Abstract
GRHL2 was implicated in regulating cancer development. Our previous study demonstrated that knockdown GRHL2 in colorectal cancer (CRC) cells inhibited cell proliferation by targeting ZEB1. It is unclear whether GRHL2 expression may have diagnostic or prognostic value in colorectal carcinoma. Additionally, how GRHL2 is associated with the clinical features of colorectal carcinoma is not known. In current study, immunohistochemistry stains were performed to examine GRHL2 in 171 colorectal cancers and paired normal colon mucosa. The prognostic value of GRHL2 was investigated in a retrospective cohort study with a five-year follow-up. The effects of GRHL2 on cell growth in vitro and in vivo were explored by GRHL2 over-expressing in HT29 and SW620 CRC cells. Further, the regulation of cell cycle and proliferation proteins by GRHL2 were assessed by flow cytometry and western blot. We found that GRHL2 was over-expressed in CRC tissues, and played an important role in CRC tumorigenesis. GRHL2 expression positively correlated with tumor size and TNM stage. Kaplan-Meier analysis showed that GRHL2 was an independent prognostic factor for both overall survival and recurrence-free survival. Ectopic over-expression of GRHL2 in CRC cell line HT29 and SW620 induced an increase of cellular proliferation in vitro and promoting tumor growth in vivo. The acquisition of GRHL2 regulated cell cycle and modulates the expression of proliferation proteins p21, p27, cyclin A and cyclin D1. Together, our findings reveal GRHL2 can be used as a novel predictive biomarker and represent a potential therapeutic target against CRC.
Keywords: GRHL2, Colorectal cancer, Prognosis, Proliferation, Cell cycle
Introduction
Colorectal cancer (CRC) is one of the most common cancers worldwide in terms of both incidence and mortality 1. The diagnosis and treatment of CRC have been improved, although, despite extensive efforts, the efficacy of surgery and chemotherapy remains unsatisfactory. Most diagnosed CRC patients present with middle or late-stage cancer. Therefore, new diagnostic and treatment strategies are urgently needed. CRC development and progression consists of mutational events in oncogenes and tumor suppressor genes 2. Thus, investigating genes associated with CRC development is crucial.
Grainyhead-like 2 (GRHL2) (also known as BOM or TFCP2L3), encoding a 325 amino acid protein, belongs to GRHL transcription factor family 3, which comprises GRHL1, GRHL2 and GRHL3 4. Grainyhead family genes have been shown to play an important role in wound healing, epidermal integrity and embryonic neural tube closure 5-7. Particularly, GRHL2 is implicated in regulating structural and functional differentiation of epithelial tissues 8-10, and controlling many physiological functions of human airway epithelium 11, 12. With regard to cancer, GRHL2 gene amplification has been noted in several tumors, including hepatocellular carcinoma 13, human oral squamous cell carcinoma 14, gastric cancer 15, breast cancer 16, 17. As such, GRHL2 has been proposed as a candidate oncogene. On the other hand, evidences have been reported for suppressing activities of GRHL2 transcription factor in tumorigenesis. Previous studies have shown that there was a tumor suppressive role of GRHL2 in gastric cancer 18. Additionally, GRHL2 could suppress EMT and restore sensitivity to anoikis in breast cancer 19. GRHL3, another member of grainyhead subfamily, was recently demonstrated to suppress squamous cell carcinoma, due to its activation of PTEN expression 20, and modulation of the miR-21 target MSH2 by RNA-binding protein DND1 21. These controversial results suggested that the role of GRHL2 was possibly tumor specific and highly dependent on its targets in different cancer cells. However, the function and precise molecular mechanism of GRHL2 in CRC were unclear and required elucidation.
In our previous study, we performed lentivirus-mediated shRNA transfection to knock-down GRHL2 gene expression in CRC cells, and we found that down-regulation of GRHL2 inhibited the proliferation of CRC cells by targeting ZEB122. However, how GRHL2 is associated with the clinical features of colorectal carcinoma is not known. In the current investigation, we examined the expression of GRHL2 in CRC tissue and its effects on cellular proliferation. Further, a retrospective cohort study with a five-year follow-up was performed to investigate the relativity between GRHL2 expression and prognosis in CRC. Our results demonstrated GRHL2 is a potent driver of CRC, as well as a prognostic marker to predict poor outcome in human CRC. In fact, GRHL2 over-expression promoted proliferation of CRC cells in vitro and enhanced the tumorigenic potential in vivo. These studies may provide new avenues of research with potential clinical implications.
Materials and methods
Patients and clinical specimens
CRC tumor specimens and paired non-tumor mucosa were collected from December 2006 to July 2008. Patients with the following criteria were excluded from participation: had received adjuvant chemotherapy or radiotherapy prior to surgery; had additional cancers diagnoses. All patients were classified according to the 7th edition of the TNM staging system 23. Postoperative adjuvant therapies were performed, according to standard schedules and doses. Patients were followed-up annually by telephone or at outpatient clinic till July, 2013 or dead. Overall survival was defined as the time elapsed from surgery to the time of patient death. Patients' death information was obtained from their family. All participating patients gave their written informed consent. This study was approved by the Ethical Committee of Shanghai Pudong Hospital.
