Abstract
Two CT features were developed to quantitatively describe lung adenocarcinomas by scoring tumor shape complexity (feature 1: convexity) and intratumor density variation (feature 2: entropy ratio) in routinely obtained diagnostic CT scans. The developed quantitative features were analyzed in two independent cohorts (cohort 1: n = 61; cohort 2: n = 47) of patients diagnosed with primary lung adenocarcinoma, retrospectively curated to include imaging and clinical data. Preoperative chest CTs were segmented semi-automatically. Segmented tumor regions were further subdivided into core and boundary sub-regions, to quantify intensity variations across the tumor. Reproducibility of the features was evaluated in an independent test-retest dataset of 32 patients. The proposed metrics showed high degree of reproducibility in a repeated experiment (concordance, CCC≥0.897; dynamic range, DR≥0.92). Association with overall survival was evaluated by Cox proportional hazard regression, Kaplan-Meier survival curves, and the log-rank test. Both features were associated with overall survival (convexity: p = 0.008; entropy ratio: p = 0.04) in Cohort 1 but not in Cohort 2 (convexity: p = 0.7; entropy ratio: p = 0.8). In both cohorts, these features were found to be descriptive and demonstrated the link between imaging characteristics and patient survival in lung adenocarcinoma.
Introduction
Lung cancer is the leading cause of cancer death in the U.S. and worldwide[1]. Despite therapeutic advances, the overall 5-year survival remains disappointingly low, at around 16%. Clinical decisions for the treatment of lung cancer are largely based on patient characteristics such as performance status, stage at diagnosis, and tumor histology. However, the clinical and biological heterogeneity within histological subtypes remain a major roadblock to successfully treatment of the disease as histologically similar tumors display a wide range of treatment response and metastatic behavior[2].
More recently, treatment strategies have begun to involve the subdivision of non-small-cell lung cancers (NSCLC) into molecular subsets based on specific driver mutational status in oncogenes and tumor suppressor genes [3,4]. Recent works have demonstrated a link between imaging features and gene expression patterns[5–8] thus highlighting the potential of imaging features to be used as independent prognostic or predictive biomarkers essential for enhancing the clinical decision making process. It is expected that the changes at the molecular level will be observable as related imaging phenomena[9]. Tumors within the same histological subtype demonstrate variable and definable imaging characteristics [10]. We propose that these characteristics can be quantified and used in addition to clinical and molecular characteristics to enhance medical decision-making process.
While complete genome profiling has not yet been adapted into the clinic, radiographic imaging is routinely performed on most patients. Computer tomography (CT) has remained an important diagnostic tool used for initial tumor assessment and staging in lung cancer. CT imaging interrogates the entire tumor ‘in situ’ in the context of its environment and can thus be used to assess the tumor globally. Additionally, it can be used to describe tumor heterogeneity and sub-regional “habitats” within the tumor[11]. Due to the increasing number of therapy options for NSCLC patients, these patient-specific prognostic biomarkers have the potential of individualizing and thus improving patient care and outcome.
NSCLC tumors are routinely characterized, using diagnostic imaging, based on their size, shape and margin morphology, and the extent of internal enhancement and necrosis. However, the terminology used in radiology to characterize the pathological findings remains subjective and the underlying data are rarely quantified; hence, we contend that their full potential to support medical decision making is underutilized. Quantifying these observations with computer assistance could provide systematic prognostic information with minimal inter- and intra-reader variability. Furthermore, quantitative data can be stored in databases, allowing these data to be mined to develop models for improved diagnosis, prognosis and prediction [12].
Although there has been an increase in research activity in the areas of lung nodule detection and classification using image processing and data mining algorithms, few investigators have pursued the development of diagnostic CT-based prognostic imaging biomarkers in NSCLC[9,13].In this work, we quantitatively analyzed diagnostic CT scans of lung adenocarcinomas to develop prognostic imaging features. In order to minimize genetic heterogeneity, we focused our research on lung adenocarcinomas, the most common histological subtype of lung cancer [14]. Diagnostic scans were collected retrospectively and augmented with patient information and clinical follow-up data, which enabled us to develop and test models to predict survival. Since the data were collected retrospectively as part of routine clinical practice, there was variability in terms of instrumentation, image acquisition and reconstruction parameters. We therefore recognized the importance of developing imaging features that were robust across the wide variability encountered in clinically-acquired diagnostic scans.
The development of the proposed imaging features was driven by the hypothesis that tumor shape and intratumor density variation reflect tumor biology and systematic quantification of these imaging characteristics can be used to describe tumor development and patient survival. Both features consistently scored tumors according to the pursued characteristics and showed prognostic behavior. Furthermore, the features were tested for reproducibility under standard patient related variations which showed high degree of reproducibility.
Materials and Methods
Ethics Statement
The University of South Florida institutional review board approved this retrospective study and waived the informed consent requirement. Data were collected and handled in accordance with the Health Insurance Portability and Accountability Act. Patient data was anonymized and de-identified prior to the analyses.
Study population and data
The protocol for this retrospective study, including the participation of the MAASTRO clinic, was approved by the Institutional Review Board (IRB). Imaging and clinical data were obtained on patients diagnosed with primary lung adenocarcinoma who were treated in the Thoracic Oncology Program at the H. Lee Moffitt Cancer Center and Research Institute and the Maastricht Radiation Oncology Clinic (MAASTRO). The Moffitt cohort (Cohort 1) included 61 patients and orthogonal MAASTRO cohort (Cohort 2) included 47 patients. Inclusion criteria encompassed patients who underwent surgical resection and had corresponding pre-surgery diagnostic CTs obtained within 60 days of the diagnosis.
For each patient, the cohorts included de-identified diagnostic pre-treatment contrast-enhanced CT scans acquired between years 2006 and 2009 as well as clinical data including demographics, diagnosis, TNM stage, and patient survival.
Clinical data were obtained from Moffitt’s cancer registry, which abstracts self-report information and clinical data from patient medical records including demographics, diagnosis, stage, and treatment. Follow-up for vital status information occurs annually through passive and active methods. Pathologic TNM staging was utilized when available and clinical stage was used if these pathologic staging was missing. For this analysis smoking status was categorized as self-report ever smoker (current or former smoker) or never smoker.
