris, France (C Bodemer PhD); Department of Pediatrics, Hospital Universitario Araba, Vitoria-Gasteiz, Spain (M P Botella MD); Manchester Centre for Genomic Medicine, Central Manchester University Hospitals NHS Foundation Trust, MAHSC, Manchester, UK (K E Chandler MD, Y J Crow); Department of Paediatric Neurology, University Hospitals of Leicester NHS Trust, Leicester, UK (L Dabydeen MRCPCH); TY Nelson Department of Neurology, and Neuroimmunology Group, the Children’s Hospital at Westmead, University of Sydney, Sydney, NSW, Australia (R C Dale PhD); Nutrition and Metabolism Unit, Hôpital Universitaire des Enfants Reine Fabiola (ULB), Brussels, Belgium (C De Laet MD); Department of Paediatric Neurology, Royal Preston Hospital, Preston, UK (C G E L De Goede FRCPCH); Pediatric Neurology Unit, Hospital Vall d’Hebron, Barcelona, Spain (M del Toro MD); Paediatric Neurology Unit, University Hospital 12 Octubre, Madrid, Spain (N Nunez Enamorado MD, R Simon MD); Department of Clinical and Experimental Sciences, Child Neurology and Psychiatry Unit, University of Brescia, Brescia, Italy (E Fazzi MD); Department of Genetics, BioCruces Health Research Institute, Hospital, Universitario Cruces, Bizkaia, Spain (B Gener MD); Institute of Biomedicine and Translational Medicine, University of Tartu, Tartu, Estonia (M Haldre BSc, P Peterson PhD, K Kisand PhD); General Neurology and Complex Motor Disorders Service, Evelina Children’s Hospital, Guy’s and St Thomas’ NHS Foundation Trust, London, UK (J-P S-M Lin PhD); Department of Paediatric Neurology, Leeds General Infirmary, Leeds, UK (J H Livingston MB ChB); Department of Neurosciences and Behavior Sciences, School of Medicine of Ribeirão Preto, University of São Paulo, São Paulo, Brazil, (C Marques Lourenco MD, W Marques Jr MD); Royal Devon and Exeter Foundation NHS Trust, Exeter, UK (P Oades FRCPCH); Section for Child Neurology, Women and Children’s Division, Oslo University Hospital, Oslo, Norway (M Rasmussen MD); Neuropediatrie, Hopital Gui de Chauliac, and INSERM U1051, INM, Montpellier, France (A Roubertie MD); Center for Genetic Medicine Research and Department of Neurology, Children’s National Medical Center, George Washington University School of Medicine, Washington, DC, USA, (J Loewenstein Schmidt CGC, A Vanderver MD); The Genetic Institute, Ha’Emek Medical Center, Afula and the Rappaport Faculty of Medicine, Technion, Haifa, Israel (S A Shalev MD, R Spiegel MD); Department of Neurology, University of Utah School of Medicine, Salt Lake City, UT, USA (K J Swoboda FACMG); Neurology Department, Royal Manchester Children’s Hospital, Manchester, UK (G Vassallo MD); Centre for Rare Diseases and Personalised Medicine and Department of Medical and Molecular Genetics, School of Clinical and Experimental Medicine, University of Birmingham, UK, and West Midlands Regional Genetics Service, Clinical Genetics Unit, Birmingham Women’s Hospital, Birmingham, UK (J Vogt MRCP); Nottingham Children’s Hospital, Nottingham University Hospitals NHS Trust, Nottingham, UK (W P Whitehouse FRCPCH); Neuropaediatrics, Department of Paediatrics, Mater Dei Hospital, Tal-Qroqq, Malta (D Soler MD); and Université Paris Descartes, EA 1833 and Hôpital Cochin, Service de Virologie, Paris, France, (F Rozenberg MD, P Lebon MD)
1,
Wilson Marques Jr
Wilson Marques Jr
1Manchester Centre for Genomic Medicine, Institute of Human Development, Faculty of Medical and Human Sciences, University of Manchester, Manchester Academic Health Sciences Centre (MAHSC), Manchester, UK (G I Rice PhD, G M A Forte MPhil, M Szynkiewicz MSc, D S Chase PhD, Y J Crow PhD); Erasme University Hospital, Universite Libre de Bruxelles (ULB), Brussels, Belgium, (A Aeby MD, C N Vilain MD, V Wermenbol MD); Medical Molecular Genetics Department, (M S Abdel-Hamid MSc, L Effat PhD), and Clinical Genetics Department, (S A Temtamy PhD, M S Aglan PhD, M S Zaki PhD, G M H Abdel-Salam PhD), Human Genetics and Genome Research Division, National Research Centre, Cairo, Egypt; Bradford Teaching Hospitals NHS Foundation Trust, Bradford, UK (S Ackroyd MB ChB); Department of Clinical Biochemistry, Lancashire Teaching Hospitals NHS Foundation Trust, Preston, UK (R Allcock FRCPath); University Hospitals of Coventry and Warwickshire, Coventry, UK (K M Bailey MRCPCH); Department of Brain and Behavioral Sciences, Unit of Child Neurology and Psychiatry, University of Pavia, Pavia, Italy, (U Balottin MD); Child Neurology and Psychiatry Unit, (U Balottin, I Olivieri MD, S Orcesi MD), and Laboratory of Experimental Neurobiology (C Cereda PhD), C. Mondino National Neurological Institute, Pavia, Italy; Praticien Hospitalier, Unité de Neurologie Pédiatrique et Centre de Référence des Maladies Neuromusculaires, Hôpital Necker-Enfants Malades, Paris, France (C Barnerias MD); Department of Pediatrics, and Department of Neurology and Neurosurgery, Montreal Children’s Hospital, McGill University Health Center, Montreal, QC, Canada (G Bernard MD); Department of Dermatology, Reference Centre for Cutaneous Rare Diseases (MAGEC), Necker-Enfants Malades APHP, Sorbonne Paris Cité, Université Paris Descartes, Institut Imagine, Paris, France (C Bodemer PhD); Department of Pediatrics, Hospital Universitario Araba, Vitoria-Gasteiz, Spain (M P Botella MD); Manchester Centre for Genomic Medicine, Central Manchester University Hospitals NHS Foundation Trust, MAHSC, Manchester, UK (K E Chandler MD, Y J Crow); Department of Paediatric Neurology, University Hospitals of Leicester NHS Trust, Leicester, UK (L Dabydeen MRCPCH); TY Nelson Department of Neurology, and Neuroimmunology Group, the Children’s Hospital at Westmead, University of Sydney, Sydney, NSW, Australia (R C Dale PhD); Nutrition and Metabolism Unit, Hôpital Universitaire des Enfants Reine Fabiola (ULB), Brussels, Belgium (C De Laet MD); Department of Paediatric Neurology, Royal Preston Hospital, Preston, UK (C G E L De Goede FRCPCH); Pediatric Neurology Unit, Hospital Vall d’Hebron, Barcelona, Spain (M del Toro MD); Paediatric Neurology Unit, University Hospital 12 Octubre, Madrid, Spain (N Nunez Enamorado MD, R Simon MD); Department of Clinical and Experimental Sciences, Child Neurology and Psychiatry Unit, University of Brescia, Brescia, Italy (E Fazzi MD); Department of Genetics, BioCruces Health Research Institute, Hospital, Universitario Cruces, Bizkaia, Spain (B Gener MD); Institute of Biomedicine and Translational Medicine, University of Tartu, Tartu, Estonia (M Haldre BSc, P Peterson PhD, K Kisand PhD); General Neurology and Complex Motor Disorders Service, Evelina Children’s Hospital, Guy’s and St Thomas’ NHS Foundation Trust, London, UK (J-P S-M Lin PhD); Department of Paediatric Neurology, Leeds General Infirmary, Leeds, UK (J H Livingston MB ChB); Department of Neurosciences and Behavior Sciences, School of Medicine of Ribeirão Preto, University of São Paulo, São Paulo, Brazil, (C Marques Lourenco MD, W Marques Jr MD); Royal Devon and Exeter Foundation NHS Trust, Exeter, UK (P Oades FRCPCH); Section for Child Neurology, Women and Children’s Division, Oslo University Hospital, Oslo, Norway (M Rasmussen MD); Neuropediatrie, Hopital Gui de Chauliac, and INSERM U1051, INM, Montpellier, France (A Roubertie MD); Center for Genetic Medicine Research and Department of Neurology, Children’s National Medical Center, George Washington University School of Medicine, Washington, DC, USA, (J Loewenstein Schmidt CGC, A Vanderver MD); The Genetic Institute, Ha’Emek Medical Center, Afula and the Rappaport Faculty of Medicine, Technion, Haifa, Israel (S A Shalev MD, R Spiegel MD); Department of Neurology, University of Utah School of Medicine, Salt Lake City, UT, USA (K J Swoboda FACMG); Neurology Department, Royal Manchester Children’s Hospital, Manchester, UK (G Vassallo MD); Centre for Rare Diseases and Personalised Medicine and Department of Medical and Molecular Genetics, School of Clinical and Experimental Medicine, University of Birmingham, UK, and West Midlands Regional Genetics Service, Clinical Genetics Unit, Birmingham Women’s Hospital, Birmingham, UK (J Vogt MRCP); Nottingham Children’s Hospital, Nottingham University Hospitals NHS Trust, Nottingham, UK (W P Whitehouse FRCPCH); Neuropaediatrics, Department of Paediatrics, Mater Dei Hospital, Tal-Qroqq, Malta (D Soler MD); and Université Paris Descartes, EA 1833 and Hôpital Cochin, Service de Virologie, Paris, France, (F Rozenberg MD, P Lebon MD)
1,
Patrick Oades
Patrick Oades
1Manchester Centre for Genomic Medicine, Institute of Human Development, Faculty of Medical and Human Sciences, University of Manchester, Manchester Academic Health Sciences Centre (MAHSC), Manchester, UK (G I Rice PhD, G M A Forte MPhil, M Szynkiewicz MSc, D S Chase PhD, Y J Crow PhD); Erasme University Hospital, Universite Libre de Bruxelles (ULB), Brussels, Belgium, (A Aeby MD, C N Vilain MD, V Wermenbol MD); Medical Molecular Genetics Department, (M S Abdel-Hamid MSc, L Effat PhD), and Clinical Genetics Department, (S A Temtamy PhD, M S Aglan PhD, M S Zaki PhD, G M H Abdel-Salam PhD), Human Genetics and Genome Research Division, National Research Centre, Cairo, Egypt; Bradford Teaching Hospitals NHS Foundation Trust, Bradford, UK (S Ackroyd MB ChB); Department of Clinical Biochemistry, Lancashire Teaching Hospitals NHS Foundation Trust, Preston, UK (R Allcock FRCPath); University Hospitals of Coventry and Warwickshire, Coventry, UK (K M Bailey MRCPCH); Department of Brain and Behavioral Sciences, Unit of Child Neurology and Psychiatry, University of Pavia, Pavia, Italy, (U Balottin MD); Child Neurology and Psychiatry Unit, (U Balottin, I Olivieri MD, S Orcesi MD), and Laboratory of Experimental Neurobiology (C Cereda PhD), C. Mondino National Neurological Institute, Pavia, Italy; Praticien Hospitalier, Unité de Neurologie Pédiatrique et Centre de Référence des Maladies Neuromusculaires, Hôpital Necker-Enfants Malades, Paris, France (C Barnerias MD); Department of Pediatrics, and Department of Neurology and Neurosurgery, Montreal Children’s Hospital, McGill University Health Center, Montreal, QC, Canada (G Bernard MD); Department of Dermatology, Reference Centre for Cutaneous Rare Diseases (MAGEC), Necker-Enfants Malades APHP, Sorbonne Paris Cité, Université Paris Descartes, Institut Imagine, Paris, France (C Bodemer PhD); Department of Pediatrics, Hospital Universitario Araba, Vitoria-Gasteiz, Spain (M P Botella MD); Manchester Centre for Genomic Medicine, Central Manchester University Hospitals NHS Foundation Trust, MAHSC, Manchester, UK (K E Chandler MD, Y J Crow); Department of Paediatric Neurology, University Hospitals of Leicester NHS Trust, Leicester, UK (L Dabydeen MRCPCH); TY Nelson Department of Neurology, and Neuroimmunology Group, the Children’s Hospital at Westmead, University of Sydney, Sydney, NSW, Australia (R C Dale PhD); Nutrition and Metabolism Unit, Hôpital Universitaire des Enfants Reine Fabiola (ULB), Brussels, Belgium (C De Laet MD); Department of Paediatric Neurology, Royal Preston Hospital, Preston, UK (C G E L De Goede FRCPCH); Pediatric Neurology Unit, Hospital Vall d’Hebron, Barcelona, Spain (M del Toro MD); Paediatric Neurology Unit, University Hospital 12 Octubre, Madrid, Spain (N Nunez Enamorado MD, R Simon MD); Department of Clinical and Experimental Sciences, Child Neurology and Psychiatry Unit, University of Brescia, Brescia, Italy (E Fazzi MD); Department of Genetics, BioCruces Health Research Institute, Hospital, Universitario Cruces, Bizkaia, Spain (B Gener MD); Institute of Biomedicine and Translational Medicine, University of Tartu, Tartu, Estonia (M Haldre BSc, P Peterson PhD, K Kisand PhD); General Neurology and Complex Motor Disorders Service, Evelina Children’s Hospital, Guy’s and St Thomas’ NHS Foundation Trust, London, UK (J-P S-M Lin PhD); Department of Paediatric Neurology, Leeds General Infirmary, Leeds, UK (J H Livingston MB ChB); Department of Neurosciences and Behavior Sciences, School of Medicine of Ribeirão Preto, University of São Paulo, São Paulo, Brazil, (C Marques Lourenco MD, W Marques Jr MD); Royal Devon and Exeter Foundation NHS Trust, Exeter, UK (P Oades FRCPCH); Section for Child Neurology, Women and Children’s Division, Oslo University Hospital, Oslo, Norway (M Rasmussen MD); Neuropediatrie, Hopital Gui de Chauliac, and INSERM U1051, INM, Montpellier, France (A Roubertie MD); Center for Genetic Medicine Research and Department of Neurology, Children’s National Medical Center, George Washington University School of Medicine, Washington, DC, USA, (J Loewenstein Schmidt CGC, A Vanderver MD); The Genetic Institute, Ha’Emek Medical Center, Afula and the Rappaport Faculty of Medicine, Technion, Haifa, Israel (S A Shalev MD, R Spiegel MD); Department of Neurology, University of Utah School of Medicine, Salt Lake City, UT, USA (K J Swoboda FACMG); Neurology Department, Royal Manchester Children’s Hospital, Manchester, UK (G Vassallo MD); Centre for Rare Diseases and Personalised Medicine and Department of Medical and Molecular Genetics, School of Clinical and Experimental Medicine, University of Birmingham, UK, and West Midlands Regional Genetics Service, Clinical Genetics Unit, Birmingham Women’s Hospital, Birmingham, UK (J Vogt MRCP); Nottingham Children’s Hospital, Nottingham University Hospitals NHS Trust, Nottingham, UK (W P Whitehouse FRCPCH); Neuropaediatrics, Department of Paediatrics, Mater Dei Hospital, Tal-Qroqq, Malta (D Soler MD); and Université Paris Descartes, EA 1833 and Hôpital Cochin, Service de Virologie, Paris, France, (F Rozenberg MD, P Lebon MD)
1,
Pärt Peterson
Pärt Peterson
1Manchester Centre for Genomic Medicine, Institute of Human Development, Faculty of Medical and Human Sciences, University of Manchester, Manchester Academic Health Sciences Centre (MAHSC), Manchester, UK (G I Rice PhD, G M A Forte MPhil, M Szynkiewicz MSc, D S Chase PhD, Y J Crow PhD); Erasme University Hospital, Universite Libre de Bruxelles (ULB), Brussels, Belgium, (A Aeby MD, C N Vilain MD, V Wermenbol MD); Medical Molecular Genetics Department, (M S Abdel-Hamid MSc, L Effat PhD), and Clinical Genetics Department, (S A Temtamy PhD, M S Aglan PhD, M S Zaki PhD, G M H Abdel-Salam PhD), Human Genetics and Genome Research Division, National Research Centre, Cairo, Egypt; Bradford Teaching Hospitals NHS Foundation Trust, Bradford, UK (S Ackroyd MB ChB); Department of Clinical Biochemistry, Lancashire Teaching Hospitals NHS Foundation Trust, Preston, UK (R Allcock FRCPath); University Hospitals of Coventry and Warwickshire, Coventry, UK (K M Bailey MRCPCH); Department of Brain and Behavioral Sciences, Unit of Child Neurology and Psychiatry, University of Pavia, Pavia, Italy, (U Balottin MD); Child Neurology and Psychiatry Unit, (U Balottin, I Olivieri MD, S Orcesi MD), and Laboratory of Experimental Neurobiology (C Cereda PhD), C. Mondino National Neurological Institute, Pavia, Italy; Praticien Hospitalier, Unité de Neurologie Pédiatrique et Centre de Référence des Maladies Neuromusculaires, Hôpital Necker-Enfants Malades, Paris, France (C Barnerias MD); Department of Pediatrics, and Department of Neurology and Neurosurgery, Montreal Children’s Hospital, McGill University Health Center, Montreal, QC, Canada (G Bernard MD); Department of Dermatology, Reference Centre for Cutaneous Rare Diseases (MAGEC), Necker-Enfants Malades APHP, Sorbonne Paris Cité, Université Paris Descartes, Institut Imagine, Paris, France (C Bodemer PhD); Department of Pediatrics, Hospital Universitario Araba, Vitoria-Gasteiz, Spain (M P Botella MD); Manchester Centre for Genomic Medicine, Central Manchester University Hospitals NHS Foundation Trust, MAHSC, Manchester, UK (K E Chandler MD, Y J Crow); Department of Paediatric Neurology, University Hospitals of Leicester NHS Trust, Leicester, UK (L Dabydeen MRCPCH); TY Nelson Department of Neurology, and Neuroimmunology Group, the Children’s Hospital at Westmead, University of Sydney, Sydney, NSW, Australia (R C Dale PhD); Nutrition and Metabolism Unit, Hôpital Universitaire des Enfants Reine Fabiola (ULB), Brussels, Belgium (C De Laet MD); Department of Paediatric Neurology, Royal Preston Hospital, Preston, UK (C G E L De Goede FRCPCH); Pediatric Neurology Unit, Hospital Vall d’Hebron, Barcelona, Spain (M del Toro MD); Paediatric Neurology Unit, University Hospital 12 Octubre, Madrid, Spain (N Nunez Enamorado MD, R Simon MD); Department of Clinical and Experimental Sciences, Child Neurology and Psychiatry Unit, University of Brescia, Brescia, Italy (E Fazzi MD); Department of Genetics, BioCruces Health Research Institute, Hospital, Universitario Cruces, Bizkaia, Spain (B Gener MD); Institute of Biomedicine and Translational Medicine, University of Tartu, Tartu, Estonia (M Haldre BSc, P Peterson PhD, K Kisand PhD); General Neurology and Complex Motor Disorders Service, Evelina Children’s Hospital, Guy’s and St Thomas’ NHS Foundation Trust, London, UK (J-P S-M Lin PhD); Department of Paediatric Neurology, Leeds General Infirmary, Leeds, UK (J H Livingston MB ChB); Department of Neurosciences and Behavior Sciences, School of Medicine of Ribeirão Preto, University of São Paulo, São Paulo, Brazil, (C Marques Lourenco MD, W Marques Jr MD); Royal Devon and Exeter Foundation NHS Trust, Exeter, UK (P Oades FRCPCH); Section for Child Neurology, Women and Children’s Division, Oslo University Hospital, Oslo, Norway (M Rasmussen MD); Neuropediatrie, Hopital Gui de Chauliac, and INSERM U1051, INM, Montpellier, France (A Roubertie MD); Center for Genetic Medicine Research and Department of Neurology, Children’s National Medical Center, George Washington University School of Medicine, Washington, DC, USA, (J Loewenstein Schmidt CGC, A Vanderver MD); The Genetic Institute, Ha’Emek Medical Center, Afula and the Rappaport Faculty of Medicine, Technion, Haifa, Israel (S A Shalev MD, R Spiegel MD); Department of Neurology, University of Utah School of Medicine, Salt Lake City, UT, USA (K J Swoboda FACMG); Neurology Department, Royal Manchester Children’s Hospital, Manchester, UK (G Vassallo MD); Centre for Rare Diseases and Personalised Medicine and Department of Medical and Molecular Genetics, School of Clinical and Experimental Medicine, University of Birmingham, UK, and West Midlands Regional Genetics Service, Clinical Genetics Unit, Birmingham Women’s Hospital, Birmingham, UK (J Vogt MRCP); Nottingham Children’s Hospital, Nottingham University Hospitals NHS Trust, Nottingham, UK (W P Whitehouse FRCPCH); Neuropaediatrics, Department of Paediatrics, Mater Dei Hospital, Tal-Qroqq, Malta (D Soler MD); and Université Paris Descartes, EA 1833 and Hôpital Cochin, Service de Virologie, Paris, France, (F Rozenberg MD, P Lebon MD)
