Abstract
No matter how many times one explores the structure of the myosin molecule, there is always something new to discover. Here, I describe the myosin mesa, a structural feature of the motor domain that has the characteristics of a binding domain for another protein, possibly myosin-binding protein C (MyBP-C). Interestingly, many well-known hypertrophic cardiomyopathy (HCM) mutations lie along this surface and may affect the putative interactions proposed here. A potential unifying hypothesis for the molecular basis of human hypertrophic cardiomyopathy is discussed here. It involves increased power output of the cardiac muscle as a result of HCM mutations causing the release of inhibition by myosin binding protein C.
Keywords: myosin, hypertrophic cardiomyopathy
Historical perspective
I was asked to give the plenary lecture at the meeting on The Dynamic Cell about the history of my laboratory’s work, what we are currently working on, and new perspectives I have. For this special issue I only briefly touch on historical aspects and focus instead on a new perspective that I hope the readership finds intriguing. It involves a structural feature of the myosin motor domain that I call the myosin mesa and the possible role it plays in hypertrophic cardiomyopathy.
My laboratory, from its inception in 1971, has been devoted to developing both in vivo and in vitro systems to understand the molecular basis of energy transduction by the myosin family of molecular motors [1–4]. Our broad interests have included the roles of myosins in non-muscle cells and the molecular basis of muscle contraction. In the first few years of my laboratory, we explored a variety of non-muscle eukaryotic organisms and began to develop new approaches, such as detergent-extraction of chick embryo fibroblasts to reveal an underlying network of actin and intermediate filaments that apparently gave the cell its shape. It was clearly the skeleton of the cell and we coined the term ‘the cytoskeleton’ (Figure 1A) [5]. But the chick embryo fibroblasts were not amenable to the type of biochemistry or genetics that we hoped to pursue, which was true for many of the other model systems we were exploring.
Figure 1. Eukaryotic cell types my laboratory has worked on.

(A) Chick embryo fibroblasts extracted with the detergent triton-X 100. The ‘cytoskeleton’ is revealed. (B) A dividing Dictyostelium myosin-II null cell in which GFP-tagged myosin was inserted to rescue cytokinesis. The amount of myosin moving to the cleavage furrow is quantified by the distribution of fluorescence intensity. (C) A human induced pluripotent stem (iPS)-cell-derived cardiomyocyte stained with phalloidin (actin, red) and a β-cardiac myosin-specific antibody (yellow). Image by Arjun Adhikari, Alexandre Ribeiro and Kristina Bezold.
One organism, Dictyostelium discoideum, rose to the forefront. Not only could we obtain large quantities of material for biochemical studies, but also in the 1980s we discovered high-efficiency homologous recombination in Dictyostelium [6], an approach that was not thought to be useful in this organism. By knocking out the single-copy muscle-like myosin II heavy chain gene in this haploid organism, we obtained the first genetic proof of the function of any molecular motor, in this case its absolute requirement for cell division in suspension cultures [6]. Importantly, those experiments also established that myosin-II is not required for cell migration as had been assumed. This led to the current view of cell migration driven by the forces of actin filament assembly [7,8]. By expressing a GFP-tagged version of the myosin II in the myosin II null cell, we rescued cytokinesis and had a visual way to monitor the cell division process (Figure 1B) [9,10]. Many years of interesting work followed, including some of the earliest mutagenesis to examine structure-function relationships of this molecular motor [11].
As powerful as the Dictyostelium system proved to be, I felt strongly that one would never fully understand how myosin worked as a biomechanical machine without an in vitro, reconstituted system for the primary function of interest, movement. After some years of establishing the necessary background for this breakthrough, in vitro motility assays for purified actin and myosin were established in the 1980s [12,13]. Having a quantitative in vitro motility assay was essential to prove that the globular head of myosin, known as subfragment-1 (S1), is the motor domain of the myosin molecule [14] and together with molecular genetic approaches, provided strong functional evidence that the light-chain-binding region of the S1 acts as a swinging lever arm during the chemomechanical coupling [15,16]. With the help of the physics of laser trapping, this assay was simplified to the single molecule level, which allowed the measurement of the step obtained when the lever arm strokes (~10 nm) as well as the intrinsic force produced by the motor (a few piconewtons) [17].
With these tools in hand, my laboratory has turned to a focus on one of the most important members of the myosin family of molecular motors, human β-cardiac myosin, the molecular motor that drives contraction of the cardiomyocyte (Figure 1C).
