Abstract
Activating RAS mutations are common in human tumors. These mutations are often markers for resistance to therapy and subsequent poor prognosis. So far, targeting the RAF-MEK-ERK and PI3K-AKT signaling pathways downstream of RAS is the only promising approach in the treatment of cancer patients harboring RAS mutations. RAL GTPase, another downstream effector of RAS, is also considered as a therapeutic option for the treatment of RAS-mutant cancers. The RAL GTPase family comprises RALA and RALB, which can have either divergent or similar functions in different tumor models. Recent studies on non-small cell lung cancer (NSCLC) have showed that different RAS mutations selectively activate specific effector pathways. This observation requires broader validation in other tumor tissue types, but if true, will provide a new approach to the treatment of RAS-mutant cancer patients by targeting specific downstream RAS effectors according to the type of RAS mutation. It also suggests that RAL GTPase inhibition will be an important treatment strategy for tumors harboring RAS glycine to cysteine (G12C) or glycien to valine (G12V) mutations, which are commonly found in NSCLC and pancreatic cancer.
Keywords: RAS, RAL, RAS mutation, RALBP1, non-small cell lung cancer
RAS GTPase overview
RAS is the most extensively studied GTPase1. It is known to regulate various cellular functions, including proliferation, survival, growth, migration, differentiation and cytoskeletal dynamics2,3,4 (Figure 1). Not coincidentally, increased activities of all of these processes are required by tumor cells to promote tumor growth. Activating oncogenic RAS mutations lead to treatment resistance in various tumor models and poor patient outcomes3,5,6,7,8,9. Three human RAS genes have been identified: HRAS, KRAS, and NRAS1,2. These encode the related proteins HRAS, KRAS and NRAS, respectively, of 189 amino acids in length1,4. KRAS exists as two isoforms, 4A and 4B, which are generated by alternative exon splicing10. These different RAS genes are well known to have differential cellular specificities and intrinsic transforming potentials3,4,10.
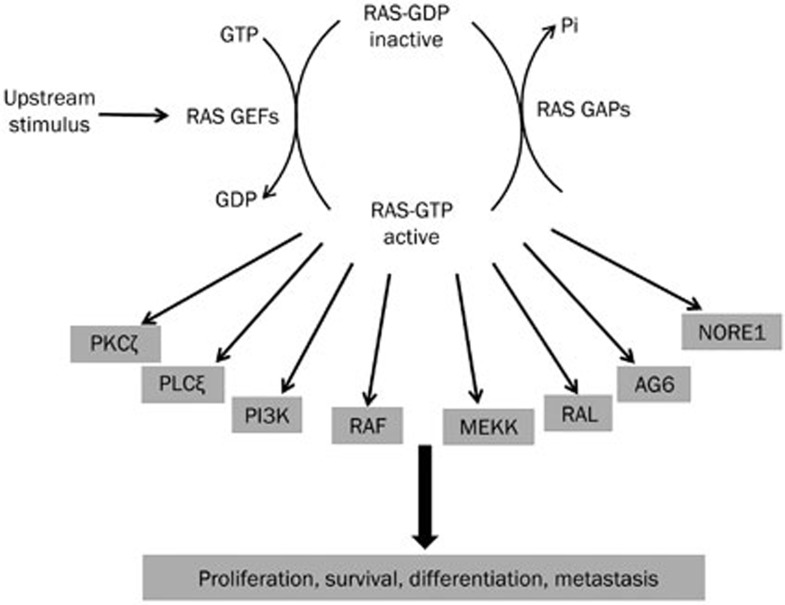
Figure 1.
RAS GTPase activation and downstream signaling. The cartoon shows the mechanism of RAS GTPase activation by upstream stimuli and numerous downstream effectors stimulated by activated RAS. RAS regulates critical cellular functions through these effectors. Constitutive RAS activation due to mutations leads to protumorigenic signaling through these effectors.
In normal cells, RAS proteins act as molecular switches for critical cellular functions. RAS proteins are activated by upstream receptors such as receptor tyrosine kinases (RTKs), G-protein coupled receptors (GPCRs) and serpentine and cytokine receptors1,4,11. RAS protein activity is tightly regulated by Guanine Nucleotide Exchange Factors (GEFs) and GTPase Activating Proteins (GAPs). Constitutive activation of the RAS protein, in which RAS is unable to hydrolyze GTP, leads to cancer and other diseases1,9,12. Activating RAS mutations have been observed in 30% of human tumors2,13; KRAS is the most commonly mutated isoform, mutated in 25%–30% of human cancers3,13. KRAS mutations are dominant in colorectal cancer (40%–45%), non-small cell lung cancer (NSCLC) (15%–40%) and pancreatic carcinoma (69%–95%)2,3 and are associated with poor patient outcomes and treatment responses in these malignancies2,3. NRAS and HRAS mutations are less common (8% and 3% of all tumors, respectively)2,13, with melanoma harboring the most NRAS mutations at 20%–30%2,13.
Activating mutations occur at codons 12, 13, and 61, which are within the GTPase catalytic domain, in all three RAS isoforms14. Approximately 80% of KRAS mutations are found in codon 12, whereas approximately 60% of NRAS mutations are found in codon 61, with 35% in codon 122,14. HRAS mutations are equally divided between codons 12 and 612,14. All of these activating mutations inhibit RAS GTPase activity by preventing GAP-stimulated GTP hydrolysis of GTP-bound activated RAS.
Approximately 20 downstream effectors bind to RAS-GTP and trigger signaling. RAS drives tumor growth via a number of prominent pathways, including the following: RAF-MEK-ERK15,16; p110 catalytic subunits (p110α, β, γ, and δ) of class I PI3K; TIAM1, a small RAC GTPase-specific GEF; RAL-specific GEFs (RALGDS, RGL, RGL2, and RGL3); and phospholipase C epsilon1,2,16.
