Abstract
Pharmaceuticals are pseudopersistent aquatic pollutants with unknown effects at environmentally relevant concentrations. Atlantic salmon (Salmo salar) were exposed to Acetaminophen: 54.77 ± 34.67; Atenolol: 11.08 ± 7.98, and Carbamazepine: 7.85 ± 0.13 μg·L−1 for 5 days. After Acetaminophen treatment, 19 proteins were differently expressed, of which 11 were significant with respect to the control group (eight up-regulated and three down-regulated). After Atenolol treatment, seven differently expressed proteins were obtained in comparison with the control, of which six could be identified (four up-regulated and two down-regulated). Carbamazepine exposure resulted in 15 differently expressed proteins compared with the control, with 10 of them identified (seven up-regulated and three down-regulated). Out of these, three features were common between Acetaminophen and Carbamazepine and one between Carbamazepine and Atenolol. One feature was common across all treatments. Principal component analysis and heat map clustering showed a clear grouping of the variability caused by the applied treatments. The obtained data suggest (1) that exposure to environmentally relevant concentrations of the pharmaceuticals alters the hepatic protein expression profile of the Atlantic salmon; and (2) the existence of treatment specific processes that may be useful for biomarker development.
Molecular approaches in ecotoxicology have greatly enhanced mechanistic understanding of the impact of aquatic pollutants in organisms. High throughput “omic” technologies, including quantitative proteomics methods such as 2D differential in-gel electrophoresis (DIGE), are now being acknowledged to be a promising tool to evaluate the effects of contaminant exposure on organisms and are becoming more widely used in ecotoxicology (1). Information on altered protein expression, including post-transcriptional modifications, can provide protein expression signatures, sets of proteins specific to different stressors, and insight into the possible mode of action (MoA)1 of chemical pollutants and their higher level toxicological effects (2–5). Contaminant specific MoAs and protein expression signatures may be used for monitorization, especially because the use of omic techniques allows earlier identification of effects than traditional endpoints. However, ecotoxicoproteomic studies are still relatively uncommon and in their infancy (6).
Human and veterinary pharmaceuticals are being released into the environment in extremely large quantities on a regular basis. Millions of prescription and nonprescription drugs are purchased and ingested by, or applied on individuals every day and eventually excreted through urine or feces, ultimately entering the effluent of wastewater treatment plants and aquatic environments (7). Frequently, sewage treatment does not affect the chemical structure, and therefore the concentrations of drugs that enter aquatic environments can be sufficient to exert toxicity on nontarget species. Most pharmaceuticals are relatively stable to avoid being biologically inactivated before carrying out their intended biological function (8). However, this stability means that they also persist and accumulate in abiotic and biotic compartments of ecosystems, potentially creating environmental problems (9). In fact, some major pharmaceutical compounds are now considered to be included in the priority list of the Water Framework Directive of the European Union (2000/60/EC). The exact effects that exposure to environmentally relevant concentrations of pharmaceuticals is having on ecosystems, biota, and also humans who may consume contaminated water or organisms are still not completely understood. Risk assessments have shown that some pharmaceuticals have the potential to cause adverse human and environmental effects from indirect exposure (8, 10–12). For the vast majority however, including most metabolites, important knowledge gaps still exist concerning long-term effects on nontarget organisms.
Some of the most pervasive groups of pharmaceuticals that are currently found in aquatic environments are analgesics, β-blockers and anti-epileptics (13–17). In this study, we have chosen Acetaminophen (APAP; analgesic), Atenolol (AT; β-blocker), and Carbamazepine (CBZ; anti-epileptic) as model compounds to assess the effects on the liver proteome of Atlantic salmon, Salmo salar, after exposure to environmentally relevant concentrations. APAP is a non-steroid anti-inflammatory and analgesic drug, used as painkiller and to reduce inflammations and fevers. The exact molecular processes are not known yet, and only a limited number of pathways have been identified (18, 19). Effects of APAP on gene and protein expression in rodents have been published elsewhere (20–22). AT is a selective β-adrenergic receptor antagonist or β-blocker for the treatment of angina, glaucoma, heart failure, high blood pressure, and other related conditions (23–25). An extensive review about the comparative physiology, pharmacology, and toxicology of β-blockers, including AT, in fish has been published recently (26). CBZ is a mood-stabilizing treatment for bipolar affective disorder. The molecular mechanisms underlying the actions of CBZ and the cause of the illness itself are unknown. However, several biochemical pathways have been postulated as possible targets of mood stabilizing drugs (27–31).
Salmonids are frequently employed in effects evaluation of environmental contaminants, and Atlantic salmon are a culturally and economically important sentinel species in many North-West European rivers. However, wild stocks of Atlantic salmon are increasingly endangered (32). A variety of ecological and climatic reasons for this have been proposed, but there is little doubt that declines in water quality are a major threat. The juvenile stages of Atlantic salmon spend a year or more in rivers, before migrating to the sea, and during these stages salmon may be particularly vulnerable to contamination by pharmaceuticals, and other chemicals, especially when river levels are low. The purpose of this study is to determine sets of proteins, in the liver of Atlantic salmon parr, whose expression is changed in response to environmentally relevant concentrations of APAP, AT, or CBZ. An ontologic analysis of the salmon proteome associated with pharmaceutical treatment can provide in-depth mechanistic information of the molecular MoA and the function of the altered proteins. Furthermore, those proteins that are highly regulated will be useful as candidates for development as biomarkers in environmental monitoring exercises, with potential for indicating exposure to specific pharmaceuticals, and thus providing an early indication of the ecological risk posed by pharmaceutical contaminant discharge.
EXPERIMENTAL PROCEDURES
Ethics Statement
All procedures were performed under license to, and in accordance with United Kingdom Home Office regulations governing animal experimentation, and following oversight by an institutional ethics review committee.
Exposure
APAP (CAS N°: 103-90-2), AT (CAS N°: 29122-68-7), and CBZ (CAS N°: 298–46-4) were purchased from Sigma. Atlantic salmon parr (approximately one year old) were purchased from the Stirling University aquaculture facility (Howietown Fish Farm, Stirling, UK). The fish were acclimated to laboratory conditions for 14 days. After that, ∼15 fish per treatment were exposed to environmentally relevant concentrations of the pharmaceuticals for 5 days under continuous flow through conditions. The concentrations were chosen based on maximum levels detected in various European freshwater bodies (13–17) and the exposure time of 5 days was selected as an approximation of low water conditions in a natural riverine environment. The experiments were carried out in duplicate and control trials were run simultaneously. A peristaltic pump supplied the compounds from daily renewed working stock solutions. Water flow through the system was adjusted to 360 L·d−1. Water samples (250 ml) were collected from each tank at days 1, 3, and 5, and stored at 4 °C not longer than 24 h until their pretreatment for posterior analysis by high performance liquid chromatography (HPLC). After 5 days of exposure, all fish were killed by a blow to the head, sexed, and final weights, tissue weights, and length were measured. Liver tissues were immediately frozen and stored at −70 °C.
