Abstract
The reversible phosphorylation of proteins on serine, threonine, and tyrosine residues is an important biological regulatory mechanism. In the context of genome integrity, signaling cascades driven by phosphorylation are crucial for the coordination and regulation of DNA repair. The two serine/threonine protein kinases ataxia telangiectasia-mutated (ATM) and Ataxia telangiectasia-mutated and Rad3-related (ATR) are key factors in this process, each specific for different kinds of DNA lesions. They are conserved across eukaryotes, mediating the activation of cell-cycle checkpoints, chromatin modifications, and regulation of DNA repair proteins. We designed a novel mass spectrometry-based phosphoproteomics approach to study DNA damage repair in Arabidopsis thaliana. The protocol combines filter aided sample preparation, immobilized metal affinity chromatography, metal oxide affinity chromatography, and strong cation exchange chromatography for phosphopeptide generation, enrichment, and separation. Isobaric labeling employing iTRAQ (isobaric tags for relative and absolute quantitation) was used for profiling the phosphoproteome of atm atr double mutants and wild type plants under either regular growth conditions or challenged by irradiation. A total of 10,831 proteins were identified and 15,445 unique phosphopeptides were quantified, containing 134 up- and 38 down-regulated ATM/ATR dependent phosphopeptides. We identified known and novel ATM/ATR targets such as LIG4 and MRE11 (needed for resistance against ionizing radiation), PIE1 and SDG26 (implicated in chromatin remodeling), PCNA1, WAPL, and PDS5 (implicated in DNA replication), and ASK1 and HTA10 (involved in meiosis).
In eukaryotes, the reversible phosphorylation of serine, threonine, and tyrosine residues within proteins is a wide-spread post-translational modification, essential for controlling a multitude of cellular processes. During the last decade, sequencing projects unexpectedly unraveled that plant genomes encode for a considerable larger number of protein kinases than the other kingdoms of life. Arabidopsis thaliana contains 1112 PKs (4% of all genes), twice the number encoded by the human genome (518 or 2% of all genes) and other plants have an even higher number of kinases (1).
Phosphatidyl inositol 3′ kinase related kinases are important players in DNA damage response (DDR)1 and crucial for genome integrity (2). Key to DNA double strand break (DSB) repair is a chain of events starting with detection of the lesion, activation of a signaling cascade, cell cycle arrest, and recruitment of the repair machinery. The cascade is triggered by the Phosphatidyl inositol 3′ kinase related kinases family kinases ataxia telangiectasia-mutated (ATM) (3) and Ataxia telangiectasia-mutated and Rad3-related (ATR) (4). Both kinases are conserved across eukaryotes. Their downstream targets have been systematically identified in yeast (5) and human cells (6, 7). Their essential role in mediating DNA repair in higher plants has been established (8–10). In Arabidopsis, loss of function mutants are viable (11); however, atm mutants are highly sensitive to genotoxic stress and have a reduced fertility. atr mutant plants have a cell-cycle checkpoint defect upon exposure to genotoxic chemicals (12). Somatic growth under nonchallenging conditions is not affected in the double mutant but plants are sterile, highlighting the role of both kinases coordinating meiotic DNA repair. In plants, systematic phosphoproteomic studies of the involved pathways have not been reported but would contribute to further elucidating the molecular mechanism of the observed phenotypes. Interestingly, plants lack clear homologs for many downstream regulatory components in the signaling cascade (e.g. CHK1, CHK2, p53, and MDC1) (13). In this context, it should be noted that DNA-PKcs (DNA-dependent protein kinase), another Phosphatidyl inositol 3′ kinase related kinases family member involved in DNA repair, has not been identified in plant genomes (14, 15), underscoring the significance of ATM and ATR as master regulators.
ATM is recruited to DSBs via its interaction with NBS1/XRS2, a member of the MRN/X complex (MRE11/RAD50/NBS1-XRS2). In plants, the detailed molecular base for ATM recruitment has remained unknown. The complex acts as damage sensor in yeast, first to be detected at DNA double strand break (DSB) sites and essential for resection of DNA (16). In all organisms analyzed, the MRN/X complex is required for genotoxic stress resistance (17). The Mre11 endonuclease activity is critical for ATM activation, likely triggered by the generation of short oligo-nucleotides (18). In higher eukaryotes, ATM activation relies on MRN binding to DSBs via MRE11, subsequent tethering of DSB ends via RAD50 and recruitment of ATM. This interaction leads to monomerisation of inactive ATM dimers, followed by autophosphorylation. The MRN subcomplex member NBS1 interacts with monomeric ATM leading to its localization in close proximity of the DSB site (19). NBS1, H2AX, the checkpoint kinase CHK2, and the trimeric replication protein A (RPA) are important downstream targets of ATM (6).
In yeast, ATR is activated by RPA coated single-stranded DNA (ssDNA) that is generated by 5′ resection mediated by MRX/N, Exo1, Sgs1, and Dna2 during DSB processing (20). Furthermore, ssDNA may become exposed because of replication fork break down during DNA replication or nucleotide excision repair (21). Exposed ssDNA is rapidly bound by RPA, attracting ATRIP, and the Rad17-RFC complex. ATRIP interacts with ATR and is essential for its activation and function (22). The Rad17-RFC complex is functional in loading the 9–1-1 protein complex (Rad9, Rad1, and Hus1) to 5′ dsDNA-ssDNA junctions, in turn stimulating ATR activity at the site of the exposed ssDNA (23). In human cells, TopBP1 is required for activation of ATR and localizing to DNA lesion sites. Rad17, TopBP1, RPA, and the checkpoint kinase CHK1 are known downstream targets of activated ATR.
The core effectors of DNA repair (e.g. RAD51), the proteins detecting DNA damage and mediating initiation of repair (e.g. MRX) and the two master regulators ATM and ATR are conserved in plants but many downstream components have diverged considerably. Yet, a comprehensive model for DDR in plants requires identification of all components to delineate the involved signaling pathways and their cross-talk with other regulatory processes.
Mass spectrometry-based methods are powerful and hypothesis-free approaches for protein characterization, enabling high-throughput studies of protein complexes (24), protein expression profiling (25), or large-scale identification of protein kinase targets (26). Also, the identification and quantification of thousands of phosphopeptides has become feasible by technological and methodological advances. As a consequence, system-wide analyses of signaling networks has become possible (27). Despite above mentioned advances, comprehensive phosphoproteomic studies remain challenging in regards to sample preparation and phosphopeptide enrichment. An additional complication in large-scale studies is imposed by the requirement for correct automatic localization of phosphorylation sites (28). Abundant metabolites make sample preparation in plants especially difficult; however, a number of large-scale phosphoproteomic studies have been reported (29–34).
Phosphorylation sites in proteins are in most cases substoichiometric. As a consequence, the comprehensive analysis requires enrichment of phosphopeptides prior to LC-MS/MS. From the large number of developed methods (35), metal-based affinity chromatography such as immobilized metal affinity chromatography (IMAC) and metal oxide affinity chromatography (MOAC) have become most widely used. Both materials have different specificities, resulting in a substantial increase of identified phosphopeptides when employed consecutively (36).
Here we delineate a methodology to identify and relatively quantify phosphorylation events proteome-wide in higher plants (Fig. 1). We demonstrate its applicability in the context of ATM and ATR dependent DNA damage repair in Arabidopsis thaliana. The approach combines filter assisted sample preparation (FASP) (37), isobaric labeling via iTRAQ, phosphopeptide enrichment using IMAC and TiO2, strong cation exchange (SCX) chromatography, followed by LC-MS/MS. We compared the relative differences between the phosphoproteomes of wild type plants with the double mutant (atm atr), studying both irradiated and nonirradiated plants. In addition, we performed an independent analysis based on peptide generation after protein precipitation.
Fig. 1.

Workflow for identification of ATM/ATR dependent and independent phosphorylations. Wild type and atm atr double mutant plants were either exposed to irradiation or grown under regular conditions. Extracted proteins were purified via FASP and labeled with iTRAQ. Phosphopeptides were enriched by consecutive application of IMAC and TiO2. Both, the phosphopeptide-enriched fraction and the flow-through of the TiO2 chromatography, were separated by SCX chromatography and fractions were analyzed by reversed phase LC-MS/MS.
All together, we identified 10,831 proteins. Four-hundred and 13 phosphoproteins are phosphorylated upon ionizing radiation, among them 108 in an ATM/ATR dependent manner. The acquired data-set represents a unique resource for plant researchers and extends the current knowledge on ATM/ATR dependent DNA repair pathways.
EXPERIMENTAL PROCEDURES
Plant Material
Arabidopsis thaliana wild type plants and mutant lines (double homozygous offspring of a double heterozygous atm atr line (atm-2: Salk_006953; (8); atr-2: Salk_032841; (12)) were in ecotype Columbia-0 (Col-0). Wild type and mutant lines were grown side-by-side under long day conditions (16 h light, 8 h dark, 21 °C; 60 to 80% humidity, 5800 LUX, 3× Philips TLD 36W, and 2× Sylvana GroLUX 36W). Homozygous atm atr mutant plants were identified by PCR using 35 cycles, standard conditions and the following primers: ATM-F1 (5′-GCTTCAAGGTTGGGCAGTTCC-3′), ATM-R1 (5′-GCAAACAGCATAACAAAACACTTCC-3′), and Lbc1 (5′-TGGACCGCTTGCTGCAACTCT-3′) and primers: atr2–1 (5′-GGATCAAGTACTACTGACTCAG-3′), atr2–3 (5′-CAACTCATTTTGAATATGAGAG-3′), and Lba1 (5′-TGGTTCACGTAGTGGGCCATCG-3′), respectively. Genotypes were validated in duplicate by PCR. For irradiation experiments, plants were exposed to 100 Gy ionizing radiation using a Co60 source (25 Gy min−1) at the developmental stage 5.10 (38). For each sample, the aerial parts of three plants were harvested 15 min after irradiation. Samples were pooled, frozen in liquid nitrogen, and stored at −80 °C.
Protein Extraction
Frozen plant tissue samples were homogenized in a liquid nitrogen-chilled cryogenic mill. For each sample, 1 ml prechilled protein extraction buffer (4% SDS (Merck), 50 mm Tris[2-carboxyethyl] phosphine (TCEP, Sigma-Aldrich), 50 mm Hepes-NaOH, pH 7.5 (Sigma-Aldrich)) was added to 170 mg of powdered plant material and incubated at 95 °C under strong agitation for 5 min. The samples were sonicated at room temperature for 1 min and incubated at 95 °C under strong agitation for further 5 min. After centrifugation (10 min, 20,000 × g at room temperature), the supernatant was cleared by a second centrifugation step.
