Abstract
Macrophage multinucleation (MM) is essential for various biological processes such as osteoclast-mediated bone resorption and multinucleated giant cell-associated inflammatory reactions. Here we study the molecular pathways underlying multinucleation in the rat through an integrative approach combining MS-based quantitative phosphoproteomics (LC-MS/MS) and transcriptome (high-throughput RNA-sequencing) to identify new regulators of MM. We show that a strong metabolic shift toward HIF1-mediated glycolysis occurs at transcriptomic level during MM, together with modifications in phosphorylation of over 50 proteins including several ARF GTPase activators and polyphosphate inositol phosphatases. We use shortest-path analysis to link differential phosphorylation with the transcriptomic reprogramming of macrophages and identify LRRFIP1, SMARCA4, and DNMT1 as novel regulators of MM. We experimentally validate these predictions by showing that knock-down of these latter reduce macrophage multinucleation. These results provide a new framework for the combined analysis of transcriptional and post-translational changes during macrophage multinucleation, prioritizing essential genes, and revealing the sequential events leading to the multinucleation of macrophages.
Macrophage multinucleation (MM)1 occurs when macrophages fuse to form a single cell containing multiple nuclei. This phenomenon plays an essential role in several homeostatic and pathological processes. In healthy subjects, macrophage multinucleation is required for the formation of osteoclasts able to resorb bone excesses and maintain bone homeostasis (1, 2). Macrophage multinucleation also plays a major role in inflammatory conditions such as sarcoidosis or tuberculosis through the formation of multinucleated giant cells (MGCs), effector cells of granulomatous reaction (3, 4).
Despite the central role of MM in various biological mechanisms, the basic knowledge of its molecular determinants and the pathways regulating multinucleation remain largely unexplored. Moreover the presence of common regulators of both MGC and osteoclast formation suggests the existence of common mechanisms for macrophage multinucleation across different cell types (5–8). Cell multinucleation was previously investigated at a transcriptional level in osteoclasts (9) and fusing rat alveolar macrophages (10). This led to the identification of novel determinants of osteoclast multinucleation (11) and macrophage fusion (10).
The Wistar-Kyoto (WKY) strain has been extensively used for its unique susceptibility to nephrotoxic nephritis (NTN), a highly reproducible and macrophage-dependent model of experimentally induced crescentic glomerulonephritis that has a large genetic component (12–14). The NTN in the WKY rat is also characterized by the formation of glomerular MGCs but the specific role of these cells in the progression of the disease remains to be determined. We observed that when the bone marrow derived macrophages (BMDMs) from the NTN-susceptible WKY rats are cultured in vitro under normal conditions, they spontaneously form MGCs after 3 days as opposed to NTN-resistant Lewis (LEW) BMDMs, which show very little fusion (Fig. 1A). We used this in vitro model of spontaneous multinucleation of BMDMs to identify a new major determinant of spontaneous macrophage multinucleation in a back-cross population derived from WKY and LEW rats (15) as well as recapitulating previously known genes (DCSTAMP (8), MMP9 (16)) as part of a macrophage gene regulatory network.
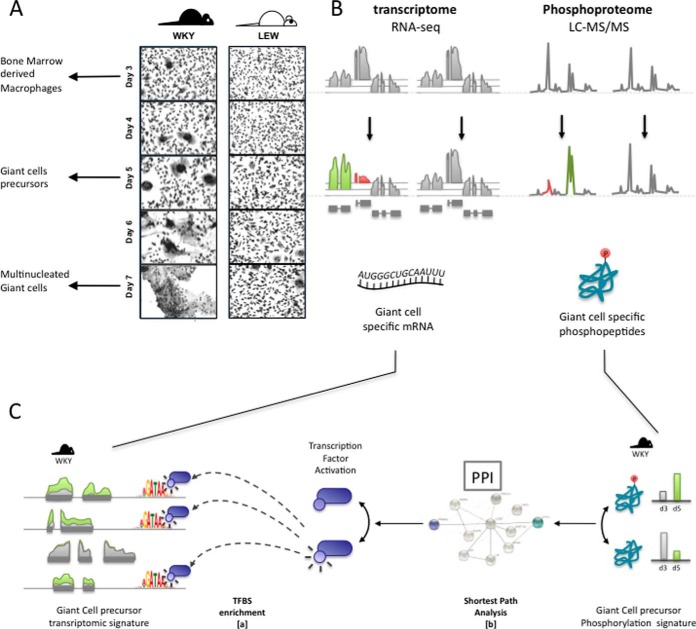
Fig. 1.
Analysis pipeline. Primary macrophages from WKY and LEW are derived from the bone marrow after 3 day of culture and show a marked phenotypic difference in multinucleation and MGC formation, A. To understand the determinants of spontaneous macrophage multinucleation, we measure the macrophage transcriptome and phospho-proteome at day 3 (mononuclear and differentiated BMDMs) and in MGC precursors (day 5). MGC precursor-specific molecular signatures are next identified by determining transcriptional changes unique to WKY day 5 BMDMs B, and by characterizing multinucleation-specific transcription factors through transcription factor binding site enrichment analysis (C, step a). Finally, multinucleation-specific transcription factors and phosphopeptides are integrated by identifying pairs in closer than random vicinity in the protein interaction network (C, step b).
Here, we developed an integrative approach to identify and characterize pathways involved in macrophage multinucleation by taking advantage of strain dependent spontaneous formation of MGCs during macrophage differentiation. We combined genome-wide and label-free phosphoproteome by LC-MS/MS and transcriptome by RNA-sequencing (RNA-seq) to pinpoint the cellular pathways as well as key regulators of transcriptomic changes occurring during macrophage multinucleation. We provide a first account of the interplay between post-translational and transcriptomic modifications occurring during macrophage multinucleation and MGC formation.
EXPERIMENTAL PROCEDURES
RNA Isolation and RNA-seq Library Preparation in Primary Macrophages
Total RNA was extracted from WKY and LEW bone-marrow derived macrophages at the indicated time points during differentiation (day 3 and day 5) using Trizol (Invitrogen) according to manufacturer's instructions with an additional purification step by on-column DNase treatment using the RNase-free DNase Kit (Qiagen, UK) to ensure elimination of any genomic DNA. The integrity and quantity of total RNA was determined using a NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific) and Agilent 2100 Bioanalyzer (Agilent Technologies, UK). One microgram of total RNA was used to generate RNA-seq libraries using TruSeq RNA sample preparation kit (Illumina, UK) according to the manufacturer's instructions. Briefly, RNA was purified and fragmented using poly-T oligo-attached magnetic beads using two rounds of purification followed by the first and second cDNA strand synthesis. Next, cDNA 3′ ends were adenylated and adapters ligated followed by 10 cycles of library amplification. Finally, the libraries were size selected using AMPue XP Beads (Beckman Coulter, UK) purified and their quality was checked using Agilent 2100 Bioanalyzer. Samples were randomized to avoid batch effects and libraries were run on a single lane per sample of the HiSeq 2000 platform (Illumina) to generate 100 bp paired-end reads. An average of 72 m reads coverage per sample was achieved (minimum 38 m).
RNA-seq Data Analysis
RNA-seq reads were aligned to the rat (rn4) reference genome using tophat 2. The average number of mapped was 67 m (minimum 36 m) corresponding to an average mapping percentage of 93%. Sequencing and mapping were quality controlled using standard tools provided in the fastQC software. Gene level read counts were computed using HT-Seq-count with “union” mode (17) and genes with less than 10 aligned reads across all samples were discarded prior to analysis leading to 15,155 genes. Clustering of samples corresponding to different conditions was done using Ward's methods based on Euclidian distance of scaled sample gene expression profile. Differential expression analyses between groups were conducted using edgeR (18) within each strain and time point using a 5% FDR for each comparison. In order to identify significant differences in gene expression variation across time-points between the WKY and LEW BMDMs, a generalized linear model was fitted on all samples including strain, time point, and the [(strain) x (time point)] interaction term. The potential interaction was then tested using a Likelihood Ratio test (comparing to a model without the interaction term).
The giant cell specific signature was defined as the set of genes that fulfilled the following criteria:
Significant interaction between strain and time point at 5% FDR;
Over-expression at day 5 compared with day 3 in WKY at 5% FDR;
No differential expression at 5% nominal p value in LEW between day 3 and day 5;
Over-expression at day 5 in WKY compared with LEW at 5% FDR.
Quantification of Macrophage Multinucleation, RNAi, and qRT-PCR
Rat bone marrow derived macrophages (BMDMs) were cultured as previously described (14). BMDMs were flushed from femur and tibia bones from rats and cultured in presence of L929-conditioned media for the indicated times in Lab-Tek chambers (Fisher Scientific, UK). To assess spontaneous MGC formation, macrophages were fixed at the indicated times using Reastain Quick Diff and MGC quantification was performed by counting the number of nuclei in 100 macrophages using light microscopy. To evaluate the effect of siRNA knockdown on macrophage multinucleation, WKY BMDMs were transfected for 48 h with siGENOME SMARTpool for either rat Lrrfip1 or Smarca4 or Dnmt1 (100 nm, GE Healthcare, UK). All transfections were performed in parallel with siGENOME nontargeting siRNA pool as the scrambled control siRNA using Dharmafect 1 (1:50, Dharmacon) as a transfection reagent in OPTIMEM medium (Invitrogen). The siRNA sequences used in the siGENOME SMARTpool for all transcripts are available upon request. All qRT-PCRs were performed with a Viaa 7 Real-Time PCR system (Life Technologies, UK). A two-step protocol was used beginning with cDNA synthesis with iScript select (Bio-Rad, UK) followed by PCR using SYBR Green Jumpstart Taq Ready Mix (Sigma). A total of 10 ng of cDNA per sample was used. All samples were amplified using a set of at least four biological replicates with three technical replicates used per sample in the PCR. Viia 7 RUO Software was used for the determination of Ct values. Results were analyzed using the comparative Ct method and each sample was normalized to the reference gene Hprt, to account for any cDNA loading differences. The primer sequences are available upon request.