Exposure Concentration Analysis
Exposure concentrations were measured as described elsewhere (33). After solid phase extraction (OASIS HLB; 60 mg, 3 ml; Waters, Milford, MA), analytes were separated under isocratic conditions with acetonitrile and 50 mm potassium dihydrogen phosphate solution. APAP and CBZ were measured using the UV signal at 250 nm, AT using the fluorescence signal at 271 nm. Compounds were identified and quantified by comparing retention times and peaks in samples and standard solution chromatograms. Limits of detection (LOD) and limits of quantification (LOQ) were calculated by using a signal-to-noise ratio of 3 and 10, respectively.
Proteomic Analysis
Frozen liver tissues were homogenized in lysis buffer (7 m urea, 2 m Thiourea, 4% CHAPS, and 30 mm Tris, pH 8.5) on ice using a glass mortar with pestle for protein solubilization. Following homogenization, the tissue lysates were centrifuged at 14,000 × g for 10 min at 4 °C to remove any insoluble particles. The supernatant was then transferred into 50 μL aliquots and stored at −80 °C until gel electrophoresis was performed. Protein concentration was determined by the method based on Bradford (Bio-Rad Protein Assay, Hercules, CA) and adjusted to 5 μg·μL−1 by dilution with lysis buffer. 24 cm long immobilized pH gradient (pH 3–11NL, GE Healthcare) IPG strips were rehydrated overnight in 450 μL DeStreak with 2.25 μL IPG buffer (pH 3–11NL) added (both GE Healthcare). After rehydration, five individual protein samples (biological replicates; 50 μg for each sample) per treatment were labeled with 400 pmol Cy3 or Cy5 minimal NHS ester dyes (GE Healthcare) allocated randomly to provide both Cy3 and Cy5 labeled samples for each treatment. A pool of material from all 20 samples was labeled with Cy2 (GE Healthcare). Different pairs of Cy3- and Cy5-labeled samples (randomly allocated and each containing 50 μg of protein) were combined and mixed with a 50-μg aliquot of the Cy2-labeled pooled standard. These mixtures were diluted 1:1 with lysis buffer containing 0.5% IPG buffer (pH 3–10) and then applied onto the IPG strips. Isoelectric focusing (IEF, first dimension) was carried out on an IPGphor system (GE Healthcare) in four stages with a ramped voltage change between each step: step and hold: 30V, 12 h; gradient 300V, 1 h; step and hold: 300V, 1 h; gradient: 1000V,1 h; step and hold: 1000V, 1 h; gradient: 8000V, 2 h; step and hold: 8000V, 8 h. Focusing was stopped after a total of 70,000 Vh accumulated. After IEF, the IPG strips were equilibrated for 15 min in equilibration buffer I (6 m urea, 2% SDS, 0.05 m Tris-Cl at pH 8.8, 50% glycerol, and 2% DTT) followed by 15 min in equilibration buffer II (same as buffer I but containing 2.5% iodacetamide instead of DTT and 0.02% bromphenol blue). The second dimensional separations were carried out on 12.5% SDS-polyacrylamide gels on the Ettan DaltSix system (GE Healthcare) at 1W/gel for one hour and subsequently at 15W/gel.
Image Analysis
Labeled proteins were visualized using the Typhoon 9000 series imager (GE Healthcare) and the Cy2, Cy3, and Cy5 components of each gel were individually imaged using excitation/emission wavelengths specific for Cy2 (488/520 nm), Cy3 (532/580 nm), and Cy5 (633/670 nm). PMT was varied in order to equalize fluorescence intensities between channels and to prevent over saturation of the signal. Background subtraction, quantitation, normalization, and first-level-of matching (within gel) was performed by Differential In-gel Analysis (DIA) using DeCyder 2D Differential Analysis Software v6.5 (GE Healthcare). For each gel, normalized spot volumes (area multiplied by density of the spot) were calculated as the ratio of each spot volume to total spot volume in the gel. Within the Biological Variation Analysis (BVA) module, each drug treatment set was combined, compared with the control treatment set (t test) and spots with a p value <0.05 returned.
In-gel Digestion and Mass Spectrometry
A preparative gel with 300 μg of protein was used for spot picking and visualized with silver staining to pick spots of interest. The gel pieces were destained and digested by trypsin (Promega, Madison, WI). Upon concentrating and desalting tryptic fragments using Millipore C18 ZipTips (Millipore, Bedford, MA), samples were mixed in a 1:1 ratio with α-cyano-4-hydroxycinnamic acid matrix (saturated solution in 50% ACN and 2.5% TFA) and spotted onto the target plate. MS/MS analyses were performed on a 4800 MALDI-TOF/TOF instrument (Applied Biosystems, Foster City, CA). Measurements were taken in the positive ion mode between 900 and 3000 m/z. Sequences were automatically acquired by scanning first in peptide mass fingerprint (MS) mode. A database search (NCBI nr) was performed, combining the results of peptide mass fingerprint (MS) with subsequent fragmentation (MS/MS) of up to twelve peptides from each spot according to the quality of the MS spectrum, using MASCOT (Version 2.0.00, release date: 19.02.2007) as a search engine. Scores greater than the given cutoff value for MS/MS fragmentation data were taken as significant (p < 0.05). Protein lists were submitted to Venn analysis (http://bioinfogp.cnb.csic.es/tools/venny/) to detect treatment specific and/or common features.
Data Evaluation by Multivariate Statistical Analyses
Multivariate analyses were performed on datasets constituted by the relative levels of all the spots that were consistently matched between the 20 gels, in order to avoid the replacement of null values by inference. Principal component analysis (PCA) was based on the obtained spots of interest where hierarchical clustering was performed on all differently expressed spots. PCA and cluster analysis were carried out using the DeCyder software package (DeCyder 2D Differential Analysis Software v6.5; GE Healthcare).
RESULTS
Exposure Concentration
The measured average concentrations of the three selected pharmaceuticals were: APAP: 54.77 ± 34.67 μg·L−1; AT: 11.08 ± 7.98 μg/L, and CBZ: 7.85 ± 0.13 μg/L. All fish survived the 5-day trial.