RAS mutation and cancer therapeutics
Devising an effective treatment strategy for patients with RAS mutations has been a major challenge2,17. However, recent attempts have been promising17,18. RNA interference is an interesting approach but has many technical hurdles, including the lack of an efficient delivery system, poor uptake and low gene silencing efficiency2. The inhibition of RAS membrane localization via the inhibition of RAS farnesylation has been investigated, but this approach has failed to materialize into a therapeutic strategy due to several limitations, including toxicity, and mainly the appearance of a compensatory mechanism via geranylgeranylation2,8,19.
So far, the only approach that has shown promise in treating cancer patients with RAS mutations is the targeting of its downstream signaling cascades such as RAF-MEK-ERK and PI3K-AKT2,8,16,19. Targeting these two pathways either separately or together is beneficial in preventing in vitro and in vivo progression of tumors harboring a RAS mutation2. Currently, clinical trials are being conducted to study the therapeutic effects of MEK and PI3K inhibitors in cancer patients harboring RAS mutations2. However, a wrinkle in this approach has appeared. Recent studies have shown that different KRAS mutations preferentially activate different downstream signaling pathways. For example, mutant KRAS with either a glycine to cysteine (G12C) or glycine to valine (G12V) mutation at codon 12 preferentially binds to RAL guanine nucleotide dissociation stimulator (RALGDS), a RAL GTPase-specific GEF, whereas KRAS harboring a glycine to aspartate mutation at codon 12 (G12D) has higher affinity for phosphatidylinositol 3-kinase (PI3K)20. These recent studies have brought to light the need to clarify the impact of such KRAS mutations on the RAL GTPase signaling pathway.
The RAL GTPase family and effectors
RAL GTPase falls under the RAS family of GTPases. RAL shares a high degree of sequence similarity with the three RAS genes, hence the name RAL (RAS-like)1. The RAL GTPase sub-family comprises the two isoforms RALA and RALB, which share high sequence homology21. Approximately 85% of the amino acid sequences of these two isoforms are identical21. RAL GTPase can be activated by six GEFs (RALGEFs), RALGDS, RGL, RGL2/Rlf, RGL3, RALGPS1, and RALGPS2, and inactivated by two GAPs, RALGAP1, and RALGAP222,23. Four RALGEFs (RALGDS, RGL, RGL2, and RGL3) are known to directly interact with the effector binding region of GTP-bound RAS and are thus important for RAS-mediated tumorigenesis22. RALGEFs and RAL play a dominant role in the RAS-mediated transformation of several different immortalized human cell lines, as well as in a RAS-driven tumor model24,25.
RAL proteins mediate various cellular activities, including filopodia formation/membrane ruffling, glycolysis, autophagy, secretion, the maintenance of polarity, apoptosis and transcription21,26 (Figure 2). Alterations to these activities can lead to tumor invasion, metastasis, altered cellular energy levels, proliferative signaling and resistance to cell death. These activities are mediated by effectors that interact with activated (GTP-bound) RAL22. Of all the RAL effectors, the most extensively studied are RALBP1 and the members of the exocyst complex Sec5 and Exo8426. Other RAL effectors include Filamin, PLD1 and ZONAB26.
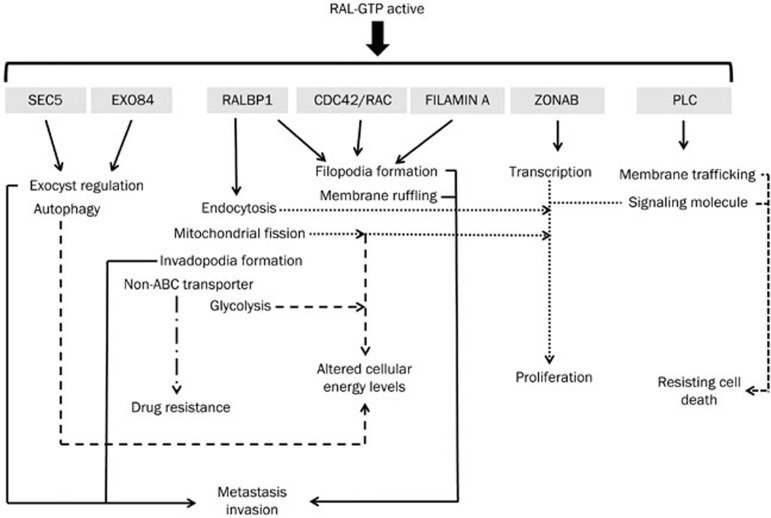
Figure 2.
RAL effectors and their functions. Upon activation, RAL GTPase regulates numerous biological processes through its effectors. Abnormal regulation of these biological processes by activated RAL leads to protumorigenic biological outcomes. The various dotted lines show the cellular processes regulated by each RAL effector and the corresponding biological outcomes.
RALBP1 has GAP activity for RAC/CDC42 proteins. In this way, RALBP1 is important for actin dynamics, filopodia formation and membrane ruffling1,26,27, thereby contributing to cancer cell adhesion, invasion and migration. RALBP1 is critical for the migration of bladder and prostate cancer cells, which is partly independent of its interaction with RAL28. RALBP1 is also important for endocytosis, including receptor-mediated endocytosis of proteins such as epidermal growth factor receptor, insulin receptor and transferrin receptor, through its interactions with POB1, Reps1 and AP229,30,31. The involvement of RALBP1 in receptor endocytosis points to the prominence of RAL in signal transduction by these receptors, which regulate various cellular processes such as survival and proliferation32,33. RALBP1 has also been identified as a non-ABC transporter that causes resistance to chemotherapeutic drugs34. RALBP1 is known to cause resistance to anthracycline derivatives such as doxorubicin and glutathione conjugates by increasing their efflux from cells in an ATP-dependent manner34,35,36. This transporter activity of RALBP1 helps to protect cancer cells from cytotoxic drugs and cellular stress34. RALBP1 also induces mitochondrial fission by promoting the phosphorylation of the GTPase DRP1 by CyclinB/CDK1 and facilitating the recruitment of DRP1 to mitochondrial membranes37. The absence of RALA-dependent RALBP1 activation leads to the improper segregation of mitochondria to daughter cells during cell division, resulting in improper cellular metabolic activity, including cellular ATP production, which assists cell growth37. RALBP1 is also known to promote invadopodia formation upon activation by RALB38. This RALBP1-dependent invadopodia formation is essential for RALB-mediated cell invasion and migration. RALBP1 ATPase function is required for invadopodia formation38.