Our current work focuses on human cardiomyopathy mutations in the cardiac contractile machinery
The human β-cardiac myosin motor is a major site in the sarcomere for hundreds of missence mutations, each thought to result in the devastating disease of the heart, hypertrophic cardiomyopathy (HCM). HCM, a monogenic inherited disease [18,19], is clinically characterized by an asymmetric thickening of the ventricular walls and a decrease in the ventricular chamber size that occurs without a predisposing cause. Systolic performance of the heart is preserved but relaxation capacity is diminished. HCM affects approximately one in 500 individuals [20–22].
There are reported to be more than 300 pathogenic mutations in β-cardiac myosin [23–25] and most of these are in the globular head domain, myosin S1. The other major site in the sarcomere for mutations is a regulatory protein known as myosin-binding protein C (MyBP-C). MyBP-C is thought to put a break on the contraction of the muscle, which can be alleviated by its phosphorylation [19,26–28]. A smaller but significant set of mutations are found in the tropomyosin–troponin (Tm–Tn) complex. Tm and the three Tn subunits, troponin T (TnT), troponin I (TnI) and troponin C (TnC) form the Ca2+ -regulatory complex of the sarcomere. The actin, myosin, Tm, the three Tns and MyBP-C constitute a seven-component system that is the fundamental-regulated contractile complex of the sarcomere (Figure 2).
Figure 2. Schematic cartoon of the seven-component regulated contractile complex.
MyBP-C is known to interact with both the myosin thick filament and the actin filament. Shown are Tm, Tn and S1, the globular head domain of myosin, which contains the myosin light chains (not shown) and is the motor domain of the molecule.
Much more is known about the Tm–Tn regulatory complex than the regulatory protein MyBP-C. There is good evidence that domains of MyBP-C near the N-terminus bind to the regulated actin filament, and the C-terminus binds to the myosin thick filament backbone (Figure 2) [26–28]. Studies also suggest binding of MyBP-C to myosin S2, the coiled-coil domain just C-terminal to the globular S1 head, as well as to the regulatory light chain of the myosin S1 [29–32]. Here, I describe features of the myosin S1 structure that may suggest binding of MyBP-C specifically to the catalytic domain (the N-terminal portion of myosin through the converter domain, but not including the lever arm; Figure 3). Such binding would have the potential of regulating many aspects of the myosin motor domain function.
Figure 3. Three views of myosin S1.

(A) A side view showing the catalytic domain oriented to illustrate the flat mesa (dashed oval), with the actin-binding region on the left and the light-chain-binding lever arm on the right. The N-terminal 25 K (light grey), 50 K (white) and C-terminal 20 K (medium grey) domains are shown, as well as the two light chains (dark grey) that are part of the lever arm. The two residues that mark the beginning and end of loop 1 (cyan) are near the edge of the mesa. (B) The mesa viewed from the top (dashed oval; view in A rotated toward you 90°). The two subdomains of the 50 K domain are marked. Loop 2 (not shown in the crystal structure) is a prominent feature of the mesa, the beginning and end of which is marked by the cyan residues. The structure shown here, and in all figures below, is a chimera of the human β-cardiac catalytic domain (MYH7; through to the converter) crystal structure (pdb id: 4DB1) with the converter and lever arm modelled in from the chicken skeletal myosin structure (MYH2) (pdb id: 2MYS). (C) The mesa viewed at an oblique angle and in surface electrostatic mode (large dashed oval). Basic patches are more raised than neighboring acidic patches. A particularly large positive cluster very rich in arginine residues (small dashed oval) is near loop 1. The beginning residues of Loops 1 and 2 (cyan) are shown. Images here and in figures below were rendered in PyMOL.
The myosin mesa, a structural feature of the myosin motor domain
The first crystal structure of the myosin motor domain was obtained by Rayment and colleagues in 1993 (33), and many important crystal structures of this domain has followed over subsequent years (eg., (34–37)). All of us in the field have examined and re-examined those structures to understand the importance of the light-chain-binding region, the 50 K upper and lower actin-binding domains, the two loops that are easily cut by proteases and so on [38–40]. But there is one feature of the myosin structure that we have all ignored. There is a particularly flat broad domain of myosin’s motor domain that is highly likely to act as a binding site for another protein, which, to my knowledge, has never been discussed before. Interestingly, it is the site of many of the well-known HCM mutations.
