Abstract
The lipid raft concept proposes that membrane environments enriched in cholesterol and sphingolipids cluster certain proteins and form platforms to integrate cell signaling. In cardiac muscle, caveolae concentrate signaling molecules and ion transporters, and play a vital role in adrenergic regulation of excitation–contraction coupling, and consequently cardiac contractility. Proteomic analysis of cardiac caveolae is hampered by the presence of contaminants that have sometimes, erroneously, been proposed to be resident in these domains. Here we present the first unbiased analysis of the proteome of cardiac caveolae, and investigate dynamic changes in their protein constituents following adrenoreceptor (AR) stimulation.
Rat ventricular myocytes were treated with methyl-β-cyclodextrin (MβCD) to deplete cholesterol and disrupt caveolae. Buoyant caveolin-enriched microdomains (BCEMs) were prepared from MβCD-treated and control cell lysates using a standard discontinuous sucrose gradient. BCEMs were harvested, pelleted, and resolubilized, then alkylated, digested, and labeled with iTRAQ reagents, and proteins identified by LC-MS/MS on a LTQ Orbitrap Velos Pro. Proteins were defined as BCEM resident if they were consistently depleted from the BCEM fraction following MβCD treatment. Selective activation of α-, β1-, and β2-AR prior to preparation of BCEMs was achieved by application of agonist/antagonist pairs for 10 min in populations of field-stimulated myocytes.
We typically identified 600–850 proteins per experiment, of which, 249 were defined as high-confidence BCEM residents. Functional annotation clustering indicates cardiac BCEMs are enriched in integrin signaling, guanine nucleotide binding, ion transport, and insulin signaling clusters. Proteins possessing a caveolin binding motif were poorly enriched in BCEMs, suggesting this is not the only mechanism that targets proteins to caveolae. With the notable exception of the cavin family, very few proteins show altered abundance in BCEMs following AR activation, suggesting signaling complexes are preformed in BCEMs to ensure a rapid and high fidelity response to adrenergic stimulation in cardiac muscle.
Caveolae are specialized invaginated lipid rafts (1), around 50–100 nm in diameter, enriched in cholesterol and sphingolipids, and characterized by the presence of caveolin and cavin proteins. The lipid environment, caveolin content, and morphology of caveolae are central to their diverse functional roles, which include coordination of signal transduction, cholesterol homeostasis, and endocytosis (2). Clustering of elements of particular signal cascades within a caveola promotes efficiency and fidelity of signaling. Although caveolae and noncaveolar rafts coexist, evidence suggests that most proteins are clustered by caveolae in the cardiac cell (3). Caveolin exists as three major isoforms: caveolin 1 and caveolin 2, which are expressed in most cell types, and caveolin 3, which is the muscle-specific isoform. Caveolins 1 and 3 are the predominant forms found in the adult cardiac myocyte (4, 5). Four members of the cavin family of related proteins exist, and all have been detected in the heart (6).
One of caveolae's best-characterized roles is as a signalosome, a compartment that brings together components of signal transduction cascades (including receptors, effectors, and targets (7)). Within caveolae, the 20-residue scaffolding domain of caveolin (CSD)1 has been proposed to interact with a complementary caveolin-binding motif (CBM) in proteins. This enables oligomeric caveolin to act as a regulatory scaffold for macromolecular signaling complex formation (8). However, the ability of this simple and commonly occurring motif to interact with caveolin (directing proteins to caveolae and regulating their activity) has recently been challenged, because it is often buried within mature proteins (9, 10). Palmitoylation of juxtamembrane cysteine residues has also been proposed to partition proteins to ordered detergent-resistant membranes such as caveolae (11).
The organization of proteins in caveolae suggests that they have a key role in regulation of signaling in the heart. We adopt the convention of the field here to assign proteins as caveolar if they are present in buoyant caveolin-containing membrane fractions obtained by sucrose gradient fractionation or in morphologically identifiable caveolae by immunogold electron microscopy. For example, α1- and β2-adrenoceptors (AR) are found exclusively in caveolae-containing membrane fractions of the adult heart (12, 13), whereas β1-AR are in both caveolar and bulk sarcolemmal fractions (14). Cardiac caveolae are also sites of enrichment of G proteins (12, 15), effectors of AR (including adenylyl cyclase V/VI, protein kinase A (RII), GRK2, phospholipase Cβ, PP2A, and eNOS (13–16)), and their downstream targets. Importantly, the distribution of receptors, effectors, and their targets is key to the efficiency and fidelity of their coupling (13, 17, 18). For example, altered β1- and β2-AR responses have been observed following cholesterol depletion (which disrupts caveolae) and severing of normal caveolin 3 interactions with a caveolin 3 CSD peptide (19, 20).
A considerable number of cardiac ion transporters are resident in cardiac caveolae: voltage-gated sodium channels (21), L-type calcium channels (16), voltage gated potassium channels (22), ATP-sensitive potassium channels (23), the sodium-calcium exchanger (24) (NCX - although this has been challenged (25)), the sodium potassium ATPase (sodium pump) (26), and the plasma membrane calcium ATPase (PMCA) (27). Physical colocalization of ion transporters in the caveolar compartment may functionally link ion flow by providing a restricted diffusional space (28) and facilitates hormonal regulation of these transporters by placing them physically adjacent to signaling molecules. For example, regulation of a subpopulation of L-type calcium channels by β2-AR requires their colocalization in caveolae (16), and regulation of the cardiac sodium pump by phospholemman (PLM) relies on phosphorylation and dephosphorylation of PLM in caveolae (29, 30). The presence of ion transporters in caveolae is likely to have functional relevance beyond signal transduction because the lipid composition of the bilayer in which an ion transporter resides will influence its activity. Membrane cholesterol modulates many aspects of ion channel function: the sodium pump, for example, is regulated by the cholesterol content of the membranes within which it resides (31).
Using traditional biochemical techniques (such as sucrose density gradient fractionation followed by Western blotting), the presence of certain proteins in cardiac caveolae has been shown to be dynamic on an acute timescale (minutes), however, no unbiased assessment of global changes in caveolar composition during signaling has been reported. PKC isoforms translocate into caveolae upon activation (32), and various G-protein coupled receptors translocate in and out of caveolae upon activation: muscarinic M2 (33) receptors into caveolae, and β2-AR out of caveolae (14, 34). Pivotally, this redistribution of proteins has been closely linked with functional responses. For example, the negative inotropic effect of the muscarinic agonist carbachol is ascribed to the ability of M2 receptors to move to caveolae and couple therein to eNOS, which is exclusively found in this compartment (33). Conversely, one suggestion for the limited functional response following β2-AR receptor stimulation is that translocation from the caveolar compartment (where it normally couples to its effector adenylyl cyclase) to clathrin-coated pits terminates its downstream effects by internalization of the receptor (14, 34).
Hence, it has been proposed that many signaling proteins and ion transporters are located in caveolae and that the distribution between caveolar and noncaveolar membranes is dynamically regulated. Although quantitative proteomics has been used to highlight the breadth of proteins localized to caveolae in nonexcitable cells (for example (35)), to date no study has used an unbiased proteomic approach to define resident proteins of cardiac caveolae, or changes in the composition of caveolae during signaling events in any cell type.
The principal challenge to overcome in using proteomics to characterize a particular membrane compartment is purifying this compartment to homogeneity. The high sensitivity of detection possible in modern proteomics means that even minor contamination of caveolar membranes with another membrane compartment leads to mis-assignment of proteins as caveolar residents. In practice, because contamination of membrane preparations cannot be reduced to zero, an alternative approach has been developed: For proteins to be considered genuinely localized to lipid rafts/caveolae, their presence must be sensitive to cholesterol depletion (36). Hence stable isotope labeling with amino acids in cell culture (SILAC) and cholesterol depletion with methyl-beta-cyclodextrin (MβCD) in noncardiac cultured cells has distinguished genuine caveolar proteins from the mitochondrial, sarco/endoplasmic reticulum, and cytosolic proteins that routinely contaminate caveolae prepared by density gradient centrifugation (35). Using such an approach, contaminating proteins appear in the caveolar fraction whether or not cholesterol is depleted with MβCD, but genuine caveolar residents are specifically depleted. This approach has refuted research that implies that mitochondrial proteins are enriched in caveolae (37, 38), including data from cardiac tissue that suggested that principal components of cardiac caveolae are mitochondrial and structural proteins (39). The current study employed a similar approach—with the modification that because cardiac myocytes cannot be maintained in culture for SILAC (because of marked phenotypic changes), we used iTRAQ-based quantitative proteomics.
Here we identify 249 high-confidence caveolar proteins in the cardiac cell and show for the first time that the caveolar proteome is remarkably stable on an acute timescale following adrenoreceptor stimulation, suggesting that signaling complexes are preformed in this microdomain.
EXPERIMENTAL PROCEDURES
Materials and Antibodies
Unless indicated otherwise, all reagents were obtained from Sigma Chemical Company (Gillingham, UK) and were of the highest grade available. Anti-clathrin heavy chain and anti-caveolin 3 were from BD Biosciences (Oxford, UK). Anti-HADHA, cavin1, GFP, and Glut4 antibodies were from Abcam (Cambridge, UK). Anti-sodium pump α2 subunit and integrin α5 were from Merck Millipore (Watford, UK). Anti-adenylyl cyclase 5/6, insulin receptor, and PKCε were from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-Giα1/2 was from Pierce Antibodies (Thermo Scientific, Rockford, IL). Anti-moesin and anti-Kir 6.2 were from the MRC Protein Phosphorylation Unit, University of Dundee. The monoclonal antibody α6F raised against the sodium pump α1 subunit by Douglas M. Fambrough were obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by the University of Iowa, Department of Biology, Iowa City, IA 52242. An antibody to PLM is described elsewhere (40). Protease and phosphatase inhibitor cocktails were from Merck Millipore and Sigma, respectively.