1,
Magnhild Rasmussen
Magnhild Rasmussen
1Manchester Centre for Genomic Medicine, Institute of Human Development, Faculty of Medical and Human Sciences, University of Manchester, Manchester Academic Health Sciences Centre (MAHSC), Manchester, UK (G I Rice PhD, G M A Forte MPhil, M Szynkiewicz MSc, D S Chase PhD, Y J Crow PhD); Erasme University Hospital, Universite Libre de Bruxelles (ULB), Brussels, Belgium, (A Aeby MD, C N Vilain MD, V Wermenbol MD); Medical Molecular Genetics Department, (M S Abdel-Hamid MSc, L Effat PhD), and Clinical Genetics Department, (S A Temtamy PhD, M S Aglan PhD, M S Zaki PhD, G M H Abdel-Salam PhD), Human Genetics and Genome Research Division, National Research Centre, Cairo, Egypt; Bradford Teaching Hospitals NHS Foundation Trust, Bradford, UK (S Ackroyd MB ChB); Department of Clinical Biochemistry, Lancashire Teaching Hospitals NHS Foundation Trust, Preston, UK (R Allcock FRCPath); University Hospitals of Coventry and Warwickshire, Coventry, UK (K M Bailey MRCPCH); Department of Brain and Behavioral Sciences, Unit of Child Neurology and Psychiatry, University of Pavia, Pavia, Italy, (U Balottin MD); Child Neurology and Psychiatry Unit, (U Balottin, I Olivieri MD, S Orcesi MD), and Laboratory of Experimental Neurobiology (C Cereda PhD), C. Mondino National Neurological Institute, Pavia, Italy; Praticien Hospitalier, Unité de Neurologie Pédiatrique et Centre de Référence des Maladies Neuromusculaires, Hôpital Necker-Enfants Malades, Paris, France (C Barnerias MD); Department of Pediatrics, and Department of Neurology and Neurosurgery, Montreal Children’s Hospital, McGill University Health Center, Montreal, QC, Canada (G Bernard MD); Department of Dermatology, Reference Centre for Cutaneous Rare Diseases (MAGEC), Necker-Enfants Malades APHP, Sorbonne Paris Cité, Université Paris Descartes, Institut Imagine, Paris, France (C Bodemer PhD); Department of Pediatrics, Hospital Universitario Araba, Vitoria-Gasteiz, Spain (M P Botella MD); Manchester Centre for Genomic Medicine, Central Manchester University Hospitals NHS Foundation Trust, MAHSC, Manchester, UK (K E Chandler MD, Y J Crow); Department of Paediatric Neurology, University Hospitals of Leicester NHS Trust, Leicester, UK (L Dabydeen MRCPCH); TY Nelson Department of Neurology, and Neuroimmunology Group, the Children’s Hospital at Westmead, University of Sydney, Sydney, NSW, Australia (R C Dale PhD); Nutrition and Metabolism Unit, Hôpital Universitaire des Enfants Reine Fabiola (ULB), Brussels, Belgium (C De Laet MD); Department of Paediatric Neurology, Royal Preston Hospital, Preston, UK (C G E L De Goede FRCPCH); Pediatric Neurology Unit, Hospital Vall d’Hebron, Barcelona, Spain (M del Toro MD); Paediatric Neurology Unit, University Hospital 12 Octubre, Madrid, Spain (N Nunez Enamorado MD, R Simon MD); Department of Clinical and Experimental Sciences, Child Neurology and Psychiatry Unit, University of Brescia, Brescia, Italy (E Fazzi MD); Department of Genetics, BioCruces Health Research Institute, Hospital, Universitario Cruces, Bizkaia, Spain (B Gener MD); Institute of Biomedicine and Translational Medicine, University of Tartu, Tartu, Estonia (M Haldre BSc, P Peterson PhD, K Kisand PhD); General Neurology and Complex Motor Disorders Service, Evelina Children’s Hospital, Guy’s and St Thomas’ NHS Foundation Trust, London, UK (J-P S-M Lin PhD); Department of Paediatric Neurology, Leeds General Infirmary, Leeds, UK (J H Livingston MB ChB); Department of Neurosciences and Behavior Sciences, School of Medicine of Ribeirão Preto, University of São Paulo, São Paulo, Brazil, (C Marques Lourenco MD, W Marques Jr MD); Royal Devon and Exeter Foundation NHS Trust, Exeter, UK (P Oades FRCPCH); Section for Child Neurology, Women and Children’s Division, Oslo University Hospital, Oslo, Norway (M Rasmussen MD); Neuropediatrie, Hopital Gui de Chauliac, and INSERM U1051, INM, Montpellier, France (A Roubertie MD); Center for Genetic Medicine Research and Department of Neurology, Children’s National Medical Center, George Washington University School of Medicine, Washington, DC, USA, (J Loewenstein Schmidt CGC, A Vanderver MD); The Genetic Institute, Ha’Emek Medical Center, Afula and the Rappaport Faculty of Medicine, Technion, Haifa, Israel (S A Shalev MD, R Spiegel MD); Department of Neurology, University of Utah School of Medicine, Salt Lake City, UT, USA (K J Swoboda FACMG); Neurology Department, Royal Manchester Children’s Hospital, Manchester, UK (G Vassallo MD); Centre for Rare Diseases and Personalised Medicine and Department of Medical and Molecular Genetics, School of Clinical and Experimental Medicine, University of Birmingham, UK, and West Midlands Regional Genetics Service, Clinical Genetics Unit, Birmingham Women’s Hospital, Birmingham, UK (J Vogt MRCP); Nottingham Children’s Hospital, Nottingham University Hospitals NHS Trust, Nottingham, UK (W P Whitehouse FRCPCH); Neuropaediatrics, Department of Paediatrics, Mater Dei Hospital, Tal-Qroqq, Malta (D Soler MD); and Université Paris Descartes, EA 1833 and Hôpital Cochin, Service de Virologie, Paris, France, (F Rozenberg MD, P Lebon MD)
1,
Agathe Roubertie
Agathe Roubertie
1Manchester Centre for Genomic Medicine, Institute of Human Development, Faculty of Medical and Human Sciences, University of Manchester, Manchester Academic Health Sciences Centre (MAHSC), Manchester, UK (G I Rice PhD, G M A Forte MPhil, M Szynkiewicz MSc, D S Chase PhD, Y J Crow PhD); Erasme University Hospital, Universite Libre de Bruxelles (ULB), Brussels, Belgium, (A Aeby MD, C N Vilain MD, V Wermenbol MD); Medical Molecular Genetics Department, (M S Abdel-Hamid MSc, L Effat PhD), and Clinical Genetics Department, (S A Temtamy PhD, M S Aglan PhD, M S Zaki PhD, G M H Abdel-Salam PhD), Human Genetics and Genome Research Division, National Research Centre, Cairo, Egypt; Bradford Teaching Hospitals NHS Foundation Trust, Bradford, UK (S Ackroyd MB ChB); Department of Clinical Biochemistry, Lancashire Teaching Hospitals NHS Foundation Trust, Preston, UK (R Allcock FRCPath); University Hospitals of Coventry and Warwickshire, Coventry, UK (K M Bailey MRCPCH); Department of Brain and Behavioral Sciences, Unit of Child Neurology and Psychiatry, University of Pavia, Pavia, Italy, (U Balottin MD); Child Neurology and Psychiatry Unit, (U Balottin, I Olivieri MD, S Orcesi MD), and Laboratory of Experimental Neurobiology (C Cereda PhD), C. Mondino National Neurological Institute, Pavia, Italy; Praticien Hospitalier, Unité de Neurologie Pédiatrique et Centre de Référence des Maladies Neuromusculaires, Hôpital Necker-Enfants Malades, Paris, France (C Barnerias MD); Department of Pediatrics, and Department of Neurology and Neurosurgery, Montreal Children’s Hospital, McGill University Health Center, Montreal, QC, Canada (G Bernard MD); Department of Dermatology, Reference Centre for Cutaneous Rare Diseases (MAGEC), Necker-Enfants Malades APHP, Sorbonne Paris Cité, Université Paris Descartes, Institut Imagine, Paris, France (C Bodemer PhD); Department of Pediatrics, Hospital Universitario Araba, Vitoria-Gasteiz, Spain (M P Botella MD); Manchester Centre for Genomic Medicine, Central Manchester University Hospitals NHS Foundation Trust, MAHSC, Manchester, UK (K E Chandler MD, Y J Crow); Department of Paediatric Neurology, University Hospitals of Leicester NHS Trust, Leicester, UK (L Dabydeen MRCPCH); TY Nelson Department of Neurology, and Neuroimmunology Group, the Children’s Hospital at Westmead, University of Sydney, Sydney, NSW, Australia (R C Dale PhD); Nutrition and Metabolism Unit, Hôpital Universitaire des Enfants Reine Fabiola (ULB), Brussels, Belgium (C De Laet MD); Department of Paediatric Neurology, Royal Preston Hospital, Preston, UK (C G E L De Goede FRCPCH); Pediatric Neurology Unit, Hospital Vall d’Hebron, Barcelona, Spain (M del Toro MD); Paediatric Neurology Unit, University Hospital 12 Octubre, Madrid, Spain (N Nunez Enamorado MD, R Simon MD); Department of Clinical and Experimental Sciences, Child Neurology and Psychiatry Unit, University of Brescia, Brescia, Italy (E Fazzi MD); Department of Genetics, BioCruces Health Research Institute, Hospital, Universitario Cruces, Bizkaia, Spain (B Gener MD); Institute of Biomedicine and Translational Medicine, University of Tartu, Tartu, Estonia (M Haldre BSc, P Peterson PhD, K Kisand PhD); General Neurology and Complex Motor Disorders Service, Evelina Children’s Hospital, Guy’s and St Thomas’ NHS Foundation Trust, London, UK (J-P S-M Lin PhD); Department of Paediatric Neurology, Leeds General Infirmary, Leeds, UK (J H Livingston MB ChB); Department of Neurosciences and Behavior Sciences, School of Medicine of Ribeirão Preto, University of São Paulo, São Paulo, Brazil, (C Marques Lourenco MD, W Marques Jr MD); Royal Devon and Exeter Foundation NHS Trust, Exeter, UK (P Oades FRCPCH); Section for Child Neurology, Women and Children’s Division, Oslo University Hospital, Oslo, Norway (M Rasmussen MD); Neuropediatrie, Hopital Gui de Chauliac, and INSERM U1051, INM, Montpellier, France (A Roubertie MD); Center for Genetic Medicine Research and Department of Neurology, Children’s National Medical Center, George Washington University School of Medicine, Washington, DC, USA, (J Loewenstein Schmidt CGC, A Vanderver MD); The Genetic Institute, Ha’Emek Medical Center, Afula and the Rappaport Faculty of Medicine, Technion, Haifa, Israel (S A Shalev MD, R Spiegel MD); Department of Neurology, University of Utah School of Medicine, Salt Lake City, UT, USA (K J Swoboda FACMG); Neurology Department, Royal Manchester Children’s Hospital, Manchester, UK (G Vassallo MD); Centre for Rare Diseases and Personalised Medicine and Department of Medical and Molecular Genetics, School of Clinical and Experimental Medicine, University of Birmingham, UK, and West Midlands Regional Genetics Service, Clinical Genetics Unit, Birmingham Women’s Hospital, Birmingham, UK (J Vogt MRCP); Nottingham Children’s Hospital, Nottingham University Hospitals NHS Trust, Nottingham, UK (W P Whitehouse FRCPCH); Neuropaediatrics, Department of Paediatrics, Mater Dei Hospital, Tal-Qroqq, Malta (D Soler MD); and Université Paris Descartes, EA 1833 and Hôpital Cochin, Service de Virologie, Paris, France, (F Rozenberg MD, P Lebon MD)
1,
Johanna Loewenstein Schmidt
Johanna Loewenstein Schmidt
1Manchester Centre for Genomic Medicine, Institute of Human Development, Faculty of Medical and Human Sciences, University of Manchester, Manchester Academic Health Sciences Centre (MAHSC), Manchester, UK (G I Rice PhD, G M A Forte MPhil, M Szynkiewicz MSc, D S Chase PhD, Y J Crow PhD); Erasme University Hospital, Universite Libre de Bruxelles (ULB), Brussels, Belgium, (A Aeby MD, C N Vilain MD, V Wermenbol MD); Medical Molecular Genetics Department, (M S Abdel-Hamid MSc, L Effat PhD), and Clinical Genetics Department, (S A Temtamy PhD, M S Aglan PhD, M S Zaki PhD, G M H Abdel-Salam PhD), Human Genetics and Genome Research Division, National Research Centre, Cairo, Egypt; Bradford Teaching Hospitals NHS Foundation Trust, Bradford, UK (S Ackroyd MB ChB); Department of Clinical Biochemistry, Lancashire Teaching Hospitals NHS Foundation Trust, Preston, UK (R Allcock FRCPath); University Hospitals of Coventry and Warwickshire, Coventry, UK (K M Bailey MRCPCH); Department of Brain and Behavioral Sciences, Unit of Child Neurology and Psychiatry, University of Pavia, Pavia, Italy, (U Balottin MD); Child Neurology and Psychiatry Unit, (U Balottin, I Olivieri MD, S Orcesi MD), and Laboratory of Experimental Neurobiology (C Cereda PhD), C. Mondino National Neurological Institute, Pavia, Italy; Praticien Hospitalier, Unité de Neurologie Pédiatrique et Centre de Référence des Maladies Neuromusculaires, Hôpital Necker-Enfants Malades, Paris, France (C Barnerias MD); Department of Pediatrics, and Department of Neurology and Neurosurgery, Montreal Children’s Hospital, McGill University Health Center, Montreal, QC, Canada (G Bernard MD); Department of Dermatology, Reference Centre for Cutaneous Rare Diseases (MAGEC), Necker-Enfants Malades APHP, Sorbonne Paris Cité, Université Paris Descartes, Institut Imagine, Paris, France (C Bodemer PhD); Department of Pediatrics, Hospital Universitario Araba, Vitoria-Gasteiz, Spain (M P Botella MD); Manchester Centre for Genomic Medicine, Central Manchester University Hospitals NHS Foundation Trust, MAHSC, Manchester, UK (K E Chandler MD, Y J Crow); Department of Paediatric Neurology, University Hospitals of Leicester NHS Trust, Leicester, UK (L Dabydeen MRCPCH); TY Nelson Department of Neurology, and Neuroimmunology Group, the Children’s Hospital at Westmead, University of Sydney, Sydney, NSW, Australia (R C Dale PhD); Nutrition and Metabolism Unit, Hôpital Universitaire des Enfants Reine Fabiola (ULB), Brussels, Belgium (C De Laet MD); Department of Paediatric Neurology, Royal Preston Hospital, Preston, UK (C G E L De Goede FRCPCH); Pediatric Neurology Unit, Hospital Vall d’Hebron, Barcelona, Spain (M del Toro MD); Paediatric Neurology Unit, University Hospital 12 Octubre, Madrid, Spain (N Nunez Enamorado MD, R Simon MD); Department of Clinical and Experimental Sciences, Child Neurology and Psychiatry Unit, University of Brescia, Brescia, Italy (E Fazzi MD); Department of Genetics, BioCruces Health Research Institute, Hospital, Universitario Cruces, Bizkaia, Spain (B Gener MD); Institute of Biomedicine and Translational Medicine, University of Tartu, Tartu, Estonia (M Haldre BSc, P Peterson PhD, K Kisand PhD); General Neurology and Complex Motor Disorders Service, Evelina Children’s Hospital, Guy’s and St Thomas’ NHS Foundation Trust, London, UK (J-P S-M Lin PhD); Department of Paediatric Neurology, Leeds General Infirmary, Leeds, UK (J H Livingston MB ChB); Department of Neurosciences and Behavior Sciences, School of Medicine of Ribeirão Preto, University of São Paulo, São Paulo, Brazil, (C Marques Lourenco MD, W Marques Jr MD); Royal Devon and Exeter Foundation NHS Trust, Exeter, UK (P Oades FRCPCH); Section for Child Neurology, Women and Children’s Division, Oslo University Hospital, Oslo, Norway (M Rasmussen MD); Neuropediatrie, Hopital Gui de Chauliac, and INSERM U1051, INM, Montpellier, France (A Roubertie MD); Center for Genetic Medicine Research and Department of Neurology, Children’s National Medical Center, George Washington University School of Medicine, Washington, DC, USA, (J Loewenstein Schmidt CGC, A Vanderver MD); The Genetic Institute, Ha’Emek Medical Center, Afula and the Rappaport Faculty of Medicine, Technion, Haifa, Israel (S A Shalev MD, R Spiegel MD); Department of Neurology, University of Utah School of Medicine, Salt Lake City, UT, USA (K J Swoboda FACMG); Neurology Department, Royal Manchester Children’s Hospital, Manchester, UK (G Vassallo MD); Centre for Rare Diseases and Personalised Medicine and Department of Medical and Molecular Genetics, School of Clinical and Experimental Medicine, University of Birmingham, UK, and West Midlands Regional Genetics Service, Clinical Genetics Unit, Birmingham Women’s Hospital, Birmingham, UK (J Vogt MRCP); Nottingham Children’s Hospital, Nottingham University Hospitals NHS Trust, Nottingham, UK (W P Whitehouse FRCPCH); Neuropaediatrics, Department of Paediatrics, Mater Dei Hospital, Tal-Qroqq, Malta (D Soler MD); and Université Paris Descartes, EA 1833 and Hôpital Cochin, Service de Virologie, Paris, France, (F Rozenberg MD, P Lebon MD)
1,
Stavit A Shalev
Stavit A Shalev
