Summary
Recent evidence supports the presence of an L-glutamyl methyltransferase(s) in eukaryotic cells, but this enzyme class has only been defined in certain prokaryotic species. Here, we characterize the human C6orf211 gene product as ‘acid residue methyltransferase-1’ (Armt1), an enzyme that specifically targets proliferating cell nuclear antigen (PCNA) in breast cancer cells, predominately methylating glutamate side chains. Armt1 homologues share structural similarities to the SAM-dependent methyltransferases, and negative regulation of activity by auto-methylation indicates a means for cellular control. Notably, shRNA-based knock down of Armt1 expression in two breast cancer cell lines altered survival in response to genotoxic stress. Increased sensitivity to UV, adriamycin, and MMS was observed in SK-Br-3 cells, while in contrast, increased resistance to these agents was observed in MCF7 cells. Together, these results lay the foundation for defining the mechanism by which this post-translational modification operates in the DNA damage response (DDR).
Introduction
Protein methyltransferases regulate important biological functions in eukaryotic cells through the post-translational modification of a wide array of targets including, but not limited to, DNA damage response mediators, DNA repair proteins and transcription factors (Grillo and Colombatto, 2005). The majority of these enzymes catalyze transfer of methyl groups from the cofactor s-adenosyl methionine (SAM) to amine containing side chains of arginine or lysine generating a methylated residue and the by-product s-adenosyl homocysteine (SAH). SAM-dependent methyltransferases (SAM-MTs) that methylate carboxylic acid groups also have been described in eukaryotic cells, and they too serve important biological roles.
Four classes of protein carboxyl methyltransferases (cSAM-MTs) have been described (Xie and Clarke, 1993). The protein iso-aspartate methyltransferases (PIMTs) are wide-spread throughout prokaryotes and eukaryotes, and these enzymes repair damaged or aging proteins. The substrates of PIMT are carboxyl groups of L-iso-aspartate or D-aspartate residues, which occur spontaneously in aging proteins. Two additional classes of cSAM-MTs are leucine carboxyl methyltransferase (LCMT) and isoprenylcysteine O-methyltransferase (ICMT). These enzymes are exclusive to eukaryotes and their substrates are the carboxyl-terminal leucine in protein phosphatase 2A (Stanevich et al., 2011) and the carboxyl-terminal isoprenylated cysteine of membrane associated proteins (Yang et al., 2011), respectively. The substrate of a fourth class of protein cSAM-MT is the carboxyl group of L-glutamate residues, but description of this type of enzyme has been limited to prokaryotic cells. Previously, we examined the post-translational state of a cancer-associated isoform of the DNA replication and repair factor proliferating cell nuclear antigen (PCNA) using LC-MS/MS (Hoelz et al., 2006). By scrutinizing the MS/MS spectra from this isoform, we identified novel carboxyl methylation (methyl esters) on several glutamate and some aspartate residues. Since our observations, glutamyl methylation has been observed on the HIV resistance protein X-DING-CD4 (Lesner et al., 2009) and aspartate and glutamate methylation has been identified and validated on ~2% of the HeLa cell proteome (Sprung et al., 2008). These findings provided compelling evidence for the existence of cSAM-MTs that target acidic residues in eukaryotic proteins and that methylation of acidic residues in PCNA may represent an unrecognized level of protein regulation in human cells.
Functionally, PCNA is essential protein and member of the DNA sliding clamp family of proteins [for reviews see (Moldovan et al., 2007; Stoimenov and Helleday, 2009)]. By tethering elements to the DNA, PCNA is required for DNA replication and DNA repair. A large and seemingly endless number of protein interactions have been observed with PCNA, which lends additional support for its function in cell cycle progression, chromatin maintenance, and programmed cell death. Interactions with PCNA therefore must be highly coordinated, and the post-translational modification (PTM) of PCNA is needed to control its functions within the cell. Ubiquitination and SUMOylation of PCNA have proven fundamental to regulation DNA damage tolerance (DDT) pathways (Hoege et al., 2002; Stelter and Ulrich, 2003). Monoubiquitination of PCNA in response to DNA damage anchors its interactions with the translesion (TLS) DNA polymerases in the error-prone branch of DDT. Interestingly, five eukaryotic TLS polymerases have been described that are capable of polymerizing (potentially incorrect) nucleotides on damaged template DNA, and the function of each polymerase appears specific to the type(s) of DNA damage encountered (Waters et al., 2009). The error-free branch of DDT requires K63-linked polyubiquitination and SUMOylation of PCNA, which supports a poorly understood template switching mechanism to bypass damage to template DNA. Phosphorylation of PCNA by the EGF receptor has also been described in breast cancer cells (Wang et al., 2006). Tyrosine phosphorylation of PCNA regulates K43-linked polyubiquitination of PCNA, which stabilizes chromatin bound PCNA by preventing its proteosomal degradation.
