Abstract
A distinct feature of human prostate cancer (PCa) is the development of osteoblastic (bone-forming) bone metastases. Metastatic growth in the bone is supported by factors secreted by PCa cells that activate signaling networks in the tumor microenvironment that augment tumor growth. To better understand these signaling networks and identify potential targets for therapy of bone metastases, we characterized the secretome of a patient-derived xenograft, MDA-PCa-118b (PCa-118b), generated from osteoblastic bone lesion. PCa-118b induces osteoblastic tumors when implanted either in mouse femurs or subcutaneously. To study signaling molecules critical to these unique tumor/microenvironment-mediated events, we performed mass spectrometry on conditioned media of isolated PCa-118b tumor cells, and identified 26 secretory proteins, such as TGF-β2, GDF15, FGF3, FGF19, CXCL1, galectins, and β2-microglobulin, which represent both novel and previously published secreted proteins. RT-PCR using human versus mouse-specific primers showed that TGFβ2, GDF15, FGF3, FGF19, and CXCL1 were secreted from PCa-118b cells. TGFβ2, GDF15, FGF3, and FGF19 function as both autocrine and paracrine factors on tumor cells and stromal cells, that is, endothelial cells and osteoblasts. In contrast, CXCL1 functions as a paracrine factor through the CXCR2 receptor expressed on endothelial cells and osteoblasts. Thus, our study reveals a complex PCa bone metastasis secretome with paracrine and autocrine signaling functions that mediate cross-talk among multiple cell types within the tumor microenvironment.
A distinct feature of human prostate cancer (PCa)1 with lethal potential is the development of metastases in bone with a bone-forming phenotype (1). This property of PCa bone metastasis suggests that PCa cells have unique interactions with cells in the bone microenvironment. Cells that are known to be present in the bone microenvironment include osteoblasts, osteoclasts, adipocytes, fibroblasts, and endothelial cells. Communication between PCa cells and each of these cells in the microenvironment is known to promote metastatic growth. This communication involves metastatic PCa cells that secrete factors to affect stromal cells in the bone microenvironment. The tumor-modified stromal cells may further alter the properties of the PCa cells to allow them to progress in the bone environment (1). Determining how secretory proteins from the metastatic PCa cells affect the PCa/stromal communication network will lead to the development of strategies to treat bone metastases.
Although men with PCa and bone metastasis most frequently present with osteoblastic bone lesions, the commonly-used PCa cell lines to study metastatic properties, for example, PC3 and C4–2B, induce osteolytic or mixed osteoblastic/osteolytic lesions, respectively, when the cells are implanted into mouse femurs or tibia (2). In contrast, the PCa-118b patient-derived xenograft (PDX), generated from an osteoblastic bone lesion of a patient with PCa and bone metastasis, shows phenotypic characteristics similar to the tumor from which it was derived, including induction of a strong osteoblastic response when implanted into femurs (3). Interestingly, PCa-118b cells are also able to induce ectopic bone formation when implanted subcutaneously (3, 4). The capacity of PCa-118b cells to induce bone formation, in which human tumor cells interact with the murine stromal microenvironment, makes this PDX an ideal model system to study tumor-microenvironment signaling pathways that create a bone-like tumor microenvironment conducive to metastatic PCa growth.
In this study, we identified secreted factors from the conditioned medium of isolated PCa-118b cells by mass spectrometry. A total of 26 secretory proteins, including cytokines and growth factors, were identified. Human- and mouse-specific PCR probes were used to identify the cells that expressed these factors. Analysis of the receptor for the corresponding secreted factor determined whether the factor exerted activities in a paracrine and/or autocrine manner. The effects of selected factors on PCa cells or stromal cells, including osteoblasts and endothelial cells, were also examined. Our studies showed that PCa-118b cells secreted multiple factors that establish an autocrine or paracrine signaling network that can mediate cross-talk among multiple cell types within the bone microenvironment.
MATERIALS AND METHODS
Materials
Generation of PCa-118b patient-derived xenograft (PDX) was described previously (3). Fingerprinting of cells isolated from PCa-118b xenografts showed that their profiles are unique as expected. MC3T3-E1 (MC4 subline), DU145 cells, and 2H11 endothelial cells were purchased from American Type Culture Collection (ATCC, Manassas, VA). PC3-mm2 cells were obtained from Dr. I. J. Fidler, M. D. Anderson Cancer Center. Rat aortic endothelial cells were described in (5). C4–2B4 PCa cells were obtained from Dr. Robert Sikes (University of Delaware). Primary mouse osteoblasts (PMO) were isolated from 2–5 day old newborn mouse calvaria as described previously (6). Accumax and Anti-Mouse MHC Class I (H-2Kd/H-2Dd) FITC were from eBioscience. PE-conjugated goat anti-human EpCAM antibody, rhBMP4, rhTGFβ2, rhGDF15, rhFGF-2, rhFGF-3, rhFGF-19, and rhCXCL1 (GROα) were from R&D Systems (Minneapolis, MN). LDN-193189, SD208, GW788388 were from AXON Medchem (Reston, VA). Antibodies against pSmad1/5, pSmad3, Smad1, Smad5, Smad3, pp44/42 MAPK, pp38MAPK, p44/42MAPK, and p38MAPK were from Cell Signaling Technology (Beverly, MA).
Tumor Cell Isolation, Conditioned Medium Preparation, and FACS Analysis
PCa-118b xenografts were generated by implanting fragments (less than 1 mm3) of tumors subcutaneously into SCID mouse. When the tumors reached 500 mm3, they were dissected, cut into small pieces, and digested with Accumax. The digested tissue was filtered through a 100 μm cell strainer and the cells were plated in tissue culture dishes in CnT-52 medium (CELLnTEC Advanced Cell System AG). After overnight incubation, the medium was removed and changed to fresh CnT-52 medium. The conditioned medium was collected at 48 h.
Fluorescence Activated Cell Sorting
Isolated PCa-118b cells were incubated with human PE-EPCAM and/or FITC-mouse MHC I (H-2Kd/H-2Dd) and FACS were done using two-color analysis. The fluorescence intensity of the cells was measured by a BD FACSCantoII flow cytometer (BD Biosciences).
Reverse Transcription and Quantitative PCR Analysis
Total RNAs were extracted from PCa-118b tumor, isolated PCa-118b cells, PC3-mm2, LNCaP, or DU145, using Trizol (Invitrogen) and the RNAs were further purified using RNeasy mini kit (Qiagen, Valencia, CA). RNAs were treated with RNase-free DNase (Qiagen) to prevent the genomic DNA contamination. The RNAs were reverse transcribed using TaqMan Reverse transcription reagent kit (Applied Biosystems, Foster City, CA). cDNA (20 ng) was used in real-time RT-PCR with SYBR Green (Applied Biosystems), using GAPDH as a control. Human- or mouse-specific PCR primers were used to determine the cell of origin of the specific secretory proteins and presence of their receptors in PCa-118b tumor. The sequences of all the PCR primers used are listed in supplemental Table S1.
Protein Identification by LC-MS/MS
Conditioned medium (10 ml) from PCa-118b cells or the control culture medium was concentrated fiftyfold in Centricon-10 (Millipore). In order to identify the maximal number of proteins, we performed LC-MS/MS on the samples that were processed in three different approaches, i.e. “in solution digestion,” “in tube-gel digestion,” and “SDS-PAGE-in gel digestion.” In the “in solution digestion” approach, the concentrated conditioned media were reduced with a final concentration of 10 mm dithiothreitol at 56 °C for 1 h and subsequently alkylated with a final concentration of 55 mm iodoacetamide at room temperature in the dark for 45 min. The proteins were then digested with trypsin, with the protein/trypsin ratio 50:1 (w/w), as previously described (7). The tryptic peptides were separated on a 12 cm × 75 μm I.D. C18 Monitor reversed-phase column (Column Engineering, Ontario, CA) containing an integrated ∼4 μm ESI tip. Peptides were eluted using a gradient starting with 100% solvent A (0.1 m acetic acid in water) to 40% solvent B (0.1 m acetic acid in acetonitrile) over 50 min, then to 70% B in 10 min (nano HPLC system, Agilent Technologies, Paolo Alto, CA). Eluted peptides were introduced into a LTQ Velos Orbitrap mass spectrometer (Thermo Electron Corporation, San Jose, CA) with a 1.9 kV electrospray voltage. Database searching was performed using peak lists created by msconvert.exe (version 2.2.3300) from the ProteoWizard package. The data were searched using Mascot Server 2.4.1 against a concatenated target + decoy UniProt human database (8). While specifying trypsin cleavage, two missed cleavages were allowed. Alkylation of cysteine by iodoacetamide was a fixed modification and oxidation of methionine was a variable modification. Mass tolerance for matching precursor ions was 7 ppm and 0.5 Da for fragment ions. The initial threshold Mascot score value for accepting individual spectra was 0.0. Based on unique (nonredundant) peptide spectrum matches, the score threshold was raised to achieve an estimated false discovery rate of 1.5%. The UniProt human database was downloaded on April 3, 2013 and decoy.pl (Matrix Science, Ltd., Boston, MA) created sequence-reversed decoys from a concatenated file that contained target (UniProt) and common contaminants sequences. There are 269,574 entries in the database.
