Abstract
The pathogenic fungus Batrachochytrium dendrobatidis (Bd) is a major conservation concern because of its role in decimating amphibian populations worldwide. We used quantitative PCR to screen 244 museum specimens from the Korean Peninsula, collected between 1911 and 2004, for the presence of Bd to gain insight into its history in Asia. Three specimens of Rugosa emeljanovi (previously Rana or Glandirana rugosa), collected in 1911 from Wonsan, North Korea, tested positive for Bd. Histology of these positive specimens revealed mild hyperkeratosis – a non-specific host response commonly found in Bd-infected frogs – but no Bd zoospores or zoosporangia. Our results indicate that Bd was present in Korea more than 100 years ago, consistent with hypotheses suggesting that Korean amphibians may be infected by endemic Asian Bd strains.
Introduction
The pathogenic fungus Batrachochytrium dendrobatidis (Bd) is linked to declines or extinctions of more than 200 amphibian species worldwide [1–3]. While Bd is widespread in Asia [4–7], no epizootic events have been reported. One potential explanation is that Asia is an origin of the global pandemic Bd lineage [5,8] (but see [9,10] for alternative hypotheses).
Bd is present throughout Asia [5–7,11–16]. Unique genotypes in Japan [5,7], China [7,11], Korea [7], and India [16,17], as well as high prevalence and low infection intensity [1,18] in Korea [3,7,19,20], support the hypothesis that some Bd strains are endemic to Asia. To fully understand Bd in Asia so that effective conservation plans can be developed, a temporal view must be included. To date, four studies have examined historical specimens in Asia [21–26], with detection in 1933 from China [25,26]. In our study, we use quantitative PCR (qPCR) and histology to screen historical amphibian specimens from Korea for the presence of Bd. These results provide a temporal perspective—going back a century—on Bd prevalence in Korea.
Material and Methods
We accessed 244 historical specimens from the Museum of Vertebrate Zoology at the University of California, Berkeley (MVZ) and the California Academy of Sciences (CAS) (S1 Table). We standardize scientific names to follow the taxonomy of AmphibiaWeb (www.amphibiaweb.org). Specimens comprise 13 of 17 species native to the Korean Peninsula and were collected between 1911 and 2004 (Table 1). We used contemporary data [6,7,14,15] (Fig. 1, Table 1) to inform our interpretation of historical data. We followed a non-invasive sampling method to detect Bd by the polymerase chain reaction (PCR) [27]. While specimens at the MVZ are susceptible to cross contamination because a jar of specimens of one species may represent several collecting trips to the same locality, CAS specimens are less so because they are stored in jars separated by species, locality, and collecting trip.