1Manchester Centre for Genomic Medicine, Institute of Human Development, Faculty of Medical and Human Sciences, University of Manchester, Manchester Academic Health Sciences Centre (MAHSC), Manchester, UK (G I Rice PhD, G M A Forte MPhil, M Szynkiewicz MSc, D S Chase PhD, Y J Crow PhD); Erasme University Hospital, Universite Libre de Bruxelles (ULB), Brussels, Belgium, (A Aeby MD, C N Vilain MD, V Wermenbol MD); Medical Molecular Genetics Department, (M S Abdel-Hamid MSc, L Effat PhD), and Clinical Genetics Department, (S A Temtamy PhD, M S Aglan PhD, M S Zaki PhD, G M H Abdel-Salam PhD), Human Genetics and Genome Research Division, National Research Centre, Cairo, Egypt; Bradford Teaching Hospitals NHS Foundation Trust, Bradford, UK (S Ackroyd MB ChB); Department of Clinical Biochemistry, Lancashire Teaching Hospitals NHS Foundation Trust, Preston, UK (R Allcock FRCPath); University Hospitals of Coventry and Warwickshire, Coventry, UK (K M Bailey MRCPCH); Department of Brain and Behavioral Sciences, Unit of Child Neurology and Psychiatry, University of Pavia, Pavia, Italy, (U Balottin MD); Child Neurology and Psychiatry Unit, (U Balottin, I Olivieri MD, S Orcesi MD), and Laboratory of Experimental Neurobiology (C Cereda PhD), C. Mondino National Neurological Institute, Pavia, Italy; Praticien Hospitalier, Unité de Neurologie Pédiatrique et Centre de Référence des Maladies Neuromusculaires, Hôpital Necker-Enfants Malades, Paris, France (C Barnerias MD); Department of Pediatrics, and Department of Neurology and Neurosurgery, Montreal Children’s Hospital, McGill University Health Center, Montreal, QC, Canada (G Bernard MD); Department of Dermatology, Reference Centre for Cutaneous Rare Diseases (MAGEC), Necker-Enfants Malades APHP, Sorbonne Paris Cité, Université Paris Descartes, Institut Imagine, Paris, France (C Bodemer PhD); Department of Pediatrics, Hospital Universitario Araba, Vitoria-Gasteiz, Spain (M P Botella MD); Manchester Centre for Genomic Medicine, Central Manchester University Hospitals NHS Foundation Trust, MAHSC, Manchester, UK (K E Chandler MD, Y J Crow); Department of Paediatric Neurology, University Hospitals of Leicester NHS Trust, Leicester, UK (L Dabydeen MRCPCH); TY Nelson Department of Neurology, and Neuroimmunology Group, the Children’s Hospital at Westmead, University of Sydney, Sydney, NSW, Australia (R C Dale PhD); Nutrition and Metabolism Unit, Hôpital Universitaire des Enfants Reine Fabiola (ULB), Brussels, Belgium (C De Laet MD); Department of Paediatric Neurology, Royal Preston Hospital, Preston, UK (C G E L De Goede FRCPCH); Pediatric Neurology Unit, Hospital Vall d’Hebron, Barcelona, Spain (M del Toro MD); Paediatric Neurology Unit, University Hospital 12 Octubre, Madrid, Spain (N Nunez Enamorado MD, R Simon MD); Department of Clinical and Experimental Sciences, Child Neurology and Psychiatry Unit, University of Brescia, Brescia, Italy (E Fazzi MD); Department of Genetics, BioCruces Health Research Institute, Hospital, Universitario Cruces, Bizkaia, Spain (B Gener MD); Institute of Biomedicine and Translational Medicine, University of Tartu, Tartu, Estonia (M Haldre BSc, P Peterson PhD, K Kisand PhD); General Neurology and Complex Motor Disorders Service, Evelina Children’s Hospital, Guy’s and St Thomas’ NHS Foundation Trust, London, UK (J-P S-M Lin PhD); Department of Paediatric Neurology, Leeds General Infirmary, Leeds, UK (J H Livingston MB ChB); Department of Neurosciences and Behavior Sciences, School of Medicine of Ribeirão Preto, University of São Paulo, São Paulo, Brazil, (C Marques Lourenco MD, W Marques Jr MD); Royal Devon and Exeter Foundation NHS Trust, Exeter, UK (P Oades FRCPCH); Section for Child Neurology, Women and Children’s Division, Oslo University Hospital, Oslo, Norway (M Rasmussen MD); Neuropediatrie, Hopital Gui de Chauliac, and INSERM U1051, INM, Montpellier, France (A Roubertie MD); Center for Genetic Medicine Research and Department of Neurology, Children’s National Medical Center, George Washington University School of Medicine, Washington, DC, USA, (J Loewenstein Schmidt CGC, A Vanderver MD); The Genetic Institute, Ha’Emek Medical Center, Afula and the Rappaport Faculty of Medicine, Technion, Haifa, Israel (S A Shalev MD, R Spiegel MD); Department of Neurology, University of Utah School of Medicine, Salt Lake City, UT, USA (K J Swoboda FACMG); Neurology Department, Royal Manchester Children’s Hospital, Manchester, UK (G Vassallo MD); Centre for Rare Diseases and Personalised Medicine and Department of Medical and Molecular Genetics, School of Clinical and Experimental Medicine, University of Birmingham, UK, and West Midlands Regional Genetics Service, Clinical Genetics Unit, Birmingham Women’s Hospital, Birmingham, UK (J Vogt MRCP); Nottingham Children’s Hospital, Nottingham University Hospitals NHS Trust, Nottingham, UK (W P Whitehouse FRCPCH); Neuropaediatrics, Department of Paediatrics, Mater Dei Hospital, Tal-Qroqq, Malta (D Soler MD); and Université Paris Descartes, EA 1833 and Hôpital Cochin, Service de Virologie, Paris, France, (F Rozenberg MD, P Lebon MD)
1,
Rogelio Simon
Rogelio Simon
1Manchester Centre for Genomic Medicine, Institute of Human Development, Faculty of Medical and Human Sciences, University of Manchester, Manchester Academic Health Sciences Centre (MAHSC), Manchester, UK (G I Rice PhD, G M A Forte MPhil, M Szynkiewicz MSc, D S Chase PhD, Y J Crow PhD); Erasme University Hospital, Universite Libre de Bruxelles (ULB), Brussels, Belgium, (A Aeby MD, C N Vilain MD, V Wermenbol MD); Medical Molecular Genetics Department, (M S Abdel-Hamid MSc, L Effat PhD), and Clinical Genetics Department, (S A Temtamy PhD, M S Aglan PhD, M S Zaki PhD, G M H Abdel-Salam PhD), Human Genetics and Genome Research Division, National Research Centre, Cairo, Egypt; Bradford Teaching Hospitals NHS Foundation Trust, Bradford, UK (S Ackroyd MB ChB); Department of Clinical Biochemistry, Lancashire Teaching Hospitals NHS Foundation Trust, Preston, UK (R Allcock FRCPath); University Hospitals of Coventry and Warwickshire, Coventry, UK (K M Bailey MRCPCH); Department of Brain and Behavioral Sciences, Unit of Child Neurology and Psychiatry, University of Pavia, Pavia, Italy, (U Balottin MD); Child Neurology and Psychiatry Unit, (U Balottin, I Olivieri MD, S Orcesi MD), and Laboratory of Experimental Neurobiology (C Cereda PhD), C. Mondino National Neurological Institute, Pavia, Italy; Praticien Hospitalier, Unité de Neurologie Pédiatrique et Centre de Référence des Maladies Neuromusculaires, Hôpital Necker-Enfants Malades, Paris, France (C Barnerias MD); Department of Pediatrics, and Department of Neurology and Neurosurgery, Montreal Children’s Hospital, McGill University Health Center, Montreal, QC, Canada (G Bernard MD); Department of Dermatology, Reference Centre for Cutaneous Rare Diseases (MAGEC), Necker-Enfants Malades APHP, Sorbonne Paris Cité, Université Paris Descartes, Institut Imagine, Paris, France (C Bodemer PhD); Department of Pediatrics, Hospital Universitario Araba, Vitoria-Gasteiz, Spain (M P Botella MD); Manchester Centre for Genomic Medicine, Central Manchester University Hospitals NHS Foundation Trust, MAHSC, Manchester, UK (K E Chandler MD, Y J Crow); Department of Paediatric Neurology, University Hospitals of Leicester NHS Trust, Leicester, UK (L Dabydeen MRCPCH); TY Nelson Department of Neurology, and Neuroimmunology Group, the Children’s Hospital at Westmead, University of Sydney, Sydney, NSW, Australia (R C Dale PhD); Nutrition and Metabolism Unit, Hôpital Universitaire des Enfants Reine Fabiola (ULB), Brussels, Belgium (C De Laet MD); Department of Paediatric Neurology, Royal Preston Hospital, Preston, UK (C G E L De Goede FRCPCH); Pediatric Neurology Unit, Hospital Vall d’Hebron, Barcelona, Spain (M del Toro MD); Paediatric Neurology Unit, University Hospital 12 Octubre, Madrid, Spain (N Nunez Enamorado MD, R Simon MD); Department of Clinical and Experimental Sciences, Child Neurology and Psychiatry Unit, University of Brescia, Brescia, Italy (E Fazzi MD); Department of Genetics, BioCruces Health Research Institute, Hospital, Universitario Cruces, Bizkaia, Spain (B Gener MD); Institute of Biomedicine and Translational Medicine, University of Tartu, Tartu, Estonia (M Haldre BSc, P Peterson PhD, K Kisand PhD); General Neurology and Complex Motor Disorders Service, Evelina Children’s Hospital, Guy’s and St Thomas’ NHS Foundation Trust, London, UK (J-P S-M Lin PhD); Department of Paediatric Neurology, Leeds General Infirmary, Leeds, UK (J H Livingston MB ChB); Department of Neurosciences and Behavior Sciences, School of Medicine of Ribeirão Preto, University of São Paulo, São Paulo, Brazil, (C Marques Lourenco MD, W Marques Jr MD); Royal Devon and Exeter Foundation NHS Trust, Exeter, UK (P Oades FRCPCH); Section for Child Neurology, Women and Children’s Division, Oslo University Hospital, Oslo, Norway (M Rasmussen MD); Neuropediatrie, Hopital Gui de Chauliac, and INSERM U1051, INM, Montpellier, France (A Roubertie MD); Center for Genetic Medicine Research and Department of Neurology, Children’s National Medical Center, George Washington University School of Medicine, Washington, DC, USA, (J Loewenstein Schmidt CGC, A Vanderver MD); The Genetic Institute, Ha’Emek Medical Center, Afula and the Rappaport Faculty of Medicine, Technion, Haifa, Israel (S A Shalev MD, R Spiegel MD); Department of Neurology, University of Utah School of Medicine, Salt Lake City, UT, USA (K J Swoboda FACMG); Neurology Department, Royal Manchester Children’s Hospital, Manchester, UK (G Vassallo MD); Centre for Rare Diseases and Personalised Medicine and Department of Medical and Molecular Genetics, School of Clinical and Experimental Medicine, University of Birmingham, UK, and West Midlands Regional Genetics Service, Clinical Genetics Unit, Birmingham Women’s Hospital, Birmingham, UK (J Vogt MRCP); Nottingham Children’s Hospital, Nottingham University Hospitals NHS Trust, Nottingham, UK (W P Whitehouse FRCPCH); Neuropaediatrics, Department of Paediatrics, Mater Dei Hospital, Tal-Qroqq, Malta (D Soler MD); and Université Paris Descartes, EA 1833 and Hôpital Cochin, Service de Virologie, Paris, France, (F Rozenberg MD, P Lebon MD)
1,
Ronen Spiegel
Ronen Spiegel
1Manchester Centre for Genomic Medicine, Institute of Human Development, Faculty of Medical and Human Sciences, University of Manchester, Manchester Academic Health Sciences Centre (MAHSC), Manchester, UK (G I Rice PhD, G M A Forte MPhil, M Szynkiewicz MSc, D S Chase PhD, Y J Crow PhD); Erasme University Hospital, Universite Libre de Bruxelles (ULB), Brussels, Belgium, (A Aeby MD, C N Vilain MD, V Wermenbol MD); Medical Molecular Genetics Department, (M S Abdel-Hamid MSc, L Effat PhD), and Clinical Genetics Department, (S A Temtamy PhD, M S Aglan PhD, M S Zaki PhD, G M H Abdel-Salam PhD), Human Genetics and Genome Research Division, National Research Centre, Cairo, Egypt; Bradford Teaching Hospitals NHS Foundation Trust, Bradford, UK (S Ackroyd MB ChB); Department of Clinical Biochemistry, Lancashire Teaching Hospitals NHS Foundation Trust, Preston, UK (R Allcock FRCPath); University Hospitals of Coventry and Warwickshire, Coventry, UK (K M Bailey MRCPCH); Department of Brain and Behavioral Sciences, Unit of Child Neurology and Psychiatry, University of Pavia, Pavia, Italy, (U Balottin MD); Child Neurology and Psychiatry Unit, (U Balottin, I Olivieri MD, S Orcesi MD), and Laboratory of Experimental Neurobiology (C Cereda PhD), C. Mondino National Neurological Institute, Pavia, Italy; Praticien Hospitalier, Unité de Neurologie Pédiatrique et Centre de Référence des Maladies Neuromusculaires, Hôpital Necker-Enfants Malades, Paris, France (C Barnerias MD); Department of Pediatrics, and Department of Neurology and Neurosurgery, Montreal Children’s Hospital, McGill University Health Center, Montreal, QC, Canada (G Bernard MD); Department of Dermatology, Reference Centre for Cutaneous Rare Diseases (MAGEC), Necker-Enfants Malades APHP, Sorbonne Paris Cité, Université Paris Descartes, Institut Imagine, Paris, France (C Bodemer PhD); Department of Pediatrics, Hospital Universitario Araba, Vitoria-Gasteiz, Spain (M P Botella MD); Manchester Centre for Genomic Medicine, Central Manchester University Hospitals NHS Foundation Trust, MAHSC, Manchester, UK (K E Chandler MD, Y J Crow); Department of Paediatric Neurology, University Hospitals of Leicester NHS Trust, Leicester, UK (L Dabydeen MRCPCH); TY Nelson Department of Neurology, and Neuroimmunology Group, the Children’s Hospital at Westmead, University of Sydney, Sydney, NSW, Australia (R C Dale PhD); Nutrition and Metabolism Unit, Hôpital Universitaire des Enfants Reine Fabiola (ULB), Brussels, Belgium (C De Laet MD); Department of Paediatric Neurology, Royal Preston Hospital, Preston, UK (C G E L De Goede FRCPCH); Pediatric Neurology Unit, Hospital Vall d’Hebron, Barcelona, Spain (M del Toro MD); Paediatric Neurology Unit, University Hospital 12 Octubre, Madrid, Spain (N Nunez Enamorado MD, R Simon MD); Department of Clinical and Experimental Sciences, Child Neurology and Psychiatry Unit, University of Brescia, Brescia, Italy (E Fazzi MD); Department of Genetics, BioCruces Health Research Institute, Hospital, Universitario Cruces, Bizkaia, Spain (B Gener MD); Institute of Biomedicine and Translational Medicine, University of Tartu, Tartu, Estonia (M Haldre BSc, P Peterson PhD, K Kisand PhD); General Neurology and Complex Motor Disorders Service, Evelina Children’s Hospital, Guy’s and St Thomas’ NHS Foundation Trust, London, UK (J-P S-M Lin PhD); Department of Paediatric Neurology, Leeds General Infirmary, Leeds, UK (J H Livingston MB ChB); Department of Neurosciences and Behavior Sciences, School of Medicine of Ribeirão Preto, University of São Paulo, São Paulo, Brazil, (C Marques Lourenco MD, W Marques Jr MD); Royal Devon and Exeter Foundation NHS Trust, Exeter, UK (P Oades FRCPCH); Section for Child Neurology, Women and Children’s Division, Oslo University Hospital, Oslo, Norway (M Rasmussen MD); Neuropediatrie, Hopital Gui de Chauliac, and INSERM U1051, INM, Montpellier, France (A Roubertie MD); Center for Genetic Medicine Research and Department of Neurology, Children’s National Medical Center, George Washington University School of Medicine, Washington, DC, USA, (J Loewenstein Schmidt CGC, A Vanderver MD); The Genetic Institute, Ha’Emek Medical Center, Afula and the Rappaport Faculty of Medicine, Technion, Haifa, Israel (S A Shalev MD, R Spiegel MD); Department of Neurology, University of Utah School of Medicine, Salt Lake City, UT, USA (K J Swoboda FACMG); Neurology Department, Royal Manchester Children’s Hospital, Manchester, UK (G Vassallo MD); Centre for Rare Diseases and Personalised Medicine and Department of Medical and Molecular Genetics, School of Clinical and Experimental Medicine, University of Birmingham, UK, and West Midlands Regional Genetics Service, Clinical Genetics Unit, Birmingham Women’s Hospital, Birmingham, UK (J Vogt MRCP); Nottingham Children’s Hospital, Nottingham University Hospitals NHS Trust, Nottingham, UK (W P Whitehouse FRCPCH); Neuropaediatrics, Department of Paediatrics, Mater Dei Hospital, Tal-Qroqq, Malta (D Soler MD); and Université Paris Descartes, EA 1833 and Hôpital Cochin, Service de Virologie, Paris, France, (F Rozenberg MD, P Lebon MD)
1,
Kathryn J Swoboda
Kathryn J Swoboda
1Manchester Centre for Genomic Medicine, Institute of Human Development, Faculty of Medical and Human Sciences, University of Manchester, Manchester Academic Health Sciences Centre (MAHSC), Manchester, UK (G I Rice PhD, G M A Forte MPhil, M Szynkiewicz MSc, D S Chase PhD, Y J Crow PhD); Erasme University Hospital, Universite Libre de Bruxelles (ULB), Brussels, Belgium, (A Aeby MD, C N Vilain MD, V Wermenbol MD); Medical Molecular Genetics Department, (M S Abdel-Hamid MSc, L Effat PhD), and Clinical Genetics Department, (S A Temtamy PhD, M S Aglan PhD, M S Zaki PhD, G M H Abdel-Salam PhD), Human Genetics and Genome Research Division, National Research Centre, Cairo, Egypt; Bradford Teaching Hospitals NHS Foundation Trust, Bradford, UK (S Ackroyd MB ChB); Department of Clinical Biochemistry, Lancashire Teaching Hospitals NHS Foundation Trust, Preston, UK (R Allcock FRCPath); University Hospitals of Coventry and Warwickshire, Coventry, UK (K M Bailey MRCPCH); Department of Brain and Behavioral Sciences, Unit of Child Neurology and Psychiatry, University of Pavia, Pavia, Italy, (U Balottin MD); Child Neurology and Psychiatry Unit, (U Balottin, I Olivieri MD, S Orcesi MD), and Laboratory of Experimental Neurobiology (C Cereda PhD), C. Mondino National Neurological Institute, Pavia, Italy; Praticien Hospitalier, Unité de Neurologie Pédiatrique et Centre de Référence des Maladies Neuromusculaires, Hôpital Necker-Enfants Malades, Paris, France (C Barnerias MD); Department of Pediatrics, and Department of Neurology and Neurosurgery, Montreal Children’s Hospital, McGill University Health Center, Montreal, QC, Canada (G Bernard MD); Department of Dermatology, Reference Centre for Cutaneous Rare Diseases (MAGEC), Necker-Enfants Malades APHP, Sorbonne Paris Cité, Université Paris Descartes, Institut Imagine, Paris, France (C Bodemer PhD); Department of Pediatrics, Hospital Universitario Araba, Vitoria-Gasteiz, Spain (M P Botella MD); Manchester Centre for Genomic Medicine, Central Manchester University Hospitals NHS Foundation Trust, MAHSC, Manchester, UK (K E Chandler MD, Y J Crow); Department of Paediatric Neurology, University Hospitals of Leicester NHS Trust, Leicester, UK (L Dabydeen MRCPCH); TY Nelson Department of Neurology, and Neuroimmunology Group, the Children’s Hospital at Westmead, University of Sydney, Sydney, NSW, Australia (R C Dale PhD); Nutrition and Metabolism Unit, Hôpital Universitaire des Enfants Reine Fabiola (ULB), Brussels, Belgium (C De Laet MD); Department of Paediatric Neurology, Royal Preston Hospital, Preston, UK (C G E L De Goede FRCPCH); Pediatric Neurology Unit, Hospital Vall d’Hebron, Barcelona, Spain (M del Toro MD); Paediatric Neurology Unit, University Hospital 12 Octubre, Madrid, Spain (N Nunez Enamorado MD, R Simon MD); Department of Clinical and Experimental Sciences, Child Neurology and Psychiatry Unit, University of Brescia, Brescia, Italy (E Fazzi MD); Department of Genetics, BioCruces Health Research Institute, Hospital, Universitario Cruces, Bizkaia, Spain (B Gener MD); Institute of Biomedicine and Translational Medicine, University of Tartu, Tartu, Estonia (M Haldre BSc, P Peterson PhD, K Kisand PhD); General Neurology and Complex Motor Disorders Service, Evelina Children’s Hospital, Guy’s and St Thomas’ NHS Foundation Trust, London, UK (J-P S-M Lin PhD); Department of Paediatric Neurology, Leeds General Infirmary, Leeds, UK (J H Livingston MB ChB); Department of Neurosciences and Behavior Sciences, School of Medicine of Ribeirão Preto, University of São Paulo, São Paulo, Brazil, (C Marques Lourenco MD, W Marques Jr MD); Royal Devon and Exeter Foundation NHS Trust, Exeter, UK (P Oades FRCPCH); Section for Child Neurology, Women and Children’s Division, Oslo University Hospital, Oslo, Norway (M Rasmussen MD); Neuropediatrie, Hopital Gui de Chauliac, and INSERM U1051, INM, Montpellier, France (A Roubertie MD); Center for Genetic Medicine Research and Department of Neurology, Children’s National Medical Center, George Washington University School of Medicine, Washington, DC, USA, (J Loewenstein Schmidt CGC, A Vanderver MD); The Genetic Institute, Ha’Emek Medical Center, Afula and the Rappaport Faculty of Medicine, Technion, Haifa, Israel (S A Shalev MD, R Spiegel MD); Department of Neurology, University of Utah School of Medicine, Salt Lake City, UT, USA (K J Swoboda FACMG); Neurology Department, Royal Manchester Children’s Hospital, Manchester, UK (G Vassallo MD); Centre for Rare Diseases and Personalised Medicine and Department of Medical and Molecular Genetics, School of Clinical and Experimental Medicine, University of Birmingham, UK, and West Midlands Regional Genetics Service, Clinical Genetics Unit, Birmingham Women’s Hospital, Birmingham, UK (J Vogt MRCP); Nottingham Children’s Hospital, Nottingham University Hospitals NHS Trust, Nottingham, UK (W P Whitehouse FRCPCH); Neuropaediatrics, Department of Paediatrics, Mater Dei Hospital, Tal-Qroqq, Malta (D Soler MD); and Université Paris Descartes, EA 1833 and Hôpital Cochin, Service de Virologie, Paris, France, (F Rozenberg MD, P Lebon MD)