In the “in tube-gel digestion” approach, the reduced and alkylated proteins were mixed with an acrylamide gel mixture and incorporated into a polyacrylamide gel as described in Cao et al. (7). After polymerization, the gel was cut into small pieces, washed sequentially with 25 mm NH4HCO3, 25 mm NH4HCO3 in 50% (v/v) acetonitrile (ACN), 100% ACN, and dried in a vacuum centrifuge. The gel pieces were then digested with 0.5 μg trypsin in 200 μl of 40 mm NH4HCO3 and 10% (v/v) ACN overnight at 37 °C. The peptides were extracted from the gel and peptide sequences determined as described above.
In the “SDS-PAGE-in gel digestion” approach, the concentrated conditioned media were separated by SDS-PAGE. The entire lane that contained the test samples was cut into 24 gel slices, whereas the lane that contained the corresponding control medium was cut into eight gel slices (see Fig. 1 in Results section). These gel slices were digested with trypsin (0.5 μg/slice) and the tryptic peptides from each gel slice were identified by mass spectrometry as described above.
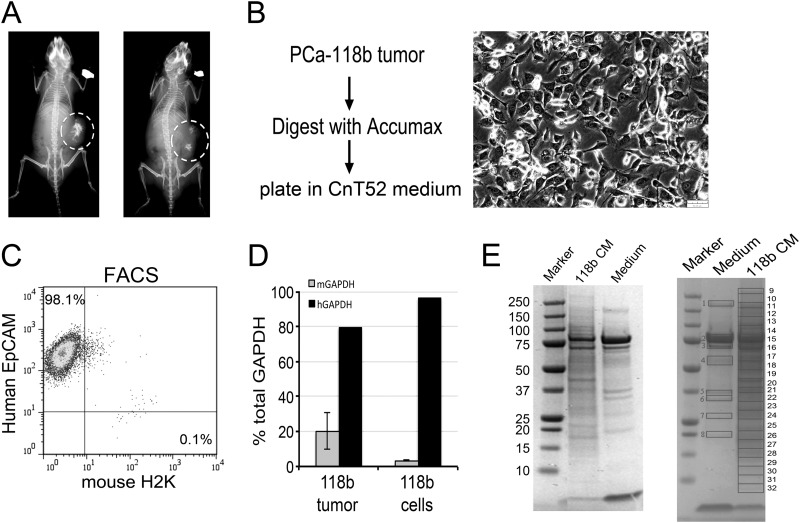
Fig. 1.
Isolation of tumor cells from the PCa-118b tumor. A, x-ray of PCa-118b tumors in SCID mice detected the presence of bone within the PCa-118b tumors. The location of the tumors is outlined by circles. B, Procedure for the isolation of PCa-118b tumor cells from the tumor. The PCa-118b tumor cells in culture exhibit cuboidal epithelial morphology. C, FACS using antibodies against human EpCAM and mouse H2K indicated the enrichment of the epithelial cell marker in the isolated tumor cell preparation. D, qRT-PCR for the relative expression of human versus mouse GAPDH in the PCa-118b tumor versus the purified PCa-118b tumor cell preparation. E, SDS-PAGE analysis of conditioned medium from the isolated PCa-118 cells. Right panel, gel slices that were used in mass spectrometry analysis.
SMAD Phosphorylation
2H11 or RAEC cells grown in 6-well plate in DMEM were serum starved overnight and then were treated with either different concentrations of BMP4 (0, 10, and 100 ng/ml), TGFβ2 (1, 10 ng/ml), or GDF15 (0.05, 0.2, 1, and 10 ng/ml) overnight, or with 10 ng/ml of BMP4, TGFβ2, and GDF15 with or without LDN-193189 (0.1 μm), SD208 (1 μm), or GW788388 (5 μm) for 16 Hrs. For PMO cells, PMO grown in αMEM medium was starved overnight and then cells were treated with 10 ng/ml of BMP4, TGFβ2, and GDF15 for 16 h. For PCa-118b cells, because the cells isolated from PCa-118b tumor are a mixture of epithelial and stromal cells, we cultured the cells in CnT-52 medium, which selectively allows epithelial but not stromal cells to survive in culture, for 2 days and then treated with LDN 193189 (0.1 μm), SD208 (1 μm), or GW788388 (5 μm) for 16 h. After the treatments, cells were harvested, lysed in SDS sample buffer (62.5 mm Tris-HCl, pH 6.8, 2% SDS, 25% (w/v) glycerol, 0.01% bromophenol blue, and 5% 2-mercaptoethanol.), resolved by SDS-PAGE and immunoblotted for p-SMAD1/5 and p-SMAD3 analysis. The same membrane was blotted with total SMAD1, 3, or 5 and actin to normalize for level of protein expression. The band intensity was quantified by Image J.
Effect of FGF3 and FGF19 on PMO Proliferation and Differentiation
PMO grown in six-well plates in Alpha MEM medium containing 10% FBS were treated with and without 10 or 50 ng/ml of FGF2, FGF3, and FGF19 for 3 days. Cells were counted using a hemocytometer. Alkaline phosphatase activity was used to monitor the effect of FGF2, FGF3, and FGF19 on PMO differentiation. In brief, cells were lysed in 0.5% Triton X-100 in Tris-buffered saline and the alkaline phosphatase activity in the cell extract was measured using p-nitrophenyl phosphate disodium salt (PNPP) as substrate (1 mg/ml in 1 m diethanolamine, pH 9.8), that was provided in the PNPP substrate kit (Fisher Scientific). The reaction product was measured at 405 nm.
p44/42 and p38 MAPK Phosphorylation
2H11 cells grown in six-well plate in DMEM medium were starved overnight and then treated with CXCL1, 10 or 50 ng/ml for 10 min. Cells were harvested in SDS gel loading buffer and Western blot analyses were performed for pp44/42 MAPK, pp38 MAPK, p44/42 MAPK, and p38 MAPK expressions.
Ethics Statement
All experimental procedures involving animals were approved by UT M D Anderson's Animal Care and Use Committee. All the experiments involving human tissue samples were approved by the UT MD Anderson Cancer Center Institutional Review Board.
Statistical Analysis
Data are expressed as the mean ± S.D. Student's t test (two-tailed, paired) was used for Statistical analyses. p values less than 0.05 were considered significant.
RESULTS
PCa-118b Xenograft Induces Ectopic Bone Formation in a Subcutaneous Site
When PCa-118b was implanted subcutaneously into SCID mice, tumor growth was observed after 4 weeks and the PCa-118b tumors exhibited ectopic bone formation (Fig. 1A). The ectopic bone was detected as dense structures in x-ray images, as either a major focus or multi-foci, with branches emanating from the central bony structure (Fig. 1A). These observations lead us to hypothesize that PCa-118b human tumor cells probably secreted factors that not only promoted tumor growth but also modulated the mouse stromal cells in the microenvironment to promote bone formation, similar to those observed in human osteoblastic bone metastases.
Culturing of Epithelial Cells from PCa-118b Xenograft In Vitro
To identify the factors that constitute the PCa-118b secretome, we separated the epithelial PCa-118b cells from stromal cells in the tumor. PCa-118b can only be maintained as a xenograft by passaging in SCID mice. Attempts to generate PCa-118b cell lines that can be passaged in vitro in culture have not been successful (unpublished observation). When tumor cells were dissociated from the PCa-118b tumor by Accumax enzyme digestion and plated onto a culture plate, cells with epithelial and stromal morphologies were observed (data not shown). To increase the proportion of epithelial cells in the tumor cell population, after dissociation cells were cultured in CnT-52 medium that selectively allows epithelial but not stromal cells to survive in culture. This method resulted in an enrichment in epithelial cells as reflected in a more cuboidal cell morphology (Fig. 1B). FACS analysis using anti-human EpCAM and anti-mouse H2K antibody was used to distinguish the proportion of human cells from mouse cells, respectively, in the cultured PCa-118b cell population. As shown in Fig. 1C, more than 98% of the isolated cells were positive for the human epithelial cell surface marker EpCAM, whereas only 0.1% of cells were derived from mouse, as evidenced by the positivity with mouse H2K cell surface marker. Real-time qRT-PCR using oligonucleotide probes specific to human versus mouse GAPDH showed that in the tumor xenograft, 80% of the cells are tumor cells, based on human-specific GAPDH primers, whereas between 10–30% of cells are stromal cells, based on mouse-specific GAPDH primers (Fig. 1D). In contrast, in the isolated epithelial cells, 98% of the cells are human cells, whereas less than 2% of cells are derived from the stromal compartment. Together, these observations indicate that the isolated PCa-118b cells contain mainly human epithelial cells.