Filter-Aided Sample Preparation (FASP)
Peptides were generated by a modified FASP protocol (37). In brief, 1 ml of plant cell lysate was mixed with 9 ml of 8 m UH-buffer (8 m urea (Amresco, Solon, OH, USA), 50 mm Hepes-NaOH, pH 8.5) containing 2 mm TCEP and loaded on an Amicon Ultra-15 centrifugal filter unit with a 30 kDa molecular weight cutoff (Millipore). All buffer exchanges were performed by centrifugation at 4000 × g at room temperature. The first buffer exchange (UH buffer) was repeated twice, followed by alkylation of proteins by addition of 40 μl of 200 mm methylmethanethiosulfonate (MMTS, Fluka) for 30 min at room temperature. MMTS was removed by performing two buffer exchanges using 8 m UH-buffer, followed by two further buffer changes using 0.8 m urea, 50 mm Triethylammonium bicarbonate (TEAB, Fluka). Trypsin (Trypsin Gold, Promega, Madison, WI) was added to 1 ml of the protein mixture at an enzyme to protein ratio of 1:100 and incubated at 37 °C for 2 h, followed by addition of an equal amount of trypsin and incubation overnight. Generated peptides were collected by centrifugation. Five-hundred microliters of 50 mm TEAB were added to the retentate and incubated, followed by isolation of peptides by centrifugation. The step was repeated and the collected samples were unified, lyophilized, and dissolved with 500 mm TEAB in a final volume of 200 μl. Two-milligrams of peptides were obtained from 170 mg of plant material as determined by UV (Nanodrop, Peqlab ND1000).
Protein Precipitation from SDS-Containing Plant Cell Lysates (Wessel-Fluegge)
One-milliliters of plant lysate was mixed with 4 ml of methanol (LC-MS Chromasolv, Fluka) and 1 ml of CHCl3 (Chromasolv Plus, Sigma-Aldrich). Three-milliliters of H2O were added and mixed again. The sample was centrifuged at 4000 × g for 30 min. The upper phase was removed and 4 ml methanol was added. The sample was centrifuged at 4000 × g for 5 min. The supernatant was removed, and the pellet was washed by two consecutive steps of suspension in methanol followed by centrifugation. The pellet was air-dried and dissolved in 250 μl 8 m urea and 50 mm TEAB, which was supported by sonication. Protein concentration was determined by a bicinchoninic acid protein assay (Pierce) to be 7.25 μg/μl. Then 140 μl of the solvent containing 1 mg protein mixture were incubated with 14 μl of 50 mm TCEP for 60 min at 30 °C, followed by alkylation with 7 μl of 200 mm MMTS for 30 min at room temperature. The sample was diluted with 50 mm TEAB to a concentration of 6 m urea, supplemented to 2 mm MgCl2 (Fluka) and 2 mm CaCl2 (Fluka) and treated with six units Turbo DNase (2 U/μl), Life Technologies, Carlsbad, CA and 800 units Benzonase (250 U/μl, purity grade I, Merck) for 30 min at 30 °C. Lys-C (Wako Pure Chemicals Industries, Osaka, Japan) was added in a ratio of 1:50. After 2 h of incubation at 30 °C, the sample was diluted to 2 m urea and the DNA digest was repeated as described above. Trypsin was added in a ratio of 1:80 and incubated for 2 h at 37 °C. The sample was further diluted to 0.8 m urea and another aliquot of trypsin was added and incubated over night at 37 °C.
iTRAQ Labeling
Each of the four different samples (FASP: 1.8 mg; protein precipitation: 1 mg) were labeled with one of the iTRAQ 4-plex reagents (wild type: iTRAQ 114; wild typeIR: iTRAQ 115; atm atr: iTRAQ 116; atm atrIR: iTRAQ 117) according to the manufacturer's instructions. A complete iTRAQ kit (AB Sciex) was used for each of the two samples. Labeling efficiency was determined by LC-MS/MS analysis of a small aliquot and the four samples were mixed in equimolar amounts. Equimolarity was validated by LC-MS/MS and the sample was freeze-dried. FASP generated samples were purified via reversed phase chromatography. In brief, the sample was dissolved in 0.1% trifluoroacetic acid (TFA, Pierce) and adjusted to a pH below two using 10% TFA. Desalting was performed with Strata-X reversed phase polymeric solid phase extraction cartridges (Phenomenex, 200 mg, Torrance, CA). Peptides were eluted with 70% acetonitrile (ACN, Chromasolv gradient grade, Sigma-Aldrich) and 0.1% formic acid (FA, Suprapur, Merck), followed by freeze-drying. Samples generated by protein precipitation were purified via SCX-chromatography using a PolySULFOETHYL A (poly LC) column (4.6 mm i.d. × 150 mm). Chromatography was performed using a steep gradient with a ternary buffer system. Starting with 100% buffer A (5 mm NaH2PO4 (Sigma), pH 2.7, 15% ACN) for 15 min, ramping to 50% buffer B (5 mm NaH2PO4, pH 2.7, 1 m NaCl (Merck), 15% ACN) and 50% buffer C (5 mm Na2HPO4 (Merck),pH 6, 15% ACN) in 5 min, followed by isocratic elution at 50% buffer B and 50% buffer C for 30 min. The fractions were freeze-dried, dissolved in 0.1% TFA, and desalted as described above.
Enrichment of Phosphopeptides by IMAC and TiO2 Chromatography
Phosphopeptides were enriched using IMAC (39) by first dissolving the dried peptide mixture in 4 ml of 40% ACN and 0.1% FA. After incubation with 140 μl of IMAC beads (Phos-Select Iron Affinity Gel, P9740, Sigma) for 60 min, the bead suspension was split and transferred to two Mobicol (MoBiTec) spin columns with a 10 μm pore filter at the outlet. The flow-through was collected by centrifugation. Beads were washed 10 times with 400 μl loading buffer and two times with 5% ACN and 0.1% FA. The first two washes were pooled with the flow-through for TiO2 chromatography and freeze-dried. Bound peptides were eluted with four times 100 μl 1 m NH4OH (Merck). The eluates of the two columns were unified and freeze-dried.
For titania enrichment (40), the dried IMAC flow-through was dissolved in 0.1% TFA, desalted as described above and again freeze-dried. The pellet was dissolved in 3 ml of TiO2 loading buffer (300 mg/ml lactic acid (Sigma-Aldrich), 80% ACN, 0.1% TFA) and incubated with 10 mg of TiO2 resin (Titansphere bulk media, 5 micron, GL Science, Tokyo, Japan) for 60 min. The beads were transferred to one Mobicol spin column and the flow-through was collected, freeze-dried and later used for protein quantification.
The TiO2 beads were washed with 8 × 500 μl of TiO2 loading buffer, 8 × 500 μl of 80% ACN, 0.1% TFA, 4 × 500 μl of 1% ACN, 0.1% TFA, 4 × 500 μl of 1% ACN, 0.1% FA. Bound peptides were eluted from the resin by addition of 4 × 100 μl 0.3 m NH4OH and 4 × 100 μl 0.7 m NH4OH, followed by freeze-drying.
SCX Chromatography
Phosphopeptides from IMAC and TiO2 enrichment were separated with SCX after combining both samples. Each of the dried samples was dissolved in 50 μl buffer A, combined and adjusted to pH < 3 with 8.5% H3PO4 (Merck). SCX was performed using an Ultimate system (Thermo Fisher Scientific) at a flow rate of 25 μl/min and a custom-made PolySULFOETHYL A (poly LC) column (5 μm particles, 300 Å, 1 mm i.d. × 250 mm). The phosphopeptides were separated by a combined salt and pH gradient. For separation, a ternary gradient was used. Starting with 100% buffer A for 10 min, followed by a linear increase to 10% buffer B and 50% buffer C in 80 min, to 25% buffer B and 50% buffer C in 10 min, 50% buffer B and 50% buffer C in 5 min, and an isocratic elution for further 15 min. The flow-through was collected as single fraction, along the shallow part of the gradient fractions were collected every minute, followed by collecting fractions every 2 min. The 94 fractions were stored at −80 °C. Two-hundred micrograms of peptides of the desalted TiO2-flow-through were separated by SCX applying the same gradient as above. Here, the total number of fractions was 98. ACN was removed by vacuum centrifugation and the samples were acidified with 0.1% TFA and analyzed by LC-MS/MS.
Samples generated by protein precipitation were separated in a slightly modified form using a steeper gradient. Starting with 100% buffer A for 20 min, followed by a linear increase to 10% buffer B and 50% buffer C in 30 min, to 25% buffer B and 50% buffer C in 10 min, to 50% buffer B and 50% buffer C in 5 min, and isocratic elution for 15 min. The flow-through was collected as one fraction, followed by one 8-min fraction, and then fractions of 2 min were collected. In total, 33 fractions were collected for further analysis.
LC-MS/MS
On-line separation was performed with an Ultimate 3000 dual gradient nano-LC system (Thermo Fisher Scientific). Peptides were loaded on a trap column (PepMap C18, 5 mm × 0.3 mm, 5 μm, 100 Å, Thermo Fisher Scientific) using 0.1% TFA at a flow rate of 25 μl/min. After a desalting step of 15 min, the trap column was switched online with the analytical column (PepMap C18, 250 mm × 0.075 mm, 3 μm, 100 Å, Thermo Fisher Scientific) using solvent A (5% ACN, 0.1% FA). Peptides generated by the FASP protocol were eluted at a flow rate of 275 nl/min applying a linear gradient from 0 to 100% B (30% ACN, 0.08% FA) in 60 min, followed by a step to 85% B and 15% C (80% ACN, 10% trifluoroethanol (Sigma-Aldrich), 0.08% FA) in 1 min, and another steep gradient to 10% B and 90% C in 4 min. For the analysis of peptides generated from the protein mixture isolated by precipitation, a 90 min dual gradient (from 0–100% B) was used.