I refer to this >20 nm2 flat area as the myosin mesa. It has the appearance of a south-western mesa landscape when viewed from the side (Figure 3A, dashed oval). When viewed from the top, the mesa can be seen to consist of all three subdomains of S1, the N-terminal 25 K domain, the actin-binding 50 K domain and a finger of the C-terminal 20 K domain (Figure 3B). The cleft between the upper and the lower 50 K domains is readily apparent and loop 2 is a central feature (Figure 3B, see cyan residues on the face of the mesa marking the beginning Ala625 and the end Phe644 of loop 2; the actual loop is flexible and does not show in the crystal structure). Loop 1, which affects the ADP release rate of the motor and therefore its velocity of movement along actin, is on the very edge of the mesa (marked by residues Arg204 and Gly212, highlighted in cyan). The converter and light-chain-binding region (myosin’s lever arm) is below the mesa in this post-stroke configuration (Figures 3A and 3B). When the mesa is viewed at an oblique angle and in surface electrostatic mode (Figure 3C, large dashed oval), it is apparent that basic patches are more raised than neighboring acidic patches. A particularly large positive cluster very rich in arginine residues is near loop 1 (Figure 3C, small dashed oval).
The cardiac myosins are fairly conserved from mouse α-cardiac to human β-cardiac in the catalytic domain. There are less than 20 major changes (although there are many more minor changes) when comparing the human β-cardiac catalytic domain to a number of non-human β-cardiac S1s and even to the most divergent mouse α-cardiac S1. Those major changes are found throughout the S1 catalytic domain (Figure 4A, red residues). There are, however, no major changes on the surface of the mesa domain (Figure 4B). Thus, this rather large surface is unusually highly conserved.
Figure 4. Major changes in residues when comparing several non-human β-cardiac myosins and mouse α-cardiac myosin (MYH6) to human β-cardiac myosin (MYH7).

(A) A view of the cartoon form of S1, looking at the face of the molecule nearly opposite to the mesa surface. The red residues mark major changes in the molecule. They are found throughout the S1; changes in the light-chain-binding region are not shown. The cyan residues mark loops 1 and 2. (B) The myosin mesa viewed from the top. Note the absence of major changes on this surface.
Such a flat >20 nm2 surface that is highly conserved is a prime candidate for an interaction site for another protein molecule. The most probable candidate for an interacting partner is MyBP-C, which is known to be in the vicinity of the myosin globular head domain in the sarcomere (Figure 2).
A further suggestion that MyBP-C may bind to the mesa comes from a comparison of the conservation of that region between cardiac S1 and skeletal S1, and between either cardiac S1 and smooth muscle S1 or cardiac S1 and non-muscle myosin II S1. Skeletal muscles, like cardiac, have MyBP-C in its sarcomere. Although the N-terminal region of skeletal MyBP-C is considerably different from its cardiac counterpart, there are only a small number of significant changes on the skeletal muscle mesa compared with the cardiac mesa, and those changes could be to better accommodate the skeletal MyBP-C isoform (compare Figures 5A and 5B; the orange residues are major changes found in human skeletal S1 compared with the human β-cardiac S1). On the other hand, neither smooth muscle nor non-muscle cells have MyBP-C regulation and there would be no reason to conserve the mesa in those cases if it were meant to be a MyBP-C-binding interface. Strikingly, both the smooth muscle myosin mesa and the non-muscle myosin II mesa are quite divergent from that of the cardiac (Figures 5C and 5D, red residues show major changes).
Figure 5. Comparison of major residue changes between human β-cardiac myosin and human skeletal myosin, smooth muscle myosin and human non-muscle myosin II.

(A) Human β-cardiac myosin (MYH7). (B) Major changes (orange residues) when compared with human skeletal muscle myosin (MYH2). (C) Major changes (red residues) when compared with human smooth muscle myosin (B1PS43). (D) Major changes (red residues) when compared with human non-muscle myosin II (MYH10).
This observation extends to the loop 2 sequences among the cardiac, skeletal, smooth and non-muscle myosin II species. Loop 2 (A625 G A D A P I E K G K G K A K K G S S F644) is much more conserved among the cardiac and skeletal species than between cardiac and smooth or cardiac and non-muscle S1. It is possible that a major role of loop 2 is to help anchor the MyBP-C to the mesa domain. It has a highly positive sequence of lysines creating a domain of positive charge, ready to engage an acidic domain on a binding partner.