Rat Ventricular Myocytes and Mouse Hearts
Calcium-tolerant adult rat ventricular myocytes (ARVM) were isolated by retrograde perfusion of collagenase in the Langendorff mode (as described in (41)). Myocytes were left to recover for 2 h at 35 °C before experiments. All drugs were applied at 35 °C. Hearts from PLM knockout (KO) animals and their wild type littermates (described in (42)) were briefly perfused in the Langendorff mode to remove contaminating blood, then snap frozen prior to homogenization.
Sucrose Gradient Fractionation of Adult Rat Ventricular Myocytes
Buoyant caveolin-enriched microdomains (BCEMs) were prepared from ARVM homogenized (5 × 6s bursts, Ultra-Turrax, with 20s intervals) and sonicated (6 × 20s bursts, 5 μm amplitude, Soniprep) in 500 mm sodium carbonate, pH 11, supplemented with 1 mm DTT, 1 mm EDTA, protease, and phosphatase inhibitors. Lysates were immediately adjusted to 45% sucrose by the addition of an equal volume of 90% w/v sucrose in (25 mm MES and 150 mm NaCl, pH 6.5) and 4 ml transferred to an ultracentrifuge tube and overlaid with 4 ml 35% sucrose and 4 ml 5% sucrose (dissolved in the same combination of salts). Following overnight centrifugation at 270,000 × g, 1 ml fractions (numbered 1 to 12) were collected. For each fraction, protein content was measured using the micro-BCA kit (Thermo Scientific), and cholesterol using Amplex red (Life Technologies, Carlsbad, CA).
Cholesterol Depletion and Loading
For cholesterol depletion, myocytes were incubated in 2% MβCD (Sigma) for 30 min with gentle mixing at 10 min intervals at room temperature prior to homogenization. Myocytes were cholesterol-loaded using MβCD that had been presaturated with cholesterol (Sigma).
Sample Preparation for iTRAQ
Membrane fractions from sucrose gradients were diluted in PBS and pelleted at 100,000 × g for 60min at 4 °C. Membranes were resuspended in 0.1% SDS, 500 mm tri-ethyl ammonium bicarbonate, reductively alkylated (dithiothreitol/iodoacetamide), tryptically digested, and labeled with iTRAQ reagents (AB Sciex, Foster City, CA) according to the manufacturer's instructions. Samples were then pooled and fractionated by strong cation exchange into 9–26 fractions, each of which was separated over a 120 min linear gradient using a UltiMate 3000 RSLCnano LC system (Dionex Corporation/Thermo Scientific) coupled to a LTQ Orbitrap Velos Pro ms system (Thermo Scientific).
Liquid Chromatography-Tandem MS (LC-MS/MS)
Trypsin digested peptides were separated using an Ultimate 3000 RSLCnano LC system (Dionex Corporation/Thermo Scientific) consisting of a HPG nano pump, an integrated column compartment, and separate thermostated autosampler. Ten microliters of sample (a total of 2 μg of peptide digest) was loaded with a constant flow of 5 μl/min onto an Acclaim PepMap 100 (C18, 100 μm × 2 cm) trap column (Dionex Corporation/Thermo Scientific). After trap enrichment peptides were eluted onto either an Acclaim PepMap100 C18 (75 μm × 15 cm, 3 μm, Dionex Corporation) column or an Acclaim PepMap RSLC (75 μm × 15 cm, nanoviper C18, 2 μm, Dionex Corporation/Thermo Scientific) column with a linear gradient of 5–40% solvent B (80% acetonitrile in 0.08% formic acid) over a 120 min with a constant flow of 300 nl/min. The UltiMate 3000 RSLC nano system was coupled to an LTQ Orbitrap Velos (Thermo Scientific) via a nano-ESI needle (Picotip Emitter, SilicaTip FS360–20-10; New Objective Inc., Woburn, MA) and nanospray flex ion source (Proxeon Biosystems/Thermo Scientific). The spray voltage was set to 1.2 kV and the temperature of the heated capillary was set to 250 °C. Full scan ms survey spectra (m/z 335–1800) in profile mode were acquired in the Orbitrap with a resolution of 60,000 after accumulation of 1e6 ions. The ten most intense peptide ions from the preview scan in the Orbitrap were fragmented by Higher-Energy Collisional Dissociation (Activation type: HCD, Normalized collision energy: 40%, Isolation width: 1.2Da, Activation time: 0.1 ms) after accumulation of 50,000 ions and analyzed in the Orbitrap at 7500 resolution. Maximal filling times were 500ms for the FT-MS full scans and 200ms for the FT-MS/MS scans. Monoisotopic precursor selection and charge state screening were enabled and all unassigned charge states as well as singly charged species were rejected. The dynamic exclusion list was restricted to a maximum of 500 entries with a maximum retention period of 45s and a relative mass window of 10 ppm. The lock mass (445.120024) option was enabled for survey scans to improve mass accuracy. Data were acquired using the XCalibur software.
Quantification and Bioinformatic Analysis
Protein identification and quantitation was performed using Proteome Discoverer (Version 1.3.0.339) with Mascot (Version 2.3.2) database searching against the International Protein Index (IPI) Rat database (Version 3.87; 39,925 sequences). Mass tolerance for peptide precursors was set at 10 ppm with fragment mass tolerance set at 0.06Da. Enzyme specificity was set to Trypsin/P with a maximum of two missed cleavages. The following fixed modifications (Carbamidomethyl (C), iTRAQ4plex (N-term) and variable modifications (Oxidation (M), Dioxidation (Met), Acetyl (N-term), Gln->pyroglu (N-term-Q), iTRAQ4plex (K), and iTRAQ 4plex (Y)) were considered. At the peptide level, the mascot ion score was applied for identification and the False Discovery Rate set to 1% with a minimum of two peptides for quantitation.
Mapping rat IPI to Human Uniprot Identifiers
In order to map outputs from Proteome Discoverer (ipi.RAT.v3.87) to the better-annotated UniProt database (release 2011_12), we used the BLAST+ executables (ftp://ftp.ncbi.nlm.nih.gov/blast/executables/blast+/) to find the closest UniProt Human ortholog to each IPI Rat entry. Python scripts (using the biopython extension (http://biopython.org)) were compiled to run the BLAST program, parse the blast output, and map the rat IPI to the human Uniprot. Two proteins were defined as homologs if they share at least 80% amino acid identity. Manual inspection was used to resolve protein homologies that could not be assigned by the automatic pipeline.
Caveolar Resident Proteins and Contaminants
Established caveolar resident proteins were identified in the UniProt database using the keyword “caveolar” and by searching the published literature. The final compiled list is reported in supplemental Table S1. Contaminant proteins were identified using the David Database (43). The proteins annotated with “mitochondrial” keyword were considered contaminants.
Adrenoreceptor Activation in Rat Ventricular Myocytes
Myocytes were field stimulated at 1 Hz throughout the experimental protocol. Antagonists were applied for 5 min before and during agonist exposure (10 min) at 35 °C. In order to achieve selective α-AR activation we used 10 μm phenylephrine in the presence of 10 μm atenolol, for β1-AR we used 100 nm isoprenaline in the presence of 100 nm ICI 118,551 and for β2-AR we used 10 μm zinterol in the presence of 300 nm CGP20712A. After treatment, myocytes were rapidly pelleted at 4 °C, and homogenized and sonicated prior to sucrose gradient fractionation.
SDS-PAGE and Western Blotting
Proteins were separated by electrophoresis on 6–20% gradient gels using the Mini Protean III system (BioRad) and transferred to PVDF membranes (Millipore) using a semidry transfer system (BioRad), which were blocked in 5% milk for minimum 1 h at room temperature. After incubation with primary antibodies overnight at 4 °C, membranes were incubated with HRP-linked secondary antibodies (GE Life Sciences) and imaged using Immobilon Chemiluminesent HRP substrate (Millipore) and a ChemiDoc XRS Imaging System (BioRad). Band densities were analyzed using the Quantity One analysis software (BioRad).
Cell Transfection and Co-immunoprecipitation
FT-293 cells expressing PLM-YFP under the control of a tetracycline-sensitive promoter were transfected with rat caveolin 3 (IMAGE clone 7103841) using lipofectamine. 24 h later cells were lyzed for immunoprecipitation of the sodium pump catalytic subunit (using monoclonal antibody C464.4, Millipore) in 2 mg/ml C12E10 in PBS supplemented with protease and phosphatase inhibitor cocktails, as well as an additional mixture of chemical phosphatase inhibitors: 5 mm sodium fluoride, 2 mm sodium orthovanadate, 2 mm sodium pyrophosphate, 2 mm sodium glycerophosphate. Immunoprecipitation was carried out as described previously (29). To ensure specificity, co-immunoprecipitation reactions were routinely immunoblotted for membrane proteins not copurifying with the immunoprecipitated protein (not shown).