Data-dependent MS/MS was performed using an LTQ-Orbitrap Velos mass spectrometer, equipped with a nano-electrospray ion source (both Thermo Fisher Scientific). For the analysis of phosphopeptides, the MS survey scan was acquired in the Orbitrap, recording a window between 350 m/z and 2000 m/z at a resolution of 60,000. The target value was 1,000,000 using a maximal fill time of 400 ms. Monoisotopic precursor selection was enabled and ion clusters with a charge higher than one were analyzed via MS/MS. Using a combined collision induced dissociation (CID)- higher-energy collisional dissociation (HCD) approach (41), the five most prominent ions were analyzed by MS/MS via CID using multistage activation in the LTQ using a target value of 10,000 and a maximal fill time of 200 ms. A normalized collision energy of 35%, a Q-value of 0.25 and an activation time of 10 ms was used. Each precursor was additionally fragmented via HCD and analyzed in the Orbitrap, using an isolation window of 1.6 m/z, a resolution of 7500, a target value of 100,000, a maximal fill time of 600 ms and a normalized collision energy of 50%. Fragmented ions were put on an exclusion list for 60 s. The lock mass option was enabled using polydimethylcyclosiloxane (m/z 445.12) for recalibration. Samples from the protein precipitation experiments and the TiO2 flow-through were analyzed in a slightly modified MS/MS set-up.
MS/MS Data Interpretation
Raw data were interpreted with Proteome Discoverer (version 1.3.0.339, Thermo Fisher Scientific, Bremen, Germany) using the search engine Mascot (Version 2.2.07, Matrix Science, UK) against The Arabidopsis thaliana protein database (TAIR10_pep_20101214, 35386 sequences), supplemented with common contaminants and concatenated with a decoy database containing the reversed sequences of the original database. The data sets from all 2D-LC-MS/MS experiments of the IMAC/TiO2-enriched sample were analyzed simultaneously. The mass tolerances were set to 5 ppm for peptide masses, 0.5 Da for CID-generated fragments and to 30 mDa for HCD spectra. Trypsin was defined as proteolytic enzyme, cleaving after lysine or arginine including cleavage before proline and three missed cleavages were allowed. Alkylation of cysteines by MMTS and iTRAQ labels on the peptide N terminus and lysines were set as fixed modification. Phosphorylation on serine, threonine, and tyrosine, oxidation of methione, acetylation on the protein N termini, and deamidation of asparagine and glutamine were set as variable modifications. The MS/MS data sets from the TiO2-flow-through were searched without allowing phosphorylation.
Peptide spectrum matches (PSMs) were filtered by requiring a minimum peptide length of eight amino acids and a search engine rank of one. Peptide confidence was set to high and proteins were grouped applying strict maximum parsimony principle. An ion score threshold was set separately for HCD and CID scans to reach a false discovery rate (FDR) of less than 0.5% as measured by the target-decoy approach. The required ion scores for the TiO2- enriched sample were 23 for CID and 11 for HCD. For the TiO2-flow-through sample, the required scores were 25 and 10, respectively.
Using Proteome Discoverer, both files were opened in a single multiconsensus report to ensure a joint protein grouping according to the maximum parsimony principle. The list of proteins was sorted by the sum of the protein ion scores, calculated by the sum of the highest scoring PSM for each unique peptide. The protein list was filtered by a FDR of 1%. By removing the respective peptides from the data set, the FDR at the PSM level was reduced to 0.05%. For the sample of the protein precipitation experiment only a dataset containing the phosphopeptide-enriched sample was generated. This dataset was filtered to 0.5% FDR for PSMs. The Mascot ion score threshold was set to 23 for CID spectra and to 13 for HCD spectra.
Phosphorylation Site Localization
To localize the phosphorylation sites in the peptides identified in the phosphopeptide-enriched sample, PhosphoRS 2.0 (42) implemented in Proteome Discoverer was applied using a cutoff of 75% for the site probability. In the majority of cases, CID and HCD spectra from the same precursor led to identical PSMs and site localizations. If both spectra resulted in different nonambiguous localizations of the phosphorylation in the phosphopeptide, it was classified as ambiguous. In cases where only one spectrum (HCD or CID) led to a nonambiguous site localization, the localization was considered nonambiguous.
iTRAQ Quantification
To quantify iTRAQ reporter ions, the integration method provided by Proteome Discoverer with a mass window of 5 mDa was used. To account for extreme regulation events, absent iTRAQ reporter ion values were replaced with a value of 100. PSMs with iTRAQ intensities below 1000 in all four channels were removed. The ratios 115/114 (WTIR/WT), 117/116 (atm atrIR/atm atr), 116/114 (atm atr/WT), and 115/117 (WTIR/atm atrIR) were calculated for each quantified PSM.
For the phosphopeptide-enriched sample, the geometric mean and standard deviations of the ratios were calculated for the repeated measurements of each unique phosphopeptide. If no peptide with localized sites was identified for a certain unique phosphopeptide sequence, then repeated measurements of the peptide with nonlocalized sites were used to calculate the geometric mean and the standard deviation.
The ratios of phosphopeptides were normalized by the corresponding protein ratios calculated from at least three quantified PSMs from the TiO2-flow-through LC-MS/MS experiments. For phosphopeptides corresponding to proteins without a sufficient relative quantitation, the original ratios are reported. To perform normalization on the protein level for the sample from the protein precipitation experiment, the nonphosphorylated peptides, which had been co-isolated in the phosphopeptide-enriched sample were used to calculate a protein ratio.
Analysis of Phosphorylation Motifs
To identify sequence motifs significantly enriched in the datasets of up- and down-regulated phosphopeptides relative to the background of all phosphopeptides quantified in this study, we utilized Motif-X (43). All quantified phosphopeptides (PhosphoRS 2.0 value > 0.75) were centered at their phosphorylated amino acid and ten positions upstream and downstream were used in the analysis (sequence context retrieved from TAIR10). If phosphorylation sites were located at the C- or N- (sub-) termini, the sequences were extended with “X” (X, any amino acid). The sequences were loaded as pre-aligned text data and all motifs were identified with a probability threshold of 1E-06 (Bonferroni corrected p value = 0.0004) and an occurrence threshold of at least three peptides.
Bioinformatic Data Analysis
Potential human orthologs of the 108 direct or indirect candidate targets of the ATM and ATR kinases were derived from the following five orthology databases using default settings: OrthoMCL DB (version V5); InParanoid (version 7.0); HomoloGene (homologene_build 65.data); Roundup (version 2.0); and Oma browser (version from march 2014).
To identify known and predicted protein–protein interactions between the 108 potential ATM and ATR target proteins, the STRING database (version 9.1) was used, applying the following settings: a medium confidence score of 0.4 was set and interactions based on either experimental evidence, curated databases, or by text-mining were scored and displayed.
A Pfam domain search was performed using Pfam 27.0 applying the standard settings
RESULTS AND DISCUSSION
Strategy for the Phosphoproteomic Analysis
We quantified DNA damage-induced changes in the phosphoproteome of Arabidopsis thaliana by employing high-resolution MS in combination with isobaric labeling using iTRAQ (Fig. 1). The wild-type scenario was compared against a homozygous atm atr double mutant. Both genotypes were analyzed under either regular conditions or DNA damage-inducing irradiation, resulting in four different samples. The double mutant was instrumental for distinguishing between ATM/ATR-dependent and -independent phosphorylation events. In order to minimize generation of artifacts by enzymatic activities outside the regular cellular context, we omitted subcellular fractionation. After FASP (37), we consecutively applied IMAC (Fe3+) and MOAC (TiO2) to the generated peptide mixtures (Fig. 1), increasing the number of identified phosphopeptides (36). In order to evaluate the performance of the applied IMAC and MOAC protocols 1% of each eluate was analyzed by LC-MS/MS (supplemental Table S1). Using IMAC, respectively TiO2 enrichment of the IMAC flow-through, 71% (IMAC) and 84% (TiO2) of the identified peptides were phosphopeptides. Of the latter, 64% were not identified after IMAC.
LC-MS/MS-based Analysis of the Proteome-wide ATM/ATR Dependent DNA Damage Response in Arabidopsis thaliana
The eluates from IMAC and TiO2 enrichment were pooled and the sample was fractionated employing SCX (94 fractions for the FASP and 33 for the precipitation method), using a combined salt/pH gradient. The results obtained for the FASP-generated samples from the LC-MS/MS analysis of all SCX-fractions are summarized in Table I and the supplemental Tables S2–S4. As expected, doubly (and higher) phosphorylated peptides were enriched in early SCX fractions, followed by singly phosphorylated and finally by nonphosphorylated peptides (Fig. 2A). We observed the enrichment of N-terminally acetylated peptides in the flow-through and in early eluting fractions (44) yielding 14% of the number of identified PSMs in these fractions. More than 60% of the identified peptides corresponded to LC-MS/MS data of single SCX fractions, demonstrating the high chromatographic resolution of the separation (Fig. 2B), as determined by analyzing the distribution of the peptides in the collected fractions. Samples were analyzed on an LTQ-Orbitrap Velos using a CID-HCD method for identification and quantification (41). Applying a FDR of 0.05%, we identified 95,356 PSMs in the phosphopeptide-enriched sample and 387,253 PSMs in the TiO2-flow-through. In most cases, PSMs corresponding to the CID and HCD spectra from the same precursor ion were redundant, reducing the relevant numbers to 56,928 and 226,495 for the phosphopeptide fractions and the flow-through, respectively. In the TiO2-flow-through sample, 85,104 peptides were identified with a FDR of 0.15%, corresponding to 9977 proteins. In the phosphopeptide-enriched sample we could identify 11,453 single, 3022 doubly, 461 triply, and 27 four times phosphorylated peptides (Fig. 2C), summing up to 14,963 phosphopeptides applying the same FDR. A total of 12,887 unique phosphorylation sites could be localized when using a conservative cutoff of 75% for the phosphoRS site probability (42). In total, 10,831 proteins were identified in the combined datasets when applying a protein FDR of 1% (supplemental Table S4).