1,
Samia A Temtamy
Samia A Temtamy
1Manchester Centre for Genomic Medicine, Institute of Human Development, Faculty of Medical and Human Sciences, University of Manchester, Manchester Academic Health Sciences Centre (MAHSC), Manchester, UK (G I Rice PhD, G M A Forte MPhil, M Szynkiewicz MSc, D S Chase PhD, Y J Crow PhD); Erasme University Hospital, Universite Libre de Bruxelles (ULB), Brussels, Belgium, (A Aeby MD, C N Vilain MD, V Wermenbol MD); Medical Molecular Genetics Department, (M S Abdel-Hamid MSc, L Effat PhD), and Clinical Genetics Department, (S A Temtamy PhD, M S Aglan PhD, M S Zaki PhD, G M H Abdel-Salam PhD), Human Genetics and Genome Research Division, National Research Centre, Cairo, Egypt; Bradford Teaching Hospitals NHS Foundation Trust, Bradford, UK (S Ackroyd MB ChB); Department of Clinical Biochemistry, Lancashire Teaching Hospitals NHS Foundation Trust, Preston, UK (R Allcock FRCPath); University Hospitals of Coventry and Warwickshire, Coventry, UK (K M Bailey MRCPCH); Department of Brain and Behavioral Sciences, Unit of Child Neurology and Psychiatry, University of Pavia, Pavia, Italy, (U Balottin MD); Child Neurology and Psychiatry Unit, (U Balottin, I Olivieri MD, S Orcesi MD), and Laboratory of Experimental Neurobiology (C Cereda PhD), C. Mondino National Neurological Institute, Pavia, Italy; Praticien Hospitalier, Unité de Neurologie Pédiatrique et Centre de Référence des Maladies Neuromusculaires, Hôpital Necker-Enfants Malades, Paris, France (C Barnerias MD); Department of Pediatrics, and Department of Neurology and Neurosurgery, Montreal Children’s Hospital, McGill University Health Center, Montreal, QC, Canada (G Bernard MD); Department of Dermatology, Reference Centre for Cutaneous Rare Diseases (MAGEC), Necker-Enfants Malades APHP, Sorbonne Paris Cité, Université Paris Descartes, Institut Imagine, Paris, France (C Bodemer PhD); Department of Pediatrics, Hospital Universitario Araba, Vitoria-Gasteiz, Spain (M P Botella MD); Manchester Centre for Genomic Medicine, Central Manchester University Hospitals NHS Foundation Trust, MAHSC, Manchester, UK (K E Chandler MD, Y J Crow); Department of Paediatric Neurology, University Hospitals of Leicester NHS Trust, Leicester, UK (L Dabydeen MRCPCH); TY Nelson Department of Neurology, and Neuroimmunology Group, the Children’s Hospital at Westmead, University of Sydney, Sydney, NSW, Australia (R C Dale PhD); Nutrition and Metabolism Unit, Hôpital Universitaire des Enfants Reine Fabiola (ULB), Brussels, Belgium (C De Laet MD); Department of Paediatric Neurology, Royal Preston Hospital, Preston, UK (C G E L De Goede FRCPCH); Pediatric Neurology Unit, Hospital Vall d’Hebron, Barcelona, Spain (M del Toro MD); Paediatric Neurology Unit, University Hospital 12 Octubre, Madrid, Spain (N Nunez Enamorado MD, R Simon MD); Department of Clinical and Experimental Sciences, Child Neurology and Psychiatry Unit, University of Brescia, Brescia, Italy (E Fazzi MD); Department of Genetics, BioCruces Health Research Institute, Hospital, Universitario Cruces, Bizkaia, Spain (B Gener MD); Institute of Biomedicine and Translational Medicine, University of Tartu, Tartu, Estonia (M Haldre BSc, P Peterson PhD, K Kisand PhD); General Neurology and Complex Motor Disorders Service, Evelina Children’s Hospital, Guy’s and St Thomas’ NHS Foundation Trust, London, UK (J-P S-M Lin PhD); Department of Paediatric Neurology, Leeds General Infirmary, Leeds, UK (J H Livingston MB ChB); Department of Neurosciences and Behavior Sciences, School of Medicine of Ribeirão Preto, University of São Paulo, São Paulo, Brazil, (C Marques Lourenco MD, W Marques Jr MD); Royal Devon and Exeter Foundation NHS Trust, Exeter, UK (P Oades FRCPCH); Section for Child Neurology, Women and Children’s Division, Oslo University Hospital, Oslo, Norway (M Rasmussen MD); Neuropediatrie, Hopital Gui de Chauliac, and INSERM U1051, INM, Montpellier, France (A Roubertie MD); Center for Genetic Medicine Research and Department of Neurology, Children’s National Medical Center, George Washington University School of Medicine, Washington, DC, USA, (J Loewenstein Schmidt CGC, A Vanderver MD); The Genetic Institute, Ha’Emek Medical Center, Afula and the Rappaport Faculty of Medicine, Technion, Haifa, Israel (S A Shalev MD, R Spiegel MD); Department of Neurology, University of Utah School of Medicine, Salt Lake City, UT, USA (K J Swoboda FACMG); Neurology Department, Royal Manchester Children’s Hospital, Manchester, UK (G Vassallo MD); Centre for Rare Diseases and Personalised Medicine and Department of Medical and Molecular Genetics, School of Clinical and Experimental Medicine, University of Birmingham, UK, and West Midlands Regional Genetics Service, Clinical Genetics Unit, Birmingham Women’s Hospital, Birmingham, UK (J Vogt MRCP); Nottingham Children’s Hospital, Nottingham University Hospitals NHS Trust, Nottingham, UK (W P Whitehouse FRCPCH); Neuropaediatrics, Department of Paediatrics, Mater Dei Hospital, Tal-Qroqq, Malta (D Soler MD); and Université Paris Descartes, EA 1833 and Hôpital Cochin, Service de Virologie, Paris, France, (F Rozenberg MD, P Lebon MD)
1,
Grace Vassallo
Grace Vassallo
1Manchester Centre for Genomic Medicine, Institute of Human Development, Faculty of Medical and Human Sciences, University of Manchester, Manchester Academic Health Sciences Centre (MAHSC), Manchester, UK (G I Rice PhD, G M A Forte MPhil, M Szynkiewicz MSc, D S Chase PhD, Y J Crow PhD); Erasme University Hospital, Universite Libre de Bruxelles (ULB), Brussels, Belgium, (A Aeby MD, C N Vilain MD, V Wermenbol MD); Medical Molecular Genetics Department, (M S Abdel-Hamid MSc, L Effat PhD), and Clinical Genetics Department, (S A Temtamy PhD, M S Aglan PhD, M S Zaki PhD, G M H Abdel-Salam PhD), Human Genetics and Genome Research Division, National Research Centre, Cairo, Egypt; Bradford Teaching Hospitals NHS Foundation Trust, Bradford, UK (S Ackroyd MB ChB); Department of Clinical Biochemistry, Lancashire Teaching Hospitals NHS Foundation Trust, Preston, UK (R Allcock FRCPath); University Hospitals of Coventry and Warwickshire, Coventry, UK (K M Bailey MRCPCH); Department of Brain and Behavioral Sciences, Unit of Child Neurology and Psychiatry, University of Pavia, Pavia, Italy, (U Balottin MD); Child Neurology and Psychiatry Unit, (U Balottin, I Olivieri MD, S Orcesi MD), and Laboratory of Experimental Neurobiology (C Cereda PhD), C. Mondino National Neurological Institute, Pavia, Italy; Praticien Hospitalier, Unité de Neurologie Pédiatrique et Centre de Référence des Maladies Neuromusculaires, Hôpital Necker-Enfants Malades, Paris, France (C Barnerias MD); Department of Pediatrics, and Department of Neurology and Neurosurgery, Montreal Children’s Hospital, McGill University Health Center, Montreal, QC, Canada (G Bernard MD); Department of Dermatology, Reference Centre for Cutaneous Rare Diseases (MAGEC), Necker-Enfants Malades APHP, Sorbonne Paris Cité, Université Paris Descartes, Institut Imagine, Paris, France (C Bodemer PhD); Department of Pediatrics, Hospital Universitario Araba, Vitoria-Gasteiz, Spain (M P Botella MD); Manchester Centre for Genomic Medicine, Central Manchester University Hospitals NHS Foundation Trust, MAHSC, Manchester, UK (K E Chandler MD, Y J Crow); Department of Paediatric Neurology, University Hospitals of Leicester NHS Trust, Leicester, UK (L Dabydeen MRCPCH); TY Nelson Department of Neurology, and Neuroimmunology Group, the Children’s Hospital at Westmead, University of Sydney, Sydney, NSW, Australia (R C Dale PhD); Nutrition and Metabolism Unit, Hôpital Universitaire des Enfants Reine Fabiola (ULB), Brussels, Belgium (C De Laet MD); Department of Paediatric Neurology, Royal Preston Hospital, Preston, UK (C G E L De Goede FRCPCH); Pediatric Neurology Unit, Hospital Vall d’Hebron, Barcelona, Spain (M del Toro MD); Paediatric Neurology Unit, University Hospital 12 Octubre, Madrid, Spain (N Nunez Enamorado MD, R Simon MD); Department of Clinical and Experimental Sciences, Child Neurology and Psychiatry Unit, University of Brescia, Brescia, Italy (E Fazzi MD); Department of Genetics, BioCruces Health Research Institute, Hospital, Universitario Cruces, Bizkaia, Spain (B Gener MD); Institute of Biomedicine and Translational Medicine, University of Tartu, Tartu, Estonia (M Haldre BSc, P Peterson PhD, K Kisand PhD); General Neurology and Complex Motor Disorders Service, Evelina Children’s Hospital, Guy’s and St Thomas’ NHS Foundation Trust, London, UK (J-P S-M Lin PhD); Department of Paediatric Neurology, Leeds General Infirmary, Leeds, UK (J H Livingston MB ChB); Department of Neurosciences and Behavior Sciences, School of Medicine of Ribeirão Preto, University of São Paulo, São Paulo, Brazil, (C Marques Lourenco MD, W Marques Jr MD); Royal Devon and Exeter Foundation NHS Trust, Exeter, UK (P Oades FRCPCH); Section for Child Neurology, Women and Children’s Division, Oslo University Hospital, Oslo, Norway (M Rasmussen MD); Neuropediatrie, Hopital Gui de Chauliac, and INSERM U1051, INM, Montpellier, France (A Roubertie MD); Center for Genetic Medicine Research and Department of Neurology, Children’s National Medical Center, George Washington University School of Medicine, Washington, DC, USA, (J Loewenstein Schmidt CGC, A Vanderver MD); The Genetic Institute, Ha’Emek Medical Center, Afula and the Rappaport Faculty of Medicine, Technion, Haifa, Israel (S A Shalev MD, R Spiegel MD); Department of Neurology, University of Utah School of Medicine, Salt Lake City, UT, USA (K J Swoboda FACMG); Neurology Department, Royal Manchester Children’s Hospital, Manchester, UK (G Vassallo MD); Centre for Rare Diseases and Personalised Medicine and Department of Medical and Molecular Genetics, School of Clinical and Experimental Medicine, University of Birmingham, UK, and West Midlands Regional Genetics Service, Clinical Genetics Unit, Birmingham Women’s Hospital, Birmingham, UK (J Vogt MRCP); Nottingham Children’s Hospital, Nottingham University Hospitals NHS Trust, Nottingham, UK (W P Whitehouse FRCPCH); Neuropaediatrics, Department of Paediatrics, Mater Dei Hospital, Tal-Qroqq, Malta (D Soler MD); and Université Paris Descartes, EA 1833 and Hôpital Cochin, Service de Virologie, Paris, France, (F Rozenberg MD, P Lebon MD)
1,
Catheline N Vilain
Catheline N Vilain
1Manchester Centre for Genomic Medicine, Institute of Human Development, Faculty of Medical and Human Sciences, University of Manchester, Manchester Academic Health Sciences Centre (MAHSC), Manchester, UK (G I Rice PhD, G M A Forte MPhil, M Szynkiewicz MSc, D S Chase PhD, Y J Crow PhD); Erasme University Hospital, Universite Libre de Bruxelles (ULB), Brussels, Belgium, (A Aeby MD, C N Vilain MD, V Wermenbol MD); Medical Molecular Genetics Department, (M S Abdel-Hamid MSc, L Effat PhD), and Clinical Genetics Department, (S A Temtamy PhD, M S Aglan PhD, M S Zaki PhD, G M H Abdel-Salam PhD), Human Genetics and Genome Research Division, National Research Centre, Cairo, Egypt; Bradford Teaching Hospitals NHS Foundation Trust, Bradford, UK (S Ackroyd MB ChB); Department of Clinical Biochemistry, Lancashire Teaching Hospitals NHS Foundation Trust, Preston, UK (R Allcock FRCPath); University Hospitals of Coventry and Warwickshire, Coventry, UK (K M Bailey MRCPCH); Department of Brain and Behavioral Sciences, Unit of Child Neurology and Psychiatry, University of Pavia, Pavia, Italy, (U Balottin MD); Child Neurology and Psychiatry Unit, (U Balottin, I Olivieri MD, S Orcesi MD), and Laboratory of Experimental Neurobiology (C Cereda PhD), C. Mondino National Neurological Institute, Pavia, Italy; Praticien Hospitalier, Unité de Neurologie Pédiatrique et Centre de Référence des Maladies Neuromusculaires, Hôpital Necker-Enfants Malades, Paris, France (C Barnerias MD); Department of Pediatrics, and Department of Neurology and Neurosurgery, Montreal Children’s Hospital, McGill University Health Center, Montreal, QC, Canada (G Bernard MD); Department of Dermatology, Reference Centre for Cutaneous Rare Diseases (MAGEC), Necker-Enfants Malades APHP, Sorbonne Paris Cité, Université Paris Descartes, Institut Imagine, Paris, France (C Bodemer PhD); Department of Pediatrics, Hospital Universitario Araba, Vitoria-Gasteiz, Spain (M P Botella MD); Manchester Centre for Genomic Medicine, Central Manchester University Hospitals NHS Foundation Trust, MAHSC, Manchester, UK (K E Chandler MD, Y J Crow); Department of Paediatric Neurology, University Hospitals of Leicester NHS Trust, Leicester, UK (L Dabydeen MRCPCH); TY Nelson Department of Neurology, and Neuroimmunology Group, the Children’s Hospital at Westmead, University of Sydney, Sydney, NSW, Australia (R C Dale PhD); Nutrition and Metabolism Unit, Hôpital Universitaire des Enfants Reine Fabiola (ULB), Brussels, Belgium (C De Laet MD); Department of Paediatric Neurology, Royal Preston Hospital, Preston, UK (C G E L De Goede FRCPCH); Pediatric Neurology Unit, Hospital Vall d’Hebron, Barcelona, Spain (M del Toro MD); Paediatric Neurology Unit, University Hospital 12 Octubre, Madrid, Spain (N Nunez Enamorado MD, R Simon MD); Department of Clinical and Experimental Sciences, Child Neurology and Psychiatry Unit, University of Brescia, Brescia, Italy (E Fazzi MD); Department of Genetics, BioCruces Health Research Institute, Hospital, Universitario Cruces, Bizkaia, Spain (B Gener MD); Institute of Biomedicine and Translational Medicine, University of Tartu, Tartu, Estonia (M Haldre BSc, P Peterson PhD, K Kisand PhD); General Neurology and Complex Motor Disorders Service, Evelina Children’s Hospital, Guy’s and St Thomas’ NHS Foundation Trust, London, UK (J-P S-M Lin PhD); Department of Paediatric Neurology, Leeds General Infirmary, Leeds, UK (J H Livingston MB ChB); Department of Neurosciences and Behavior Sciences, School of Medicine of Ribeirão Preto, University of São Paulo, São Paulo, Brazil, (C Marques Lourenco MD, W Marques Jr MD); Royal Devon and Exeter Foundation NHS Trust, Exeter, UK (P Oades FRCPCH); Section for Child Neurology, Women and Children’s Division, Oslo University Hospital, Oslo, Norway (M Rasmussen MD); Neuropediatrie, Hopital Gui de Chauliac, and INSERM U1051, INM, Montpellier, France (A Roubertie MD); Center for Genetic Medicine Research and Department of Neurology, Children’s National Medical Center, George Washington University School of Medicine, Washington, DC, USA, (J Loewenstein Schmidt CGC, A Vanderver MD); The Genetic Institute, Ha’Emek Medical Center, Afula and the Rappaport Faculty of Medicine, Technion, Haifa, Israel (S A Shalev MD, R Spiegel MD); Department of Neurology, University of Utah School of Medicine, Salt Lake City, UT, USA (K J Swoboda FACMG); Neurology Department, Royal Manchester Children’s Hospital, Manchester, UK (G Vassallo MD); Centre for Rare Diseases and Personalised Medicine and Department of Medical and Molecular Genetics, School of Clinical and Experimental Medicine, University of Birmingham, UK, and West Midlands Regional Genetics Service, Clinical Genetics Unit, Birmingham Women’s Hospital, Birmingham, UK (J Vogt MRCP); Nottingham Children’s Hospital, Nottingham University Hospitals NHS Trust, Nottingham, UK (W P Whitehouse FRCPCH); Neuropaediatrics, Department of Paediatrics, Mater Dei Hospital, Tal-Qroqq, Malta (D Soler MD); and Université Paris Descartes, EA 1833 and Hôpital Cochin, Service de Virologie, Paris, France, (F Rozenberg MD, P Lebon MD)
1,
Julie Vogt
Julie Vogt