Table 1. Comparison of modern and historical specimen data.
| Species | Modern samples | Historical samples | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Bd + | Total | % Bd + | ZE | 1910–1929 | 1930–1949 | 1950–1969 | 1970–1989 | 1990–2004 | TOTAL | |
| Bombina orientalis | 62 | 373 | 16.6% | 13.85 | 0 | 2 | 17 | 0 | 0 | 19 |
| Bufo gargarizans | 3 | 93 | 3.2% | 20.00 | 0 | 0 | 2 | 0 | 0 | 2 |
| Bufo stejnegeri | 2 | 36 | 5.6% | - | 0 | 0 | 0 | 0 | 0 | 0 |
| Hyla japonica | 62 | 420 | 14.8% | 28.07 | 0 | 0 | 48 | 0 | 0 | 48 |
| Hyla suweonensis | 0 | 4 | 0% | - | 0 | 0 | 0 | 0 | 0 | 0 |
| Hynobius leechii | 59 | 123 | 48.0% | 1.50 | 0 | 0 | 0 | 20 | 18 | 38 |
| Hynobius quelpaertenses | 46 | 117 | 39.3% | - | 0 | 0 | 0 | 0 | 0 | 0 |
| Hynobius yangii | 0 | 6 | 0% | - | 0 | 0 | 0 | 0 | 1 | 1 |
| Kaloula borealis | 0 | 20 | 0% | - | 0 | 0 | 4 | 0 | 0 | 4 |
| Karsenia koreana | 3 | 9 | 33.3% | 2.67 | 0 | 0 | 0 | 0 | 5 | 5 |
| Onychodactylus fischeri | 0 | 22 | 0% | - | 0 | 0 | 0 | 1 | 5 | 6 |
| Pelophylax chosenicus | 1 | 13 | 7.7% | - | 0 | 0 | 1 | 0 | 0 | 1 |
| Pelophylax nigromaculatus | 5 | 168 | 3.0% | - | 0 | 0 | 13 | 2 | 0 | 15 |
| Rana (Lithobates) catesbeiana | 28 | 102 | 27.5% | 305.36 | 0 | 0 | 0 | 0 | 0 | 0 |
| Rana coreana | 11 | 60 | 18.3% | - | 0 | 0 | 18 | 0 | 0 | 18 |
| Rana dybowskii | 46 | 255 | 18.1% | 5.17 | 0 | 0 | 0 | 5 | 0 | 5 |
| Rana huarenensis | 2 | 14 | 14.3% | - | 0 | 0 | 0 | 0 | 0 | 0 |
| Rana sp. | 0 | 1 | 0% | - | 0 | 0 | 33 | 0 | 0 | 33 |
| Rugosa emeljanovi | 19 | 98 | 19.4% | 14.12 | 7* | 0 | 42 | 0 | 0 | 49 |
| TOTAL | 349 | 1,934 | 18.0% | 61.18 | 7 | 2 | 178 | 28 | 29 | 244 |
Historical samples grouped into ~20 year categories. Of all historical samples tested, only three samples of Rugosa emeljanovi (previously Rana or Glandirana rugosa) were positive for Batrachochytrium dendrobatidis (Bd). ZE denotes the estimated Bd zoospore genomic equivalents on each swab. Data for modern samples are compiled from previous publications [6,7,14,15].
* Three of these historical samples tested positive for Bd.
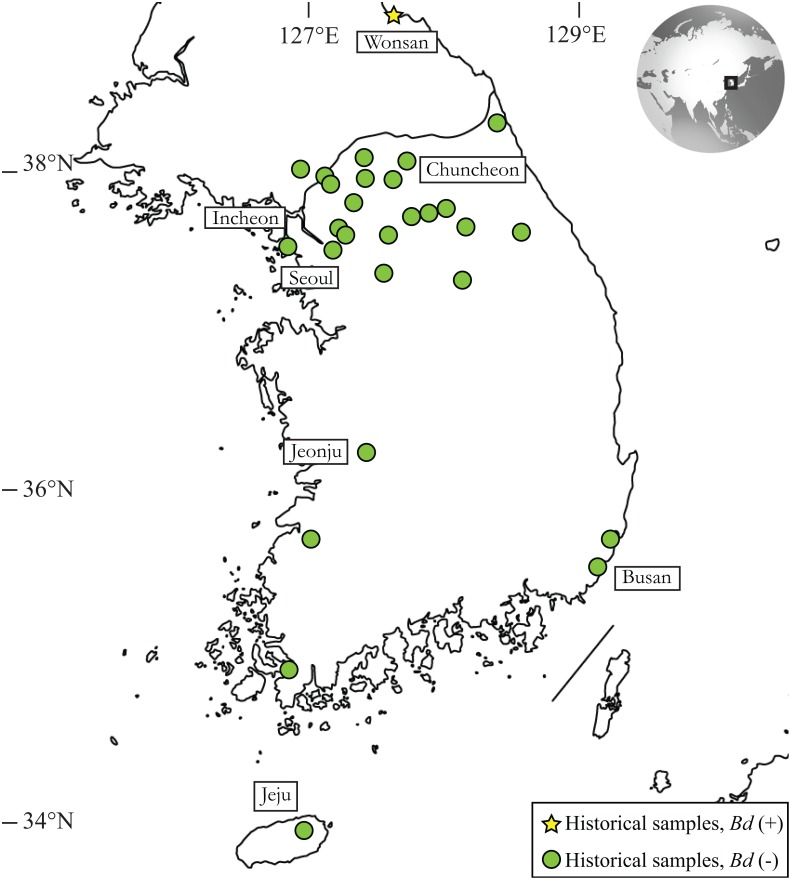
Fig 1. Sampling of historical specimens across the Korean Peninsula used in this study.