Immunohistochemical staining
Immunohistochemistry (IHC) stains were performed using formalin-fixed, paraffin-embedded tissue sections of tissue blocks, according to the well-established protocols 24. The primary antibody used was the rabbit monoclonal anti-GRHL2 antibody (HPA004820, Sigma, St. Louis, MO, USA) at 1: 300 dilutions. Negative controls were stained with IgG as primary antibody. We scored the GRHL2 IHC staining based on the proportion of cell staining and the staining intensity. The percentage of positive cells was divided into five grades (percentage scores) : <10 % (0), 10-25 % (1), 25-50 % (2), 50-75 % (3) and>75 % (4). The intensity of staining was divided into four grades (intensity scores): no staining (0), weak staining (1), moderate staining (2) and strong staining (3). GRHL2 staining positivity was determined by the following formula: overall score = percentage score × intensity score. The overall score ≤ 3 was defined as negative, and > 3 was defined as positive. GRHL2 staining was scored by two independent researchers, blinded to the clinical characteristics of the patients.
Cell lines and cell culture
Human colorectal cancer cells HCT116, SW116, LoVo, SW480 and Caco2 were preserved in our institute. These cells were grown in RPMI-1640(Corning Cellgro®, Manassas, VA, USA) medium containing 10 % fetal bovine serum (FBS, Corning Cellgro®). HT29 and SW620 cells were obtained from the American Type Culture Collection (ATCC), grown in McCoy's 5A (for HT29) and Leibovitz's L-15 medium (for SW620) supplemented with 10% (v/v) FBS (Corning Cellgro®), penicillin and streptomycin (GIBCO BRL, Gaithersburg, MD, USA). All the cells were cultured at 37℃ in a humidified atmosphere of 5% CO2.
Generation of stable cell lines by lentiviral transduction
For GRHL2 over-expression, human GRHL2 full length cDNA was amplified, introduced into the LV5 vector, and cloned into the NotI/BamHI sites of the V2251-2 plasmid (GenePharma, Shanghai, China). HT29 and SW620 cells infected with lentiviral vectors expressing GRHL2 or empty vector were performed for 12 h in the presence of 5μg/ml polybrene (Santa Cruz, Dallas, Texas, USA). Infected cells were selected for more than 2 weeks by incubation with puromycin (5μg/ml for HT29, 3μg/ml for SW620), followed by qRT-PCR and western blot analysis to verify GRHL2 expression. Stable transfectant cells were maintained in medium containing 1 µg/ml puromycin.
Quantitative real-time PCR
Total RNA extracts were prepared from cell cultures using Trizol (Invitrogen, Grand Island, NY, USA) according to the manufacturer's protocol. cDNA was amplified using a reverse transcriptional kit (Promega, Madison, WI, USA). The qRT-PCR was carried out using cDNA as a template and Universal PCR Master Mix (Applied Biosystems, Carlsbad, CA, USA) on an Applied Biosystems 7900HT sequence detection system (Applied Biosystems, Foster City, CA, USA). Primers used for qRT-PCR analysis were as follows: GRHL2: GGAAATCTAGCCCTGGGTTTG (forward) and TCAGGGAGGAACGCACTGA (reverse). GAPDH: AAGGTGAA-GGTCGGAGTCAAC (forward) and GGGGTCATTGATGGCAACAATA (reverse). The relative amount of mRNA was normalized using GAPDH as endogenous control.
Western blot
Whole cellular or tissue proteins were extracted with RIPA lysis buffer (Solarbio, Beijing, China) containing 0.2 mM phenylmethylsulfonyl fluoride (PMSF), according to standard methods. Protein concentrations were determined by the Bradford assay (Bio-Rad Laboratories, Hercules, CA, USA). Western blot was performed via established protocols 25. Primary antibodies used in this study were as follows: GRHL2 (1:300, Cat. HPA004820, Sigma); cyclin A (1:500, Cat. sc-596), cyclin D1 (1:500, Cat. sc-753), p21 (1:500, Cat. sc-397), p27 (1:500, Cat. sc-528) were from Santa Cruz biotechnology. The secondary antibodies used were horseradish peroxidase (HRP)-conjugated anti-rabbit (1:1000, Cat. 7074) and anti-mouse (1:1000, Cat. 7076) from Cell Signaling Technology; Primary antibody against GAPDH (1:5000, Cat. sc-365062, Santa Cruz, USA) was used as a loading control.
Cell proliferation assay
Cell proliferation assays were performed using a Cell Counting Kit-8 (CCK-8; cat.CK04, DOJINDO, Kumamoto, Japan) following the manufacturer's protocol. Parental, Vector and GRHL2+ cells were seeded in a 96-well plate (3000 cells per well) and cultured for 24, 48, 72, 96 and 120 h. Then, 10 μl of the CCK-8 solution was added to each well of the plate and incubated for another 3 hours. The absorbance at 450 nm was measured with a microplate reader.
Colony formation assay
Parental, Vector and GRHL2+ cells (1000 cells per well) were plated at low densities in six-well plates and incubated for 14 days with the medium replaced every 3 days. Cells were washed twice with PBS, fixed and then stained with 1% crystal violet staining solution. The number of colonies containing more than 50 cells was counted under a microscope.
Flow cytometry
Vector and GRHL2+ cells were fixed in 70% ice-cold ethanol overnight at 4℃, and then incubated with 100 ug/mL RNase at 37℃ for 20 min. After staining with propidium iodide (50 ug/mL, Roche Applied Science, Basel, Switzerland), the cells were subjected to fluorescence activated cell sorting (FACS) on a FACScan (Beckman Instruments, Fullerton, CA, USA). The cell populations were quantified using the Modfit software (Becton-Dickinson).
Experimental in vivo animal model
Male BALB/c nu/nu nude mice (Institute of Zoology Chinese Academy of Sciences), 4-week-old, were housed at a specific pathogen-free environment in the Animal Laboratory Unit. Mice (6 mice per group) were subcutaneously implanted with Vector and GRHL2+ cells (1 × 106 in 0.1 ml of sterilized PBS buffer). Tumor growth were determined by measuring the size (mm3, V = (Width2 × Length) / 2) every 4 days using calipers 26. The mice were euthanized after 32 days post-inoculation, and tumor were excised for analyzing. Mice received humane care and the study protocols comply with the Institution's guidelines and animal research laws.