Protein Expression Analysis
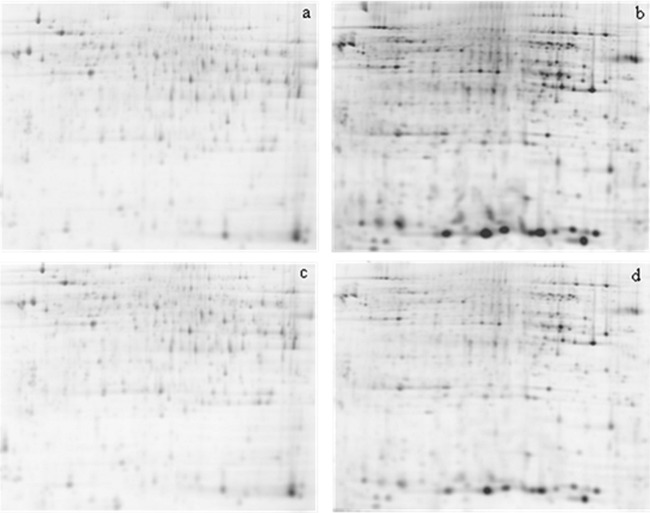
In order to identify sets of proteins that respond to different pharmaceutical exposure in Atlantic salmon, we compared the liver proteome of the four experimental groups (CTRL, APAP, AT, and CBZ) using 2D-DIGE. One representative gel for each experimental condition is shown in Fig. 1A–1D. A total of 53 significantly differently expressed spots were acquired in a reproducible way across all gels and after statistical analysis (t test, p ≤ 0.05) significant differences in several spots were detected between the experimental groups. For the APAP treatment, 19 spots were differently expressed, of which 11 were significant with respect to the control group. Eight of these 11 spots were up-regulated, whereas only three were down-regulated. After AT treatment, seven differently expressed spots were obtained in comparison with the control of which six could be identified. Four of them were up-regulated and two down-regulated. CBZ exposure resulted in 15 differently expressed spots compared with the control, with 10 of them identified. In this last treatment, seven features were up-regulated and three down-regulated. Tables I, II, and III show the results of protein spot identification for the APAP, AT, and CBZ treatment, respectively. The 78kDa glucose regulated protein observed in the AT treatment (Table II) is the feature with the highest absolute change, down-regulated 2.469-fold. Three features were common between different treatments (Fig. 2): the phosphoglycerate kinase 1 (gi 197631857) was up-regulated in all three treatments (APAP: 1.722; AT: 1.307, and CBZ: 1.754 and 1.491). Two other features were represented both in the APAP as well as in the CBZ treatment: acetyl-CoA acetyltransferase, cytosolic (gi 213513638), which was up-regulated during the APAP treatment 1.412 times and during the CBZ treatment 1.279 times, and the glyceraldehyde-3-phosphate dehydrogenase (gi 209737954), which was up-regulated during the APAP treatment 1.484 times, whereas during the CBZ treatment it was down-regulated 1.704 times. Obtained fold changes throughout all the treatments were relatively low, and absolute values comprised between 1.2 (threshold setting) and 2.5.
Fig. 1.
Representative 2D DIGE gel images for the three treatments a) APAP, b) AT, c) CBZ and d) control. Each multiplexed gel was charged with 50 μg of protein per dye.
Table I. Identification of significantly altered proteins after APAP treatment by Mass Fingerprint approach.
| # Master spot | Putative identification | Homology to protein (Genebank) | Protein scorea | Protein score C.I. % | Protein MW | Protein PI | Peptide count | Total ion score | Total ion C.I. % | T-test value | Fold change |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 556 | Transketolase | Salmo salar | 114 | 100 | 68896.6 | 6.18 | 15 | 24 | 0 | 0.0151 | 1.978 |
| gi 213511480 | |||||||||||
| 899 | ATP synthase H+ transporting mitochondrial F1 complex beta | Salmo salar | 105 | 99.998 | 52910.6 | 4.87 | 11 | 33 | 56.937 | 0.0061 | 1.639 |
| gi 198285477 | |||||||||||
| 952 | Elongation factor 1-gamma | Salmo salar | 232 | 100 | 34931.8 | 9.19 | 11 | 164 | 100 | 0.0081 | −1.414 |
| gi 213515528 | |||||||||||
| 1052 | Phosphoglycerate kinase 1 | Salmo salar | 315 | 100 | 44917.1 | 8.31 | 11 | 257 | 100 | 0.0004 | −1.722 |
| gi 197631857 | |||||||||||
| 1053 | RNA polymerase beta subunit protein | Rhododendro n Simsii | 73 | 97.18 | 156861.6 | 9.05 | 18 | – | – | 0.0458 | −1.534 |
| gi 290489550 | |||||||||||
| 1061 | unknown | Arabidopsis thaliana | 69 | 92.582 | 8922.7 | 9.3 | 6 | – | – | 0.0055 | −1.423 |
| gi 116830485 | |||||||||||
| 1122 | Acetyl-CoA acetyltransferase, cytosolic | Salmo salar | 243 | 100 | 41722 | 8.07 | 8 | 196 | 100 | 0.0024 | −1.412 |
| gi 213513638 | |||||||||||
| 1177 | OsmC-like family protein | Dichelobacter nodosus VCS1703A | 73 | 97.428 | 16299 | 5.22 | 8 | – | – | 0.0359 | −1.649 |
| gi 146329847 | |||||||||||
| 1284 | Glyceraldehyde-3-phosphate dehydrogenase-1 | Salmo salar | 468 | 100 | 36097.4 | 7.12 | 21 | 238 | 100 | 0.0309 | −1.51 |
| gi 209737954 | |||||||||||
| 1408 | Caspase-3 | Salmo salar | 94 | 99.975 | 31376.4 | 5.97 | 10 | – | – | 0.0109 | 1.541 |
| gi 213513742 | |||||||||||
| 1788 | Peptidyl-prolyl cis-trans isomerase B precursor | Salmo salar | 325 | 100 | 23993.6 | 9.18 | 11 | 222 | 100 | 0.004 | −1.484 |
| gi 209735348 |
a Mascot score: Probability Based Mowse Score: Ions score is −10*Log(P), where P is the probability that the observed match is a random event. Protein scores greater than 83 are significant (p < 0.05). MALDI-TOF-TOF peak lists were searched against a NCBInr 10997816 protein database using MASCOT TM software with the following settings: Type of search: Sequence Query; Enzyme: Trypsin; Fixed modifications: Carbamidomethyl (C); Variable modifications: Oxidation (M); Mass values: MONOISOTOPIC; Protein Mass: Unrestricted; Peptide Mass Tolerance: ± 100 ppm; Fragment Mass Tolerance: ± 0.2 Da; Max Missed Cleavages: 1; Instrument type: MALDI-TOF-TOF.