Macrophage Phosphoproteome and Proteome: Cell Lysis, Protein Digestion Peptide, and Phosphopeptide Extraction
Cells in the flasks were washed twice with cold PBS supplemented with phosphatase inhibitors (1 mm Na3VO4 and 1 mm NaF) and lyzed in urea lysis buffer (8 m Urea in 20 mm HEPES, pH 8.0) supplemented with phosphatase inhibitors (1 mm Na3VO4; 1 mm NaF; 1 mm β-glycerol-phosphate; 2.5 mm Na4P2O7; and 1 μm okadaic acid). Cell extracts were then scraped from the flasks, transfer to 2 ml low protein binding eppendorf tubes and further homogenized by sonication (three pulses of 15 s). Insoluble material was removed by centrifugation at 20,000 × g for 5 min at 4 °C and proteins in the supernatants were quantified by Bradford. A total amount of 500 μg and 200 μg of protein/sample were reduced and alkylated by sequential incubation with 10 mm DTT and 10 mm Iodoacetamide for 15 min at room temperature for phosphopeptides and whole peptides, respectively. 20 mm HEPES, pH 8.0, was used to reduce urea concentration to 2 m and proteins were digested overnight at 37 °C with Immobilized tosyl-lysine chloromethyl ketone (TLCK)–trypsin [20 p-toluenesulfonyl-l-arginine methyl ester (TAME) units/mg]. Digestion was stopped by adding trifluoroacetic acid (TFA) to 1% final concentration and trypsin beads were removed by centrifugation. Peptide mixtures were then desalted by reverse phase chromatographic cartridges (Oasis HLB, Waters, Milford, MA), washed, and dried.
For quantitative phosphoproteomics, phosphopeptides were enriched following a procedure previously described, with some modifications (19). Briefly, sample volumes after elution from Oasis cartridges were adjusted to 1 ml with glycolic acid solution (1 m Glycolic acid; 80% ACN; and 5% TFA) and 50 μl of TiO2 beads (50% slurry in 1% TFA) were added to the peptide mixture. After 5 min incubation at RT with rotation, TiO2 slurry was packed into preconditioned empty spin tips by centrifugation. Spin tips were conditioned with 200 μl of 100% ACN. TiO2 beads were sequentially washed with 200 μl of glycolic acid solution, 100 mm Ammonium acetate in 25% ACN, and 1% ACN. Phosphopeptides were then eluted four times with 50 μl of 5% NH4OH in 1% ACN. Following the removal of the insoluble material by centrifugation from the eluents, supernatants were snap frozen in dry ice for 15 min. Samples were dried in a speed-vac and stored at −80 °C for mass spectrometry. For quantitative proteomics, mass spectrometry was applied without TiO2 enrichment only on WKY and LEW BMDM whole peptide extracts (analyzed in technical duplicates) at the peak of multinucleation. For quantitative phosphoproteomics, each of the four samples were provided in biological triplicate (in single technical replicate) for mass spectrometry analysis.
Mass Spectrometry
Phosphopeptide and whole peptide dried extracts were resuspended in 14 μl of reconstitution buffer (0.1% TFA containing 20 nm of an enolase digest) and 4 μl (phosphopeptides) and 5 μl (peptides) were loaded in a LC-MS/MS system. This consists of a nanoflow liquid chromatography (nanoLC, Ultimate 3000, Thermo Scientific) system coupled online to an LTQ-Orbitrap Velos mass spectrometer (Thermo Scientific). The nanoLC system delivered a flow of 8 μl/min (loading) onto a trap column (Thermo Scientific Acclaim Pepmap 100 with dimensions 100 μm internal diameter and 2 cm length, C18 reverse phase material with 5 μm diameter beads and 100Å pore size) in 98% water, 2% acetonitrile, and 0.1% TFA. Peptides were then eluted on-line to an analytical column (Thermo Scientific Acclaim Pepmap RSLC with dimensions 75 μm internal diameter and 25 cm length, C18 reverse phase material with 2 μm diameter beads and 100Å pore size) and separated using a gradient with conditions: initial 5 min with 4% B (96% A), then 120 min gradient 4–35% B, then 10 min isocratic at 100% B, then 5 min isocratic at 4% B (solvent A: 98% water, 2% acetonitrile, and 0.1% formic acid; solvent B: 20% water, 80% acetonitrile, and 0.1% formic acid).
For phosphopeptide quantification, a LTQ-Orbitrap Velos mass spectrometer acquired full scan survey spectra (m/z 350–1500) with a 15,000 resolution at m/z 400. A maximum of the seven (phosphopeptides) most abundant multiply-charged ions registered in each survey spectrum were selected in a data-dependent manner, fragmented by collision induced dissociation (multi-stage activation enabled) with a normalized collision energy of 35% and scan in the LTQ (m/z 50–2000). This produced a duty cycle of 2.4 s. A dynamic exclusion was enabled with the exclusion list restricted to 500 entries, exclusion duration of 60 s and mass window of 10 ppm. Chromatographic peaks were about 30 s at the base, which ensured at least 10 data points per extracted ion chromatogram (XIC).
For whole protein quantification, a Q Exactive mass spectrometer acquired full scan survey spectra (m/z 400–2000) with 70,000 resolution at m/z 400 was used. A maximum of the 12 most abundant multiply-charged ions registered in each survey spectrum were selected in a data-dependent manner, fragmented by higher-energy collision induced dissociation (HCD) with a normalized collision energy of 28%. This produced a duty cycle of ∼1.5 s. MS/MS resolution was 17,500.
Phosphopeptide and Peptide (Protein) Identification
For phosphopeptide identification, Mascot Distiller v2.4.3.1 was used to smoothen and centroid the MS/MS data and Mascot v2.4.1 search engine was used to match peaks to peptides in proteins present in the SwissProt Database (SwissProt_2013Jan.fasta) restricted to rattus norvegicus entries (7,853 sequences) (20). The process was automated with Mascot Daemon v2.4, mass tolerance was set to 10 ppm and 600 millimass units for precursor and fragment ions, respectively. Phosphorylation on Ser, Thr, and Tyr; PyroGlu on N-terminal Glu; and oxidation of Met were allowed in the search as variable modifications and carbamidomethyl Cys as fixed modification. Trypsin (cleaves C-terminal to Arg and Lys residues provided there is not a Pro C-term to the Arg/Lys residue) was selected as the digestion enzyme and two missed cleavages were allowed. Sites of modification are reported when they had delta scores >10. Delta scores were calculated as previously described (21). Otherwise the site of modification was deemed to be ambiguous; in such cases phosphopeptides are reported as the start-end residues within the protein sequence. Results from Mascot searches were deposited into the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository (22) with the dataset identifier Project accession PXD001269 and Project DOI: 10.6019/PXD001269. Phosphopeptides showing a Mascot expectancy of <0.05 (∼2% false discovery rate as assessed by searches against a decoy database) were placed in a database of peptides quantifiable by MS. In cases where a given peptide sequence matched more than one database entry, all such entries were listed in the final result table (supplemental Table S1). Pescal software (19, 23) was used to construct XICs of peptides included in the database across all the samples being compared. Enolase peptides spiked in all samples were used as reference points along chromatograms to align retention times. The XIC windows were 7 ppm and 2 min.
For quantitative proteomics, raw data files were uploaded onto Progenesis QI for Proteomics software (Nonlinear Dynamics, 2014, version: 2.0.5387.52102). Chromatographic alignment (with additional manual manipulation), data normalization, and peak picking were performed by Progenesis QI. Mascot server (version 2.5.0) was used for peptide/protein identification as searched against the Uniprot Swissprot rattus norvegicus FASTA (downloaded June 6th 2014) which contained 7914 sequences. Mass tolerance was set to 5 ppm and 25 millimass units for precursor and fragment ions, respectively. Deamidation of Asn and Gln, PyroGlu on N-terminal Glu, and oxidation of Met were allowed in the search as variable modifications and carbamidomethyl on Cys as fixed modification. Trypsin was selected as the digestion enzyme and two missed cleavages were permitted. All peptides used in quantification were exported from Mascot at identity threshold with p value 0.05. The overall search FDR was 1.3%.