Table I. Numbers of identified and quantified spectra, peptides and proteins in the FASP-generated phosphopeptide-enriched and TiO2-flow-through sample.
| Number of | Phospho-enriched | TiO2-flow-through |
|---|---|---|
| PSMs (0.05% FDR) | 95,356 | 387,253 |
| Identified HCD-spectra (0.03% FDR) | 51,032 | 190,183 |
| Identified CID-spectra (0.07% FDR) | 44,324 | 197,070 |
| Nonredundant PSMsa (0.08% FDR) | 56,928 | 226,818 |
| Unique quantification eventsb (0.08% FDR) | 50,940 | 209,831 |
| Unique peptidesc (0.15% FDR) | 21,495 | 85,104 |
| Unique phosphopeptidesc | 14,963 | n.a. |
| Unique phosphopeptidesc with localized sitesd | 14,084 | n.a. |
| Unique peptidesc quantifiede(0.16% FDR) | 20,249 | 81,905 |
| Unique phosphopeptidesc quantifiede | 13,974 | n.a. |
| Proteins (1%FDR) | 5,819 | 9,977 |
| Proteins found in both samples | 10,831 | |
| Proteins quantifiedf | 7,915 | |
n.a., nonapplicable, as the spectra of the TiO2-flow-through sample were not searched against phosphorylation.
a CID/HCD triggered from same precursor.
b Nonredundant PSMs with at least one iTRAQ channel >1000.
c Unique based on amino acid sequence, distinguishing only acetylation and phosphorylation and number and position of phospho-sites.
d Unique phosphopeptide with localized sites contains all identified sites with a pRS probability > 75%.
e Unique peptides were only used for quantification, if at least 1 iTRAQ channel was > 1000.
f Protein ratios were only calculated, if at least 3 unique quantification events were detected.
Fig. 2.
Numbers of peptides identified after phosphopeptide enrichment. Following FASP and phosphopeptide enrichment samples were fractionated by SCX chromatography and analyzed by LC-MS/MS. A, Diagram showing the distribution of nonphosphorylated, singly and higher phosphorylated, and N-terminally acetylated peptides within each of the 94 collected fractions. The combined salt- and pH-gradient is indicated as a solid and dashed line, respectively. B, Pie chart diagram depicting the number of fractions in which a unique peptide was found. C, The phosphorylation status of the identified peptides of all combined fractions is depicted. D, The unique phosphopeptides identified with the two different approaches (FASP and Wessel/Fluegge (W/F)) and with unambiguously localized phospho-sites are compared with those deposited in the PhosPhAt 4.0 database.
Limited Influence of Irradiation on Protein Abundance
A total number of 50,940 PSMs of the phosphopeptide-enriched sample and 209,831 PSMs of the TiO2 flow-through had sufficient reporter ion intensities for quantification. When requiring at least three quantified PSMs, the data acquired from the TiO2-flow-through led to the quantification of 7915 proteins (supplemental Table S4). We observed only subtle differences between overall protein abundances in the four samples, with less than 0.5% of all proteins differing in their abundance by a factor of two when comparing any of the four different conditions with each other.
The time frame for sample collection was chosen to monitor the immediate response at the level of phosphorylations; however, up- or down-regulated proteins might additionally offer interesting clues about the biological response to irradiation induced stress. Applying a confidence interval of 4σ, 18 proteins in the atm atrIR sample were significantly more abundant compared with both the WTIR sample and the double mutant without irradiation (supplemental Table S4). Sixteen of them are GO (Gene Ontology)-annotated to be involved in general stress and defense response. Seven of these proteins are in the subcategory of response to reactive oxygen species (ROS). Among the proteins involved in general stress and defense response are PR1, PR2, PR5 (PR4 would be in the same group but is up-regulated within a confidence interval of 3σ), PNP-A, GSTF6, and GSTF7, all factors reported to be involved in pathogen defense (45, 46). Our data is consistent with previous observations of ATM involvement in stress and defense response (47, 48).
Twenty-seven proteins are less abundant in the atm atr double mutant when compared with the wild type in both the irradiated and nonirradiated samples. Among these 27 proteins are five LEA (Late Embryogenesis Abundant) proteins, fitting to the observed phenotype of a slight delay in development in the atm atr double mutant plants.
Ionizing Radiation Increased Phosphorylation of Specific Target Proteins
First, iTRAQ ratios of phosphopeptides were normalized to the respective changes in protein abundance (49). In summary, we could normalize 9782 phosphopeptides from a total number of 13,974 quantified peptides (supplemental Table S5). In a preceding experiment, the sample preparation included an initial protein precipitation step instead of FASP. In this experiment, we could quantify 5553 phosphopeptides (supplemental Table S6). In combination with slightly higher amounts of starting material, FASP processing led to significantly higher numbers of identified and quantified phosphopeptides; however, some peptides were only detected in the sample generated by protein precipitation (Fig. 2D). Combining both data sets a total number of 15,445 unique phosphopeptides could be quantified (supplemental Table S5).
To determine the overlap of the phosphopeptides identified in this study with already published phosphopeptides, all unique peptides with nonambiguous localized sites from the FASP and Wessel/Fluegge dataset were compared with the phosphopeptides provided as “full dataset experimental sites” at the PhosPhAt 4.0 Arabidopsis Protein Phosphorylation Site Database (http://phosphat.mpimp-golm.mpg.de/downloads.html). The comparison was made based on the phosphopeptide sequence considering the localized phosphosite. The dataset of this study increases the already annotated phosphopeptides by 11,910 entries, which represents an increase by 116% (Fig. 2D).
We chose a conservative cutoff defining a minimum ratio of 1.8 for regulated phosphorylation sites. For the important class of ATM/ATR and ionizing radiation (IR) dependent phosphorylation sites (Fig. 3, first column), both of the ratios WTIR/WT (iTRAQ channels 115/114) and WTIR/atm atrIR (iTRAQ channels 115/117) had to be at least 1.8. For the irradiation-induced and ATM/ATR independent phosphorylation events (Fig. 3, second column), we used the same cutoff applied to the ratios WTIR/WT (iTRAQ channels 115/114) and atm atrIR/atm atr (iTRAQ channels 117/116). We tested the cutoff by applying it to the nonphosphorylated peptides co-isolated in the phosphopeptide-enriched samples, assuming that they should be unaffected. Only 0.05% of the nonphosphorylated peptides passed this cutoff in any of the above mentioned combinations. Of the 15,445 quantified phosphopeptides, we identified 604 phosphopeptides up-regulated in response to irradiation. Of these, 134 phosphopeptides, grouping to 108 proteins, were more abundant in response to irradiation only in wild-type plants, but not in atm atr mutant plants, therefore representing potential direct or indirect targets of the ATM and ATR kinases (Fig. 3 and supplemental Table S7). Additionally, we identified 12 additional potential targets of ATM and ATR (supplemental Table S7, last sheet) applying relaxed filter criteria but requiring manual validation of spectra.
Fig. 3.

Overview of relative phosphopeptide abundances. Normalized and quantified phosphopeptides were grouped into five different categories depending on the relative abundance in the four different samples as measured by their iTRAQ ratios. The first row depicts phosphopeptides that are up-regulated following exposure to IR and dependent on ATM/ATR. The second row depicts phosphopeptides enriched after exposure to IR, but independent of ATM/ATR. Phosphorylated S/T-Q motifs are only present in peptides with higher abundance depending on both, IR exposure and ATM and ATR (first row).
From the 134 peptides, 84 (62.7%) were phosphorylated on a SQ or TQ motif. In yeast and mammals, this motif is the preferred target of both ATM and ATR (5, 7, 50). Among the regulated target sites in our data set, 84% of the 50 strongest up-regulated phosphopeptides contain S/T-Q sites, suggesting that the corresponding sites in plants are also direct substrates. In addition to confirming the S/T-Q motif, “Motif-X” analysis (43) additionally identified a SE motif in 16 of the ATM/ATR and irradiation dependent phosphorylation sites (Fig. 4). Although the body of evidence points to SQ and TQ being direct target sites of ATM and ATR (5, 7, 50, 51), phosphorylation on SE sites might be indirectly regulated. The SE motif was only found among those up-regulated phosphorylation sites that depend on both irradiation and ATM/ATR. The core SE motif and its environment match the acidic consensus target motif of casein kinase 2 (CK2) (52). CK2 is another important player in cellular signal transduction, including DNA stress response, phosphorylating important downstream targets (53). Although it remains to be proven that Arabidopsis CK2 is recognizing these motifs in an ATM/ATR dependent manner, it is intriguing that plants expressing a dominant negative version of CK2 are more sensitive to ionizing radiation (54).
Fig. 4.
Motif X analysis of significantly up-regulated phosphopeptides. A, Three consensus motifs are significantly enriched in the dataset of irradiation induced and ATM/ATR dependent up-regulated phosphopeptides. Five motifs are found in the dataset corresponding to ATM/ATR independent up-regulated phosphopeptides. The enrichment was calculated against the background of all quantified phosphopeptides with localized sites and is shown in the last column of the table. (*** - Bonferroni corrected p value = 0.0004) B, Graphical representation of the amino acid environment of the identified motifs shown in panel A.
We furthermore found motifs around irradiation-induced regulated phosphorylation sites, which were ATM and ATR independent (Fig. 4). The predominant motifs found were SP (230 residues of a total of 397 phosphorylated serine residues are embedded in a SP motif), either as a stand-alone motif or embedded in different neighboring consensus amino acids, and TP (62 residues of a total of 97 phosphorylated threonines reside in a TP motif).
We furthermore found mitogen activated protein kinases 3, 6, and 11 (the identified peptide for MAPK 11 also matches MAPK 4) with elevated double-phosphorylations on T#EY# in its activation loop (55). MAPKs have been identified as key signal transducers in various signaling cascades, involving pathogen defense, abiotic stress, and response to ionizing radiation (56, 57). Importantly, MAPKs phosphorylate proteins on consensus TP and SP target sites (the latter has also been defined as a minimal consensus sequence of cyclin dependent kinases) (58). Together, these results suggest a parallel and ATM/ATR independent signaling cascade following ionizing radiation.
We also identified 71 phosphopeptides that appear to be down-regulated in response to irradiation. Of these, 38 peptides were down-regulated in the wild typeIR, but not in the atm atrIR sample. However, in several of these peptides, we detected up-regulation of the same site in combination with another S/T-Q site. The detection of the down-regulation of the peptide could therefore be a consequence of the up-regulation of the same site in combination with the second S/T-Q site.
Analysis of ATM/ATR Target Proteins
In a first step, we grouped the proteins with up-regulated phosphorylation events depending on ionizing radiation and ATM/ATR according to their GO annotations (supplemental Table S8). The 30 peptides with the most prominent irradiation and ATM/ATR dependent phosphorylation events are depicted in Table II.
Table II. The top 30 phosphopeptides (out of 132) according to the strongest increase of ATM and ATR dependent phosphorylation following treatment with ionizing radiation (sorted by decreasing WTIR/WT ratios). Unambigously assigned phosphosites are marked in red, for one singly phosphorylated peptide with ambiguous site assignment possible acceptor sites are marked in blue. In two cases the peptide spans the protein N-terminus, which was found to be acetylated (marked in green).