Statistical Analysis
IBM SPSS Statistical software (version 19.0) was utilized for statistical analysis. Correlations between GRHL2 expression in CRC tissues and clinicopathological parameters were analyzed using the Pearson's Chi-square (χ2) test. Overall survival (OS) and recurrence-free survival (RFS) were evaluated with the Kaplan-Meier method and differences were compared by log-rank test. Cox's proportional hazards modeling of factors potentially related to survival were conducted to calculate hazard ratios (HR). Differences were compared using a two-tailed Student's t test. All P-values were determined from 2-tailed tests and differences with a P-value<0.05 were considered to be statistically significant.
Results
GRHL2 expression was increased in CRC tissues comparing to the paired non-tumor tissues and varied in CRC cell lines
To gain insights into the role of GRHL2 in colorectal tumorigenesis, we examined its expression in primary CRC and paired normal mucosal tissues by IHC staining. Representative staining results were shown in Figure 1. IHC staining indicated that GRHL2 was predominantly presented in the nucleus in CRC cells and normal cells in the paired normal colorectal mucosa. Among 171 cases, 106(62.0%) were positive for GRHL2 expression (Fig. 1D-F), while 65 (38.0%) were negative for GRHL2 expression (Fig. 1C), only 74/171 (43.2%) matched normal mucosa were detected with positive GRHL2 expression (Fig. 1B). Collectively, GRHL2 levels were significantly higher in CRC tissues than non-tumor tissues (P=0.001, Table 1). This indicated that GRHL2 might facilitate tumorigenesis in colon.
Figure 1.
GRHL2 expression was increased in colorectal cancer (CRC) tissues and varied in CRC cell lines. (A) GRHL2-negative normal mucosa epithelium adjacent to carcinoma. (B) Nuclear staining of GRHL2 in normal epithelium adjacent to cancer. (C) GRHL2-negative cancer tissue. (D) Moderate positive nuclear stain for GRHL2 in cancer tissue. (E-F) Strong nuclear staining for GRHL2 in CRC tissue. The scale bar indicates 100 um. Original Magnification: × 200 (A-D, F), × 100 (E). (G) The expression of GRHL2 gene transcripts in a panel of human CRC cell lines were detected by qRT-PCR. The relative amount of mRNA was normalized using GAPDH as endogenous control. (H) GRHL2 protein levels were detected by western blot, with GAPDH as control.
Table 1.
Expression of GRHL2 in colorectal cancer tissues and matched non-tumor mucosa
| Total | GRHL2(-) | GRHL2(+) | P value | |
|---|---|---|---|---|
| tumor specimens | 171 | 65 (38%) | 106 (62%) | 0.001* |
| non-tumor mucosa | 171 | 97 (57%) | 74 (43%) |
*Statistically significant
To investigate whether there was heterogeneity in the expression of GRHL2 in different CRC cell lines, qRT-PCR (Fig. 1G) and western blot (Fig. 1H) were performed to assess GRHL2 gene transcripts and proteins levels. GRHL2 expression was varied in seven different CRC cell lines. Five of which showed a high expression, including HCT116, LoVo, HT29, SW480 and Caco2 cells. Low level expression of GRHL2 was found in SW620 and SW1116 cells.
GRHL2 expression correlated with different clinicopathologic parameters
Considering the level of GRHL2 in CRC tissues was significantly higher than that in non-tumor tissues, the correlations between GRHL2 expression and clinicopathologic parameters in CRC were analyzed by Pearson's Chi-square. We subdivided the CRC cases in two groups according to GRHL2 levels (negative and positive). Correlation analysis demonstrated that the positive expression of GRHL2 was correlated with the large tumor size, later clinical stage in CRCs (p<0.05), but was not significantly correlated to gender, tumor location, cell differentiation (Table 2). GRHL2 level was higher in advanced tumor stages III+IV than tumor stages I+II (69.2% vs. 53.8%, p =0.037). Larger tumor size was found with higher GRHL2 expression (67.9% vs. 51.6%, p =0.035). Our data revealed that GRHL2 was highly expressed in the more aggressive tumors and had a correlation with the well-established prognostic evaluation factors. Hence, GRHL2 could be used as a potential unfavorable prognostic biomarker for CRC.
Table 2.
