Abstract
Specific electrostatics (i.e. salt-bridge) includes both local and non-local interactions that contribute to the overall stability of proteins. It has been shown that a salt-bridge could either be buried or exposed, networked or isolated, hydrogen-bonded or nonhydrogen bonded, in secondary-structure or in coil, formed by single or multiple bonds. Further it could also participates either in intra- or inter-dipole interactions with preference in orientation either for basic residue at N-terminal (orientation-I) or acidic residue at N-terminal (orientation-II). In this context SBION2 is unique in that it reports above mentioned binary items in excel format along with details on intra and inter-dipole interactions and orientations. These results are suitable for post run statistical analyses involving large datasets. Reports are also made on protein-protein interactions, intervening residue distances and general residue specific salt-bridge details. A ready to use compact supplementary table is also produced. The program runs in three alternative modes. Each mode works on any number of structure files with any number of chains at any given atomic distance of ion-pair. Thus SBION2 provides intricate details on salt-bridges and finds application in structural bioinformatics.
Availability
SBION2 is freely available at http://sourceforge.net/projects/sbion2/ for academic users
Keywords: salt-bridge, intra helical, bond multiplicity, core, surface, networked, isolated
Background
Salt-bridges (SBs) (Figure 1C), bonds between oppositely charged side chain atoms of acidic (acceptor atoms, red residue in Figure 1C) and basic (donor atoms, blue residues in Figure 1C) residues that are within 4~ distance, contribute to the overall stability of native state of proteins [1]. Software for determination of atomic intra- and inter-chain SBs and their secondary structural distribution are now available for single input of user choice per run [2] and for multiple structure files per run along with additional details on networked SB and missing residues information [1]. However, further development is sought for details on residue specific SB (but not atomic ones as above) as to whether it is in core or surface of protein, isolated or networked, single or multiple (bonds ≥ 2) bonded, hydrogen or non hydrogen bonded, local or non-local (if local, i→(i+n) typing) and intra- or inter-helical/strand/coil. Each of this binary condition is known to modulate energetic contribution of SBs [3]. Focus may also be paid on the fact that helix and strand are permanent dipole in protein and thus determination of orientation for intra type SBs (i.e. basic residue is at N-terminal [orientation-I] or acidic residue is at Nterminal [Orientation-II]) would be useful in understanding structural stability as well as packing [3]. In this end SBION2 has been the first of its kind which not only display all the above details in systematic manner along with others [1] but also store similar kind of data in excel format for comprehensive post run statistical analyses involving a large number of structural files.
Figure 1.
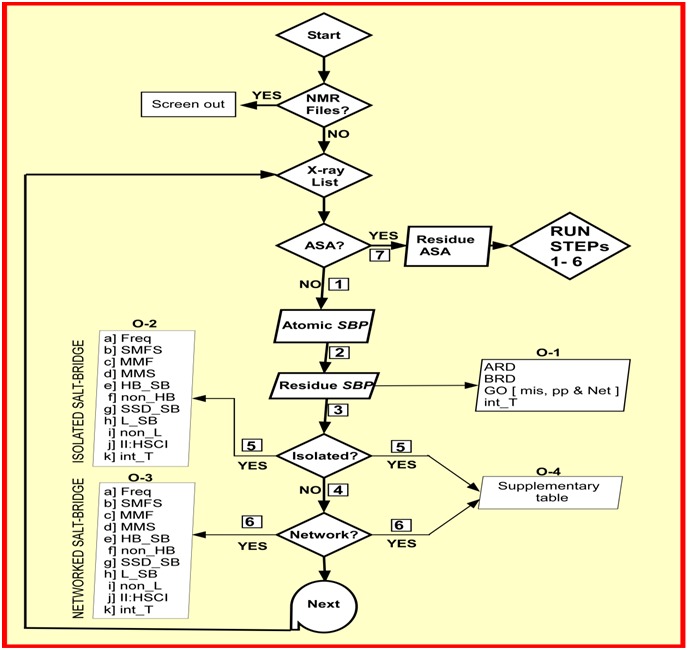
Flowchart of the functioning of the program SBION2. Upon start of the program it screens out NMR files if they are there in the working directory followed by making a list of X-ray structure files. The program verify for accessibility calculation program (ASA) in specified directory. If absent (i.e. NO), the program obtains atomic salt bridge pairs (step-1) and then converts them in residue specific salt bridge pairs (step-2). At this level four outputs are redirected (O-1). Output details of isolated (O-2) and networked salt-bridges (O-3) are separately reported (STEPs 3 to 6). A supplementary table (O-4) is prepared which includes both isolated and networked salt-bridge information (STEPs 5 and 6). If ASA (STEP 7) is YES, SBION2 performs step 1 through 6 that include core and surface location information of salt bridges along with earlier ones. Freq: Frequency; SMFS: Single bond multiplicity frequency and sum; MMF: Multiple bond multiplicity frequency; MMS: Multiple bond multiplicity sum; HB_SB: Hydrogen bonded salt-bridge frequency; non_HB: non-Hydrogen bonded salt-bridge frequency; SSD_SB: secondary structure (helix, sheet and coil) distribution of salt-bridge; L_SB: local salt-bridges; non_L: non-local salt bridges; II:_HSCI: intra- and interhelix, sheet and coil interactions; int_T: intervening residue table.