Analysis of PCa-118 Conditioned Medium by Mass Spectrometry
The conditioned medium (CM) was collected from these enriched PCa-118b epithelial cells cultured in CnT-52 medium after 48 h. The PCa-118b-CM and the control CnT-52 medium, which contains bovine pituitary extract, was concentrated by Centricon centrifugation, analyzed on SDS-PAGE, and stained by Coomassie blue. As shown in Fig. 1E, in addition to bands observed in the CnT-52 medium, additional protein bands were detected in the PCa-118b-CM (left panel). To maximize protein identification, proteins in PCa-118b-CM were subjected to different protein digestion strategies, including “in solution digestion,” “in tube-gel digestion” (7), and “SDS-PAGE-in gel digestion.” The results from the “in solution digestion” approach are shown in supplemental Table S2 (control, Ctr_30) and supplemental Table S3 (sample 118b_10). The results from “in tube-gel digestion” are shown in supplemental Table S4 (Ctr) and supplemental Table S5 (sample 118b). In the “SDS-PAGE-in gel digestion” method, proteins were separated in SDS-PAGE followed by cutting the proteins bands for in gel trypsinization (Fig. 1E, right panel). There are differences in the SDS-PAGE protein profiles between the control media and the conditioned media (Fig. 1E, left panel). Protein bands in the control media are mainly from the components of the pituitary extract supplements in the CnT-52 medium. Some components in the pituitary extract are likely consumed by the tumor cells during the 2-day culturing, resulting in differences in their intensity compared with control media. A total of 32 gel slices from control CnT-52 medium and PCa-118b-CM were subjected to mass spectrometry analyses. The results from “SDS-PAGE-in gel digestion” are shown in supplemental Table S6.
We focused our analysis only on the secretory proteins that are differentially detected in the PCa-118b-CM compared with CnT52 medium. A total of 26 secretory proteins were identified in the PCa-118b-CM from the combination of three mass spectrometry analyses (Table I). Of these, 21 were identified from the “in solution digestion” and 19 from the “in tube-gel digestion” approach, with 14 proteins in common from both analyses. The “SDS-PAGE-in gel digestion” yielded only nine out of the 26 secretory proteins, likely because of the limited amount of total proteins that were loaded onto the SDS-PAGE. All of those proteins identified from “SDS-PAGE-in gel digestion” were also found by the other two approaches. The peptide sequences of the 26 secretory proteins identified by mass spectrometry are listed in supplemental Table S7. For single peptide identifications, annotated spectra are shown as supplemental Figs. S1–S8.
Table I. Secretory factors identified by mass spectrometry.
| Category | Accession number | Protein name | Peptide# | Protein score | % Seq. cover | Method | Peptide # | Protein score | % Seq. cover | Method |
|---|---|---|---|---|---|---|---|---|---|---|
| IGF family | ||||||||||
| Q9NP10 | IGF1 | 1 | 35.99 | 16.5 | In solution | |||||
| Q6UW32 | IGFL1 | 1 | 35.87 | 9.1 | In solution | |||||
| Q6ZSD0 | IGFBP3 | 1 | 34.82 | 5.6 | In solution | |||||
| Q8WX77 | IGFBPL1 | 1 | 35.85 | 4.3 | In solution | |||||
| TGFβ family | ||||||||||
| P61812 | TGFβ2a | 1 | 31.02 | 4.6 | In solution | 1 | 85.47 | 3.9 | In tube-gel | |
| Q99988 | GDF15a | 6 | 162.49 | 26.6 | In solution | 11 | 375.04 | 44.8 | In tube-gel | |
| FGF family | ||||||||||
| P11487 | FGF3 | 1 | 63.74 | 4.2 | In solution | |||||
| O95750 | FGF19 | 1 | 25.17 | 7.4 | In solution | |||||
| Q14512 | FGFBP1a | 5 | 238.69 | 16.2 | In solution | 5 | 204.3 | 28.2 | In tube-gel | |
| Other growth factors | ||||||||||
| P36955 | PEDFa | 2 | 108.46 | 9.1 | In solution | 4 | 208.54 | 14.8 | In tube-gel | |
| P61916 | epididymal secretory protein E1 | 1 | 58.66 | 9.2 | In solution | 2 | 89.46 | 12.5 | In tube-gel | |
| Cytokines | ||||||||||
| P09341 | CXCL1 | 1 | 25.9 | 11.2 | in tube-gel | |||||
| P21741 | midkine | 3 | 130.84 | 32.7 | In solution | 7 | 245.84 | 33.3 | In tube-gel | |
| P14174 | MIFa | 1 | 64.14 | 7.8 | In solution | 2 | 92.9 | 11.3 | In tube-gel | |
| Galectin | ||||||||||
| P09382 | Galectin-1 | 1 | 44.07 | 7.4 | In solution | 2 | 71.3 | 13.3 | In tube-gel | |
| P17931 | Galectin-3a | 7 | 255.85 | 34.8 | In solution | 8 | 179.9 | 28 | In tube-gel | |
| Q08380 | Galectin-3-binding proteina | 5 | 200.5 | 7.9 | In tube-gel | |||||
| Neuronal | ||||||||||
| P10645 | Chromogranin A | 2 | 72.94 | 8.5 | In tube-gel | |||||
| P55145 | mesencephalic astrocyte-derived neurotrophic factor | 2 | 83.7 | 16.8 | In tube-gel | |||||
| Protease inhibitors | ||||||||||
| Q6FGX5 | TIMP1a | 1 | 45.34 | 6.7 | In solution | 1 | 35.02 | 30 | In tube-gel | |
| P16035 | TIMP2 | 1 | 47.21 | 3.6 | In solution | 6 | 166.88 | 25.5 | In tube-gel | |
| Miscellaneous | ||||||||||
| P61769 | β2-microglobulina | 2 | 60.93 | 23.8 | In solution | 3 | 68.62 | 18.9 | In tube-gel | |
| B7Z351 | Osteopontin | 1 | 47.99 | 13.5 | In solution | 2 | 98.94 | 10.3 | In tube-gel | |
| Q6X4U4 | SOSTDC1 | 1 | 60.38 | 5.3 | In solution | |||||
| Q9H3U7 | SMOC2 | 1 | 35.42 | 1.8 | In tube-gel | |||||
| O94907 | DKK1 | 4 | 191.01 | 15.8 | In solution | 2 | 109.96 | 10.2 | In tube-gel |
a Proteins also identified by SDS-PAGE-in gel digestion.
Based on their biological activities, the secretory proteins were divided into nine groups, including IGF, TGFβ, FGF family proteins, other growth factors, cytokines, galectins, neuronal related proteins, protease inhibitors, and miscellaneous (Table I). We have previously used cytokine arrays to characterize the cytokines in the PCa-118b-CM (4), and some of the cytokines identified by cytokine arrays are also found by mass spectrometry. Three proteins, that is, CXCL1, MIF, and TIMP2, were detected by both mass spectrometry and cytokine antibody array. As mass spectrometry is not as sensitive as antibody array, these results suggest that CXCL1, MIF, and TIMP2 are present at higher abundance than the other cytokines in the PCa-118b-CM. On the other hand, BMP-4 was detected in the cytokine array but not by mass spectrometry. Together, the combined data provide a more complete list of secretory factors found in the PCa-118b secretome.
Expression of TGFβ and GDF Family Genes in PCa-118b Tumor Cells
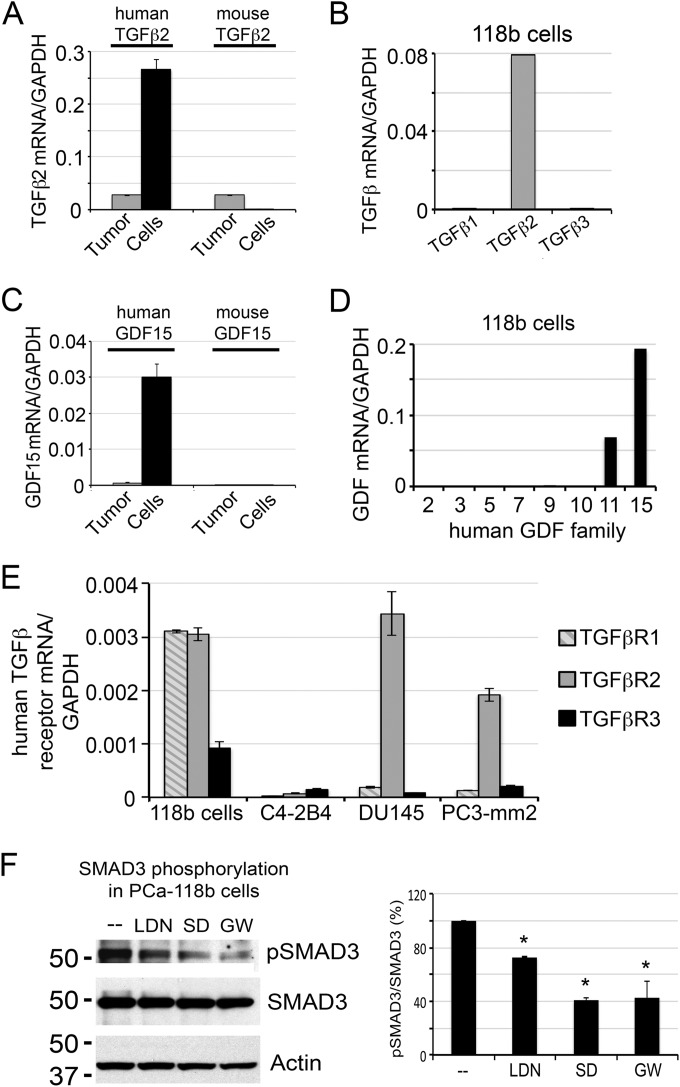
We first examined the TGFβ family proteins as they have been shown to have pleiotropic effects on multiple cell types. To distinguish whether TGFβ2 is expressed from PCa-118b tumor cells or tumor-associated stromal cells, oligonucleotides specific for human or mouse TGFβ family proteins were used in qRT-PCR of mRNA from PCa-118b tumor or isolated PCa-118b epithelial cells. Only human-specific oligonucleotides produced strong signals in isolated PCa-118b epithelial cells (Fig. 2A), indicating that TGFβ2 is mainly secreted from PCa-118b tumor cells. The TGFβ family contains three TGFβ isoforms. Consistent with the mass spectrometry data (Table I), qRT-PCR showed that only TGFβ2 is highly expressed, whereas messages for TGFβ1 and TGFβ3 were low to nondetectable in PCa-118b cells (Fig. 2B). These observations suggest that TGFβ2 is the major TGFβ family protein expressed in PCa-118b cells. Our results are similar to those observed by Dallas et al. (9), which showed that TGFβ2 is the major TGFβ isoform produced by prostate cancer.