RESULTS
Acute Cholesterol Depletion Removes Caveolar Resident Proteins from Buoyant Membranes
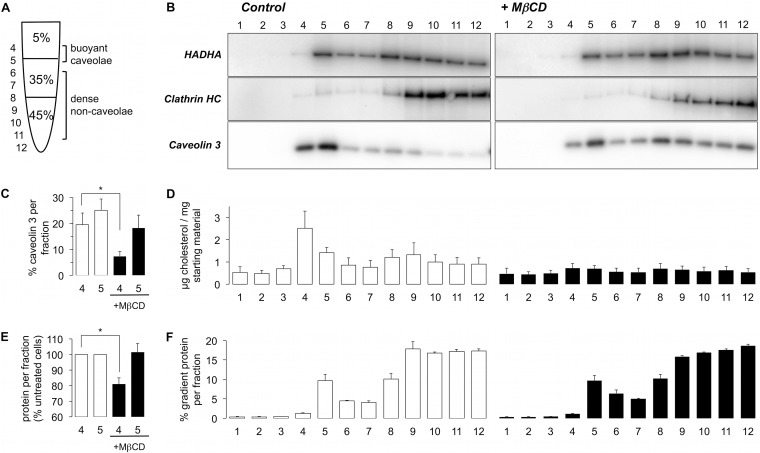
We prepared BCEMs from control, cholesterol-depleted, and cholesterol-loaded adult rat ventricular myocytes using density gradient centrifugation (Fig. 1A). BCEMs were retrieved from gradient fractions 4 and 5 (Fig. 1B). Cholesterol loading was without effect on the distribution of caveolar (caveolin 3), noncaveolar (clathrin heavy chain), or mitochondrial (HADHA) marker proteins (not shown). Cholesterol depletion significantly attenuated the purification of BCEMs in gradient fraction 4, but was largely without influence on the distribution of noncaveolar sarcolemmal or mitochondrial membranes (Fig. 1B). We typically observed a 3-fold reduction in caveolin 3 in gradient fraction 4 following cholesterol depletion (Fig. 1C). Interestingly, although the distribution of cholesterol throughout the sucrose gradient was profoundly reduced by MβCD treatment (as expected, Fig. 1D), the gross distribution of cellular protein throughout the gradient was largely unaltered (Fig. 1F). A significant peak of cellular protein was observed in gradient fraction 5 regardless of the cholesterol status of the myocytes from which the gradient was prepared. This fraction also contained a significant amount of mitochondrial protein (Fig. 1B). Because the cardiac mitochondrial compartment quantitatively dwarves the lipid raft compartment, gradient fraction 5 was not used for analysis. We typically recovered 50–100 μg cellular protein in gradient fraction 4, which represented ∼50% of the buoyant caveolin 3-containing membranes, 20% of all caveolin 3 on the gradient, but less than 2% of the protein loaded onto the gradient, and this was significantly reduced following cholesterol depletion (Fig. 1E). We compared this fraction from control and MβCD treated myocytes.
Fig. 1.
Depletion of caveolin 3 and cholesterol, but not bulk sarcolemmal or mitochondrial proteins from buoyant membranes following MβCD treatment of adult rat ventricular myocytes (n = 5 per group for all measurements, mean ± S. E. shown). A, Buoyant membranes were prepared using a discontinuous sucrose gradient. B, Caveolae were isolated from control (left) and MβCD-treated (right) adult rat ventricular myocytes. Fractions were immunoblotted for the proteins indicated. C, Quantitative assessment of the depletion of caveolin 3 from gradient fractions 4 and 5 following MβCD treatment. D, Quantitative assessment of the distribution of cholesterol in all gradient fractions (control left, MβCD right). E, Depletion of protein from gradient fractions 4 and 5 following MβCD treatment. F, Distribution of protein for all gradient fractions (control left, MβCD right).
iTRAQ Analysis of the Cardiac BCEM Proteome
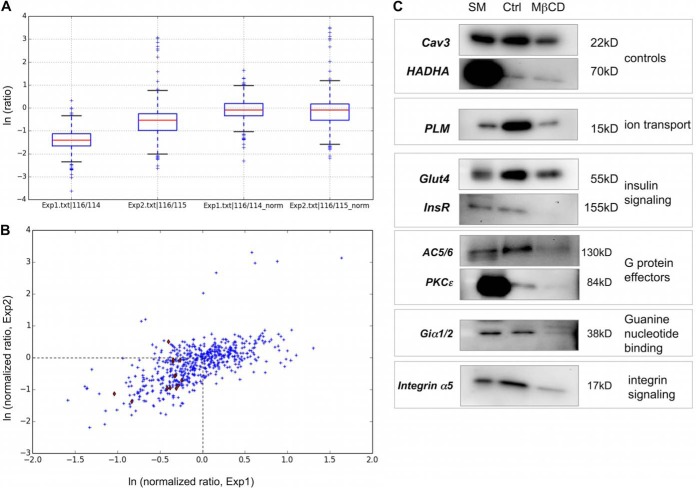
Gradient fractions 4 from control and MβCD treated rat ventricular myocytes were compared using iTRAQ. A full list of proteins identified with iTRAQ ratios from two independent experiments is available in supplemental Table S1. For analysis, rat IPI identifiers were mapped to human UniProt orthologs using a custom built dictionary. We found that MβCD treatment depleted both established caveolar proteins and contaminants (e.g. mitochondrial proteins) from gradient fraction 4, but that annotated caveolar proteins were consistently more depleted (i.e. a lower iTRAQ ratio). The extent of depletion was not consistent between experiments, so in order to account for experiment-to-experiment variability, we divided the iTRAQ ratio values from each replicate by the average iTRAQ ratio for that experiment. Having normalized in this way, potential caveolar residents are defined as having a normalized iTRAQ ratio less than one (natural logarithm (ratio) less than zero in Fig. 2A and Fig. 2B), and the caveolar proteome was defined as those proteins with a normalized ratio below one in 2/2 experiments. There was good reproducibility between the two experiments (Fig. 2B). Although a considerable number of contaminant proteins were selected from individual experiments using this approach, the final list of 249 high confidence caveolar proteins generated (supplemental Table S1) includes 34 annotated mitochondrial proteins, of which the majority (27/34) are either erroneously annotated as mitochondrial or not entirely mitochondrial in location. Hypergeometric distribution indicates the probability that assignment to caveolae is random is 0.015.
Fig. 2.
iTraq analysis of control and MβCD treated BCEMs. A, Distribution of iTraq ratios from two independent experiments before (left) and after (right) normalization. B, After normalization, iTRAQ ratios show good reproducibility between experiments. Proteins annotated as caveolar residents in Uniprot are highlighted in red. The caveolar proteome was defined as the boxed area. C, Candidate caveolar proteins were verified by immunoblotting fraction 4 purified from control and MβCD treated cells. SM = gradient starting material.
Candidate caveolar proteins were verified by immunoblotting gradient fraction 4 from control and MβCD treated cells for representative members of the ion transport (the Na pump accessory subunit phospholemman, PLM), insulin signaling (Glut4, insulin receptor), G protein effector (adenylyl cyclase 5/6, PKCε), G protein (Giα1/2), and integrin signaling (integrin α5 light chain) functional clusters (Fig. 2C). We assessed the enrichment of the caveolar binding motif (CBM, ΦXΦXXXXΦ or ΦXXXXΦXXΦ where Φ is an aromatic amino acid (44)) in our caveolar data set. Thirty-seven percent (13407/36272) of proteins in the human proteome contain this motif, whereas it is found in 45% (111/249) of high-confidence caveolar proteins.
Phospholemman and the Na Pump in Cardiomyocyte BCEMs
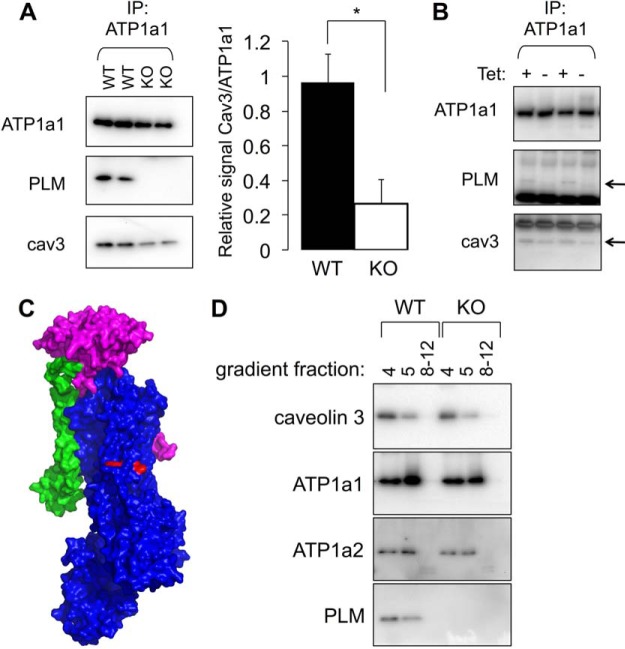
As an exemplar of a cardiac caveolar resident protein, we investigated the relationship between caveolin 3 and the sodium pump catalytic α and regulatory subunit phospholemman (PLM (30)). The Na pump catalytic α subunit interacts with caveolins through a conserved CBM close to its intracellular amino terminus (45). In nonexcitable cells, this interaction results in the Na pump regulating the endocytic trafficking and stabilizing the surface membrane pool of caveolin 1 (45). Surprisingly, we found that co-immunoprecipitation of caveolin 3 with the Na pump α1 subunit was substantially reduced in homogenates from PLM KO compared with WT ventricles (Fig. 3A). We also assessed co-immunoprecipitation of transfected caveolin 3 with Na pump α1 subunit in a cell line engineered to express PLM-YFP under the control of a tetracycline sensitive promoter (46). Induction of PLM-YFP increased the copurification of caveolin 3 with Na pump α1 subunit (Fig. 3B), suggesting that the presence of PLM influences the affinity of caveolin for the pump, despite the relatively large distance between the sites of interaction of these proteins on the pump α subunit (Fig. 3C). We therefore examined whether the localization of the Na pump to cardiomyocyte caveolae was modified in PLM KO compared with WT hearts using sucrose gradient fractionation (Fig. 3D). Despite the apparent influence of PLM on the affinity of Na pump α subunit for caveolin 3, the distribution of neither α1 nor α2 subunits to buoyant caveolar membranes was altered in PLM KO hearts, suggesting the modulation of α subunit affinity for caveolin 3 by PLM does not influence its recruitment to caveolae.