Tumor and lung segmentation
Patient CT scans were segmented to identify lung fields and tumors. The delineation of the lung fields was carried out using single click ensemble algorithm developed using Lung Tumor Analysis (LuTA) tool within the Definiens Developer XD© (Munich, Germany) software platform. Target lesions were volumetrically segmented using semiautomatic approach. The resident radiologist (over 2 years of experience) oversaw the segmentation boundaries on the CT slices. Performing semi-automatic segmentation not only decreased user interaction and eliminated the need for a manually drawn boundary, but also provided robust, reproducible and consistent delineation of the tumor region across the CT slices. We have previously demonstrated that the single click ensemble segmentation algorithm reduced inter-observer variability while capturing the intricacies and important details of the tumor boundary[15].
Algorithms for image feature extraction and quantification of the segmented tumor regions were implemented in MATLAB (Mathworks, Natick, MA).
Convexity morphological feature
Convexity algorithm (S1 Fig.) was developed to quantify shape variation of the tumor border. Irregularities along tumor perimeter can result from intratumor heterogeneity and differences in growth patterns, interaction with the surrounding environment and spiculations, multiple finger-like projections into the parenchyma which are generally considered to be a poor prognostic indicator[14,16].
The convexity of the tumor border was calculated as the ratio of the areas contained within a) the tumor perimeter to b) the calculated convex hull (Fig. 1A). The convex hull was computed by defining the smallest convex polygon enclosing a planar tumor region of interest (ROI) using the implementation of QuickHull algorithm[17]. Using tumor segmentation mask for a given CT slice as input, the divide and conquer approach automatically computed a convex hull vector of pixel locations. Intrinsically the ratio between the area of the tumor mask and its convex hull described the amount of substantial protuberances and depressions along the tumor border.
Fig 1. Convexity feature was developed to quantify tumor shape.
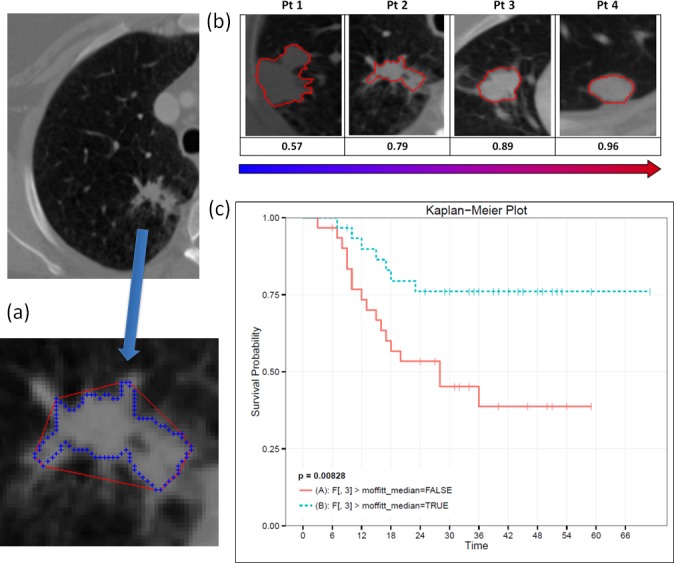
Convexity is computed as a ratio of tumor border (blue) to convex hull (red) (a). Convexity feature tracks the change in tumor morphology (b). Convexity is predictive of patient overall survival when dichotomized at the median value (c).
In order to account for all sequential slices containing tumor ROI, the calculation was performed individually on each slice and a mean score was computed using all slices for the tumor. A convexity score of one corresponded to a shape that does not present with any concavities along its perimeter. Convexity scores less than one measure the degree of deviation from a convex shape.
Since tumor pressing against the pleural wall can compromise the appearance of tumor morphology, automatic pleural attachment detection was incorporated into the convexity feature algorithm. For each CT slice, perimeters from segmented tumor and lung regions were extracted (S2 Fig.). Pleural attachment was determined if perimeters overlapped. If for a particular slice more than half of the tumor perimeter pixels were also in the lung perimeter, its convexity score was eliminated from the mean score calculation. This step allowed us to account for significant cases of shape smoothing that resulted from tumor pressing against the pleural wall.
Entropy Ratio of intratumor intensity variation feature
Entropy ratio feature (S1 Fig.) was developed to score heterogeneity of pixel attenuation coefficients across the tumor. The entropy filter is the implementation of Shannon entropy. In the image processing context, entropy is the measure of variation computed on the pixel histogram distribution within a given ROI. It is defined as where p i represents the probability (normalized frequency) of the given intensity value i. In our study we used a 256 intensity bins.
Segmented tumor mask was subdivided into two distinct regions: core and boundary. The subdivision was driven by the hypothesis that these distinct regions reflect unique, spatially explicit biological processes, e.g. necrosis in the core and proliferation along the periphery, and should therefore be assessed separately. Tumor growth and interaction with the surrounding microenvironment has been shown to lead to intratumor changes observable in radiographic scans[5]. In addition, the introduced spatial constraint helped account for edge effect manifesting itself in higher changes in intensities at the tumor interface which would otherwise skew the calculations had the summary statistics across the entire tumor ROI been applied.
Core and boundary masks were generated automatically from the original tumor segmentation using a series of morphological operations, erosion and dilation (S3 Fig.). Two disk-shaped structural elements with radii of 3 or 5 pixels were used. A binary segmentation mask for each tumor slice was first dilated using a disk with a 3 pixel radius. This produced a dilated mask which was then eroded using a disk with a 5 pixel radius to generate the core region mask. The ‘doughnut shape’ mask for the boundary region was produced by subtracting the core mask from the dilated mask (S3 Fig.).