Fig. 2.
TGFβ2 and GDF15 are expressed from tumor cells and function as autocrine factors. A, qRT-PCR for the expression of TGF-β2 in PCa-118b tumor and isolated PCa-118b cells using human- and mouse-specific primers showed that TGF-β2 is mainly expressed from PCa-118b cells. B, qRT-PCR for the expression of TGF-β1, β2, and β3 in isolated PCa-118b cells using human-specific primers showed that PCa-118b cells only expressed TGFβ2. C, qRT-PCR for the expression of GDF15 in PCa-118b tumor and isolated PCa-118b cells using human and mouse-specific primers showed that GDF15 is expressed from PCa-118b cells. D, qRT-PCR for the expression of GDF family genes detected only GDF15 and GDF11 messages in isolated PCa-118b cells. E, Expression of TGF-β receptors in isolated PCa-118b cells and C4–2B4, DU145, and PC3-mm2 PCa cell lines. F, Inhibition of Smad3 phosphorylation by LDN193189, an inhibitor of BMP type 1 receptors, and by TGFβ receptor inhibitors SD208 and GW788388 (left panel). The averages of pSMAD3 to total SMAD3 ratio from two experiments are shown (right panel). *, p < 0.05
GDF15 (also known as MIC-1) is one of the GDF family proteins, and a member of the TGFβ super family. Detection of GDF15 by mass spectrometry suggests that GDF15 is likely expressed in relatively high abundance. To examine whether the GDF15 detected in the PCa-118b-CM is from tumor cells or stromal cells, we used human and mouse-specific primers for GDF15 mRNA analysis. Human GDF primers detected high levels of GDF15 in the mRNA from enriched PCa-118b cells, but much less in RNA prepared from PCa-118b tumor (Fig. 2C). These results suggest that GDF15 is expressed mainly by the PCa-118b tumor cells. To distinguish which members of the GDF family are being expressed, we employed a GDF primer array (TaqMan Array human TGFB pathway) representing eight GDF family genes for qRT-PCR. This analysis detected GDF11 in addition to GDF15 in PCa-118b cells, with less GDF11 transcripts relative to those of GDF15 (Fig. 2D). The low levels of GDF11 transcripts may explain, in part, why we did not detect GDF11 protein in the mass spectrometry analysis. These observations indicate that GDF15 is the major GDF family protein expressed in PCa-118b tumor cells.
Expression of TGFβ Receptors in PCa-118b and Other PCa Cell Lines
Next, we examined whether TGFβ2 and GDF15 secreted from PCa-118b cells function as autocrine or paracrine factors in the PCa-118b tumor. Because secretory proteins function through cell surface receptors, we examined the expression of TGFβ receptors in PCa-118b epithelial and stromal cells. The receptors for TGFβ family proteins include TGFβR1, TGFβR2, and TGFβR3. However, the receptors for GDF family proteins are unknown. Analysis of TGFβ receptors using TaqMan PCR array showed that PCa-118b cells expressed transcripts for all three TGFβ receptors (data not shown). We further used qRT-PCR to examine the expression of TGFβR1, TGFβR2, and TGFβR3 on RNAs prepared from PCa-118b tumor cells as well as C4–2B4, DU145, and PC3-mm2 PCa cell lines. As shown in Fig. 2E, PCa-118b expressed RNA transcripts for all three TGFβ receptors, suggesting that TGFβ2 can function as an autocrine factor in PCa-118b cells through its TGFβ receptors. The expression of TGFβ receptors in other PCa cells was used as comparison.
To examine whether TGFβ2 functions as an autocrine factor in PCa-118b cells, we treated isolated PCa-118b cells with TGFβ receptor 1 inhibitors, SD208 (10–12) or GW788388 (13, 14), or BMP type 1 receptors ALK2 and ALK3 inhibitor, LDN-193189 (3, 15, 16), and examined their effects on TGFβ2 signaling via Smad3 phosphorylation. As shown in Fig. 2F, there were significant reductions (about 60%) in the level of Smad3 phosphorylation by SD208 or GW788388. Interestingly, LDN-193189 also led to a small but significant decrease (about 25%) of Smad3 phosphorylation (Fig. 2F). These observations indicate that TGFβ2 functions as an autocrine factor in PCa-118b cells.
Effect of TGFβ2, GDF15, and BMP4 on Endothelial Cells
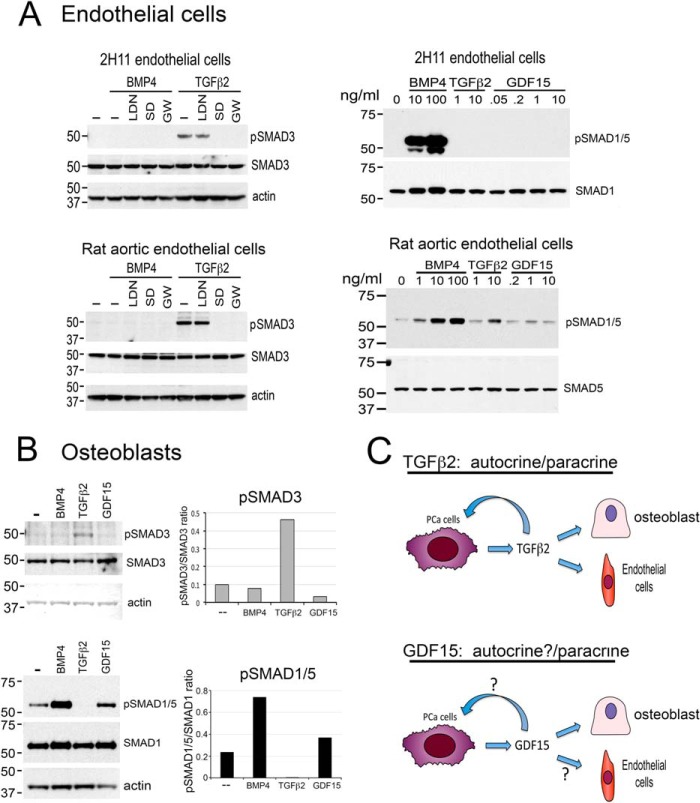
Next, we examined whether TGFβ2 and GDF15 function as paracrine factors on stromal cells present in the tumor microenvironment. Endothelial cells in the tumor stroma play a critical role in tumor angiogenesis. Two endothelial cell lines, including the mouse 2H11 cell line and the rat aortic endothelial (RAEC) cell line, were treated with TGFβ2 or GDF15 and cellular response via Smad3 phosphorylation was examined. As shown in Fig. 3A (left panels), treatment of 2H11 or RAEC cells with 10 ng/ml TGFβ2 stimulated Smad3 phosphorylation, and this effect was inhibited by SD208 (10–12) and GW788388 (13, 14), but not by LDN-193189 (3, 15, 16), indicating that TGFβ2 increases Smad3 phosphorylation through the TGFβR1 receptor. As expected, BMP4 did not stimulate Smad3 phosphorylation in 2H11 or RAEC cells (Fig. 3A). In contrast, no effect of GDF15 on SMAD3 phosphorylation was observed in either 2H11 or RAEC cells (data not shown). As the receptor and signaling pathways are not clear for GDF15, we further examined the effect of GDF15 on SMAD1/5 phosphorylation in 2H11 or RAEC cells. BMP4, which is known to stimulate SMAD1/5 phosphorylation, was used as a control. As shown in Fig. 3A (right panels), treatment of 2H11 or RAEC cells with various doses of GDF15 did not show a significant effect on SMAD1/5 phosphorylation. In contrast, BMP4 induces strong Smad1/5 phosphorylation in these cells and TGFβ2 stimulates SMAD1/5 phosphorylation in a dose-dependent manner in rat aortic endothelial cells. These results indicate that TGFβ2 functions as a paracrine factor on endothelial cells, but the effect of GDF15 on endothelial cells is not clear.
Fig. 3.