Table II. Identification of significantly altered proteins after AT treatment by Mass Fingerprint approach.
| # Master spot | Putative identification | Homology to protein (Genebank) | Protein score | Protein score C.I. % | Protein MW | Protein PI | Peptide count | Total ion score | Total ion C.I. % | T-test value | Fold change |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 105 | Elongation factor 2 | Salmo salar | 212 | 100 | 96406.4 | 6.19 | 21 | 76 | 99.995 | 0.0462 | −1.748 |
| gi 223647986 | |||||||||||
| 265 | 78kDa glucose/regulated protein Salmo salar | Salmo salar | 538 | 100 | 72539.3 | 5 | 28 | 288 | 100 | 0.0042 | 2.469 |
| gi 213511032 | |||||||||||
| 813 | GL16441 | Drosophila persimilis | 76 | 98.619 | 21738.9 | 10.33 | 10 | – | – | 0.0121 | −1.498 |
| gi 195164237 | |||||||||||
| 1052 | Phosphoglycerate kinase 1 | Salmo salar | 315 | 100 | 44917.1 | 8.31 | 11 | 257 | 100 | 0.0002 | −1.307 |
| gi 197631857 | |||||||||||
| 1539 | Branched chain amino acid aminotransferase | Zunongwangia profunda SM-A87 | 73 | 97.244 | 40105.2 | 5.63 | 11 | – | – | 0.0359 | 1.423 |
| gi 295134966 | |||||||||||
| 1782 | hypothetical protein | Salmo salar | 69 | 92.751 | 6919.7 | 9.4 | 4 | 29 | 0 | 0.0414 | −1.526 |
| gi 281416447 |
Mascot score: Probability Based Mowse Score: Ions score is −10*Log(P), where P is the probability that the observed match is a random event. Protein scores greater than 83 are significant (p < 0.05). MALDI-TOF-TOF peak lists were searched against a NCBInr 10997816 protein database using MASCOT TM software with the following settings: Type of search: Sequence Query; Enzyme: Trypsin; Fixed modifications: Carbamidomethyl (C); Variable modifications: Oxidation (M); Mass values: MONOISOTOPIC; Protein Mass: Unrestricted; Peptide Mass Tolerance: ± 100 ppm; Fragment Mass Tolerance: ± 0.2 Da; Max Missed Cleavages: 1; Instrument type: MALDI-TOF-TOF.
Table III. Identification of significantly altered proteins after CBZ treatment by Mass Fingerprint approach.
| # Master spot | Putative identification | Homology to protein (Genebank) | Protein score | Protein score C.I. % | Protein MW | Protein PI | Peptide count | Total ion score | Total ion C.I. % | T-test value | Fold change |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 272 | Elongation factor 2 | Salmo salar | |||||||||
| gi 223647986 | 212 | 100 | 96406.4 | 6.19 | 21 | 76 | 99.995 | 0.0087 | −1.644 | ||
| 395 | Contactin 1a precursor | Danio rerio | |||||||||
| gi 136256388 | 70 | 94.749 | 115044.4 | 6.19 | 15 | – | – | 0.0168 | −1.576 | ||
| 404 | TenA family transcriptional activator | Marinomonas sp. MWYL1 | |||||||||
| gi 152997829 | 71 | 95.632 | 25638.9 | 5.16 | 9 | – | – | 0.0285 | −1.652 | ||
| 559 | Transketolase | Salmo salar | |||||||||
| gi 213511480 | 165 | 100 | 68896.6 | 6.18 | 20 | – | – | 0.0406 | 1.445 | ||
| 1020 | Phosphoglycerate kinase 1 | Salmo salar | |||||||||
| gi 197631857 | 315 | 100 | 44917.1 | 8.31 | 11 | 257 | 100 | 0.0272 | −1.754 | ||
| 1027 | Fumarylacetoacetase | Osmerus mordax | |||||||||
| gi 225706644 | 71 | 95.829 | 49875.9 | 6.27 | 5 | 53 | 99.485 | 0.0211 | 1.87 | ||
| 1052 | Phosphoglycerate kinase 1 | Salmo salar | |||||||||
| gi 197631857 | 315 | 100 | 44917.1 | 8.31 | 11 | 257 | 100 | 0.0272 | −1.754 | ||
| 1061 | unknown | Arabidopsis thaliana | |||||||||
| gi 116830485 | 69 | 92.582 | 8922.7 | 9.3 | 6 | – | – | 0.0281 | −1.206 | ||
| 1122 | Acetyl-CoA acetyltransferase, cytosolic | Salmo salar | |||||||||
| gi 213513638 | 243 | 100 | 41722 | 8.07 | 8 | 196 | 100 | 0.0057 | −1.279 | ||
| 1323 | Glyceraldehyde-3-phosphate dehydrogenase | Salmo salar | |||||||||
| gi 209737954 | 468 | 100 | 36097.4 | 7.12 | 21 | 238 | 100 | 0.0006 | 1.704 |
Mascot score: Probability Based Mowse Score: Ions score is −10*Log(P), where P is the probability that the observed match is a random event. Protein scores greater than 83 are significant (p < 0.05). MALDI-TOF-TOF peak lists were searched against a NCBInr 10997816 protein database using MASCOT TM software with the following settings: Type of search: Sequence Query; Enzyme: Trypsin; Fixed modifications: Carbamidomethyl (C); Variable modifications: Oxidation (M); Mass values: MONOISOTOPIC; Protein Mass: Unrestricted; Peptide Mass Tolerance: ± 100 ppm; Fragment Mass Tolerance: ± 0.2 Da; Max Missed Cleavages: 1; Instrument type: MALDI-TOF-TOF.
Fig. 2.
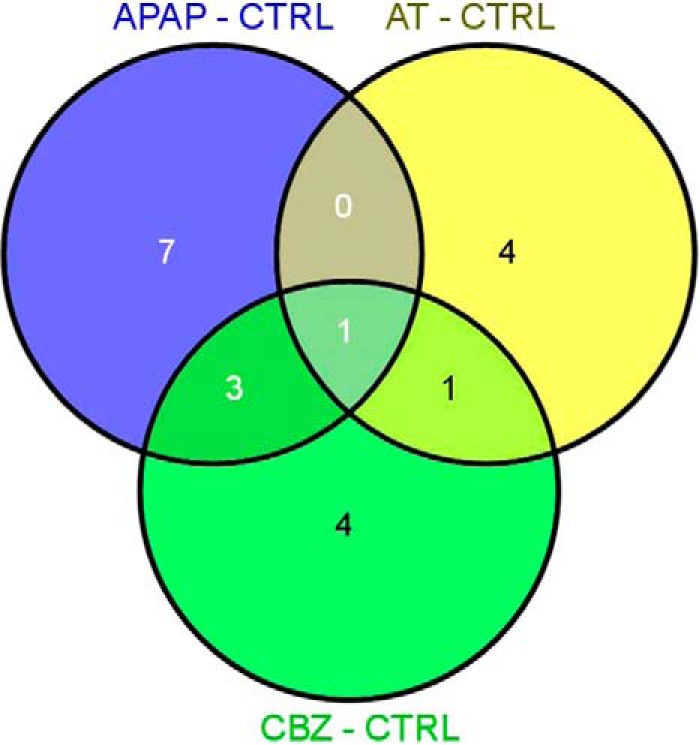
Venn diagram representation of differentially expressed proteins for each treatment in comparison to the control. Individuals were exposed to 54.77 ± 34.67, 11.08 ± 7.98 and 7.85 ± 0.13 μg·L−1 APAP, AT and CBZ, respectively for 5 days. Venn diagram shows the overlaps of differentially expressed proteins based on at least a 1.2-fold filter change with a p = 0.05.