Here, we report that a novel human gene (C6orf211) product is a first-in-class eukaryotic cSAM-MT that is capable of methylating PCNA. This protein, which we’ve termed ‘acidic residue methyltransferase 1’ (Armt1), shows significant structural similarities to class I SAM-MTs (Schubert et al., 2003) and its activity appears to be tightly regulated in human cells. Knockdown of Armt1 expression alters DNA damage survival rates in breast cancer cells linking its function to the DNA damage response. Opposing survival phenotypes support a complex role for Armt1 in the DNA damage response and indicate a dependence on factor(s) that are differentially expressed between these two cell types. In addition to uncovering another level of protein regulation in eukaryotic cells, our identification of Armt1 uncovers a potentially powerful target to selectively kill cancer cells.
Results
PCNA is the substrate of a carboxyl methyltransferase in human cells
Our previous identification of methylation on several acid residues in PCNA was surprising because a eukaryotic methyltransferase capable of catalyzing this reaction was unknown. To determine whether carboxyl methylation was an enzymatic phenomenon we investigated breast cancer cell extracts for carboxyl methyltransferase activity (Figure 1A). Using a vapor diffusion assay we were not only able to detect SAM-MT activity, but were also able to distinguish cSAM-MT activity (Murray and Clarke, 1984). After assaying extracts with 3H-methyl-labeled SAM, the reactions were equilibrated with base and spotted onto filter paper placed in the neck of a scintillation vial. Hydrolysis of carboxyl methylation then generates volatile methanol which evaporates from the extracts and diffuses into scintillation fluid. The amount of radioactivity in the fluid is therefore proportional to cSAM-MT activity of the extracts. A small but significant amount of activity was observed when breast cancer cell extracts were assayed by vapor diffusion (Figure 1A). This activity was specific to the extracts and sensitive to heat denaturation indicating the presence of cSAM-MT enzymes and substrates in the extracts. To investigate whether PCNA was a substrate for a cSAM-MT in breast cancer cells, we added increasing amounts of exogenous PCNA to the assays and examined its impact on activity (Figure 1B). As a result of PCNA addition, a large and dose-dependent increase in cSAM-MT activity was observed. When equivalent amounts of a non-specific protein, bovine serum albumin (BSA), were added to the assays, an increase in cSAM-MT activity was not observed. These data indicated that a cSAM-MT was present in human cell extracts and was capable of modifying acidic residues in PCNA.
Figure 1. A carboxyl methyltransferase targets PCNA in human cells.
(A) MDA-MB-468 breast cancer cell extracts possess SAM-dependent carboxyl methyltransferase activities. Mean counts from vapor diffusion assays are presented ± SD and compared using student’s t-test. (B) PCNA (2, 5, or 10 μg) or bovine serum albumin (BSA) were added to breast cancer cell extracts and mean counts from vapor diffusion assay presented ± SD and results compared using student’s t-test. (C) PCNA-dependent methyltransferase activity was enriched and resolved by 2D-PAGE. The position of the C6orf211 gene product in the gel is noted with an arrow. (D) Proteins identified by LC-MS/MS in activity enriched fractions classified by cellular functions. See also Figure S1.
C6orf211 encodes a DUF 89 protein containing a conserved SAM-MT structural fold
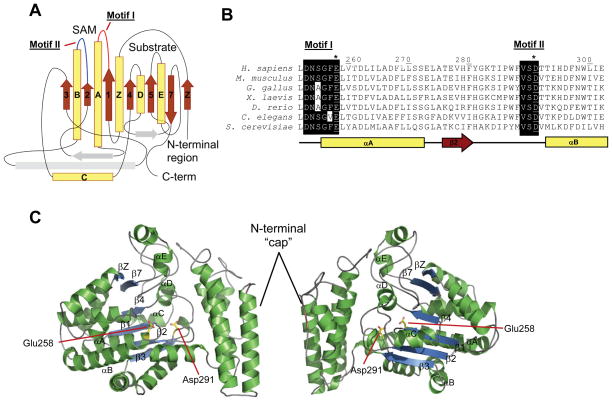
To identify the cSAM-MT responsible for modifying PCNA we fractionated cell extracts and enriched for enzyme activity. Following protein precipitation with 30% ammonium sulfate, activity was further enriched by phenyl Sepharose chromatography. Active fractions were then separated by gel filtration chromatography prior to other chromatographic steps. However, additional chromatographic attempts yielded no activity. This apparent loss of activity at steps of higher enrichment prevented us from isolating the enzyme to near homogeneity, so we closely examined enriched fractions displaying PCNA-directed cSAM-MT activity for the presence of a potential cSAM-MT. Individual polypeptides present in the active gel filtration fractions were separated by two-dimensional polyacrylamide electrophoresis (2D-PAGE), and the polypeptides present in the gel were subsequently excised, proteolytically digested and identified by LC-MS/MS (Figures 1C & D). Previously identified methyltransferases were not observed in the active fractions, so the identified proteins were classified according to their cellular function (Figure 1D). Aiding identification of the methyltransferase in question is that, in general and despite having high sequence divergence, SAM-MTs contain an evolutionarily conserved Rossman-like structural fold. The Rossman-like SAM-MT fold is composed of a core ‘α-β-α’ sandwich of six parallel β-strands and a C-terminal antiparallel β-strand, flanked by five α-helices, in addition to a variable N-terminal cap region (Martin and McMillan, 2002). Blast-based sequence alignments, together with secondary structure prediction and fold recognition using the I-TASSER server (Zhang, 2008), revealed that one isolate in the 2D-PAGE gel (Figure 1C), the product of an uncharacterized human gene C6orf211, likely contained a SAM-MT fold (Figure 2A).