Differential Phosphorylation Analysis
Peak height intensity XIC values were log transformed and normalized to using PQN normalization (24). For each pair of condition, differential expression was assessed both quantitatively and qualitatively. Only peptides with at least two observations in each condition were considered for the quantitative analysis. Quantitative assessment of differential phosphorylation was assessed using a slightly modified version of the framework proposed in (25). Briefly, in keeping with (25), we assume that peptide expression is missing with a probability that depends on both the true expression of the peptide (data is missing when the true expression is under a peptide-dependent threshold ci) and the sample specific probability of missing data (sample dependent-probability πl to be missing at random). Similarly to (25), the censoring threshold ci is set for each peptide at the minimal observed a value across all samples (Maximum likelihood estimator) and the πl are estimated by modeling the occurrence of missing values as a function of the mean expression of the peptides. However, opposite to Karpievitch, for each individual l we model the probability of not observing a peptide as a parametric function of the mean μ of the peptide in all samples from the same condition:
where πl represents the probability of missingness at random (probability of missing expression measurement for a peptide with infinitely high expression). We then estimate the parameters πl ∈[0,1], al and bl>0 via likelihood maximization. The use of a parametric fit to estimate the probability of missing values rather than the nonparametric estimate proposed by Karpievitch et al. is justified here by the fact the nonparametric estimate led to unrealistic values (πl < 0) because of the unconstrained interpolation that was performed. Our estimate on the other hand, enforces strictly positive values of πl thus allowing more reliable estimates of the missing-at-random probability πl.
p values resulting from the quantitative models were adjusted for multiple testing using Benjamini-Hochberg correction and a 5% FDR threshold as used for significance.
Differential Protein Abundance
For each biological replicate, values of protein abundance exported from Mascot were averaged between technical replicates to yield the final measures of protein abundance. Protein abundances were then log transformed (adding an offset of 1 to avoid infinite values) and samples intensities were scaled to have identical total intensity. For each protein, differential protein abundance between WKY and LEW was assessed using a t test.
Giant-cell Specific Phosphorylation Signature
Giant-cell specific phosphorylation signature was defined based on peptides that were detected in at least two samples in all conditions. In order to improve power of the enrichment analysis (see below) we used nonconservative statistical thresholds to identify giant-cell specific phosphorylation signature. Although this approach might yield increased false positives, this will not bias the subsequent enrichment analysis because false positives will by definition occur at random. In detail, phosphopeptides were considered as exhibiting giant-cell specific signature if they fulfilled any of the two following set of criteria (see also supplemental Fig. S4):
- WKY specific difference between time points
- Differentially phosphorylated in WKY between day 3 and day 5 at a 5% FDR;
- Not differentially phosphorylated in LEW between day 3 and day 5 at 5% nominal p value;
- Differentially phosphorylated in WKY compared with LEW at day 5 at 5% nominal p value.
- Day 5 specific difference between strains
- Differentially phosphorylated in WKY compared with LEW at day 5 at a 5% FDR;
- Not differentially phosphorylated WKY compared with LEW at day 3 at 5% nominal p value;
- Differentially phosphorylated in WKY compared with LEW at day 5 at 5% nominal p value.
In both cases the third criteria iii) was used as a mean to remove phosphopeptides exhibiting the wrong trends rather than as a statistical test, hence the decision not to account for multiple testing and use nominal p values < 0.05.
In addition to the peptides selected by the procedure described above, we included phospho-peptides that were considered specifically absent/present in MGC (i.e. in WKY at D5). Specifically, a phosphorylated peptide was declared as detected above background when it was quantified and showed an log2 (intensity) (i.e. area under curve) above eight (empirically determined). Peptides where then considered as specifically present in MGC if they were detected above background in all three WKY D5 samples and in none of the other sample. Conversely, they were considered as specifically absent from MGC if they showed the opposite pattern (see supplemental Fig. S5).
Phosphopeptide Enrichment Analysis
Enrichment analysis of phosphopeptides was carried out by DAVID (26) using UniprotKb IDs and taking all phosphoproteins (retrieved from UniProtKb using keywords Phosphoprotein [KW-0597] and Organism: Rat [10116]) as a reference set.
Transcription Factor Binding Site Enrichment
One-hundred and thirty-one transcription factors binding sites (TFBS) motifs were retrieved from the Jaspar Core vertebrate database and a Transcription Factor (TF) to gene binding affinity was computed for each gene on the genome. For each gene, the gene promoter was defined as a 1000 bp wide window centered on the transcription start site (TSS) and the promoter sequence was extracted from the rn4 reference genome sequence.
When more than one TSS existed for a gene, only the promoter sequence around the first TSS was used to avoid biasing affinities toward genes with multiple TSS. Each promoter was then scored using TRAP (27) and PASTAA (28) was used to compute enrichment TFBS site among the genes identified from of the giant cell specific transcriptome analysis. For LRRFIP1, a motif was built from the consensus sequence given in (29) by fixing an arbitrary 90% probability on each nucleotide of the consensus sequence and equal probability for all other nucleotides.
Functional Enrichment of Predicted Targets of Transcription Factors
To assess the functional enrichment of the targets of each MGC specific transcription factor, we considered the top 1000 genes (genome wide) whose promoter had the highest binding affinity to the transcription factor according to TRAP predictions of transcription factor binding affinity (using a 1000 bp promoter around the TSS). Among these 1000 putative targets, we extracted the subset that was specifically up-regulated in WKY at day 5, and considered these genes as putative targets of the MGC specific transcription factors. We then used DAVID to test functional enrichment of these genes.
PPI Network Analysis
We used 118,363 known physical interactions from the Biogrid database (Human PPI network, version 3.2, as retrieved on October 31st 2013) (removing the >9000 ubiquitination interactions involving UBC) to construct a physical interaction network and searched for each phospho-peptide the 10 shortest paths linking it to the transcriptions factors whose targets are up-regulated during MGC formation. We defined topological proximity between a phospho-peptide and a transcription factor as the average length of the 10 shortest path between them.
All phosphopeptide-transcription factor pairs were then ranked by topological proximity, and a null distribution of topological proximity was assessed by computing topological proximity between 1000 random pairs of protein. A p value of significance was then computed for each protein pair, and protein pairs were considered as potentially interacting when p < 0.01.
FDR among the predicted interactions was estimated by computing the ratio of the estimated number of false positive to the number of positive for various p value threshold (supplemental Fig. S7A). The estimated FDR for a p = 0.01 threshold was of 15%.
To further ensure the selected interacting pairs were not biased toward proteins with high number of interactors, we used a second permutation strategy, testing separately for each member of a pair, whether it was closer to its interactor in the PPI network than to a randomly chosen protein. Specifically, a null distribution of topological proximity was assessed separately for each transcription factor by computing topological proximity between 200 random proteins and the TF. A TF-centered p value (PTF) was then computed for each phosphoprotein-TF pair indicating the probability that a particular genes is involved in TF activation.
Symmetrically, a null distribution of topological proximity was assessed separately for each phosphoprotein, and a phosphoprotein-centered p value (PPhospho) was computed for each pair indicating the likelihood that a particular gene is activated by a phosphoprotein. All phosphopeptide-transcription factor pairs with PTF<0.05 and PPhospho<0.05 were considered as significant (supplemental Figs. S7B, S7C, and S8).
Ethic Statement
Rats were culled by asphyxiation with CO2 and cervical dislocation and the bones were isolated immediately for primary macrophage culture. All procedures were performed in accordance with the United Kingdom Animals (Scientific Procedures) Act, 1986.
RESULTS
Study Design for Combining Transcriptional and Phosphoproteomic Analyses During Macrophage Multinucleation
We first identified MGC-specific transcriptomic and phosphoproteomic signatures by focusing on critical time points during multinucleation in fusion-competent WKY BMDMs. We compared the transcriptome and phosphoproteome profiles of WKY rats BMDMs with LEW BMDMs, which present a markedly reduced MGC formation (Fig. 1A and 1B). In the WKY rat BMDMs, macrophage fusion events start to appear after 3 days of culture. At day 5 the multinucleation process is well underway with formation of MGCs and further culture of these cells lead to almost complete fusion of all cells in the plate at day 7 (Fig. 1A). In order to capture the dynamics of molecular events underlying of MGC formation, we have quantified the phosphopeptides by LC-MS/MS and mRNA by RNA-seq at day 3 (mononucleated macrophages) and day 5 in fusing macrophages. (Fig. 1A and 1B). We have then investigated transcription factors (TF) underlying the transcriptional changes through TF motif enrichment analysis (Fig. 1C, step [a]). We combined the phosphoproteomics and transcriptional data by using a k-shortest-path strategy similar to ((30)) to investigate the link between differentially abundant phosphopeptides and TFs driving transcriptional changes measured by RNA-seq during macrophage multinucleation (Fig. 1C, step [b]). Briefly, our k-shortest-path strategy measures the relatedness of a phosphoprotein to a given TF, as a function of the average distance between them in the protein–protein interaction network (see Experimental Procedures). The proposed integrative approach, thus allows the identification of novel regulators of macrophage multinucleation both at the transcriptional and cell-signaling levels.