Correlation of GRHL2 with different clinicopathologic parameters
| Parameters | GRHL2 expression | ||||
|---|---|---|---|---|---|
| All cases | Negative | Positive | P value | ||
| Age | <60 | 77 | 30(39.0%) | 47(61.0%) | 0.974 |
| ≧60 | 94 | 35(37.2%) | 59(62.8%) | ||
| Gender | Male | 97 | 40(41.2%) | 57(58.8%) | 0.323 |
| Female | 74 | 25(33.8%) | 49(66.2%) | ||
| Histology | Tubular | 136 | 57(41.9%) | 79(58.1%) | 0.060 |
| Mucinous | 28 | 7 (25.0%) | 21(75.0%) | ||
| Papillary | 7 | 1 (14.3%) | 6 (85.7%) | ||
| Site | Rectum & sigmoid | 112 | 38(33.9%) | 74(66.1%) | 0.053 |
| Right colon | 40 | 21(52.5%) | 19(47.5%) | ||
| Left colon | 19 | 6(31.6%) | 13(68.4%) | ||
| Size status | <3cm | 62 | 30(48.4%) | 32(51.6%) | 0.035* |
| ≧3cm | 109 | 35(32.1%) | 74(67.9%) | ||
| Stage status | I+ II | 80 | 37(46.3%) | 43(53.7%) | 0.037* |
| III+IV | 91 | 28(30.8%) | 63(69.2%) | ||
*Statistically significant
GRHL2 expression was associated with poor clinical outcomes
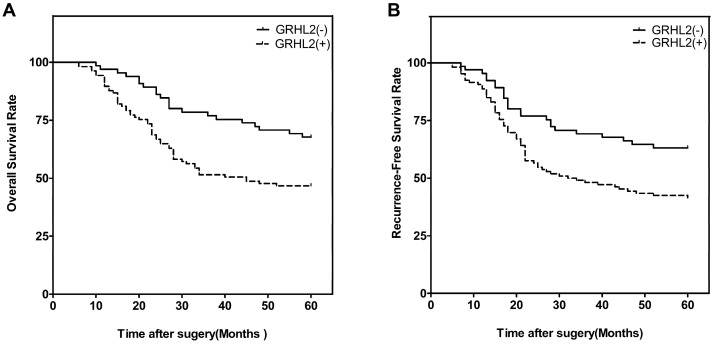
We next investigated the correlation of GRHL2 expression with patient survival. At the end of the follow-up period (60 months), 93 of the 171 patients were alive, 78 patients were dead. Patients were then divided into two groups: GRHL2 (+) and GRHL2 (-). A Kaplan-Meier survival analysis revealed that GRHL2 up-regulation was associated with a lower OS rate (p=0.003, Fig. 2A) and RFS (p=0.006, Fig. 2B). Further multivariate Cox regression analysis determined GRHL2 positive-expression was an independent prognostic factor for the poor OS (HR=0.545, p=0.018, Table 3) and lower RFS (HR=0.601, p=0.036, Table 3) in CRC patients. Thus, GRHL2 could be a potential biomarker indicating poor outcome in CRC patient.
Figure 2.
GRHL2 expression was associated with poor clinical outcomes. (A) Kaplan-Meier survival analysis of OS of CRCs according to the expression level of GRHL2 protein. GRHL2 up-regulation was associated with a lower OS, compared with GRHL2-negative CRC patients (p=0.003, log-rank test). (B) Kaplan-Meier survival analysis of RFS of CRC patients according to the expression level of GRHL2. Positive expression were associated with a shorter RFS (p=0.006, log-rank test).
Table 3.
Univariate and multivariate analysis of clinicopathlogical parameters with overall survival (OS ) and recurrence-free survival(RFS) by Cox proportional hazards regression.
| Factors | Univeriate | Multivariate | ||||
|---|---|---|---|---|---|---|
| HR | 95%CI | P value | HR | 95%CI | P value | |
| OS | ||||||
| Age | 0.993 | 0.972-1.015 | 0.525 | |||
| Gender | 0.903 | 0.575-1.418 | 0.658 | |||
| Histology | 1.020 | 0.760-1.370 | 0.894 | |||
|
Site Size |
1.151 1.036 |
0.878-1.508 0.654-1.640 |
0.309 0.880 |
|||
| Clinical stage | 1.792 | 1.415-2.270 | 0.000* | 1.728 | 1.365-2.187 | 0.000* |
| GRHL2 | 0.481 | 0.291-0.793 | 0.004* | 0.545 | 0.329-0.902 | 0.018* |
| RFS | ||||||
| Age | 1.000 | 0.980-1.021 | 0.995 | |||
| Gender | 0.934 | 0.608-1.433 | 0.753 | |||
| Histology | 0.927 | 0.706-1.217 | 0.585 | |||
|
Site Size |
1.185 1.031 |
0.912-1.539 0.663-1.601 |
0.205 0.894 |
|||
| Clinical stage | 1.936 | 1.530-2.450 | 0.000* | 1.879 | 1.484-2.379 | 0.000* |
| GRHL2 | 0.522 | 0.326-0.837 | 0.007* | 0.601 | 0.374-0.967 | 0.036* |
*Statistically significant
GRHL2 promoted cell proliferation of CRC cell lines in vitro
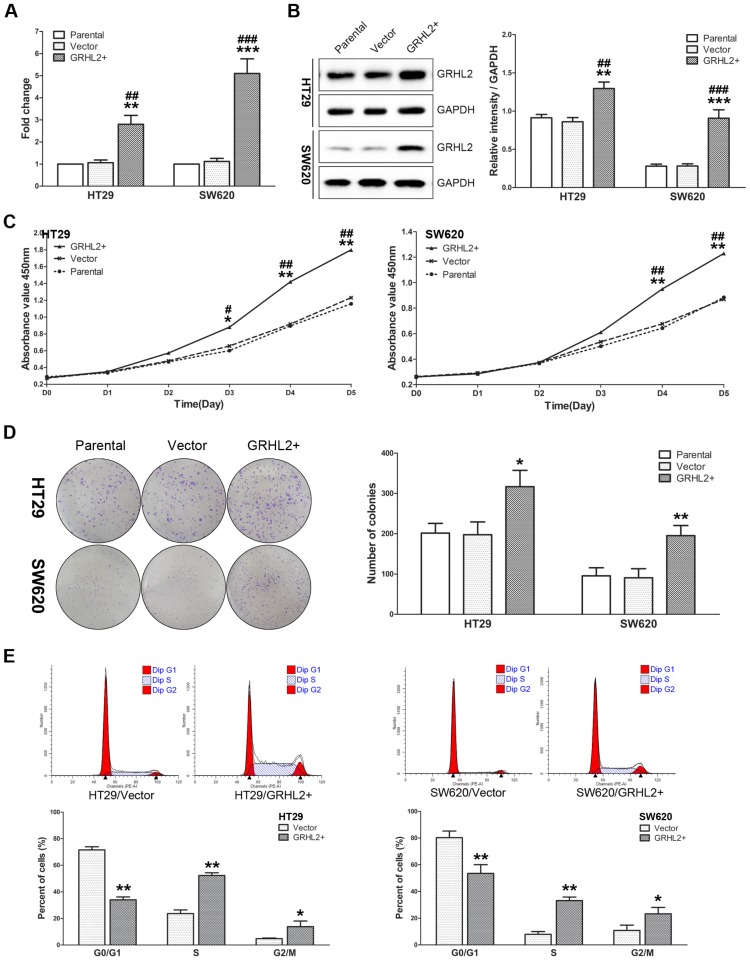
Given that GRHL2 was positively correlated with tumor stage in CRC clinical specimens, we sought to further investigate its influence on cell proliferation in vitro. We utilized human CRC cells HT29 and SW620 to establish the model stably over-expressing exogenous human GRHL2 by lentivirus-mediated experiments. Both qRT-PCR (Fig. 3A) and western blot analysis (Fig. 3B) demonstrated that HT29/GRHL2+ cells (p<0.01) and SW620/GRHL2+ (p<0.001) had a significant increase of GRHL2 expression relative to their empty vector transfected control and parental control cells.