Multivariate Analyses
Data were analyzed by two different multivariate analysis methods using the DeCyder-Extended Data Analysis (EDA) module. Principal component analysis (PCA) was applied to visualize the differences in expression patterns in the different spot maps from treated and untreated organisms (Fig. 3).
Fig. 3.
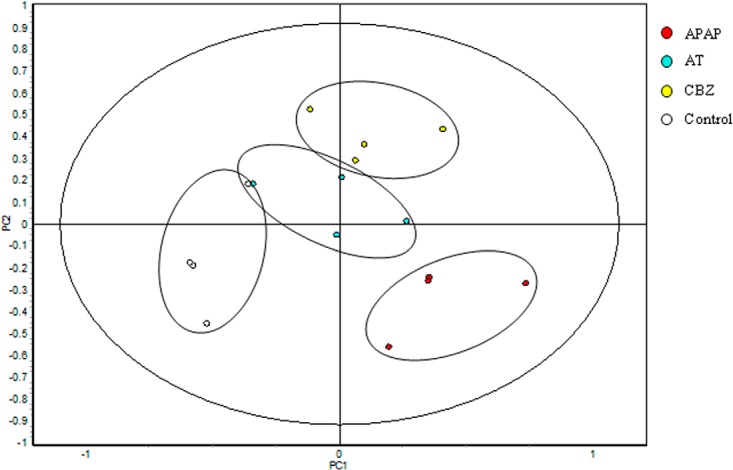
Principal component analysis (PCA) of the protein spot maps. PC#1 represents the variability because of the applied treatments; PC#2 represents the variability because of biological and technical variations between samples. The proteins that are included in the PCA were present in at least 75% of the spot maps and passed the filter of the one-way ANOVA (p < 0.01) test.
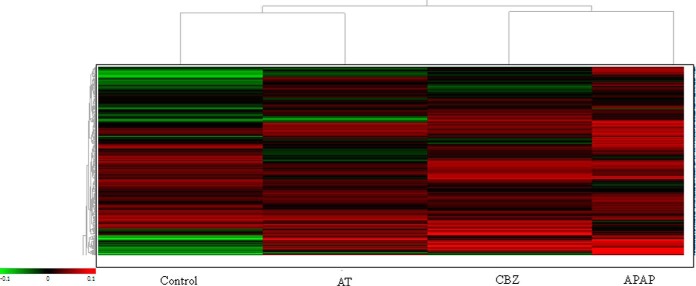
By analyzing the first two PCs a total separation of the different spot maps into the four different groups is observed, with the treatments APAP and CBZ more separated from each other than the AT and Control group. These latter treatments, although also clearly separated, show a slight overlap and are positioned closer to the coordinate intersection than the other two treatments. The control group was separated from the rest of the treatments in the first component (PC1) occupying the most negative position. The rest of the treatments are more placed toward positive values, with a clear vertical separation (PC2). The second component (PC2) separated the rest of the experimental groups that were also clustered individually, although a slight overlap between the treatments AT and CBZ could be observed. Also, a pattern analysis generating a heat map was carried out (Fig. 4) with manual base set where including those spots that were present in > 75% of the spot maps. Unassigned spots were removed and the applied normalization comprised scaling corresponding to the subtraction of the internal reference from the control. This resulted in 1273 spots that were represented in the heat map. The pattern analysis revealed a hierarchical clustering where the control group and the APAP treatment showed the most pronounced difference. Control and AT, as well as APAP and CBZ showed a closer relationship to each other than to the rest, forming, in both groups, two separate clusters.
Fig. 4.
Hierarchical cluster analysis. Heat map showing the separation of the different experimental groups taking into account all altered proteins present in at least 75% of the spot maps and filtered by one-way ANOVA (p < 0.01). Every colored box represents a protein that is up-regulated (red) or down-regulated (green) in a certain treatment. The brighter the color, the more intense is the change.
DISCUSSION
Because of their wide distribution and pseudo persistence, an increasing number of studies are focusing on the toxicological effects of exposure to pharmaceuticals in mammalian (34–37) and fish model species (26, 38–39). To better understand the effects of these potentially hazardous chemicals on ecologically relevant fish species, the present study aimed at evaluating the toxicity of short-term exposure to three highly environmentally prevalent pharmaceuticals in the liver of the Atlantic salmon parr, Salmo salar, a candidate sentinel species. We showed that short-term, subacute exposure to the three pharmaceuticals induced significant alterations in the hepatic protein expression profile. To date, effects of exposure to pharmaceuticals at environmentally relevant concentrations on nontarget organisms in general and, especially in Atlantic salmon remain largely unknown. Experiments with both mammals and teleost fish have shown high 50% lethal concentration (LC50) values (exposure concentration that produced mortality in 50% of the exposed organisms), suggesting that acutely exposed animals can tolerate quite high levels of these pharmaceuticals (40). In the present study, the tested concentrations were chosen according to environmental levels reported in the literature (13–17) and no mortality was observed. Using a proteomic analysis as means of evaluation of low level subacute effects, the present study reported a number of proteins displaying significant changes in abundance following exposure to the selected pharmaceuticals and of which most of them were successfully identified and validated through MASCOT from primary sequence databases using Peptide Mass Fingerprint, sequence query and MS/MS ion search. The function of these proteins may provide new clues on the molecular mechanisms by which APAP, AT, and CBZ induce effects in liver tissue and help to identify effective biomarkers of this kind of pollutants.
Regarding PCA (Fig. 3), the horizontal distance (PC#1) between the treated and control group is slightly larger than the distance between samples within one group, indicating that the most important difference between groups is because of the application of different treatments. The vertical distance (PC#2) between samples in the plot represents biological and technical variation. Proteins from individual samples were extracted and processed for each gel, so differences in resistance or sensitivity toward adverse conditions are to be expected. Additionally, technical variation induced by protein extraction, labeling, separation, spot picking, and digestion procedures may also provide a source of variation within a treatment group. The spot maps with the largest PC#1 values between each other are those who presented most differences in expression patterns. These are in our case the maps from the APAP and the Control treatments, whereas the AT and CBZ treatment present the least distance and lowest PC#1 value. This is in concordance with the fact that most significantly different proteins (this study) and also transcripts (41) have been found after APAP treatment where the supplied dose was 10 and 5 times higher than the one for CBZ and AT as found in the environment. Moreover, the target organ for APAP is the liver, the test tissue employed in this experiment, whereas the target tissues for AT and CBZ are the heart and the brain, respectively. Nevertheless, and although CBZ and AT are not pharmaceutically targeted to liver, these pharmaceuticals induced changes in hepatic protein expression in previous studies, such as hepatic cytochrome P450 subfamily members in the case of CBZ (42, 43, 44) or general hepatic dysfunction in the case of AT (45).