Figure 2. The C6orf211 protein possesses a SAM-dependent methyltransferase fold.
(A) I-Tasser predicted folds of C6orf211 suggest a SAM-MT fold. (B) Alignment of C6orf211 homologs identified conservation in predicted SAM binding site. (C) Two 180° views of a threaded model of C6orf211 based on 3PT1.pdb structure, central β sheets and α-helices of the SAM-MT core fold are labelled. Conserved active site glutamate 258 and aspartate 291 highlighted as yellow sticks. Structural images were produced using PyMOL (www.pymol.org). See also Figures S1 and S2.
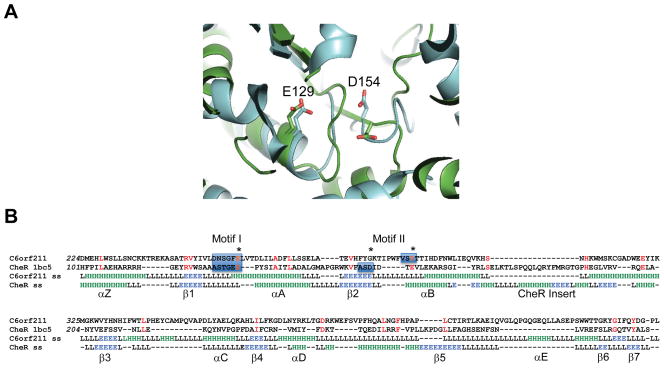
C6orf211 belongs to the domain of unknown function (DUF) 89 family of uncharacterized proteins, present in both eukaryota and archaea. Structure-based sequence alignments of C6orf211 with the four DUF89 protein homologues, as previously determined by structural genomics efforts, revealed a predicted conservation of secondary structures (Figure S1). Of these homologs, the closest structural neighbor to human C6orf211 was the biochemically uncharacterized DUF89 protein from budding yeast, YMR027W (PDB code: 3PT1), which shared 29% sequence identity. This suggested that the DUF89 family of proteins likely represents a subset of the SAM-MT domain, as the 7-stranded central β-sheet of the DUF89 structure differs slightly from the classical SAM-MT fold by having a β-strand occurring before β1, which we term βZ, and the loss of β-strand 6 (Figures 2A, 2C & S1). The noted sequence divergence of SAM-MT family proteins extends to their active sites; but, the occurrence of two SAM-binding motifs is generally well-conserved. Motif I is present in the loop regions connecting the first β-strand (β1) and second α-helix (αA), while motif II is located in the loop connecting the second β-strand (β2) and third α-helix (αB). Together, motifs I and II help form the SAM binding pocket. Sequence alignments of several eukaryotic C6orf211 proteins revealed that motifs I and II sequences are well conserved, and thus, likely essential for protein function (Figure 2B). Notably, one of the closest homologues to the DUF89 family with known structure and catalytic activity is the bacterial glutamyl cMTase CheR. Despite showing limited sequence conservation to other SAM-MTs in its motif I and II sequences, the SAM binding pocket of CheR possesses conserved acidic residues that are responsible for hydrogen bonding with SAH (Djordjevic and Stock, 1997). These acidic residues are conserved in DUF89 proteins including yeast YMR027W, and in human C6orf211, are residues Glu258 and Asp291 (Figure 2 & S2). Sequence alignments and structural superimpositions reveal that the first acidic active site residue is conserved between S. cerevisiae YMR027W (3PT1.pdb) and S. typhimurium CheR (1BC5.pdb) (Figure 3). A second acidic residue is in a structurally equivalent position, but it occurs at the end of a loop insert after β-strand 2 in the DUF89 sequences that includes C6orf211. The equivalent residue in CheR occurs at the end of β-strand 2. Human C6orf211 additionally shares homology to the human methyltransferase 10 domain containing protein (Figure S3A), although SAM binding in the active site of this latter protein does not require the well conserved acidic residues (Wu H., 2006). Sequence analyses also suggested a second C6orf211-like DUF89 domain in the human genome, occurring in the C-terminus of Pantothenate kinase 4 (PNK4; Figure S3B). The N-terminal kinase domain of PNK4 lacks an essential catalytic residue, and thus, the C-terminal C6orf211-like/DUF89 domain could instead be key to its poorly defined cellular function. As far as we are aware, this is the first prediction of structural and functional commonalties between C6orf211, the DUF89 protein family and methyltransferases that include the bacterial glutamyl cSAM-MT CheR.