Sec5 and Exo84 are members of the exocyst complex and are responsible for targeting various secretory vesicles to specific regions of the cellular membrane39,40. This regulation of vesicular trafficking controls various complex cellular functions such as polarized membrane formation, cytokinesis, tight junction formation, and tumor cell invasion41,42,43,44. RALB also interacts with Sec5 to activate the IκB kinase TBK121,45. Upon chronic RALB activation, the RALB/Sec5 effector complex, through TBK1, promotes cancer cell survival and RALB-mediated protection from apoptosis45. A separate RALB/Exo84 complex activates autophagosome assembly under starvation conditions via ULK1 and Beclin1-VPS3421,46. Thus, RALB can act as a nutrient sensor by promoting autophagosome formation upon nutrient deprivation46. The RALB/Exo84 complex is able to maintain the balance between cellular growth and maintenance based on nutrient availability through autophagocytosis46. On the other hand, the RALA/Exo84 complex is known to regulate the morphology and polarized migration of neuronal progenitor cells. Loss of the RALA/Exo84 interaction hampers polarity in postnatal migratory neuroblasts and embryonic neurons47. Thus, RALA/Exo84 can promote cellular migration by altering cell polarization47.
RAL also interacts with ZONAB to regulate cellular transcription, which gives rise to cellular proliferative signaling48. Upon its activation by RAS, RALA is known to complex with ZONAB and inhibit its transcription-repressing activity, thereby promoting the transcription of genes required for cellular proliferation and tumorigenesis48. RALA is known to bind Filamin to regulate actin cross-linking, which ultimately results in membrane ruffling and filopodia formation49. RALA is required for CDC42-mediated filopodia formation with the help of Filamin, which aids in cell invasion, adhesion and migration49. PLD1 is another important effector of RAL GTPase; PLD1 plays a role in membrane trafficking and as a signaling molecule50. PLD1 is responsible for RALA-mediated mTOR pathway activation51. RALA acts as a nutrient sensor and promotes protumorigenic signaling through mTOR with the help of PLD1.
RAL in cancer
Even though RALA and RALB have significant amino acid sequence homology, they also have some different cellular functions. RALA is important for tumor growth, whereas RALB is important for tumor metastasis and invasion21,25,52,53,54. However, it has also been reported a number of times that both isoforms also have redundant functions in cancer24,54,55. Increased expression and activation of the RAL isoforms are observed in various tumor types compared with normal tissues, as well as in high-stage and high-grade tumors, signifying their importance in tumorigenesis55,56,57,58,59. In most cancer models, RALA has been established as the dominant RAL isoform and the main driver of tumor progression and metastasis.
In melanoma and non-small cell lung cancer (NSCLC), RALA and RALB seem to have redundant effects on tumor growth24,55,59,60. In melanoma, RALA is the dominant isoform regulating tumor growth59. However, the RAL effectors responsible for melanoma tumor growth have not been studied59. Male et al showed that RALA is important for in vitro and in vivo growth and invasion of NSCLC cells using only the A549 cell line as a model60. The role of RALB in growth and invasion of the A549 NSCLC cell line was not investigated in this study60. RALBP1 was established as the primary RALA effector in A549 cells60. Our lab used a panel of 14 NSCLC cell lines and showed that RALA and RALB had redundant functions in NSCLC tumor growth and that RALBP1 is the primary effector driving this phenotype55. Our lab further developed the first known RAL inhibitors and showed that these small molecule drugs inhibited the growth of NSCLC cell lines in vitro and in vivo by limiting the binding of RALBP1 to both RALA and RALB61. In pancreatic cancer, RALA is important for tumor growth, whereas RALA and RALB are both required for invasion54. The RALBP1-RAL interaction was attributed to pancreatic tumor growth and invasion54. Phosphorylation of RALA at S194 by Aurora-A has been shown to induce pancreatic cancer growth62. This study showed that RALA phosphorylation assists in the RALA-RALBP1 interaction and that this interaction drives pancreatic tumor growth62. RALA and RALB have also been shown to provide protection against ionizing radiation via the effectors RALBP1 and Sec5 in pancreatic cancer cell lines63.
Both RAL isoforms have important functions in the growth and metastasis of bladder cancer28,64,65. RALA is important for the progression of hepatocellular carcinoma, colorectal cancer, chronic myeloid leukemia, squamous cell carcinoma of the skin and nerve sheath tumors57,66,67,68,69. RALA is also pro-migratory, assisting in the metastasis of prostate cancer to bone and aiding in the invasion of squamous cell carcinoma of the skin and renal cancers28,68,70,71. RALB, however, is important for the invasion and migration of bladder cancer, oral squamous cell carcinoma, B cells and multiple myeloma cells53,72,73. RALBP1 has been identified as the main RAL effector in the majority of these tumors. RAL-driven colorectal cancer growth has been attributed to RALBP1, Sec5, and Exo8466. Sec5 and Exo84 have been identified as important RAL effectors in squamous cell carcinoma of the skin68. The phosphorylation of RALB at S198 by protein kinase C (PKC) is an important driver of bladder cancer growth and metastasis via promotion of the RALB-RALBP1 interaction65. A transcriptional signature was recently developed to report cellular RAL activity levels 74. This RAL signature was predictive of cancer patient outcomes, indicating for the first time that the genes and pathways regulated by RAL are important for tumor formation and progression in cancer patients 74.