Fig. 3.
Phospholemman and the Na pump in cardiomyocyte caveolae. A, The sodium pump catalytic subunit was immunoprecipitated from ventricular homogenates from PLM WT and KO hearts, and copurifying proteins immunoblotted as shown. In the absence of PLM, substantially less caveolin 3 copurifies with the pump (n = 5 hearts per group, mean ± S.E. shown). B, Copurification of transfected caveolin 3 with endogenous pump α1 subunit in FT-293 cells is enhanced when PLM-YFP expression is induced. PLM and caveolin 3 are marked with an arrow, resolved from the immunoprecipitating antibody that cross-reacts with the secondary antibody. C, Position of the Na pump α subunit CBM (red) relative to the associated PLM (green) in the Na pump crystal structure (α subunit shown in blue, β subunit in magenta). D, Sucrose density gradient fractionation of PLM WT and KO ventricular homogenates reveals no difference in the distribution of sodium pump α1 or α2 subunit to buoyant (4, 5) or dense (8–12) membranes in the absence of PLM.
Quantitative Changes in Caveolar Content Following Adrenoreceptor Activation
Signaling through cardiac AR is among the best-characterized pathways in the cardiovascular system, and is a high-priority research area given the substantial remodeling of AR pathways during the development of cardiac hypertrophy and failure (47). A proportion of the β1-, and the majority of the α- and β2-AR signal pass through cardiac caveolae. We therefore investigated changes in the composition of gradient fraction 4 following activation of each receptor subtype in field-stimulated myocytes using 4-plex iTRAQ in 4 independent experiments. Individual experimental results are provided in Supplemental Table S2. Agonists were applied to field-stimulated cells for 10 min. This time point was selected as the optimum one for studying caveolar dynamics based on the temporal characteristics of cAMP generation and kinase activation. We have previously reported sustained increases in cAMP, substrate phosphorylation, and contractility (for β1-AR stimulation (48)), sustained generation of cAMP (for β2-AR stimulation (20)), and sustained substrate phosphorylation (for α-AR stimulation (41)) following agonist application for 5–10 min. We also observed phosphorylation of PLM at Ser68 (a protein kinase A and C site) following 10 min agonist application in all treatment groups (not shown), confirming adequate receptor activation.
For analysis, we found significant experiment-to-experiment variability in the absolute iTRAQ ratios for particular proteins. This is likely the result of iTRAQ ratio compression (49), where other labeled peptide ions are co-isolated during isolation of the iTRAQ-labeled precursor ion for fragmentation. The iTRAQ ratios calculated are influenced by the co-isolated peptides, thus causing variability. Ratio compression is dependent on sample complexity, so samples were divided into nine fractions (Experiments 3 and 4, supplemental Table S2) and 23–26 fractions (Experiments 5 and 6, supplemental Table S2) prior to analysis in an attempt to improve replicate variability. Data for each particular AR agonist were therefore merged from four experiments, and the iTRAQ ratios converted to a z-score to identify proteins up- (z-score >0.6) or down- (z-score <−0.6) regulated in a minimum of three out of four experiments. Proteins not present in the 249 high-confidence caveolar resident proteins were excluded. Hence, we defined a list of proteins that were recruited into or expelled from cardiomyocyte caveolae following activation of α-, β1-, or β2-AR (supplemental Table 3A–D, summarized in Table I). Of particular interest is cavin 1 (polymerase 1 and transcript release factor, PTRF), which is recruited to caveolae following β1 and β2-AR activation (Table I, supplemental Table S3A and S3B).
Table I. Selected proteins recruited into (+) or expelled from (-) cardiomyocyte caveolae following activation of α-, β1-, or β2-AR. A full list of proteins showing altered abundance is in supplementary Tables S3D and S3E.
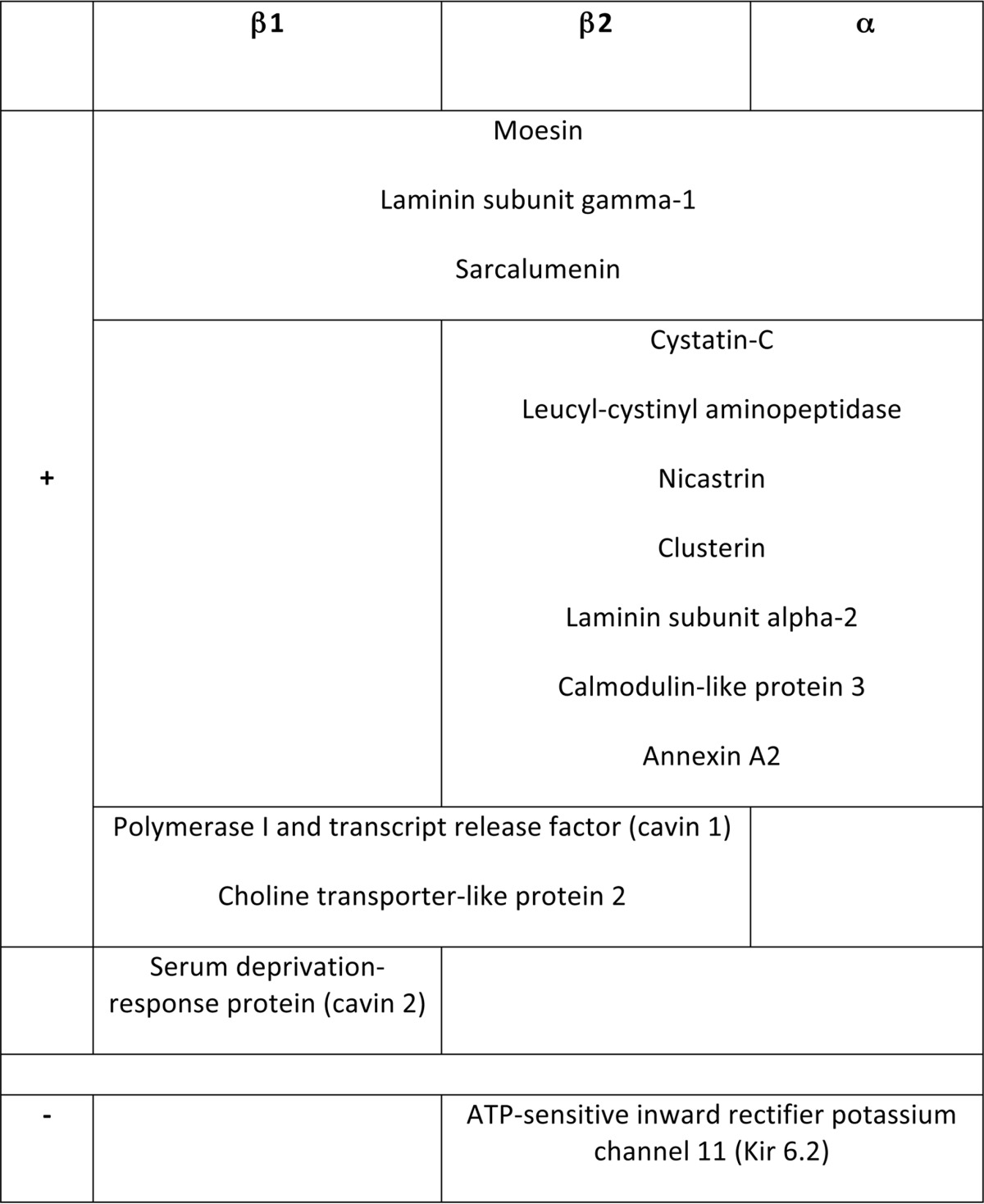
Cavin 1 Redistributes to Caveolae Following Activation of Ventricular β1 or β2 Adrenoceptors
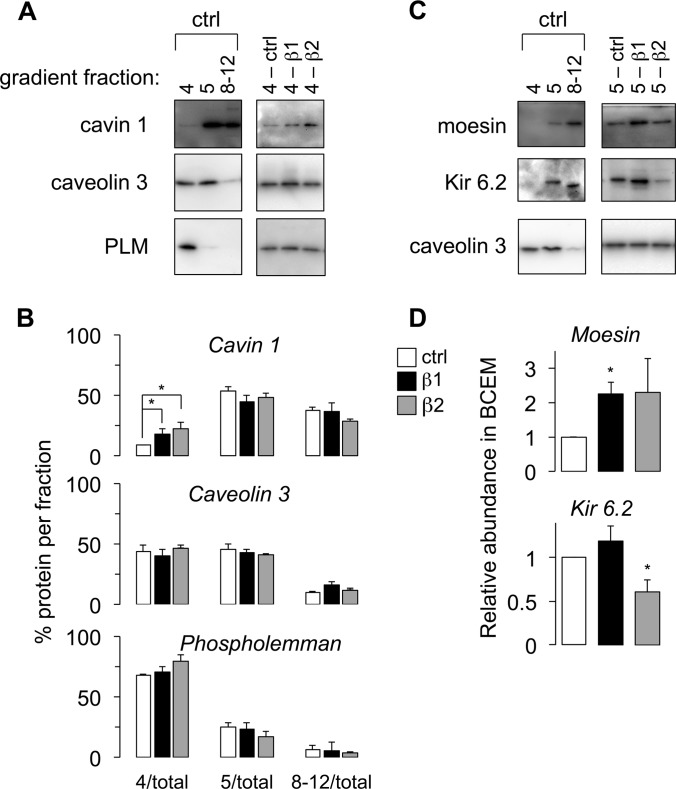
Functional clustering of proteins showing increased and decreased abundance following AR activation is presented in supplemental Table S3D and S3E, respectively. We focused on validating the redistribution of proteins following activation of β1 and β2 AR in an independent replication cohort. Freshly prepared gradient fractions from field stimulated β1- and β2-AR agonist treated myocytes were immunoblotted for cavin 1, caveolin 3, PLM, moesin, and the ATP-sensitive potassium channel pore-forming subunit Kir 6.2 (Fig. 4). Although the role of the cavin family in caveolae is still the subject of some debate, it is generally agreed that cavins stabilize the caveolin oligomer and assist in the formation of caveolae (50), which may facilitate signal platform formation therein. Western blotting confirmed that activation of β1- and β2-AR in field-stimulated myocytes leads to recruitment of cavin 1 into buoyant caveolar membranes, particularly gradient fraction 4 (Fig. 4A), but is without effect on the distribution of either caveolin 3 or PLM (Fig. 4B). Neither moesin nor Kir 6.2 were detectable in gradient fraction 4 (Fig. 4C). We observed translocation of moesin into gradient fraction 5 following β1-AR activation, whereas Kir 6.2 abundance in gradient fraction 5 decreased after β2-AR activation (Fig. 4D).