Specimens were collected between 1911 and 2004. Three specimens of Rugosa emeljanovi (previously Rana or Glandirana rugosa), collected from Wonsan, North Korea in 1911, tested qPCR positive for Batrachochytrium dendrobatidis (Bd) infection.
Each specimen was rinsed with 70% EtOH and then swabbed (MW113, Medical Wire and Equipment, Corsham, UK) 30 times across its dorsal and ventral surfaces. Swabs were stored dry in 1.5 mL microcentrifuge tubes at 4° C until processed. Prior to extraction, swabs were dried in a Spin Vac (Savant Instruments, Farmingdale, NY, USA) to remove EtOH. Extraction was performed using 40 μL of Prepman Ultra (Applied Biosystems, Carlsbad, CA, USA) [19,20] and diluted 1:10 with 0.25 × TE buffer. We analyzed each sample in duplicate using 5 μL of the diluted DNA extract. Universal DNA standards from the global pandemic lineage (provided by A.S. Hyatt) were included to calibrate the qPCR (0.1, 1.0, 10, and 100 zoospore equivalents per reaction). Negative controls were included during extraction and qPCR to detect contamination. Samples were run on an Applied Biosystems 7300 Real-Time PCR thermocycler. A specimen was considered Bd-positive if one or both qPCR replicates were positive. We calculated the number of zoospores in terms of Zswab (i.e., estimated Bd zoospore genomic equivalents on each swab) [1,3] by multiplying qPCR results by 80 to account for sample dilution (40 μL Prepman × 10 dilution / 5 μL for reaction = 80).
Three additional tests were performed to check Bd-positive samples. First, we ran two more qPCR replicates from the original DNA extract. Next, the positive specimen and all other specimens in the same jar were swabbed again and processed using the same qPCR protocol as above (duplicate runs per sample). Lastly, skin samples of qPCR positive specimens were screened for Bd using histological methods [4]. Histology was conducted at the Wildlife Disease Laboratories at San Diego Zoo (by AP) on full-thickness skin (4 × 4 mm) excised from the ventral pelvic area (n = 2) and webbing between rear digits (n = 4) from each specimen. Past studies showed these areas are likely to harbor Bd infection [8]. Samples were routinely processed for paraffin embedding, sectioned at 5 to 6 μm, and stained with hematoxylin and eosin [9]. In all, 120 serial sections were examined from each skin sample for the presence of Bd thalli, zoosporangia, and associated lesions.
Results
From qPCR assays, 241 samples scored negative and three samples (CAS32672, CAS33676, CAS33678) positive with low levels of Bd amplification (Table 2). Four to six qPCR reactions (2–4 from first swab, 2 from second swab) were run for each positive sample. For the first round of qPCR, a single reaction of sample CAS32672 was positive (Zswab = 0.008). The second round of qPCR from the same swab extract yielded the same result, with a single positive out of two reactions (Zswab = 0.385). From qPCR of a new swab extraction, one of two reactions was positive (Zswab = 0.772). Of re-swabbed specimens from the same jar (same species, same collecting trip), two additional positive samples were discovered: CAS33676 (Zswab = 0.242) and CAS33678 (Zswab = 1.016). Attempts to sequence these PCR products were unsuccessful, probably due to the low DNA quantity. Histological analysis of skin from the three specimens did not reveal the presence of Bd thalli or zoosporangia, but showed evidence of mild parakeratotic hyperkeratosis.