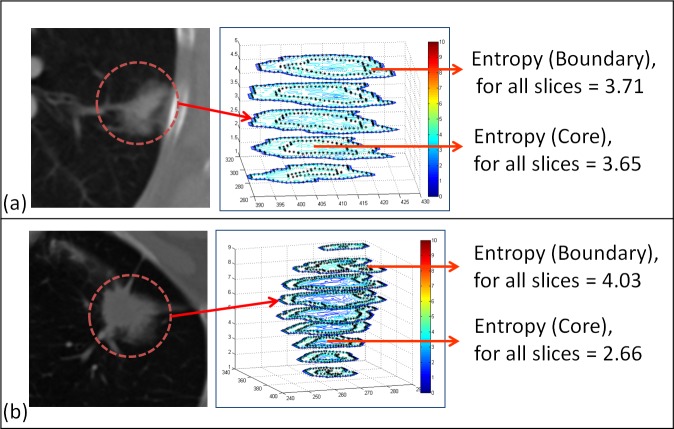
Prior to entropy filtering, each CT slice containing tumor was converted to its normalized grayscale equivalent. Binning Hounsfield units into 256 discrete intensity bins allowed the method to normalize tumor intensity ranges and to emphasize the differences in intensity gradient over small local pixel intensity variations. For each pixel within the tumor, an entropy coefficient was computed in its 7-by-7 pixel surrounding neighborhood. The mean of the coefficients was computed for pixels in the boundary and core regions as ensemble scores to represent them (Fig. 2). Entropy ratio feature was computed by dividing entropy score of the boundary region by the entropy score of the core region in order to characterize the contrast between them.
Fig 2. Entropy ratio was developed to quantify intensity variations across the tumor.
While some tumors present with consistent mean entropy across the core and the boundary (a), others have a distinct difference in the values (b).
Image feature reproducibility analysis. Since feature stability is an important trait for a biomarker, we tested the developed metrics against typical patient variations using the Reference Image Database to Evaluate Therapy Response (RIDER) dataset. The RIDER project was a National Cancer Center (NCI) sponsored project to create consensus across institutions and help in harmonization of the quantitative features. The dataset was downloaded from the NBIA National Biomedical Imaging Archive [18]. Unenhanced thoracic CT images for 32 patients in the test-retest dataset (baseline and follow-up) were acquired within 15 minutes of each other, using identical CT scanner and imaging protocol[18]. GE Medical Systems Lightspeed 16 or VCT scanner with 16/ 64 detectors in 28/4 patients were used, respectively. The dataset was matched with equal number of early and late stage tumors with equal mixture of men and women in the study.
Tumors were delineated by a semi-automated segmentation tool with boundary markings finalized by a radiologist. Convexity and entropy ratio features were extracted independently from baseline and follow-up scans of the RIDER dataset and consistency of the assigned score was examined. For each proposed feature, concordance correlation coefficient (CCC) was computed to quantify reproducibility between two scans performed on each patient. The concordance correlation coefficient measures deviation from the 45 degree line which has been shown to be superior to the Pearson correlation coefficient for comparing repeated experiments[19]. We then computed the Dynamic Range (DR) for each feature, which is defined as the average difference between measurements to the observed biological (inter-patient) range in the data set. The method has been first proposed and used to find informative feature set [20].
Statistical analyses
The imaging feature data, demographic information, and vital status data were merged into a single file for subsequent statistical analyses using Stata/MP 12.1 (StataCorp LP, College Station, TX). Student’s t-test and ANOVA were used to test for differences in imaging features by the demographic features and imaging parameters. A correlation matrix was used to assess dependency between the imaging features. Survival analyses were performed using Cox proportional hazard regression and Kaplan-Meier survival curves; statistical significance was computed using the log-rank test. The imaging features were dichotomized into two groups using the median score value.
Results
Demographics and imaging parameters by imaging biomarkers
Table 1 captures the variability of key clinical and imaging parameters for Cohort 1. This variability is representative of clinical applications that rely on patient imaging captured during the course of clinical care rather than for research purposes.
Table 1. Distribution of study population demographics and imaging parameters by imaging biomarkers in Cohort 1.
| Imaging biomarkers | ||||||||
|---|---|---|---|---|---|---|---|---|
| Entropy ratio | Tumor volume | Convexity | ||||||
| Characteristic 1 | No. | (%) | Mean | (SD) | Mean | (SD) | Mean | (SD) |
| Overall | 61 | (100.0) | 1.41 | (0.26) | 7884.6 | (11205.9) | 0.87 | (0.07) |
| Demographics | ||||||||
| Age at diagnosis | ||||||||
| < 65 | 20 | (32.8) | 1.42 | (0.05) | 11436.4 | (3651.2) | 0.87 | (0.02) |
| ≥ 65 | 41 | (67.2) | 1.40 | (0.04) | 6152.0 | (1129.7) | 0.86 | (0.01) |
| P-value | 0.834 | 0.084 | 0.829 | |||||
| Gender | ||||||||
| Female | 30 | (49.2) | 1.35 | (0.04) | 6467.9 | (1462.0) | 0.87 | (0.1) |
| Male | 31 | (50.8) | 1.46 | (0.05) | 9255.6 | (2444.3) | 0.87 | (0.1) |
| P-value | 0.102 | 0.336 | 0.995 | |||||
| Stage | ||||||||
| Stage I | 25 | (41.0) | 1.36 | (0.27) | 5875.2 | (10830.4) | 0.87 | (0.07) |
| Stage II | 19 | (31.2) | 1.44 | (0.24) | 9791.5 | (10927.1) | 0.86 | (0.09) |
| Stages III and IV | 17 | (27.8) | 1.42 | (0.25) | 8708.2 | (12217.9) | 0.87 | (0.07) |
| P-value | 0.590 | 0.493 | 0.817 | |||||
| Imaging Parameters | ||||||||
| Voltage, KvP | ||||||||
| 120 | 57 | (93.4) | 1.42 | (0.26) | 8187.2 | (11510.8) | 0.87 | (0.08) |
| 130 or 140 | 4 | (6.6) | 1.22 | (0.19) | 3572.3 | (3406.5) | 0.85 | (0.03) |