Figure 3.
Over-expression of GRHL2 promoted proliferation of CRC cell lines in vitro. (A) A qRT-PCR analysis of GRHL2 gene expression revealed a striking up-regulation of exogenous GRHL2 expression in HT29 (HT29/GRHL2+) and SW620 (SW620/GRHL2+) cells compared to Parental and Vector cells. (B) In HT29/GRHL+ and SW620/GRHL2 cells, there were a marked increase in the expression of GRHL2, verified by western blot. Immunoblotting results are typical blots from 3 experiments. Densitometric analysis was expressed relative to the loading control, GAPDH. (C) Effect of GRHL2 on cellular proliferation was measured by CCK-8 assay. HT29/GRHL+ and SW620/GRHL2 cells showed higher proliferation than control cells. (D) GRHL2 over-expression resulted in a significant increase in colony formation in GRHL2+ cell. (E) Representative charts for cell-cycle distribution in Vector and GRHL2 cells. Results are mean ± SD (3-5 experiments). *Relative to Vector control; #relative to Parental control. * p<0.05, ** p<0.01, *** p<0.001, # p<0.05, # p<0.01, ### p<0.001.
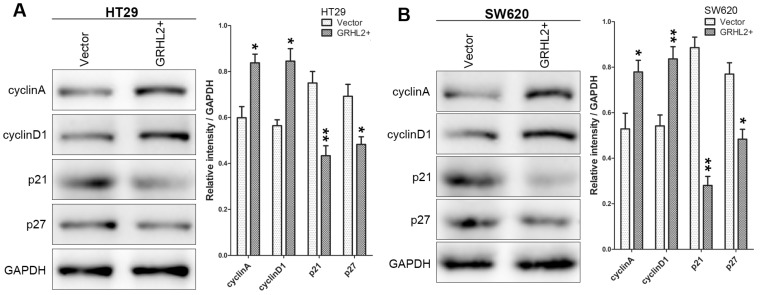
To assess the effects of GRHL2 on cell growth, CCK-8 assays were performed. Cells transfected with GRHL2 demonstrated a significant increase in proliferation rate relative to vector control or parental cells. A significant higher proliferation rate was observed in HT29/GRHL2+ and SW620/GRHL2+ cells (Fig. 3C). GRHL2 over-expression resulted in a significant increase in colony formation in HT29 and SW620 cells (Fig. 3D). We then examined cell-cycle distribution in GRHL2-over-expressed cells. The percentage of cells in S phase and G2/M phase was increased in HT29/GRHL2+ and SW620/GRHL2+ cells, while reduced in G1 phase comparing to vector control cells (Fig. 3E). This indicated that the growth-promoted effect of GRHL2 may partly due to the alteration of cell cycle distribution. Furthermore, we assessed the proliferation markers (such as cyclin A, cyclin D1, p21, p27) expression by western blot analysis. Consistently, over-expressing GRHL2 resulted in a significant increase of the expression of cyclin A and cyclin D1, while attenuating p21 and p27 levels (Fig. 4A&B). Collectively, GRHL2 expression promoted cellular proliferation and tumor growth in vitro.
Figure 4.
Over-expression of GRHL2 modulated the expression of proliferation marker proteins. The proliferation marker proteins were assessed by western blot analysis. Over-expression GRHL2 in HT29 (A) and SW620 (B) cells significant increased the expression of cyclin A and cyclin D1, while decreasing the levels of p21 and p27. Immunoblotting results are typical from 3 experiments and the densitometry is mean ± SD (3-5 experiments) *Relative to Vector control * p<0.05, ** p<0.01.