There is a striking correlation with the locations of well-known HCM mutations and the β-cardiac myosin mesa
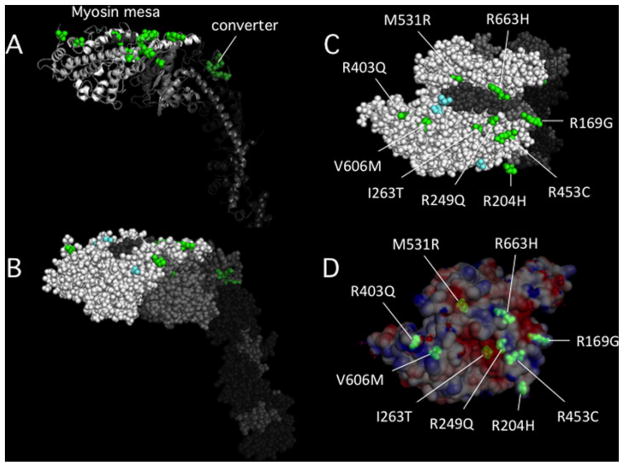
My colleague Kathy Ruppel and I are leading an effort to understand at the molecular level how missense mutations in human β-cardiac myosin lead to hypertrophic cardiomyopathy (HCM). There has been no unifying hypothesis as to why HCM mutations lead to hypercontractility of the muscle. To develop a better understanding of the molecular basis of this hypercontractility, we are focusing on fifteen key HCM mutations that have been well-documented to be the cause of the disease, given the number of separate families who show correlation of the disease with the mutation as well as family genetic histories that emphasize the link between the mutation and the disease [18,19,23,41]. Thus, we are focusing on R169G, A199V, R204H, R249Q, I263T, R403Q, R453C, M531R, G584R, V606M, R663H, G716R, R719W, R723G and G741R. M531R, although only documented in one family, is a left ventricular non-compaction mutant myosin that appears to be hypercontractile in our studies and is therefore included. G716R, R719W, R723G and G741R are in the converter domain (Figures 6A and 6B; mutations denoted by dark-green residues), but where are the 11 others? Strikingly, they are almost all on the surface of the myosin mesa (Figure 6A, mutations denoted by bright-green residues). Nine are visible on the mesa surface in a space-filling model (Figure 6C and 6D) whereas the remaining two are just below the mesa surface, only slightly buried (Figure 6A). Note also that six of the nine surface residues that are HCM mutations are all arginine residues that are changed to less charged residues, suggesting disruption of a positively charged domain that could be involved in binding another protein (possibly MyBP-C) that carries a negatively charged complementary surface. This positively-charged (blue) arginine-rich region is right at the surface of the mesa, while the negatively-charged (red) neighboring regions, possibly important players in the effects of the HCM mutations, are somewhat buried in small pockets (Figure 6D).
Figure 6. The locations of 15 HCM mutations in the myosin catalytic domain.
(A) Cartoon image view showing two groups of HCM mutations. Four mutations are in the converter domain (dark green) and 11 are on or very near the myosin mesa (bright green). Fourteen of the mutations were chosen because they have been well-documented to be the cause of HCM in families carrying these mutations. The fifteenth is M531R, which, although only documented in one family, is a left ventricular non-compaction mutant myosin that appears to be hypercontractile in our studies. (B) The same view as in (A) except in all sphere representation. (C) The top view of the mesa showing that nine of the 11 mutations are on the mesa surface (the other two are just below the surface). (D) The same view as in (C) except the surface charge distribution is shown. M531R and I263T are slightly below the surface in acidic pockets, while the remainder of the mutations are right at the surface and most are arginine residues, 5 of which form a particularly large domain of positive charge on the lower right (see also Figure 3C, small dashed oval).
This distinctive pattern of the HCM mutations is not seen for dilated cardiomyopathy (DCM) mutations. The DCM mutations are not highly localized to the human β-cardiac S1 mesa domain (Figure 7).
Figure 7. The locations of fourteen DCM mutations in the myosin catalytic domain.