Table 2. Quantitiative PCR (qPCR) results of frog specimens that screened positive for Batrachochytrium dendrobatidis (Bd) infection.
| Specimen # | Swab 1 | Swab 2 | ||||
|---|---|---|---|---|---|---|
| Reaction 1 | Reaction 2 | Reaction 3 | Reaction 4 | Reaction 1 | Reaction 2 | |
| CAS32672 | 0 | 0.008 | 0.385 | 0 | 0 | 0.772 |
| CAS33676 | 0 | 0 | - | - | 0.242 | 0 |
| CAS33678 | 0 | 0 | - | - | 0 | 1.016 |
Values of positive qPCR reactions are calculated as Zswab (estimated Bd zoospore genomic equivalents on each swab) by multiplying qPCR results by 80 to account for sample dilution (40 μL Prepman × 10 dilution / 5 μL for reaction = 80).
Discussion
We discovered three Bd-positive individuals out of 244 historical specimens (1.2%) from the Korean Peninsula. All positives were of Rugosa emeljanovi (previously Rana or Glandirana rugosa) from Wonsan, North Korea collected in 1911. Infection intensities ranged between 0.008 and 1.016 Zswab (Table 2). Histological inspection of the positive samples revealed mild hyperkeratosis, which would be expected if subjects had been infected by Bd. These results demonstrate that Bd was present in Korea in the early 1900s, consistent with hypotheses suggesting the existence of endemic Bd strains in Asia [7].
Recent surveys in South Korea found a Bd prevalence of 18% [7] and average infection intensities for species ranging between 305.36 zoospore equivalents in the introduced American bullfrog (Rana [Lithobates] catesbeiana) and 1.50 zoospore equivalents in the Korean salamander (Hynobius leechii) (Table 1). Modern samples of Rugosa emeljanovi showed a Bd prevalence of 19.4% and an infection intensity of 14.12 zoospore equivalents (Table 1), values greater than historical samples in our study (prevalence = 6.1%, infection intensity = 0.008–1.016 Zswab; Table 2). The inconsistencies revealed when comparing historical and modern data may be attributable to three factors: 1) increased Bd prevalence in recent times by either an endemic strain or emergence of a new strain (e.g. [28]), 2) reduced detection in historical samples due to degraded DNA or the presence of PCR inhibitors [29], or 3) sampling error associated with the availability of historical specimens. Our data cannot differentiate among these possibilities. Although we know that Bd was present in Korea in 1911, we do not know its status between 1911 and present time. Further study involving time-calibrated genetic analyses and additional historical specimens is needed to determine how Bd spread through Korea over time.
We minimized the possibility of false positives for the three specimens collected in 1911 by running replicate tests on each sample using two different methods—qPCR and histology. All tests consistently yielded the same result—low Bd infection loads in the three specimens. The robustness of our conclusions is boosted by a recent study that found low probabilities of false positives due to contamination using the same qPCR assay that we used [18]. Although uncertainty exists as to how the museum specimens were handled in the past, we undertook precautions to minimize sample contamination by continually sterilizing equipment and the workspace. Additionally, specimens from CAS were stored separately by species, locality, and collecting trip, further reducing the possibility of cross-contamination. In the jar of the three positive samples, four other frogs consistently yielded negative results for Bd in all trials.
For qPCR, positive and negative controls were run throughout the procedure, and neither produced results that would indicate a failed reaction or contamination. One concern is the DNA standards used for qPCR (a requirement to calibrate DNA quantification) are a source of contamination. We expect contamination of samples by DNA standards to be random. However, repeated runs of positive samples consistently gave positive results. This PCR assay has been applied extensively since 2004 without any reports of false positives [3,19,20].