Figure 3. Structural similarities of the C6orf211 pocket with the SAM binding pocket of CheR.
(A) Structural superimpostions of S. cerevisiae protein YMR027W (3PT1.pdb) in cyan and S. typhimurium CheR (Uniprot code: P07801, PDB code: 1BC5.pdb) in green, revealing two acidic residues (E129 and D154 in CheR) in both proteins in similar positions within the active site. (B) Structure-based sequence alignment of human C6orf211 with S. typhimurium CheR. Conserved residues highlighted in red, stars indicate active site acidic residues, motifs I and II are highlighted with blue boxes. The first active site glutamate is conserved, the second, structurally equivalent acid residue occurs after a loop insert in C6orf211. I-Tasser predicted secondary structure shown for C6orf211 together with 1BC5.pdb secondary structure as defined by DSSP, green H indicates helix, blue E indicates strand and L is loop/coil. The conserved secondary structure elements in common with the core SAM-MT fold and the CheR insert are labeled. See also Figure S3.
The product of C6orf211, Armt1, carboxyl methylates PCNA
The identification of C6orf211 in protein fractions enriched for PCNA-dependent cSAM-MT activity and its structural similarities to the bacterial cSAM-MT CheR was intriguing because it supported a novel protein cSAM-MT function for this uncharacterized gene product. To establish the human C6orf211 gene as encoding a cSAM-MT, we expressed, purified and examined the recombinant protein for cSAM-MT activity directed towards PCNA (Figure 4). Using the vapor diffusion assay, we were able to detect cSAM-MT activity in the presence of purified recombinant PCNA (Figure 4B). This confirmed that the C6orf211 gene product was a cSAM-MT, and we designated it Armt1. In addition to the activity observed in the presence of PCNA, significant activity also was observed in the absence of PCNA. Although this was a surprising result, it did support an auto-methylation function for Armt1, and it was possible that Armt1 was regulating its activity perhaps by modifying key active site acidic residues (Figure 2). Negative regulation of Armt1 activity by self-methylation was congruent with observations made during initial isolation of the enzyme (Figure 1). In this instance, purification of Armt1 would support auto-methylation, and this could explain the apparent loss of activity observed with more highly enriched fractions. To investigate Armt1 activity further, we used an alternative assay that detects loss of adenine absorbance resulting from the enzyme-coupled degradation of the by-product of the methyltransferase reaction, SAH. Initially, attempts to detect SAH production with purified Armt1 were unsuccessful (Figure 4C). We then attempted to remove potentially inhibitory methylation on Armt1 prior to assaying.
Figure 4. C6orf211 codes for Armt1 - a carboxyl methyltransferase capable of modifying PCNA.
(A) Recombinant His-tagged C6orf211 was expressed in insect cells, isolated by Ni2+ Sepharose chromatography prior to 10% SDS-PAGE and colloidal Coomassie blue staining. (B) C6orf211 (0.2μg) was assayed by vapor diffusion in absence and presence of PCNA (2 μg). Mean background (PCNA alone) subtracted counts are presented ± SD and comparisons made using student’s t-test. (C) Self methylation restricts activity. 2 μg of untreated C6orf211 or C6orf211 pretreated at 37°C for 90 min in the presence of 10 μM sinefungin were assayed for methyltransferase activity. Mean background (PCNA alone) subtracted absorbances from three replicate assays are presented. (D) PCNA is a target of C6orf211 methyltransferase activity. Mean background (PCNA alone) subtracted activities from three independent assays in the absence and presence of increasing amounts of PCNA are presented. (E) Pre-steady state kinetics support an inhibitory role for Armt1 self-methylation.
Compared to most other PTMs, including amine methylation, carboxyl methylation is highly unstable and spontaneously hydrolyzes to an unmodified residue within minutes under basic or physiologic conditions (Kim and Paik, 1976). We exploited this lability and removed the majority of carboxyl methylation by pretreating Armt1 in a pH 8 solution. To prevent re-methylation of the hydrolyzed residues in Armt1, we added the cSAM-MT inhibitor sinefungin. As a result of pretreatment, we consistently observed significant levels of SAM-MT activity above background (Figure 4C). Consistent with the results generated using the vapor diffusion assay, we detected Armt1 auto-methylation using this assays, but only after pretreatment. These results support the ability of Armt1 to negatively regulate its activity. To confirm PCNA as a target of Armt1, we investigated SAM-MT activity after the addition of increasing amounts of recombinant PCNA (Figure 4D). A dose-dependent increase in activity was observed following addition of PCNA, which further supported it as a substrate of Armt1. Pre-steady state or burst kinetics were also observed early in these assays (Figure 4E). The burst phase represents enzyme-substrate complex formation, and in the presence of Armt1 alone the burst phase closely matched enzyme concentration. At the end of the burst phase and when auto-methylation of Armt1 was near complete, the reaction likely shifted to an equilibrium between enzyme methylation and inhibition followed by spontaneous methylation hydrolysis and enzyme re-activation. a dose-dependent increase in the burst phase also was observed upon addition of the substrate PCNA (Figure 4E). Unlike in the presence of Armt1 alone, where all activity contributes to inhibition of enzymatic activity, methylation of the substrate PCNA would not contribute to enzyme inhibition resulting in a concentration dependent increase in the burst phase. Thus, based on the activities observed with the recombinant human protein, we confirm our structural conclusions and identify the DUF89 family member and product of the uncharacterized human gene C6orf211 gene as a novel cSAM-MT that methylates both itself and PCNA.