(A) Cartoon image of DCM mutations (orange) showing the same view as in Figure 6A. Most are not on the myosin mesa. The DCM mutations were chosen on the basis of high likelihood of being causative of DCM [45]. They are I201T, A223T, R237W, G245E, I248F, R369Q, D469Y, I524V, E525K, S532P, I533V, R567H, N597K and F764L. (B) The view from the other side of the S1 compared to the view in A. (C) The top view of the mesa showing that 4 of the 14 mutations are on the mesa surface, and all in a clustered linear sequence. (D) The same view as in C except the surface charge distribution is shown.
There are many more HCM mutations reported in the literature, but for the most part they have not been firmly identified to be causative of the disease by genetic analyses or numbers of independent HCM-diseased families carrying the mutation. It will be interesting to see how many truly causative mutations fall into the class of HCM mutations focused on here, which I will call the myosin mesa HCM family. It is likely that there will be additional categories of myosin HCM mutations that effect power output in different mechanistic ways, the converter HCMs being possibly its own distinct class. It is possible, however, that all myosin HCM mutations, including the converter mutations, have the primary effect of releasing the inhibitory response of MyBP-C, resulting in hypercontractility.
Conclusion and future perspectives
In a recent review I described the importance of the fundamental parameters that determine the power output of muscle contraction [42]. The power output is the product of force and velocity of contraction. The ensemble force (Fensemble) of the contractile sarcomere is determined by the intrinsic force of a single myosin molecule (f) and the duty ratio, the fraction of the myosin heads in the sarcomere that are in a strongly-bound force-producing state at any moment. The duty ratio for those heads available to interact with the actin is ts/tc, where ts is the strongly-bound state time of myosin on the actin and tc is the total cycle time of the actin-activated myosin chemomechanical cycle. Thus the ensemble force in the sarcomere is
where Nt is the total number of myosin heads functionally available to interact with actin.
The potential binding of MyBP-C to the myosin mesa could affect any of the fundamental parameters that determine power output. A most likely effect, however, might be to reduce Nt by keeping some of the myosin heads out of play, thus reducing the Fensemble. The concept of MyBP-C removing myosin heads from the functioning pool of heads in the sarcomere has been suggested previously, with evidence for binding to S2 and the RLC (for reviews, see [30,43]).
A simple hypothesis is that the myosin mesa class of myosin HCM mutations, and possibly the corresponding HCM missense mutations on the domain(s) of the MyBP-C that putatively interacts with the myosin mesa, reduce the affinity of the MyBP-C for the myosin head, releasing those heads to now be involved in the contractile process. This would result in hypercontractility of the muscle, which is characteristic of HCM clinically. Since the myosin motor domain is highly allosteric and there may be other binding sites on S1 for MyBP-C, this molecular mechanism for HCM hypercontractility may extend to HCM mutations not on the mesa, and thus this mechanism may be a unifying hypothesis.
It is noteworthy that there are acidic patches on several of the MyBP-C domains that could act as a binding interface to the basic patch(s) on the myosin mesa. Of particular note is the negatively-charged cardiac-specific insert of the C5 domain.
One might expect that there could be other mutations at the myosin mesa that would strengthen the affinity of MyBP-C for the S1 and therefore remove more heads than normal from the pool of interacting heads in the sarcomere. Such mesa mutations would lead to hypocontractility, which is characteristic of dilated cardiomyopathy. Interestingly, 4 out of 14 DCM mutations that we are analyzing (I524V, E525K, S532P, and I533V) are in a linear cluster on the myosin mesa (Figure 7C and 7D), perhaps poised to bind MyBP-C more strongly.
My laboratory currently has a major focus on the changes in biochemical and biomechanical parameters in HCM mutant forms of human β-cardiac myosin. Our studies have used both the two-component human system actin and myosin and the six-component reconstituted human system, actin, myosin, Tm, TnC, TnI and TnT. From the considerations described here, it is possible that many, if not all, of the HCM mutations in the main body of the motor domain alter significantly the effects of the putative MyBP-C interaction with the myosin S1 surface. Thus, reconstitution of the seven-component system will be necessary for fully understanding the effects of the HCM mutations.
Similarly, the effects of small-molecule activators and inhibitors on the human cardiac ventricular system (eg., [44]) will probably have to be understood at the seven-component level in molecular studies to correlate effects seen in the reconstituted system with those observed in skinned fibres, cardiomyocytes, the whole organ and animals. Indeed, MyBP-C may be a particularly good target for a small molecule therapeutic approach.