Difficulties of extracting and amplifying DNA from preserved specimens, especially those exposed to formalin, have been well documented [21,23] (but see [18]). There is greater success with ethanol-preserved specimens and PCR success appears negatively correlated with exposure time to formalin [30]. Therefore, the preservation history of a specimen should be considered when making conclusions using such data. The three positive samples and four negative samples stored in the same jar were collected by Joseph C. Thompson (also known as Victor Kuhne), preserved in ethanol and, to the best of curators’ knowledge, never exposed to formalin (J. Vindum, pers. comm.). The known preservation history of the three positive samples gives us greater confidence that they are true positives.
Histological analyses of the three Bd-positive specimens showed mild hyperkeratosis—a thickened stratum corneum. Hyperkeratosis is a non-specific host response seen in a variety of skin diseases, but associated with Bd infection [4]. Although no Bd organisms were found, histological examination of the skin is known to be an insensitive method for detection of Bd, especially with low-level or subclinical infections [19,31]. Evidence of Bd is often found in the stratum corneum and sloughed skin, so swabbing itself may have physically removed zoosporangia and destroyed histological evidence of positive infection. Histological results are consistent with qPCR results that detected low zoospore counts. Similarly, contemporary samples of native Korean amphibian species reveal low levels of Bd infection, with an average of 1.50 to 28.07 zoospore equivalents per species (Table 1).
Our results need to be interpreted within an historical anthropogenic context. Korea was relatively isolated from other countries until the late 1800s, when several international treaties opened ports to trade. Wonsan, the site of the three positive specimens, was opened in 1880 to trade with Japan, Russia, China, Germany, and Great Britain [32,33]. The Bd-positive samples were collected in 1911, which raises the possibility that Bd was introduced into Korea during the late 1800s or early 1900s from one of the trade partner countries. Japan and China are the most likely sources, as trade with these countries was the highest [32]. We cannot determine whether Bd detected in our study is native or introduced, but if introduced, its likely origin is still Asia.
Two major hypotheses have been proposed to explain the pattern of Bd in amphibian populations: 1) Bd is a novel pathogen that infects and kills naïve hosts [28], and 2) Bd is an endemic pathogen that infects but does not kill hosts in its native range because of a stable pathogen-host equilibrium [1]. The evolutionary history of Bd may be complex [10], with both endemic and novel strains existing in a region. Amphibian hosts exposed to Bd may acquire resistance [34], while Bd-naïve hosts may be more susceptible to the invasive fungus [35,36]. Understanding the geographic origins of Bd is thus important in interpreting the effects on hosts and in determining the best way to conserve threatened species. Worldwide, the earliest records of Bd are 1894 from Brazil [17], 1933 from Cameroon [37] and China [26], 1934 from Kenya [38], and 1938 from South Africa [39]. Our study pushes the presence of Bd in Asia back to 1911. All these studies point toward longer, potentially independent histories of Bd in some parts of the world. Continued testing of historical specimens coupled with repeated surveys in present-day populations are needed to give a perspective on the dynamic pathogen-host relationship and the differing effects of endemic and introduced Bd strains.
Supporting Information
Shaded cells for latitude/longitude indicate that data have been added or modified based on detailed verbatim locality data. Taxonomy was changed to follow AmphibiaWeb (www.amphibiaweb.org). Under preservation type, the preservatives used for specimen preparation followed by that used for storage are given. Preservation method of specimens was assumed to be formalin/EtOH unless we had information stating otherwise. CAS = California Academy of Sciences, MVZ = Museum of Vertebrate Zoology.