Transcription Factor Driven Networks Underlying the Transcriptional Changes in Multinucleating Macrophages
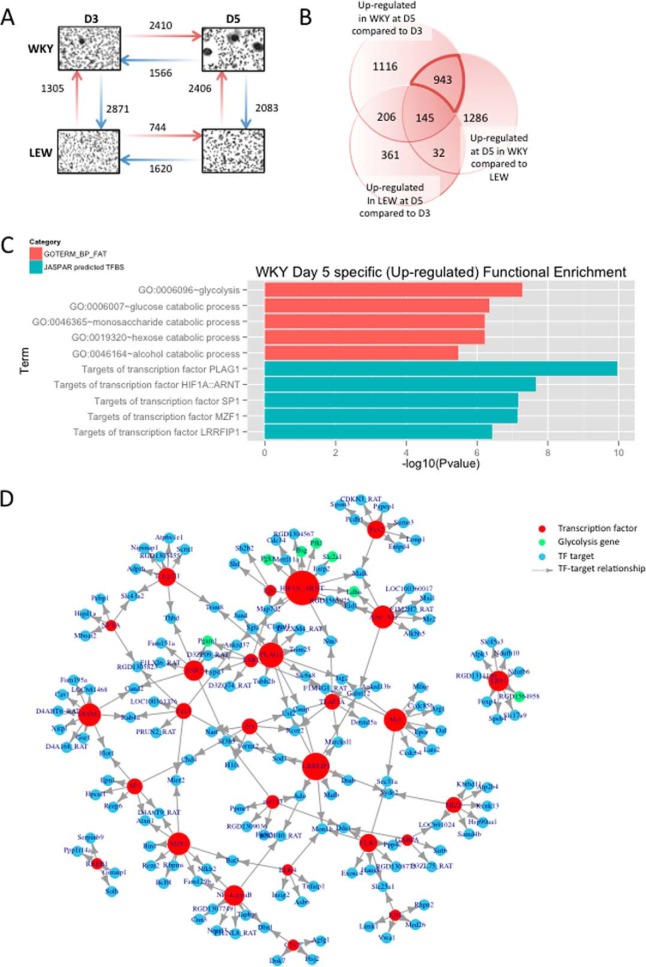
Clustering and principal component analysis of RNA-seq profiles revealed major differences in the transcriptome of WKY and LEW BMDMs as well as between the onset of fusion (day 3) and during early stage of MGC formation (day 5) in the WKY rat. The difference in the LEW transcriptome between day 3 (D3) and day 5 (D5), however, was of smaller magnitude (supplemental Fig. S1A and S1B). We performed differential expression analysis between D3 and D5 in both strains and observed a stronger change of the transcriptional program measured by RNA-seq in WKY than in LEW (3976 differentially expressed genes versus 2364 in LEW, supplemental Table S2); this was mostly because of WKY-specific over-expression of genes at D5 during the early stage of multinucleated giant cell formation (Fig. 2A). We first investigated the functional enrichment of differentially expressed genes over the time course (D3 versus D5 in both strains) and between the rat strains (independently of the time course, supplemental Fig. S1C and S1D). We then focused on the identification of the putative regulators of macrophage multinucleation by filtering for transcripts specifically up-regulated in early MGC formation. To this aim, we extracted the subset of 943 genes whose expression was showing WKY-specific increase during multinucleation (Fig. 2B and see Experimental Procedures). Functional enrichment analysis of these MGC-specific genes showed a marked enrichment for glycolysis (p < 10−7, false discovery rate (FDR) = 0.0002%) and its components (glucose, monosaccharide, and hexose catabolic processes) as well as response to hypoxia (p < 10−3, FDR = 2.6%) (Fig. 2C, supplemental Table S3).
Fig. 2.
Transcriptomic signature of macrophage multinucleation uncovers a role of HIF1-mediated glycolysis in MGCs. A, The number of up (red arrow) and down (blue arrow) regulated transcripts between each pair of condition (strain of rat and time of culture). B, the overlap of genes up-regulated at D5 in both strains and genes up-regulated in WKY at D5 and and highlighting of 943 MGC precursor specific genes. C, GO and TFBS enrichment for the set of MGC specific genes. D, the network of TF found in the enrichment analysis (red) with MGC specific genes that among their top 200 predicted targets (blue). Node size reflects number of neighbors. Glycolysis genes and are highlighted in green. E, transcript level expression of the top 3 TF found in the Enrichment analysis.
Transcription factor binding site enrichment analysis revealed that the MGC-specific up-regulated genes were most significantly enriched for transcription factor targets of PLAG1 (p < 1.1 × 10−10, FDR<3.6 × 10−8), and HIF-1 complex (p < 2.2 × 10−8, FDR<1.5 × 10−6) (Fig. 2C and supplemental Table S4). We then reconstructed the network of enriched TFs and their target genes, which suggested that the effect on glycolytic genes was mostly mediated by HIF1 (Fig. 2D). All enrichments were robust to the use of more stringent significance thresholds (1% FDR) for the definition of MGC-specific genes.
Macrophage Multinucleation is Associated with Increased HIF1-Mediated Glycolysis
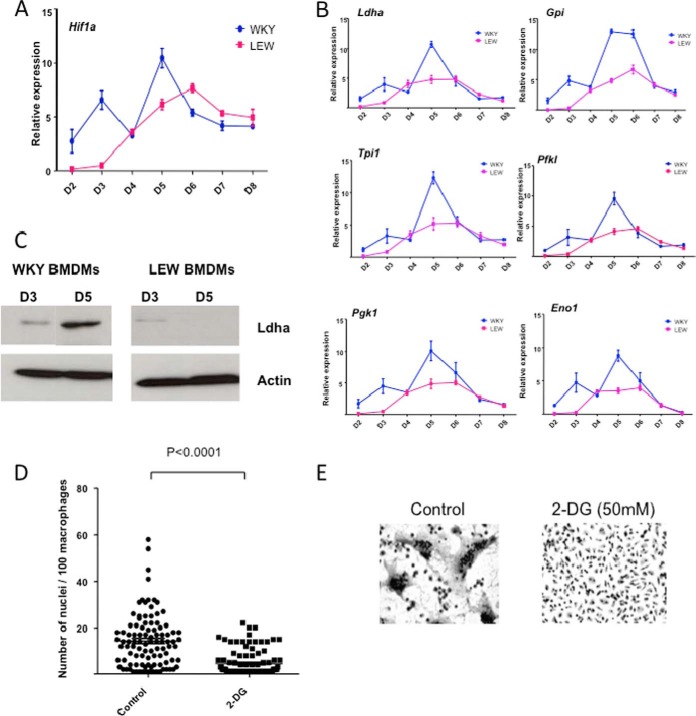
HIF1 is constitutively expressed in macrophages (supplemental Fig. S2). Given the previously established regulatory role of HIF1 in hypoxia-mediated glycolysis (reviewed in (31)), we hypothesized that HIF-mediated glycolysis is up-regulated in WKY during MGC formation and is required for spontaneous MGC formation. qRT-PCR analysis showed transient mRNA expression of Hif1a (Fig. 3A) as well as its glycolytic targets (Ldha, Gpi, tpi1, Pfkl, Eno1, and Pgk1) peaking at early stage of MGC formation in WKY BMDMs (Fig. 3B). Given the key role played by LDHA in HIF1-mediated glycolysis (32), we next measured LDHA protein levels as readout of HIF1-mediated glycolysis. LDHA over-expression specific to the fusion-competent WKY BMDMs at day 5 was confirmed at the protein level by Western blot analysis (Fig. 3C). We then inhibited cellular glycolysis by addition of 2-deoxy-d-glucose (2-DG) to the WKY BMDM at the onset of fusion (Day 3) and observed decreased expression of glycolytic genes (supplemental Fig. S3A), decreased protein amounts of LDHA (supplemental Fig. S3B) as well as decreased formation of MGC at day 5 (Fig. 3D and 3E), suggesting a role of HIF-mediated glycolysis during MGC formation in macrophages.
Fig. 3.
HIF1 mediated glycolysis is essential for macrophage multinucleation. A, expression of HIF1a in macrophages from WKY and LEW across time points. B, expression of HIF1a targets Ldha, Gpi, Tpi1, Pfkl, Pgk1, and Eno1 in macrophages from WKY and LEW and across time points. C, Western blot of Ldha at day 3 and day 5 in WKY and LEW, respectively. D and E, the effect of 2 Deoxy glucose treatment on multinucleation of WKY BMDMs.