TGFβ2 and GDF15 exert paracrine effects by stimulating SMAD phosphorylation in endothelial cells and osteoblasts. A, Left panel, TGFβ2 stimulates SMAD3 phosphorylation in 2H11 endothelial cells and rat aortic endothelial cells. The signaling is inhibited by SD208 and GW788388, two TGFβ receptor 1 inhibitors, but not by LDN193189, an inhibitor of BMP type 1 receptors ALK2 and ALK3. Right panel, TGFβ2 stimulates SMAD1/5 phosphorylation in a dose-dependent manner in rat aortic endothelial cells, but not in 2H11 cells. GDF15 did not affect SMAD1/5 phosphorylation in both 2H11 and rat aortic endothelial cells. B, TGFβ2 stimulates SMAD3, but not SMAD1/5, phosphorylation in osteoblasts. GDF15 stimulates SMAD1/5, but not SMAD3, phosphorylation in osteoblasts. C, Role of TGFβ2 and GDF15 in PCa-118b tumor. Based on receptor expression and SMAD signaling, TGFβ2 functions as an autocrine as well as a paracrine factor in PCa-118b tumor. GDF15 can function as a paracrine factor on osteoblasts. Whether GDF15 can function as an autocrine factor is as yet unclear.
Effect of TGFβ2 and GDF15 on Osteoblasts
We further examined whether TGFβ2 and GDF15 can function as paracrine factors on osteoblasts present in the PCa-118b tumor microenvironment. Primary mouse osteoblasts isolated from newborn mouse calvaria were treated with or without 10 ng/ml TGFβ2 or GDF15 and their effects on SMAD3 or SMAD1/5 phosphorylation were measured. Treatment with BMP4 was used as a control. As shown in Fig. 3B (upper panel), TGFβ2 induced SMAD3 phosphorylation in primary osteoblasts, whereas GDF15 did not. In contrast, TGFβ2 did not affect SMAD1/5 phosphorylation, whereas GDF15 showed a slight, but detectable, increase in SMAD1/5 phosphorylation relative to controls in osteoblasts (Fig. 3B, lower panel). BMP4 treatment led to SMAD1/5 but not SMAD3 phosphorylation as expected. Together, these studies showed that TGFβ2 exhibits both autocrine and paracrine signalings on both tumor cells and stromal cells, whereas GDF15 may have paracrine signaling on osteoblasts (Fig. 3C). Wakchoure et al. (17) reported that GDF15/MIC-1 has a slight stimulatory effect on the early steps of osteoblast differentiation. It is possible that GDF15/MIC-1 signals through SMAD1/5 to stimulate osteoblast differentiation.
FGF3 and FGF19 Function as both Autocrine and Paracrine Factors
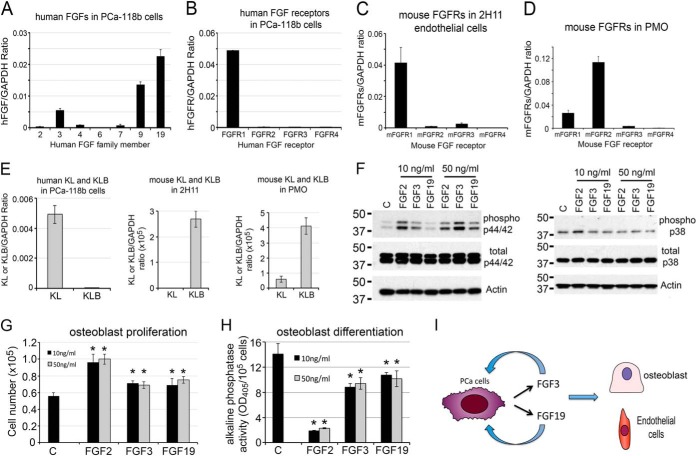
We next examined the FGF signaling axis. Among the 22 FGF family growth factors, FGF3 and FGF19 were the two members identified in the PCa118b-CM by mass spectrometry (Table I). FGF1–10, 16–18, 20, and 22 are classical FGFs, whereas FGF19, 21, and 23 are considered endocrine FGFs (18). Thus, PCa-118b expresses both classical and endocrine FGFs. qRT-PCR of RNA from PCa-118b detected the messages for FGF 3 and 19 (Fig. 4A), confirming the results from mass spectrometry analysis. qRT-PCR also detected the message for FGF9, which has been reported previously (3).
Fig. 4.
FGF3 and FGF19 function as both autocrine and paracrine factors in PCa-118b tumor. A, qRT-PCR for the expression of human FGF family genes detected FGF3, 9, and 19 messages in isolated PCa-118b cells. B, qRT-PCR for the expression of FGF receptors in PCa-118b cells showed that PCa-118b cells mainly expressed FGFR1. C, qRT-PCR using mouse-specific primers for the expression of FGF receptors in 2H11 endothelial cells showed that 2H11 cells expressed FGFR1. D, qRT-PCR using mouse-specific primers detected FGFR1 and FGFR2 in mouse osteoblasts. E, qRT-PCR for the expression of human KL and KLB in PCa-118b cells, mouse KL and KLB in 2H11 endothelial cells, and mouse KL and KLB in PMO. F, Effects of FGF3 and FGF19 on p44/42 and p38 phosphorylation. G, Stimulation of osteoblast proliferation by FGF3 and FGF19, as measured by increases in cell numbers over 3 days. *, p < 0.05. H, Effects of FGF2, FGF3, and FGF19 on osteoblast differentiation, as measured by alkaline phosphatase activity over 3 days. *, p < 0.05. I, FGF3 and FGF19 function as both autocrine and paracrine factors based on receptor expression and their effects on osteoblasts.
To determine whether FGFs function as an autocrine factor on PCa cells or paracrine factor on stromal cells, we examined the expression of FGF receptors in PCa cells and several stromal cell lines by qRT-PCR. There are four major FGF receptors, FGFR1 to FGFR4, which mediate classic FGF functions. The endocrine FGF19, on the other hand, requires α-Klotho and β-Klotho in addition to FGF receptors for their biological activity (18). To identify the cell type that expresses the receptors for FGF3, qRT-PCR using primers specific to human or mouse FGF receptors was performed. We found that FGFR1, but not FGFR2–4, is mainly expressed in PCa118b tumor cells (Fig. 4B). Previous studies by Yang et al. (19) have shown that FGFR1 is essential for PCa progression and metastasis in a mouse model of PCa. Thus, FGF3 likely regulates PCa-118b cell activity through FGFR1. In the stromal cells, mouse FGFR-specific primers detected FGFR1 as the dominant FGFR receptor in 2H11 endothelial cells (Fig. 4C), whereas FGFR2 is the major FGFR in primary osteoblasts (Fig. 4D). These results indicate that FGF3 secreted from PCa-118b cells likely mediates both autocrine and paracrine signaling activities on tumor cells and stromal cells.
Because FGF19 is an endocrine FGF and requires Klotho co-receptors for its function, qRT-PCR using primers specific to human or mouse α-Klotho and β-Klotho were performed to identify cells that may respond to FGF19. As shown in Fig. 4E, PCa-118b cells expressed mainly α-Klotho (KL), whereas mouse endothelial cells and osteoblasts expressed mainly β-Klotho (KLB). These results suggest that FGF19 may have effects on both PCa-118b cells and stromal cells. FGF family proteins were previously shown to stimulate the phosphorylation of p44/42 MAPK (20). To examine whether FGF3 and FGF19 have an effect on osteoblasts, primary mouse osteoblasts were treated with 10 ng/ml of FGF2, FGF3, or FGF19. As shown in Fig. 4F, FGF2 or FGF3 treatments led to an increase in the phosphorylation of p44/42 MAPK. FGF19, at a higher concentration (50 ng/ml), also stimulated p44/42 MAPK phosphorylation (Fig. 4F). In contrast, FGF2, 3, or 19 did not have a significant effect on p38 MAPK phosphorylation in mouse osteoblasts (Fig. 4F).
The functional roles of FGF19 in PCa cells have recently been reported (21). However, the roles of FGF3 and FGF19 on osteoblasts are not known. FGF2 and FGF9 have previously been shown to stimulate osteoblast proliferation (3, 4, 22). Thus, the effects of FGF3 and FGF19 on osteoblast proliferation were examined. FGF2 was used as a positive control. We found that FGF3 and FGF19 both stimulated osteoblast proliferation (Fig. 4G), albeit with weaker activity when compared with FGF2. FGF2 is known to increase osteoblast proliferation accompanied with inhibition of osteoblast differentiation, as reflected in a decrease in alkaline phosphatase activity (Fig. 4H). Similarly, both FGF3 and FGF19 decrease alkaline phosphatase activity (Fig 4H), but with much weaker activity when compared with FGF2. Together, these findings suggest that FGF3 and FGF19 secreted from PCa-118b cells exert both autocrine and paracrine effects on tumor cells and stromal cells (Fig. 4I).