Acknowledgments
I thank Kathy Ruppel for constant discussions involving cardiomyopathies and her key role in our cardiomyopathy programme. I also thank Arjun Adhikari, Alexandre Ribeiro and Kristina Bezold for the cardiomyocyte image for Figure 1(C), and Carol Cho for her help with figure 3(C).
Funding
This work was supported by the National Institutes of Health [grant numbers GM33289 and HL117138].
Abbreviations
- DCM
dilated cardiomyopathy
- HCM
hypertrophic cardiomyopathy
- MyBP-C
myosin-binding protein C
- S1
subfragment 1
- Tm
tropomyosin
- Tn
troponin
- TnC
troponin C
- TnI
troponin I
- TnT
troponin T
References
- 1.Spudich JA. How molecular motors work. Nature. 1994;372:515–518. doi: 10.1038/372515a0. [DOI] [PubMed] [Google Scholar]
- 2.Spudich JA. The myosin swinging cross-bridge model. Nat Rev Mol Cell Biol. 2001;2:387–392. doi: 10.1038/35073086. [DOI] [PubMed] [Google Scholar]
- 3.Spudich JA. Molecular motors: forty years of interdisciplinary research. Mol Biol Cell. 2011;22:3936–3939. doi: 10.1091/mbc.E11-05-0447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Spudich JA. One path to understanding energy transduction in biological systems. Nat Med. 2012;18:1478–1482. doi: 10.1038/nm.2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown S, Levinson W, Spudich JA. Cytoskeletal elements of chick embryo fibroblasts revealed by detergent extraction. J Supramol Struct. 1976;5:119–130. doi: 10.1002/jss.400050203. [DOI] [PubMed] [Google Scholar]
- 6.De Lozanne A, Spudich JA. Disruption of the Dictyostelium myosin heavy chain gene by homologous recombination. Science. 1987;236:1086–1091. doi: 10.1126/science.3576222. [DOI] [PubMed] [Google Scholar]
- 7.Pollard TD. Cellular motility powered by actin filament assembly and disassembly. Harvey Lect. 2002;98:1–17. [PubMed] [Google Scholar]
- 8.Pollard TD, Cooper JA. Actin, a central player in cell shape and movement. Science. 2009;326:1208–1212. doi: 10.1126/science.1175862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moores SL, Sabry JH, Spudich JA. Myosin dynamics in live Dictyostelium cells. Proc Natl Acad Sci USA. 1996;93:443–446. doi: 10.1073/pnas.93.1.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zang JH, Cavet G, Sabry JH, Wagner P, Moores SL, Spudich JA. On the role of myosin-II in cytokinesis: division of Dictyostelium cells under adhesive and nonadhesive conditions. Mol Biol Cell. 1997;8:2617–2629. doi: 10.1091/mbc.8.12.2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ruppel KM, Spudich JA. Structure-function analysis of the motor domain of myosin. Annu Rev Cell Dev Biol. 1996;12:543–573. doi: 10.1146/annurev.cellbio.12.1.543. [DOI] [PubMed] [Google Scholar]
- 12.Spudich JA, Kron SJ, Sheetz MP. Movement of myosin-coated beads on oriented filaments reconstituted from purified actin. Nature. 1985;315:584–586. doi: 10.1038/315584a0. [DOI] [PubMed] [Google Scholar]
- 13.Kron SJ, Spudich JA. Fluorescent actin filaments move on myosin fixed to a glass surface. Proc Natl Acad Sci USA. 1986;83:6272–6276. doi: 10.1073/pnas.83.17.6272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Toyoshima YY, Kron SJ, McNally EM, Niebling KR, Toyoshima C, Spudich JA. Myosin subfragment-1 is sufficient to move actin filaments in vitro. Nature. 1987;328:536–539. doi: 10.1038/328536a0. [DOI] [PubMed] [Google Scholar]
- 15.Uyeda TQ, Abramson PD, Spudich JA. The neck region of the myosin motor domain acts as a lever arm to generate movement. Proc Natl Acad Sci. 1996;93:4459–4464. doi: 10.1073/pnas.93.9.4459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shih WM, Gryczynski Z, Lakowicz JR, Spudich JA. A FRET-based sensor reveals large ATP hydrolysis-induced conformational changes and three distinct states of the molecular motor myosin. Cell. 2000;102:683–694. doi: 10.1016/s0092-8674(00)00090-8. [DOI] [PubMed] [Google Scholar]
