Abstract
Objective: The human T cell transcription factor-4 (TCF4) interacts functionally with β-catenin in the Wnt signaling pathway, whose deregulation is involved in the tumorigenesis of various types of cancers. Recent studies showed that TCF4 mRNAs were subject to alternative splicing, which was proposed to be important in regulating transactivational properties of the corresponding protein isoforms. Here we investigated the splicing isoforms and the roles of TCF4 in human esophageal squamous cell carcinoma.
Methods: RT-PCR and subsequent cloning and sequencing were applied to identify the splicing isoforms. Western blotting and realtime PCR were used to analyze the expression of TCF4. Knockdown of TCF4 was achieved with siRNA and stable transfection of expression vectors was performed.
Results: Our results showed there were a lot of different isoforms of TCF4 mRNA both in human esophageal cancers and cell line. Further, knockdown of TCF4E isoform expression in EC109 cells inhibited the cell growth, while overexpression of TCF4M isoform did not alter its transcription activity. Moreover, sixteen potential binding proteins of TCF4 were preliminarily identified by mass spectrometry.
Conclusions: Our data suggested that deregulation of TCF4 isoforms may contribute to the tumorigenesis of ESCC.
Keywords: TCF4, colon cancer, esophageal squamous cell carcinoma, alternative splicing, tumorigenesis
Introduction
Esophageal cancer is one of the leading causes of cancer-related death, and its incidence is increasing year by year in the world 1. Compared with other populations, esophageal squamous cell carcinoma (ESCC) is the most common subtype of esophageal cancer in Chinese population, whose incidence is much higher than esophageal adenocarcinomas 2. As a typical cancer, esophageal cancer is thought to be derived from multistep genetic and epigenetic alterations, but the molecular mechanisms have not been convincingly identified. Molecular studies of human esophageal tumors have identified numerous genetic abnormalities associated with DNA replication and repair, cell proliferation and apoptosis, cell cycle control, oncogenes and tumor suppressor genes (TSG) 3. Among them, abnormalities of transcription factors and coactivators have attracted many interests. Many TSG such p53, p21and p16 can function as transcription factors by binding to cis-acting elements, which lead to activation of its substrates.
Wnt/β-catenin signaling pathway consists of some transcription coactivators such as β-catenin and T cell factors (TCFs). This signaling pathway has been reported to control a variety of functions in various types of cells. During tissue development and regeneration, Wnt/β-catenin signals are well-controlled to ensure the proper balance between proliferation and differentiation 4,5. Activation of Wnt/β-catenin signaling pathway is thought to be associated with cell proliferation, as aberrant expressions of Wnt components are found in many types of cancers. Evidences indicate that abnormal activation of the Wnt pathway plays an important role in cancer progression. The hallmark of the Wnt signaling pathway is the accumulation of β-catenin in the cytoplasm, which then translocates to the nucleus to trigger the β-catenin/TCF transcriptional machinery, and finally upregulates target genes.
Recently, accumulation of nuclear and cytoplasmic β-catenin has also been observed in ESCC 6,7. Under normal conditions, the protein level of cytoplasmic β-catenin is very low because it is phosphorylated and degraded by a multiprotein complex. In the canonical Wnt/β-catenin pathway, the TCFs factors (including lymphoid enhancer-binding factor (LEF1), TCF1, TCF3 and TCF4) transduce Wnt/β-catenin signals to activate downstream target genes. Besides adenomatous polyposis coli (APC) protein, glycogen synthase kinase 3β (GSK 3β), casein kinase I and Axin, overexpression of T-cell lymphomas-1 (FRAT1) or End-binding protein 1 (EB1) can also lead to activation of β-catenin/TCF pathway in ESCC 8,9. Activated β-catenin/TCF pathway can upregulate STAT3 expression in ESCC at transcription level, which indicates a crosstalk between these signaling pathways 10. To upregulate its targets, TCF plays an important role in the transcription machinery.
TCF4, a member of the large family of evolutionary conserved HMG-box-containing proteins, can bind to DNA motif of 5'(A/T)(A/T)CAAAG3'. Its transcription activity is activated by nuclear β-catenin in Wnt signal pathway. However, other studies showed that TCF/LEF factors could also act as transcriptional repressors of Wnt/β-catenin target genes in the absence of β-catenin. Other proteins were shown to act as corepressors in this process, particularly CtBP that binds to the C-terminal end of TCF4 protein 11-13. Recent results showed that TCF mRNAs were subject to alternative splicing (AS), which has been proposed to be important in regulating transactivational properties of the corresponding protein isoforms. Different transcription isoforms of TCF4 have been found in various types of cancer including colorectal cancer, brain and renal carcinoma 14-17. Three regions of TCF4 gene are most frequently found to be alternative spliced. The first one is exon 4, which encodes a 23 amino acid protein domain without known function. The second one is exon 8, which has splice donor at the 3'-end to yield TCF4N isoform 18. This isoform, functions as a dominant negative factor in Wnt signaling, contains of the N-terminal part of TCF4 including the β-catenin binding domain but lacks the HMG-box. The other is exon 13~16, which extensively spliced in various combinations to generate TCF4L, TCFM and TCF4S isoforms with highly diverging C-termini.