Hierarchical Cluster Analysis result representation as heat map (Fig. 4) visualizes the pattern of expression of proteins across treatments as compared with the internal standard. The trees to the left of the heat map and above the heat map show the relationships between the proteins and between the treatments, respectively. Proteins more distantly placed within branches show greatest differences in expression patterns between treatments. All three drug treatments varied significantly from the expression pattern observed in the control (with respect to the internal standard), with the largest difference occurring between the Control and APAP treatment. This is consistent with the PCA, as observed above (Fig. 3).
Expression of Proteins Involved in Energy Metabolism
In the present study, the proteins transketolase, mitochondrial ATP synthase, acetyl-CoA acetyltransferase, phosphoglycerate kinase 1, and glyceraldehydes-3-phosphate dehydrogenase were modulated by low level exposure within all the three treatments. All these enzymes play important roles in energy metabolism. Several ATP synthase mitochondrial precursors, a CoA isomerase precursor, as well as several heat shock proteins have also shown to be altered in the liver proteome of APAP treated mice (21), indicating similar responses in these different vertebrates. However, the doses applied to mice in these previous experiments were significantly higher that may explain the higher total number of altered proteins. One of the first responses of adaptation to stress in fish is the mobilization of energetic reserves, including the activation of liver glycogenolysis and gluconeogenesis in order to maintain a continuous supply of glucose to essential organs such as brain and muscle (46). In all three treatments phosphoglycerate kinase 1, an enzyme of the gluconeogenic and glycolytic pathways is increased. In APAP glyceraldehyde-3-phosphate dehydrogenase (GAPDH), another gluconoegenic/glycolytic enzyme is also increased, although this enzyme is decreased in CBZ. Transketolase, important for NADPH production in the pentose phosphate shunt, is also increased in APAP and CBZ. At the same time, we observed a 1.4 and 1.3-fold abundance increase of acetyl-CoA acetyltransferase in organisms exposed to APAP or CBZ. Acetyl-CoA acetyltransferase is a major enzyme of ketone body production in liver, and increases suggest responses to stressors, as well as, gluconeogenesis and, thus, glucose production. Overall these results suggest an ametabolic shift from glycolysis to gluconeogenesis fueled by an increase in NADPH production and concurrent with up-regulation of the ketogenic pathway producing metabolic fuel for the liver and peripheral tissues (47). However, as no significant changes in direct lipid metabolic enzymes or pathways were observed we did not see any evidence for mobilization of energy stores. The 1.639-fold decrease in ATP synthase H+ transporting mitochondrial F1 complex beta (gi 198285477) protein concentration upon APAP treatment may indicate mitochondrial toxicity and a loss of cellular energy production in line with mitochondrial damage. Previous studies indicated that mitochondrial permeability transition (MPT) is the principal mechanism in APAP-induced injury, with a potential to open the transition pore (48). MPT is recently focused as a mechanism for drug-induced hepatocyte injury (49–53) and the observed up-regulations of different energy related features could be a compensatory mechanism. Notably induction of MPT is a major signal for activation of apoptotic cell death pathways, and the major executioner of apoptosis, caspase 3, was also increased after APAP treatment.
Other Proteins
OsmC-like family proteins: in bacteria, this kind of proteins have shown to be induced by different types of stress, particularly high osmotic pressure and starvation (54). The test specimens employed in our experiments were salmon parr, which were about one month before smoltification, still adapted to fresh water. However, undergoing adaptation processes may have been already under way as the process of smoltification implies an adaptation from fresh to salt water with the accompanying physiological changes required. It is possible that some osmotic mechanism was already working in the fish. However, this feature was only detected in one (APAP) of the three treatments which were carried out under identical conditions, where its expression was down-regulated by 1.65 (p = 0.0359) indicating an interaction of APAP with normal osmotic adaptation processes in the liver.
Elongation factor: In eukaryotes, peptide chain elongation is mediated by elongation factors EF1 and EF2. EF2, which was up-regulated in our AT and CBZ treatments, catalyzes the translocation of peptidyl-tRNA on the ribosome. Elongation factors are highly conserved among different species and may be involved in functions other than protein synthesis, such as organization of the mitotic apparatus, signal transduction, developmental regulation, aging and transformation (55). EF2 can be modulated by reversible phosphorylation. Increased levels of phosphorylated EF2 reduce elongation rates presumably because phosphorylated EF2 fails to bind the ribosomes. Treatment of mammalian cells with agents that raise the cytoplasmic Ca2+ and cAMP levels reduce elongation rates by activating the kinase responsible for phosphorylating EF2. One of the therapeutic actions of CBZ is a result of its interaction with the adenylyl cyclase system and the consequent reduction of intracellular cAMP levels (56, 57). Possibly, at least after the CBZ treatment, the expression of EF2 has been induced to compensate for the reduction in elongation rates because of activation of the kinase responsible for phosphorylating EF2. In relation to AT, it was shown that beta-adrenergic receptor blockers can inhibit β adrenoreceptor-mediated cAMP accumulation in living cardiac rat cells (58).
TenA family transcriptional activator: this protein is a heme oxygenase related feature that was significantly changed under the CBZ treatment. Effects of CBZ on heme oxidase, glutathione-S-transferase and cytochrome P450 3A-like have been observed in the crustacean T. platyurus and the cnidarian H. attenuate, and lipid peroxidation was reduced in both organisms suggesting redox activity of the lipophilic CBZ molecule (59). In a related study where we analyzed the effects of the exposure trials on the liver transcriptome, we observed altered electron carrier activity and heme binding activity after CBZ treatment by Blast2Go analysis within the top 10 GO terms related to molecular function (41). In human, CBZ is well known to have various hematological toxic effects, such as aplastic anemia, leukopenia, eosinophilia, agranulocytosis and thrombocytopenia with often selective failure of red cell production during CBZ monotherapy (60).