- 17.Finer JT, Simmons RM, Spudich JA. Single myosin molecule mechanics: piconewton forces and nanometre steps. Nature. 1994;368:113–119. doi: 10.1038/368113a0. [DOI] [PubMed] [Google Scholar]
- 18.Geisterfer-Lowrance AA, Kass S, Tanigawa G, Vosberg HP, McKenna W, Seidman CE, Seidman JG. A molecular basis for familial hypertrophic cardiomyopathy: a beta cardiac myosin heavy chain gene missense mutation. Cell. 1990;62:999–1006. doi: 10.1016/0092-8674(90)90274-i. [DOI] [PubMed] [Google Scholar]
- 19.Seidman CE, Seidman JG. Hypertrophic cardiomyopathy. In: Scriver CR, Beaudet AL, Valle D, Sly WS, Childs KW, Vogelstein B, editors. The Metabolic and Molecular Bases of Inherited Disease. McGraw-Hill; 2000. pp. 5532–5452. [Google Scholar]
- 20.Harvey PA, Leinwand LA. Cellular mechanisms of cardiomyopathy. J Cell Biol. 2011;194:355–365. doi: 10.1083/jcb.201101100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maron BJ. Braunwald’s Heart Disease. Elsevier Inc; 2010. Hypertrophic cardiomyopathy; pp. 1582–1594. [Google Scholar]
- 22.Maron BJ, Gardin JM, Flack JM, Gidding SS, Kurosaki TT, Bild DE. Prevalence of hypertrophic cardiomyopathy in a general population of young adults: echocardiographic analysis of 4111 subjects in the CARDIA study. Circulation. 1995;92:785–789. doi: 10.1161/01.cir.92.4.785. [DOI] [PubMed] [Google Scholar]
- 23.Buvoli M, Hamady M, Leinwand LA, Knight R. Bioinformatics assessment of β-myosin mutations reveals myosin’s high sensitivity to mutations. Trends Cardiovasc Med. 2008;18:141–149. doi: 10.1016/j.tcm.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seidman CE, Seidman JG. Identifying sarcomere gene mutations in hypertrophic cardiomyopathy: a personal history. Circ Res. 2011;108:743–750. doi: 10.1161/CIRCRESAHA.110.223834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Walsh R, Rutland C, Thomas R, Loughna S. Cardiomyopathy: a systematic review of disease-causing mutations in myosin heavy chain 7 and their phenotypic manifestations. Cardiology. 2010;115:49–60. doi: 10.1159/000252808. [DOI] [PubMed] [Google Scholar]
- 26.Kensler RW, Shaffer JF, Harris SP. Binding of the N-terminal fragment C0-C2 of cardiac MyBP-C to cardiac F-actin. J Struct Biol. 2011;174:44–51. doi: 10.1016/j.jsb.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Dijk SJ, Bezold KL, Harris SP. Earning stripes: myosin binding protein-C interactions with actin. Pflugers Archiv. 2014;466:445–450. doi: 10.1007/s00424-013-1432-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chow ML, Shaffer JF, Harris SP, Dawson JF. Altered interactions between cardiac myosin binding protein-C and alpha-cardiac actin variants associated with cardiomyopathies. Arch Biochem Biophys. 2014;550–551:28–32. doi: 10.1016/j.abb.2014.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harris SP, Rostkova E, Gautel M, Moss RL. Binding of myosin binding protein-C to myosin subfragment S2 affects contractility independent of a tether mechanism. Circ Res. 2004;95:930–936. doi: 10.1161/01.RES.0000147312.02673.56. [DOI] [PubMed] [Google Scholar]
- 30.Kampourakis T, Yan Z, Gautel M, Sun YB, Irving M. Myosin binding protein-C activates thin filaments and inhibits thick filaments in heart muscle cells. Proc Natl Acad Sci USA. 2014;111:18763–18768. doi: 10.1073/pnas.1413922112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pfuhl M, Gautel M. Structure, interactions and function of the N-terminus of cardiac myosin binding protein C (MyBP-C): who does what, with what, and to whom? J Muscle Res Cell Motil. 2012;33:83–94. doi: 10.1007/s10974-012-9291-z. [DOI] [PubMed] [Google Scholar]