As mentioned above, aberrant expression of Wnt/β-catenin signal pathway components was found in ESCC, the alterative splicing of TCF4 may contribute to carcinogenesis and tumor progress. To better understand its roles in human carcinogenesis, we analyzed the alterative splicing forms of TCF4 in human ESCC by RT-PCR, and subsequently sequencing the clones. Our results revealed the different isoforms of TCF4 mRNA both in human esophageal cancers and cell lines.
Materials and Methods
Materials
CCK-8 and PI staining kits were purchased from Beyotime Biotechnology Inc. (Nantong, China). Primary antibodies against TCF4 and GAPDH were purchased from Cell Signaling Technology (Beverly, USA). Lipofectamine 2000 and Trizol were purchased from Invitrogen (Carlsbad, USA). Small interference RNA duplex (siRNA) was synthesized by Shanghai GenePharma (Shanghai, China). Experssion constructs of TCF4E and TCF4M were obtained from FulenGen (Guangzhou, China).
Patients and samples
Cancerous and non-cancerous tissues were obtained by surgical resection from patients with esophageal cancer at the Southwest Hospital, Chongqing, China. Histological classification was based on postoperative histological examination. Non-cancerous tissues were located in the esophagus at least 5 cm away from the tumor. Tissues were applied for protein and RNA isolation. This study was approved by the institutional review board of Third Military Medical University.
Cell culture
Human esophageal cancer cell line EC109, KYSE150 and KYSE450 were obtained from Shanghai Cell Bank (http://www.ctcccas.ac.cn/xibao) and cultured in Dulbecco modified Eagle medium containing 100 U/ml of penicillin, 100 μg/ml of streptomycin, and 10% fetal bovine serum. Materials for cell culture were obtained from Thermo Fisher Scientific (Shanghai, China).
RNA isolation, reverse transcription polymerase chain reaction (RT-PCR) and cloning of PCR products
Total RNAs were extracted from tissues and cell culture with Trizol reagent. RNA concentration was quantified with a NanoDrop spectrophotometer. An amount of 1 µg total RNA was reverse transcribed to cDNA according to the manufacturer's directions (Roche (China) Ltd., Shanghai, China). Regular and realtime PCR was performed as previously described 19. The PCR conditions were: initial denaturation at 95°C for 5 min; 32 cycles of denaturation at 95°C for 20 s; annealing at 60°C for 30 s and elongation at 72°C for 30 s. PCR products were detected by electrophoresis on 6-10% polyacrylamide gels, or on 2% agarose gels. DNA fragments were isolated from the agarose gels with DNA and Gel Purification Kit (Bodatake, China). PCR products were further sequenced or subcloned into T-vector and individual clone was picked for sequencing.
For realtime PCR, all primers were list in Table 2, and the amplification conditions were similar to that of regular RT-PCR. Relative gene expression of each sample was normalized to the expression level of GAPDH and was presented as fold increase to that of normal control.
Table 2.
sequence of real-time RT-PCR primers
| Gene Name | Sequence | Product (bp) |
|---|---|---|
| CyclinD1 | F: GGATGCTGGAGGTCTGCGAGGAA | 358 |
| R: GCGTGTTTGCGGATGATCTGTTTGT | ||
| MMP7 | F: TTTAGAAGCCAAACTCAAGGA | 218 |
| R: GATCCACTGTAATATGCGGTAA | ||
| GAPDH | F: GGGAAGGTGAAGGTCGGAGTC | 232 |
| R: CCTGGAAGATGGTGATGGGAT |
Proteins samples and Western blotting
Protein samples from tissues and cell cultures were analyzed by Western blotting.
Tissues or cell pellets were homogenized in ice-cold RIPA buffer containing complete protease inhibitors cocktail (Roche (China) Ltd., Shanghai, China). The extraction mixture was then centrifuged at 12000g at 4℃ for 20 min and the protein concentration in the supernatant was determined by Bradford assay. Western blotting was performed as previously described 19. The expression of TCF4 protein was normalized by that of GAPDH.
Cell cycle analysis
Exponentially growing cells in 6-well plate were transfected with siRNA or TCF4E construct using Lipofectamine 2000 according to the manufacturer's directions. Two days later, cells were harvested and washed with ice-cold PBS and fixed in 70% ethanol. Before flow cytometric analysis, DNA content was labeled with propidium iodide in the presence of the RNase (1mg/ml) for 30 min at room temperature. Then the proportions of cells in G1, S and G2/M phases were analyzed on a FACSort (Becton Dickinson, San Jose, CA) with Cell Quest software (version 313).
Knockdown of TCF4
EC109 cells in 6-well plate were transfected with siRNA against TCF4 or control siRNA. Target sequences of TCF4 siRNA were listed in Supplementary Material: Table S1. Cells were harvested and protein samples were prepared for Western blotting 48h after transfection. For cell growth assays, EC109 cells in 96-well plate were treated with siRNA for 48h and then stained with CCK-8.
Immunoprecipitation and mass spectrometry (MS)
EC109 cells were harvested and washed in ice-cold PBS. Then cells were suspended in RIPA buffer (P0013D, Beyotime, China) containing protease inhibitor cocktail and phosphatase inhibitor cocktail (Roche, China), and incubated on ice for 60 min with brief vortex (10 seconds) every 3 mins. After centrifuged (2 000×g) for 10 min, the supernatant was collected and mixed with anti-TCF4 antibody, and then rotated overnight at 4°C. After protein-A/G-agarose beads (Beyotime, China) were added, the mixtures were further rotated for 2h at 4°C. Then the beads were washing with lysis buffer for 5 times. The samples of immunoprecipitated proteins were collected by centrifugation, eluted, and analyzed by mass spectrometry.