1Manchester Centre for Genomic Medicine, Institute of Human Development, Faculty of Medical and Human Sciences, University of Manchester, Manchester Academic Health Sciences Centre (MAHSC), Manchester, UK (G I Rice PhD, G M A Forte MPhil, M Szynkiewicz MSc, D S Chase PhD, Y J Crow PhD); Erasme University Hospital, Universite Libre de Bruxelles (ULB), Brussels, Belgium, (A Aeby MD, C N Vilain MD, V Wermenbol MD); Medical Molecular Genetics Department, (M S Abdel-Hamid MSc, L Effat PhD), and Clinical Genetics Department, (S A Temtamy PhD, M S Aglan PhD, M S Zaki PhD, G M H Abdel-Salam PhD), Human Genetics and Genome Research Division, National Research Centre, Cairo, Egypt; Bradford Teaching Hospitals NHS Foundation Trust, Bradford, UK (S Ackroyd MB ChB); Department of Clinical Biochemistry, Lancashire Teaching Hospitals NHS Foundation Trust, Preston, UK (R Allcock FRCPath); University Hospitals of Coventry and Warwickshire, Coventry, UK (K M Bailey MRCPCH); Department of Brain and Behavioral Sciences, Unit of Child Neurology and Psychiatry, University of Pavia, Pavia, Italy, (U Balottin MD); Child Neurology and Psychiatry Unit, (U Balottin, I Olivieri MD, S Orcesi MD), and Laboratory of Experimental Neurobiology (C Cereda PhD), C. Mondino National Neurological Institute, Pavia, Italy; Praticien Hospitalier, Unité de Neurologie Pédiatrique et Centre de Référence des Maladies Neuromusculaires, Hôpital Necker-Enfants Malades, Paris, France (C Barnerias MD); Department of Pediatrics, and Department of Neurology and Neurosurgery, Montreal Children’s Hospital, McGill University Health Center, Montreal, QC, Canada (G Bernard MD); Department of Dermatology, Reference Centre for Cutaneous Rare Diseases (MAGEC), Necker-Enfants Malades APHP, Sorbonne Paris Cité, Université Paris Descartes, Institut Imagine, Paris, France (C Bodemer PhD); Department of Pediatrics, Hospital Universitario Araba, Vitoria-Gasteiz, Spain (M P Botella MD); Manchester Centre for Genomic Medicine, Central Manchester University Hospitals NHS Foundation Trust, MAHSC, Manchester, UK (K E Chandler MD, Y J Crow); Department of Paediatric Neurology, University Hospitals of Leicester NHS Trust, Leicester, UK (L Dabydeen MRCPCH); TY Nelson Department of Neurology, and Neuroimmunology Group, the Children’s Hospital at Westmead, University of Sydney, Sydney, NSW, Australia (R C Dale PhD); Nutrition and Metabolism Unit, Hôpital Universitaire des Enfants Reine Fabiola (ULB), Brussels, Belgium (C De Laet MD); Department of Paediatric Neurology, Royal Preston Hospital, Preston, UK (C G E L De Goede FRCPCH); Pediatric Neurology Unit, Hospital Vall d’Hebron, Barcelona, Spain (M del Toro MD); Paediatric Neurology Unit, University Hospital 12 Octubre, Madrid, Spain (N Nunez Enamorado MD, R Simon MD); Department of Clinical and Experimental Sciences, Child Neurology and Psychiatry Unit, University of Brescia, Brescia, Italy (E Fazzi MD); Department of Genetics, BioCruces Health Research Institute, Hospital, Universitario Cruces, Bizkaia, Spain (B Gener MD); Institute of Biomedicine and Translational Medicine, University of Tartu, Tartu, Estonia (M Haldre BSc, P Peterson PhD, K Kisand PhD); General Neurology and Complex Motor Disorders Service, Evelina Children’s Hospital, Guy’s and St Thomas’ NHS Foundation Trust, London, UK (J-P S-M Lin PhD); Department of Paediatric Neurology, Leeds General Infirmary, Leeds, UK (J H Livingston MB ChB); Department of Neurosciences and Behavior Sciences, School of Medicine of Ribeirão Preto, University of São Paulo, São Paulo, Brazil, (C Marques Lourenco MD, W Marques Jr MD); Royal Devon and Exeter Foundation NHS Trust, Exeter, UK (P Oades FRCPCH); Section for Child Neurology, Women and Children’s Division, Oslo University Hospital, Oslo, Norway (M Rasmussen MD); Neuropediatrie, Hopital Gui de Chauliac, and INSERM U1051, INM, Montpellier, France (A Roubertie MD); Center for Genetic Medicine Research and Department of Neurology, Children’s National Medical Center, George Washington University School of Medicine, Washington, DC, USA, (J Loewenstein Schmidt CGC, A Vanderver MD); The Genetic Institute, Ha’Emek Medical Center, Afula and the Rappaport Faculty of Medicine, Technion, Haifa, Israel (S A Shalev MD, R Spiegel MD); Department of Neurology, University of Utah School of Medicine, Salt Lake City, UT, USA (K J Swoboda FACMG); Neurology Department, Royal Manchester Children’s Hospital, Manchester, UK (G Vassallo MD); Centre for Rare Diseases and Personalised Medicine and Department of Medical and Molecular Genetics, School of Clinical and Experimental Medicine, University of Birmingham, UK, and West Midlands Regional Genetics Service, Clinical Genetics Unit, Birmingham Women’s Hospital, Birmingham, UK (J Vogt MRCP); Nottingham Children’s Hospital, Nottingham University Hospitals NHS Trust, Nottingham, UK (W P Whitehouse FRCPCH); Neuropaediatrics, Department of Paediatrics, Mater Dei Hospital, Tal-Qroqq, Malta (D Soler MD); and Université Paris Descartes, EA 1833 and Hôpital Cochin, Service de Virologie, Paris, France, (F Rozenberg MD, P Lebon MD)
1,
Vanessa Wermenbol
Vanessa Wermenbol
1Manchester Centre for Genomic Medicine, Institute of Human Development, Faculty of Medical and Human Sciences, University of Manchester, Manchester Academic Health Sciences Centre (MAHSC), Manchester, UK (G I Rice PhD, G M A Forte MPhil, M Szynkiewicz MSc, D S Chase PhD, Y J Crow PhD); Erasme University Hospital, Universite Libre de Bruxelles (ULB), Brussels, Belgium, (A Aeby MD, C N Vilain MD, V Wermenbol MD); Medical Molecular Genetics Department, (M S Abdel-Hamid MSc, L Effat PhD), and Clinical Genetics Department, (S A Temtamy PhD, M S Aglan PhD, M S Zaki PhD, G M H Abdel-Salam PhD), Human Genetics and Genome Research Division, National Research Centre, Cairo, Egypt; Bradford Teaching Hospitals NHS Foundation Trust, Bradford, UK (S Ackroyd MB ChB); Department of Clinical Biochemistry, Lancashire Teaching Hospitals NHS Foundation Trust, Preston, UK (R Allcock FRCPath); University Hospitals of Coventry and Warwickshire, Coventry, UK (K M Bailey MRCPCH); Department of Brain and Behavioral Sciences, Unit of Child Neurology and Psychiatry, University of Pavia, Pavia, Italy, (U Balottin MD); Child Neurology and Psychiatry Unit, (U Balottin, I Olivieri MD, S Orcesi MD), and Laboratory of Experimental Neurobiology (C Cereda PhD), C. Mondino National Neurological Institute, Pavia, Italy; Praticien Hospitalier, Unité de Neurologie Pédiatrique et Centre de Référence des Maladies Neuromusculaires, Hôpital Necker-Enfants Malades, Paris, France (C Barnerias MD); Department of Pediatrics, and Department of Neurology and Neurosurgery, Montreal Children’s Hospital, McGill University Health Center, Montreal, QC, Canada (G Bernard MD); Department of Dermatology, Reference Centre for Cutaneous Rare Diseases (MAGEC), Necker-Enfants Malades APHP, Sorbonne Paris Cité, Université Paris Descartes, Institut Imagine, Paris, France (C Bodemer PhD); Department of Pediatrics, Hospital Universitario Araba, Vitoria-Gasteiz, Spain (M P Botella MD); Manchester Centre for Genomic Medicine, Central Manchester University Hospitals NHS Foundation Trust, MAHSC, Manchester, UK (K E Chandler MD, Y J Crow); Department of Paediatric Neurology, University Hospitals of Leicester NHS Trust, Leicester, UK (L Dabydeen MRCPCH); TY Nelson Department of Neurology, and Neuroimmunology Group, the Children’s Hospital at Westmead, University of Sydney, Sydney, NSW, Australia (R C Dale PhD); Nutrition and Metabolism Unit, Hôpital Universitaire des Enfants Reine Fabiola (ULB), Brussels, Belgium (C De Laet MD); Department of Paediatric Neurology, Royal Preston Hospital, Preston, UK (C G E L De Goede FRCPCH); Pediatric Neurology Unit, Hospital Vall d’Hebron, Barcelona, Spain (M del Toro MD); Paediatric Neurology Unit, University Hospital 12 Octubre, Madrid, Spain (N Nunez Enamorado MD, R Simon MD); Department of Clinical and Experimental Sciences, Child Neurology and Psychiatry Unit, University of Brescia, Brescia, Italy (E Fazzi MD); Department of Genetics, BioCruces Health Research Institute, Hospital, Universitario Cruces, Bizkaia, Spain (B Gener MD); Institute of Biomedicine and Translational Medicine, University of Tartu, Tartu, Estonia (M Haldre BSc, P Peterson PhD, K Kisand PhD); General Neurology and Complex Motor Disorders Service, Evelina Children’s Hospital, Guy’s and St Thomas’ NHS Foundation Trust, London, UK (J-P S-M Lin PhD); Department of Paediatric Neurology, Leeds General Infirmary, Leeds, UK (J H Livingston MB ChB); Department of Neurosciences and Behavior Sciences, School of Medicine of Ribeirão Preto, University of São Paulo, São Paulo, Brazil, (C Marques Lourenco MD, W Marques Jr MD); Royal Devon and Exeter Foundation NHS Trust, Exeter, UK (P Oades FRCPCH); Section for Child Neurology, Women and Children’s Division, Oslo University Hospital, Oslo, Norway (M Rasmussen MD); Neuropediatrie, Hopital Gui de Chauliac, and INSERM U1051, INM, Montpellier, France (A Roubertie MD); Center for Genetic Medicine Research and Department of Neurology, Children’s National Medical Center, George Washington University School of Medicine, Washington, DC, USA, (J Loewenstein Schmidt CGC, A Vanderver MD); The Genetic Institute, Ha’Emek Medical Center, Afula and the Rappaport Faculty of Medicine, Technion, Haifa, Israel (S A Shalev MD, R Spiegel MD); Department of Neurology, University of Utah School of Medicine, Salt Lake City, UT, USA (K J Swoboda FACMG); Neurology Department, Royal Manchester Children’s Hospital, Manchester, UK (G Vassallo MD); Centre for Rare Diseases and Personalised Medicine and Department of Medical and Molecular Genetics, School of Clinical and Experimental Medicine, University of Birmingham, UK, and West Midlands Regional Genetics Service, Clinical Genetics Unit, Birmingham Women’s Hospital, Birmingham, UK (J Vogt MRCP); Nottingham Children’s Hospital, Nottingham University Hospitals NHS Trust, Nottingham, UK (W P Whitehouse FRCPCH); Neuropaediatrics, Department of Paediatrics, Mater Dei Hospital, Tal-Qroqq, Malta (D Soler MD); and Université Paris Descartes, EA 1833 and Hôpital Cochin, Service de Virologie, Paris, France, (F Rozenberg MD, P Lebon MD)
1,
William P Whitehouse
William P Whitehouse
1Manchester Centre for Genomic Medicine, Institute of Human Development, Faculty of Medical and Human Sciences, University of Manchester, Manchester Academic Health Sciences Centre (MAHSC), Manchester, UK (G I Rice PhD, G M A Forte MPhil, M Szynkiewicz MSc, D S Chase PhD, Y J Crow PhD); Erasme University Hospital, Universite Libre de Bruxelles (ULB), Brussels, Belgium, (A Aeby MD, C N Vilain MD, V Wermenbol MD); Medical Molecular Genetics Department, (M S Abdel-Hamid MSc, L Effat PhD), and Clinical Genetics Department, (S A Temtamy PhD, M S Aglan PhD, M S Zaki PhD, G M H Abdel-Salam PhD), Human Genetics and Genome Research Division, National Research Centre, Cairo, Egypt; Bradford Teaching Hospitals NHS Foundation Trust, Bradford, UK (S Ackroyd MB ChB); Department of Clinical Biochemistry, Lancashire Teaching Hospitals NHS Foundation Trust, Preston, UK (R Allcock FRCPath); University Hospitals of Coventry and Warwickshire, Coventry, UK (K M Bailey MRCPCH); Department of Brain and Behavioral Sciences, Unit of Child Neurology and Psychiatry, University of Pavia, Pavia, Italy, (U Balottin MD); Child Neurology and Psychiatry Unit, (U Balottin, I Olivieri MD, S Orcesi MD), and Laboratory of Experimental Neurobiology (C Cereda PhD), C. Mondino National Neurological Institute, Pavia, Italy; Praticien Hospitalier, Unité de Neurologie Pédiatrique et Centre de Référence des Maladies Neuromusculaires, Hôpital Necker-Enfants Malades, Paris, France (C Barnerias MD); Department of Pediatrics, and Department of Neurology and Neurosurgery, Montreal Children’s Hospital, McGill University Health Center, Montreal, QC, Canada (G Bernard MD); Department of Dermatology, Reference Centre for Cutaneous Rare Diseases (MAGEC), Necker-Enfants Malades APHP, Sorbonne Paris Cité, Université Paris Descartes, Institut Imagine, Paris, France (C Bodemer PhD); Department of Pediatrics, Hospital Universitario Araba, Vitoria-Gasteiz, Spain (M P Botella MD); Manchester Centre for Genomic Medicine, Central Manchester University Hospitals NHS Foundation Trust, MAHSC, Manchester, UK (K E Chandler MD, Y J Crow); Department of Paediatric Neurology, University Hospitals of Leicester NHS Trust, Leicester, UK (L Dabydeen MRCPCH); TY Nelson Department of Neurology, and Neuroimmunology Group, the Children’s Hospital at Westmead, University of Sydney, Sydney, NSW, Australia (R C Dale PhD); Nutrition and Metabolism Unit, Hôpital Universitaire des Enfants Reine Fabiola (ULB), Brussels, Belgium (C De Laet MD); Department of Paediatric Neurology, Royal Preston Hospital, Preston, UK (C G E L De Goede FRCPCH); Pediatric Neurology Unit, Hospital Vall d’Hebron, Barcelona, Spain (M del Toro MD); Paediatric Neurology Unit, University Hospital 12 Octubre, Madrid, Spain (N Nunez Enamorado MD, R Simon MD); Department of Clinical and Experimental Sciences, Child Neurology and Psychiatry Unit, University of Brescia, Brescia, Italy (E Fazzi MD); Department of Genetics, BioCruces Health Research Institute, Hospital, Universitario Cruces, Bizkaia, Spain (B Gener MD); Institute of Biomedicine and Translational Medicine, University of Tartu, Tartu, Estonia (M Haldre BSc, P Peterson PhD, K Kisand PhD); General Neurology and Complex Motor Disorders Service, Evelina Children’s Hospital, Guy’s and St Thomas’ NHS Foundation Trust, London, UK (J-P S-M Lin PhD); Department of Paediatric Neurology, Leeds General Infirmary, Leeds, UK (J H Livingston MB ChB); Department of Neurosciences and Behavior Sciences, School of Medicine of Ribeirão Preto, University of São Paulo, São Paulo, Brazil, (C Marques Lourenco MD, W Marques Jr MD); Royal Devon and Exeter Foundation NHS Trust, Exeter, UK (P Oades FRCPCH); Section for Child Neurology, Women and Children’s Division, Oslo University Hospital, Oslo, Norway (M Rasmussen MD); Neuropediatrie, Hopital Gui de Chauliac, and INSERM U1051, INM, Montpellier, France (A Roubertie MD); Center for Genetic Medicine Research and Department of Neurology, Children’s National Medical Center, George Washington University School of Medicine, Washington, DC, USA, (J Loewenstein Schmidt CGC, A Vanderver MD); The Genetic Institute, Ha’Emek Medical Center, Afula and the Rappaport Faculty of Medicine, Technion, Haifa, Israel (S A Shalev MD, R Spiegel MD); Department of Neurology, University of Utah School of Medicine, Salt Lake City, UT, USA (K J Swoboda FACMG); Neurology Department, Royal Manchester Children’s Hospital, Manchester, UK (G Vassallo MD); Centre for Rare Diseases and Personalised Medicine and Department of Medical and Molecular Genetics, School of Clinical and Experimental Medicine, University of Birmingham, UK, and West Midlands Regional Genetics Service, Clinical Genetics Unit, Birmingham Women’s Hospital, Birmingham, UK (J Vogt MRCP); Nottingham Children’s Hospital, Nottingham University Hospitals NHS Trust, Nottingham, UK (W P Whitehouse FRCPCH); Neuropaediatrics, Department of Paediatrics, Mater Dei Hospital, Tal-Qroqq, Malta (D Soler MD); and Université Paris Descartes, EA 1833 and Hôpital Cochin, Service de Virologie, Paris, France, (F Rozenberg MD, P Lebon MD)
1,
Doriette Soler
Doriette Soler
1Manchester Centre for Genomic Medicine, Institute of Human Development, Faculty of Medical and Human Sciences, University of Manchester, Manchester Academic Health Sciences Centre (MAHSC), Manchester, UK (G I Rice PhD, G M A Forte MPhil, M Szynkiewicz MSc, D S Chase PhD, Y J Crow PhD); Erasme University Hospital, Universite Libre de Bruxelles (ULB), Brussels, Belgium, (A Aeby MD, C N Vilain MD, V Wermenbol MD); Medical Molecular Genetics Department, (M S Abdel-Hamid MSc, L Effat PhD), and Clinical Genetics Department, (S A Temtamy PhD, M S Aglan PhD, M S Zaki PhD, G M H Abdel-Salam PhD), Human Genetics and Genome Research Division, National Research Centre, Cairo, Egypt; Bradford Teaching Hospitals NHS Foundation Trust, Bradford, UK (S Ackroyd MB ChB); Department of Clinical Biochemistry, Lancashire Teaching Hospitals NHS Foundation Trust, Preston, UK (R Allcock FRCPath); University Hospitals of Coventry and Warwickshire, Coventry, UK (K M Bailey MRCPCH); Department of Brain and Behavioral Sciences, Unit of Child Neurology and Psychiatry, University of Pavia, Pavia, Italy, (U Balottin MD); Child Neurology and Psychiatry Unit, (U Balottin, I Olivieri MD, S Orcesi MD), and Laboratory of Experimental Neurobiology (C Cereda PhD), C. Mondino National Neurological Institute, Pavia, Italy; Praticien Hospitalier, Unité de Neurologie Pédiatrique et Centre de Référence des Maladies Neuromusculaires, Hôpital Necker-Enfants Malades, Paris, France (C Barnerias MD); Department of Pediatrics, and Department of Neurology and Neurosurgery, Montreal Children’s Hospital, McGill University Health Center, Montreal, QC, Canada (G Bernard MD); Department of Dermatology, Reference Centre for Cutaneous Rare Diseases (MAGEC), Necker-Enfants Malades APHP, Sorbonne Paris Cité, Université Paris Descartes, Institut Imagine, Paris, France (C Bodemer PhD); Department of Pediatrics, Hospital Universitario Araba, Vitoria-Gasteiz, Spain (M P Botella MD); Manchester Centre for Genomic Medicine, Central Manchester University Hospitals NHS Foundation Trust, MAHSC, Manchester, UK (K E Chandler MD, Y J Crow); Department of Paediatric Neurology, University Hospitals of Leicester NHS Trust, Leicester, UK (L Dabydeen MRCPCH); TY Nelson Department of Neurology, and Neuroimmunology Group, the Children’s Hospital at Westmead, University of Sydney, Sydney, NSW, Australia (R C Dale PhD); Nutrition and Metabolism Unit, Hôpital Universitaire des Enfants Reine Fabiola (ULB), Brussels, Belgium (C De Laet MD); Department of Paediatric Neurology, Royal Preston Hospital, Preston, UK (C G E L De Goede FRCPCH); Pediatric Neurology Unit, Hospital Vall d’Hebron, Barcelona, Spain (M del Toro MD); Paediatric Neurology Unit, University Hospital 12 Octubre, Madrid, Spain (N Nunez Enamorado MD, R Simon MD); Department of Clinical and Experimental Sciences, Child Neurology and Psychiatry Unit, University of Brescia, Brescia, Italy (E Fazzi MD); Department of Genetics, BioCruces Health Research Institute, Hospital, Universitario Cruces, Bizkaia, Spain (B Gener MD); Institute of Biomedicine and Translational Medicine, University of Tartu, Tartu, Estonia (M Haldre BSc, P Peterson PhD, K Kisand PhD); General Neurology and Complex Motor Disorders Service, Evelina Children’s Hospital, Guy’s and St Thomas’ NHS Foundation Trust, London, UK (J-P S-M Lin PhD); Department of Paediatric Neurology, Leeds General Infirmary, Leeds, UK (J H Livingston MB ChB); Department of Neurosciences and Behavior Sciences, School of Medicine of Ribeirão Preto, University of São Paulo, São Paulo, Brazil, (C Marques Lourenco MD, W Marques Jr MD); Royal Devon and Exeter Foundation NHS Trust, Exeter, UK (P Oades FRCPCH); Section for Child Neurology, Women and Children’s Division, Oslo University Hospital, Oslo, Norway (M Rasmussen MD); Neuropediatrie, Hopital Gui de Chauliac, and INSERM U1051, INM, Montpellier, France (A Roubertie MD); Center for Genetic Medicine Research and Department of Neurology, Children’s National Medical Center, George Washington University School of Medicine, Washington, DC, USA, (J Loewenstein Schmidt CGC, A Vanderver MD); The Genetic Institute, Ha’Emek Medical Center, Afula and the Rappaport Faculty of Medicine, Technion, Haifa, Israel (S A Shalev MD, R Spiegel MD); Department of Neurology, University of Utah School of Medicine, Salt Lake City, UT, USA (K J Swoboda FACMG); Neurology Department, Royal Manchester Children’s Hospital, Manchester, UK (G Vassallo MD); Centre for Rare Diseases and Personalised Medicine and Department of Medical and Molecular Genetics, School of Clinical and Experimental Medicine, University of Birmingham, UK, and West Midlands Regional Genetics Service, Clinical Genetics Unit, Birmingham Women’s Hospital, Birmingham, UK (J Vogt MRCP); Nottingham Children’s Hospital, Nottingham University Hospitals NHS Trust, Nottingham, UK (W P Whitehouse FRCPCH); Neuropaediatrics, Department of Paediatrics, Mater Dei Hospital, Tal-Qroqq, Malta (D Soler MD); and Université Paris Descartes, EA 1833 and Hôpital Cochin, Service de Virologie, Paris, France, (F Rozenberg MD, P Lebon MD)