Fig. 4.
Quantitative changes in caveolar content following β-AR activation (n = 3 per group, mean ± S. E. shown). A, Representative Western blot analysis of cavin 1, caveolin 3, and PLM distribution to buoyant membranes following activation of β1- and β2-AR. B, Mean data indicate that cavin 1, but not cav 3 or PLM, relocalizes to caveolae. C, Representative Western blot analysis of moesin, Kir 6.2, and caveolin 3 distribution in control cells (left) and after β-AR activation in BCEM (right). D, Mean data indicate recruitment of moesin to cardiac BCEM after β1-AR activation, and ejection of Kir 6.2 after β2-AR activation.
DISCUSSION
Analysis of the Caveolar Interaction Network
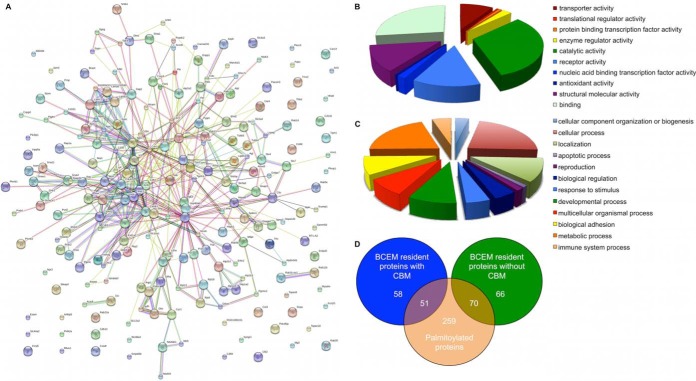
The caveolar interaction network was visualized using MINT (51) (Fig. 5A). Panther analysis (52, 53) of the molecular function of high-confidence caveolar resident proteins is presented in Fig. 5B, with detailed analysis in supplemental Table S1. The biological processes with which caveolar residents are involved is presented in Fig. 5C. We also used functional clustering with DAVID (43) to identify particular functional groups enriched in cardiac caveolae (supplemental Table S1).
Fig. 5.
Analysis of caveolar interaction network. A, Molecular Interaction Network visualization of 249 high confidence caveolar resident proteins in ventricular muscle. B, Panther analysis of annotated molecular function for 249 high confidence caveolar residents. C, Panther analysis of annotated biological process for high confidence caveolar residents. D, Overlap between cardiac palmitoyl proteome and cardiac caveolar proteome: over 50% of caveolar proteins are palmitoylated.
As expected, caveolin and cavin proteins featured in the list of high-confidence caveolar residents. Caveolin 1, 2, and 3 and cavins 1, 2, and 4 were depleted by MβCD in 2/2 experiments, however, only caveolin 1β, caveolin 3, and cavins 1 and 2 are classified as caveolar residents after filtering (see Limitations, below). Ion channels and transporters are heavily represented in cardiac caveolae, with subunits of both voltage activated sodium and calcium channels present, highlighting their likely importance in action potential propagation and excitation–contraction coupling. The cardiac caveolar L type calcium channels have also been linked with excitation–transcription coupling (54). In addition, P-type ATP-dependent transporters including sodium pump subunits, a proton ATPase, and a phospholipid flippase, and ATP independent transporters including the Na/K/Cl and K/Cl cotransporters, aquaporin 1, Glut4, and the Cl/HCO3 exchanger are all present. Although we routinely detect it in BCEM by immunoblotting (not shown), the sodium calcium exchanger fails to be classified as BCEM resident in this study because it was not detected in one of the two MβCD depletion experiments.
GTP binding proteins are understandably highly enriched in cardiac BCEMs. We detected α, β, and γ subunits of several heterotrimeric G proteins (including αs, αo, αi2, and α11), as well as H-ras and multiple ras-related small G proteins (Supplemental Table S1). Signaling molecules downstream of G proteins, including PI4 kinase, inositol-1,4,5-trisphosphate 5-phosphatase, and the phosphoinositide phosphatase SAC1 are also present, highlighting the role of caveolae in both the initiation and termination of signaling events.
The large number of proteins classified either as structural or binding molecules emphasizes the role of caveolae in the assembly of macro-molecular signaling complexes. Although we did not detect G-protein coupled receptors (discussed below), a considerable number of receptors were detected, particularly those involved in cell–cell and cell–extracellular matrix interactions (for example, cadherins). The presence of laminins, integrins, and components of the integrin signaling pathway (Lyn kinase, moesin) is also notable. It is perhaps curious that such adhesion molecules are localized to a surface membrane domain whose architecture will not facilitate interaction with either the extracellular matrix or adjacent cells. Their presence may reflect the fact that caveolae internalize and degrade/recycle such receptors, but is also likely relevant to the role of integrins in mechanosignaling (55): caveolar deformation during, for example, stretch would facilitate integrin binding to ligands in the extracellular matrix.
The need for cholesterol depletion to define the caveolar proteome is emphasized by the presence of 268 annotated mitochondrial proteins out of 981 unique proteins identified in gradient fraction 4 from control and MβCD-treated ventricular myocytes (27%, supplemental Table S1). The strength of the cholesterol depletion approach is demonstrated by the elimination of the vast majority of these false positives simply by setting appropriate depletion thresholds in two independent experiments. The seven annotated mitochondrial proteins in our high confidence caveolar proteome (3%) are the largest group of false positives that also include a small number of sarcoplasmic reticulum resident proteins. Hence the two quantitatively largest membrane compartments in cardiac muscle are almost entirely eliminated from our caveolar proteome.
Failure to Enrich the Caveolin Binding Motif in Caveolar Proteins
The CBM is present in 37% of proteins in the human proteome, and is rather poorly enriched (45%) in our high-confidence caveolar data set. This lends support to the argument that although this motif is clearly an important determinant of the caveolar localization of certain proteins (for example the sodium pump (45) and see (9) for discussion), its presence cannot be the only determinant of caveolar content (9, 10). We have recently defined the cardiac palmitoyl proteome (Howie et al., manuscript under preparation, Uniprot IDs are listed in supplemental Table S1). Because palmitoylation is proposed to partition proteins to detergent-resistant membranes such as caveolae (11), we investigated the overlap between the cardiac palmitoyl proteome and our high-confidence caveolar data set (Fig. 5D). Forty-nine percent (121/249) of caveolar proteins are palmitoylated, meaning palmitoylation is a better prediction of localization to BCEMs than the presence of a CBM. Interestingly, we identified no palmitoylating (DHHC-containing palmitoyl acyl transferases) or depalmitoylating (thioesterases) enzymes in BCEMs, which implies that palmitoylation outside these domains directs proteins to caveolae (possibly in the secretory pathway) and that depalmitoylation may require internalization. Notably 73% (179/249) of caveolar proteins either contain a CBM or are palmitoylated (or both), meaning the presence of only 27% of proteins in BCEMs cannot be accounted for by these mechanisms. This raises the possibility that other mechanisms recruit proteins to caveolae—possibly other lipid modifications, or protein–protein interactions with caveolar residents. Our data also suggest that the use of peptides that interfere with the interaction between CBM and CSD as a means to disrupt signaling through BCEM will not target all BCEM residents.
Multiple Caveolar Subpopulations in Ventricular Muscle?
The physical space (50–100 nm) available in a single caveola is not sufficient to accommodate phospholipids, the caveolin oligomer, cavins, as well as the 249 high confidence caveolar proteins identified in this investigation. Hence, our study strengthens the argument that caveolae are a nonhomogenous population in the cardiac myocyte (56, 57). Because physical colocalization facilitates functional colocalization, an important question for future investigations is what subpopulations of caveolae exist in ventricular muscle, and which proteins are present therein?