3 Results
3.1 TCF4 expression in esophageal cancer cell lines
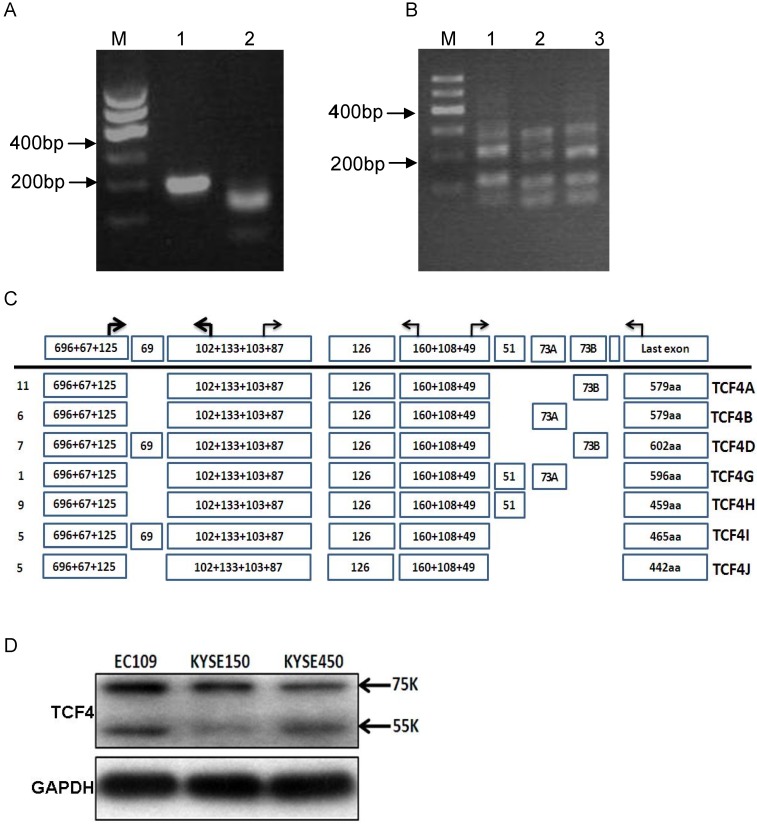
TCF4N isoform, which can function as a dominant negative factor in Wnt signaling, was reported to be existed in mouse tissues except embryonic stem cells 18. However, it was not clear whether TCF4N variants also occur in human cell types. We first analyzed the TCF4N variant ESCC cells. Its existence was assessed by using specific primer located in exon 7(P2F) and putative exon 8(P2R). As expected, TCF4N variant was found in mouse liver and spleen. Similarly, RT-PCR product was also detected in EC109 cell, and was further confirmed by digestion with AluI. Moreover, sequencing analysis showed that TCF4N variant was expressed in human ESCC cells (Fig. 1A and Supplementary Material: Fig. S1A). Then, we determined the status of exon 4 in the transcript variants. As shown in Fig.1B, PCR products with the corresponding size were detected in both cell lines, which were further confirmed by subcloning and sequencing (Data not shown). However, the relative proportions of transcripts containing or lacking exon 4 varied (Fig. S1B). Next, we detected the transcripts generated by alternative splicing of exon 13-16. PCR products were much complex by using primer pairs located in exons 12 and 17 (Fig. 1B). More than 3 types of products were detected, with different abundance. The main products were further confirmed by cloning and sequencing.
Figure 1.
TCF4 expression in esophageal cancer cell line EC109. (A) Identification of TCF4N isoform in EC109 cells. Total RNA was isolated from EC109 cells and RT-PCR was performed as described in Materials and Methods. Lane1: RT-PCR product; lane2: RT-PCR product digested with AluI. (B) RT-PCR analysis the splicing of exon 13~16 with primer P3F and P3R in EC109 (lane 1), KESE150 (lane 2) and KYSE450 (lane 3). (C) Transcript variants spanning exon 3 and exons 17 in EC109 cells were identified by RT-PCR and subsequent cloning and sequencing. Numbers of clones were list before each isoform variant. (D) TCF4 protein expression was detected by Western blotting in esophageal cancer cell lines.
To exactly identify all the transcript variants spanning exon 3 to exons 17 in EC109 cells, primer pair located in exon 3(P1F) and exon 17(P3R) was applied to amplify the main body of all the transcript variants, which generate a chief product (Fig.S1C). Then the product was cloned and sequenced. We finally got the sequences of 44 clones. Each variant and its relative abundance were list in Fig.1C. Interestingly, exon 14 and 15 were mutually exclusive, and exon 16 was not detected in any clone.
To determine the protein expression of TCF4 in esophageal cancer cell line, total protein from cell line EC109, KYSE150 and KYSE450 was detected by Western blotting with specific antibody, which recognized the N-terminal of TCF4. As shown in Fig. 1D, two main bands were found, with molecular weight of about 75 and 55Ku, which matched TCF4E isoforms and TCF4M respectively.