CXCL1 Functions as a Paracrine Factor
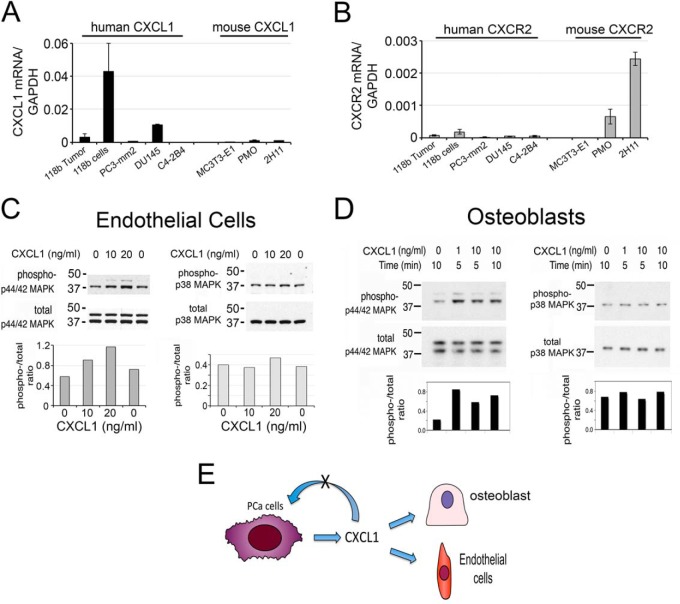
CXCL1 was detected in PCa-118b conditioned medium by mass spectrometry (Table I) as well as by cytokine array (4). Quantitative RT-PCR using oligonucleotide probes specific to human or mouse CXCL1 showed that CXCL1 is secreted from PCa-118b cells as only human-specific primers, but not mouse-specific primers, detected the CXCL1 message in PCa-118b cells (Fig. 5A). Interestingly, among several PCa cell lines, PCa-118b expressed the highest levels of CXCL1 compared with PC3-mm2 and DU145 cells (Fig. 5A).
Fig. 5.
CXCL1 secreted from PCa-118b cells functions as a paracrine factor on osteoblasts and endothelial cells. A, CXCL1 is expressed from PCa-118b tumor cells as well as DU145 cells. B, CXCR2, the CXCL1 receptor, is expressed in PMO and 2H11 endothelial cells, but not in human PCa cells. C, CXCL1 induces p44/42 but not p38MAPK phosphorylation in 2H11 endothelial cells. D, CXCL1 induces p44/42 but not p38MAPK phosphorylation in osteoblasts. E, CXCL1 functions as a paracrine factor based on receptor expression and p44/42 phosphorylation.
To determine whether CXCL1 functions as an autocrine factor on PCa cells or paracrine factor on stromal cells, we next examined the expression of the CXCL1 receptor, CXCR2, in PCa cells and several stromal cell lines by qRT-PCR. In contrast to the ligand CXCL1, qRT-PCR using human-specific CXCR2 primers did not detect the CXCR2 message in PCa-118b, PC3-mm2, DU145 and C4–2B4 cells (Fig. 5B). On the other hand, qRT-PCR using mouse-specific primers for CXCR2 showed that CXCR2 is expressed in primary mouse osteoblasts and immortalized mouse endothelial cells 2H11 (Fig. 5B). Because CXCL1 is secreted from tumor cells while its receptor CXCR2 is mainly present in stromal cells, these observations suggest that CXCL1 likely functions as a paracrine factor on tumor-associated stromal cells. Treatment of mouse endothelial cells 2H11 with CXCL1 showed a dose-dependent increase in the phosphorylation of p44/42 MAPK, but not p38 MAPK (Fig. 5C), suggesting that CXCL1 may play a role in tumor angiogenesis. This is consistent with a previous report by Moore et al. (23) that showed that CXCL1 played a role in mediating tumorigenicity of DU145 PCa cells through an induction of angiogenesis.
To examine whether CXCL1 has an effect on osteoblasts, primary mouse osteoblasts were treated with CXCL1. As shown in Fig. 5D, CXCL1 stimulated p42/44MAPK phosphorylation at either 1 or 10 ng/ml for 5 or 10 min in mouse osteoblasts. In contrast, CXCL1 did not have an effect on p38 phosphorylation in osteoblasts (Fig. 5D). These observations suggest that CXCL1 may elicit paracrine functions on osteoblasts. Previously studies have shown that CXCL1 enhanced osteoblast differentiation, but not proliferation (4). Together, based on receptor expression and cellular signaling, we establish that CXCL1 serves as a paracrine factor to modulate osteoblast and endothelial cell activity (Fig. 5E).
DISCUSSION
We have identified 26 secretory proteins in the secretome of the osteogenic PCa-118b tumor and examined five of these proteins for their expression, signal transduction, and function on multiple cell types within the tumor microenvironment. Our study demonstrated that the cross-talk between PCa cells and their microenvironment can be mediated via some of these secreted factors. We showed that FGF3, FGF19, and TGFβ2 exert both autocrine and paracrine effects on tumor cells and stromal cells, whereas GDF15 and CXCL1 have mainly paracrine effects on stromal cells. Thus, our secretome analysis reveals paracrine and autocrine networks involving multiple cell types present in the PCa-118b tumor microenvironment. Characterization of factors secreted from the tumor-associated stromal cells should provide information on how the stromal cells in turn influence the growth and survival of PCa cells. Based on the results from this study, we proposed a model for the multi-factor and multi-cell type tumor-microenvironment signaling networks in the PCa-118b tumor (Fig. 6). We envisage that human PCa-118b cells secrete multiple factors to modulate the activities of various stromal cells in bone, including endothelial cells, osteoblasts, and fibroblasts. Stromal cells in turn may secrete factors, which are yet to be identified, to further modulate the human PCa tumor cells. These observations may explain why single modality treatments in the clinical setting frequently produce modest effects in the control of disease progression in bone and suggest that therapy strategies that tackle both tumor cells and relevant stromal cells will be critical in achieving therapeutic effect.
Fig. 6.
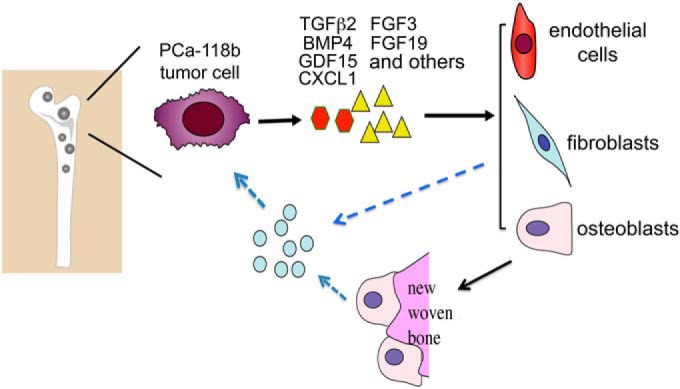
Model for the multi-factor and multi-cell type tumor-microenvironment signaling networks in PCa-118b tumor. We propose that human PCa-118b cells, which elicit strong osteoblastic foci formation in bone, secrete multiple factors to modulate the activities of stromal cells in the bone microenvironment, including endothelial cells, osteoblasts, and fibroblasts. Stromal cells in turn may secrete factors, which are yet to be identified, to further modulate the human PCa tumor cells.
A distinct feature of human PCa bone metastasis is a strong bone-forming phenotype and there is evidence that PCa-induced bone formation supports PCa progression in bone marrow (3, 4). Thus, there is a strong interest in identifying factors secreted by PCa cells that mediate the osteoblastic response. Our previous study has shown that PCa-118b secretes FGF9, which is involved in ectopic bone formation (3). In this study, we further showed that the PCa-118b cells also secretes FGF3 and endocrine FGF19 and both factors can stimulate osteoblast proliferation, although with a much weaker potency when compared with FGF2. Together, PCa-118b secretes multiple FGF family proteins that stimulate osteoblast proliferation. In addition to proliferation, an increase in osteoblast differentiation, a critical process leading to bone matrix secretion and mineralization, is also essential for bone formation. Using a cytokine array, we have previously shown that PCa-118b secretes BMP4, one of the most potent osteoblast differentiation factors (4). Together, the expression of FGF3, 9, 19, and BMP4 likely contribute to the strong osteoblastic phenotype observed in the PCa-118b tumors. Previous studies using inhibitors that block BMP signaling (4) or neutralizing antibody against FGF9 (3) have resulted in a reduction in osteogenesis in PCa-118b tumor. It is likely that targeting multiple FGF ligands and BMP4 activity are needed to inhibit PCa-118b tumor induced osteogenesis.
In addition to osteogenesis, the FGF/FGFR axis has been shown to play an important role in PCa initiation and progression. All four FGF receptors are expressed in human PCa (20). Yang et al. (19) showed that FGFR1 is essential for PCa progression and metastasis in a mouse model of PCa. Consistently, we found that PCa-118b mainly expresses FGFR1. Interestingly, we found that PCa-118b expresses the endocrine FGF19 and the α-Klotho (KL) co-receptor. As Feng et al. (21) showed that FGF19 promotes the growth, invasion, adhesion, and colony formation of PCa cells in vitro, we suggest that FGF19 plays an autocrine role in enhancing the tumorigenicity of PCa-118b cells. We also showed that the 2H11 endothelial cells and primary mouse osteoblasts express the β-Klotho (KLB) co-receptor, and addition of exogenous FGF19 to primary mouse osteoblasts increases osteoblast proliferation. Our observations raise the possibility that endocrine FGF19 exerts not only autocrine effects on tumor cells but also paracrine effects on the tumor microenvironment. Collectively, these results suggest that the FGF19/FGFR signaling axis plays an important role in epithelial-stromal interactions in PCa bone metastasis.