Armt1 regulates the DNA damage response in breast cancer cells
To explore a cellular function for Armt1, we knocked down C6orf211 gene expression in two breast cancer cell lines with lentiviral shRNA. SK-Br-3 and MCF7 cells stably expressing either non-targeting control (shCon) or C6orf211 targeting shRNA (shArmt1) were selected, and a 70% reduction in expression was observed in SK-Br-3 cells, and an 85% reduction was observed in MCF7 cells (Figure S4). The SK-Br-3 and MCF7 knockdown cell lines were then assayed for clonogenic survival, and no significant differences in damage-free survival were observed in these cell lines after Armt1 knockdown (data not shown). However, when the knock down cell lines were exposed to different kinds of DNA damage, Armt1 expression significantly affected cell survival (Figure 5). Armt1 knockdown sensitized SK-Br-3 cells to UV, adriamycin (doxorubicin) and methyl methanesulfonate (MMS). SK-Br-3 cells expressing alternative shRNA constructs displayed similar phenotypes discounting off-target effects (Figure S4C). Conversely, reduced Armt1 expression in MCF7 cells generated the opposite phenotype (Figure 5B). In response to UV, adriamycin and MMS, reduced Armt1 expression produced a damage-resistant phenotype in MCF7 cells, and similar results were observed with other shRNAs (Figure S4D). These results strongly support a role for Armt1 activity, and likely the methylation of acidic residues in PCNA, in the cellular response to DNA damage. These results also suggest that reduction of Armt1 activity can affect survival by sensitizing some cell types to DNA damage while creating resistance in others. Targeting Armt1 may therefore enhance selective killing of tumor cells with DNA damaging cytotoxic agents.
Figure 5. Armt1 Functions in the DNA damage response.
Armt1 differentially regulates survival in SK-Br-3 (A) and MCF7 (B) cells. Cells stably expressing either control (shCon) or Armt1 targeting (shArmt1) shRNA were exposed to DNA damage and survival was assessed by clonogenic assay. Mean colony numbers from three replicates were normalized to the untreated controls and are presented ± SEM. See also Figure S4.
Discussion
Here, we provide the original description of Armt1, a first-in-class eukaryotic methyltransferase encoded by the uncharacterized gene C6orf211. Structurally, Armt1 belongs to the ‘DUF89’ family, and of the four DUF89 structures that have been determined to date, all possess conserved and strong structural similarities to key active site residues of the SAM-MT fold (Martin and McMillan, 2002). Based on these structural observations, activities for the DUF89 family of proteins have been previously proposed, but these studies lacked biochemical and cellular analyses. For example, in 2010 the structure of the S. cerevisiae Armt1 homolog was deposited into the protein data bank by a structural genomics group (PDB code: 3PT1). After soaking the crystals with 6-fructophosphate, the depositors found the molecule in the central pocket leading them to postulate it as novel carbohydrate phosphatase. Instead, we observe that the yeast structure belongs to the SAM-MT domain family, readily docks the cofactor SAM (data not shown), and our in-depth characterization of carboxyl methyltransferase activity with the human homolog supports this domain as a SAM-MT fold. In these studies we not only detected methyltransferase activity in the presence of PCNA, but also in its absence. Self-methylation of Armt1 appears to generate negative feedback that limits its activity.
In addition to enzymatic analyses, the peer reviewed literature also supports C6orf211 gene function in processes alternative to metabolism. Mec1ATR-dependent up-regulation of the C6orf211 homologue YMR027W in S. cerevisiae, for example, was observed following MMS treatment (Gasch et al., 2001). Yeast Armt1 was also implicated in homologous recombination repair in a study examining spontaneous Rad52 foci formation (Alvaro et al., 2007). In this study, knockouts of YMR027W were among a group of knockout cells that displayed the highest rates of spontaneous Rad52 foci formation indicating either an increase in DNA damage and/or reduction in DNA repair. YMR027W knockout cells actually formed higher Rad52 foci rates than knockouts of the homologous recombination and repair (HRR) protein Rad51 and the RecQ helicase SGS1. Equivalent Rad52 foci formation rates were observed in knockouts of the mismatch repair gene MLH1 and the DNA repair HRR genes RAD54, and RAD57. Correspondingly, human C6orf211 has been closely linked to cancer. In breast cancer, C6orf211 was observed to be tightly co-expressed with ESR1, the gene encoding the estrogen receptor (ER) (Dunbier et al., 2011). siRNA knockdown of C6orf211 expression reduced breast cancer cell proliferation, which was independent of estrogen. C6orf211 gene expression also positively correlated with proliferation metagene expression in 354 breast tumors. In another study, a small nucleotide polymorphism (SNP) in close proximity to C6orf211 was identified as a positive indicator of susceptibility to chronic myeloid leukemia (Kim et al., 2011). Here we define the product of C6orf211 as a novel protein cSAM-MT that functions in the DNA damage response. We identify that Armt1 modifies PCNA and we propose that methylation of this essential DNA clamp is, at least in part, responsible for the alterations to survival observed in Armt1 knockdown cells following DNA damage.