3.2 TCF4 expression in human esophageal cancer
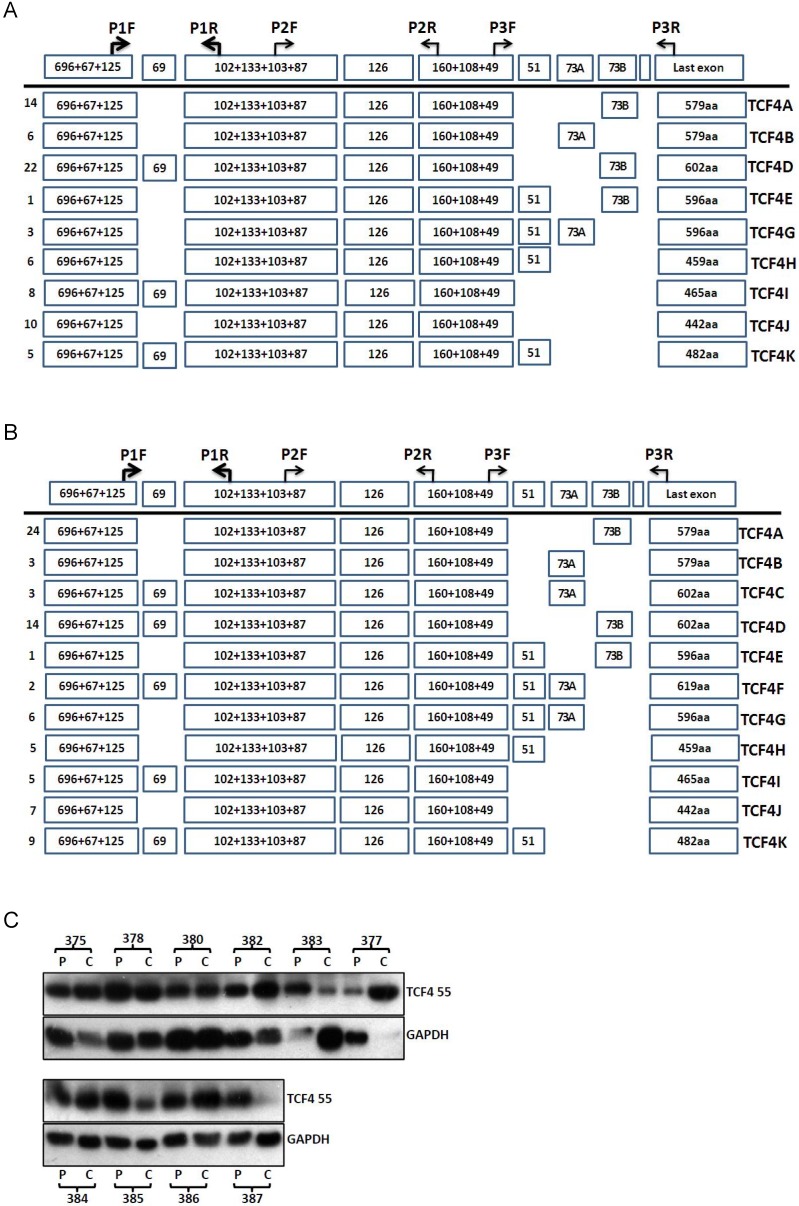
Then, we detected the transcript variants of TCF4 in pooled samples of esophageal cancer tissues and the adjacent normal tissues. As same as that in esophageal cancer cell line, TCF4N isoform was detected in these two group samples. Compared with that of adjacent normal tissues, the expression level of TCF4N was downregulated in esophageal cancer tissues (Supplementary Material: Fig.S2). As for exon 4, it was alternative spliced both in cancer and normal tissues. Again, the PCR products from cancer and normal tissues sample by amplifying with primers P1F and P3R were cloned and sequenced. Totally 75 and 79 clones were verified from cancer and adjacent tissues. These identified isoforms showed some characters similar with that observed in cancer cell line. As shown in Fig.2, the most frequent isoform in both groups encoded TCF4E isoform.
Figure 2.
TCF4 expression in esophageal cancer and adjacent tissues. Total RNA was isolated from tissues and RT-PCR with primer set of P1F and P3R was performed as described in Materials and Methods. The PCR products from cancer tissues (A) and adjacent tissues (B) were subcloned and individual clones was further sequenced to identify all the transcript variants spanning exon 3 to exons 17. Numbers of clones were list before each isoform variant. Total proteins were isolated from esophageal cancer and adjacent tissues and further detected by Western blotting (C). P: adjacent tissues, C: cancer tissues.
The protein level of TCF4 was also detected by westen blotting (Fig.2C). Although many splicing isoforms were detected at mRNA level, only TCF4M (about 55ku) was detected at protein level in both groups. The protein isoforms did not match with that of mRNA isoform, and the reason of this discrepancy was not clear. To understand the role of TCF4 in esophageal cancer, we compared the protein expression of TCF4 between cancer tissues and its matched adjacent tissues. Among ten pairs of tissues, the protein level of TCF4 in cancer tissues was increased in five pairs of tissues (5/10), and decreased in 3 pairs (3/10). This increase suggested a role of TCF4 in esophageal tumorigenesis.