Dependency of RAL activity on RAS mutation
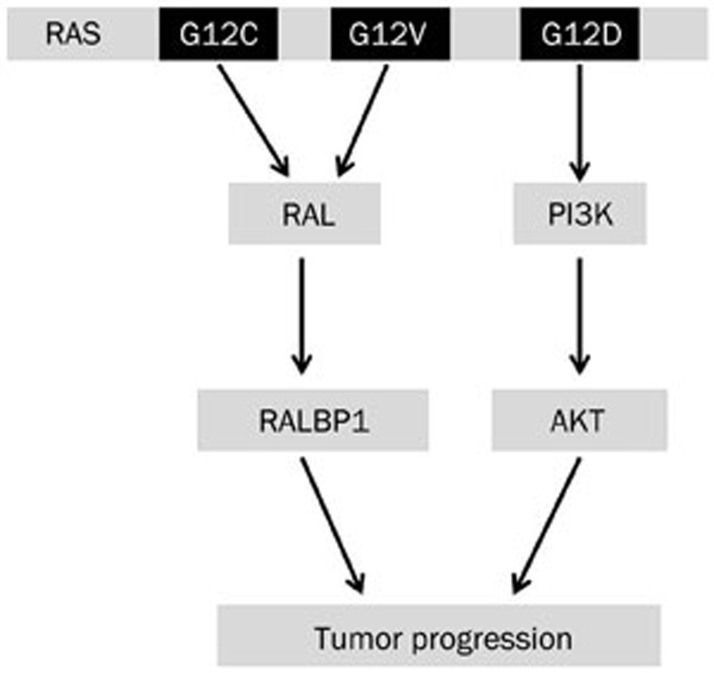
Recent studies on NSCLC patient samples showed that patients carrying a glycine to cysteine (G12C) or a glycine to valine (G12V) amino acid mutation at codon 12 of KRAS have worse outcomes compared with patients carrying other KRAS mutations or wildtype KRAS20. Interestingly, it was observed that NSCLC cell lines carrying the KRAS G12C or G12V mutation have higher RAL GTPase activities and lower PI3K/AKT or RAF-MEK-ERK pathway activation compared with cell lines carrying other KRAS mutations (Figure 3) or wild-type KRAS20. It is known that KRAS homodimer formation is important for RAL GTPase activation via RALGDS binding to KRAS75. Ihle et al used extensive molecular modeling to show that in case of the KRAS G12C mutation, the smaller cysteine residue allows for KRAS homodimer formation, RALGDS binding and subsequent RAL activation20. Another common KRAS mutation at codon 12, the glycine to aspartate (G12D) mutation, was shown to predominantly activate the PI3K/AKT pathway20 (Figure 3). Aspartate is a bulkier amino acid than glycine, G12D mutation prevents KRAS homodimer formation via steric hindrance; thus, RAL activation is prevented20. Since KRAS directly binds to PI3K, this pathway can be activated irrespective of KRAS dimer formation20. Thus, different KRAS mutations help select for different downstream signaling pathways based on the properties of the oncogenic amino acid.
Figure 3.
RAS mutations and activated pathways. Specific RAS mutations are known to activate specific downstream signaling pathways in non-small cell lung cancers.
Another study on a large group of Korean NSCLC patients showed that KRAS mutations are less common in this patient population compared with patients from Western countries76. This observation is consistent with other studies showing that the frequencies of KRAS mutations vary with ethnicity77,78,79,80. G12D was the most frequent KRAS transition mutation found in the Korean patient population; this is consistent with other studies conducted on Asian NSCLC patients76. The study on the Korean patients also showed high RALA membrane expression in 80% of the patient samples and high RALB membrane and cytoplasmic expression in 57% of the samples from patients carrying a KRAS mutation76. Since RAL membrane expression is associated with RAL activity, we can speculate that patients with KRAS mutations have high RAL activity55. The authors showed that there was no difference in RAL expression between patients harboring KRAS transition mutations, including G12D, compared to patients carrying KRAS transversion mutations such as G12C and G12V76. Similarly, AKT was also not differentially expressed between patients carrying KRAS transition and transversion mutations76. This study suggests that although different downstream pathways may be activated by specific KRAS mutations, the expression of these downstream signaling molecules may not change as a function of these specific KRAS mutations.
Using in vitro and in vivo functional assays, our lab established that both RAL isoforms are required for the growth of NSCLC cells carrying the KRAS G12C mutation compared with cells carrying other KRAS mutations and wild-type KRAS55. We used a gene knockdown approach on multiple NSCLC cell lines that carry either wild-type KRAS or different KRAS mutations at codon 1255. We showed that the loss of RALA and RALB massively inhibited anchorage-dependent and anchorage-independent growth of NSCLC cell lines carrying the KRAS G12C mutation55. The loss of RALA and RALB also inhibited in vivo tumor growth with the NSCLC cell line H2122, which carries the G12C mutation55. NSCLC cell lines that are wild-type for KRAS or that carry other mutations showed moderate to no inhibition in anchorage-dependent and anchorage-independent growth assays upon the loss of the RAL GTPases55. Our study also showed that overexpression of the KRAS G12C mutant in the KRAS-wild-type cell line H2228 increased the activation of RAL and the dependency of the cells on RAL GTPases for anchorage-independent growth55. Thus, we used detailed functional analysis to validate that the KRAS G12C mutation indeed drives tumorigenesis, predominantly via RAL GTPases. In non-smoking NSCLC patients, the probability of having a G12C or G12V KRAS mutation at codon 12 is 9% or 28%, respectively, whereas in smokers, this probability is 47% or 24%, respectively20,81. Since KRAS G12C and G12V are the most common RAS mutations found in NSCLC, these studies point to the importance of RAL in driving tumor growth downstream of KRAS in NSCLC.