Phosphoproteome Analysis in Multinucleating Macrophages
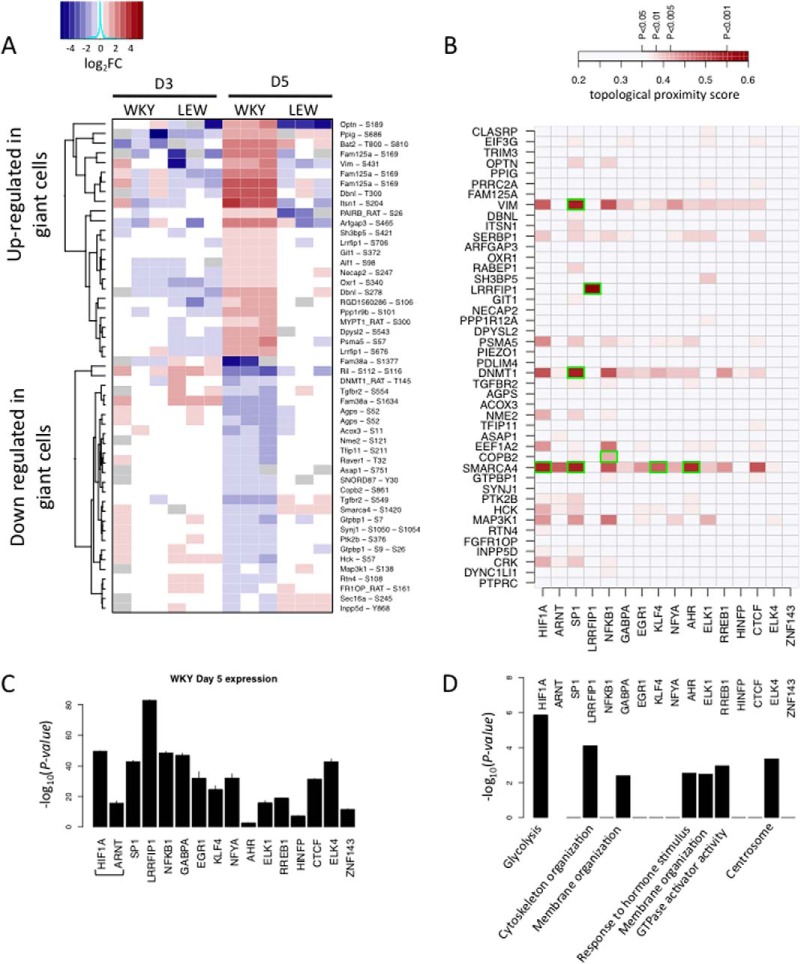
In order to investigate the mechanisms upstream the transcriptional regulation underlying the spontaneous multinucleation observed in WKY BMDMs, we characterized phosphorylation sites showing specific phosphorylation patterns during MGC formation. We performed titanium dioxide enrichment in protein lysates to isolate phosphopeptides (19) from LEW and WKY in BMDM after 3 and 5 days of culture, respectively. We then used LC-MS/MS to quantify over 2000 highly abundant phosphopeptides, among which 1006 were reliably detected in at least one condition (see Experimental Procedures). We first focused on a subset of 425 phosphopeptides reliably detected in all conditions, for quantitative analysis of differential phosphoprotein abundance (supplemental Table S5and Table S6). Out of the 425 phosphopeptides, we identified 128 differentially phosphorylated peptides at 5% FDR level between LEW and WKY macrophages at day 5, but only 17 at day 3, therefore suggesting multinucleation-specific phosphorylations patterns. Similar to the RNA-seq analysis, we further identified a set of 50 peptides specifically phosphorylated or dephosphorylated in MGC (i.e. in WKY at day 5; see supplemental Fig. S4 and Experimental Procedures). The expression of this phosphopeptide set across all conditions is shown in Fig. 4A. We further searched for specific absence/presence patterns in MGC formation and identified six phosphopeptides that were either specifically present or absent in WKY macrophages at D5 (supplemental Fig. S5A).
Fig. 4.
Phosphoproteome reprogramming and prediction of new regulators of MGC transcriptomic reprogramming via shortest path analysis. A, the heatmap of phosphopeptide expression across all conditions for the set of peptides harboring MGC specific phosphorylation pattern. For each phospho-peptides, abundance is shown relative to the median across all samples and in log2 scale. Gray spots represent missing quantification of the peptide. B, topological proximity score between MGC specific phospho-peptides and transcription factors, based on 10-shortest path analysis. Stronger color indicates higher topological proximity in the protein–protein interaction network. Only the scores that are significant at a 5% nominal p value are displayed (pairs among the top 5% closest pairs of the PPI network). MGC specific phosphoprotein-transcription factors pairs that have a direct connection in the PPI network are highlighted in green. C, expression (FPKM) of transcription factors with over-represented targets among MGC specific transcripts. D, the strongest over-represented Gene Ontology among the targets of each transcription factor. Bar height reflects -log10 p values of enrichment.
For the large majority of differentially phosphorylated sites, mRNAs encoding the proteins showed no change in expression, suggesting post-translational modification rather than transcriptional regulation of the peptides (supplemental Fig. S5B). This observation was further supported by the low fold changes in protein expression, observed when comparing protein abundance in WKY and LEW at the peak of multinucleation (supplemental Fig. S5C and supplemental Table S6 and Table S7). Functional enrichment analysis of phosphoproteins showing MGC-specific phosphorylation patterns did not reveal any significant enrichment at a 5% FDR threshold. Interestingly however, we noticed three poly-phosphate inositol phosphatases among the set of differentially phosphorylated sites, which were specifically down-phosphorylated in MGC precursors (supplemental Fig. S6A–6F) This observation was supported by a marginal trend toward enrichment for poly-phosphate inositol related phosphatases (p = 0.02, FDR = 18%). These findings are consistent with the previously reported role of poly-phosphates inositol phosphorylation in RANKL-mediated osteoclast multinucleation (33, 34), (summarized in supplemental Fig. S6G).
Integrative Analysis of TF-driven Networks and Phosphoproteome Data in Multinucleating Macrophages
In order to investigate how the observed changes in the phosphoproteome can underlie the transcriptomic reprogramming of macrophages during multinucleation and characterize the resulting regulatory effects, we examined the molecular links between differentially phosphorylated peptides and transcription factors using known interaction networks. We focused on expressed transcription factors showing specific change in activity during macrophage multinucleation (WKY D5, supplemental Fig. S2). We used a k-shortest path algorithm (35) to find the 10 shortest path linking phosphoprotein-transcription factor pair in the protein–protein interaction network. Firstly, the length of the 10 shortest paths was used to rank pairs according to their topological proximity in the protein–protein interaction network (Fig. 4B). We then used a two way random sampling strategy to test each pair for higher-than-random proximity in the network topology (see Experimental Procedures and supplemental Figs. S7B–7C). This approach led to the identification of 25 putative phosphoprotein-TF relationships, including six direct phosphoprotein-TF interactions (supplemental Fig. S8). These interactions are likely to be indicative of putative regulators of transcription factors showing increased activity during macrophage multinucleation (i.e. differentially expressed TFs targets during macrophage multinucleation). Our shortest path analysis identified LRRFIP1 as the only transcription factor undergoing direct phosphorylation during MM. LRRFIP1 was also the most strongly expressed among MGC specific TFs (Fig. 4C), and its predicted target genes showed functional enrichment for cytoskeleton organization (Fig. 4D, supplemental Table S7, see Experimental Procedures). This led us to prioritize LRRFIP1 for further analyses. In addition, this analysis identified DNMT1 and SMARCA4 as phosphoproteins that were predicted to be functionally related to a large number of TFs showing differential activity during multinucleation (after accounting for possible connectivity bias, see supplemental Fig. S8), which were followed up through validation experiments. Of note, Vimentin (VIM) was also identified by our shortest path analysis as a MGC specific phosphoprotein, in close proximity to a large number of MGC specific TFs. However, a strong decrease in the significance of Vimentin was observed when accounting for connectivity bias in the shortest path analysis (supplemental Figs. S7C and S8), suggesting that the significance of could be driven by its high connectivity alone (257 interactors in Biogrid). This observation led us to discard Vimentin as a potential candidate for follow-up studies.
Identification and Validation of Novel Determinants of Macrophage Multinucleation
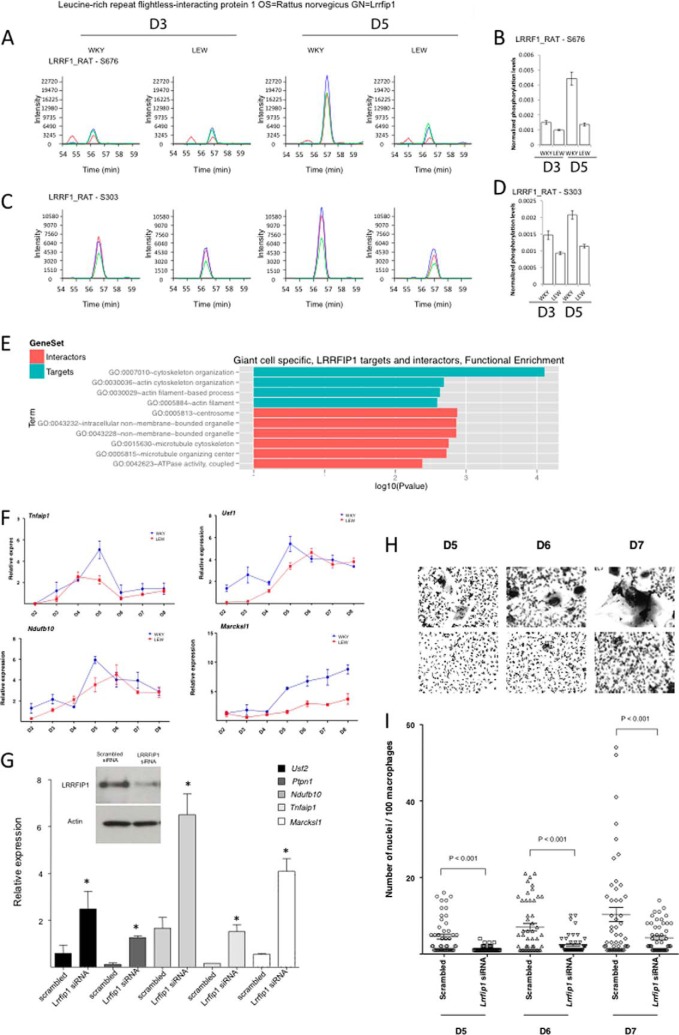
Leucine rich repeat (in FLII) interacting protein 1 (LRRFIP1) is a transcriptional repressor binding GC-rich motif sequences previously described as a regulator of Toll-Like Receptor response and Nf-kappaB activity (36). LRRFIP1 showed MGC-specific differential phosphorylation (Figs. 4A and 5A–5D) and enrichment for predicted targets in the MGC-specific genes assessed by RNA-seq (p < 3.8 × 10−7, FDR<9.9 × 10−6, Fig. 2C). Moreover, the MGC-specific up-regulated transcripts that are LRRFIP1-targets were enriched for cytoskeleton organization (p < 0.0002, FDR<0.3%, Fig. 5E), a key biological process associated with macrophage fusion and multinucleation events (37) and showed a strong over-expression at day 5 (Fig. 5F). We therefore hypothesized that phosphorylation of LRRFIP1 might play a role in macrophage multinucleation through a down-regulation of its repressor activity. To test this hypothesis, we knocked-down LRRFIP1 expression in fusion-competent WKY BMDMs at the onset of fusion (i.e. at D3) and observed an up-regulation of its transcriptional targets (Fig. 5G) associated with a marked decrease in macrophage multinucleation (Fig. 5H and 5I). Although these data validate the role of LRRFIP1 in macrophage multinucleation, the decrease of multinucleation following LRRFIP1 knock down suggests that the role of LRRFIP1 in multinucleation is independent of the transcriptional control of its target genes.