The closest previously characterized functional neighbour to Armt1 is the bacterial glutamyl cSAM-MT, CheR. By methylating specific glutamate residues in chemotaxis receptors, CheR modulates intracellular receptor interactions and signal transduction events that cause the bacterium to swim towards nutrients. Methylation of chemotaxis receptors by CheR was found to act as gain-control allowing the bacterium to adapt receptor output across a broad spectrum of ligand concentrations (Levit and Stock, 2002). We have demonstrated that, in addition to methylating the DNA replication and repair factor PCNA, Armt1 functions to regulate the DDR in human cells. Armt1 function in the DDR also depends on cell type, and opposite survival phenotypes were identified in SK-Br-3 and MCF7 cells. The reason for the different survival phenotypes is currently unclear, but the background genetics of these cell lines likely hold the keys to these observations. One important difference between these cells is status p53 an important mediator of the DDR in human cells. MCF7 cells, for example, express wild-type p53 and can induce expression of responsive genes in the DDR. In contrast, SK-Br-3 cells express mutant p53 that is incapable of inducing gene expression in the DDR (Runnebaum et al., 1991). MCF7 cells also lack the pro-apoptotic factor caspase 3 (Janicke et al., 1998), which could alter survival in the DDR. Our functional description of this uncharacterized human gene product as a methyltransferase and its ability to differential regulate cell survival in the DDR also implicates Armt1 as potentially powerful target for anti-cancer therapy. Future research will help determine whether modulation of this novel signalling pathway will be of clinical utility in selecting certain tumor cells to cytotoxic therapies.
Experimental Procedures
Cell culture
MCF7, MDA MB468, and SKBr3 cells were obtained from ATCC and maintained in DMEM or McCoys 5A supplemented with 10% FBS and antibiotics at 37°C with 5% CO2. Cells were exposed to UV-C (254 nm) using a Spectrolinker (Spectronics) and exposed to MMS and Adriamycin (Sigma) at the indicated concentrations for 4 h. Armt1 was cloned into a baculovirus vector used to infect T.ni insect cells (Allele Biotech) at an MOI of 1:3. T.ni cells were maintained at 27°C in serum free media (Allele Biotech) and harvested 72 hours after infection.
Assays
Vapor diffusion assays were carried out basically as described (Murray and Clarke, 1984). Briefly, whole cell extracts (100 μg) were incubated in the presence of 1.5 μM [3H-methyl]-SAM (PerkinElmer) and 7 μM unlableled SAM (Sigma-Aldrich) in 50 mM Tris buffer, pH 7.5 for 1 hour at 37° C. An equal volume of a 200 mM NaOH and 2% SDS solution was added, mixed, and the mixture immediately spotted onto filter paper placed in the neck of a scintillation vial above the fluid. Vials were capped and incubated overnight at room temperature. Volatile 3H-methanol was measured by scintillation counting (Beckman). The SAM265 Methyltransferase Assay (GBiosciences) performed according the manufacturer’s instruction. Briefly, loss of adenine absorbance resulting from the degradation of SAH was monitored with a microplate reader (BioTek). Clonogenic survival and host cell reactivation assays were performed as previously described (Koch-Paiz et al., 2004; Birger et al., 2003) Clonogenic assays were performed by exposing the cells to DNA damage followed by seeding onto 6 cm tissue culture dishes in triplicate. Cells were fixed with 70% ethanol after 10–14 d and stained with crystal violet. Colonies with >30 cells were scored.