PLM and the Na Pump in Cardiomyocyte Caveolae
The Na pump catalytic α subunit interacts with caveolins via a CBM at its intracellular amino terminus (Fig. 3C) (45). Surprisingly, we found the presence of PLM in both adult hearts and an engineered cell line enhanced the interaction between the pump α1 subunit and caveolin 3, despite the relatively large distance between PLM and this CBM in the pump complex. As well as modulating the kinetic properties of the pump, PLM amplifies phosphatidylserine (PS) interactions with the pump α subunit, probably by promoting PS binding close to the α subunit's ninth transmembrane domain (58). Cavins (50, 59–61) and caveolins (62) also bind PS, which has been found to cluster in caveolar regions of the cell surface membrane (63). Our finding that the pump is identically distributed to BCEMs regardless of the presence of PLM suggests that the principal determinant of its sarcolemmal distribution in cardiac muscle is the presence of the CBM rather than phospholipid interactions. Nonetheless, this highlights another potential means by which localization to caveolae can be regulated—through modifying the ability of a particular protein to interact with either caveolins or the phospholipids enriched in caveolae.
Dynamic Regulation of the Cavin Content of Caveolae
Dynamic changes in certain caveolar components may be necessary to ensure a high fidelity and temporally distinct response to AR stimulation in cardiac muscle. Cavin 1 is lost from caveolae during stretch-induced mechanical stress (64), which is associated with flattening of morphologically-identifiable caveolae (5). Cavins stabilize the caveolin oligomer, and cavin 1 is necessary and sufficient for the formation of caveolae in the presence of caveolins (50). Our finding that cavin 1 migrates into caveolar fraction 4 following activation of β1- or β2-AR in field-stimulated ventricular myocytes is the first report that the cavin content of muscle caveolae may be dynamically regulated. We also observe cavin 2 translocation into BCEM following β1-AR activation. We speculate that this translocation represents a mechanism to stabilize caveolae during the additional mechanical stress associated with positive inotropy in vivo; this will be the subject of future investigations. Our finding that the cavin content of ventricular caveolae can be increased, and that a significant pool of cavin 1 is present in nonbuoyant membranes from sucrose gradients (Fig. 4), implies a noncaveolar pool of cavin 1 exists in cardiac muscle. The recent report that prolonged (30 min) α1-AR activation causes cavin 4 translocation out of caveolae into the cytosol (65) is in agreement with our finding that the cavin content of caveolae may be dynamically regulated. Although cavin 4 failed to be classified as BCEM resident (discussed below), it is recruited into BCEM membranes following both β1- and α1-AR activation (supplemental Table S3A and S3C), suggesting that this dissociation from caveolae following prolonged α1-AR activation (65) is preceded by recruitment into caveolae.
With respect to the molecular mechanisms underlying cavin recruitment to caveolae following β-AR activation, it is noteworthy that all cavins are phosphorylated at multiple sites (66), and all are phosphorylated in response to β-AR stimulation in the murine heart in vivo (67). Of particular significance in the context of β-AR signaling pathways is the presence of a serine in a conserved PKA consensus phosphorylation site in the coiled-coil region of cavins 1 (ser 118), 2 (ser 122), and 4 (ser 97), which has been found phosphorylated in cavin 1 in phosphopeptide proteomic screens (68). Notably, we detect recruitment of all cavins except cavin 3 (which lacks the necessary upstream arg to possess a PKA consensus phosphorylation site at the homologous position, ser 89) to caveolae following β1-AR stimulation, although cavin 4 is not included in supplemental Table S3A because it is excluded from the list of high-confidence caveolar residents (see Limitations, below). That said, PKA is not the only effector of β-AR signaling and it has recently been found that cavin 1 is acutely phosphorylated following β1-AR stimulation in the murine heart in vivo at the C-terminal serines 367, 389, and 391, which lie in casein kinase 2 and Ca-calmodulin dependent protein kinase 2 (CaMKII) consensus motifs (66). CaMKII is a well-established mediator of β-AR signaling in cardiac muscle, being activated as a result of the increased Ca cycling following AR activation. In preliminary experiments (not shown) we fail to see translocation of cavin 1 into caveolae following AR activation in the absence of field stimulation, which implies a role for CaMKII rather than PKA in this translocation.
Caveolae Protein Dynamics in Ventricular Muscle
In this study, agonists were applied to field-stimulated myocytes for 10 min. Using Western blotting, other researchers report receptor/effector translocation following a similar period of signaling pathway stimulation: phorbol ester and endothelin −1 induced translocation of protein kinase C isoforms and ERK is seen within 5 min and plateaus at 15 min (32); isoprenaline induced β2-AR translocation is detected at 10 min (14, 34); carbachol induced M2 receptor translocation is seen within 15 min (33). GRK-dependent phosphorylation of the β2 receptor is proposed to underlie β2 AR translocation from caveolae (34), and protein kinase A dependent phosphorylation has also been shown to have a role in promoting movement of this receptor (69). Because we have previously established 10 min as the time required for the stable peak response in terms of a change in phosphorylation of the caveolar resident protein PLM following application of submaximal concentrations of AR agonists (41), we considered 10 min an optimum time point.
Although we did not observe redistribution of receptors or kinases to cardiac caveolae, a number of classes of proteins do translocate in and out of BCEM following AR activation, which are described in detail in supplemental Table S3D and S3E. A notable number of secreted (laminins, cystatin C) and cell adhesion (integrins, moesin) proteins are found recruited into caveolae. We were unable to detect secreted proteins in our sucrose gradient fractions by western blotting (not shown), which questions the biological significance of these particular changes, however moesin is recruited into caveolae following β1-AR activation. Because moesin facilitates the interaction between the cytoskeleton and cell membranes (70), this again suggests a mechanism to stabilize caveolae during mechanical stress. The translocation of the pore-forming subunit of the cardiac ATP-sensitive potassium channel (Kir 6.2) out of cardiac caveolae following β2-AR activation implies a functional link between these two that has not hitherto been reported.
Limitations: False Negatives in the Caveolar Proteome
As discussed above, this study is not without limitations; of particular relevance are false negatives. Notably, we are unable to detect low abundance proteins such as G protein coupled receptors in our proteomic experiments, despite the apparent ease with which they are detectable by Western blotting of BCEMs (for example (14, 33, 34)).
In addition, some known caveolar proteins do not reach the threshold for inclusion in our list of high confidence caveolar proteins, and are therefore also false negatives in the list of residents and proteins that redistribute to/from caveolae with AR stimulation. Included in the list of false negatives for both categories is cavin 4, despite it being readily detected in fraction 4 in 2/2 experiments (supplemental Table S1) and enriched in fraction 4 following β1-AR stimulation in 3/4 experiments. The same is true of PKA RII regulatory subunit, which is depleted from gradient fraction 4 following β1-AR activation. Interestingly, cavin 4 has been reported to be cytosolic in native cardiac and skeletal muscle (71, 72), as well as being unequivocally localized to caveolae in these tissues (61). Our cholesterol-depletion approach is best tailored toward identifying proteins whose predominant location is caveolar. Proteins present in both caveolae and a noncaveolar compartment that contaminates the buoyant membranes will be less depleted from the buoyant membranes by MβCD (because this only depletes the caveolar fraction), and hence may fail to be defined as caveolar. This may account for our failure to define cavin 4 as caveolar, among others.
Supplementary Material
Footnotes
Author contributions: M.L.J.A., S.C.C., and W.F. designed research; K.J.W. and W.C. performed research; M.T., W.C., D.L., and W.F. analyzed data; S.C.C. and W.F. wrote the paper.
* This work was supported by grants from the British Heart Foundation (PG/10/93/28650 to WF & SCC, PG/12/6/29366 to WF & MLJA).
 This article contains supplemental Tables S1 to S3.
This article contains supplemental Tables S1 to S3.
1 The abbreviations used are:
- CSD
- caveolin scaffolding domain
- BCEM
- buoyant caveolin-enriched microdomain
- CaMKII
- Calcium-calmodulin dependent protein kinase 2
- CBM
- caveolin binding motif
- MβCD
- methyl-β-cyclodextrin.