Fig. 5.
LRRFIP1, a new regulator of MGC formation. A and C, extracted ion chromatograms of the two LRRFIP1 phosphopeptides exhibiting differential phosphorylation across time points in both strains. On each graph the blue, green and red curve show the intensity of the three main isotopic peaks. B and D, show the associated quantification of normalized phosphoprotein abundance. E, represents GO enrichment of LRRFIP1 direct interactors (salmon) and predicted targets specifically up-regulated in MGC (turquoise). F, shows the expression of LRRFIP1 predicted targets over time in both WKY and LEW during multinucleation. G, the effect of LRRFIP1 knockdown on its predicted targets. H, and I highlight the role of LRRFIP1 in macrophage fusion by showing the reduction of macrophage multinucleation following LRRFIP1 Knock down.
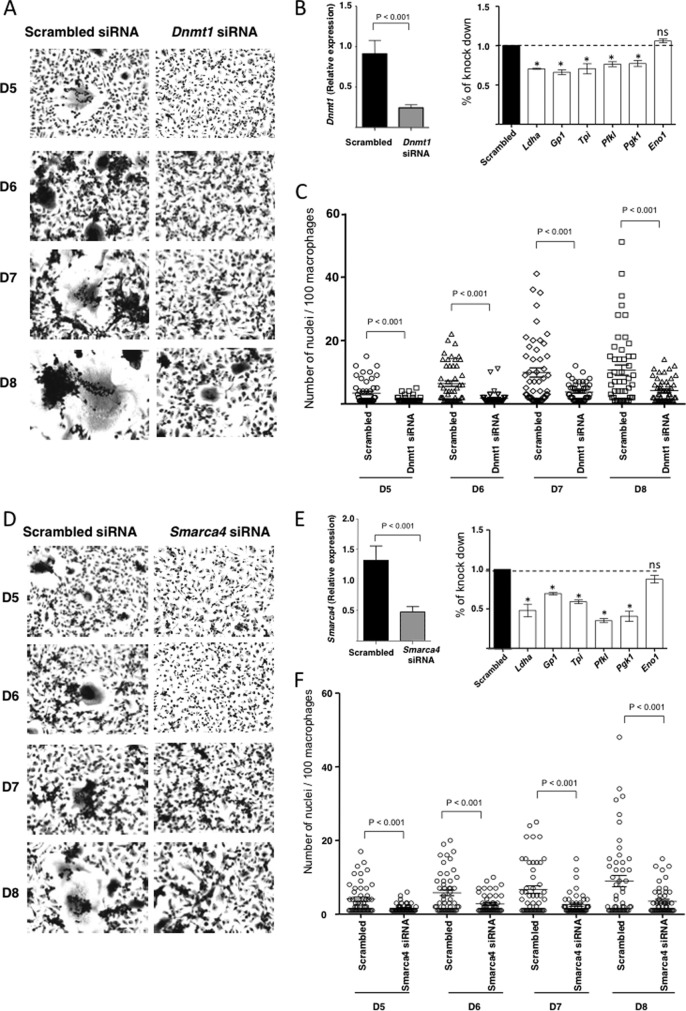
In addition to LRRFIP1, our integrated analysis of TFs-networks and phosphoproteome data in multinucleating macrophages also revealed SMARCA4 and DNMT1 as potential regulators of the transcriptional program in these cells (Fig. 4B). Both phosphoproteins showed close interactions with more than seven TFs active during multinucleation (including HIF1), suggesting a potential role in the regulation of the multinucleation transcriptional program (Fig. 4B and supplemental Fig. S8). SMARCA4 is a transcriptional activator also known as BRG1, previously shown to regulate the expression of the well-established macrophage multinucleation marker, CD44 (10), whereas DNMT1 is a DNA methyl-transferase involved in transcriptional repression via methylation of transcription factor binding site (38). We therefore hypothesized that SMARCA4 and DNMT1 were required for HIF1-mediated transcriptomic activation of glycolysis and more generally for the transcriptomic reprogramming occurring during macrophage multinucleation. To test this hypothesis, we performed siRNAs mediated knock-down of SMARCA4 and DNMT1 in fusion competent WKY macrophages at the onset of fusion (D3), and observed a reduction of transcription of glycolytic enzymes (Ldha, GP1, Tpi, Pfkl, and Pgk1) at day 5 together with a significant reduction of multinucleation (Fig. 6A-F). For SMARCA4 but not DNMT1, monitoring of LDHA levels following knock down, confirmed the reduction at the protein level (supplemental Fig. S9).
Fig. 6.
Effect of DNMT1 and SMARCA4 knock-down on glycolysis and multinucleation. A, B, and C, show the effect of siRNA knock down of DNMT1 on multinucleation and glycolysis in WKY macrophages. A comparison of cell cultures over time and quantifications of multinucleation are shown in A and C. B, the effect of knock down of DNMT1 on glylotyic gene expression at day 5. D, E, and F, show the effect of siRNA knock down of SMARCA4 on multinucleation and glycolysis in WKY macrophages. A comparison of cell cultures over time and quantifications of multinucleation are shown in D and F. C shows the effect of knock down of SMARCA4 on glylotyic gene expression at day 5.
Taken together our integrative analysis of transcriptional and phosphoproteomics data in multinucleating macrophages revealed complex cellular pathways as well as key regulators (HIF1A, LRRFIP1, SMARCA4, and DNMT1) underlying macrophage multinucleation and MGC formation.
DISCUSSION
In the present study, we have taken advantage of a spontaneous multinucleation occurring in the BMDMs of the inbred WKY rat and employed an integrative strategy to identify new pathways involved in macrophage multinucleation. We show that early MGC formation is regulated at metabolic and cell signaling levels. We further demonstrate that, by combining RNA-seq and phosphoproteome, novel determinants of macrophage multinucleation can be identified.
At the metabolic level, we showed that macrophage multinucleation required activation of HIF1-mediated glycolytic transcription, mediated partly by dephosphorylation of SMARCA4 and DNMT1. HIF1 has been reported previously as a potential target in osteoporosis because of its effect on osteoclast mediated bone resorption and osteoclast activity (39), suggesting the hypothesis that HIF1-mediated effects in multinucleation might also be relevant for the downstream osteoclast activity and bone resorption. In cancer, a similar activation of HIF1 under normoxic conditions has been described as the Warburg effect (40). This observation suggests some shared metabolic features between giant cell formation and tumorigenesis. Our integrative framework also identified PSMA5, MAP3K1, and EEF1A2 as putative regulators of HIF1 activation. PSMA5 is a component of the proteasome complex, that is responsible of down-regulation of HIF1 activity under normoxic condition through fast degradation of HIF1A (41), whereas MAP3K1 is an activator of the JNK kinase pathway that leads to activation of HIF1 and Nf-kappaB (42).
At a cell signaling level, we observed significant changes in phosphorylation of ARF GTPases (43, 44) as well as several phosphatidyl-inositol and inositol phosphatase during early multinucleation in macrophages. Phosphatidyl-inositol (PI) phosphorylation has also been linked to cytoskeletal regulation and membrane trafficking (45) and PI kinase such as PI3K have been previously linked to osteoclast differentiation (33). Our finding corroborates the already proposed role of PI3K in osteoclast differentiation and further suggests that increased osteoclast formation is maintained by down-regulation of the activity of PI phosphatases Ipp5d, Ipp5e, and Synj1.