GRHL2 increased the tumorigenic potential of CRC cell line in vivo
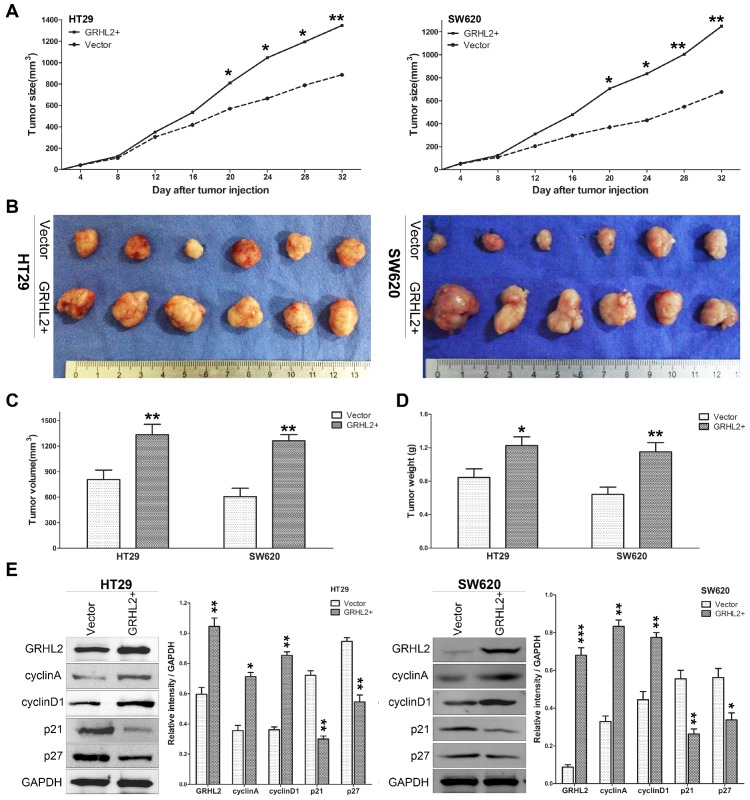
Since GRHL2 plays an oncogenic role in vitro, it was very important to assess its effect in vivo. Studies were performed utilizing the xenograft model by injecting Vector and GRHL2 cells subcutaneously into the nude mice, respectively. As demonstrated in Figure 5, over-expression of GRHL2 in HT29 and SW620 cells promoted tumor growth (Fig. 5A), compared to the Vector cells. Notably, the average volume and weight of tumors excised after 32 days post-inoculation in GRHL2+ groups were markedly larger and heavier than those in Vector groups (Fig. 4B-D). Furthermore, we also assessed the proliferation markers expression in mice tumor masses by western blot analysis. Similar to the observation in vitro, GRHL2+ groups had an increase in the expression of cyclin A and cyclin D1, while a decrease in the levels of p21 and p27 (Fig. 5E). Altogether, these data clearly demonstrated that GRHL2 expression promoted tumor growth in vivo.
Figure 5.
Over-expression of GRHL2 increased the tumorigenic potential of CRC cell lines in vivo. (A) Tumor growth curves demonstrated over-expression of GRHL2 promoted tumor growth. Tumor sizes were examined every 4 days after implantation of Vector cells and GRHL2+ cells. (B) The tumor masses originated from GRHL+ cells were larger than tumors from Vector cells. (C) As compared with Vector controls, the average tumor volume of the GRHL2+ group was markedly larger. (D) The average weight of primary tumors originated from GRHL2 groups was heavier than that from Vector groups. (E) GRHL2over-expression significantly increased the expression of the proliferation markers cyclin A and cyclin D1, while decreasing the levels of p21 and p27. Each data point represents the mean ± SD (3-5 experiments). Densitometric analysis is expressed relative to the loading control, GAPDH. *Relative to Vector control * p<0.05, ** p<0.01, *** p<0.001.
Discussion
The precise molecular mechanisms underlying CRC tumorigenesis is crucial, as it can help us to develop novel therapeutic strategies for CRC patients 27. In recent studies, GRHL2 aroused great interest and was selected as a candidate gene since it has been reported to be involved in several malignant tumors 13-17. GRHL2 is a transcriptional regulator of proliferation and differentiation in epithelial cells, both during development and tumorigenesis. Although GRHL2 was demonstrated to act as an oncogene in several tumors, its role in CRC remains unclear. Herein, we examined GRHL2 expression in CRC and paired normal colorectal epithelium, and found that GRHL2 was over-expressed in CRC. We gave the first evidence that GRHL2 plays a pivotal role in CRC tumorigenesis. The characteristics observed in patients with CRC were recapitulated in cells and mouse xenograft models. Over-expressing GRHL2 in HT29 and SW620 cells induced an increase in cellular proliferation, and promoting tumor growth in vivo. To date, however, evidence has been reported for tumor-suppressing activities of GRHL2 transcription factor in tumorigenesis 18. These conflicting data suggests that GRHL2 may have dual functions depending on the cell background and cancer type 28.
The abilities to predict outcome and to identify key players in biological mechanisms that lead to poor outcome are two important objectives in cancer research. In this study, we included 171 CRC specimens with various TNM stages (I-IV) to investigate the clinical potential of GRHL2 alone as an independent prognostic factor. Our study demonstrated that GRHL2 expression positively correlated with large tumor size, later TNM stage. Intriguingly, Kaplan-Meier analysis showed that CRC patients with GRHL2-positive tumors had significantly worse prognosis in both OS and RFS. In agreement with our study, using a large public microarray database, Xiang 16 found that higher expression levels of GRHL2 was significantly correlated with a shorter relapse-free survival interval. In hepatocellular carcinoma, GRHL2 was demonstrated to be an independent predictive marker for HCC recurrence 13. Thus, our study further confirmed that GRHL2 could be used as an unfavorable prognostic biomarker for CRC patients.