3.3 Inhibition of TCF4E expression in EC109 cells
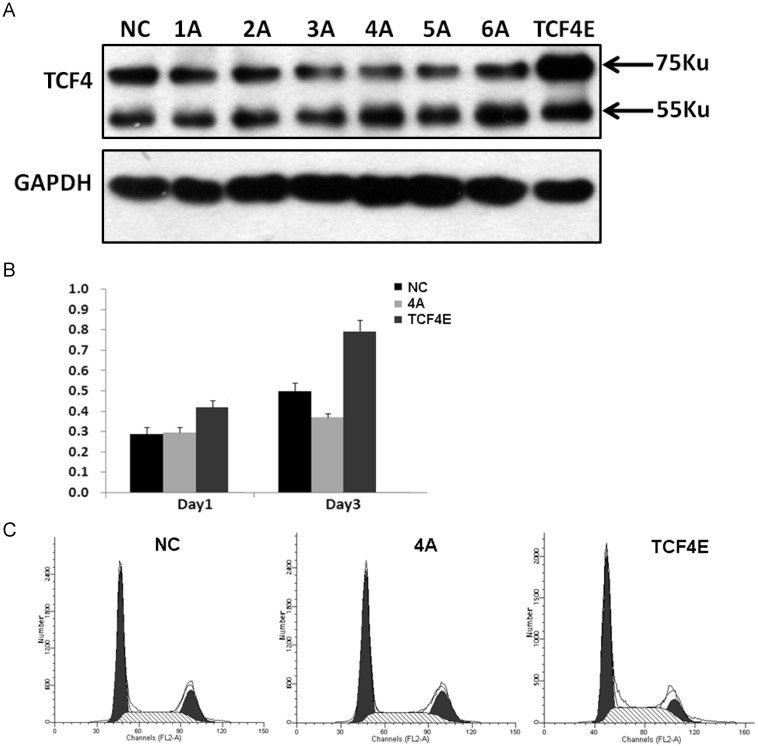
Although various types of TCF splicing isoforms were found in EC109 cells, its exact biological function of each individual remains unknown, especially for those in-frame alternative spliced exons. At protein level, TCF4E was the most frequent isoform in EC109 cell as detected by western blotting. This isoform contains most functional domains such as DNA binding domain, β-catenin binding domain and CTBP. In order to determine its effects on cell growth in EC109 cells, gene expression of TCF4E knockdown by RNAi with different target site. As shown in Fig.3A, gene expression of TCF4E isoform was significantly inhibited, while TCF4M isoform was not affected. Then, its effect on cell growth was determined by CCK-8, which showed that cell growth was inhibited as TCF4E isoform was inhibited. Simultaneously, cell cycle distribution was also altered by TCF4E knockdown, with decreased S-phase and increased G2 phase (Fig.3C and Supplementary Material: Fig.S3). These data suggested TCF4E isoform promote cell growth in EC109 cells.
Figure 3.
Funtional analysis of TCF4E variant in EC109 cells. Knockdown of TCF4E isoform in EC109 cell by siRNA (1A to 6A, and negative control) was confirmed by Western blotting. Overexpression of TCF4E was performed by transfection of TCF4E construct into EC109 cells (A). Cell growth (B) and cell cycle distribution (C) of EC109 cells was determined by CCK-8 and FACS as TCF4E expression was altered. NC: negative control.
3.4 Over-expression of TCF4M isoform does not alter its transcription activity
The most important role of TCF4 was to function as a transcription factor. It binds to its target promoters with its DNA bingding domain and activates its transcription. Compared with TCF4E isoform, TCF4M lacks CtBP domain which can co-operate with other regulation proteins. As TOPflash reporter contains TCF4 binding site, it was extensively applied to detect TCF4 transcription activity. Transient expression of TCF4M plasmid was confirmed western blotting (Supplementary Material: Fig.S4). Then its effect on transcription activity was determined by TOPflash reporter. As shown if Fig.4A, augment of transcription activity was not observed, although the gene expression of TCF4M isoform was greatly increased. It was reported that β-catenin is needed to activate TCF4 activity. Constitutively active β-catenin expressing vector was co-transfected into EC109 cells with TCF4M. However, β-catenin did not significantly change the luciferase expression. Moreover, other canonical Wnt targets such as MMP7 and cyclinD1 were not altered by TCF4M (Fig.4B). This suggested other regulative proteins, which were lacked in EC109 cells, are needed to activate β-catenin/TCF4 pathway.
Figure 4.
Funtional analysis of TCF4M overexpression in EC109 cells. TCF4M construct and TOPflash reporter were co-transfected into EC109 cells, combined with or without β-catenin expression vector. The transcription activity was determined by relative luciferase activity (A). Total RNA was isolated for realtime PCR to detect the gene expression of CyclinD1 and MMP7 (B). N: empty vector.
3.5 Proteins interact with TCF4 protein in EC109 cells
As a transcription factor, TCF4 binds to its target promoter. Its chromatin occupancy was revealed by chromatin-immunoprecipitation combined with massively DNA sequencing in several studies 20-22. To further understand the physiological functions of TCF4, we employed immunoprecipitation followed by mass spectrometry to identify the binding proteins. The immunoprecipitated samples were subjected to mass spectrometry and retrieved to identify the candidate proteins. Sixteen proteins were identified (Table 3). Among these proteins, several histone components were found to potentially interact with TCF4, which was coincident with the findings of its chromatin occupancy. Other most frequent identified proteins are some kinases including serine/threonine-protein kinase and myosin light chain kinase. These proteins need further verification to convince their interacting with TCF4.
Table 3.