TGFβ family proteins are known to have pleotropic effects on multiple cell types. We found that the PCa-118b tumor secretes high levels of TGFβ2, which are able to activate TGFβ signaling pathways in endothelial cells and osteoblasts. Recent work of Carstens et al. (24) showed that the invasive prostate tumor was associated with the stroma that exhibited increased TGF-β signaling. In addition, they showed that elevated stromal TGF-β signaling, as reflected in Smad2+ nuclei, was associated with higher Gleason scores in human biopsies (24). Because TGFβ is one of the factors that can drive epithelial cells toward mesenchymal phenotype, we suggest that TGFβ2 secreted from PCa-118b cells functions as an autocrine factor that leads to epithelial-to-mesenchymal transition of tumor cells as well as a paracrine factor to modulate the properties of tumor-associated stromal cells including fibroblast, endothelial cells, and osteoblasts. Together, these activities contribute to the metastatic potential of PCa cells.
GDF15 is one of the TGFβ family proteins up-regulated during PCa progression and its expression has been correlated with bone metastasis (25). Although a significant increase in the serum levels of GDF15 has been observed in many cancer types, the role of GDF15 in cancer has been controversial (26, 27). In PCa, Senapati et al. (28) reported that overexpression of GDF15 induces metastasis of PC3 human PCa cells. In the TRAMP mouse model, Husaini et al. (29) reported that TRAMP mice with increased expression of GDF15 showed increased metastases, although the tumors exhibited a slower growth rate. In normal skeletal development, recombinant GDF15 has been shown to induce cartilage formation and promote early stage of endochondral bone formation (30). Wakchoure et al. (17) also showed that GDF15/MIC-1 has a slight stimulatory effect on the early steps of osteoblast differentiation. Consistently, we found that GDF15 treatment led to a small increase in SMAD1/5 phosphorylation in primary mouse osteoblasts. Together, these observations suggest that GDF15 may have effects on the metastatic and the osteogenic potential of PCa-118b cells.
CXCL1 has been known to promote prostate tumor growth through the stimulation of tumor angiogenesis (23). In this study, we also showed that CXCL1 stimulates Erk1/2 phosphorylation in osteoblasts, suggesting a role in osteoblast proliferation and/or differentiation. Our observations are consistent with the studies by Bischoff et al. (31), which showed that mCXCR(−/−) mice have decreased amounts of trabecular and cortical long bone, suggesting that CXCR2 plays a role in maintaining normal bone homeostasis. Previous studies have shown that the CXCL1/CXCR2 axis plays a major role in neutrophil function, and that CXCR2 is the major mediator of neutrophil migration to sites of inflammation (32). CXCL1 has also been shown to increase tumor growth by increasing pancreatic stromal fibroblast to express connective tissue growth factor (33). Together, CXCL1 may contribute to metastatic PCa progression in bone by affecting both tumor angiogenesis and the interactions between PCa cells and multiple cell types present in the bone microenvironment.
Although the secretome of the isolated human epithelial cells from patient-derived xenograft as described here provides important insights into the cross-talk between PCa cells and their microenvironment, the epithelial cells from a patient-derived xenograft may differ from those in human tumors. Therefore, in the future, it would be important to identify the secretome from freshly sorted human epithelial cells or after co-culturing them with murine stromal cells.
In our study, we used three approaches in sample preparation, that is, in-solution digestion, in tube-gel digestion, and SDS-PAGE-in gel digestion, for identifying proteins in PCa-118b conditioned medium by mass spectrometry. Our results showed that “in tube-gel digestion” generated the most peptides, whereas the “SDS-PAGE in gel digestion” generated the least peptides from mass spectrometry analysis. A total of 501 and 748 proteins were identified from “in-solution digestion” and “in tube-gel digestion,” respectively. Among the 26 secretory proteins, 14 were found in both “in-solution digestion” and “in tube-gel digestion” approaches (Table I). Seven of them were only found in “in solution digestion” and five were only found “in tube-gel digestion” (Table I). In “SDS-PAGE-in gel digestion” approach, only nine of the 26 secretory proteins were found (Table I). The possible explanation is that highly glycosylated proteins and proteins with hydrophobic domains may negatively influence trypsin digestion, resulting in their limited detection by subsequent mass spectrometric analysis (34). The “in tube-gel digestion” method was shown to increase the detection of hydrophobic proteins and proteins with a high degree of post-translational modification (7, 35, 36). The “SDS-PAGE-in gel digestion” method is limited by the amount of proteins that are loaded onto the gel. The low number of peptides identified by this method may be caused by detection sensitivity.
In conclusion, metastatic PCa cells are in close communication with their microenvironment that may support PCa cells' progression in the distant metastatic site. The identification of PCa-118b secreted factors within the secretome allows us to begin to dissect the contribution of each factor on PCa-118b tumorigenesis. Understanding the unique communication networks between PCa cells and the various components of the tumor microenvironment will allow us to design novel therapeutic strategies that target both the tumor cells and their interactions within the tumor microenvironment in bone.
Supplementary Material
Footnotes
Author contributions: Y.L. and S.L. designed research; Y.L., M.S.G., and D.J. performed research; Y.L., M.S.G., D.J., J.G.C., C.L., L.Y., G.E.G., S.N.M., and S.L. analyzed data; Y.L. and S.L. wrote the paper.
* This work was supported by Cancer Prevention and Research Institute of Texas (CPRIT RP110327), National Institutes of Health Grant RO1 174798, P50-CA140388, US Department of Defense Grant PC093132, and an award from the Prostate Cancer Foundation, a Cancer Center Support grant 2P30CA016647 from the National Cancer Institute, and the European Union FP7-PEOPLE-2012-IAPP- proposal No. 324400 - GlycoMet. This research is also in part conducted using the Rhode Island NSF/EPSCoR Proteomics Shared Resource Facility, which is supported in part by the National Science Foundation EPSCoR Grant No. 1004057, National Institutes of Health Grant No. 1S10RR020923, a Rhode Island Science and Technology Advisory Council grant, and the Division of Biology and Medicine, Brown University.
 This article contains supplemental Figs. S1 to S8 and Tables S1 to S7.
This article contains supplemental Figs. S1 to S8 and Tables S1 to S7.
1 The abbreviations used are:
- PCa
- prostate cancer
- CM
- conditioned medium.
REFERENCES
- 1. Logothetis C. J., Lin S. H. (2005) Osteoblasts in prostate cancer metastasis to bone. Nat. Rev. Cancer 5, 21–28 [DOI] [PubMed] [Google Scholar]
- 2. Hall C. L., Bafico A., Dai J., Aaronson S. A., Keller E. T. (2005) Prostate cancer cells promote osteoblastic bone metastases through Wnts. Cancer Res. 65, 7554–7560 [DOI] [PubMed] [Google Scholar]
- 3. Li Z. G., Mathew P., Yang J., Starbuck M. W., Zurita A. J., Liu J., Sikes C., Multani A. S., Efstathiou E., Lopez A., Wang J., Fanning T. V., Prieto V. G., Kundra V., Vazquez E. S., Troncoso P., Raymond A. K., Logothetis C. J., Lin S. H., Maity S., Navone N. M. (2008) Androgen receptor-negative human prostate cancer cells induce osteogenesis in mice through FGF9-mediated mechanisms. J. Clin. Invest. 118, 2697–2710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lee Y. C., Cheng C. J., Bilen M. A., Lu J. F., Satcher R. L., Yu-Lee L. Y., Gallick G. E., Maity S. N., Lin S. H. (2011) BMP4 promotes prostate tumor growth in bone through osteogenesis. Cancer Res. 71, 5194–5203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lee S. H., Kunz J., Lin S. H., Yu-Lee L. Y. (2007) 16-kDa prolactin inhibits endothelial cell migration by down-regulating the Ras-Tiam1-Rac1-Pak1 signaling pathway. Cancer Res. 67, 11045–11053 [DOI] [PubMed] [Google Scholar]
- 6. Lin S. H., Cheng C. J., Lee Y. C., Ye X., Tsai W. W., Kim J., Pasqualini R., Arap W., Navone N. M., Tu S. M., Hu M., Yu-Lee L. Y., Logothetis C. J. (2008) A 45-kDa ErbB3 secreted by prostate cancer cells promotes bone formation. Oncogene 27, 5195–5203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cao L., Clifton J. G., Reutter W., Josic D. (2013) Mass spectrometry-based analysis of rat liver and hepatocellular carcinoma Morris hepatoma 7777 plasma membrane proteome. Anal. Chem. 85, 8112–8120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Elias J. E., Gygi S. P. (2007) Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 4, 207–214 [DOI] [PubMed] [Google Scholar]