The deregulation of cell proliferation is a prerequisite for carcinogenesis and cancer progression 29. The achievement of the proliferation follows from the activation/inactivation of signaling pathways regulating this process 30. In the context of cancer, this occurs owing to both the activation of oncogenes and the inactivation of tumor suppressors regulating proliferation 31. The association between proliferation and poor prognosis has been established 32. Previous studies suggest that GRHL2 is involved in cell proliferation. GRHL2 knocking-down markedly repressed hepatoma cells growth 13. Loss of GRHL2 induced a striking reduction in cell proliferation in breast cancer 28. Importantly, GRHL2 appears to be a critical factor for hTERT expression and telomerase activity, GRHL2 over-expression prevented the senescence-associated loss of telomerase activity and enhanced cellular proliferation in oral carcinoma 14. Also, GRHL2 was shown to significantly promote tumor growth and metastasis in breast cancer 16. In the current study, we demonstrated that over-expressing GRHL2 promotes cell cycle progression, as documented by the significant enrichment of the cell population in the S and G2/M phase. Consistently, modulation of GRHL2 levels resulted in significant changes in the expression of the proliferation marker proteins in vitro and in vivo.
In conclusion, we identified GRHL2, as an oncogene, was frequently over-expressed in CRC tissues and cell lines. In fact, high GRHL2 level correlates with poorer prognosis in CRC. GRHL2 over-expression was demonstrated to promote CRC progression both in vitro and in vivo. Furthermore, GRHL2 could regulate cell cycle and modulate cell cycle proteins. In fact, our current results indicate that GRHL2 can be used as a novel predictive biomarker and potential therapeutic target for CRC.
Acknowledgments
We thank Yan Shen for help of performing the mouse xenografts experiment. This work was supported by the Pudong Hospital Funds for Distinguished Young Scientists (201407Y).
References
- 1.Siegel R, Desantis C, Jemal A. Colorectal cancer statistics, 2014. CA: a cancer journal for clinicians. 2014;64(2):104–117. doi: 10.3322/caac.21220. [DOI] [PubMed] [Google Scholar]
- 2.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61(5):759–767. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- 3.Wilanowski T, Tuckfield A, Cerruti L, O'Connell S, Saint R, Parekh V, Tao J, Cunningham JM, Jane SM. A highly conserved novel family of mammalian developmental transcription factors related to Drosophila grainyhead. Mechanisms of development. 2002;114(1-2):37–50. doi: 10.1016/s0925-4773(02)00046-1. [DOI] [PubMed] [Google Scholar]
- 4.Auden A, Caddy J, Wilanowski T, Ting SB, Cunningham JM, Jane SM. Spatial and temporal expression of the Grainyhead-like transcription factor family during murine development. Gene expression patterns: GEP. 2006;6(8):964–970. doi: 10.1016/j.modgep.2006.03.011. [DOI] [PubMed] [Google Scholar]
- 5.Narasimha M, Uv A, Krejci A, Brown NH, Bray SJ. Grainy head promotes expression of septate junction proteins and influences epithelial morphogenesis. Journal of cell science. 2008;121(Pt 6):747–752. doi: 10.1242/jcs.019422. [DOI] [PubMed] [Google Scholar]
- 6.Ting SB, Wilanowski T, Cerruti L, Zhao LL, Cunningham JM, Jane SM. The identification and characterization of human Sister-of-Mammalian Grainyhead (SOM) expands the grainyhead-like family of developmental transcription factors. The Biochemical journal. 2003;370(Pt 3):953–962. doi: 10.1042/BJ20021476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pyrgaki C, Liu A, Niswander L. Grainyhead-like 2 regulates neural tube closure and adhesion molecule expression during neural fold fusion. Developmental biology. 2011;353(1):38–49. doi: 10.1016/j.ydbio.2011.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boglev Y, Wilanowski T, Caddy J, Parekh V, Auden A, Darido C, Hislop NR, Cangkrama M, Ting SB, Jane SM. The unique and cooperative roles of the Grainy head-like transcription factors in epidermal development reflect unexpected target gene specificity. Developmental biology. 2011;349(2):512–522. doi: 10.1016/j.ydbio.2010.11.011. [DOI] [PubMed] [Google Scholar]
- 9.Werth M, Walentin K, Aue A, Schonheit J, Wuebken A, Pode-Shakked N, Vilianovitch L, Erdmann B, Dekel B, Bader M. et al. The transcription factor grainyhead-like 2 regulates the molecular composition of the epithelial apical junctional complex. Development. 2010;137(22):3835–3845. doi: 10.1242/dev.055483. [DOI] [PubMed] [Google Scholar]
- 10.Senga K, Mostov KE, Mitaka T, Miyajima A, Tanimizu N. Grainyhead-like 2 regulates epithelial morphogenesis by establishing functional tight junctions through the organization of a molecular network among claudin3, claudin4, and Rab25. Molecular biology of the cell. 2012;23(15):2845–2855. doi: 10.1091/mbc.E12-02-0097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gao X, Vockley CM, Pauli F, Newberry KM, Xue Y, Randell SH, Reddy TE, Hogan BL. Evidence for multiple roles for grainyheadlike 2 in the establishment and maintenance of human mucociliary airway epithelium. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(23):9356–9361. doi: 10.1073/pnas.1307589110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Varma S, Cao Y, Tagne JB, Lakshminarayanan M, Li J, Friedman TB, Morell RJ, Warburton D, Kotton DN, Ramirez MI. The transcription factors Grainyhead-like 2 and NK2-homeobox 1 form a regulatory loop that coordinates lung epithelial cell morphogenesis and differentiation. The Journal of biological chemistry. 2012;287(44):37282–37295. doi: 10.1074/jbc.M112.408401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tanaka Y, Kanai F, Tada M, Tateishi R, Sanada M, Nannya Y, Ohta M, Asaoka Y, Seto M, Shiina S. et al. Gain of GRHL2 is associated with early recurrence of hepatocellular carcinoma. Journal of hepatology. 2008;49(5):746–757. doi: 10.1016/j.jhep.2008.06.019. [DOI] [PubMed] [Google Scholar]