| P-value | 0.124 | 0.431 | 0.761 | |||||
| Convolution kernel | ||||||||
| B30F | 8 | (13.1) | 1.47 | (0.24) | 3314.3 | (4500.1) | 0.845 | (0.13) |
| B40f | 19 | (31.2) | 1.31 | (0.15) | 11752.4 | 1(5712.3) | 0.837 | (0.07) |
| B41F | 22 | (36.1) | 1.46 | (0.25) | 6487.3 | (7959.1) | 0.889 | (0.05) |
| Other 2 | 12 | (19.7) | 1.44 | 0.37) | 7369.0 | (10095.2) | 0.884 | (0.08) |
| P-value | 0.222 | 0.269 | 0.101 | |||||
| Slice thickness, mm | ||||||||
| <5.0 | 18 | (29.5) | 1.47 | (0.26) | 9836.8 | 0.844 | (0.10) | |
| ≥ 5.0 | 43 | (70.5) | 1.38 | (0.25) | 7067.4 | (11301.7) | 0.875 | (0.06) |
| P-value | 0.181 | 0.383 | 0.142 | |||||
| Pixel resolution 3 , mm | ||||||||
| < 0.6926 | 20 | (32.8) | 1.44 | (0.25) | 8013.6 | (10733.4) | 0.875 | (0.06) |
| ≥ 0.6926 to < 0.7785 | 20 | (32.8) | 1.36 | (0.21) | 10414.8 | (14356.6) | 0.878 | (0.09) |
| ≥ 0.7785 | 21 | (34.4) | 1.49 | (0.29) | 5352.1 | (7636.5) | 0.845 | (0.09) |
| P-value | 0.529 | 0.357 | 0.290 | |||||
1 96.7% (No. = 59) of this study population were ever smokers and 96.7% (No. = 59) were White race
2 Other includes B30s, B41s, B70s, CHST, FC01, FC13, LUNG, and STANDARD
3 Distribution based on the tertile values
There were no statistically significant differences in the imaging biomarkers by age, gender, stage, or by the imaging parameters including KVP, convolution kernel, slice thickness, or pixel resolution (Table 1). We however observed significant differences for both imaging features by stage (Convexity: p = 0.017; Entropy Ratio: p = 0.002) and for entropy ratio feature, by convolution kernel (p = 0.026) in Cohort 2 (Table A in S1 File).
Convexity and Entropy Ratio features reproducibility confirmed by Test-Retest analyses
Concordance correlation coefficients (CCC) and dynamic range (DR), averaged over the volume of the tumor, are summarized in Table B in S1 File. CCCs and DR for the features extracted from a single center slice of the tumor, matched by a radiologist between test and retest scans, are summarized in Table C in S1 File. Convexity feature showed high concordance in the test-retest experiment (>0.88) in both single slice and volumetric evaluations. Entropy ratio had a low CCC when averaged over the volume of the tumor. While entropy of the core and boundary regions had high CCC (>0.81), the concordance metric penalized their ratio as entropy ratio scores showed unproportional variation to the 45° line. The third column of Table B in S1 File shows the statistics of the absolute percent difference between test and retest feature values and it should be noted that the mean difference for entropy ratio feature is approximately 1.69%. Both convexity and entropy ratio showed high level of concordance (>0.99) and dynamic range (>0.96) in single slice evaluation (Table C in S1 File).
The repeatability of the features confirms their ability to consistently score tumor characteristics with respect to variability in the repeated experiments (S4 Fig.). The tumors in the RIDER set are a diverse population of mixed stages and histology. The Bland-Altman plot for the computed features (S5 Fig.) shows close bound (95% confidence) for the individual cases; larger test-retest variations can be attributed to the above discussed challenges.
Overall Survival of Convexity Feature
A lower convexity score was reflective of a more irregular shape and expected worse survival. Higher convexity scores describe convex shapes with fewer irregularities along the boundary. Fig. 1B demonstrates tumor shape morphology ordered according to the computed convexity score. Fig. 1C shows that high (> median) and low convexity separated patients with good and poor overall survival time (p = 0.008). Convexity values for the Cohort 1 ranged from 0.57 to 0.97, with the median value = 0.89.
Overall Survival of Entropy Ratio Feature
Median score were used to discriminate between low and high entropy groups. Entropy measures for the entire segmented region were not statistically significant (p = 0.28) with respect to overall survival. Evaluating entropy within the boundary region only was also not significant (p = 0.96). Furthermore, it appeared that high entropy values in the border region skewed the performance of the feature when calculated across the entire tumor region. While restricting the calculation of entropy to the core region was not statistically significant (p = 0.059), tumors with high (>median) core entropy tended to associate with worse overall survival. Entropy of the core may capture important intratumor characteristics such as necrosis and heterogeneity. While entropies of core and boundary regions were not statistically significant independently, their ratio was associated with overall survival (p = 0.04, Fig. 3A). Tumors for which the entropy was consistent throughout the core and boundary regions (i.e. ratio<1.41; S6 Fig.) were associated with better survival outcome and tumors presenting with larger (>1.41) ratios (S6 Fig.) were correlated with worse overall survival.
Fig 3. Entropy ratio between the core and border regions of the tumor is predictive of patient survival.
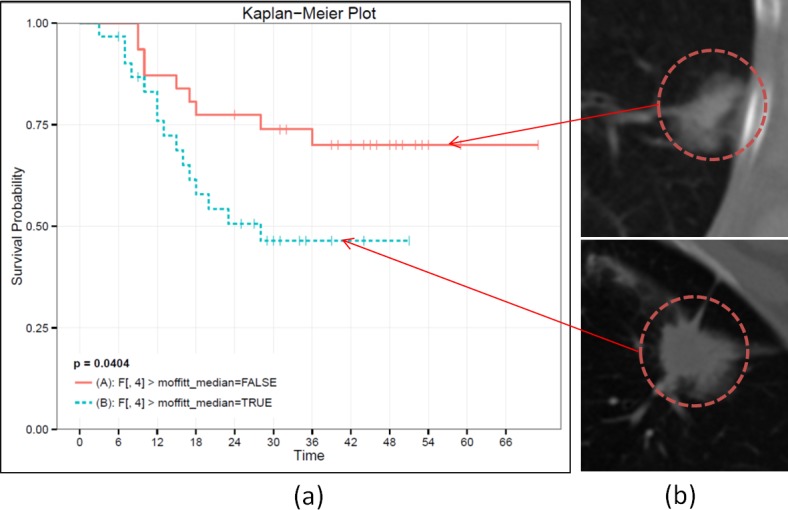
The tumors in the two prognostic groups (a) did not appear significantly different in the CT scans (b).