Protein Chemistry
Whole cell extracts were generated with MPer containing protease inhibitor cocktail (Pierce), and 1 mM DTT. Chromatography was performed using a Biologic DuoFlow FPLC (BioRad). MDA MB468 whole cell extracts were subjected to 30 % NH4SO4 precipitation for 2 h on ice. Precipitates were clarified by centrifugation prior to passage over a 5 ml Phenyl Sepharose HP (HiTrap) column (GE Biosciences) in 20 mM phosphate buffer, pH 7.0. Active fractions were eluted with a linear gradient of NH4SO4 from 30 to 0% in 20 mM phosphate buffer, pH 7.0. Fractions were desalted with Protein Desalting Spin Columns (Pierce) and carboxyl methyltransferase activity assayed using the vapor diffusion assay in the presence of 2 μg of PCNA, as described. Active fractions were combined and separated on a Superdex S200 gel filtration column (GE Biosciences) in 50 mM Tris, 150 mM NaCl, 10% glycerol, and 1 mM DTT, pH 7.5. Active fractions were acetone precipitated prior to 2D-PAGE and colloidal Coomassie blue staining (Candiano et al., 2004). SDS-PAGE, 2D-PAGE, protein identification and sequencing were performed as previously described (Hoelz et al., 2006). Recombinant PCNA was expressed as a 6 x His-tagged fusion using a pET303/CT-His (InVitrogen) vector or a calmodulin binding peptide (CBP) fusion using the pDual expression system (Stratagene) and purified with Ni2+ Sepharose (GE Biosciences) or Calmodulin agarose (Stratagene), respectively.
Supplementary Material
Acknowledgments
We thank A. Travesa for manuscript review, R. VanBeneden for manuscript review and the generous use of laboratory space, A. Loehrer and M. Diallo for experimental support, T. Kunkel helpful discussions, R. Freund for providing the GST-p21 plasmid, and J. Yates, III for providing the microspray stage designs. This work was supported Eastern Maine Healthcare Systems (D.J.H.), the Maine Cancer Foundation Grant (D.J.H.), generous support from Sue and Joe Cyr from Old Town, ME (D.J.H.), DOD BCRP DAMD17-02-1-0467 (D.J.H.) and DAMD17-99-1-9273 (D.J.H.), USAMRMC W81XWH-10-2-0014 (J. Hock; Project PI: D.J.H.), NIH 5R01CA121289-03 (L.H.M.), and NIH AR059968-03 (J.J.P.P.)
Footnotes
Author Contributions
D.J.H. conceived the project, analyzed and carried out experiments, and wrote the manuscript and provided financial support. J.J.P.P. directed and carried out experiments, analyzed data, wrote the manuscript and provided financial support. G.B.D. and A.E.A designed and carried out experiments. L.E.D. carried out experiments and critically reviewed the manuscript. L.H.M. provided scientific advice and supported the research.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alvaro D, Lisby M, Rothstein R. Genome-wide analysis of Rad52 foci reveals diverse mechanisms impacting recombination. PLoS genetics. 2007;3:e228. doi: 10.1371/journal.pgen.0030228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candiano G, Bruschi M, Musante L, Santucci L, Ghiggeri GM, Carnemolla B, Orecchia P, Zardi L, Righetti PG. Blue silver: a very sensitive colloidal Coomassie G-250 staining for proteome analysis. Electrophoresis. 2004;25:1327–1333. doi: 10.1002/elps.200305844. [DOI] [PubMed] [Google Scholar]
- Djordjevic S, Stock AM. Crystal structure of the chemotaxis receptor methyltransferase CheR suggests a conserved structural motif for binding S-adenosylmethionine. Structure. 1997;5:545–558. doi: 10.1016/s0969-2126(97)00210-4. [DOI] [PubMed] [Google Scholar]
- Dunbier AK, Anderson H, Ghazoui Z, Lopez-Knowles E, Pancholi S, Ribas R, Drury S, Sidhu K, Leary A, Martin LA, et al. ESR1 is co-expressed with closely adjacent uncharacterised genes spanning a breast cancer susceptibility locus at 6q25.1. PLoS genetics. 2011;7:e1001382. doi: 10.1371/journal.pgen.1001382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasch AP, Huang M, Metzner S, Botstein D, Elledge SJ, Brown PO. Genomic expression responses to DNA-damaging agents and the regulatory role of the yeast ATR homolog Mec1p. Molecular biology of the cell. 2001;12:2987–3003. doi: 10.1091/mbc.12.10.2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grillo MA, Colombatto S. S-adenosylmethionine and protein methylation. Amino acids. 2005;28:357–362. doi: 10.1007/s00726-005-0197-6. [DOI] [PubMed] [Google Scholar]
- Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419:135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- Hoelz DJ, Arnold RJ, Dobrolecki LE, Abdel-Aziz W, Loehrer AP, Novotny MV, Schnaper L, Hickey RJ, Malkas LH. The discovery of labile methyl esters on proliferating cell nuclear antigen by MS/MS. Proteomics. 2006;6:4808–4816. doi: 10.1002/pmic.200600142. [DOI] [PubMed] [Google Scholar]