Ninty-three phosphoproteins and 60 of the subgroup containing at least one phosphorylated S/T-Q site were annotated to be localized to the nucleus. These annotations correspond well to the nuclear localization of both ATM and ATR. All 18 proteins with a phosphorylated SE motif were also annotated with a nuclear localization.
Forty-eight of the 108 proteins are associated with biological processes of nitrogen compound metabolism (Supplemental Table S8). This cluster encompasses processes related to DNA and RNA metabolism, which is anticipated to undergo dynamic changes following genotoxic stress. Along the same lines, 16 proteins are related to the biological process of chromosome organization and 34 to the biological process of RNA metabolic processes. In the GO category describing molecular functions, 33 proteins are annotated to be DNA binding. Not surprisingly, most of the subcategories are related to DDR, with actions taking place in the nucleus.
The relevance of the data-set is substantiated by the identification of phosphorylation events on proteins that have already been found to be crucial for resistance against ionizing radiation in plants, such as LIG4, MRE11, or UVH3, factors implicated in chromatin remodeling such as PIE1, CHR4, SDG26, PA200, FVE/MSI4, and REF6, implicated in DNA replication such as PCNA1, WAPL, PDS5, RAD21.3, and RHL1, proteins involved in meiosis such as ASK1, MRE11, PIE1, and HTA10 and further factors involved in transcriptional control such as WRKY1, HON4, GTE4, GTE5, and NF-YC11 (Fig. 5 and supplemental Table S7).
Fig. 5.
Graphical representation of ATM/ATR dependent phosphorylation targets clustered according to GO-annotation terms. Selected phosphoproteins carrying irradiation induced and ATM/ATR dependent up-regulated phosphosites are grouped according to their biological function and color-coded according to their regulatory ratio.
Among afore mentioned proteins is ATLIG4 (AT5G57160), well described for its essential function in the nonhomologous end joining DSB repair pathway (59). Phosphorylation on S1100 (YTES#QQR) was 78-fold enriched upon IR in an ATM/ATR dependent manner. Another crucial DNA repair factor, MRE11 (AT5G54260), being involved in both, nonhomologous end joining and homologous recombination DNA repair pathways (17, 60), was also found to be phosphorylated upon IR in an ATM/ATR dependent manner (S413, SEANIDDS#ER, 19-fold enrichment). We hypothesize that MRE11 is phosphorylated by an elusive downstream kinase and signal mediator of ATM/ATR, as suggested by the presence of a SE motif.
UVH3 (AT3G28030) (IGILHDT#DS#QNER, 15-fold enrichment) is related to the yeast Rad2 and to the human XPG protein and is an endonuclease required for nucleotide excision repair. The corresponding Arabidopsis mutant plants displays increased sensitivity to ultraviolet (UV) light, H2O2, and ionizing radiation and a premature senescence phenotype (61).
We furthermore found that H2AX (AT1G08880; AT1G54690) is phosphorylated upon IR in an ATM/ATR dependent manner on S139 (GDIGSAS#QEF, 80-fold enrichment). Phosphorylation of H2AX (collectively referred as γ-H2AX), including the phosphorylation on S139 (62) has been described as cellular marker specific for DSBs, crucial for recruitment of DNA damage repair proteins and as a target of ATM. Additionally, H2A.7 (AT5G27670) (KSTASSS#QAEK, 88-fold enrichment and MESS#QATTK, fivefold enrichment) and H2A.10 (AT1G51060) (KTGASKPS#AEDD, eightfold enrichment) were found to be phosphorylated in an irradiation- and ATM/ATR dependent manner. Although the role of H2A.7 is a novel finding, the importance of H2A.10 during meiosis and for centromere function has already been recognized (63). The observed histone modifications likely relate to chromatin dynamics following genotoxic stress. Consequentially, we also detected increased phosphorylation in several chromatin modifiers, such as FVE/MSI4/RbA48p (AT2G19520) (EDS#QTPSSQQQSDVK, 24-fold enrichment; EDSQTPSS#QQQSDVK, fourfold enrichment), a core component of chromatin remodeling complexes and important for flower development, chromatin silencing and DNA repair (64). We also identified the chromatin remodeler SDG26/ASHH1 (AT1G76710) as a target of irradiation- and ATM/ATR dependent phosphorylation (LLSQNS#QEDSSPK, threefold enrichment). SDG26 contains a SET domain and is a lysine methyltransferase enzyme related to the Trithorax Group (TrxG) proteins and again implicated in control of flowering time and DNA repair (64). Another chromatin remodeling factor identified in the given study is the PIE1 protein (AT3G12810) (AVITTSS#QEDDTDVLDDVK, fivefold enrichment), the catalytic partner of the Swi2/Snf2-related (SWR1) complex. The SWR1 complex has been found to be important for H2A.Z histone variant deposition and in a broader sense for DNA damage resistance and meiotic recombination (65, 66). We also identified up-regulated phosphopeptides of the catalytic partner of a nucleosome remodeler complex, CHR4 (AT5G44800) (VGSSS#QTTK, fourfold enrichment) known to be needed for recombination in plants and DNA repair in Drosophila and humans (67, 68). Another chromatin remodeling factor, REF6 (AT3G48430) (DT#QETLSDAER, tenfold enrichment), is a H3K27me3 demethylase, related to metazoan H3K9me2/3 and H3K36me2/3 demethylases and important for derepression of gene expression by releasing the H3K27me3 bound LHP1 protein, the Arabidopsis homolog of HP1 (69).
We identified several proteins, which are involved in sister chromatid cohesion, such as the Arabidopsis homologs of WAPL (AT1G11060) (LNS#QEESSNR,130-fold enrichment) and one of the PDS5 homologs (AT5G47690) (VEGDKS#QVGEEK, eightfold enrichment) as direct ATM/ATR targets following genotoxic stress. It is worth to highlight, that of the many phosphorylated residues identified on peptides corresponding to WAPL (three P-sites identified) and PDS5 (28 P-sites identified), only those embedded in SQ motifs were strongly up-regulated. Arabidopsis WAPL has a crucial role in meiosis (70). In other model organisms, WAPL and PDS5 have been described to be important for chromatin structure, chromosome segregation and gene expression, also in the context of genotoxic stress (71, 72). Additionally we identified one of the RAD21 homologs (part of the cohesin complex) as an ATM/ATR target (RAD21.3/SYN 4, AT5G16270, SQIDENVNT#QNEPEEER, fivefold enrichment) (73). Another protein factor that is important during DNA synthesis and that we identified as an indirect target of ATM/ATR is PCNA1 (AT1G07370) (IEEEEDT#NP, twofold enrichment). ATM has been found to physically interact with PCNA to regulate DNA synthesis in human cell culture (74).
We also identified transcriptional regulators as targets of ATM/ATR following genotoxic stress. For instance, the general transcription factor NF-YC11 (AT3G12480) as a potential direct target (VPDYGHS#QGQGHGDVTMDDR; 146-fold enrichment) (75) and also the transcription factor WRKY1 (AT2G04880) as a potential indirect target (AVGTLES#EEQKPK, twofold enrichment). WRKY1 has been shown previously to be transcriptionally up-regulated by IR and has putative binding sites in promoter regions of DNA repair genes (e.g. RAD51, AtCOM1) (76). An unusual transcriptional regulator is the protein DAYSLEEPER (AT3G42170), related to transposase-like proteins (77). It is essential for normal plant development and we found ATM/ATR and IR dependent up-regulation of a corresponding peptide (MEVYNDDTEMRS#PET#QPIK, 15-fold enrichment).
One of the strongest up-regulated phosphopeptides belongs to the proteasome activating factor PA200 (AT3G13330) (S#QDLGGDVDDR, 34-fold enrichment), which is known to be involved in DNA repair (78). Another factor involved in protein turn-over and in many different biological processes, including meiosis, is the SCF complex partner SKP1/ASK1 (AT1G75950) for which we found an up-regulated phosphopeptide depending on IR and ATM/ATR (NDFT#PEEEEEVRR, twofold enrichment) (79–81). We also found the Prp19-like E3 ubiquitin ligases MAC3A (AT1G04510) (IFGLPDDDNTEDS#AQDS; IFGLPDDDNTEDSAQDS#, sixfold enrichment) and MAC3B (AT2G33340) (ANVDDDS#AQDS; ANVDDDSAQDS#, sevenfold enrichment). In plants they are important for resistance against pathogen infection (82). More importantly, in human cell culture Prp19 is involved in DDR by directly binding to RPA at ssDNA lesion sites, promoting RPA ubiquitylation and further promote ATR activation (83, 84).
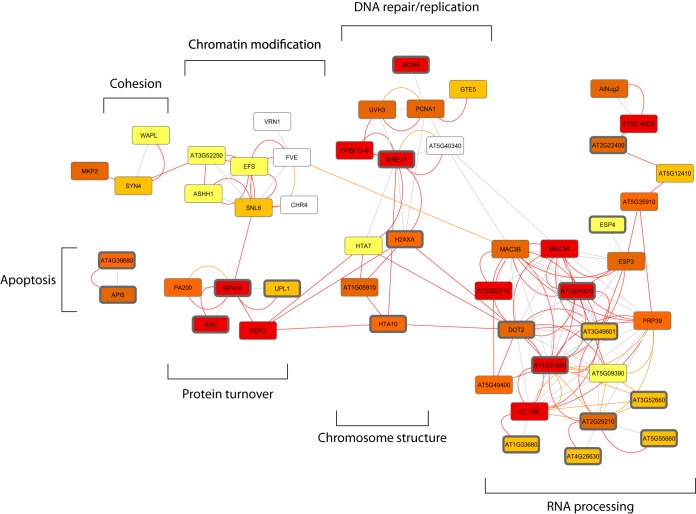
Looking at the data from a more systemic angle allows depicting potential communalities between different organisms. Sixty-nine human orthologs of the 108 plant ATM/ATR targets protein were identified using five different programs (Supplemental Table S9) and the information has been superimposed on a STRING network, visualizing the clusters and relations of the found ATM/ATR target proteins. Several of the human orthologs (26 of 69) have also been found to be affected by differential phosphorylation in DDR in other screens. The conserved proteins cluster in different groups involved in RNA processing, DNA repair and replication, chromosome structure, chromatin modifications, cohesion, and apoptosis (Fig. 6). Notably, none of these processes is related to signal transduction. This underlines that the factors that transduce the DNA damage signal are not conserved in plants, whereas those that detect the damage and elicit the signaling cascade (ATM and ATR) and those that implement the actual DNA repair are conserved. As outlined above, the canonical signal transducer kinases CHK1 and CHK2, that act downstream of ATM and ATR in humans, have not been found in plants. This indicates a dramatic difference of the “wiring” of the plant DDR. The plant specific transcription activator SOG1 has been recognized as a direct and immediate connection between DNA damage and transcriptional changes (15). It is a direct target of ATM and the genes that depend on ATM, ATR, and SOG1 are largely identical, indicating that SOG1 exclusively integrates ATM and ATR mediated DNA damage signals (85, 86). Phosphorylated SOG1 peptides have not been identified in the given screen because of an adverse tryptic cleavage pattern. Though not related by sequence, SOG1 has been defined to have an analogous function of human p53, which is not present in plant genomes (87). p53 is a transcriptional activator and downstream target of (among others) ATM, ATR, CHK1, and CHK2 (88). It is a key integrator of DNA damage signaling (and other stresses) and needed for the expression of many genes that encode DNA repair proteins and cell cycle and apoptosis regulators (89).