- 14.Kang X, Chen W, Kim RH, Kang MK, Park NH. Regulation of the hTERT promoter activity by MSH2, the hnRNPs K and D, and GRHL2 in human oral squamous cell carcinoma cells. Oncogene. 2009;28(4):565–574. doi: 10.1038/onc.2008.404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheng L, Wang P, Yang S, Yang Y, Zhang Q, Zhang W, Xiao H, Gao H, Zhang Q. Identification of genes with a correlation between copy number and expression in gastric cancer. BMC medical genomics. 2012;5:14. doi: 10.1186/1755-8794-5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xiang X, Deng Z, Zhuang X, Ju S, Mu J, Jiang H, Zhang L, Yan J, Miller D, Zhang HG. Correction: grhl2 determines the epithelial phenotype of breast cancers and promotes tumor progression. PloS one. 2013. 8(4) [DOI] [PMC free article] [PubMed]
- 17.Yang X, Vasudevan P, Parekh V, Penev A, Cunningham JM. Bridging cancer biology with the clinic: relative expression of a GRHL2-mediated gene-set pair predicts breast cancer metastasis. PloS one. 2013;8(2):e56195. doi: 10.1371/journal.pone.0056195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xiang J, Fu X, Ran W, Chen X, Hang Z, Mao H, Wang Z. Expression and role of grainyhead-like 2 in gastric cancer. Medical oncology. 2013;30(4):714. doi: 10.1007/s12032-013-0714-5. [DOI] [PubMed] [Google Scholar]
- 19.Cieply B, Riley Pt, Pifer PM, Widmeyer J, Addison JB, Ivanov AV, Denvir J, Frisch SM. Suppression of the epithelial-mesenchymal transition by Grainyhead-like-2. Cancer research. 2012;72(9):2440–2453. doi: 10.1158/0008-5472.CAN-11-4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Darido C, Georgy SR, Wilanowski T, Dworkin S, Auden A, Zhao Q, Rank G, Srivastava S, Finlay MJ, Papenfuss AT. et al. Targeting of the tumor suppressor GRHL3 by a miR-21-dependent proto-oncogenic network results in PTEN loss and tumorigenesis. Cancer cell. 2011;20(5):635–648. doi: 10.1016/j.ccr.2011.10.014. [DOI] [PubMed] [Google Scholar]
- 21.Bhandari A, Gordon W, Dizon D, Hopkin AS, Gordon E, Yu Z, Andersen B. The Grainyhead transcription factor Grhl3/Get1 suppresses miR-21 expression and tumorigenesis in skin: modulation of the miR-21 target MSH2 by RNA-binding protein DND1. Oncogene. 2013;32(12):1497–1507. doi: 10.1038/onc.2012.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Quan Y, Jin R, Huang A, Zhao H, Feng B, Zang L, Zheng M. Downregulation of GRHL2 inhibits the proliferation of colorectal cancer cells by targeting ZEB1. Cancer biology & therapy. 2014;15(7):878–887. doi: 10.4161/cbt.28877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Edge SB, Compton CC. The American Joint Committee on Cancer: the 7th edition of the AJCC cancer staging manual and the future of TNM. Annals of surgical oncology. 2010;17(6):1471–1474. doi: 10.1245/s10434-010-0985-4. [DOI] [PubMed] [Google Scholar]
- 24.Yu W, Li Q, Yao Y, Quan Y, Zhang Y. Two Novel 30K Proteins Overexpressed in Baculovirus System and Their Antiapoptotic Effect in Insect and Mammalian Cells. International journal of genomics. 2013;2013:323592. doi: 10.1155/2013/323592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kovacevic Z, Fu D, Richardson DR. The iron-regulated metastasis suppressor, Ndrg-1: identification of novel molecular targets. Biochimica et biophysica acta. 2008;1783(10):1981–1992. doi: 10.1016/j.bbamcr.2008.05.016. [DOI] [PubMed] [Google Scholar]
- 26.Jacob D, Davis J, Fang B. Xenograftic tumor models in mice for cancer research, a technical review. Gene Ther Mol Biol. 2004;8:213–219. [Google Scholar]
- 27.Colussi D, Brandi G, Bazzoli F, Ricciardiello L. Molecular pathways involved in colorectal cancer: implications for disease behavior and prevention. International journal of molecular sciences. 2013;14(8):16365–16385. doi: 10.3390/ijms140816365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Werner S, Frey S, Riethdorf S, Schulze C, Alawi M, Kling L, Vafaizadeh V, Sauter G, Terracciano L, Schumacher U. et al. Dual roles of the transcription factor grainyhead-like 2 (GRHL2) in breast cancer. The Journal of biological chemistry. 2013;288(32):22993–23008. doi: 10.1074/jbc.M113.456293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 30.Duronio RJ, Xiong Y. Signaling pathways that control cell proliferation. Cold Spring Harbor perspectives in biology. 2013;5(3):a008904. doi: 10.1101/cshperspect.a008904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alkarain A, Slingerland J. Deregulation of p27 by oncogenic signaling and its prognostic significance in breast cancer. Breast cancer research: BCR. 2004;6(1):13–21. doi: 10.1186/bcr722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Starmans MH, Krishnapuram B, Steck H, Horlings H, Nuyten DS, van de Vijver MJ, Seigneuric R, Buffa FM, Harris AL, Wouters BG. et al. Robust prognostic value of a knowledge-based proliferation signature across large patient microarray studies spanning different cancer types. British journal of cancer. 2008;99(11):1884–1890. doi: 10.1038/sj.bjc.6604746. [DOI] [PMC free article] [PubMed] [Google Scholar]