REFERENCES
- 1. Simons K., Ikonen E. (1997) Functional rafts in cell membranes. Nature 387, 569–572 [DOI] [PubMed] [Google Scholar]
- 2. Harvey R. D., Calaghan S. C. (2012) Caveolae create local signaling domains through their distinct protein content, lipid profile, and morphology. J. Mol. Cell. Cardiol. 52, 366–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Morris J. B., Huynh H., Vasilevski O., Woodcock E. A. (2006) Alpha1-adrenergic receptor signaling is localized to caveolae in neonatal rat cardiomyocytes. J. Mol. Cell. Cardiol. 41, 17–25 [DOI] [PubMed] [Google Scholar]
- 4. Head B. P., Patel H. H., Roth D. M., Murray F., Swaney J. S., Niesman I. R., Farquhar M. G., Insel P. A. (2006) Microtubules and actin microfilaments regulate lipid raft/caveolae localization of adenylyl cyclase signaling components. J. Biol. Chem. 281, 26391–26399 [DOI] [PubMed] [Google Scholar]
- 5. Kozera L., White E., Calaghan S. (2009) Caveolae act as membrane reserves which limit mechanosensitive I (Cl,swell) channel activation during swelling in the rat ventricular myocyte. PLoS One 4, e8312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hansen C. G., Shvets E., Howard G., Riento K., Nichols B. J. (2013) Deletion of cavin genes reveals tissue-specific mechanisms for morphogenesis of endothelial caveolae. Nature Communications 4, 1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Razani B., Woodman S. E., Lisanti M. P. (2002) Caveolae: from cell biology to animal physiology. Pharmacol. Rev. 54, 431–467 [DOI] [PubMed] [Google Scholar]
- 8. Sargiacomo M., Scherer P. E., Tang Z., Kubler E., Song K. S., Sanders M. C., Lisanti M. P. (1995) Oligomeric structure of caveolin: implications for caveolae membrane organization. Proc. Natl. Acad. Sci. U.S.A. 92, 9407–9411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Byrne D. P., Dart C., Rigden D. J. (2012) Evaluating caveolin interactions: do proteins interact with the caveolin scaffolding domain through a widespread aromatic residue-rich motif? PLoS One 7, e44879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Collins B. M., Davis M. J., Hancock J. F., Parton R. G. (2012) Structure-based reassessment of the caveolin signaling model: do caveolae regulate signaling through caveolin-protein interactions? Dev. Cell 23, 11–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Levental I., Lingwood D., Grzybek M., Coskun U., Simons K. (2010) Palmitoylation regulates raft affinity for the majority of integral raft proteins. Proc. Natl. Acad. Sci. U.S.A. 107, 22050–22054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fujita T., Toya Y., Iwatsubo K., Onda T., Kimura K., Umemura S., Ishikawa Y. (2001) Accumulation of molecules involved in alpha1-adrenergic signal within caveolae: caveolin expression and the development of cardiac hypertrophy. Cardiovasc. Res. 51, 709–716 [DOI] [PubMed] [Google Scholar]
- 13. Head B. P., Patel H. H., Roth D. M., Lai N. C., Niesman I. R., Farquhar M. G., Insel P. A. (2005) G-protein-coupled receptor signaling components localize in both sarcolemmal and intracellular caveolin-3-associated microdomains in adult cardiac myocytes. J. Biol. Chem. 280, 31036–31044 [DOI] [PubMed] [Google Scholar]
- 14. Rybin V. O., Xu X., Lisanti M. P., Steinberg S. F. (2000) Differential targeting of β-adrenergic receptor subtypes and adenylyl cyclase to cardiomyocyte caveolae: a mechanism to functionally regulate the cAMP signaling pathway. J. Biol. Chem. 275, 41447–41457 [DOI] [PubMed] [Google Scholar]
- 15. Rybin V. O., Pak E., Alcott S., Steinberg S. F. (2003) Developmental changes in beta2-adrenergic receptor signaling in ventricular myocytes: the role of Gi proteins and caveolae microdomains. Mol. Pharmacol. 63, 1338–1348 [DOI] [PubMed] [Google Scholar]
- 16. Balijepalli R. C., Foell J. D., Hall D. D., Hell J. W., Kamp T. J. (2006) Localization of cardiac L-type Ca(2+) channels to a caveolar macromolecular signaling complex is required for beta(2)-adrenergic regulation. Proc. Natl. Acad. Sci. U.S.A. 103, 7500–7505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Calaghan S., White E. (2006) Caveolae modulate excitation–contraction coupling and beta2-adrenergic signaling in adult rat ventricular myocytes. Cardiovasc. Res. 69, 816–824 [DOI] [PubMed] [Google Scholar]
- 18. Calaghan S., Kozera L., White E. (2008) Compartmentalization of cAMP-dependent signaling by caveolae in the adult cardiac myocyte. J. Mol. Cell. Cardiol. 45, 88–92 [DOI] [PubMed] [Google Scholar]
- 19. Agarwal S. R., MacDougall D. A., Tyser R., Pugh S. D., Calaghan S. C., Harvey R. D. (2011) Effects of cholesterol depletion on compartmentalized cAMP responses in adult cardiac myocytes. J. Mol. Cell. Cardiol. 50, 500–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Macdougall D. A., Agarwal S. R., Stopford E. A., Chu H., Collins J. A., Longster A. L., Colyer J., Harvey R. D., Calaghan S. (2012) Caveolae compartmentalize beta2-adrenoceptor signals by curtailing cAMP production and maintaining phosphatase activity in the sarcoplasmic reticulum of the adult ventricular myocyte. J. Mol. Cell. Cardiol. 52, 388–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yarbrough T. L., Lu T., Lee H. C., Shibata E. F. (2002) Localization of cardiac sodium channels in caveolin-rich membrane domains: regulation of sodium current amplitude. Circ. Res. 90, 443–449 [DOI] [PubMed] [Google Scholar]
- 22. Maguy A., Hebert T. E., Nattel S. (2006) Involvement of lipid rafts and caveolae in cardiac ion channel function. Cardiovasc. Res. 69, 798–807 [DOI] [PubMed] [Google Scholar]
- 23. Garg V., Jiao J., Hu K. (2009) Regulation of ATP-sensitive K+ channels by caveolin-enriched microdomains in cardiac myocytes. Cardiovasc. Res. 82, 51–58 [DOI] [PubMed] [Google Scholar]
- 24. Bossuyt J., Taylor B. E., James-Kracke M., Hale C. C. (2002) Evidence for cardiac sodium–calcium exchanger association with caveolin-3. FEBS Lett. 511, 113–117 [DOI] [PubMed] [Google Scholar]
- 25. Cavalli A., Eghbali M., Minosyan T. Y., Stefani E., Philipson K. D. (2007) Localization of sarcolemmal proteins to lipid rafts in the myocardium. Cell Calcium 42, 313–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Liu L., Askari A. (2006) Beta-subunit of cardiac Na+-K+-ATPase dictates the concentration of the functional enzyme in caveolae. Am. J. Physiol. Cell Physiol. 291, C569–C578 [DOI] [PubMed] [Google Scholar]
- 27. Hammes A., Oberdorf-Maass S., Rother T., Nething K., Gollnick F., Linz K. W., Meyer R., Hu K., Han H., Gaudron P., Ertl G., Hoffmann S., Ganten U., Vetter R., Schuh K., Benkwitz C., Zimmer H. G., Neyses L. (1998) Overexpression of the sarcolemmal calcium pump in the myocardium of transgenic rats. Circ. Res. 83, 877–888 [DOI] [PubMed] [Google Scholar]
- 28. Verdonck F., Mubagwa K., Sipido K. R. (2004) [Na(+)] in the subsarcolemmal “fuzzy” space and modulation of [Ca(2+)](i) and contraction in cardiac myocytes. Cell Calcium 35, 603–612 [DOI] [PubMed] [Google Scholar]
- 29. Wypijewski K. J., Howie J., Reilly L., Tulloch L. B., Aughton K. L., McLatchie L. M., Shattock M. J., Calaghan S. C., Fuller W. (2013) A separate pool of cardiac phospholemman that does not regulate or associate with the sodium pump: multimers of phospholemman in ventricular muscle. J. Biol. Chem. 288, 13808–13820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fuller W., Tulloch L. B., Shattock M. J., Calaghan S. C., Howie J., Wypijewski K. J. (2013) Regulation of the cardiac sodium pump. Cell Mol. Life Sci. 70, 1357–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cornelius F., Turner N., Christensen H. R. (2003) Modulation of Na,K-ATPase by phospholipids and cholesterol. II. Steady-state and presteady-state kinetics. Biochemistry 42, 8541–8549 [DOI] [PubMed] [Google Scholar]
- 32. Rybin V. O., Xu X., Steinberg S. F. (1999) Activated protein kinase C isoforms target to cardiomyocyte caveolae : stimulation of local protein phosphorylation. Circ. Res. 84, 980–988 [DOI] [PubMed] [Google Scholar]
- 33. Feron O., Smith T. W., Michel T., Kelly R. A. (1997) Dynamic targeting of the agonist-stimulated m2 muscarinic acetylcholine receptor to caveolae in cardiac myocytes. J. Biol. Chem. 272, 17744–17748 [DOI] [PubMed] [Google Scholar]
- 34. Ostrom R. S., Gregorian C., Drenan R. M., Xiang Y., Regan J. W., Insel P. A. (2001) Receptor number and caveolar colocalization determine receptor coupling efficiency to adenylyl cyclase. J. Biol. Chem. 276, 42063–42069 [DOI] [PubMed] [Google Scholar]
- 35. Foster L. J., De Hoog C. L., Mann M. (2003) Unbiased quantitative proteomics of lipid rafts reveals high specificity for signaling factors. Proc. Natl. Acad. Sci. U.S.A. 100, 5813–5818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zheng Y. Z., Foster L. J. (2009) Contributions of quantitative proteomics to understanding membrane microdomains. J. Lipid Res. 50, 1976–1985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zheng Y. Z., Berg K. B., Foster L. J. (2009) Mitochondria do not contain lipid rafts, and lipid rafts do not contain mitochondrial proteins. J. Lipid Res. 50, 988–998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bini L., Pacini S., Liberatori S., Valensin S., Pellegrini M., Raggiaschi R., Pallini V., Baldari C. T. (2003) Extensive temporally regulated reorganization of the lipid raft proteome following T-cell antigen receptor triggering. Biochem. J. 369, 301–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Banfi C., Brioschi M., Wait R., Begum S., Gianazza E., Fratto P., Polvani G., Vitali E., Parolari A., Mussoni L., Tremoli E. (2006) Proteomic analysis of membrane microdomains derived from both failing and nonfailing human hearts. Proteomics 6, 1976–1988 [DOI] [PubMed] [Google Scholar]