Methodology
A detailed flow-chart of functioning of SBION2 is presented in Figure 1. It runs in three different modes such as mode-1, mode-2 and mode-3 (Figure 2A). First two modes are for extraction of information on surface and core location of SBs by two alternative paths. Mode-3 utilizes different path (Figure 1). If the program is called with option 1 or 2, upon listing of PDB files it checks for existence of ASA analytical software in the local machine in specified directory (http:// sourceforge.net /projects/sbion2/ files/ README.txt/ download) for extraction of surface and core location of salt bridges along with others (see below). Alternatively, if it is called with option 3 (Figure 1), it performs step 1 through 6 (Figure 2A) for extraction of a total of 27 outputs of which 4 are on residue specific general outputs (Figure 1: list O-1), 11 are on isolated SB (list O-2), 11 are on networked SB (list O-3) and one is for supplementary table (O-4). Output lists in O-2 and O-3 are designed for binary comparison as well as post run statistical analysis and that in O-4 is for ready use of SB results. The run is repeated the times the number of PDB files present in the current directory.
Figure 2.
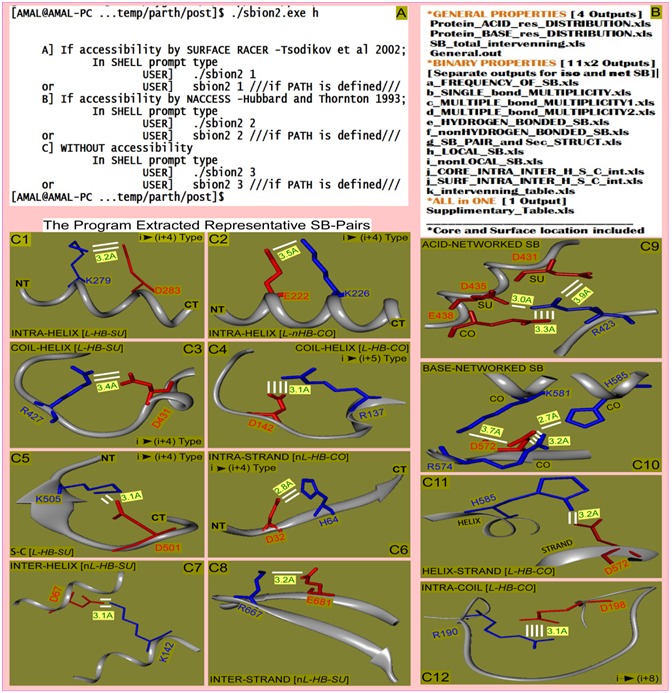
SBION2 execution (A), its outputs (B) and display of few of its output details on salt-bridges (C: 1 through 12) are presented. Execution of the program with option “h” shows three modes of its operation (A). A total of 27 kinds of outputs relevant for presentation and statistical calculation are obtained (B). The program extracts salt-bridge interactions such as INTRA-HELIX (C1 and C2), COIL-HELIX (C3 and C4), STRAND-COIL (C5), INTRA-STRAND (C6), INTER-HELIX (C7), INTER-STRAND (C8), ACID-NETWORK (C9), BASE-NETWORK (C10), HELIX-STRAND (C11) and LOCAL-COIL (C12) are shown with orientation-I (C1 and C3 with basic residue is at N-terminal) and orientation-II (C2 and C4 with acidic residue is at N-terminal) for representative candidate SB pairs. Bond multiplicity (1, 2, 4 etc; white line), Average distance (Green letter in yellow shade), local/non-local (L/nL) and residue separations (i →i+n) are highlighted.
Program input:
Users need to input PDB files, ion-pair distance, ASA and Local/non-Local threshold (http://sourceforge.net /projects/ sbion2/). Crystal structures mixed with NMR files are screen out. Execution of the program allows user to avail three different modes (identified as 1/2/3) (Figure 2A). For mode 1 and 2, user needs to provide ion-pair distance (for salt bridge, the distance is ≤ 4 ~) and ASA threshold (20 to 25) and local/non-Local boundary (≥5 and ≤9). However, for mode 3, only ion-pair distance and local/non-local threshold are necessary.
Program output:
SBION2, like our earlier developments [1, 4], is meant for automated analysis of single or multiple input(s). The program possesses many new features over the existing ones. Firstly it lists results in binary items of which 11 are for isolated (Figure 1, O-2) and 11 are for networked (Figure 1, O-3) SBs. Secondly, within a given chain of protein, SBs could either be intrahelical/ strand/coil or inter-helical/strand/coil with i → (i+n) type of connections. The program reports these result in excel files (Figure 1, O-4). Representative from these SBs are presented in Figure 2 (C1 through C12). Finally, the program also performs analyses during scanning of topology of structures and reports them as general outputs (Figure 1, O-1). While the first item is useful for post run statistical analyses involving a large databases the last two items are in ready to use form.
Caveats and future development:
Program is written in AWK programming language which can preferably run in any C shell UNIX prompt including CYGWIN and also be made work in B shell LINUX and WINDOWS environment. Presently we are actively engaged in developing web interface to integrate SBION2 and other related software tools of our laboratory such as SBION [1] and PHYSICO [4] such that their availability could reach to academic users within an unique web service.
Acknowledgments
We thankfully acknowledge the computational facility Laboratory of the Department of Biotechnology, The University of Burdwan.
Footnotes
Citation:Gupta et al, Bioinformation 11(1): 039-042 (2015)
References
- 1.Gupta PS, et al. Bioinformation. 2014;10:164. doi: 10.6026/97320630010164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gurusaran M, et al. IUCrJ. 2013;1:74. doi: 10.1107/S2052252513031485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rose GD, et al. Science. 1994;20:1126. doi: 10.1126/science.8178170. [DOI] [PubMed] [Google Scholar]
- 4.Gupta PS, et al. Bioinformation. 2014;10:105. doi: 10.6026/97320630010105. [DOI] [PMC free article] [PubMed] [Google Scholar]