1,
Ivana Olivieri
Ivana Olivieri
1Manchester Centre for Genomic Medicine, Institute of Human Development, Faculty of Medical and Human Sciences, University of Manchester, Manchester Academic Health Sciences Centre (MAHSC), Manchester, UK (G I Rice PhD, G M A Forte MPhil, M Szynkiewicz MSc, D S Chase PhD, Y J Crow PhD); Erasme University Hospital, Universite Libre de Bruxelles (ULB), Brussels, Belgium, (A Aeby MD, C N Vilain MD, V Wermenbol MD); Medical Molecular Genetics Department, (M S Abdel-Hamid MSc, L Effat PhD), and Clinical Genetics Department, (S A Temtamy PhD, M S Aglan PhD, M S Zaki PhD, G M H Abdel-Salam PhD), Human Genetics and Genome Research Division, National Research Centre, Cairo, Egypt; Bradford Teaching Hospitals NHS Foundation Trust, Bradford, UK (S Ackroyd MB ChB); Department of Clinical Biochemistry, Lancashire Teaching Hospitals NHS Foundation Trust, Preston, UK (R Allcock FRCPath); University Hospitals of Coventry and Warwickshire, Coventry, UK (K M Bailey MRCPCH); Department of Brain and Behavioral Sciences, Unit of Child Neurology and Psychiatry, University of Pavia, Pavia, Italy, (U Balottin MD); Child Neurology and Psychiatry Unit, (U Balottin, I Olivieri MD, S Orcesi MD), and Laboratory of Experimental Neurobiology (C Cereda PhD), C. Mondino National Neurological Institute, Pavia, Italy; Praticien Hospitalier, Unité de Neurologie Pédiatrique et Centre de Référence des Maladies Neuromusculaires, Hôpital Necker-Enfants Malades, Paris, France (C Barnerias MD); Department of Pediatrics, and Department of Neurology and Neurosurgery, Montreal Children’s Hospital, McGill University Health Center, Montreal, QC, Canada (G Bernard MD); Department of Dermatology, Reference Centre for Cutaneous Rare Diseases (MAGEC), Necker-Enfants Malades APHP, Sorbonne Paris Cité, Université Paris Descartes, Institut Imagine, Paris, France (C Bodemer PhD); Department of Pediatrics, Hospital Universitario Araba, Vitoria-Gasteiz, Spain (M P Botella MD); Manchester Centre for Genomic Medicine, Central Manchester University Hospitals NHS Foundation Trust, MAHSC, Manchester, UK (K E Chandler MD, Y J Crow); Department of Paediatric Neurology, University Hospitals of Leicester NHS Trust, Leicester, UK (L Dabydeen MRCPCH); TY Nelson Department of Neurology, and Neuroimmunology Group, the Children’s Hospital at Westmead, University of Sydney, Sydney, NSW, Australia (R C Dale PhD); Nutrition and Metabolism Unit, Hôpital Universitaire des Enfants Reine Fabiola (ULB), Brussels, Belgium (C De Laet MD); Department of Paediatric Neurology, Royal Preston Hospital, Preston, UK (C G E L De Goede FRCPCH); Pediatric Neurology Unit, Hospital Vall d’Hebron, Barcelona, Spain (M del Toro MD); Paediatric Neurology Unit, University Hospital 12 Octubre, Madrid, Spain (N Nunez Enamorado MD, R Simon MD); Department of Clinical and Experimental Sciences, Child Neurology and Psychiatry Unit, University of Brescia, Brescia, Italy (E Fazzi MD); Department of Genetics, BioCruces Health Research Institute, Hospital, Universitario Cruces, Bizkaia, Spain (B Gener MD); Institute of Biomedicine and Translational Medicine, University of Tartu, Tartu, Estonia (M Haldre BSc, P Peterson PhD, K Kisand PhD); General Neurology and Complex Motor Disorders Service, Evelina Children’s Hospital, Guy’s and St Thomas’ NHS Foundation Trust, London, UK (J-P S-M Lin PhD); Department of Paediatric Neurology, Leeds General Infirmary, Leeds, UK (J H Livingston MB ChB); Department of Neurosciences and Behavior Sciences, School of Medicine of Ribeirão Preto, University of São Paulo, São Paulo, Brazil, (C Marques Lourenco MD, W Marques Jr MD); Royal Devon and Exeter Foundation NHS Trust, Exeter, UK (P Oades FRCPCH); Section for Child Neurology, Women and Children’s Division, Oslo University Hospital, Oslo, Norway (M Rasmussen MD); Neuropediatrie, Hopital Gui de Chauliac, and INSERM U1051, INM, Montpellier, France (A Roubertie MD); Center for Genetic Medicine Research and Department of Neurology, Children’s National Medical Center, George Washington University School of Medicine, Washington, DC, USA, (J Loewenstein Schmidt CGC, A Vanderver MD); The Genetic Institute, Ha’Emek Medical Center, Afula and the Rappaport Faculty of Medicine, Technion, Haifa, Israel (S A Shalev MD, R Spiegel MD); Department of Neurology, University of Utah School of Medicine, Salt Lake City, UT, USA (K J Swoboda FACMG); Neurology Department, Royal Manchester Children’s Hospital, Manchester, UK (G Vassallo MD); Centre for Rare Diseases and Personalised Medicine and Department of Medical and Molecular Genetics, School of Clinical and Experimental Medicine, University of Birmingham, UK, and West Midlands Regional Genetics Service, Clinical Genetics Unit, Birmingham Women’s Hospital, Birmingham, UK (J Vogt MRCP); Nottingham Children’s Hospital, Nottingham University Hospitals NHS Trust, Nottingham, UK (W P Whitehouse FRCPCH); Neuropaediatrics, Department of Paediatrics, Mater Dei Hospital, Tal-Qroqq, Malta (D Soler MD); and Université Paris Descartes, EA 1833 and Hôpital Cochin, Service de Virologie, Paris, France, (F Rozenberg MD, P Lebon MD)
1,
Simona Orcesi
Simona Orcesi
1Manchester Centre for Genomic Medicine, Institute of Human Development, Faculty of Medical and Human Sciences, University of Manchester, Manchester Academic Health Sciences Centre (MAHSC), Manchester, UK (G I Rice PhD, G M A Forte MPhil, M Szynkiewicz MSc, D S Chase PhD, Y J Crow PhD); Erasme University Hospital, Universite Libre de Bruxelles (ULB), Brussels, Belgium, (A Aeby MD, C N Vilain MD, V Wermenbol MD); Medical Molecular Genetics Department, (M S Abdel-Hamid MSc, L Effat PhD), and Clinical Genetics Department, (S A Temtamy PhD, M S Aglan PhD, M S Zaki PhD, G M H Abdel-Salam PhD), Human Genetics and Genome Research Division, National Research Centre, Cairo, Egypt; Bradford Teaching Hospitals NHS Foundation Trust, Bradford, UK (S Ackroyd MB ChB); Department of Clinical Biochemistry, Lancashire Teaching Hospitals NHS Foundation Trust, Preston, UK (R Allcock FRCPath); University Hospitals of Coventry and Warwickshire, Coventry, UK (K M Bailey MRCPCH); Department of Brain and Behavioral Sciences, Unit of Child Neurology and Psychiatry, University of Pavia, Pavia, Italy, (U Balottin MD); Child Neurology and Psychiatry Unit, (U Balottin, I Olivieri MD, S Orcesi MD), and Laboratory of Experimental Neurobiology (C Cereda PhD), C. Mondino National Neurological Institute, Pavia, Italy; Praticien Hospitalier, Unité de Neurologie Pédiatrique et Centre de Référence des Maladies Neuromusculaires, Hôpital Necker-Enfants Malades, Paris, France (C Barnerias MD); Department of Pediatrics, and Department of Neurology and Neurosurgery, Montreal Children’s Hospital, McGill University Health Center, Montreal, QC, Canada (G Bernard MD); Department of Dermatology, Reference Centre for Cutaneous Rare Diseases (MAGEC), Necker-Enfants Malades APHP, Sorbonne Paris Cité, Université Paris Descartes, Institut Imagine, Paris, France (C Bodemer PhD); Department of Pediatrics, Hospital Universitario Araba, Vitoria-Gasteiz, Spain (M P Botella MD); Manchester Centre for Genomic Medicine, Central Manchester University Hospitals NHS Foundation Trust, MAHSC, Manchester, UK (K E Chandler MD, Y J Crow); Department of Paediatric Neurology, University Hospitals of Leicester NHS Trust, Leicester, UK (L Dabydeen MRCPCH); TY Nelson Department of Neurology, and Neuroimmunology Group, the Children’s Hospital at Westmead, University of Sydney, Sydney, NSW, Australia (R C Dale PhD); Nutrition and Metabolism Unit, Hôpital Universitaire des Enfants Reine Fabiola (ULB), Brussels, Belgium (C De Laet MD); Department of Paediatric Neurology, Royal Preston Hospital, Preston, UK (C G E L De Goede FRCPCH); Pediatric Neurology Unit, Hospital Vall d’Hebron, Barcelona, Spain (M del Toro MD); Paediatric Neurology Unit, University Hospital 12 Octubre, Madrid, Spain (N Nunez Enamorado MD, R Simon MD); Department of Clinical and Experimental Sciences, Child Neurology and Psychiatry Unit, University of Brescia, Brescia, Italy (E Fazzi MD); Department of Genetics, BioCruces Health Research Institute, Hospital, Universitario Cruces, Bizkaia, Spain (B Gener MD); Institute of Biomedicine and Translational Medicine, University of Tartu, Tartu, Estonia (M Haldre BSc, P Peterson PhD, K Kisand PhD); General Neurology and Complex Motor Disorders Service, Evelina Children’s Hospital, Guy’s and St Thomas’ NHS Foundation Trust, London, UK (J-P S-M Lin PhD); Department of Paediatric Neurology, Leeds General Infirmary, Leeds, UK (J H Livingston MB ChB); Department of Neurosciences and Behavior Sciences, School of Medicine of Ribeirão Preto, University of São Paulo, São Paulo, Brazil, (C Marques Lourenco MD, W Marques Jr MD); Royal Devon and Exeter Foundation NHS Trust, Exeter, UK (P Oades FRCPCH); Section for Child Neurology, Women and Children’s Division, Oslo University Hospital, Oslo, Norway (M Rasmussen MD); Neuropediatrie, Hopital Gui de Chauliac, and INSERM U1051, INM, Montpellier, France (A Roubertie MD); Center for Genetic Medicine Research and Department of Neurology, Children’s National Medical Center, George Washington University School of Medicine, Washington, DC, USA, (J Loewenstein Schmidt CGC, A Vanderver MD); The Genetic Institute, Ha’Emek Medical Center, Afula and the Rappaport Faculty of Medicine, Technion, Haifa, Israel (S A Shalev MD, R Spiegel MD); Department of Neurology, University of Utah School of Medicine, Salt Lake City, UT, USA (K J Swoboda FACMG); Neurology Department, Royal Manchester Children’s Hospital, Manchester, UK (G Vassallo MD); Centre for Rare Diseases and Personalised Medicine and Department of Medical and Molecular Genetics, School of Clinical and Experimental Medicine, University of Birmingham, UK, and West Midlands Regional Genetics Service, Clinical Genetics Unit, Birmingham Women’s Hospital, Birmingham, UK (J Vogt MRCP); Nottingham Children’s Hospital, Nottingham University Hospitals NHS Trust, Nottingham, UK (W P Whitehouse FRCPCH); Neuropaediatrics, Department of Paediatrics, Mater Dei Hospital, Tal-Qroqq, Malta (D Soler MD); and Université Paris Descartes, EA 1833 and Hôpital Cochin, Service de Virologie, Paris, France, (F Rozenberg MD, P Lebon MD)
1,
Mona S Aglan
Mona S Aglan
1Manchester Centre for Genomic Medicine, Institute of Human Development, Faculty of Medical and Human Sciences, University of Manchester, Manchester Academic Health Sciences Centre (MAHSC), Manchester, UK (G I Rice PhD, G M A Forte MPhil, M Szynkiewicz MSc, D S Chase PhD, Y J Crow PhD); Erasme University Hospital, Universite Libre de Bruxelles (ULB), Brussels, Belgium, (A Aeby MD, C N Vilain MD, V Wermenbol MD); Medical Molecular Genetics Department, (M S Abdel-Hamid MSc, L Effat PhD), and Clinical Genetics Department, (S A Temtamy PhD, M S Aglan PhD, M S Zaki PhD, G M H Abdel-Salam PhD), Human Genetics and Genome Research Division, National Research Centre, Cairo, Egypt; Bradford Teaching Hospitals NHS Foundation Trust, Bradford, UK (S Ackroyd MB ChB); Department of Clinical Biochemistry, Lancashire Teaching Hospitals NHS Foundation Trust, Preston, UK (R Allcock FRCPath); University Hospitals of Coventry and Warwickshire, Coventry, UK (K M Bailey MRCPCH); Department of Brain and Behavioral Sciences, Unit of Child Neurology and Psychiatry, University of Pavia, Pavia, Italy, (U Balottin MD); Child Neurology and Psychiatry Unit, (U Balottin, I Olivieri MD, S Orcesi MD), and Laboratory of Experimental Neurobiology (C Cereda PhD), C. Mondino National Neurological Institute, Pavia, Italy; Praticien Hospitalier, Unité de Neurologie Pédiatrique et Centre de Référence des Maladies Neuromusculaires, Hôpital Necker-Enfants Malades, Paris, France (C Barnerias MD); Department of Pediatrics, and Department of Neurology and Neurosurgery, Montreal Children’s Hospital, McGill University Health Center, Montreal, QC, Canada (G Bernard MD); Department of Dermatology, Reference Centre for Cutaneous Rare Diseases (MAGEC), Necker-Enfants Malades APHP, Sorbonne Paris Cité, Université Paris Descartes, Institut Imagine, Paris, France (C Bodemer PhD); Department of Pediatrics, Hospital Universitario Araba, Vitoria-Gasteiz, Spain (M P Botella MD); Manchester Centre for Genomic Medicine, Central Manchester University Hospitals NHS Foundation Trust, MAHSC, Manchester, UK (K E Chandler MD, Y J Crow); Department of Paediatric Neurology, University Hospitals of Leicester NHS Trust, Leicester, UK (L Dabydeen MRCPCH); TY Nelson Department of Neurology, and Neuroimmunology Group, the Children’s Hospital at Westmead, University of Sydney, Sydney, NSW, Australia (R C Dale PhD); Nutrition and Metabolism Unit, Hôpital Universitaire des Enfants Reine Fabiola (ULB), Brussels, Belgium (C De Laet MD); Department of Paediatric Neurology, Royal Preston Hospital, Preston, UK (C G E L De Goede FRCPCH); Pediatric Neurology Unit, Hospital Vall d’Hebron, Barcelona, Spain (M del Toro MD); Paediatric Neurology Unit, University Hospital 12 Octubre, Madrid, Spain (N Nunez Enamorado MD, R Simon MD); Department of Clinical and Experimental Sciences, Child Neurology and Psychiatry Unit, University of Brescia, Brescia, Italy (E Fazzi MD); Department of Genetics, BioCruces Health Research Institute, Hospital, Universitario Cruces, Bizkaia, Spain (B Gener MD); Institute of Biomedicine and Translational Medicine, University of Tartu, Tartu, Estonia (M Haldre BSc, P Peterson PhD, K Kisand PhD); General Neurology and Complex Motor Disorders Service, Evelina Children’s Hospital, Guy’s and St Thomas’ NHS Foundation Trust, London, UK (J-P S-M Lin PhD); Department of Paediatric Neurology, Leeds General Infirmary, Leeds, UK (J H Livingston MB ChB); Department of Neurosciences and Behavior Sciences, School of Medicine of Ribeirão Preto, University of São Paulo, São Paulo, Brazil, (C Marques Lourenco MD, W Marques Jr MD); Royal Devon and Exeter Foundation NHS Trust, Exeter, UK (P Oades FRCPCH); Section for Child Neurology, Women and Children’s Division, Oslo University Hospital, Oslo, Norway (M Rasmussen MD); Neuropediatrie, Hopital Gui de Chauliac, and INSERM U1051, INM, Montpellier, France (A Roubertie MD); Center for Genetic Medicine Research and Department of Neurology, Children’s National Medical Center, George Washington University School of Medicine, Washington, DC, USA, (J Loewenstein Schmidt CGC, A Vanderver MD); The Genetic Institute, Ha’Emek Medical Center, Afula and the Rappaport Faculty of Medicine, Technion, Haifa, Israel (S A Shalev MD, R Spiegel MD); Department of Neurology, University of Utah School of Medicine, Salt Lake City, UT, USA (K J Swoboda FACMG); Neurology Department, Royal Manchester Children’s Hospital, Manchester, UK (G Vassallo MD); Centre for Rare Diseases and Personalised Medicine and Department of Medical and Molecular Genetics, School of Clinical and Experimental Medicine, University of Birmingham, UK, and West Midlands Regional Genetics Service, Clinical Genetics Unit, Birmingham Women’s Hospital, Birmingham, UK (J Vogt MRCP); Nottingham Children’s Hospital, Nottingham University Hospitals NHS Trust, Nottingham, UK (W P Whitehouse FRCPCH); Neuropaediatrics, Department of Paediatrics, Mater Dei Hospital, Tal-Qroqq, Malta (D Soler MD); and Université Paris Descartes, EA 1833 and Hôpital Cochin, Service de Virologie, Paris, France, (F Rozenberg MD, P Lebon MD)
1,
Maha S Zaki
Maha S Zaki