Our integrative analysis of phosphoproteome and transcriptome using the k-shortest path analysis identified three novel regulators of macrophage multinucleation, LRRFIP1, DNMT1, and SMARCA4. We showed that LRRFIP1 acts as a repressor of the cytoskeleton re-organization occurring during macrophage multinucleation and suggested that its phosphorylation is responsible for the down-regulation of its transcriptional activity during MGC formation. The decrease in multinucleation that we observed after knock-down of LRRFIP1 indicates that LRRFIP1 is still active during multinucleation independently of its transcriptional repressor activity. Interestingly, beside its transcriptional repressor activity, LRRFIP1 was also shown to play a role in transduction of TLR4 signaling by competing with FLII for binding to MYD88, leading to activation of Nf-Kappa B (46). Hence, phosphorylation of LRRFIP1 may act by switching LRRFIP1 from a transcriptional repressor activity to a signaling role in interaction with MYD88 leading to giant cell formation via Nf-KappaB activation. SMARCA4 (also known as brahma-related gene-1, Brg1) is a mammalian homolog of SWI/SNF chromatin-remodeling factor subunits that can regulate both transcriptional activation and repression. SMARCA4 is mutated or deleted in numerous cancer cell lines (47–49), leading to the altered expression of genes that influence cell proliferation and metastasis. Interestingly, the promoter of CD44, a previously established macrophage multinucleation marker (10), is hypermethylated in cells that have a genetic deletion of SMARCA4 (50). In keeping with this, the de-phosphorylation of DNMT1, a DNA (cytosine-5-)-methyltransferase 1, suggests modification of its enzymatic activity that could result in aberrant methylation levels in the DNMT1-target promoters during multinucleation. The role of DNA methylation in macrophage multinucleation is unknown and future studies will be pivotal in establishing the methylation-driven transcriptional control of macrophage multinucleation. Beyond the identification of novel determinants of multinucleation, the specific effect of phosphorylation sites on the functional properties of these and on cell fusion preceding macrophage multinucleation remain to be determined. In summary, we integrated transcriptome and phosphoproteome data to investigate regulatory networks underlying multinucleation and prioritized new candidate genes and pathways underlying multinucleation of macrophages.
Supplementary Material
Footnotes
Author contributions: M.R., E.P., and J.B. designed research; M.R. performed research; C.M. and P.R.C. contributed new reagents or analytic tools; M.R., J.K., and C.M. analyzed data; M.R., P.R.C., E.P., and J.B. wrote the paper; J.K., A.K., and J.B. performed experiments; P.K.S. performed the rna-seq mapping and quality-control; A.M. and P.F. did phosphoprotein experiments and quantification.
* This work was supported by the Medical Research Council (MR/M004716/1 to J.B.) and by Kidney Research UK (RP9/2013 to J.B.). We also acknowledge funding from European Community's Seventh Framework Programme (FP7/2007-2013) under grant agreement no. HEALTH-F4-2010-241504 (EURATRANS).
 This article contains supplemental Figs. S1 to S9 and Tables S1 to S9.
This article contains supplemental Figs. S1 to S9 and Tables S1 to S9.
Conflict of interest: The authors declare that they have no conflict of interest.
Contributions: JB, EP, and MR designed the study. PS performed the RNA-seq mapping and quality-control. AM, PC, and PF did the phosphoprotein experiments and quantification. MR performed the statistical and integrative analyses. JB, JHK, and AK performed the lab experiments and analysed the data with CM. MR, EP, and JB wrote the manuscript with contributions from all authors.
1 The abbreviations used are:
- MM
- macrophage multinucleation
- MGCs
- multinucleated giant cells
- NTN
- nephrotoxic nephritis
- BMDM
- bone marrow derived macrophages
- FDR
- false discovery rate
- TF
- transcription factor
- 2-DG
- 2 Deoxy glucose
- PI
- Phosphatidyl-inositol.
REFERENCES
- 1. Teitelbaum S. L. (2000) Bone resorption by osteoclasts. Science 289, 1504–1508 [DOI] [PubMed] [Google Scholar]
- 2. Tonna E. A. (1963) Origin of osteoclats from the fusion of phagocytes. Nature 200, 226–227 [DOI] [PubMed] [Google Scholar]
- 3. Brodbeck W. G., Anderson J. M. (2009) Giant cell formation and function. Curr. Opin. Hematol. 16, 53–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Van Maarsseveen T. C. M. T., Vos W., van Diest P. J. (2009) Giant cell formation in sarcoidosis: cell fusion or proliferation with nondivision? Clin. Exp. Immunol. 155, 476–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Helming L., Gordon S. (2009) Molecular mediators of macrophage fusion. Trends Cell Biol. 19, 514–522 [DOI] [PubMed] [Google Scholar]
- 6. Peng Q., Malhotra S., Torchia J. A., Kerr W. G., Coggeshall K. M., Humphrey M. B. (2010) TREM2- and DAP12-dependent activation of PI3K requires DAP10 and is inhibited by SHIP1. Sci. Signal. 3, ra38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Vignery A. (2005) Macrophage fusion the making of osteoclasts and giant cells. J. Exp. Med. 202, 337–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhang C., Dou C., Xu J., Dong S. (2014) DC-STAMP, the key fusion-mediating molecule in osteoclastogenesis. J. Cell Physiol 229, 1330–1335 [DOI] [PubMed] [Google Scholar]
- 9. Rho J., Altmann C. R., Socci N. D., Merkov L., Kim N., So H., Lee O., Takami M., Brivanlou A. H., Choi Y. (2002) Gene expression profiling of osteoclast differentiation by combined suppression subtractive hybridization (SSH) and cDNA microarray analysis. DNA Cell Biol. 21, 541–549 [DOI] [PubMed] [Google Scholar]
- 10. Cui W., Ke J. Z., Zhang Q., Ke H.-Z., Chalouni C., Vignery A. (2006) The intracellular domain of CD44 promotes the fusion of macrophages. Blood 107, 796–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lee S.-H., Rho J., Jeong D., Sul J.-Y., Kim T., Kim N., Kang J.-S., Miyamoto T., Suda T., Lee S.-K., Pignolo R. J., Koczon-Jaremko B., Lorenzo J., Choi Y. (2006) v-ATPase V0 subunit d2-deficient mice exhibit impaired osteoclast fusion and increased bone formation. Nat. Med. 12, 1403–1409 [DOI] [PubMed] [Google Scholar]
- 12. Aitman T. J., Dong R., Vyse T. J., Norsworthy P. J., Johnson M. D., Smith J., Mangion J., Roberton-Lowe C., Marshall A. J., Petretto E., Hodges M. D., Bhangal G., Patel S. G., Sheehan-Rooney K., Duda M., Cook P. R., Evans D. J., Domin J., Flint J., Boyle J. J., Pusey C. D., Cook H. T. (2006) Copy number polymorphism in Fcgr3 predisposes to glomerulonephritis in rats and humans. Nature 439, 851–855 [DOI] [PubMed] [Google Scholar]
- 13. Behmoaras J., Bhangal G., Smith J., McDonald K., Mutch B., Lai P. C., Domin J., Game L., Salama A., Foxwell B. M., Pusey C. D., Cook H. T., Aitman T. J. (2008) Jund is a determinant of macrophage activation and is associated with glomerulonephritis susceptibility. Nat. Genet. 40, 553–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Behmoaras J., Smith J., D'Souza Z., Bhangal G., Chawanasuntoropoj R., Tam F. W. K., Pusey C. D., Aitman T. J., Cook H. T. (2010) Genetic loci modulate macrophage activity and glomerular damage in experimental glomerulonephritis. J. Am. Soc. Nephrol. JASN 21, 1136–1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kang H., Kerloc'h A., Rotival M., Xu X., Zhang Q., D'Souza Z., Kim M., Scholz J. C., Ko J.-H., Srivastava P. K., Genzen J. R., Cui W., Aitman T. J., Game L., Melvin J. E., Hanidu A., Dimock J., Zheng J., Souza D., Behera A. K., Nabozny G., Cook H. T., Bassett J. H. D., Williams G. R., Li J., Vignery A., Petretto E., Behmoaras J. (2014) Kcnn4 is a regulator of macrophage multinucleation in bone homeostasis and inflammatory disease. Cell Rep. 8, 1210–1224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. MacLauchlan S., Skokos E. A., Meznarich N., Zhu D. H., Raoof S., Shipley J. M., Senior R. M., Bornstein P., Kyriakides T. R. (2009) Macrophage fusion, giant cell formation, and the foreign body response require matrix metalloproteinase 9. J. Leukoc. Biol. 85, 617–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Anders S., Pyl P. T., Huber W. (2015) HTSeq–A Python framework to work with high-throughput sequencing data. Bioinformatics 15, 166–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Robinson M. D., McCarthy D. J., Smyth G. K. (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Montoya A., Beltran L., Casado P., Rodríguez-Prados J.-C., Cutillas P. R. (2011) Characterization of a TiO2 enrichment method for label-free quantitative phosphoproteomics. Methods 54, 370–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Perkins D. N., Pappin D. J., Creasy D. M., Cottrell J. S. (1999) Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20, 3551–3567 [DOI] [PubMed] [Google Scholar]