TCF4 interacting proteins identified by mass spectrometry
| Protein symbol | mRNA | Description |
|---|---|---|
| ACTA_HUMAN | NM_001141945.1 | Actin, aortic smooth muscle |
| ACTB_HUMAN | NM_001101.3 | Actin, cytoplasmic 1 |
| ACTBL_HUMAN | NM_001017992.3 | Beta-actin-like protein 2 |
| ALBU_HUMAN | NM_000477.5 | Serum albumin |
| ARF4_HUMAN | NM_001660.3 | ADP-ribosylation factor 4 |
| ARHG1_HUMAN | NM_004706.3 | Rho guanine nucleotide exchange factor 1 |
| COQ6_HUMAN | NM_182476.2 | Ubiquinone biosynthesis monooxygenase COQ6 |
| CU129_HUMAN | ND | Uncharacterized protein C21orf129 |
| H2A1B_HUMAN | NM_003513.2 | Histone H2A type 1-B/E |
| H2B1B_HUMAN | NM_021062.2 | Histone H2B type 1-B |
| H4_HUMAN | NM_175054.2 | Histone H4 |
| MOS_HUMAN | NM_005372.1 | Proto-oncogene serine/threonine-protein kinase mos |
| MRCKG_HUMAN | NM_017525.2 | Serine/threonine-protein kinase MRCK gamma |
| MYLK2_HUMAN | NM_033118.3 | Myosin light chain kinase 2 |
| NEK10_HUMAN | NM_199347.2 | Serine/threonine-protein kinase |
| XPO2_HUMAN | NM_001256135.1 | Exportin-2 |
4 Discussion
Wnt/β-catenin signaling pathway has been recognized to control a variety of functions and properties in various types of cells. One of its main effects in a given cell is to change the gene expression at the transcriptional level. Many target genes of this pathway are subject to accurate control. The downstream molecules of Wnt/β-catenin signaling pathway include various types of transcription coactivators such as TCF/LEF, cAMP response element-binding protein (CREB)-binding protein/E1A binding protein p300 (CBP/p300), and Pygopus 2, which transduce Wnt/β-catenin signals to activate downstream target genes, thereby initiating transcription of multiple factors that promote cell growth 23.
Previous studies showed that human TCF4 gene was alternative spliced in some types of cancers including colon cancer, hepatocellular carcinoma, renal carcinoma and brain tumor. AS of this gene was also found in other species such as mouse and zebrafish 24,25. Moreover, its splicing was found to be associated with type 2 diabetes 26-28. However, there is no report on ESCC regarding the AS of TCF4 and its significance in cell proliferation. In this study, we analyzed the TCF4 isoforms both in vivo and in vitro. Compared with the TCF4 splicing isoforms in other studies, we have identified 11 different TCF4 isoforms by sequencing code region generated from complete mRNA clones derived ESCC cancer and its adjacent tissues. Our results showed some differences with other studies. First, exon 16 was not included in any splicing isoform, while it was identified in other types of cancer 14. Exon 16', which was described in brain tumours 15, was not found in our samples. Second, exon 14 and 15 were mutually exclusive. However, these two exons, which both contain 73bp, could co-exist in a same isoform. These results suggested a tumor-specific splicing of TCF4 in human cells. Moreover, we convinced the existence of TCF4N variant in human cells, as this isoform has only been reported in mouse but not in human cells. This isoform consists of the N-terminal part of TCF4 including the β-catenin binding domain. As it lacks the HMG-box, TCF4N inhibits the activation of β-catenin-dependent promoter. We found the TCF4N expression was downregulated in esophageal cancer tissues, compared with that of adjacent tissues. As Wnt/β-catenin was activated in esophageal cancer, downregulation of TCF4N might contribute to tumorigenesis.
According to recent nomenclature of TCF4 variants, the protein products are classified as 3 groups: TCF4E, TCF4M and TCF4S 24. Transcripts which code for TCF4 protein isoform containing a complete C-clamp and the two CtBP-binding motifs, represent the E-type similar to the TCF1E variant, and transcripts which give rise to TCF4 protein with a partial C-clamp form the type 'S' group, while splice products coding for TCF4 lacking all of the C-clamp constitute the M-type. The protein products encoded by the splicing variants in this study are grouped as TCF4E and TCF4M, which was coincident with the result of Western blotting in EC109 cells (Fig.1). However, only TCF4M was detected in human tissue samples. The reasons remain unknown. Maybe the TCF4 isoforms are translated with different efficiency, which suggested a post-transciptional regulation of TCF4 isoforms.
The TCF4 protein can act as either transcriptional activators or repressors according to its association with other proteins. Transcriptional activation is driven partially by β-catenin which directly binding with TCF4 through its N-ternimal domains, while the repression is mediated by co-repressors such as Groucho in the absence of nuclear β-catenin 29. Therefore, the transcriptional activity of TCF4 is much dependent on its interacting proteins. By using MS to detect the TCF4 immunoprecipitation from EC109 cells, 16 proteins were preliminarily identified. As TCF4 located in specific DNA motifs, several histone components were identified from the TCF4 immunoprecipitation. This might suggest that TCF4 binds partial via histones. Other potential binding proteins are some kinases including serine/threonine-protein kinase and myosin light chain kinase. MOS is a serine/threonine kinase that activates the MAP kinase cascade through direct phosphorylation of the MAP kinase activator MEK, while NEK10 mediates G2/M cell cycle arrest and MEK autoactivation 30,31. The functions of these kinases are related to cell cycle control. However, these proteins need to be further convinced.