- 40. Fuller W., Eaton P., Bell J. R., Shattock M. J. (2004) Ischemia-induced phosphorylation of phospholemman directly activates rat cardiac Na/K-ATPase. FASEB J. 18, 197–199 [DOI] [PubMed] [Google Scholar]
- 41. Fuller W., Howie J., McLatchie L. M., Weber R. J., Hastie C. J., Burness K., Pavlovic D., Shattock M. J. (2009) FXYD1 phosphorylation in vitro and in adult rat cardiac myocytes: threonine 69 is a novel substrate for protein kinase C. Am. J. Physiol. Cell Physiol. 296, C1346–C1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bell J. R., Kennington E., Fuller W., Dighe K., Donoghue P., Clark J. E., Jia L. G., Tucker A. L., Moorman J. R., Marber M. S., Eaton P., Dunn M. J., Shattock M. J. (2008) Characterization of the phospholemman knockout mouse heart: depressed left ventricular function with increased Na-K-ATPase activity. Am. J. Physiol. Heart Circ. Physiol. 294, H613–H621 [DOI] [PubMed] [Google Scholar]
- 43. Sherman B. T., Huang da W., Tan Q., Guo Y., Bour S., Liu D., Stephens R., Baseler M. W., Lane H. C., Lempicki R. A. (2007) DAVID Knowledgebase: a gene-centered database integrating heterogeneous gene annotation resources to facilitate high-throughput gene functional analysis. BMC Bioinformatics 8, 426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Couet J., Li S., Okamoto T., Ikezu T., Lisanti M. P. (1997) Identification of peptide and protein ligands for the caveolin-scaffolding domain. Implications for the interaction of caveolin with caveolae-associated proteins. J. Biol. Chem. 272, 6525–6533 [DOI] [PubMed] [Google Scholar]
- 45. Cai T., Wang H., Chen Y., Liu L., Gunning W. T., Quintas L. E., Xie Z. J. (2008) Regulation of caveolin-1 membrane trafficking by the Na/K-ATPase. J. Cell Biol. 182, 1153–1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tulloch L. B., Howie J., Wypijewski K. J., Wilson C. R., Bernard W. G., Shattock M. J., Fuller W. (2011) The inhibitory effect of phospholemman on the sodium pump requires its palmitoylation. J. Biol. Chem. 286, 36020–36031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Koitabashi N., Kass D. A. (2012) Reverse remodeling in heart failure–mechanisms and therapeutic opportunities. Nat. Rev. Cardiology 9, 147–157 [DOI] [PubMed] [Google Scholar]
- 48. Calaghan S. C., White E., Colyer J. (1998) Coordinated changes in cAMP, phosphorylated phospholamban, Ca2+, and contraction following beta-adrenergic stimulation of rat heart. Pflugers Arch. 436, 948–956 [DOI] [PubMed] [Google Scholar]
- 49. Ow S. Y., Salim M., Noirel J., Evans C., Rehman I., Wright P. C. (2009) iTRAQ underestimation in simple and complex mixtures: “the good, the bad, and the ugly.” J. Proteome Res. 8, 5347–5355 [DOI] [PubMed] [Google Scholar]
- 50. Hill M. M., Bastiani M., Luetterforst R., Kirkham M., Kirkham A., Nixon S. J., Walser P., Abankwa D., Oorschot V. M., Martin S., Hancock J. F., Parton R. G. (2008) PTRF-Cavin, a conserved cytoplasmic protein required for caveola formation and function. Cell 132, 113–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Licata L., Briganti L., Peluso D., Perfetto L., Iannuccelli M., Galeota E., Sacco F., Palma A., Nardozza A. P., Santonico E., Castagnoli L., Cesareni G. (2012) MINT, the molecular interaction database: 2012 update. Nucleic Acids Res. 40, D857–D861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Mi H., Muruganujan A., Casagrande J. T., Thomas P. D. (2013) Large-scale gene function analysis with the PANTHER classification system. Nat. Protoc. 8, 1551–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Mi H., Muruganujan A., Thomas P. D. (2013) PANTHER in 2013: modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res. 41, D377–D386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Makarewich C. A., Correll R. N., Gao H., Zhang H., Yang B., Berretta R. M., Rizzo V., Molkentin J. D., Houser S. R. (2012) A caveolae-targeted L-type Ca(2)+ channel antagonist inhibits hypertrophic signaling without reducing cardiac contractility. Circ. Res. 110, 669–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hannigan G. E., Coles J. G., Dedhar S. (2007) Integrin-linked kinase at the heart of cardiac contractility, repair, and disease. Circ. Res. 100, 1408–1414 [DOI] [PubMed] [Google Scholar]
- 56. Shibata E. F., Brown T. L., Washburn Z. W., Bai J., Revak T. J., Butters C. A. (2006) Autonomic regulation of voltage-gated cardiac ion channels. J. Cardiovasc. Electrophysiol. 1, S34–S42 [DOI] [PubMed] [Google Scholar]
- 57. Sampson L. J., Davies L. M., Barrett-Jolley R., Standen N. B., Dart C. (2007) Angiotensin II-activated protein kinase C targets caveolae to inhibit aortic ATP-sensitive potassium channels. Cardiovasc. Res. 76, 61–70 [DOI] [PubMed] [Google Scholar]
- 58. Mishra N. K., Peleg Y., Cirri E., Belogus T., Lifshitz Y., Voelker D. R., Apell H. J., Garty H., Karlish S. J. (2011) FXYD proteins stabilize Na,K-ATPase: amplification of specific phosphatidylserine-protein interactions. J. Biol. Chem. 286, 9699–9712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Gustincich S., Vatta P., Goruppi S., Wolf M., Saccone S., Della Valle G., Baggiolini M., Schneider C. (1999) The human serum deprivation response gene (SDPR) maps to 2q32-q33 and codes for a phosphatidylserine-binding protein. Genomics 57, 120–129 [DOI] [PubMed] [Google Scholar]
- 60. Izumi Y., Hirai S., Tamai Y., Fujise-Matsuoka A., Nishimura Y., Ohno S. (1997) A protein kinase Cdelta-binding protein SRBC whose expression is induced by serum starvation. J. Biol. Chem. 272, 7381–7389 [DOI] [PubMed] [Google Scholar]
- 61. Bastiani M., Liu L., Hill M. M., Jedrychowski M. P., Nixon S. J., Lo H. P., Abankwa D., Luetterforst R., Fernandez-Rojo M., Breen M. R., Gygi S. P., Vinten J., Walser P. J., North K. N., Hancock J. F., Pilch P. F., Parton R. G. (2009) MURC/Cavin-4 and cavin family members form tissue-specific caveolar complexes. J. Cell Biol. 185, 1259–1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wanaski S. P., Ng B. K., Glaser M. (2003) Caveolin scaffolding region and the membrane binding region of SRC form lateral membrane domains. Biochemistry 42, 42–56 [DOI] [PubMed] [Google Scholar]
- 63. Fairn G. D., Schieber N. L., Ariotti N., Murphy S., Kuerschner L., Webb R. I., Grinstein S., Parton R. G. (2011) High-resolution mapping reveals topologically distinct cellular pools of phosphatidylserine. J. Cell Biol. 194, 257–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Sinha B., Koster D., Ruez R., Gonnord P., Bastiani M., Abankwa D., Stan R. V., Butler-Browne G., Vedie B., Johannes L., Morone N., Parton R. G., Raposo G., Sens P., Lamaze C., Nassoy P. (2011) Cells respond to mechanical stress by rapid disassembly of caveolae. Cell 144, 402–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Ogata T., Naito D., Nakanishi N., Hayashi Y. K., Taniguchi T., Miyagawa K., Hamaoka T., Maruyama N., Matoba S., Ikeda K., Yamada H., Oh H., Ueyama T. (2014) MURC/Cavin-4 facilitates recruitment of ERK to caveolae and concentric cardiac hypertrophy induced by alpha1-adrenergic receptors. Proc. Natl. Acad. Sci. U.S.A. 111, 3811–3816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Hansen C. G., Nichols B. J. (2010) Exploring the caves: cavins, caveolins, and caveolae. Trends Cell Biol. 20, 177–186 [DOI] [PubMed] [Google Scholar]
- 67. Lundby A., Andersen M. N., Steffensen A. B., Horn H., Kelstrup C. D., Francavilla C., Jensen L. J., Schmitt N., Thomsen M. B., Olsen J. V. (2013) In vivo phosphoproteomics analysis reveals the cardiac targets of beta-adrenergic receptor signaling. Sci. Signal. 6, rs11. [DOI] [PubMed] [Google Scholar]
- 68. Wang B., Malik R., Nigg E. A., Korner R. (2008) Evaluation of the low-specificity protease elastase for large-scale phosphoproteome analysis. Anal. Chem. 80, 9526–9533 [DOI] [PubMed] [Google Scholar]
- 69. Zamah A. M., Delahunty M., Luttrell L. M., Lefkowitz R. J. (2002) Protein kinase A-mediated phosphorylation of the beta 2-adrenergic receptor regulates its coupling to Gs and Gi. Demonstration in a reconstituted system. J. Biol. Chem. 277, 31249–31256 [DOI] [PubMed] [Google Scholar]
- 70. Turunen O., Wahlstrom T., Vaheri A. (1994) Ezrin has a COOH-terminal actin-binding site that is conserved in the ezrin protein family. J. Cell Biol. 126, 1445–1453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Ogata T., Ueyama T., Isodono K., Tagawa M., Takehara N., Kawashima T., Harada K., Takahashi T., Shioi T., Matsubara H., Oh H. (2008) MURC, a muscle-restricted coiled-coil protein that modulates the Rho/ROCK pathway, induces cardiac dysfunction, and conduction disturbance. Mol. Cell. Biol. 28, 3424–3436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Tagawa M., Ueyama T., Ogata T., Takehara N., Nakajima N., Isodono K., Asada S., Takahashi T., Matsubara H., Oh H. (2008) MURC, a muscle-restricted coiled-coil protein, is involved in the regulation of skeletal myogenesis. Am. J. Physiol. Cell Physiol. 295, C490–C498 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.