Representative CT slices of the tumors from the two prognostic groups, as defined by larger than median entropy ratio of the core and boundary regions are shown in Fig. 3B. When visually inspected by resident radiologists, tumors from these two prognostic groups were not described as being prognostically different. In fact, the tumor at the top in Fig. 3B might be considered to have worse prognosis due to its attachment to the pleural wall[21], which is a known negative prognostic indicator.
Cox Proportional Hazards Models
When the imaging biomarkers were analyzed independently in separate univariate models for overall survival (Table 2) in Cohort 1, we noted statistically significant associations with tumor volume (HR = 2.59; 95% CI 1.06–6.29) and convexity (HR = 0.34; 95% CI 0.14–0.82), and a borderline significant association with entropy ratio (HR = 2.19; 95% CI 0.94–5.08). Age, gender, and stage were significantly associated with survival in the univariable models. When all three imaging biomarkers were included in a stepwise forward selection model (see methods), tumor volume dropped out while convexity (HR = 0.32; 95% CI 0.13–0.78) and entropy ratio (HR = 2.33; 95% CI 1.00–5.45) remained. Convexity and entropy ratio remained statistically significant in the final multivariable model adjusting for age, gender, and stage. The final model was replicated in Cohort 2. Although the hazard ratios for entropy and convexity were not statistically significant, the point estimates trended in the same direction.
Table 2. Cox Proportional Hazards Models for Overall Survival.
| Cohort 1 (N = 62) | Cohort 2 (N = 47) | |||||||
|---|---|---|---|---|---|---|---|---|
| Covariates 1 | Unadjusted HR (95% CI) 2 | P-value | Multivariable HR (95% CI) 3 | P-value | Multivariable HR (95% CI) 4 | P-value | Multivariable HR (95% CI) 4 | P-value |
| Entropy ratio | 2.19 (0.94–5.08) | 0.07 | 2.33 (1.00–5.45) | 0.05 | 2.36 (1.00–5.58) | 0.05 | 1.24 (0.53–2.87) | 0.62 |
| Tumor volume | 2.59 (1.06–6.29) | 0.04 | — | — | — | — | — | — |
| Convexity | 0.34 (0.14–0.82) | 0.02 | 0.32 (0.13–0.78) | 0.01 | 0.31 (0.12–0.78) | 0.01 | 0.82 (0.33–2.03) | 0.67 |
| Age | 1.22 (0.50–2.96) | 0.67 | — | — | 1.11 (0.44–2.83) | 0.82 | 1.54 (0.60–3.97) | 0.36 |
| Gender | 1.25 (0.55–2.85) | 0.60 | — | — | 1.76 (0.73–4.23) | 0.21 | 1.49 (0.65–3.46) | 0.35 |
| Stage | 1.52 (0.92–2.53) | 0.11 | — | — | 1.45 (0.84–2.49) | 0.18 | 2.73 (1.36–5.48) | < 0.01 |
Abbreviations: Hazard Ratio, HR; Confidence Intervals, CI.
Statistically significant hazard ratios (p < 0.05) are shown in bold.
1 The imaging features are dichotomized at their respective median values and age is dichotomized at 65 years
2 Each imaging biomarker is analyzed independently in separate univariate models. The unadjusted HRs represent the main effects of each covariate.
3 Based on forward selection, only two imaging biomarkers are included in the model but excluded age, gender, and stage.
4 Only two imaging biomarkers are included in the model in addition to age, gender, and stage
Imaging features s performance in an independent cohort
The distributions of the computed feature scores were compared between two independent cohorts: Cohort 1 and Cohort 2 (Fig. 4). The convexity descriptor had a similar range in both cohorts (Fig. 4A); however, Cohort 1 (green) distribution was skewed towards rounder and more convex tumor shapes. In either of the cohorts, low convexity corresponded to more irregular tumor shapes, while high convexity scores were indicative of rounder shapes with fewer concavities and irregularities along the tumor perimeter.
Fig 4. Histogram of the two imaging features across cohorts.
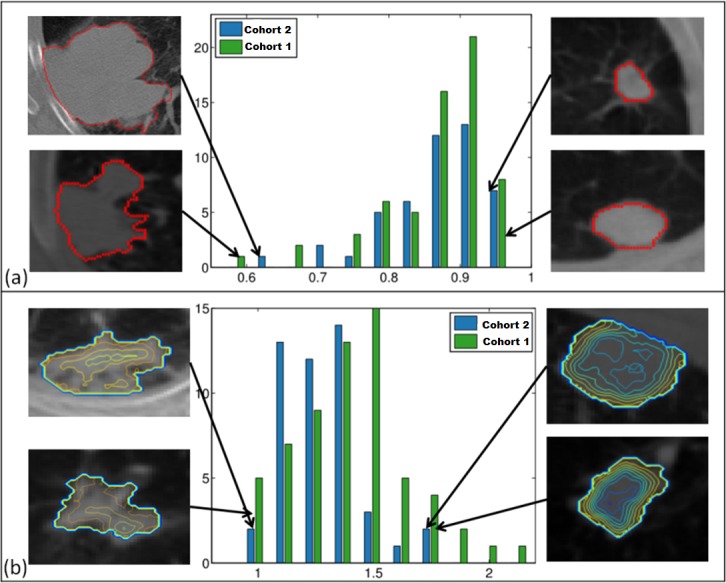
Convexity (a) shows similar range across cohorts (training-green, test-blue) However, training cohort is enriched with round tumors. The range of values for entropy ratio feature (b) is larger in training cohort. Both convexity (a) and entropy ratio (b) consistently capture targeted tumor characteristics in both cohorts.
Histograms of entropy ratio scores were different between the cohorts (Fig. 4B). Contour plots of entropy coefficients in the tumor regions were generated to visualize intratumor entropy differences in extreme imaging phenotypes. The phenotypes described by the low value entropy ratio (Fig. 4B, left) were similar across both cohorts. Contour plot profile corresponding to high entropy ratio (Fig. 4B, right) was consistent for both cohorts, as well.