The number of significantly differentially expressed features we obtained in our three exposure experiments was slightly lower or at the lower threshold of the average amount found in proteomic studies carried out in human and rodents (61). In a study compiling the results from about 200 proteomic experiments in human and rodents, these authors found that a typical published 2D-based expression proteomics experiment features 400–1500 spots and reports between 10 and 40 identified up- or down-regulated proteins which in many cases are repeated across the whole range of examined studies (61). Two features of their top 15 protein list from humans and rodents were also present in one or more of our identified protein lists: elongation factors and ATP synthase beta subunit. In relation with the microarray studies carried out within the same experimental trial, we can observe that tissue samples from the same individuals have shown to induce far more features at transcriptome level than at protein level. The number of differently expressed features per treatment we detected in these experiments was significantly higher (order of several hundred) than the differently expressed proteins detected in this study (41). Whereas at transcriptome level the main induced pathways were also those related to energy metabolism, genes belonging to other processes were altered, in many cases similar to the responses observed in mammalian studies of similar treatments. These are for instance inflammatory responses after APAP treatment, iron ion related processes after AT treatment and Ca2+ channel activity after CBZ treatment. However, when analyzing the same tissues at protein level, the main effect was alteration of proteins belonging to energy related processes (41). The simultaneous measurement of thousands of proteins in a cell has high potential impact in toxicology, as cellular effects and thus modes of action are more relevant at protein than at mRNA level. However, as all proteins have different properties (mass, isoelectric point, solubility, stability, etc.), and may exist in multiple forms, the accurate measurement of thousands of proteins in a sample is a very complicated task. Several approaches have been followed to study the problem of the complexity of the cellular proteome and the difficulty to isolate homogeneous samples for this type of studies (62). It is important to realize that the relationship between mRNA and protein content is heavily dependent on time after treatment, cellular localization as well as stability of the molecules (63).
The lack of obvious congruence in differently expressed features after transcriptomic and proteomic analysis, and the results observed by Petrak et al. (61) that the same proteins seem to be observed after widely varying treatments, raises the concern that with current proteomic techniques, only the highly abundant and soluble proteins are detected giving rise to technical artifacts, limitations or biases of the method. However, the combination of techniques, transcriptomic and proteomic, offers the opportunity to reliably quantify expression changes and identify previously unknown features which can be useful tools in ecotoxicology, if followed by further studies to confirm potential as biomarkers of contaminant specific contamination.
Environmental Relevance
Traditional risk assessment is based on the derivation of no observed effect concentrations (NOECs) from laboratory derived dose-response curves for comparison with measured or estimated exposure concentrations. Although proteomic data does not provide additional information for the derivation of NOECs, the function of the altered proteins provides in-depth mechanistic information of the molecular processes related with the exposure to environmental contaminants. In addition, proteins that are found to be specifically induced by one (group of) contaminant could be used as a base to develop contaminant specific biomarkers for monitoring environmental exposure in wildlife.
It is yet to be determined if the exposure to the selected concentration ranges would directly produce an adverse response, as proteome change alone does not necessarily equate to toxicity or adversity. An ecologically relevant adverse response is the impairment of functional capacity (morphology, development, lifespan, growth) because of an insult that exceeds an organism's homeostatic range and is likely to have consequences finally at community and ecosystem level. However, energy used for alteration of the proteome and metabolic adaptation in response to contamination will not be available for other critical physiological processes, and may thus have indirect adverse responses and consequent knock-on ecosystem effects.
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (64) via the PRIDE partner repository with the dataset identifier PXD001354.
Supplementary Material
Acknowledgments
We thank Dr Alberto Pascual Bravo, Raquel Gómez Díaz, and Dr Antonio Romero Ruíz from the Institute for Biomedicine (IBIS-CSIC) in Seville, Spain for their valuable help with the preparative gels and bioinformatics.
Footnotes
Author contributions: M.H. and M.L. designed research; M.H. performed research; E.A., I.A., J.S., and M.L. contributed new reagents or analytic tools; M.H., E.A., I.A., and J.S. analyzed data; M.H. wrote the paper.
* This work was supported by a Marie Curie Fellowship to MH (Proposal N° EIF-039691-SALMONPHARM, FP6-2005-Mobility-5).
 This article contains supplemental Files S1 to S4.
This article contains supplemental Files S1 to S4.
1 The abbreviations used are:
- MoA
- Mode of Action
- APAP
- Acetaminophen
- AT
- Atenolol
- CBZ
- Carbamazepine
- CSIC
- Spanish National Council for Scientific Research
- CAS N°
- Chemical Abstracts Service Number
- LOD
- Limits of detection
- LOQ
- limits of quantification
- DIA
- Differential In-gel Analysis
- BVA
- Biological Variation Analysis
- NCBI
- National Center for Biotechnology Information
- MASCOT
- software search engine that uses mass spectrometry data to identify proteins from primary sequence databases
- PCA
- Principal Component Analysis
- CTRL
- Control
- EDA
- Extended Data Analysis
- PC
- Principal Components
- LC50
- 50% Lethal Concentration
- GAPDH
- glyceraldehydes-3-phosphate dehydrogenase
- MPT
- mitochondrial permeability transition
- EF
- Elongation Factor
- cAMP
- 3′-5′-cyclic adenosine monophosphate
- NOEC
- no observed effect concentration.