(DOCX)
Acknowledgments
We thank Jim McGuire (MVZ) and Jens Vindum (CAS) for access to specimen collections, Mi-Sook Min for access to Korean data, and Matthew Fujita for comments on the manuscript. We also thank Alan Leviton, David Wake, and Ted Papenfuss for discussions on museum preservation methods.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
The research was supported by the National Research Foundation of Korea (NRF) funded by the government of the Republic of Korea (MSIP) (grants 2010-0002767, 2012R1A1A2044449 and 2012K1A2B1A03000496 to BW), Seoul National University (Brain Fusion Program, Brain Korea 21 Program, and New Faculty Resettlement Fund grants to BW), and the National Science Foundation (grant 1258133 to VTV). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Briggs CJ, Knapp RA, Vredenburg VT (2010) Enzootic and epizootic dynamics of the chytrid fungal pathogen of amphibians. Proc Natl Acad Sci USA 107: 9695–9700. 10.1073/pnas.0912886107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Fisher MC, Garner TW, Walker SF (2009) Global emergence of Batrachochytrium dendrobatidis and amphibian chytridiomycosis in space, time, and host. Annu Rev Microbiol 63: 291–310. 10.1146/annurev.micro.091208.073435 [DOI] [PubMed] [Google Scholar]
- 3. Vredenburg VT, Knapp RA, Tunstall TS, Briggs CJ (2010) Dynamics of an emerging disease drive large-scale amphibian population extinctions. Proc Natl Acad Sci USA 107: 9689–9694. 10.1073/pnas.0914111107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pessier A, Nichols D, Longcore J, Fuller M (1999) Cutaneous chytridiomycosis in poison dart frogs (Dendrobates spp.) and White's tree frogs (Litoria caerulea). J Vet Diag Invest 11: 194–199. [DOI] [PubMed] [Google Scholar]
- 5. Goka K, Yokoyama J, Une Y, Kuroki T, Suzuki K, et al. (2009) Amphibian chytridiomycosis in Japan: distribution, haplotypes and possible route of entry into Japan. Mol Ecol 18: 4757–4774. 10.1111/j.1365-294X.2009.04384.x [DOI] [PubMed] [Google Scholar]
- 6. Swei A, Rowley JJL, Rödder D, Diesmos MLL, Diesmos AC, et al. (2011) Is chytridiomycosis an emerging infectious disease in Asia? PLoS ONE 6: e23179 10.1371/journal.pone.0023179.t004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bataille A, Fong JJ, Cha M, Wogan GOU, Baek H-J, et al. (2013) Genetic evidence for a high diversity and wide distribution of endemic strains of the pathogenic chytrid fungus Batrachochytrium dendrobatidis in wild Asian amphibians. Mol Ecol 22: 4196–4209. 10.1111/mec.12385 [DOI] [PubMed] [Google Scholar]
- 8. Puschendorf R, Bolaños F (2006) Detection of Batrachochytrium dendrobatidis in Eleutherodactylus fitzingeri: effects of skin sample location and histologic stain. J Wildl Dis 42: 301–306. [DOI] [PubMed] [Google Scholar]
- 9. Lillie RD, Fullmer HM (1976) Histopathologic Technic and Practical Histochemistry. 4th ed New York: McGraw-Hill. [Google Scholar]
- 10. Rosenblum EB, James TY, Zamudio KR, Poorten TJ, Ilut D, et al. (2013). Complex history of the amphibian-killing chytrid fungus revealed with genome resequencing data. Proc Natl Acad Sci U S A 110: 9385–9390. 10.1073/pnas.1300130110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bai C, Liu X, Fisher MC, Garner TWJ, Li Y (2012) Global and endemic Asian lineages of the emerging pathogenic fungus Batrachochytrium dendrobatidis widely infect amphibians in China. Diversity and Distributions 18: 307–318. 10.1111/j.1472-4642.2011.00878.x [DOI] [Google Scholar]