Survival analyses were carried out on Cohort 2 strictly in accordance with the previously established procedure for Cohort 1. This is a very stringent test of reproducibility, as the populations and acquisition conditions were different between the cohorts. Based on the median splits derived from Cohort 1 (convexity = 0.89, and entropy ratio = 1.4), in Cohort 2 neither convexity (p = 0.7) nor entropy ratio (p = 0.8) were associated with overall survival. In Cohort 2, convexity feature was statistically significant with respect to survival (p = 0.008) in univariate Cox proportional hazards regression analysis.
The developed tumor descriptors consistently and objectively scored and ordered tumors according to their shape (S7 Fig.) and intratumor intensity variation (S8 Fig.) in both datasets.
Discussion
The aim of this study was to develop objective and robust quantitative imaging descriptors that were associated with patient survival. The developed CT features quantified lung adenocarcinomas based on their shape and intratumor intensity variations. The features systematically scored tumors and identified imaging phenotypes which exhibited survival differences in two independent cohorts. The features were extracted from routinely obtained CT images and were reproducible and stable despite the inherent clinical image acquisition variability.
Outcomes after resection in early-stage NSCLC are poor, with 30–50% of the deaths due to distant recurrence[22]. Current decision support for the use of adjuvant chemotherapy following surgery is ill-informed. Gene signatures [2] have been developed to improve risk stratification of early stage NSCLC patients and while an improvement, the approach suffers from sampling bias, as to all biopsy-dependent approaches. Incorporating additional knowledge from individual patients’ scans into the post-operative decision making can potentially identify distinct phenotypes of the disease and serve as a noninvasive approach to identify high-risk patients who may benefit from additional post-surgical treatment such as adjuvant chemotherapy. In addition, imaging features can be used as surrogates for tumor specific factors when limited pathological specimens are obtained from diagnostic biopsies [21]. Furthermore, we believe that inferring tumor biological activity and aggressiveness from minimally-invasive imaging techniques can inform precision medicine strategies longitudinally where serial biopsies may be too difficult to achieve.
In order to avoid data sparsity, we recognized the importance of keeping the number of imaging features reasonably low and reflective of the cohort. It has been recommended to keep a rather stringent ratio between the number of patients in each prognostic class and the features above 10[23]. In order to adequately relay tumor characteristics while coping with dimensionality restrictions, preference should be given to hypothesis-driven features predicated on available biomarkers and tumor biology associations[24].
Owning to the absence of a validation cohort with matching clinical and imaging parameters, we could not adequately explore the prognostic behavior of the developed features. Validation of imaging features derived from retrospectively assembled cohorts presents an interesting challenge as the field continues to mature. It is essential to refute spurious findings by validating imaging biomarkers in independent, previously unseen cohorts. However, the lack of ‘golden standard’ for testing derived features is often coupled with a number of confounding factors stemming from imaging variability and heterogeneity within patient cohort. It is therefore important to recognize that differences in patient population, quality and manner of data acquisition and subsequent processing can result in validation cohorts that are not adequate for fair assessment. This also highlights the significance and merits of hypothesis-driven imaging feature design which can serve as additional validation criteria in an independent cohort. In our study we have taken the effort to develop image based metrics to quantify observable tumor characteristics.
Additional exploration of the prognostic utility was carried out in an independent cohort (Cohort 2) from a partnering institution located on a different continent, with inherent differences in patient population. In both cohorts, extreme tumor phenotypes were identified based on the distribution of the computed feature scores (Table D in S1 File). Patient subpopulations were chosen using the opposite extreme quartiles of the convexity score distribution (<0.8 and >0.9) from Cohort 1. Survival difference between the two phenotypes was statistically significant for Cohort 2 (p = 0.02; Cohort 1: p = 0.06). Similarly, two subpopulations were identified using low (<1.23) and high (>1.5) entropy ratio scores from Cohort 1. The difference between the subpopulations was statistically significant for Cohort 1 (15 patients vs. 19 patients, p = 0.04). However, due to the differences in entropy score distributions (Fig. 4B), there were not enough patients (n = 2) in Cohort 2 to represent the phenotype with high (>1.5) entropy ratio scores. The survival significance found in the entire Cohort 1 and in Cohort 2, using quartile split, is promising.
Our study had several limitations. The cohort sample sizes were small which limited our ability to perform extensive stratified analyses. Additionally, the two cohorts are likely not comparable since we have observed significant differences for the imaging features by stage and convolution kernel in Cohort 2 but no difference by stage (or any other characteristic) in Cohort 1. While patients in Cohort 1 were surgical candidates, predominantly stages I and II, Cohort 2 consisted of radiotherapy planning patients with more advanced stages of the disease. Furthermore, the overall survival trend for the cohorts differed (S9 Fig.). The differences between the two cohorts may explain why our multivariate model did not replicate in Cohort 2 (Table 2). Standardization of image protocols remains a challenge in image biomarker development[25]. Variability in image acquisition and reconstruction parameters is inherent to retrospective imaging studies. Although this variability can be a significant limitation in studies, ultimately for the computed tomographic biomarkers to be adopted into clinical practice and utilized across multiple imaging centers, these biomarkers must be stable in the presence of image acquisition variability[26]. The size of our cohorts and imaging parameter variability are equivalent to other recently published studies [7,13,27,28]. The strength of our study was the development of imaging features that were descriptive and reproducible using retrospectively acquired clinical scans.
Our results suggest that quantitative imaging biomarkers can be used as an additional diagnostic tool in management of lung adenocarcinomas. Although more work is needed to determine clinical utility, it is clear that these descriptors are capable of quantifying and consistently ranking key tumor characteristics. Imaging biomarkers, combined with RECIST measurements and laboratory test results, will in the future be the de facto standard of a decision support pipeline employed to personalize and optimize treatment protocols.
Supporting Information
Convexity (a) and entropy ratio (b) algorithms are described.
(TIF)
The lung perimeter is outlined with green unfilled circles and tumor perimeter—with red filled circles.