- 32.Ratti J, Rostkova E, Gautel M, Pfuhl M. Structure and interactions of myosin-binding protein C domain C0: cardiac-specific regulation of myosin at its neck? J Biol Chem. 2011;286:12650–12658. doi: 10.1074/jbc.M110.156646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rayment I, Rypniewski WR, Schmidt-Base K, Smith R, Tomchick DR, Benning MM, Winkelmann DA, Wesenberg G, Holden HM. Three-dimensional structure of myosin subfragment-1: a molecular motor. Science. 1993;261:50–58. doi: 10.1126/science.8316857. [DOI] [PubMed] [Google Scholar]
- 34.Dominguez R, Freyzon Y, Trybus KM, Cohen C. Crystal structure of a vertebrate smooth muscle myosin motor domain and its complex with the essential light chain: visualization of the pre-power stroke state. Cell. 1998;94:559–571. doi: 10.1016/s0092-8674(00)81598-6. [DOI] [PubMed] [Google Scholar]
- 35.Houdusse A, Kalabokis VN, Himmel D, Szent-Gyorgyi AG, Cohen C. Atomic structure of scallop myosin subfragment S1 complexed with MgADP: a novel conformation of the myosin head. Cell. 1999;97:459–470. doi: 10.1016/s0092-8674(00)80756-4. [DOI] [PubMed] [Google Scholar]
- 36.Houdusse A, Szent-Gyorgyi AG, Cohen C. Three conformational states of scallop myosin S1. Proc Natl Acad Sci USA. 2000;97:11238–11243. doi: 10.1073/pnas.200376897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Coureux PD, Sweeney HL, Houdusse A. Three myosin V structures delineate essential features of chemomechanical transduction. EMBO J. 2004;23:4527–4537. doi: 10.1038/sj.emboj.7600458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Holmes KC. The molecular basis of cross-bridge function. Adv Exp Med Biol. 2005;565:13–22. doi: 10.1007/0-387-24990-7_2. discussion 23, 359–369. [DOI] [PubMed] [Google Scholar]
- 39.Geeves MA, Holmes KC. The molecular mechanism of muscle contraction. Adv Protein Chem. 2005;71:161–193. doi: 10.1016/S0065-3233(04)71005-0. [DOI] [PubMed] [Google Scholar]
- 40.Sweeney HL, Houdusse A. Structural and functional insights into the myosin motor mechanism. Annu Rev Biophys. 2010;39:539–557. doi: 10.1146/annurev.biophys.050708.133751. [DOI] [PubMed] [Google Scholar]
- 41.Pan S, Caleshu CA, Dunn KE, Foti MJ, Moran MK, Soyinka O, Ashley EA. Cardiac structural and sarcomere genes associated with cardiomyopathy exhibit marked intolerance of genetic variation. Circ Cardiovasc Genet. 2012;5:602–610. doi: 10.1161/CIRCGENETICS.112.963421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Spudich JA. Hypertrophic and dilated cardiomyopathy: four decades of basic research on muscle lead to potential therapeutic approaches to these devastating genetic diseases. Biophys J. 2014;106:1236–1249. doi: 10.1016/j.bpj.2014.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moss RL, Fitzsimons DP, Ralphe JC. Cardiac MyBP-C Regulates the Rate and Force of Contraction in Mammalian Myocardium. Circ Res. 2015;116:183–192. doi: 10.1161/CIRCRESAHA.116.300561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Malik FI, Hartman JJ, Elias KA, Morgan BP, Rodriguez H, Brejc K, Anderson RL, Sueoka SH, Lee KH, Finer JT, Sakowicz R, Baliga R, Cox DR, Garard M, Godinez G, Kawas R, Kraynack E, Lenzi D, Lu PP, Muci A, Niu C, Qian X, Pierce DW, Pokrovskii M, Suehiro I, Sylvester S, Tochimoto T, Valdez C, Wang W, Katori T, Kass DA, Shen YT, Vatner SF, Morgans DJ. Cardiac myosin activation: a potential therapeutic approach for systolic heart failure. Science. 2011;331:1439–1443. doi: 10.1126/science.1200113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pugh TJ, Kelly MA, Gowrisankar S, Hynes E, Seidman MA, Baxter SM, Bowser M, Harrison B, Aaron D, Mahanta LM, Lakdawala NK, McDermott G, White ET, Rehm HL, Lebo M, Funke BH. The landscape of genetic variation in dilated cardiomyopathy as surveyed by clinical DNA sequencing. Genet Med. 2014;16:601–608. doi: 10.1038/gim.2013.204. [DOI] [PubMed] [Google Scholar]