1Manchester Centre for Genomic Medicine, Institute of Human Development, Faculty of Medical and Human Sciences, University of Manchester, Manchester Academic Health Sciences Centre (MAHSC), Manchester, UK (G I Rice PhD, G M A Forte MPhil, M Szynkiewicz MSc, D S Chase PhD, Y J Crow PhD); Erasme University Hospital, Universite Libre de Bruxelles (ULB), Brussels, Belgium, (A Aeby MD, C N Vilain MD, V Wermenbol MD); Medical Molecular Genetics Department, (M S Abdel-Hamid MSc, L Effat PhD), and Clinical Genetics Department, (S A Temtamy PhD, M S Aglan PhD, M S Zaki PhD, G M H Abdel-Salam PhD), Human Genetics and Genome Research Division, National Research Centre, Cairo, Egypt; Bradford Teaching Hospitals NHS Foundation Trust, Bradford, UK (S Ackroyd MB ChB); Department of Clinical Biochemistry, Lancashire Teaching Hospitals NHS Foundation Trust, Preston, UK (R Allcock FRCPath); University Hospitals of Coventry and Warwickshire, Coventry, UK (K M Bailey MRCPCH); Department of Brain and Behavioral Sciences, Unit of Child Neurology and Psychiatry, University of Pavia, Pavia, Italy, (U Balottin MD); Child Neurology and Psychiatry Unit, (U Balottin, I Olivieri MD, S Orcesi MD), and Laboratory of Experimental Neurobiology (C Cereda PhD), C. Mondino National Neurological Institute, Pavia, Italy; Praticien Hospitalier, Unité de Neurologie Pédiatrique et Centre de Référence des Maladies Neuromusculaires, Hôpital Necker-Enfants Malades, Paris, France (C Barnerias MD); Department of Pediatrics, and Department of Neurology and Neurosurgery, Montreal Children’s Hospital, McGill University Health Center, Montreal, QC, Canada (G Bernard MD); Department of Dermatology, Reference Centre for Cutaneous Rare Diseases (MAGEC), Necker-Enfants Malades APHP, Sorbonne Paris Cité, Université Paris Descartes, Institut Imagine, Paris, France (C Bodemer PhD); Department of Pediatrics, Hospital Universitario Araba, Vitoria-Gasteiz, Spain (M P Botella MD); Manchester Centre for Genomic Medicine, Central Manchester University Hospitals NHS Foundation Trust, MAHSC, Manchester, UK (K E Chandler MD, Y J Crow); Department of Paediatric Neurology, University Hospitals of Leicester NHS Trust, Leicester, UK (L Dabydeen MRCPCH); TY Nelson Department of Neurology, and Neuroimmunology Group, the Children’s Hospital at Westmead, University of Sydney, Sydney, NSW, Australia (R C Dale PhD); Nutrition and Metabolism Unit, Hôpital Universitaire des Enfants Reine Fabiola (ULB), Brussels, Belgium (C De Laet MD); Department of Paediatric Neurology, Royal Preston Hospital, Preston, UK (C G E L De Goede FRCPCH); Pediatric Neurology Unit, Hospital Vall d’Hebron, Barcelona, Spain (M del Toro MD); Paediatric Neurology Unit, University Hospital 12 Octubre, Madrid, Spain (N Nunez Enamorado MD, R Simon MD); Department of Clinical and Experimental Sciences, Child Neurology and Psychiatry Unit, University of Brescia, Brescia, Italy (E Fazzi MD); Department of Genetics, BioCruces Health Research Institute, Hospital, Universitario Cruces, Bizkaia, Spain (B Gener MD); Institute of Biomedicine and Translational Medicine, University of Tartu, Tartu, Estonia (M Haldre BSc, P Peterson PhD, K Kisand PhD); General Neurology and Complex Motor Disorders Service, Evelina Children’s Hospital, Guy’s and St Thomas’ NHS Foundation Trust, London, UK (J-P S-M Lin PhD); Department of Paediatric Neurology, Leeds General Infirmary, Leeds, UK (J H Livingston MB ChB); Department of Neurosciences and Behavior Sciences, School of Medicine of Ribeirão Preto, University of São Paulo, São Paulo, Brazil, (C Marques Lourenco MD, W Marques Jr MD); Royal Devon and Exeter Foundation NHS Trust, Exeter, UK (P Oades FRCPCH); Section for Child Neurology, Women and Children’s Division, Oslo University Hospital, Oslo, Norway (M Rasmussen MD); Neuropediatrie, Hopital Gui de Chauliac, and INSERM U1051, INM, Montpellier, France (A Roubertie MD); Center for Genetic Medicine Research and Department of Neurology, Children’s National Medical Center, George Washington University School of Medicine, Washington, DC, USA, (J Loewenstein Schmidt CGC, A Vanderver MD); The Genetic Institute, Ha’Emek Medical Center, Afula and the Rappaport Faculty of Medicine, Technion, Haifa, Israel (S A Shalev MD, R Spiegel MD); Department of Neurology, University of Utah School of Medicine, Salt Lake City, UT, USA (K J Swoboda FACMG); Neurology Department, Royal Manchester Children’s Hospital, Manchester, UK (G Vassallo MD); Centre for Rare Diseases and Personalised Medicine and Department of Medical and Molecular Genetics, School of Clinical and Experimental Medicine, University of Birmingham, UK, and West Midlands Regional Genetics Service, Clinical Genetics Unit, Birmingham Women’s Hospital, Birmingham, UK (J Vogt MRCP); Nottingham Children’s Hospital, Nottingham University Hospitals NHS Trust, Nottingham, UK (W P Whitehouse FRCPCH); Neuropaediatrics, Department of Paediatrics, Mater Dei Hospital, Tal-Qroqq, Malta (D Soler MD); and Université Paris Descartes, EA 1833 and Hôpital Cochin, Service de Virologie, Paris, France, (F Rozenberg MD, P Lebon MD)
1,
Ghada M H Abdel-Salam
Ghada M H Abdel-Salam
1Manchester Centre for Genomic Medicine, Institute of Human Development, Faculty of Medical and Human Sciences, University of Manchester, Manchester Academic Health Sciences Centre (MAHSC), Manchester, UK (G I Rice PhD, G M A Forte MPhil, M Szynkiewicz MSc, D S Chase PhD, Y J Crow PhD); Erasme University Hospital, Universite Libre de Bruxelles (ULB), Brussels, Belgium, (A Aeby MD, C N Vilain MD, V Wermenbol MD); Medical Molecular Genetics Department, (M S Abdel-Hamid MSc, L Effat PhD), and Clinical Genetics Department, (S A Temtamy PhD, M S Aglan PhD, M S Zaki PhD, G M H Abdel-Salam PhD), Human Genetics and Genome Research Division, National Research Centre, Cairo, Egypt; Bradford Teaching Hospitals NHS Foundation Trust, Bradford, UK (S Ackroyd MB ChB); Department of Clinical Biochemistry, Lancashire Teaching Hospitals NHS Foundation Trust, Preston, UK (R Allcock FRCPath); University Hospitals of Coventry and Warwickshire, Coventry, UK (K M Bailey MRCPCH); Department of Brain and Behavioral Sciences, Unit of Child Neurology and Psychiatry, University of Pavia, Pavia, Italy, (U Balottin MD); Child Neurology and Psychiatry Unit, (U Balottin, I Olivieri MD, S Orcesi MD), and Laboratory of Experimental Neurobiology (C Cereda PhD), C. Mondino National Neurological Institute, Pavia, Italy; Praticien Hospitalier, Unité de Neurologie Pédiatrique et Centre de Référence des Maladies Neuromusculaires, Hôpital Necker-Enfants Malades, Paris, France (C Barnerias MD); Department of Pediatrics, and Department of Neurology and Neurosurgery, Montreal Children’s Hospital, McGill University Health Center, Montreal, QC, Canada (G Bernard MD); Department of Dermatology, Reference Centre for Cutaneous Rare Diseases (MAGEC), Necker-Enfants Malades APHP, Sorbonne Paris Cité, Université Paris Descartes, Institut Imagine, Paris, France (C Bodemer PhD); Department of Pediatrics, Hospital Universitario Araba, Vitoria-Gasteiz, Spain (M P Botella MD); Manchester Centre for Genomic Medicine, Central Manchester University Hospitals NHS Foundation Trust, MAHSC, Manchester, UK (K E Chandler MD, Y J Crow); Department of Paediatric Neurology, University Hospitals of Leicester NHS Trust, Leicester, UK (L Dabydeen MRCPCH); TY Nelson Department of Neurology, and Neuroimmunology Group, the Children’s Hospital at Westmead, University of Sydney, Sydney, NSW, Australia (R C Dale PhD); Nutrition and Metabolism Unit, Hôpital Universitaire des Enfants Reine Fabiola (ULB), Brussels, Belgium (C De Laet MD); Department of Paediatric Neurology, Royal Preston Hospital, Preston, UK (C G E L De Goede FRCPCH); Pediatric Neurology Unit, Hospital Vall d’Hebron, Barcelona, Spain (M del Toro MD); Paediatric Neurology Unit, University Hospital 12 Octubre, Madrid, Spain (N Nunez Enamorado MD, R Simon MD); Department of Clinical and Experimental Sciences, Child Neurology and Psychiatry Unit, University of Brescia, Brescia, Italy (E Fazzi MD); Department of Genetics, BioCruces Health Research Institute, Hospital, Universitario Cruces, Bizkaia, Spain (B Gener MD); Institute of Biomedicine and Translational Medicine, University of Tartu, Tartu, Estonia (M Haldre BSc, P Peterson PhD, K Kisand PhD); General Neurology and Complex Motor Disorders Service, Evelina Children’s Hospital, Guy’s and St Thomas’ NHS Foundation Trust, London, UK (J-P S-M Lin PhD); Department of Paediatric Neurology, Leeds General Infirmary, Leeds, UK (J H Livingston MB ChB); Department of Neurosciences and Behavior Sciences, School of Medicine of Ribeirão Preto, University of São Paulo, São Paulo, Brazil, (C Marques Lourenco MD, W Marques Jr MD); Royal Devon and Exeter Foundation NHS Trust, Exeter, UK (P Oades FRCPCH); Section for Child Neurology, Women and Children’s Division, Oslo University Hospital, Oslo, Norway (M Rasmussen MD); Neuropediatrie, Hopital Gui de Chauliac, and INSERM U1051, INM, Montpellier, France (A Roubertie MD); Center for Genetic Medicine Research and Department of Neurology, Children’s National Medical Center, George Washington University School of Medicine, Washington, DC, USA, (J Loewenstein Schmidt CGC, A Vanderver MD); The Genetic Institute, Ha’Emek Medical Center, Afula and the Rappaport Faculty of Medicine, Technion, Haifa, Israel (S A Shalev MD, R Spiegel MD); Department of Neurology, University of Utah School of Medicine, Salt Lake City, UT, USA (K J Swoboda FACMG); Neurology Department, Royal Manchester Children’s Hospital, Manchester, UK (G Vassallo MD); Centre for Rare Diseases and Personalised Medicine and Department of Medical and Molecular Genetics, School of Clinical and Experimental Medicine, University of Birmingham, UK, and West Midlands Regional Genetics Service, Clinical Genetics Unit, Birmingham Women’s Hospital, Birmingham, UK (J Vogt MRCP); Nottingham Children’s Hospital, Nottingham University Hospitals NHS Trust, Nottingham, UK (W P Whitehouse FRCPCH); Neuropaediatrics, Department of Paediatrics, Mater Dei Hospital, Tal-Qroqq, Malta (D Soler MD); and Université Paris Descartes, EA 1833 and Hôpital Cochin, Service de Virologie, Paris, France, (F Rozenberg MD, P Lebon MD)
1,
Adeline Vanderver
Adeline Vanderver
1Manchester Centre for Genomic Medicine, Institute of Human Development, Faculty of Medical and Human Sciences, University of Manchester, Manchester Academic Health Sciences Centre (MAHSC), Manchester, UK (G I Rice PhD, G M A Forte MPhil, M Szynkiewicz MSc, D S Chase PhD, Y J Crow PhD); Erasme University Hospital, Universite Libre de Bruxelles (ULB), Brussels, Belgium, (A Aeby MD, C N Vilain MD, V Wermenbol MD); Medical Molecular Genetics Department, (M S Abdel-Hamid MSc, L Effat PhD), and Clinical Genetics Department, (S A Temtamy PhD, M S Aglan PhD, M S Zaki PhD, G M H Abdel-Salam PhD), Human Genetics and Genome Research Division, National Research Centre, Cairo, Egypt; Bradford Teaching Hospitals NHS Foundation Trust, Bradford, UK (S Ackroyd MB ChB); Department of Clinical Biochemistry, Lancashire Teaching Hospitals NHS Foundation Trust, Preston, UK (R Allcock FRCPath); University Hospitals of Coventry and Warwickshire, Coventry, UK (K M Bailey MRCPCH); Department of Brain and Behavioral Sciences, Unit of Child Neurology and Psychiatry, University of Pavia, Pavia, Italy, (U Balottin MD); Child Neurology and Psychiatry Unit, (U Balottin, I Olivieri MD, S Orcesi MD), and Laboratory of Experimental Neurobiology (C Cereda PhD), C. Mondino National Neurological Institute, Pavia, Italy; Praticien Hospitalier, Unité de Neurologie Pédiatrique et Centre de Référence des Maladies Neuromusculaires, Hôpital Necker-Enfants Malades, Paris, France (C Barnerias MD); Department of Pediatrics, and Department of Neurology and Neurosurgery, Montreal Children’s Hospital, McGill University Health Center, Montreal, QC, Canada (G Bernard MD); Department of Dermatology, Reference Centre for Cutaneous Rare Diseases (MAGEC), Necker-Enfants Malades APHP, Sorbonne Paris Cité, Université Paris Descartes, Institut Imagine, Paris, France (C Bodemer PhD); Department of Pediatrics, Hospital Universitario Araba, Vitoria-Gasteiz, Spain (M P Botella MD); Manchester Centre for Genomic Medicine, Central Manchester University Hospitals NHS Foundation Trust, MAHSC, Manchester, UK (K E Chandler MD, Y J Crow); Department of Paediatric Neurology, University Hospitals of Leicester NHS Trust, Leicester, UK (L Dabydeen MRCPCH); TY Nelson Department of Neurology, and Neuroimmunology Group, the Children’s Hospital at Westmead, University of Sydney, Sydney, NSW, Australia (R C Dale PhD); Nutrition and Metabolism Unit, Hôpital Universitaire des Enfants Reine Fabiola (ULB), Brussels, Belgium (C De Laet MD); Department of Paediatric Neurology, Royal Preston Hospital, Preston, UK (C G E L De Goede FRCPCH); Pediatric Neurology Unit, Hospital Vall d’Hebron, Barcelona, Spain (M del Toro MD); Paediatric Neurology Unit, University Hospital 12 Octubre, Madrid, Spain (N Nunez Enamorado MD, R Simon MD); Department of Clinical and Experimental Sciences, Child Neurology and Psychiatry Unit, University of Brescia, Brescia, Italy (E Fazzi MD); Department of Genetics, BioCruces Health Research Institute, Hospital, Universitario Cruces, Bizkaia, Spain (B Gener MD); Institute of Biomedicine and Translational Medicine, University of Tartu, Tartu, Estonia (M Haldre BSc, P Peterson PhD, K Kisand PhD); General Neurology and Complex Motor Disorders Service, Evelina Children’s Hospital, Guy’s and St Thomas’ NHS Foundation Trust, London, UK (J-P S-M Lin PhD); Department of Paediatric Neurology, Leeds General Infirmary, Leeds, UK (J H Livingston MB ChB); Department of Neurosciences and Behavior Sciences, School of Medicine of Ribeirão Preto, University of São Paulo, São Paulo, Brazil, (C Marques Lourenco MD, W Marques Jr MD); Royal Devon and Exeter Foundation NHS Trust, Exeter, UK (P Oades FRCPCH); Section for Child Neurology, Women and Children’s Division, Oslo University Hospital, Oslo, Norway (M Rasmussen MD); Neuropediatrie, Hopital Gui de Chauliac, and INSERM U1051, INM, Montpellier, France (A Roubertie MD); Center for Genetic Medicine Research and Department of Neurology, Children’s National Medical Center, George Washington University School of Medicine, Washington, DC, USA, (J Loewenstein Schmidt CGC, A Vanderver MD); The Genetic Institute, Ha’Emek Medical Center, Afula and the Rappaport Faculty of Medicine, Technion, Haifa, Israel (S A Shalev MD, R Spiegel MD); Department of Neurology, University of Utah School of Medicine, Salt Lake City, UT, USA (K J Swoboda FACMG); Neurology Department, Royal Manchester Children’s Hospital, Manchester, UK (G Vassallo MD); Centre for Rare Diseases and Personalised Medicine and Department of Medical and Molecular Genetics, School of Clinical and Experimental Medicine, University of Birmingham, UK, and West Midlands Regional Genetics Service, Clinical Genetics Unit, Birmingham Women’s Hospital, Birmingham, UK (J Vogt MRCP); Nottingham Children’s Hospital, Nottingham University Hospitals NHS Trust, Nottingham, UK (W P Whitehouse FRCPCH); Neuropaediatrics, Department of Paediatrics, Mater Dei Hospital, Tal-Qroqq, Malta (D Soler MD); and Université Paris Descartes, EA 1833 and Hôpital Cochin, Service de Virologie, Paris, France, (F Rozenberg MD, P Lebon MD)
1,
Kai Kisand
Kai Kisand
1Manchester Centre for Genomic Medicine, Institute of Human Development, Faculty of Medical and Human Sciences, University of Manchester, Manchester Academic Health Sciences Centre (MAHSC), Manchester, UK (G I Rice PhD, G M A Forte MPhil, M Szynkiewicz MSc, D S Chase PhD, Y J Crow PhD); Erasme University Hospital, Universite Libre de Bruxelles (ULB), Brussels, Belgium, (A Aeby MD, C N Vilain MD, V Wermenbol MD); Medical Molecular Genetics Department, (M S Abdel-Hamid MSc, L Effat PhD), and Clinical Genetics Department, (S A Temtamy PhD, M S Aglan PhD, M S Zaki PhD, G M H Abdel-Salam PhD), Human Genetics and Genome Research Division, National Research Centre, Cairo, Egypt; Bradford Teaching Hospitals NHS Foundation Trust, Bradford, UK (S Ackroyd MB ChB); Department of Clinical Biochemistry, Lancashire Teaching Hospitals NHS Foundation Trust, Preston, UK (R Allcock FRCPath); University Hospitals of Coventry and Warwickshire, Coventry, UK (K M Bailey MRCPCH); Department of Brain and Behavioral Sciences, Unit of Child Neurology and Psychiatry, University of Pavia, Pavia, Italy, (U Balottin MD); Child Neurology and Psychiatry Unit, (U Balottin, I Olivieri MD, S Orcesi MD), and Laboratory of Experimental Neurobiology (C Cereda PhD), C. Mondino National Neurological Institute, Pavia, Italy; Praticien Hospitalier, Unité de Neurologie Pédiatrique et Centre de Référence des Maladies Neuromusculaires, Hôpital Necker-Enfants Malades, Paris, France (C Barnerias MD); Department of Pediatrics, and Department of Neurology and Neurosurgery, Montreal Children’s Hospital, McGill University Health Center, Montreal, QC, Canada (G Bernard MD); Department of Dermatology, Reference Centre for Cutaneous Rare Diseases (MAGEC), Necker-Enfants Malades APHP, Sorbonne Paris Cité, Université Paris Descartes, Institut Imagine, Paris, France (C Bodemer PhD); Department of Pediatrics, Hospital Universitario Araba, Vitoria-Gasteiz, Spain (M P Botella MD); Manchester Centre for Genomic Medicine, Central Manchester University Hospitals NHS Foundation Trust, MAHSC, Manchester, UK (K E Chandler MD, Y J Crow); Department of Paediatric Neurology, University Hospitals of Leicester NHS Trust, Leicester, UK (L Dabydeen MRCPCH); TY Nelson Department of Neurology, and Neuroimmunology Group, the Children’s Hospital at Westmead, University of Sydney, Sydney, NSW, Australia (R C Dale PhD); Nutrition and Metabolism Unit, Hôpital Universitaire des Enfants Reine Fabiola (ULB), Brussels, Belgium (C De Laet MD); Department of Paediatric Neurology, Royal Preston Hospital, Preston, UK (C G E L De Goede FRCPCH); Pediatric Neurology Unit, Hospital Vall d’Hebron, Barcelona, Spain (M del Toro MD); Paediatric Neurology Unit, University Hospital 12 Octubre, Madrid, Spain (N Nunez Enamorado MD, R Simon MD); Department of Clinical and Experimental Sciences, Child Neurology and Psychiatry Unit, University of Brescia, Brescia, Italy (E Fazzi MD); Department of Genetics, BioCruces Health Research Institute, Hospital, Universitario Cruces, Bizkaia, Spain (B Gener MD); Institute of Biomedicine and Translational Medicine, University of Tartu, Tartu, Estonia (M Haldre BSc, P Peterson PhD, K Kisand PhD); General Neurology and Complex Motor Disorders Service, Evelina Children’s Hospital, Guy’s and St Thomas’ NHS Foundation Trust, London, UK (J-P S-M Lin PhD); Department of Paediatric Neurology, Leeds General Infirmary, Leeds, UK (J H Livingston MB ChB); Department of Neurosciences and Behavior Sciences, School of Medicine of Ribeirão Preto, University of São Paulo, São Paulo, Brazil, (C Marques Lourenco MD, W Marques Jr MD); Royal Devon and Exeter Foundation NHS Trust, Exeter, UK (P Oades FRCPCH); Section for Child Neurology, Women and Children’s Division, Oslo University Hospital, Oslo, Norway (M Rasmussen MD); Neuropediatrie, Hopital Gui de Chauliac, and INSERM U1051, INM, Montpellier, France (A Roubertie MD); Center for Genetic Medicine Research and Department of Neurology, Children’s National Medical Center, George Washington University School of Medicine, Washington, DC, USA, (J Loewenstein Schmidt CGC, A Vanderver MD); The Genetic Institute, Ha’Emek Medical Center, Afula and the Rappaport Faculty of Medicine, Technion, Haifa, Israel (S A Shalev MD, R Spiegel MD); Department of Neurology, University of Utah School of Medicine, Salt Lake City, UT, USA (K J Swoboda FACMG); Neurology Department, Royal Manchester Children’s Hospital, Manchester, UK (G Vassallo MD); Centre for Rare Diseases and Personalised Medicine and Department of Medical and Molecular Genetics, School of Clinical and Experimental Medicine, University of Birmingham, UK, and West Midlands Regional Genetics Service, Clinical Genetics Unit, Birmingham Women’s Hospital, Birmingham, UK (J Vogt MRCP); Nottingham Children’s Hospital, Nottingham University Hospitals NHS Trust, Nottingham, UK (W P Whitehouse FRCPCH); Neuropaediatrics, Department of Paediatrics, Mater Dei Hospital, Tal-Qroqq, Malta (D Soler MD); and Université Paris Descartes, EA 1833 and Hôpital Cochin, Service de Virologie, Paris, France, (F Rozenberg MD, P Lebon MD)