KRAS mutations are also frequently found in pancreatic and colorectal cancers. Approximately 90% of pancreatic cancer patients have a KRAS mutation at codon 12, with glycine to valine, aspartate or arginine (G to V, D, or A) mutations being the most frequent3,13. In pancreatic cancers, RAL GTPase was shown to be required for in vivo metastatic growth of tumors driven by the KRAS G12V mutation54, pointing to the dependency of these cancers that carry this specific KRAS mutations on the RAL pathway. KRAS mutations at codon 12, predominantly glycine to aspartate or valine (G to D or V) mutations, are also frequent in colorectal cancers13. However, the role of RAL in colorectal cancer with respect to different KRAS mutations is unclear66. Higher RAL GTPase activation was also observed in bladder cancer cell lines carrying the G12V HRAS mutation compared with cell lines that either are wild-type for RAS or carry other RAS mutations56. Bladder cancers have a low to medium RAS mutation frequency, but 60% of the total RAS (either KRAS or HRAS) mutations present are G12V mutations, pointing to a potential dependence on the RAL signaling pathway in the majority of RAS-mutant tumors13. Based on the currently available information in NSCLC, the dependency of pancreatic, colorectal and bladder tumors carrying specific RAS mutations on RAL or other downstream pathways requires investigation.
Conclusion
Drugs against the PI3K/AKT and RAF-MEK-ERK pathways are used for the treatment of patients carrying RAS mutations. RAL GTPase has not been explored as a treatment target for cancer patients harboring RAS mutations. The only reason for this is the lack of RAL GTPase inhibitors. The recent discovery that different RAS mutations specifically activate different downstream signaling pathways stresses the need for personalized treatment strategies for patients that are stratified based on their RAS mutation status. These recent findings denote that RAL GTPase is a potential drug target for the treatment of cancer patients carrying specific RAS mutations. For example, we expect that NSCLC patients carrying KRAS G12C or G12V mutations would respond to treatment with a RAL inhibitor, whereas patients carrying KRAS G12D mutations should be treated with a PI3K inhibitor. This recent observation, predominantly made in NSCLC, also calls for a comprehensive investigation in other tumor types to validate that specific RAS mutations drive specific signaling pathways. We expect that targeted therapy against RAL GTPase will be vital in the treatment of cancer patients because KRAS G12C and G12V mutations are abundant in NSCLC as well as colorectal and pancreatic cancers.
Abbreviations
NSCLC, non-small cell lung cancer; GEF, guanine nucleotide exchange factor; GAP, GTPase-activating proteins; RTK, receptor tyrosine kinase; PI3K, phosphatidylinositol 3-kinase.
References
- Takai Y, Sasaki T, Matozaki T. Small GTP-binding proteins. Physiol Rev. 2001;81:153–208. doi: 10.1152/physrev.2001.81.1.153. [DOI] [PubMed] [Google Scholar]
- Takashima A, Faller DV. Targeting the RAS oncogene. Expert Opin Ther Targets. 2013;17:507–31. doi: 10.1517/14728222.2013.764990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chetty R, Govender D. Gene of the month: KRAS. J Clin Pathol. 2013;66:548–50. doi: 10.1136/jclinpath-2013-201663. [DOI] [PubMed] [Google Scholar]
- Symons M, Takai Y. Ras GTPases: singing in tune. Sci STKE. 2001;2001:pe1. doi: 10.1126/stke.2001.68.pe1. [DOI] [PubMed] [Google Scholar]
- Tan C, Du X. KRAS mutation testing in metastatic colorectal cancer. World J Gastroenterol. 2012;18:5171–80. doi: 10.3748/wjg.v18.i37.5171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin P, Leighl NB, Tsao MS, Shepherd FA. KRAS mutations as prognostic and predictive markers in non-small cell lung cancer. J Thorac Oncol. 2013;8:530–42. doi: 10.1097/JTO.0b013e318283d958. [DOI] [PubMed] [Google Scholar]
- di Magliano MP, Logsdon CD. Roles for KRAS in pancreatic tumor development and progression. Gastroenterology. 2013;144:1220–9. doi: 10.1053/j.gastro.2013.01.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacco E, Spinelli M, Vanoni M. Approaches to Ras signaling modulation and treatment of Ras-dependent disorders: a patent review (2007 — present) Expert Opin Ther Pat. 2012;22:1263–87. doi: 10.1517/13543776.2012.728586. [DOI] [PubMed] [Google Scholar]
- Ward AF, Braun BS, Shannon KM. Targeting oncogenic Ras signaling in hematologic malignancies. Blood. 2012;120:3397–406. doi: 10.1182/blood-2012-05-378596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosten M, Dhawahir A, Sum EY, Urosevic J, Lechuga CG, Esteban LM, et al. Genetic analysis of Ras signalling pathways in cell proliferation, migration and survival. EMBO J. 2010;29:1091–104. doi: 10.1038/emboj.2010.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matozaki T, Nakanishi H, Takai Y. Small G-protein networks: their crosstalk and signal cascades. Cell Signal. 2000;12:515–24. doi: 10.1016/s0898-6568(00)00102-9. [DOI] [PubMed] [Google Scholar]
- Cherfils J, Zeghouf M. Regulation of small GTPases by GEFs, GAPs, and GDIs. Physiol Rev. 2013;93:269–309. doi: 10.1152/physrev.00003.2012. [DOI] [PubMed] [Google Scholar]