(TIF)
Original tumor segmented ROI (a) is subdivided into core and boundary sub-regions (c). Tumor ROI (b, red) is dilated (b, pink) and eroded (b, green). Subtracting the core mask (b, bottom, green contour) from the dilated region forms the boundary mask (b, bottom, pink contour).
(TIF)
Representative slices from tumors with high (a,b) and low (c,d) convexity scores are displayed. Tumor scores were computed on baseline scans: 0.54(a) and 0.92(c) and follow-up scans: 0.6 (c) and 0.91 (d).
(TIF)
The dotted lines show 95% confidence limit for the features in the sample set.
(TIF)
Two classes of tumors were identified: tumors with low entropy ratio between core and boundary regions (a) and tumors with high entropy ratio between core and boundary regions (b).
(TIF)
Convexity feature consistently scored tumor shape in both cohorts (Cohort 1: a; Cohort 2: b).
(TIF)
Tumors were ordered based on their entropy ratio score and showed similar patterns of entropy coefficient distribution in both cohorts. Entropy ratio feature consistently scored intratumor density variation in both cohorts (Cohort 1: a; Cohort 2: b).
(TIF)
Overall survival trends for Cohort1 (a) and Cohort 2 (b) are different.
(TIF)
Table B, Concordance correlation coefficient (CCC) and dynamic range for CT features in the test-retest experiment, averages over tumor volume. Table C, Feature Reproducibility in test-retest analyses using matching center slice on the CT image. Table D, Clinical parameters and feature scores for Cohort 1 patients.
(DOCX)
Data Availability
Quantitative features data for Cohort 1 has been added to Suppl. materials. Cohort 1 deidentified image data will be available via TCIA, upon publication. The dataset can be found at: https://public.cancerimagingarchive.net/ncia/login.jsf Using Search, under Collection(s), the dataset is available as LungCT-Diagnosis, and the DOI is https://wiki.cancerimagingarchive.net/x/VIYiAQ. Patient scan and clinical data for Cohort 2 is de-identified and can be made available upon request. Please contact Olya.Grove@moffitt.org, to establish a data transfer protocol, for images.
Funding Statement
Funding provided by National Cancer Institute: http://www.cancer.gov/, 1.NIH/NCI U01CA143062, Radiomics of NSCLC; 2. NIH/NCI P50CA119997, Radiomics of Lung Cancer Screening. Florida Biomedical Research Programs: http://researchtest.magnet.fsu.edu/newsletters/2013/July/documents/2013-2014%20SUMMER%20CALL%20King%20Program.pdf, King Team Science Grant 2KT01. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Siegel R, Naishadham D, Jemal A (2013) Cancer statistics, 2013. CA Cancer J Clin 63: 11–30. 10.3322/caac.21166 [DOI] [PubMed] [Google Scholar]
- 2. Kratz JR, He J, Van Den Eeden SK, Zhu ZH, Gao W, et al. (2012) A practical molecular assay to predict survival in resected non-squamous, non-small-cell lung cancer: development and international validation studies. Lancet 379: 823–832. 10.1016/S0140-6736(11)61941-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ding L, Getz G, Wheeler DA, Mardis ER, McLellan MD, et al. (2008) Somatic mutations affect key pathways in lung adenocarcinoma. Nature 455: 1069–1075. 10.1038/nature07423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Peifer M, Fernandez-Cuesta L, Sos ML, George J, Seidel D, et al. (2012) Integrative genome analyses identify key somatic driver mutations of small-cell lung cancer. Nat Genet 44: 1104–1110. 10.1038/ng.2396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rutman AM, Kuo MD (2009) Radiogenomics: creating a link between molecular diagnostics and diagnostic imaging. Eur J Radiol 70: 232–241. 10.1016/j.ejrad.2009.01.050 [DOI] [PubMed] [Google Scholar]
- 6. Segal E, Sirlin CB, Ooi C, Adler AS, Gollub J, et al. (2007) Decoding global gene expression programs in liver cancer by noninvasive imaging. Nat Biotechnol 25: 675–680. [DOI] [PubMed] [Google Scholar]
- 7. Gevaert O, Xu J, Hoang CD, Leung AN, Xu Y, et al. (2012) Non-small cell lung cancer: identifying prognostic imaging biomarkers by leveraging public gene expression microarray data—methods and preliminary results. Radiology 264: 387–396. 10.1148/radiol.12111607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Diehn M, Nardini C, Wang DS, McGovern S, Jayaraman M, et al. (2008) Identification of noninvasive imaging surrogates for brain tumor gene-expression modules. Proc Natl Acad Sci U S A 105: 5213–5218. 10.1073/pnas.0801279105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jaffe CC (2012) Imaging and genomics: is there a synergy? Radiology 264: 329–331. 10.1148/radiol.12120871 [DOI] [PubMed] [Google Scholar]
- 10. Rees JH, Smirniotopoulos JG, Jones RV, Wong K (1996) Glioblastoma multiforme: radiologic-pathologic correlation. Radiographics 16: 1413–1438; quiz 1462–1413. [DOI] [PubMed] [Google Scholar]
- 11. Gatenby RA, Grove O, Gillies RJ (2013) Quantitative imaging in cancer evolution and ecology. Radiology 269: 8–15. 10.1148/radiol.13122697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lambin P, Rios-Velazquez E, Leijenaar R, Carvalho S, van Stiphout RG, et al. (2012) Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer 48: 441–446. 10.1016/j.ejca.2011.11.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ganeshan B, Panayiotou E, Burnand K, Dizdarevic S, Miles K (2012) Tumour heterogeneity in non-small cell lung carcinoma assessed by CT texture analysis: a potential marker of survival. Eur Radiol 22: 796–802. 10.1007/s00330-011-2319-8 [DOI] [PubMed] [Google Scholar]
- 14. Honda T, Kondo T, Murakami S, Saito H, Oshita F, et al. (2013) Radiographic and pathological analysis of small lung adenocarcinoma using the new IASLC classification. Clin Radiol 68: e21–26. 10.1016/j.crad.2012.09.002 [DOI] [PubMed] [Google Scholar]