In summary, we revealed the alterative splicing isoforms of TCF4 gene both in human esophageal cancers and cell lines. The gene functions of splicing isoform TCF4E and TCF4M were analyzed in EC109 cells, and TCF4 potential binding proteins were preliminarily identified by MS. Our data suggested that deregulation of TCF4 isoform may contribute to the tumorigenesis of ESCC.
Supplementary Material
Table S1, Figures S1 - S4.
Table 1.
sequence of TCF4 primers
| Primer Name | Sequence | Product (bp) |
|---|---|---|
| P1F | AAGAAGCGGCCAAGAGGCAAGAT | 299/368 |
| P1R | GTACGTGATAAGAGGCGTGAGGG | |
| P2F | ATCGCCTGGCACCGTAGGACAAA | 424 |
| P2R | CCGCCCAAGGATCTGGTTGATGG | |
| P3F | GCGGGATAACTATGGAAAGAAGAA | 95/146/168/219 |
| P3R | CCTTGTATGTAGCGAACGCACTT | |
| TCF4NF | CCCCGTATTACCCACTATCGC | 179 |
| TCF4NR | GGCCCCGGTTCTTGGAACTCA |
Acknowledgments
This work was supported by a grant from the Chongqing Natural Science Foundation (CSTC2011BB5022).
Author contributions
BZ: conceived and designed the experiments. GH, XG, XC, YW, CL: performed the experiments. BZ, YW: analyzed the data. BZ, GH: drafted the manuscript. All authors approved the final version of the manuscript.
Abbreviations
- AS
alternative splicing
- ESCC
esophageal squamous cell carcinoma
- CREB
cAMP response element-binding protein
- GSK 3β
glycogen synthase kinase 3β
- HMG
high mobilty group
- LEF1
lymphoid enhancer-binding factor
- MS
mass spectrometry
- TCFs
T cell factors.
References
- 1.Thallinger CM, Kiesewetter B, Raderer M, Hejna M. Pre- and postoperative treatment modalities for esophageal squamous cell carcinoma. Anticancer Res. 2012;32:4609–27. [PubMed] [Google Scholar]
- 2.Lin Y, Totsuka Y, He Y. et al. Epidemiology of esophageal cancer in Japan and China. J Epidemiol. 2013;23:233–42. doi: 10.2188/jea.JE20120162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Denlinger CE, Thompson RK. Molecular basis of esophageal cancer development and progression. Surg Clin North Am. 2012;92:1089–103. doi: 10.1016/j.suc.2012.07.002. [DOI] [PubMed] [Google Scholar]
- 4.Zhang Y, Goss AM, Cohen ED. et al. A Gata6-Wnt pathway required for epithelial stem cell development and airway regeneration. Nat Genet. 2008;40:862–70. doi: 10.1038/ng.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Katoh M, Katoh M. WNT signaling pathway and stem cell signaling network. Clin Cancer Res. 2007;13:4042–5. doi: 10.1158/1078-0432.CCR-06-2316. [DOI] [PubMed] [Google Scholar]
- 6.Zhang G, Zhou X, Xue L. et al. Accumulation of cytoplasmic beta-catenin correlates with reduced expression of E-cadherin, but not with phosphorylated Akt in esophageal squamous cell carcinoma: immunohistochemical study. Pathol Int. 2005;55:310–7. doi: 10.1111/j.1440-1827.2005.01840.x. [DOI] [PubMed] [Google Scholar]
- 7.Lv J, Cao XF, Ji L. et al. Association of β-catenin, Wnt1, Smad4, Hoxa9, and Bmi-1 with the prognosis of esophageal squamous cell carcinoma. Med Oncol. 2012;29:151–60. doi: 10.1007/s12032-010-9816-5. [DOI] [PubMed] [Google Scholar]
- 8.Wang Y, Liu S, Zhu H. et al. FRAT1 overexpression leads to aberrant activation of beta-catenin/TCF pathway in esophageal squamous cell carcinoma. Int J Cancer. 2008;123:561–8. doi: 10.1002/ijc.23600. [DOI] [PubMed] [Google Scholar]
- 9.Yan S, Zhou C, Zhang W. et al. beta-Catenin/TCF pathway upregulates STAT3 expression in human esophageal squamous cell carcinoma. Cancer Lett. 2008;271:85–97. doi: 10.1016/j.canlet.2008.05.035. [DOI] [PubMed] [Google Scholar]
- 10.Wang Y, Zhou X, Zhu H. et al. Overexpression of EB1 in human esophageal squamous cell carcinoma (ESCC) may promote cellular growth by activating beta-catenin/TCF pathway. Oncogene. 2005;24:6637–45. doi: 10.1038/sj.onc.1208819. [DOI] [PubMed] [Google Scholar]
- 11.Valkenburg KC, Graveel CR, Zylstra-Diegel CR. et al. Wnt/β-catenin Signaling in Normal and Cancer Stem Cells. Cancers (Basel) 2011;3:2050–79. doi: 10.3390/cancers3022050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou C, Liu S, Zhou X. et al. Overexpression of human pituitary tumor transforming gene (hPTTG), is regulated by beta-catenin /TCF pathway in human esophageal squamous cell carcinoma. Int J Cancer. 2005;113:891–8. doi: 10.1002/ijc.20642. [DOI] [PubMed] [Google Scholar]
- 13.Deng YZ, Chen PP, Wang Y. et al. Connective tissue growth factor is overexpressed in esophageal squamous cell carcinoma and promotes tumorigenicity through beta-catenin-T-cell factor/Lef signaling. J Biol Chem. 2007;282:36571–81. doi: 10.1074/jbc.M704141200. [DOI] [PubMed] [Google Scholar]