- 12. Kusrini M, Skerratt L, Garland S, Berger L, Endarwin W (2008) Chytridiomycosis in frogs of Mount Gede Pangrango, Indonesia. Dis Aquat Org 82: 187–194. 10.3354/dao01981 [DOI] [PubMed] [Google Scholar]
- 13. Savage AE, Grismer LL, Anuar S, Onn CK, Grismer JL, et al. (2011) First record of Batrachochytrium dendrobatidis infecting four frog families from Peninsular Malaysia. EcoHealth 8: 121–128. 10.1007/s10393-011-0685-y [DOI] [PubMed] [Google Scholar]
- 14. Yang H, Baek H, Speare R, Webb R, Park S, et al. (2009) First detection of the amphibian chytrid fungus Batrachochytrium dendrobatidis in free-ranging populations of amphibians on mainland Asia: survey in South Korea. Dis Aquat Org 86: 9–13. 10.3354/dao02098 [DOI] [PubMed] [Google Scholar]
- 15. Jeong A, Yang HJ, Baek H-J, Ko YM, Lee H, et al. (2010) Distribution of the amphibians infected by chytrid fungus (Bd) in South Korea. Korean Journal of Herpetology 2: 9–15. [Google Scholar]
- 16. Dahanukar N, Krutha K, Paingankar MS, Padhye AD, Modak N, et al. (2013) Endemic Asian chytrid strain infection in threatened and endemic anurans of the northern Western Ghats, India. PLoS ONE 8: e77528 10.1371/journal.pone.0077528.t001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rodriguez D, Becker CG, Pupin NC, Haddad CFB, Zamudio KR (2014) Long-term endemism of two highly divergent lineages of the amphibian-killing fungus in the Atlantic Forest of Brazil. Mol Ecol 23: 774–787. 10.1111/mec.12615 [DOI] [PubMed] [Google Scholar]
- 18. Richards-Hrdlicka KL (2012) Extracting the amphibian chytrid fungus from formalin-fixed specimens. Method Ecol Evol 3: 842–849. 10.1111/j.2041-210X.2012.00228.x [DOI] [Google Scholar]
- 19. Boyle D, Boyle D, Olsen V, Morgan J, Hyatt A (2004) Rapid quantitative detection of chytridiomycosis (Batrachochytrium dendrobatidis) in amphibian samples using real-time Taqman PCR assay. Dis Aquat Org 60: 141–148. [DOI] [PubMed] [Google Scholar]
- 20. Hyatt A, Boyle D, Olsen V, Boyle D, Berger L, et al. (2007) Diagnostic assays and sampling protocols for the detection of Batrachochytrium dendrobatidis . Dis Aquat Org 73: 175–192. [DOI] [PubMed] [Google Scholar]
- 21. Stuart BL, Dugan K, Allard M, Kearney M (2006) Extraction of nuclear DNA from bone of skeletonized and fluid-preserved museum specimens. Syst Biodivers 4: 13–136. 17118181 [Google Scholar]
- 22. Ouellet M, Mikaelian I, Pauli BD, Rodrigue J, Green DM (2005) Historical evidence of widespread chytrid Infection in North American amphibian populations. Cons Biol 19: 1431–1440. 10.1111/j.1523-1739.2005.00108.x [DOI] [Google Scholar]
- 23. Wandeler P, Hoeck PEA, Keller LF (2007) Back to the future: museum specimens in population genetics. Trends Ecol Evol 22: 634–642. 10.1016/j.tree.2007.08.017 [DOI] [PubMed] [Google Scholar]
- 24. McLeod D, Sheridan J, Jiraungkoorskul W, Khonsue W (2008) A survey for chytrid fungus in Thai amphibians. Raff Bull Zool 56: 199–204. [Google Scholar]
- 25. Zeng Z, Bai S, Zhu Y (2011) Genetic differentiation of the pathogen of Batrachochytrium dendrobatidis in toads. J Econ Anim 15: 160–163. [Google Scholar]
- 26. Zhu W, Bai C, Wang S, Soto-Azat C, Li X, et al. (2014) Retrospective survey of museum specimens reveals historically widespread presence of Batrachochytrium dendrobatidis in China. EcoHealth: 241–250. 10.1007/s10393-013-0894-7 [DOI] [PubMed] [Google Scholar]