- 9. Dallas S. L., Zhao S., Cramer S. D., Chen Z., Peehl D. M., Bonewald L. F. (2005) Preferential production of latent transforming growth factor beta-2 by primary prostatic epithelial cells and its activation by prostate-specific antigen. J. Cell Physiol. 202, 361–370 [DOI] [PubMed] [Google Scholar]
- 10. Uhl M., Aulwurm S., Wischhusen J., Weiler M., Ma J. Y., Almirez R., Mangadu R., Liu Y. W., Platten M., Herrlinger U., Murphy A., Wong D. H., Wick W., Higgins L. S., Weller M. (2004) SD-208, a novel transforming growth factor beta receptor I kinase inhibitor, inhibits growth and invasiveness and enhances immunogenicity of murine and human glioma cells in vitro and in vivo. Cancer Res. 64, 7954–7961 [DOI] [PubMed] [Google Scholar]
- 11. Zhou L., Nguyen A. N., Sohal D., Ying Ma J., Pahanish P., Gundabolu K., Hayman J., Chubak A., Mo Y., Bhagat T. D., Das B., Kapoun A. M., Navas T. A., Parmar S., Kambhampati S., Pellagatti A., Braunchweig I., Zhang Y., Wickrema A., Medicherla S., Boultwood J., Platanias L. C., Higgins L. S., List A. F., Bitzer M., Verma A. (2008) Inhibition of the TGF-beta receptor I kinase promotes hematopoiesis in MDS. Blood 112, 3434–3443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ge R., Rajeev V., Ray P., Lattime E., Rittling S., Medicherla S., Protter A., Murphy A., Chakravarty J., Dugar S., Schreiner G., Barnard N., Reiss M. (2006) Inhibition of growth and metastasis of mouse mammary carcinoma by selective inhibitor of transforming growth factor-beta type I receptor kinase in vivo. Clin. Cancer Res. 12, 4315–4330 [DOI] [PubMed] [Google Scholar]
- 13. Petersen M., Thorikay M., Deckers M., van Dinther M., Grygielko E. T., Gellibert F., de Gouville A. C., Huet S., ten Dijke P., Laping N. J. (2008) Oral administration of GW788388, an inhibitor of TGF-beta type I and II receptor kinases, decreases renal fibrosis. Kidney Int. 73, 705–715 [DOI] [PubMed] [Google Scholar]
- 14. Gellibert F., de Gouville A. C., Woolven J., Mathews N., Nguyen V. L., Bertho-Ruault C., Patikis A., Grygielko E. T., Laping N. J., Huet S. (2006) Discovery of 4-{4-[3-(pyridin-2-yl)-1H-pyrazol-4-yl]pyridin-2-yl}-N-(tetrahydro-2H- pyran-4-yl)benzamide (GW788388): a potent, selective, and orally active transforming growth factor-beta type I receptor inhibitor. J. Med. Chem. 49, 2210–2221 [DOI] [PubMed] [Google Scholar]
- 15. Cuny G. D., Yu P. B., Laha J. K., Xing X., Liu J. F., Lai C. S., Deng D. Y., Sachidanandan C., Bloch K. D., Peterson R. T. (2008) Structure-activity relationship study of bone morphogenetic protein (BMP) signaling inhibitors. Bioorg. Med. Chem. Lett. 18, 4388–4392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yu P. B., Deng D. Y., Lai C. S., Hong C. C., Cuny G. D., Bouxsein M. L., Hong D. W., McManus P. M., Katagiri T., Sachidanandan C., Kamiya N., Fukuda T., Mishina Y., Peterson R. T., Bloch K. D. (2008) BMP type I receptor inhibition reduces heterotopic [corrected] ossification. Nat. Med. 14, 1363–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wakchoure S., Swain T. M., Hentunen T. A., Bauskin A. R., Brown D. A., Breit S. N., Vuopala K. S., Harris K. W., Selander K. S. (2009) Expression of macrophage inhibitory cytokine-1 in prostate cancer bone metastases induces osteoclast activation and weight loss. Prostate 69, 652–661 [DOI] [PubMed] [Google Scholar]
- 18. Itoh N., Ornitz D. M. (2011) Fibroblast growth factors: from molecular evolution to roles in development, metabolism and disease. J. Biochem. 149, 121–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yang F., Zhang Y., Ressler S. J., Ittmann M. M., Ayala G. E., Dang T. D., Wang F., Rowley D. R. (2013) FGFR1 is essential for prostate cancer progression and metastasis. Cancer Res. 73, 3716–3724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Feng S., Shao L., Yu W., Gavine P., Ittmann M. (2012) Targeting fibroblast growth factor receptor signaling inhibits prostate cancer progression. Clin. Cancer Res. 18, 3880–3888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Feng S., Dakhova O., Creighton C. J., Ittmann M. (2013) Endocrine fibroblast growth factor FGF19 promotes prostate cancer progression. Cancer Res. 73, 2551–2562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lee Y. C., Huang C. F., Murshed M., Chu K., Araujo J. C., Ye X., deCrombrugghe B., Yu-Lee L. Y., Gallick G. E., Lin S. H. (2010) Src family kinase/abl inhibitor dasatinib suppresses proliferation and enhances differentiation of osteoblasts. Oncogene 29, 3196–3207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Moore B. B., Arenberg D. A., Stoy K., Morgan T., Addison C. L., Morris S. B., Glass M., Wilke C., Xue Y. Y., Sitterding S., Kunkel S. L., Burdick M. D., Strieter R. M. (1999) Distinct CXC chemokines mediate tumorigenicity of prostate cancer cells. Am. J. Pathol. 154, 1503–1512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Carstens J. L., Shahi P., Van Tsang S., Smith B., Creighton C. J., Zhang Y., Seamans A., Seethammagari M., Vedula I., Levitt J. M., Ittmann M. M., Rowley D. R., Spencer D. M. (2014) FGFR1-WNT-TGF-beta signaling in prostate cancer mouse models recapitulates human reactive stroma. Cancer Res. 74, 609–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Selander K. S., Brown D. A., Sequeiros G. B., Hunter M., Desmond R., Parpala T., Risteli J., Breit S. N., Jukkola-Vuorinen A. (2007) Serum macrophage inhibitory cytokine-1 concentrations correlate with the presence of prostate cancer bone metastases. Cancer Epidemiol. Biomarkers Prev. 16, 532–537 [DOI] [PubMed] [Google Scholar]
- 26. Mimeault M., Batra S. K. (2010) Divergent molecular mechanisms underlying the pleiotropic functions of macrophage inhibitory cytokine-1 in cancer. J. Cell. Physiol. 224, 626–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Vanhara P., Hampl A., Kozubik A., Soucek K. (2012) Growth/differentiation factor-15: prostate cancer suppressor or promoter? Prostate Cancer Prostatic Dis. 15, 320–328 [DOI] [PubMed] [Google Scholar]
- 28. Senapati S., Rachagani S., Chaudhary K., Johansson S. L., Singh R. K., Batra S. K. (2010) Overexpression of macrophage inhibitory cytokine-1 induces metastasis of human prostate cancer cells through the FAK-RhoA signaling pathway. Oncogene 29, 1293–1302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Husaini Y., Qiu M. R., Lockwood G. P., Luo X. W., Shang P., Kuffner T., Tsai V. W., Jiang L., Russell P. J., Brown D. A., Breit S. N. (2012) Macrophage inhibitory cytokine-1 (MIC-1/GDF15) slows cancer development but increases metastases in TRAMP prostate cancer prone mice. PloS One 7, e43833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Paralkar V. M., Vail A. L., Grasser W. A., Brown T. A., Xu H., Vukicevic S., Ke H. Z., Qi H., Owen T. A., Thompson D. D. (1998) Cloning and characterization of a novel member of the transforming growth factor-beta/bone morphogenetic protein family. J. Biol. Chem. 273, 13760–13767 [DOI] [PubMed] [Google Scholar]
- 31. Bischoff D. S., Sakamoto T., Ishida K., Makhijani N. S., Gruber H. E., Yamaguchi D. T. (2011) CXC receptor knockout mice: characterization of skeletal features and membranous bone healing in the adult mouse. Bone 48, 267–274 [DOI] [PubMed] [Google Scholar]
- 32. Cacalano G., Lee J., Kikly K., Ryan A. M., Pitts-Meek S., Hultgren B., Wood W. I., Moore M. W. (1994) Neutrophil and B cell expansion in mice that lack the murine IL-8 receptor homolog. Science 265, 682–684 [DOI] [PubMed] [Google Scholar]
- 33. Ijichi H., Chytil A., Gorska A. E., Aakre M. E., Bierie B., Tada M., Mohri D., Miyabayashi K., Asaoka Y., Maeda S., Ikenoue T., Tateishi K., Wright C. V., Koike K., Omata M., Moses H. L. (2011) Inhibiting Cxcr2 disrupts tumor-stromal interactions and improves survival in a mouse model of pancreatic ductal adenocarcinoma. J. Clin. Invest. 121, 4106–4117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Speers A. E., Wu C. C. (2007) Proteomics of integral membrane proteins–theory and application. Chem. Rev. 107, 3687–3714 [DOI] [PubMed] [Google Scholar]
- 35. Cao R., He Q., Zhou J., He Q., Liu Z., Wang X., Chen P., Xie J., Liang S. (2008) High-throughput analysis of rat liver plasma membrane proteome by a nonelectrophoretic in-gel tryptic digestion coupled with mass spectrometry identification. J. Proteome Res. 7, 535–545 [DOI] [PubMed] [Google Scholar]
- 36. Lu X., Zhu H. (2005) Tube-gel digestion: a novel proteomic approach for high throughput analysis of membrane proteins. Mol. Cell. Proteomics 4, 1948–1958 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.