- Fernandez-Medarde A, Santos E. Ras in cancer and developmental diseases. Genes Cancer. 2011;2:344–58. doi: 10.1177/1947601911411084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prior IA, Lewis PD, Mattos C. A comprehensive survey of Ras mutations in cancer. Cancer Res. 2012;72:2457–67. doi: 10.1158/0008-5472.CAN-11-2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malumbres M, Barbacid M. RAS oncogenes: the first 30 years. Nat Rev Cancer. 2003;3:459–65. doi: 10.1038/nrc1097. [DOI] [PubMed] [Google Scholar]
- Rajalingam K, Schreck R, Rapp UR, Albert S. Ras oncogenes and their downstream targets. Biochim Biophys Acta. 2007;1773:1177–95. doi: 10.1016/j.bbamcr.2007.01.012. [DOI] [PubMed] [Google Scholar]
- Ostrem JM, Peters U, Sos ML, Wells JA, Shokat KM. K-Ras (G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature. 2013;503:548–51. doi: 10.1038/nature12796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patgiri A, Yadav KK, Arora PS, Bar-Sagi D. An orthosteric inhibitor of the Ras-Sos interaction. Nat Chem Biol. 2011;7:585–7. doi: 10.1038/nchembio.612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downward J. Targeting RAS signalling pathways in cancer therapy. Nat Rev Cancer. 2003;3:11–22. doi: 10.1038/nrc969. [DOI] [PubMed] [Google Scholar]
- Ihle NT, Byers LA, Kim ES, Saintigny P, Lee JJ, Blumenschein GR, et al. Effect of KRAS oncogene substitutions on protein behavior: implications for signaling and clinical outcome. J Natl Cancer Inst. 2012;104:228–39. doi: 10.1093/jnci/djr523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashatus DF. Ral GTPases in tumorigenesis: emerging from the shadows. Exp Cell Res. 2013;319:2337–42. doi: 10.1016/j.yexcr.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feig LA. Ral-GTPases: approaching their 15 minutes of fame. Trends Cell Biol. 2003;13:419–25. doi: 10.1016/s0962-8924(03)00152-1. [DOI] [PubMed] [Google Scholar]
- Vigil D, Martin TD, Williams F, Yeh JJ, Campbell SL, Der CJ. Aberrant overexpression of the Rgl2 Ral small GTPase-specific guanine nucleotide exchange factor promotes pancreatic cancer growth through Ral-dependent and Ral-independent mechanisms. J Biol Chem. 2010;285:34729–40. doi: 10.1074/jbc.M110.116756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peschard P, McCarthy A, Leblanc-Dominguez V, Yeo M, Guichard S, Stamp G, et al. Genetic deletion of RALA and RALB small GTPases reveals redundant functions in development and tumorigenesis. Curr Biol. 2012;22:2063–8. doi: 10.1016/j.cub.2012.09.013. [DOI] [PubMed] [Google Scholar]
- Lim KH, Baines AT, Fiordalisi JJ, Shipitsin M, Feig LA, Cox AD, et al. Activation of RalA is critical for Ras-induced tumorigenesis of human cells. Cancer Cell. 2005;7:533–45. doi: 10.1016/j.ccr.2005.04.030. [DOI] [PubMed] [Google Scholar]
- Bodemann BO, White MA. Ral GTPases and cancer: linchpin support of the tumorigenic platform. Nat Rev Cancer. 2008;8:133–40. doi: 10.1038/nrc2296. [DOI] [PubMed] [Google Scholar]
- Ikeda M, Ishida O, Hinoi T, Kishida S, Kikuchi A. Identification and characterization of a novel protein interacting with Ral-binding protein 1, a putative effector protein of Ral. J Biol Chem. 1998;273:814–21. doi: 10.1074/jbc.273.2.814. [DOI] [PubMed] [Google Scholar]
- Wu Z, Owens C, Chandra N, Popovic K, Conaway M, Theodorescu D. RalBP1 is necessary for metastasis of human cancer cell lines. Neoplasia. 2010;12:1003–12. doi: 10.1593/neo.101080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi A, Urano T, Goi T, Feig LA. An Eps homology (EH) domain protein that binds to the Ral-GTPase target, RalBP1. J Biol Chem. 1997;272:31230–4. doi: 10.1074/jbc.272.50.31230. [DOI] [PubMed] [Google Scholar]
- Nakashima S, Morinaka K, Koyama S, Ikeda M, Kishida M, Okawa K, et al. Small G protein Ral and its downstream molecules regulate endocytosis of EGF and insulin receptors. EMBO J. 1999;18:3629–42. doi: 10.1093/emboj/18.13.3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jullien-Flores V, Mahe Y, Mirey G, Leprince C, Meunier-Bisceuil B, Sorkin A, et al. RLIP76, an effector of the GTPase Ral, interacts with the AP2 complex: involvement of the Ral pathway in receptor endocytosis. J Cell Sci. 2000;113:2837–44. doi: 10.1242/jcs.113.16.2837. [DOI] [PubMed] [Google Scholar]
- Wang Y, Pennock S, Chen X, Wang Z. Endosomal signaling of epidermal growth factor receptor stimulates signal transduction pathways leading to cell survival. Mol Cell Biol. 2002;22:7279–90. doi: 10.1128/MCB.22.20.7279-7290.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resat H, Ewald JA, Dixon DA, Wiley HS. An integrated model of epidermal growth factor receptor trafficking and signal transduction. Biophys J. 2003;85:730–43. doi: 10.1016/s0006-3495(03)74516-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awasthi S, Cheng J, Singhal SS, Saini MK, Pandya U, Pikula S, et al. Novel function of human RLIP76: ATP-dependent transport of glutathione conjugates and doxorubicin. Biochemistry. 2000;39:9327–34. doi: 10.1021/bi992964c. [DOI] [PubMed] [Google Scholar]
- Drake KJ, Singhal J, Yadav S, Nadkar A, Pungaliya C, Singhal SS, et al. RALBP1/RLIP76 mediates multidrug resistance. Int J Oncol. 2007;30:139–44. [PubMed] [Google Scholar]
- Singhal SS, Singhal J, Nair MP, Lacko AG, Awasthi YC, Awasthi S. Doxorubicin transport by RALBP1 and ABCG2 in lung and breast cancer. Int J Oncol. 2007;30:717–25. [PubMed] [Google Scholar]