- 27. Cheng TL, Rovito SM, Wake DB, Vredenburg VT (2011) Coincident mass extirpation of neotropical amphibians with the emergence of the infectious fungal pathogen Batrachochytrium dendrobatidis . Proc Natl Acad Sci USA 108: 9502–9507. 10.1073/pnas.1105538108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lips K, Diffendorfer J, Mendelson J III, Sears M (2008) Riding the wave: reconciling the roles of disease and climate change in amphibian declines. PLoS Biol 6: e72 10.1371/journal.pbio.0060072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hall LM, Willcox MS, Jones DS (1997) Association of enzyme inhibition with methods of museum skin preparation. BioTechniques 22: 928–&. [DOI] [PubMed] [Google Scholar]
- 30. Zimmermann J, Hajibabaei M, Blackburn DC, Hanken J, Cantin E, et al. (2008) DNA damage in preserved specimens and tissue samples: a molecular assessment. Front Zool 5: 18 10.1186/1742-9994-5-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mendelson JR III, Jones MEB, Pessier AP, Toledo G, Kabay EH, et al. (2014) On the timing of an epidemic of amphibian chytridiomycosis in the highlands of Guatemala. South Am J Herpetol 9: 151–153. 10.2994/SAJH-D-14-00021.1 [DOI] [Google Scholar]
- 32. Larsen KW (2006) Trade, dependency, and colonialism: foreign trade and Korea's regional integration In: Armstrong CK, Rozman G, Kim SS, Kotin S, editors. Korea at the Center. Armonk, New York: M.E. Sharpe; 51–69. [Google Scholar]
- 33. Lukin A (2006) Russian views of Korea, China, and the regional order in northeast Asia In: Armstrong CK, Rozman G, Kim SS, Kotin S, editors. Korea at the Center. Armonk, New York: M.E. Sharpe; 15–34. [Google Scholar]
- 34. McMahon TA, Sears BF, Venesky MD, Bessler SM, Brown JM, et al. (2014) Amphibians acquire resistance to live and dead fungus overcoming fungal immunosuppression. Nature 511: 224–227. 10.1038/nature13491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Garner T, Perkins M, Govindarajulu P, Seglie D, Walker S, et al. (2006) The emerging amphibian pathogen Batrachochytrium dendrobatidis globally infects introduced populations of the North American bullfrog, Rana catesbeiana . Biol Lett 2: 455–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Farrer RA, Weinert LA, Bielby J, Garner TWJ, Balloux F, et al. (2011) Multiple emergences of genetically diverse amphibian-infecting chytrids include a globalized hypervirulent recombinant lineage. Proc Natl Acad Sci U S A 108: 18732–18736. 10.1073/pnas.1111915108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Soto-Azat C, Clarke BT, Fisher MC, Walker SF, Cunningham AA (2010) Widespread historical presence of Batrachochytrium dendrobatidis in African pipid frogs. Divers Distrib 16: 126–131. 10.1111/j.1472-4642.2009.00618.x [DOI] [Google Scholar]
- 38. Vredenburg VT, Felt SA, Morgan EC, McNally SVG, Wilson S, et al. (2013) Prevalence of Batrachochytrium dendrobatidis in Xenopus collected in Africa (1871–2000) and in California (2001–2010). PLoS ONE 8: e63791 10.1371/journal.pone.0063791.t003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Weldon C, Preez Du L, Hyatt A, Muller R, Speare R (2004) Origin of the amphibian chytrid fungus. Emerg Infect Dis 10: 2100–2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Shaded cells for latitude/longitude indicate that data have been added or modified based on detailed verbatim locality data. Taxonomy was changed to follow AmphibiaWeb (www.amphibiaweb.org). Under preservation type, the preservatives used for specimen preparation followed by that used for storage are given. Preservation method of specimens was assumed to be formalin/EtOH unless we had information stating otherwise. CAS = California Academy of Sciences, MVZ = Museum of Vertebrate Zoology.
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.