Fig. 6.
Many plant ATM/ATR targets are functionally interconnected and are related to human orthologs. The 108 proteins identified as direct and indirect targets of ATM and ATR were entered to STRING version 9.1 to identify known and predicted protein–protein interactions. About half of the proteins could be connected by either experimental evidence (red lines), by interactions based on curated databases (orange lines) or by text-mining (gray lines). The node colors indicate, with how many out of five used programs a human ortholog could be identified. No color: 0, light yellow: 1, dark yellow: 2, orange: 3, brown: 4, dark red: 5. Candidates with orthologs being affected by differential phosphorylation in response to DNA damage in other screens are marked with dark gray edges (see also supplemental Table S9).
Although transcriptional changes are important for DDR (e.g. to synthesize effective DNA repair proteins like AtRAD51 (90), the full range of physiological changes cannot be explained by transcriptional changes alone, especially considering the many targets of the CHK1 and CHK2 kinases in humans and other nonplant organisms. In our screen we found two kinases following a “Pfam domain” search that are phosphorylated in an ATM/ATR and IR dependent manner: (1) An unknown protein with a canonical phospho-kinase motif (encoded by At2g07180). It is highly conserved in plants but has no clear relatives outside the plant kingdom. It is phosphorylated on a non-S/TQ residue (S#VTLYEASSDSQGTR, threefold enrichment), shows low ubiquitous and strong expression in pollen. It is strongly up-regulated upon genotoxic and other stress conditions (91); (2) The dual function protein kinase/phosphatase VTC3 (encoded by At2g40860), which has been found to be phosphorylated on a S/TQ residue (TSNS#QS#DTDGADIIK, twofold enrichment). This protein shows strong ubiquitous expression (91) and is involved in maintaining the ascorbic acid pool, important to counteract oxidative stress (92). Furthermore the finding of a dominant “SE” motif (16 sites), embedded in acidic residues, indicates that also in plants ATM and ATR initiate a signaling cascade involving transducer kinases. As written above, it is tempting to speculate that the corresponding kinase is the multimeric CK2 kinase, which has been shown to be involved in DDR in plants and various organisms. We hypothesize that in plants CHK1 and CHK2 have been substituted by other kinases like the modular CK2, VTC3, and the kinase encoded by At2g07180.
Our data-set also pictures dynamic features of the Arabidopsis phosphoproteome that do not depend on ATM and ATR. Of the 604 phosphopeptides up-regulated upon irradiation, 470 peptides were ATM/ATR independent (Fig. 3; supplemental Table S7). In the corresponding 310 proteins, only 1.7% of the phosphorylations occurred on S/T-Q sites. Furthermore, only 164 have a GO-annotation of nuclear localization and many of the phosphorylation events will not be related to DDR.
CONCLUSIONS
Here we present the first large-scale phosphoproteomic study, quantitatively dissecting DNA damage repair mechanisms in plants, focusing on the master regulators, ATM and ATR. As outlined above, we found many factors directly involved in DNA repair and chromatin remodeling. Although many of the proteins have already been known for their importance to maintain genome integrity, we identified several of them as direct or indirect targets of ATM and ATR. The canonical “S/T-Q” sites previously identified as the minimal consensus site for ATM and ATR kinases in other model organisms (51) also appears to be the preferential target in higher plants. This statement is supported by our data demonstrating that that 62.7% of all phosphorylation events induced by irradiation and depending on ATM and ATR occurred within the S/T-Q motif, but only 1.7% of the irradiation induced and ATM/ATR independent phosphorylation events. Moreover, the proteins corresponding to the experimentally identified ATM/ATR targets are strongly enriched for S/T-Q sites with an average of 6.0 S/T-Q sites/protein compared with 2.6 S/T-Q sites/protein in the Arabidopsis proteome. We also find a significant enrichment of S/T-Q cluster domains (SCD – defined by at least three S/T-Q sites within a window of 100 amino acids (51)) with about 42% of the ATM/ATR target proteins in our data-set that harbor at least one phosphorylated S/T-Q site also harbor at least one SCD compared with only 12% of all Arabidopsis proteins that contain a least one S/T-Q site also harbor at least one SCD.
Interestingly, phosphorylation of SE motifs was also significantly up-regulated in an irradiation and ATM/ATR dependent manner. As written above, the SE motif has been described as a consensus motif of Casein Kinase 2 (CK2) (52). We speculate that CK2 may be one of the still elusive transducer kinases acting downstream of ATM and ATR. This idea is supported by the fact that CK2 has been implicated in DNA repair in mammalians, targeting known DNA repair proteins like MRE11, XRCC1, or p53 (93). Furthermore, CK2-defective plants show decreased homologous recombination and are more sensitive to IR (54). Further two candidates to act as transducer kinases have been identified in the screen (see above) and all three kinases and their potential roles in DNA damage signaling are currently investigated.
Taken together, this study presents a very large proteome and phospho-proteome data-sets obtained in the plant Arabidopsis thaliana. We demonstrate the (few) changes of relative protein abundance 15 min after plants have been exposed to IR. More importantly, we identified, in a quantitative manner, the fast changes of the phosphorylation status of about 461 proteins following a pulse of IR. Utilizing the atm atr double mutant plant line enabled us to attribute 134 of these changes to be ATM and ATR dependent. Importantly, our work revealed 66 novel targets of ATM/ATR in higher plants and we anticipate that these findings will stimulate further research to understand the regulation of DNA repair in higher plants.
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository (94) with the dataset identifier PXD000033.
Supplementary Material
Footnotes
Author contributions: P.S. and K.M. designed research; E.R. and M.H. performed research; E.R., M.H., T.K., P.P., M.N., and J.Y. analyzed data; E.R., M.H., T.K., P.S., and K.M. wrote the paper.
* This work was supported by EUFP7/2007-2014 grant KBBE-2009-222883 (http://cordis.europa.eu/fp7/home_en.html) (KM and PS) and No 262067 PRIME-XS (KM) and by the Austrian Science Fund FWF SFB F3402 (KM and PS), P 24685-B24 (KM) and TRP 308-N15 (KM).
 This article contains supplemental Tables S1 to S9.
This article contains supplemental Tables S1 to S9.
1 The abbreviations used are:
- DDR
- DNA damage response
- ACN
- acetonitrile
- ATM
- ataxia telangiectasia-mutated
- ATR
- ataxia telangiectasia-mutated and Rad3-related
- CID
- collision induced dissociation
- DSB
- DNA double strand break
- FA
- formic acid
- FASP
- filter assisted sample preparation
- FDR
- false discovery rate
- HCD
- higher-energy collisional dissociation
- IMAC
- immobilized metal affinity chromatography
- IR
- ionizing radiation
- iTRAQ
- isobaric tags for relative and absolute quantification
- MOAC
- metal oxide affinity chromatography
- PSM
- peptide spectrum match
- SCX
- strong cation exchange chromatography
- TCEP
- tris[2-carboxyethyl] phosphine
- TEAB
- triethylammonium bicarbonate.