- 15. Gu Y, Kumar V, Hall LO, Goldgof DB, Li CY, et al. (2013) Automated Delineation of Lung Tumors from CT Images Using a Single Click Ensemble Segmentation Approach. Pattern Recognit 46: 692–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Prokop M (2001) Image Analysis In: Prokop M, Galanski M, Van Der Molen AJ, Schaefer-Prokop C, editors. Spiral and Multislice Computed Tomography of the Body. 1 ed: Thieme; pp. 1104. [Google Scholar]
- 17. Barber CB, Dobkin DP, Huhdanpaa HT (1996) The Quickhull Algorithm for Convex Hulls. ACM Transactions on Mathematical Software 22: 469–483. [Google Scholar]
- 18. Zhao B, James LP, Moskowitz CS, Guo P, Ginsberg MS, et al. (2009) Evaluating variability in tumor measurements from same-day repeat CT scans of patients with non-small cell lung cancer. Radiology 252: 263–272. 10.1148/radiol.2522081593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lin LI (1989) A concordance correlation coefficient to evaluate reproducibility. Biometrics 45: 255–268. [PubMed] [Google Scholar]
- 20. Balagurunathan Y, Gu Y, Wang H, Kumar V, Grove O, et al. (2014) Reproducibility and Prognosis of Quantitative Features Extracted from CT Images. Transl Oncol 7: 72–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shultz DB, Trakul N, Abelson JA, Murphy JD, Maxim PG, et al. (2014) Imaging features associated with disease progression after stereotactic ablative radiotherapy for stage I non-small-cell lung cancer. Clin Lung Cancer 15: 294–301 e293. 10.1016/j.cllc.2013.12.011 [DOI] [PubMed] [Google Scholar]
- 22. Kelsey CR, Marks LB, Hollis D, Hubbs JL, Ready NE, et al. (2009) Local recurrence after surgery for early stage lung cancer: an 11-year experience with 975 patients. Cancer 115: 5218–5227. 10.1002/cncr.24625 [DOI] [PubMed] [Google Scholar]
- 23. Miles KA, Ganeshan B, Griffiths MR, Young RC, Chatwin CR (2009) Colorectal cancer: texture analysis of portal phase hepatic CT images as a potential marker of survival. Radiology 250: 444–452. 10.1148/radiol.2502071879 [DOI] [PubMed] [Google Scholar]
- 24. Yang X, Knopp MV (2011) Quantifying tumor vascular heterogeneity with dynamic contrast-enhanced magnetic resonance imaging: a review. J Biomed Biotechnol 2011: 732848 10.1155/2011/732848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kuo MD, Jamshidi N (2014) Behind the numbers: decoding molecular phenotypes with radiogenomics-guiding principles and technical considerations. Radiology 270: 320–325. 10.1148/radiol.13132195 [DOI] [PubMed] [Google Scholar]
- 26. Rose CJ, Mills SJ, O'Connor JP, Buonaccorsi GA, Roberts C, et al. (2009) Quantifying spatial heterogeneity in dynamic contrast-enhanced MRI parameter maps. Magn Reson Med 62: 488–499. 10.1002/mrm.22003 [DOI] [PubMed] [Google Scholar]
- 27. Nair VS, Barnett PG, Ananth L, Gould MK, Veterans Affairs Solitary Nodule Accuracy Project Cooperative Studies G (2010) PET scan 18F-fluorodeoxyglucose uptake and prognosis in patients with resected clinical stage IA non-small cell lung cancer. Chest 137: 1150–1156. 10.1378/chest.09-2356 [DOI] [PubMed] [Google Scholar]
- 28. Ravanelli M, Farina D, Morassi M, Roca E, Cavalleri G, et al. (2013) Texture analysis of advanced non-small cell lung cancer (NSCLC) on contrast-enhanced computed tomography: prediction of the response to the first-line chemotherapy. Eur Radiol 23: 3450–3455. 10.1007/s00330-013-2965-0 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Convexity (a) and entropy ratio (b) algorithms are described.
(TIF)
The lung perimeter is outlined with green unfilled circles and tumor perimeter—with red filled circles.
(TIF)
Original tumor segmented ROI (a) is subdivided into core and boundary sub-regions (c). Tumor ROI (b, red) is dilated (b, pink) and eroded (b, green). Subtracting the core mask (b, bottom, green contour) from the dilated region forms the boundary mask (b, bottom, pink contour).
(TIF)
Representative slices from tumors with high (a,b) and low (c,d) convexity scores are displayed. Tumor scores were computed on baseline scans: 0.54(a) and 0.92(c) and follow-up scans: 0.6 (c) and 0.91 (d).
(TIF)
The dotted lines show 95% confidence limit for the features in the sample set.
(TIF)
Two classes of tumors were identified: tumors with low entropy ratio between core and boundary regions (a) and tumors with high entropy ratio between core and boundary regions (b).
(TIF)
Convexity feature consistently scored tumor shape in both cohorts (Cohort 1: a; Cohort 2: b).
(TIF)
Tumors were ordered based on their entropy ratio score and showed similar patterns of entropy coefficient distribution in both cohorts. Entropy ratio feature consistently scored intratumor density variation in both cohorts (Cohort 1: a; Cohort 2: b).
(TIF)
Overall survival trends for Cohort1 (a) and Cohort 2 (b) are different.
(TIF)
Table B, Concordance correlation coefficient (CCC) and dynamic range for CT features in the test-retest experiment, averages over tumor volume. Table C, Feature Reproducibility in test-retest analyses using matching center slice on the CT image. Table D, Clinical parameters and feature scores for Cohort 1 patients.
(DOCX)
Data Availability Statement
Quantitative features data for Cohort 1 has been added to Suppl. materials. Cohort 1 deidentified image data will be available via TCIA, upon publication. The dataset can be found at: https://public.cancerimagingarchive.net/ncia/login.jsf Using Search, under Collection(s), the dataset is available as LungCT-Diagnosis, and the DOI is https://wiki.cancerimagingarchive.net/x/VIYiAQ. Patient scan and clinical data for Cohort 2 is de-identified and can be made available upon request. Please contact Olya.Grove@moffitt.org, to establish a data transfer protocol, for images.