- Janicke RU, Sprengart ML, Wati MR, Porter AG. Caspase-3 is required for DNA fragmentation and morphological changes associated with apoptosis. The Journal of biological chemistry. 1998;273:9357–9360. doi: 10.1074/jbc.273.16.9357. [DOI] [PubMed] [Google Scholar]
- Kim DH, Lee ST, Won HH, Kim S, Kim MJ, Kim HJ, Kim SH, Kim JW, Kim HJ, Kim YK, et al. A genome-wide association study identifies novel loci associated with susceptibility to chronic myeloid leukemia. Blood. 2011;117:6906–6911. doi: 10.1182/blood-2011-01-329797. [DOI] [PubMed] [Google Scholar]
- Lesner A, Shilpi R, Ivanova A, Gawinowicz MA, Lesniak J, Nikolov D, Simm M. Identification of X-DING-CD4, a new member of human DING protein family that is secreted by HIV-1 resistant CD4(+) T cells and has anti-viral activity. Biochemical and biophysical research communications. 2009;389:284–289. doi: 10.1016/j.bbrc.2009.08.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levit MN, Stock JB. Receptor methylation controls the magnitude of stimulus-response coupling in bacterial chemotaxis. The Journal of biological chemistry. 2002;277:36760–36765. doi: 10.1074/jbc.M204325200. [DOI] [PubMed] [Google Scholar]
- Martin JL, McMillan FM. SAM (dependent) I AM: the S-adenosylmethionine-dependent methyltransferase fold. Current opinion in structural biology. 2002;12:783–793. doi: 10.1016/s0959-440x(02)00391-3. [DOI] [PubMed] [Google Scholar]
- Moldovan GL, Pfander B, Jentsch S. PCNA, the maestro of the replication fork. Cell. 2007;129:665–679. doi: 10.1016/j.cell.2007.05.003. [DOI] [PubMed] [Google Scholar]
- Murray ED, Jr, Clarke S. Synthetic peptide substrates for the erythrocyte protein carboxyl methyltransferase. Detection of a new site of methylation at isomerized L-aspartyl residues. The Journal of biological chemistry. 1984;259:10722–10732. [PubMed] [Google Scholar]
- Runnebaum IB, Nagarajan M, Bowman M, Soto D, Sukumar S. Mutations in p53 as potential molecular markers for human breast cancer. Proceedings of the National Academy of Sciences of the United States of America. 1991;88:10657–10661. doi: 10.1073/pnas.88.23.10657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert HL, Blumenthal RM, Cheng X. Many paths to methyltransfer: a chronicle of convergence. Trends in biochemical sciences. 2003;28:329–335. doi: 10.1016/S0968-0004(03)00090-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprung R, Chen Y, Zhang K, Cheng D, Zhang T, Peng J, Zhao Y. Identification and validation of eukaryotic aspartate and glutamate methylation in proteins. Journal of proteome research. 2008;7:1001–1006. doi: 10.1021/pr0705338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanevich V, Jiang L, Satyshur KA, Li Y, Jeffrey PD, Li Z, Menden P, Semmelhack MF, Xing Y. The structural basis for tight control of PP2A methylation and function by LCMT-1. Molecular cell. 2011;41:331–342. doi: 10.1016/j.molcel.2010.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelter P, Ulrich HD. Control of spontaneous and damage-induced mutagenesis by SUMO and ubiquitin conjugation. Nature. 2003;425:188–191. doi: 10.1038/nature01965. [DOI] [PubMed] [Google Scholar]
- Stoimenov I, Helleday T. PCNA on the crossroad of cancer. Biochemical Society transactions. 2009;37:605–613. doi: 10.1042/BST0370605. [DOI] [PubMed] [Google Scholar]
- Wang SC, Nakajima Y, Yu YL, Xia W, Chen CT, Yang CC, McIntush EW, Li LY, Hawke DH, Kobayashi R, et al. Tyrosine phosphorylation controls PCNA function through protein stability. Nature cell biology. 2006;8:1359–1368. doi: 10.1038/ncb1501. [DOI] [PubMed] [Google Scholar]
- Waters LS, Minesinger BK, Wiltrout ME, D’Souza S, Woodruff RV, Walker GC. Eukaryotic translesion polymerases and their roles and regulation in DNA damage tolerance. Microbiology and molecular biology reviews : MMBR. 2009;73:134–154. doi: 10.1128/MMBR.00034-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, MJ, Zeng H, Loppnau P, Weigelt J, Sundstrom M, Arrowsmith CH, Edwards AM, Bochkarev A, Plotnikov AN. PDB submission. 2006. The crystal structure of human methyltransferase 10 domain containing protein. [Google Scholar]
- Xie H, Clarke S. Methyl esterification of C-terminal leucine residues in cytosolic 36-kDa polypeptides of bovine brain. A novel eucaryotic protein carboxyl methylation reaction. The Journal of biological chemistry. 1993;268:13364–13371. [PubMed] [Google Scholar]
- Yang J, Kulkarni K, Manolaridis I, Zhang Z, Dodd RB, Mas-Droux C, Barford D. Mechanism of isoprenylcysteine carboxyl methylation from the crystal structure of the integral membrane methyltransferase ICMT. Molecular cell. 2011;44:997–1004. doi: 10.1016/j.molcel.2011.10.020. [DOI] [PubMed] [Google Scholar]
- Zhang Y. I-TASSER server for protein 3D structure prediction. BMC bioinformatics. 2008;9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.