- Kashatus DF, Lim KH, Brady DC, Pershing NL, Cox AD, Counter CM. RALA and RALBP1 regulate mitochondrial fission at mitosis. Nat Cell Biol. 2011;13:1108–15. doi: 10.1038/ncb2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neel NF, Rossman KL, Martin TD, Hayes TK, Yeh JJ, Der CJ. The RalB small GTPase mediates formation of invadopodia through a GTPase-activating protein-independent function of the RalBP1/RLIP76 effector. Mol Cell Biol. 2012;32:1374–86. doi: 10.1128/MCB.06291-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugihara K, Asano S, Tanaka K, Iwamatsu A, Okawa K, Ohta Y. The exocyst complex binds the small GTPase RalA to mediate filopodia formation. Nat Cell Biol. 2002;4:73–8. doi: 10.1038/ncb720. [DOI] [PubMed] [Google Scholar]
- Moskalenko S, Henry DO, Rosse C, Mirey G, Camonis JH, White MA. The exocyst is a Ral effector complex. Nat Cell Biol. 2002;4:66–72. doi: 10.1038/ncb728. [DOI] [PubMed] [Google Scholar]
- Spiczka KS, Yeaman C. Ral-regulated interaction between Sec5 and paxillin targets Exocyst to focal complexes during cell migration. J Cell Sci. 2008;121:2880–91. doi: 10.1242/jcs.031641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shipitsin M, Feig LA. RalA but not RalB enhances polarized delivery of membrane proteins to the basolateral surface of epithelial cells. Mol Cell Biol. 2004;24:5746–56. doi: 10.1128/MCB.24.13.5746-5756.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazelett CC, Sheff D, Yeaman C. RalA and RalB differentially regulate development of epithelial tight junctions. Mol Biol Cell. 2011;22:4787–800. doi: 10.1091/mbc.E11-07-0657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascone I, Selimoglu R, Ozdemir C, Del Nery E, Yeaman C, White M, et al. Distinct roles of RalA and RalB in the progression of cytokinesis are supported by distinct RalGEFs. EMBO J. 2008;27:2375–87. doi: 10.1038/emboj.2008.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien Y, Kim S, Bumeister R, Loo YM, Kwon SW, Johnson CL, et al. RalB GTPase-mediated activation of the IkappaB family kinase TBK1 couples innate immune signaling to tumor cell survival. Cell. 2006;127:157–70. doi: 10.1016/j.cell.2006.08.034. [DOI] [PubMed] [Google Scholar]
- Bodemann BO, Orvedahl A, Cheng T, Ram RR, Ou YH, Formstecher E, et al. RalB and the exocyst mediate the cellular starvation response by direct activation of autophagosome assembly. Cell. 2011;144:253–67. doi: 10.1016/j.cell.2010.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das A, Gajendra S, Falenta K, Oudin MJ, Peschard P, Feng S, et al. RalA promotes a direct exocyst-Par6 interaction to regulate polarity in neuronal development. J Cell Sci. 2014;127:686–99. doi: 10.1242/jcs.145037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel P, Aronheim A, Kavanagh E, Balda MS, Matter K, Bunney TD, et al. RalA interacts with ZONAB in a cell density-dependent manner and regulates its transcriptional activity. EMBO J. 2005;24:54–62. doi: 10.1038/sj.emboj.7600497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta Y, Suzuki N, Nakamura S, Hartwig JH, Stossel TP. The small GTPase RalA targets filamin to induce filopodia. Proc Natl Acad Sci U S A. 1999;96:2122–8. doi: 10.1073/pnas.96.5.2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Luo JQ, Urano T, Frankel P, Lu Z, Foster DA, et al. Involvement of Ral GTPase in v-Src-induced phospholipase D activation. Nature. 1995;378:409–12. doi: 10.1038/378409a0. [DOI] [PubMed] [Google Scholar]
- Xu L, Salloum D, Medlin PS, Saqcena M, Yellen P, Perrella B, et al. Phospholipase D mediates nutrient input to mammalian target of rapamycin complex 1 (mTORC1) J Biol Chem. 2011;286:25477–86. doi: 10.1074/jbc.M111.249631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien Y, White MA. RAL GTPases are linchpin modulators of human tumour-cell proliferation and survival. EMBO Rep. 2003;4:800–6. doi: 10.1038/sj.embor.embor899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oxford G, Owens CR, Titus BJ, Foreman TL, Herlevsen MC, Smith SC, et al. RalA and RalB: antagonistic relatives in cancer cell migration. Cancer Res. 2005;65:7111–20. doi: 10.1158/0008-5472.CAN-04-1957. [DOI] [PubMed] [Google Scholar]
- Lim KH, O'Hayer K, Adam SJ, Kendall SD, Campbell PM, Der CJ, et al. Divergent roles for RalA and RalB in malignant growth of human pancreatic carcinoma cells. Curr Biol. 2006;16:2385–94. doi: 10.1016/j.cub.2006.10.023. [DOI] [PubMed] [Google Scholar]
- Guin S, Ru Y, Wynes MW, Mishra R, Lu X, Owens C, et al. Contributions of KRAS and RAL in non-small-cell lung cancer growth and progression. J Thorac Oncol. 2013;8:1492–501. doi: 10.1097/JTO.0000000000000007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SC, Oxford G, Baras AS, Owens C, Havaleshko D, Brautigan DL, et al. Expression of ral GTPases, their effectors, and activators in human bladder cancer. Clin Cancer Res. 2007;13:3803–13. doi: 10.1158/1078-0432.CCR-06-2419. [DOI] [PubMed] [Google Scholar]
- Ezzeldin M, Borrego-Diaz E, Taha M, Esfandyari T, Wise AL, Peng W, et al. RalA signaling pathway as a therapeutic target in hepatocellular carcinoma (HCC) Mol Oncol. 2014;8:1043–53. doi: 10.1016/j.molonc.2014.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Terai K, Peng W, Rouyanian A, Liu J, Roby KF, et al. The role of RalA in biology and therapy of ovarian cancer Oncotarget 2013. Dec 10. [Epub ahead of print]
- Zipfel PA, Brady DC, Kashatus DF, Ancrile BD, Tyler DS, Counter CM. Ral activation promotes melanoma genesis. Oncogene. 2010;29:4859–64. doi: 10.1038/onc.2010.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Male H, Patel V, Jacob MA, Borrego-Diaz E, Wang K, Young DA, et al. Inhibition of RalA signaling pathway in treatment of non-small cell lung cancer. Lung Cancer. 2012;77:252–9. doi: 10.1016/j.lungcan.2012.03.007. [DOI] [PubMed] [Google Scholar]