REFERENCES
- 1. Dissmeyer N., Schnittger A. (2011) Guide to the book plant kinases. Methods Mol. Biol. 779, 3–5 [DOI] [PubMed] [Google Scholar]
- 2. Lovejoy C. A., Cortez D. (2009) Common mechanisms of PIKK regulation. DNA Rep. 8, 1004–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Greenwell P. W., Kronmal S. L., Porter S. E., Gassenhuber J., Obermaier B., Petes T. D. (1995) TEL1, a gene involved in controlling telomere length in S. cerevisiae, is homologous to the human ataxia telangiectasia gene. Cell 82, 823–829 [DOI] [PubMed] [Google Scholar]
- 4. Weinert T. A., Kiser G. L., Hartwell L. H. (1994) Mitotic checkpoint genes in budding yeast and the dependence of mitosis on DNA replication and repair. Genes Dev. 8, 652–665 [DOI] [PubMed] [Google Scholar]
- 5. Smolka M. B., Albuquerque C. P., Chen S.-h., Zhou H. (2007) Proteome-wide identification of in vivo targets of DNA damage checkpoint kinases. Proc. Natl. Acad. Sci. U.S.A. 104, 10364–10369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Matsuoka S., Ballif B. A., Smogorzewska A., McDonald E. R., 3rd, Hurov K. E., Luo J., Bakalarski C. E., Zhao Z., Solimini N., Lerenthal Y., Shiloh Y., Gygi S. P., Elledge S. J. (2007) ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science 316, 1160–1166 [DOI] [PubMed] [Google Scholar]
- 7. Stokes M. P., Rush J., MacNeill J., Ren J. M., Sprott K., Nardone J., Yang V., Beausoleil S. A., Gygi S. P., Livingstone M., Zhang H., Polakiewicz R. D., Comb M. J. (2007) Profiling of UV-induced ATM/ATR signaling pathways. Proc. Natl. Acad. Sci. U.S.A. 104, 19855–19860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Garcia V., Bruchet H., Camescasse D., Granier F., Bouchez D., Tissier A. (2003) AtATM is essential for meiosis and the somatic response to DNA damage in plants. Plant Cell 15, 119–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Friesner J. D., Liu B., Culligan K., Britt A. B. (2005) Ionizing radiation-dependent gamma-H2AX focus formation requires ataxia telangiectasia mutated and ataxia telangiectasia mutated and Rad3-related. Mol. Biol. Cell 16, 2566–2576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Culligan K. M., Britt A. B. (2008) Both ATM and ATR promote the efficient and accurate processing of programmed meiotic double-strand breaks. Plant J. 55, 629–638 [DOI] [PubMed] [Google Scholar]
- 11. Culligan K. M., Robertson C. E., Foreman J., Doerner P., Britt A. B. (2006) ATR and ATM play both distinct and additive roles in response to ionizing radiation. Plant J. 48, 947–961 [DOI] [PubMed] [Google Scholar]
- 12. Culligan K., Tissier A., Britt A. (2004) ATR regulates a G2-phase cell-cycle checkpoint in Arabidopsis thaliana. Plant Cell 16, 1091–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cools T., De Veylder L. (2009) DNA stress checkpoint control and plant development. Curr. Opin. Plant Biol. 12, 23–28 [DOI] [PubMed] [Google Scholar]
- 14. Templeton G. W., Moorhead G. B. (2005) The phosphoinositide-3-OH-kinase-related kinases of Arabidopsis thaliana. EMBO Rep. 6, 723–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yoshiyama K., Sakaguchi K., Kimura S. (2013) DNA damage response in plants: conserved and variable response compared to animals. Biology 2, 1338–1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lisby M., Barlow J. H., Burgess R. C., Rothstein R. (2004) Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell 118, 699–713 [DOI] [PubMed] [Google Scholar]
- 17. Bundock P., Hooykaas P. (2002) Severe developmental defects, hypersensitivity to DNA-damaging agents, and lengthened telomeres in Arabidopsis MRE11 mutants. Plant Cell 14, 2451–2462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jazayeri A., Balestrini A., Garner E., Haber J. E., Costanzo V. (2008) Mre11-Rad50-Nbs1-dependent processing of DNA breaks generates oligonucleotides that stimulate ATM activity. EMBO J. 27, 1953–1962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wyman C., Kanaar R. (2006) DNA double-strand break repair: all's well that ends well. Annu. Rev. Genet. 40, 363–383 [DOI] [PubMed] [Google Scholar]
- 20. Gobbini E., Cesena D., Galbiati A., Lockhart A., Longhese M. P. (2013) Interplays between ATM/Tel1 and ATR/Mec1 in sensing and signaling DNA double-strand breaks. DNA Repair 12, 791–799 [DOI] [PubMed] [Google Scholar]
- 21. Zeman M. K., Cimprich K. A. (2013) Causes and consequences of replication stress. Nat. Cell Biol. 16, 2–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zou L., Elledge S. J. (2003) Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 300, 1542–1548 [DOI] [PubMed] [Google Scholar]
- 23. Zou L., Cortez D., Elledge S. J. (2002) Regulation of ATR substrate selection by Rad17-dependent loading of Rad9 complexes onto chromatin. Genes Dev. 16, 198–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kocher T., Superti-Furga G. (2007) Mass spectrometry-based functional proteomics: from molecular machines to protein networks. Nat. Methods 4, 807–815 [DOI] [PubMed] [Google Scholar]
- 25. Walther T. C., Mann M. (2010) Mass spectrometry-based proteomics in cell biology. J. Cell Biol. 190, 491–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Schulze W. X. (2010) Proteomics approaches to understand protein phosphorylation in pathway modulation. Curr. Opin. Plant Biol. 13, 280–287 [DOI] [PubMed] [Google Scholar]
- 27. Choudhary C., Mann M. (2010) Decoding signaling networks by mass spectrometry-based proteomics. Nat. Rev. Mol. Cell Biol. 11, 427–439 [DOI] [PubMed] [Google Scholar]
- 28. Chalkley R. J., Clauser K. R. (2013) Modification site localization scoring: strategies and performance. Mol. Cell. Proteomics 11, 3–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rose C. M., Venkateshwaran M., Volkening J. D., Grimsrud P. A., Maeda J., Bailey D. J., Park K., Howes-Podoll M., den Os D., Yeun L. H., Westphall M. S., Sussman M. R., Ane J. M., Coon J. J. (2012) Rapid phosphoproteomic and transcriptomic changes in the rhizobia-legume symbiosis. Mol. Cell. Proteomics 11, 724–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lan P., Li W. F., Wen T. N., Schmidt W. (2012) Quantitative phosphoproteome profiling of iron-deficient Arabidopsis roots. Plant Physiol. 159, 403–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mayank P., Grossman J., Wuest S., Boisson-Dernier A., Roschitzki B., Nanni P., Nuhse T., Grossniklaus U. (2012) Characterization of the phosphoproteome of mature Arabidopsis pollen. Plant J. 72, 89–101 [DOI] [PubMed] [Google Scholar]
- 32. Meyer L. J., Gao J. J., Xu D., Thelen J. J. (2012) Phosphoproteomic analysis of seed maturation in Arabidopsis, rapeseed, and soybean. Plant Physiol. 159, 517–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kline K. G., Barrett-Wilt G. A., Sussman M. R. (2010) In planta changes in protein phosphorylation induced by the plant hormone abscisic acid. Proc. Natl. Acad. Sci. U.S.A. 107, 15986–15991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhang H. T., Zhou H. J., Berke L., Heck A. J. R., Mohammed S., Scheres B., Menke F. L. H. (2013) Quantitative phosphoproteomics after auxin-stimulated lateral root induction identifies an SNX1 protein phosphorylation site required for growth. Mol. Cell. Proteomics 12, 1158–1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nilsson C. L. (2012) Advances in quantitative phosphoproteomics. Anal. Chem. 84, 735–746 [DOI] [PubMed] [Google Scholar]
- 36. Thingholm T. E., Jensen O. N., Robinson P. J., Larsen M. R. (2008) SIMAC (sequential elution from IMAC), a phosphoproteomics strategy for the rapid separation of monophosphorylated from multiply phosphorylated peptides. Mol. Cell. Proteomics 7, 661–671 [DOI] [PubMed] [Google Scholar]
- 37. Wisniewski J. R., Zougman A., Nagaraj N., Mann M. (2009) Universal sample preparation method for proteome analysis. Nat. Methods 6, U359–U360 [DOI] [PubMed] [Google Scholar]
- 38. Boyes D. C., Zayed A. M., Ascenzi R., McCaskill A. J., Hoffman N. E., Davis K. R., Gorlach J. (2001) Growth stage-based phenotypic analysis of Arabidopsis: a model for high throughput functional genomics in plants. Plant Cell 13, 1499–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Villen J., Gygi S. P. (2008) The SCX/IMAC enrichment approach for global phosphorylation analysis by mass spectrometry. Nat. Protoc. 3, 1630–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bodenmiller B., Mueller L. N., Mueller M., Domon B., Aebersold R. (2007) Reproducible isolation of distinct, overlapping segments of the phosphoproteome. Nat. Methods 4, 231–237 [DOI] [PubMed] [Google Scholar]
- 41. Kocher T., Pichler P., Schutzbier M., Stingl C., Kaul A., Teucher N., Hasenfuss G., Penninger J. M., Mechtler K. (2009) High precision quantitative proteomics using iTRAQ on an LTQ Orbitrap: a new mass spectrometric method combining the benefits of all. J. Proteome Res. 8, 4743–4752 [DOI] [PubMed] [Google Scholar]
- 42. Taus T., Koecher T., Pichler P., Paschke C., Schmidt A., Henrich C., Mechtler K. (2011) Universal and confident phosphorylation site localization using phosphoRS. J. Proteome Res. 10, 5354–5362 [DOI] [PubMed] [Google Scholar]
- 43. Schwartz D., Gygi S. P. (2005) An iterative statistical approach to the identification of protein phosphorylation motifs from large-scale data sets. Nat. Biotech. 23, 1391–1398 [DOI] [PubMed] [Google Scholar]
- 44. Mohammed S., Heck A. J. R. (2011) Strong cation exchange (SCX) based analytical methods for the targeted analysis of protein post-translational modifications. Curr. Opin. Biotechnol. 22, 9–16 [DOI] [PubMed] [Google Scholar]
- 45. Meier S., Bastian R., Donaldson L., Murray S., Bajic V., Gehring C. (2008) Co-expression and promoter content analyses assign a role in biotic and abiotic stress responses to plant natriuretic peptides. BMC Plant Biol. 8, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Dubreuil-Maurizi C., Vitecek J., Marty L., Branciard L., Frettinger P., Wendehenne D., Meyer A. J., Mauch F., Poinssot B. (2011) Glutathione deficiency of the Arabidopsis mutant pad2–1 affects oxidative stress-related events, defense gene expression, and the hypersensitive response. Plant Physiol. 157, 2000–2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Barzilai A., Rotman G., Shiloh Y. (2002) ATM deficiency and oxidative stress: a new dimension of defective response to DNA damage. DNA Repair 1, 3–25 [DOI] [PubMed] [Google Scholar]
- 48. Guo Z., Kozlov S., Lavin M. F., Person M. D., Paull T. T. (2010) ATM activation by oxidative stress. Science 330, 517–521 [DOI] [PubMed] [Google Scholar]