- 14.Shiina H, Igawa M, Breault J. et al. The human T-cell factor-4 gene splicing isoforms, Wnt signal pathway, and apoptosis in renal cell carcinoma. Clin Cancer Res. 2003;9:2121–32. [PubMed] [Google Scholar]
- 15.Howng SL, Huang FH, Hwang SL. et al. Differential expression and splicing isoform analysis of human Tcf-4 transcription factor in brain tumors. Int J Oncol. 2004;25:1685–92. [PubMed] [Google Scholar]
- 16.Tsedensodnom O, Koga H, Rosenberg SA. et al. Identification of T-cell factor-4 isoforms that contribute to the malignant phenotype of hepatocellular carcinoma cells. Exp Cell Res. 2011;317:920–31. doi: 10.1016/j.yexcr.2011.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duval A, Rolland S, Tubacher E. et al. The human T-cell transcription factor-4 gene: structure, extensive characterization of alternative splicings, and mutational analysis in colorectal cancer cell lines. Cancer Res. 2000;60:3872–9. [PubMed] [Google Scholar]
- 18.Kennell JA, O'Leary EE, Gummow BM. et al. T-cell factor 4N (TCF-4N), a novel isoform of mouse TCF-4, synergizes with beta-catenin to coactivate C/EBPalpha and steroidogenic factor 1 transcription factors. Mol Cell Biol. 2003;23:5366–75. doi: 10.1128/MCB.23.15.5366-5375.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu Y, He G, Wang Y. et al. MCM-2 is a therapeutic target of Trichostatin A in colon cancer cells. Toxicol Lett. 2013;221:23–30. doi: 10.1016/j.toxlet.2013.05.643. [DOI] [PubMed] [Google Scholar]
- 20.Hatzis P, van der Flier LG, van Driel MA. et al. Genome-wide pattern of TCF7L2/TCF4 chromatin occupancy in colorectal cancer cells. Mol Cell Biol. 2008;28:2732–44. doi: 10.1128/MCB.02175-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yochum GS, McWeeney S, Rajaraman V. et al. Serial analysis of chromatin occupancy identifies beta-catenin target genes in colorectal carcinoma cells. Proc Natl Acad Sci U S A. 2007;104:3324–9. doi: 10.1073/pnas.0611576104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gougelet A, Torre C, Veber P. et al. T-cell factor 4 and β-catenin chromatin occupancies pattern zonal liver metabolism in mice. Hepatology. 2014;59:2344–57. doi: 10.1002/hep.26924. [DOI] [PubMed] [Google Scholar]
- 23.Ring A, Kim YM, Kahn M. Wnt/Catenin signaling in adult stem cell physiology and disease. Stem Cell Rev. 2014;10:512–25. doi: 10.1007/s12015-014-9515-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.AS Weise A, Bruser K, Elfert S. et al. Alternative splicing of Tcf7l2 transcripts generates protein variants with differential promoter-binding and transcriptional activation properties at Wnt/beta-catenin targets. Nucleic Acids Res. 2010;38:1964–81. doi: 10.1093/nar/gkp1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Young RM, Reyes AE, Allende ML. Expression and splice variant analysis of the zebrafish tcf4 transcription factor. Mech Dev. 2002;117:269–73. doi: 10.1016/s0925-4773(02)00180-6. [DOI] [PubMed] [Google Scholar]
- 26.Le Bacquer O, Shu L, Marchand M. et al. TCF7L2 splice variants have distinct effects on beta-cell turnover and function. Hum Mol Genet. 2011;20:1906–15. doi: 10.1093/hmg/ddr072. [DOI] [PubMed] [Google Scholar]
- 27.Hansson O, Zhou Y, Renström E. et al. Molecular function of TCF7L2: Consequences of TCF7L2 splicing for molecular function and risk for type 2 diabetes. Curr Diab Rep. 2010;10:444–51. doi: 10.1007/s11892-010-0149-8. [DOI] [PubMed] [Google Scholar]
- 28.Prokunina-Olsson L, Kaplan LM, Schadt EE. et al. Alternative splicing of TCF7L2 gene in omental and subcutaneous adipose tissue and risk of type 2 diabetes. PLoS One. 2009;4:e7231. doi: 10.1371/journal.pone.0007231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cuilliere-Dartigues P, El-Bchiri J, Krimi A. et al. TCF-4 isoforms absent in TCF-4 mutated MSI-H colorectal cancer cells colocalize with nuclear CtBP and repress TCF-4-mediated transcription. Oncogene. 2006;25:4441–8. doi: 10.1038/sj.onc.1209471. [DOI] [PubMed] [Google Scholar]
- 30.Moniz LS, Stambolic V. Nek10 mediates G2/M cell cycle arrest and MEK autoactivation in response to UV irradiation. Mol Cell Biol. 2011;31:30–42. doi: 10.1128/MCB.00648-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kalejs M, Ivanov A, Plakhins G. et al. Upregulation of meiosis-specific genes in lymphoma cell lines following genotoxic insult and induction of mitotic catastrophe. BMC Cancer. 2006;6:6. doi: 10.1186/1471-2407-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1, Figures S1 - S4.