- Yan C, Liu D, Li L, Wempe MF, Guin S, Khanna M, et al. Discovery and characterization of small molecules that target the GTPase Ral. Nature. 2014;515:443–7. doi: 10.1038/nature13713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim KH, Brady DC, Kashatus DF, Ancrile BB, Der CJ, Cox AD, et al. Aurora-A phosphorylates, activates, and relocalizes the small GTPase RalA. Mol Cell Biol. 2010;30:508–23. doi: 10.1128/MCB.00916-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd AR, 3rd, Snider JL, Martin TD, Graboski SF, Der CJ, Cox AD. Ras-related small GTPases RalA and RalB regulate cellular survival after ionizing radiation. Int J Radiat Oncol Biol Phys. 2010;78:205–12. doi: 10.1016/j.ijrobp.2010.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SC, Theodorescu D. The Ral GTPase pathway in metastatic bladder cancer: key mediator and therapeutic target. Urol Oncol. 2009;27:42–7. doi: 10.1016/j.urolonc.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Owens C, Chandra N, Conaway MR, Brautigan DL, Theodorescu D. Phosphorylation of RalB is important for bladder cancer cell growth and metastasis. Cancer Res. 2010;70:8760–9. doi: 10.1158/0008-5472.CAN-10-0952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin TD, Samuel JC, Routh ED, Der CJ, Yeh JJ. Activation and involvement of Ral GTPases in colorectal cancer. Cancer Res. 2011;71:206–15. doi: 10.1158/0008-5472.CAN-10-1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Li Y, Luo X, Fei J. Inhibition of small GTPase RalA regulates growth and arsenic-induced apoptosis in chronic myeloid leukemia (CML) cells. Cell Signal. 2012;24:1134–40. doi: 10.1016/j.cellsig.2012.01.016. [DOI] [PubMed] [Google Scholar]
- Sowalsky AG, Alt-Holland A, Shamis Y, Garlick JA, Feig LA. RalA function in dermal fibroblasts is required for the progression of squamous cell carcinoma of the skin. Cancer Res. 2011;71:758–67. doi: 10.1158/0008-5472.CAN-10-2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodempudi V, Yamoutpoor F, Pan W, Dudek AZ, Esfandyari T, Piedra M, et al. Ral overactivation in malignant peripheral nerve sheath tumors. Mol Cell Biol. 2009;29:3964–74. doi: 10.1128/MCB.01153-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin J, Pollock C, Tracy K, Chock M, Martin P, Oberst M, et al. Activation of the RalGEF/Ral pathway promotes prostate cancer metastasis to bone. Mol Cell Biol. 2007;27:7538–50. doi: 10.1128/MCB.00955-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Zhang Y, Kim WJ, Daaka Y. PGE2 promotes renal carcinoma cell invasion through activated RalA. Oncogene. 2013;32:1408–15. doi: 10.1038/onc.2012.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada M, Miki T, Iwai S, Shimizu H, Yura Y. Involvement of RhoA and RalB in geranylgeranyltransferase I inhibitor-mediated inhibition of proliferation and migration of human oral squamous cell carcinoma cells. Cancer Chemother Pharmacol. 2011;68:559–69. doi: 10.1007/s00280-010-1520-9. [DOI] [PubMed] [Google Scholar]
- de Gorter DJ, Reijmers RM, Beuling EA, Naber HP, Kuil A, Kersten MJ, et al. The small GTPase Ral mediates SDF-1-induced migration of B cells and multiple myeloma cells. Blood. 2008;111:3364–72. doi: 10.1182/blood-2007-08-106583. [DOI] [PubMed] [Google Scholar]
- Smith SC, Baras AS, Owens CR, Dancik G, Theodorescu D. Transcriptional signatures of Ral GTPase are associated with aggressive clinicopathologic characteristics in human cancer. Cancer Res. 2012;72:3480–91. doi: 10.1158/0008-5472.CAN-11-3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Hofer F, Martin GS, Kim SH. Structural basis for the interaction of Ras with RalGDS. Nat Struct Biol. 1998;5:422–6. doi: 10.1038/nsb0698-422. [DOI] [PubMed] [Google Scholar]
- Kim EY, Kim A, Kim SK, Kim HJ, Chang J, Ahn CM, et al. KRAS oncogene substitutions in Korean NSCLC patients: clinical implication and relationship with pAKT and RalGTPases expression. Lung Cancer. 2014;85:299–305. doi: 10.1016/j.lungcan.2014.04.012. [DOI] [PubMed] [Google Scholar]
- Bae NC, Chae MH, Lee MH, Kim KM, Lee EB, Kim CH, et al. EGFR, ERBB2, and KRAS mutations in Korean non-small cell lung cancer patients. Cancer Genet Cytogenet. 2007;173:107–13. doi: 10.1016/j.cancergencyto.2006.10.007. [DOI] [PubMed] [Google Scholar]
- Kim YT, Kim TY, Lee DS, Park SJ, Park JY, Seo SJ, et al. Molecular changes of epidermal growth factor receptor (EGFR) and KRAS and their impact on the clinical outcomes in surgically resected adenocarcinoma of the lung. Lung Cancer. 2008;59:111–8. doi: 10.1016/j.lungcan.2007.08.008. [DOI] [PubMed] [Google Scholar]
- Soung YH, Lee JW, Kim SY, Seo SH, Park WS, Nam SW, et al. Mutational analysis of EGFR and K-RAS genes in lung adenocarcinomas. Virchows Arch. 2005;446:483–8. doi: 10.1007/s00428-005-1254-y. [DOI] [PubMed] [Google Scholar]
- Jang TW, Oak CH, Chang HK, Suo SJ, Jung MH. EGFR and KRAS mutations in patients with adenocarcinoma of the lung. Korean J Intern Med. 2009;24:48–54. doi: 10.3904/kjim.2009.24.1.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts PJ, Stinchcombe TE. KRAS mutation: should we test for it, and does it matter. J Clin Oncol. 2013;31:1112–21. doi: 10.1200/JCO.2012.43.0454. [DOI] [PubMed] [Google Scholar]