- 49. Wu R. H., Dephoure N., Haas W., Huttlin E. L., Zhai B., Sowa M. E., Gygi S. P. (2011) Correct interpretation of comprehensive phosphorylation dynamics requires normalization by protein expression changes. Mol. Cell. Proteomics 10, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bennetzen M. V., Larsen D. H., Bunkenborg J., Bartek J., Lukas J., Andersen J. S. (2010) Site-specific phosphorylation dynamics of the nuclear proteome during the DNA damage response. Mol. Cell. Proteomics 9, 1314–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Traven A., Heierhorst J. (2005) SQ/TQ cluster domains: concentrated ATM/ATR kinase phosphorylation site regions in DNA-damage-response proteins. Bioessays 27, 397–407 [DOI] [PubMed] [Google Scholar]
- 52. Pinna L. A. (2002) Protein kinase CK2: a challenge to canons. J. Cell Sci. 115, 3873–3878 [DOI] [PubMed] [Google Scholar]
- 53. Litchfield D. W. (2003) Protein kinase CK2: structure, regulation and role in cellular decisions of life and death. Biochem. J. 369, 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Moreno-Romero J., Armengot L., Marques-Bueno M. M., Britt A., Martinez M. C. (2012) CK2-defective Arabidopsis plants exhibit enhanced double-strand break repair rates and reduced survival after exposure to ionizing radiation. Plant J. 71, 627–638 [DOI] [PubMed] [Google Scholar]
- 55. Cargnello M., Roux P. P. (2011) Activation and function of the MAPKs and their substrates, the MAPK-activated protein kinases. Microbiol. Mol. Biol. Rev. 75, 50–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Beckers G. J. M., Jaskiewicz M., Liu Y. D., Underwood W. R., He S. Y., Zhang S. Q., Conrath U. (2009) Mitogen-activated protein kinases 3 and 6 are required for full priming of stress responses in Arabidopsis thaliana. Plant Cell 21, 944–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Umezawa T., Sugiyama N., Takahashi F., Anderson J. C., Ishihama Y., Peck S. C., Shinozaki K. (2013) Genetics and phosphoproteomics reveal a protein phosphorylation network in the abscisic acid signaling pathway in Arabidopsis thaliana. Sci. Signal. 6, 13. [DOI] [PubMed] [Google Scholar]
- 58. Lee T. Y., Bretana N. A., Lu C. T. (2011) PlantPhos: using maximal dependence decomposition to identify plant phosphorylation sites with substrate site specificity. BMC Bioinformatics 12, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. van Attikum H., Bundock P., Overmeer R. M., Lee L. Y., Gelvin S. B., Hooykaas P. J. J. (2003) The Arabidopsis AtLIG4 gene is required for the repair of DNA damage, but not for the integration of Agrobacterium T-DNA. Nucleic Acids Res. 31, 4247–4255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Puizina J., Siroky J., Mokros P., Schweizer D., Riha K. (2004) Mre11 deficiency in Arabidopsis is associated with chromosomal instability in somatic cells and Spo11-dependent genome fragmentation during meiosis. Plant Cell 16, 1968–1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Liu Z. R., Hall J. D., Mount D. W. (2001) Arabidopsis UVH3 gene is a homolog of the Saccharomyces cerevisiae RAD2 and human XPG DNA repair genes. Plant J. 26, 329–338 [DOI] [PubMed] [Google Scholar]
- 62. Rogakou E. P., Pilch D. R., Orr A. H., Ivanova V. S., Bonner W. M. (1998) DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 273, 5858–5868 [DOI] [PubMed] [Google Scholar]
- 63. Dong Q. H., Han F. P. (2012) Phosphorylation of histone H2A is associated with centromere function and maintenance in meiosis. Plant J. 71, 800–809 [DOI] [PubMed] [Google Scholar]
- 64. Campi M., D'Andrea L., Emiliani J., Casati P. (2012) Participation of chromatin-remodeling proteins in the repair of ultraviolet-B-damaged DNA. Plant Physiol. 158, 981–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Mizuguchi G., Shen X. T., Landry J., Wu W. H., Sen S., Wu C. (2004) ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science 303, 343–348 [DOI] [PubMed] [Google Scholar]
- 66. Choi K., Zhao X., Kelly K. A., Venn O., Higgins J. D., Yelina N. E., Hardcastle T. J., Ziolkowski P. A., Copenhaver G. P., Franklin F. C. H., McVean G., Henderson I. R. (2013) Arabidopsis meiotic crossover hot spots overlap with H2A.Z nucleosomes at gene promoters. Nat. Genet. 45, 1327–1336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Shaked H., Avivi-Ragolsky N., Levy A. A. (2006) Involvement of the Arabidopsis SWI2/SNF2 chromatin remodeling gene family in DNA damage response and recombination. Genetics 173, 985–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Stanley F. K. T., Moore S., Goodarzi A. A. (2013) CHD chromatin remodeling enzymes and the DNA damage response. Mutat. Res. Fundam. Mol. Mech. Mutagen. 750, 31–44 [DOI] [PubMed] [Google Scholar]
- 69. Lu F. L., Cui X., Zhang S. B., Jenuwein T., Cao X. F. (2011) Arabidopsis REF6 is a histone H3 lysine 27 demethylase. Nat. Genet. 43, U715–U144 [DOI] [PubMed] [Google Scholar]
- 70. De K., Sterle L., Krueger L., Yang X. H., Makaroff C. A. (2014) Arabidopsis thaliana WAPL is essential for the prophase removal of cohesin during meiosis. PLoS Genet. 10, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Losada A., Yokochi T., Hirano T. (2005) Functional contribution of Pds5 to cohesin-mediated cohesion in human cells and Xenopus egg extracts. J. Cell Sci. 118, 2133–2141 [DOI] [PubMed] [Google Scholar]
- 72. Tedeschi A., Wutz G., Huet S., Jaritz M., Wuensche A., Schirghuber E., Davidson I. F., Tang W., Cisneros D. A., Bhaskara V., Nishiyama T., Vaziri A., Wutz A., Ellenberg J., Peters J. M. (2013) Wapl is an essential regulator of chromatin structure and chromosome segregation. Nature 501, 564–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. da Costa-Nunes J. A., Bhatt A. M., O'Shea S., West C. E., Bray C. M., Grossniklaus U., Dickinson H. G. (2006) Characterization of the three Arabidopsis thaliana RAD21 cohesins reveals differential responses to ionizing radiation. J. Exp. Bot. 57, 971–983 [DOI] [PubMed] [Google Scholar]
- 74. Gamper A. M., Choi S., Matsumoto Y., Banerjee D., Tomkinson A. E., Bakkenist C. J. (2012) ATM protein physically and functionally interacts with proliferating cell nuclear antigen to regulate DNA synthesis. J. Biol. Chem. 287, 12445–12454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Leyva-Gonzalez M. A., Ibarra-Laclette E., Cruz-Ramirez A., Herrera-Estrella L. (2012) Functional and transcriptome analysis reveals an acclimatization strategy for abiotic stress tolerance mediated by Arabidopsis NF-YA family members. Plos One 7, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Chen I. P., Haehnel U., Altschmied L., Schubert I., Puchta H. (2003) The transcriptional response of Arabidopsis to genotoxic stress – a high-density colony array study (HDCA). Plant J. 35, 771–786 [DOI] [PubMed] [Google Scholar]
- 77. Bundock P., Hooykaas P. (2005) An Arabidopsis hAT-like transposase is essential for plant development. Nature 436, 282–284 [DOI] [PubMed] [Google Scholar]
- 78. Ustrell V., Hoffman L., Pratt G., Rechsteiner M. (2002) PA200, a nuclear proteasome activator involved in DNA repair. EMBO J. 21, 3516–3525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Gray W. M., del Pozo J. C., Walker L., Hobbie L., Risseeuw E., Banks T., Crosby W. L., Yang M., Ma H., Estelle M. (1999) Identification of an SCF ubiquitin-ligase complex required for auxin response in Arabidopsis thaliana. Genes Dev. 13, 1678–1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Yang M., Hu Y., Lodhi M., McCombie W. R., Ma H. (1999) The Arabidopsis SKP1-LIKE1 gene is essential for male meiosis and may control homolog separation. Proc. Natl. Acad. Sci. U.S.A. 96, 11416–11421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Yang X., Timofejeva L., Ma H., Makaroff C. A. (2006) The Arabidopsis SKP1 homolog ASK1 controls meiotic chromosome remodeling and release of chromatin from the nuclear membrane and nucleolus. J. Cell Sci. 119, 3754–3763 [DOI] [PubMed] [Google Scholar]
- 82. Monaghan J., Xu F., Gao M., Zhao Q., Palma K., Long C., Chen S., Zhang Y., Li X. (2009) Two Prp19-like u-box proteins in the MOS4-associated complex play redundant roles in plant innate immunity. PLoS Pathog. 5, e1000526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Lu X., Legerski R. J. (2007) The Prp19/Pso4 core complex undergoes ubiquitylation and structural alterations in response to DNA damage. Biochem. Biophys. Res. Commun. 354, 968–974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Marechal A., Li J. M., Ji X. Y., Wu C. S., Yazinski S. A., Nguyen H. D., Liu S. Z., Jimenez A. E., Jin J. P., Zou L. (2014) PRP19 transforms into a sensor of RPA-ssDNA after DNA damage and drives ATR activation via a ubiquitin-mediated circuitry. Mol. Cell 53, 235–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Yoshiyama K. O., Kobayashi J., Ogita N., Ueda M., Kimura S., Maki H., Umeda M. (2013) ATM-mediated phosphorylation of SOG1 is essential for the DNA damage response in Arabidopsis. EMBO Rep. 14, 817–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Yoshiyama K., Conklin P. A., Huefner N. D., Britt A. B. (2009) Suppressor of gamma response 1 (SOG1) encodes a putative transcription factor governing multiple responses to DNA damage. Proc. Natl. Acad. Sci. U.S.A. 106, 12843–12848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Yoshiyama K. O., Kimura S., Maki H., Britt A. B., Umeda M. (2014) The role of SOG1, a plant-specific transcriptional regulator, in the DNA damage response. Plant Signal. Behav. 9, e28889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Dai C., Gu W. (2010) p53 post-translational modification: deregulated in tumorigenesis. Trends Mol. Med. 16, 528–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Carvajal L. A., Manfredi J. J. (2013) Another fork in the road-life or death decisions by the tumour suppressor p53. EMBO Rep. 14, 414–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Culligan K. M., Robertson C. E., Foreman J., Doerner P., Britt A. B. (2006) ATR and ATM play both distinct and additive roles in response to ionizing radiation. Plant J. 48, 947–961 [DOI] [PubMed] [Google Scholar]
- 91. Winter D., Vinegar B., Nahal H., Ammar R., Wilson G. V., Provart N. J. (2007) An “electronic fluorescent pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS One 2, e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Conklin P. L., DePaolo D., Wintle B., Schatz C., Buckenmeyer G. (2013) Identification of Arabidopsis VTC3 as a putative and unique dual function protein kinase::protein phosphatase involved in the regulation of the ascorbic acid pool in plants. J. Exp. Bot. 64, 2793–2804 [DOI] [PubMed] [Google Scholar]
- 93. Guerra B., Iwabuchi K., Issinger O.-G. (2014) Protein kinase CK2 is required for the recruitment of 53BP1 to sites of DNA double-strand break induced by radiomimetic drugs. Cancer Lett. 345, 115–123 [DOI] [PubMed] [Google Scholar]
- 94. Vizcaino J. A., Cote R. G., Csordas A., Dianes J. A., Fabregat A., Foster J. M., Griss J., Alpi E., Birim M., Contell J., O'Kelly G., Schoenegger A., Ovelleiro D., Perez-Riverol Y., Reisinger F., Rios D., Wang R., Hermjakob H. (2013) The Proteomics Identifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res. 41, D1063–D1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.