1,
Flore Rozenberg
Flore Rozenberg
1Manchester Centre for Genomic Medicine, Institute of Human Development, Faculty of Medical and Human Sciences, University of Manchester, Manchester Academic Health Sciences Centre (MAHSC), Manchester, UK (G I Rice PhD, G M A Forte MPhil, M Szynkiewicz MSc, D S Chase PhD, Y J Crow PhD); Erasme University Hospital, Universite Libre de Bruxelles (ULB), Brussels, Belgium, (A Aeby MD, C N Vilain MD, V Wermenbol MD); Medical Molecular Genetics Department, (M S Abdel-Hamid MSc, L Effat PhD), and Clinical Genetics Department, (S A Temtamy PhD, M S Aglan PhD, M S Zaki PhD, G M H Abdel-Salam PhD), Human Genetics and Genome Research Division, National Research Centre, Cairo, Egypt; Bradford Teaching Hospitals NHS Foundation Trust, Bradford, UK (S Ackroyd MB ChB); Department of Clinical Biochemistry, Lancashire Teaching Hospitals NHS Foundation Trust, Preston, UK (R Allcock FRCPath); University Hospitals of Coventry and Warwickshire, Coventry, UK (K M Bailey MRCPCH); Department of Brain and Behavioral Sciences, Unit of Child Neurology and Psychiatry, University of Pavia, Pavia, Italy, (U Balottin MD); Child Neurology and Psychiatry Unit, (U Balottin, I Olivieri MD, S Orcesi MD), and Laboratory of Experimental Neurobiology (C Cereda PhD), C. Mondino National Neurological Institute, Pavia, Italy; Praticien Hospitalier, Unité de Neurologie Pédiatrique et Centre de Référence des Maladies Neuromusculaires, Hôpital Necker-Enfants Malades, Paris, France (C Barnerias MD); Department of Pediatrics, and Department of Neurology and Neurosurgery, Montreal Children’s Hospital, McGill University Health Center, Montreal, QC, Canada (G Bernard MD); Department of Dermatology, Reference Centre for Cutaneous Rare Diseases (MAGEC), Necker-Enfants Malades APHP, Sorbonne Paris Cité, Université Paris Descartes, Institut Imagine, Paris, France (C Bodemer PhD); Department of Pediatrics, Hospital Universitario Araba, Vitoria-Gasteiz, Spain (M P Botella MD); Manchester Centre for Genomic Medicine, Central Manchester University Hospitals NHS Foundation Trust, MAHSC, Manchester, UK (K E Chandler MD, Y J Crow); Department of Paediatric Neurology, University Hospitals of Leicester NHS Trust, Leicester, UK (L Dabydeen MRCPCH); TY Nelson Department of Neurology, and Neuroimmunology Group, the Children’s Hospital at Westmead, University of Sydney, Sydney, NSW, Australia (R C Dale PhD); Nutrition and Metabolism Unit, Hôpital Universitaire des Enfants Reine Fabiola (ULB), Brussels, Belgium (C De Laet MD); Department of Paediatric Neurology, Royal Preston Hospital, Preston, UK (C G E L De Goede FRCPCH); Pediatric Neurology Unit, Hospital Vall d’Hebron, Barcelona, Spain (M del Toro MD); Paediatric Neurology Unit, University Hospital 12 Octubre, Madrid, Spain (N Nunez Enamorado MD, R Simon MD); Department of Clinical and Experimental Sciences, Child Neurology and Psychiatry Unit, University of Brescia, Brescia, Italy (E Fazzi MD); Department of Genetics, BioCruces Health Research Institute, Hospital, Universitario Cruces, Bizkaia, Spain (B Gener MD); Institute of Biomedicine and Translational Medicine, University of Tartu, Tartu, Estonia (M Haldre BSc, P Peterson PhD, K Kisand PhD); General Neurology and Complex Motor Disorders Service, Evelina Children’s Hospital, Guy’s and St Thomas’ NHS Foundation Trust, London, UK (J-P S-M Lin PhD); Department of Paediatric Neurology, Leeds General Infirmary, Leeds, UK (J H Livingston MB ChB); Department of Neurosciences and Behavior Sciences, School of Medicine of Ribeirão Preto, University of São Paulo, São Paulo, Brazil, (C Marques Lourenco MD, W Marques Jr MD); Royal Devon and Exeter Foundation NHS Trust, Exeter, UK (P Oades FRCPCH); Section for Child Neurology, Women and Children’s Division, Oslo University Hospital, Oslo, Norway (M Rasmussen MD); Neuropediatrie, Hopital Gui de Chauliac, and INSERM U1051, INM, Montpellier, France (A Roubertie MD); Center for Genetic Medicine Research and Department of Neurology, Children’s National Medical Center, George Washington University School of Medicine, Washington, DC, USA, (J Loewenstein Schmidt CGC, A Vanderver MD); The Genetic Institute, Ha’Emek Medical Center, Afula and the Rappaport Faculty of Medicine, Technion, Haifa, Israel (S A Shalev MD, R Spiegel MD); Department of Neurology, University of Utah School of Medicine, Salt Lake City, UT, USA (K J Swoboda FACMG); Neurology Department, Royal Manchester Children’s Hospital, Manchester, UK (G Vassallo MD); Centre for Rare Diseases and Personalised Medicine and Department of Medical and Molecular Genetics, School of Clinical and Experimental Medicine, University of Birmingham, UK, and West Midlands Regional Genetics Service, Clinical Genetics Unit, Birmingham Women’s Hospital, Birmingham, UK (J Vogt MRCP); Nottingham Children’s Hospital, Nottingham University Hospitals NHS Trust, Nottingham, UK (W P Whitehouse FRCPCH); Neuropaediatrics, Department of Paediatrics, Mater Dei Hospital, Tal-Qroqq, Malta (D Soler MD); and Université Paris Descartes, EA 1833 and Hôpital Cochin, Service de Virologie, Paris, France, (F Rozenberg MD, P Lebon MD)
1,
Pierre Lebon
Pierre Lebon
1Manchester Centre for Genomic Medicine, Institute of Human Development, Faculty of Medical and Human Sciences, University of Manchester, Manchester Academic Health Sciences Centre (MAHSC), Manchester, UK (G I Rice PhD, G M A Forte MPhil, M Szynkiewicz MSc, D S Chase PhD, Y J Crow PhD); Erasme University Hospital, Universite Libre de Bruxelles (ULB), Brussels, Belgium, (A Aeby MD, C N Vilain MD, V Wermenbol MD); Medical Molecular Genetics Department, (M S Abdel-Hamid MSc, L Effat PhD), and Clinical Genetics Department, (S A Temtamy PhD, M S Aglan PhD, M S Zaki PhD, G M H Abdel-Salam PhD), Human Genetics and Genome Research Division, National Research Centre, Cairo, Egypt; Bradford Teaching Hospitals NHS Foundation Trust, Bradford, UK (S Ackroyd MB ChB); Department of Clinical Biochemistry, Lancashire Teaching Hospitals NHS Foundation Trust, Preston, UK (R Allcock FRCPath); University Hospitals of Coventry and Warwickshire, Coventry, UK (K M Bailey MRCPCH); Department of Brain and Behavioral Sciences, Unit of Child Neurology and Psychiatry, University of Pavia, Pavia, Italy, (U Balottin MD); Child Neurology and Psychiatry Unit, (U Balottin, I Olivieri MD, S Orcesi MD), and Laboratory of Experimental Neurobiology (C Cereda PhD), C. Mondino National Neurological Institute, Pavia, Italy; Praticien Hospitalier, Unité de Neurologie Pédiatrique et Centre de Référence des Maladies Neuromusculaires, Hôpital Necker-Enfants Malades, Paris, France (C Barnerias MD); Department of Pediatrics, and Department of Neurology and Neurosurgery, Montreal Children’s Hospital, McGill University Health Center, Montreal, QC, Canada (G Bernard MD); Department of Dermatology, Reference Centre for Cutaneous Rare Diseases (MAGEC), Necker-Enfants Malades APHP, Sorbonne Paris Cité, Université Paris Descartes, Institut Imagine, Paris, France (C Bodemer PhD); Department of Pediatrics, Hospital Universitario Araba, Vitoria-Gasteiz, Spain (M P Botella MD); Manchester Centre for Genomic Medicine, Central Manchester University Hospitals NHS Foundation Trust, MAHSC, Manchester, UK (K E Chandler MD, Y J Crow); Department of Paediatric Neurology, University Hospitals of Leicester NHS Trust, Leicester, UK (L Dabydeen MRCPCH); TY Nelson Department of Neurology, and Neuroimmunology Group, the Children’s Hospital at Westmead, University of Sydney, Sydney, NSW, Australia (R C Dale PhD); Nutrition and Metabolism Unit, Hôpital Universitaire des Enfants Reine Fabiola (ULB), Brussels, Belgium (C De Laet MD); Department of Paediatric Neurology, Royal Preston Hospital, Preston, UK (C G E L De Goede FRCPCH); Pediatric Neurology Unit, Hospital Vall d’Hebron, Barcelona, Spain (M del Toro MD); Paediatric Neurology Unit, University Hospital 12 Octubre, Madrid, Spain (N Nunez Enamorado MD, R Simon MD); Department of Clinical and Experimental Sciences, Child Neurology and Psychiatry Unit, University of Brescia, Brescia, Italy (E Fazzi MD); Department of Genetics, BioCruces Health Research Institute, Hospital, Universitario Cruces, Bizkaia, Spain (B Gener MD); Institute of Biomedicine and Translational Medicine, University of Tartu, Tartu, Estonia (M Haldre BSc, P Peterson PhD, K Kisand PhD); General Neurology and Complex Motor Disorders Service, Evelina Children’s Hospital, Guy’s and St Thomas’ NHS Foundation Trust, London, UK (J-P S-M Lin PhD); Department of Paediatric Neurology, Leeds General Infirmary, Leeds, UK (J H Livingston MB ChB); Department of Neurosciences and Behavior Sciences, School of Medicine of Ribeirão Preto, University of São Paulo, São Paulo, Brazil, (C Marques Lourenco MD, W Marques Jr MD); Royal Devon and Exeter Foundation NHS Trust, Exeter, UK (P Oades FRCPCH); Section for Child Neurology, Women and Children’s Division, Oslo University Hospital, Oslo, Norway (M Rasmussen MD); Neuropediatrie, Hopital Gui de Chauliac, and INSERM U1051, INM, Montpellier, France (A Roubertie MD); Center for Genetic Medicine Research and Department of Neurology, Children’s National Medical Center, George Washington University School of Medicine, Washington, DC, USA, (J Loewenstein Schmidt CGC, A Vanderver MD); The Genetic Institute, Ha’Emek Medical Center, Afula and the Rappaport Faculty of Medicine, Technion, Haifa, Israel (S A Shalev MD, R Spiegel MD); Department of Neurology, University of Utah School of Medicine, Salt Lake City, UT, USA (K J Swoboda FACMG); Neurology Department, Royal Manchester Children’s Hospital, Manchester, UK (G Vassallo MD); Centre for Rare Diseases and Personalised Medicine and Department of Medical and Molecular Genetics, School of Clinical and Experimental Medicine, University of Birmingham, UK, and West Midlands Regional Genetics Service, Clinical Genetics Unit, Birmingham Women’s Hospital, Birmingham, UK (J Vogt MRCP); Nottingham Children’s Hospital, Nottingham University Hospitals NHS Trust, Nottingham, UK (W P Whitehouse FRCPCH); Neuropaediatrics, Department of Paediatrics, Mater Dei Hospital, Tal-Qroqq, Malta (D Soler MD); and Université Paris Descartes, EA 1833 and Hôpital Cochin, Service de Virologie, Paris, France, (F Rozenberg MD, P Lebon MD)
1,
Yanick J Crow
Yanick J Crow
1Manchester Centre for Genomic Medicine, Institute of Human Development, Faculty of Medical and Human Sciences, University of Manchester, Manchester Academic Health Sciences Centre (MAHSC), Manchester, UK (G I Rice PhD, G M A Forte MPhil, M Szynkiewicz MSc, D S Chase PhD, Y J Crow PhD); Erasme University Hospital, Universite Libre de Bruxelles (ULB), Brussels, Belgium, (A Aeby MD, C N Vilain MD, V Wermenbol MD); Medical Molecular Genetics Department, (M S Abdel-Hamid MSc, L Effat PhD), and Clinical Genetics Department, (S A Temtamy PhD, M S Aglan PhD, M S Zaki PhD, G M H Abdel-Salam PhD), Human Genetics and Genome Research Division, National Research Centre, Cairo, Egypt; Bradford Teaching Hospitals NHS Foundation Trust, Bradford, UK (S Ackroyd MB ChB); Department of Clinical Biochemistry, Lancashire Teaching Hospitals NHS Foundation Trust, Preston, UK (R Allcock FRCPath); University Hospitals of Coventry and Warwickshire, Coventry, UK (K M Bailey MRCPCH); Department of Brain and Behavioral Sciences, Unit of Child Neurology and Psychiatry, University of Pavia, Pavia, Italy, (U Balottin MD); Child Neurology and Psychiatry Unit, (U Balottin, I Olivieri MD, S Orcesi MD), and Laboratory of Experimental Neurobiology (C Cereda PhD), C. Mondino National Neurological Institute, Pavia, Italy; Praticien Hospitalier, Unité de Neurologie Pédiatrique et Centre de Référence des Maladies Neuromusculaires, Hôpital Necker-Enfants Malades, Paris, France (C Barnerias MD); Department of Pediatrics, and Department of Neurology and Neurosurgery, Montreal Children’s Hospital, McGill University Health Center, Montreal, QC, Canada (G Bernard MD); Department of Dermatology, Reference Centre for Cutaneous Rare Diseases (MAGEC), Necker-Enfants Malades APHP, Sorbonne Paris Cité, Université Paris Descartes, Institut Imagine, Paris, France (C Bodemer PhD); Department of Pediatrics, Hospital Universitario Araba, Vitoria-Gasteiz, Spain (M P Botella MD); Manchester Centre for Genomic Medicine, Central Manchester University Hospitals NHS Foundation Trust, MAHSC, Manchester, UK (K E Chandler MD, Y J Crow); Department of Paediatric Neurology, University Hospitals of Leicester NHS Trust, Leicester, UK (L Dabydeen MRCPCH); TY Nelson Department of Neurology, and Neuroimmunology Group, the Children’s Hospital at Westmead, University of Sydney, Sydney, NSW, Australia (R C Dale PhD); Nutrition and Metabolism Unit, Hôpital Universitaire des Enfants Reine Fabiola (ULB), Brussels, Belgium (C De Laet MD); Department of Paediatric Neurology, Royal Preston Hospital, Preston, UK (C G E L De Goede FRCPCH); Pediatric Neurology Unit, Hospital Vall d’Hebron, Barcelona, Spain (M del Toro MD); Paediatric Neurology Unit, University Hospital 12 Octubre, Madrid, Spain (N Nunez Enamorado MD, R Simon MD); Department of Clinical and Experimental Sciences, Child Neurology and Psychiatry Unit, University of Brescia, Brescia, Italy (E Fazzi MD); Department of Genetics, BioCruces Health Research Institute, Hospital, Universitario Cruces, Bizkaia, Spain (B Gener MD); Institute of Biomedicine and Translational Medicine, University of Tartu, Tartu, Estonia (M Haldre BSc, P Peterson PhD, K Kisand PhD); General Neurology and Complex Motor Disorders Service, Evelina Children’s Hospital, Guy’s and St Thomas’ NHS Foundation Trust, London, UK (J-P S-M Lin PhD); Department of Paediatric Neurology, Leeds General Infirmary, Leeds, UK (J H Livingston MB ChB); Department of Neurosciences and Behavior Sciences, School of Medicine of Ribeirão Preto, University of São Paulo, São Paulo, Brazil, (C Marques Lourenco MD, W Marques Jr MD); Royal Devon and Exeter Foundation NHS Trust, Exeter, UK (P Oades FRCPCH); Section for Child Neurology, Women and Children’s Division, Oslo University Hospital, Oslo, Norway (M Rasmussen MD); Neuropediatrie, Hopital Gui de Chauliac, and INSERM U1051, INM, Montpellier, France (A Roubertie MD); Center for Genetic Medicine Research and Department of Neurology, Children’s National Medical Center, George Washington University School of Medicine, Washington, DC, USA, (J Loewenstein Schmidt CGC, A Vanderver MD); The Genetic Institute, Ha’Emek Medical Center, Afula and the Rappaport Faculty of Medicine, Technion, Haifa, Israel (S A Shalev MD, R Spiegel MD); Department of Neurology, University of Utah School of Medicine, Salt Lake City, UT, USA (K J Swoboda FACMG); Neurology Department, Royal Manchester Children’s Hospital, Manchester, UK (G Vassallo MD); Centre for Rare Diseases and Personalised Medicine and Department of Medical and Molecular Genetics, School of Clinical and Experimental Medicine, University of Birmingham, UK, and West Midlands Regional Genetics Service, Clinical Genetics Unit, Birmingham Women’s Hospital, Birmingham, UK (J Vogt MRCP); Nottingham Children’s Hospital, Nottingham University Hospitals NHS Trust, Nottingham, UK (W P Whitehouse FRCPCH); Neuropaediatrics, Department of Paediatrics, Mater Dei Hospital, Tal-Qroqq, Malta (D Soler MD); and Université Paris Descartes, EA 1833 and Hôpital Cochin, Service de Virologie, Paris, France, (F Rozenberg MD, P Lebon MD)
1