- 21. Savitski M. M., Lemeer S., Boesche M., Lang M., Mathieson T., Bantscheff M., Kuster B. (2011) Confident phosphorylation site localization using the Mascot Delta Score. Mol. Cell. Proteomics 10, M110.003830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Vizcaíno J. A., Côté R. G., Csordas A., Dianes J. A., Fabregat A., Foster J. M., Griss J., Alpi E., Birim M., Contell J., O'Kelly G., Schoenegger A., Ovelleiro D., Pérez-Riverol Y., Reisinger F., Ríos D., Wang R., Hermjakob H. (2013) The PRoteomics IDEntifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res. 41, D1063–D1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cutillas P. R., Vanhaesebroeck B. (2007) Quantitative profile of five murine core proteomes using label-free functional proteomics. Mol. Cell. Proteomics 6, 1560–1573 [DOI] [PubMed] [Google Scholar]
- 24. Frank Dieterle A. R. (2006) Probabilistic quotient normalization as robust method to account for dilution of complex biological mixtures. Application in 1H NMR metabonomics. Anal. Chem. 78, 4281–4290 [DOI] [PubMed] [Google Scholar]
- 25. Karpievitch Y., Stanley J., Taverner T., Huang J., Adkins J. N., Ansong C., Heffron F., Metz T. O., Qian W.-J., Yoon H., Smith R. D., Dabney A. R. (2009) A statistical framework for protein quantitation in bottom-up MS-based proteomics. Bioinforma. Oxf. Engl. 25, 2028–2034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Huang D. W., Sherman B. T., Lempicki R. A. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 [DOI] [PubMed] [Google Scholar]
- 27. Roider H. G., Kanhere A., Manke T., Vingron M. (2007) Predicting transcription factor affinities to DNA from a biophysical model. Bioinformatics 23, 134–141 [DOI] [PubMed] [Google Scholar]
- 28. Roider H. G., Manke T., O'Keeffe S., Vingron M., Haas S. A. (2009) PASTAA: identifying transcription factors associated with sets of co-regulated genes. Bioinforma. Oxf. Engl. 25, 435–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Reed A. L., Yamazaki H., Kaufman J. D., Rubinstein Y., Murphy B., Johnson A. C. (1998) Molecular cloning and characterization of a transcription regulator with homology to gc-binding factor. J. Biol. Chem. 273, 21594–21602 [DOI] [PubMed] [Google Scholar]
- 30. Shih Y.-K., Parthasarathy S. (2012) A single source k-shortest paths algorithm to infer regulatory pathways in a gene network. Bioinformatics 28, i49–i58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Semenza G. L. (2013) HIF-1 mediates metabolic responses to intratumoral hypoxia and oncogenic mutations. J. Clin. Invest. 123, 3664–3671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Semenza G. L., Roth P. H., Fang H. M., Wang G. L. (1994) Transcriptional regulation of genes encoding glycolytic enzymes by hypoxia-inducible factor 1. J. Biol. Chem. 269, 23757–23763 [PubMed] [Google Scholar]
- 33. Mandal C. C., Ghosh Choudhury G., Ghosh-Choudhury N. (2009) Phosphatidylinositol 3 kinase/Akt signal relay cooperates with smad in bone morphogenetic protein-2-induced colony stimulating factor-1 (CSF-1) expression and osteoclast differentiation. Endocrinology 150, 4989–4998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Moon J. B., Kim J. H., Kim K., Youn B. U., Ko A., Lee S. Y., Kim N. (2012) Akt induces osteoclast differentiation through regulating the GSK3β/NFATc1 signaling cascade. J. Immunol.188, 163–169 [DOI] [PubMed] [Google Scholar]
- 35. Yen J. Y. (1971) Finding the k shortest loopless paths in a network. Manag. Sci. 17, 712–716 [Google Scholar]
- 36. Lee S. Y., Lee S. Y., Choi Y. (1997) TRAF-interacting Protein (TRIP): A novel component of the Tumor Necrosis Factor Receptor (TNFR)- and CD30-TRAF signaling complexes that inhibits TRAF2-mediated NF-?B activation. J. Exp. Med. 185, 1275–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. DeFife K. M., Jenney C. R., Colton E., Anderson J. M. (1999) Cytoskeletal and adhesive structural polarizations accompany IL-13-induced human macrophage fusion. J. Histochem. Cytochem. 47, 65–74 [DOI] [PubMed] [Google Scholar]
- 38. Robertson K. D., Ait-Si-Ali S., Yokochi T., Wade P. A., Jones P. L., Wolffe A. P. (2000) DNMT1 forms a complex with Rb, E2F1, and HDAC1 and represses transcription from E2F-responsive promoters. Nat. Genet. 25, 338–342 [DOI] [PubMed] [Google Scholar]
- 39. Miyauchi Y., Sato Y., Kobayashi T., Yoshida S., Mori T., Kanagawa H., Katsuyama E., Fujie A., Hao W., Miyamoto K., Tando T., Morioka H., Matsumoto M., Chambon P., Johnson R. S., Kato S., Toyama Y., Miyamoto T. (2013) HIF1α is required for osteoclast activation by estrogen deficiency in postmenopausal osteoporosis. Proc. Natl. Acad. Sci. U.S.A. 110, 16568–16573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Palsson-McDermott E. M., O'Neill L. A. J. (2013) The Warburg effect then and now: from cancer to inflammatory diseases. BioEssays News Rev. Mol. Cell Dev. Biol. 35, 965–973 [DOI] [PubMed] [Google Scholar]
- 41. Lee J.-W., Bae S.-H., Jeong J.-W., Kim S.-H., Kim K.-W. (2004) Hypoxia-inducible factor (HIF-1)alpha: its protein stability and biological functions. Exp. Mol. Med. 36, 1–12 [DOI] [PubMed] [Google Scholar]
- 42. Ikeda F., Matsubara T., Tsurukai T., Hata K., Nishimura R., Yoneda T. (2008) JNK/c-Jun signaling mediates an anti-apoptotic effect of RANKL in osteoclasts. J. Bone Miner. Res. Off. J. Am. Soc. Bone Miner. Res. 23, 907–914 [DOI] [PubMed] [Google Scholar]
- 43. Chavrier P., Goud B. (1999) The role of ARF and Rab GTPases in membrane transport. Curr. Opin. Cell Biol. 11, 466–475 [DOI] [PubMed] [Google Scholar]
- 44. Randazzo P. A., Hirsch D. S. (2004) Arf GAPs: multifunctional proteins that regulate membrane traffic and actin remodelling. Cell. Signal. 16, 401–413 [DOI] [PubMed] [Google Scholar]
- 45. Gillooly D. J., Simonsen A., Stenmark H. (2001) Phosphoinositides and phagocytosis. J. Cell Biol. 155, 15–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Dai P., Jeong S. Y., Yu Y., Leng T., Wu W., Xie L., Chen X. (2009) Modulation of TLR signaling by multiple MyD88-interacting partners including leucine-rich repeat Fli-I-interacting proteins. J. Immunol. 182, 3450–3460 [DOI] [PubMed] [Google Scholar]
- 47. Jelinic P., Mueller J. J., Olvera N., Dao F., Scott S. N., Shah R., Gao J., Schultz N., Gonen M., Soslow R. A., Berger M. F., Levine D. A. (2014) Recurrent SMARCA4 mutations in small cell carcinoma of the ovary. Nat. Genet. 46, 424–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ramos P., Karnezis A. N., Craig D. W., Sekulic A., Russell M. L., Hendricks W. P. D., Corneveaux J. J., Barrett M. T., Shumansky K., Yang Y., Shah S. P., Prentice L. M., Marra M. A., Kiefer J., Zismann V. L., McEachron T. A., Salhia B., Prat J., D'Angelo E., Clarke B. A., Pressey J. G., Farley J. H., Anthony S. P., Roden R. B. S., Cunliffe H. E., Huntsman D. G., Trent J. M. (2014) Small cell carcinoma of the ovary, hypercalcemic type, displays frequent inactivating germline and somatic mutations in SMARCA4. Nat. Genet. 46, 427–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Witkowski L., Carrot-Zhang J., Albrecht S., Fahiminiya S., Hamel N., Tomiak E., Grynspan D., Saloustros E., Nadaf J., Rivera B., Gilpin C., Castellsagué E., Silva-Smith R., Plourde F., Wu M., Saskin A., Arseneault M., Karabakhtsian R. G., Reilly E. A., Ueland F. R., Margiolaki A., Pavlakis K., Castellino S. M., Lamovec J., Mackay H. J., Roth L. M., Ulbright T. M., Bender T. A., Georgoulias V., Longy M., Berchuck A., Tischkowitz M., Nagel I., Siebert R., Stewart C. J. R., Arseneau J., McCluggage W. G., Clarke B. A., Riazalhosseini Y., Hasselblatt M., Majewski J., Foulkes W. D. (2014) Germline and somatic SMARCA4 mutations characterize small cell carcinoma of the ovary, hypercalcemic type. Nat. Genet. 46, 438–443 [DOI] [PubMed] [Google Scholar]
- 50. Banine F., Bartlett C., Gunawardena R., Muchardt C., Yaniv M., Knudsen E. S., Weissman B. E., Sherman L. S. (2005) SWI/SNF chromatin-remodeling factors induce changes in DNA methylation to promote transcriptional activation. Cancer Res. 65, 3542–3547 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.