REFERENCES
- 1. Sanchez B. C., Ralston-Hooper K., Sepúlveda M. S. (2011) Review of recent proteomic applications in aquatic toxicology. Environ. Toxicol. Chem. 30, 274–282 [DOI] [PubMed] [Google Scholar]
- 2. Martyniuk C. J., Popesku J. T., Chown B., Denslow N. D., Trudeau V. L. (2012) Quantitative proteomics in teleost fish: insights and challenges for neuroendocrine and neurotoxicology research. Gen. Comp. Endocr. 176, 314–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Martyniuk C. J., Alvarez S., Denslow N. D. (2012) DIGE and iTRAQ as biomarker discovery tools in aquatic toxicology. Ecotox. Environ. Saf. 76, 3–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gillardin V., Silvestre F., Dieu M., Delaive E., Raes M., Thomé J. P., Kestemont P. (2009) Protein expression profiling in the African clawed frog Xenopus laevis tadpoles exposed to the polychlorinated biphenyl mixture aroclor 1254. Mol. Cell. Proteomics 8, 596–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dortsa J., Kestemonta P., Marchanda P.-A., D'Hollanderb W., Thézenasc M.-L., Raesc M., Silvestre F. (2011) Ecotoxicoproteomics in gills of the sentinel fish species, Cottus gobio, exposed to perfluorooctane sulfonate (PFOS). Aquat. Toxicol. 103, 1–8 [DOI] [PubMed] [Google Scholar]
- 6. Viarengo A., Lowe D., Bolognesi C., Fabbri E., Koehler A. (2007) The use of biomarkers in biomonitoring: a 2-tier approach assessing the level of pollutant-induced stress syndrome in sentinel organisms. Comp. Biochem. Physiol. C. Toxicol. Pharmacol. 146, 281–300 [DOI] [PubMed] [Google Scholar]
- 7. Boxall A., Breton R. (2003) Pharmaceuticals and personal care products in the environment: regulatory drivers and research needs. QSAR Comb. Sci. 22, 399–409 [Google Scholar]
- 8. Halling-Sorensen B., Nielsen S. N., Lansky P. F., Ingerslev F., Holten Lutzhoft H. C., Jorgensen S. E. (1998) Occurrence, fate, and effects of pharmaceutical substances in the environment- a review. Chemosphere 36, 357–393 [DOI] [PubMed] [Google Scholar]
- 9. European Environment Agency (2010). Pharmaceuticals in the environment. Results of an EEA workshop. EEA Technical report No 1/2010 [Google Scholar]
- 10. Harder B. (2003) Extracting estrogens. Sci. News 164, 67–68 [Google Scholar]
- 11. Webb S, Ternes T., Gilbert M., Olejniczak K. (2003) Indirect human exposure to pharmaceuticals via drinking water. Toxicol. Lett. 142, 157–167 [DOI] [PubMed] [Google Scholar]
- 12. Cleuvers M. (2004) Mixture toxicity of the anti-inflammatory drugs diclofenac, ibuprofen, naproxen, and acetylsalicylic acid. Ecotox. Environ. Saftey 59, 309–315 [DOI] [PubMed] [Google Scholar]
- 13. Buser H.-R., Mullere M. D., Theobald N. (1998) Occurrence of the pharmaceutical clofibric acid and the herbicide mecoprop in various Swiss Lakes and in the North Sea. Environ. Sci. Technol. 32, 188–192 [Google Scholar]
- 14. Ternes T. A. (1998) Occurrence of pharmaceuticals in German sewage treatment plants and rivers. Water Res. 32, 3245–3260 [Google Scholar]
- 15. Ternes T. A., Stumpf M., Mueller J., Haberer K., Wilken R.-D., Servos M. (1999) Behavior and occurrence of estrogens in municipal sewage treatment plants—I. investigations in Germany, Canada, and Brazil. Sci. Total Environ. 225, 81–90 [DOI] [PubMed] [Google Scholar]
- 16. Kümmerer K. (2001) Pharmaceuticals in the environment: emission of pharmaceuticals, diagnostic aids, and disinfectants into wastewater by hospitals in relation to other sources—a review. Chemosphere 45, 957–969 [DOI] [PubMed] [Google Scholar]
- 17. Kolpin D. W., Furlong E. T., Meyer M. T., Thurman E. M., Zaugg S. D., Barber L. B., Buxton H. T. (2002) Pharmaceuticals, hormones, and other organic wastewater contaminants in US streams 1999–2000: a national reconnaissance. Environ. Sci. Technol. 36, 1202–1211 [DOI] [PubMed] [Google Scholar]
- 18. Salminen W. F., Voellmy R., Roberts S. M. (1997) Protection against hepatotoxicity by a single dose of amphetamine: the potential role of heat shock protein. Toxicol. Appl. Pharm. 147, 247–258 [DOI] [PubMed] [Google Scholar]
- 19. Liu J., Liu Y., Hartley D., Klaassen C. D., Shehin-Johnson S. E., Lucas A., Cohen S. D. (1999) Metallothionein-I/II knockout mice are sensitive to acetaminophen-induced hepatotoxicity. J. Pharmacol. Exp. Ther. 289, 580–586 [PubMed] [Google Scholar]
- 20. Reilly T. P., Bourdi M., Brady J. N., Pise-Masison C. A., Radonovich M. F., George J. W., Pohl L. R. (2001) Expression profiling of acetaminophen liver toxicity in mice using microarray technology. Biochem. Biophys. Res. Comm. 282, 321–328 [DOI] [PubMed] [Google Scholar]
- 21. Ruepp S. U., Tonge R. P., Shaw J., Wallis N., Pognan F. (2002) Genomics and proteomics analysis of acetaminophen toxicity in mouse liver. Toxicol. Sci. 65, 135–150 [DOI] [PubMed] [Google Scholar]
- 22. Jaeschke H., Bajt M. L. (2006) Intracellular signaling mechanisms of acetaminophen-induced liver cell death. Toxicol. Sci. 89, 31–41 [DOI] [PubMed] [Google Scholar]
- 23. Black J. W., Stephenson J. S. (1962) Pharmacology of a new adrenergic beta-receptor blocking compound. Lancet 2, 311–314 [DOI] [PubMed] [Google Scholar]
- 24. Bowman W. C., Rand M. J. (1980) Textbook of Pharmacology, second ed Blackwell, Oxford, 2368 pp. [Google Scholar]
- 25. Toda N. (2003) Vasodilating β-adrenoceptor blockers as cardiovascular therapeutics. Pharmacol. Therapeut. 100, 215–234 [DOI] [PubMed] [Google Scholar]
- 26. Owen S. F., Giltrow E., Huggett D. B., Hutchinson T. H., Saye J. A., Winter M. J., Sumpter J. P. (2007) Comparative physiology, pharmacology and toxicology of β-blockers: Mammals versus fish. Aquat. Toxicol. 82 145–162 [DOI] [PubMed] [Google Scholar]
- 27. Berridge M. J., Downes C. P., Hanley M. R. (1989) Neural and developmental actions of lithium: a unifying hypothesis. Cell 59, 411–419 [DOI] [PubMed] [Google Scholar]
- 28. Klein P. S., Melton D. A. (1996) A molecular mechanism for the effect of lithium on development. Proc. Nat. Acad. Sci.U.S.A. 93, 8455–8459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lucas F. R., Salinas P. C. (1997) WNT-7a induces axonal remodeling and increases synapsin I levels in cerebellar neurons. Dev. Biol. 192, 31–44 [DOI] [PubMed] [Google Scholar]
- 30. Beutler A. S., Li S., Nicol R., Walsh M. J. (2005) Carbamazepine is an inhibitor of histone deacetylases. Life Sci. 76, 3107–3115 [DOI] [PubMed] [Google Scholar]
- 31. Lee H. J., Rao J. S., Rapoport S. I., Bazinet R. P. (2007) Antimanic therapies target brain arachidonic acid signaling: lessons learned about the regulation of brain fatty acid metabolism. Prostag. Leukotr. Ess. 77, 239–246 [DOI] [PubMed] [Google Scholar]
- 32. Taggart J. B., Bron J. E., Martin S. A. M., Seear P. J., Høyheim B., Talbot R., Villeneuve L., Sweeney G. E., Houlihan D. F., Secombes C. J., Tocher D. R., Teale A. J. (2008) A description of the origins, design, and performance of the TRAITS / SGP Atlantic salmon (Salmo salar L.) cDNA microarray. J. Fish Biol. 72, 2071–2094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Santos J. L., Aparicio I., Alonso E., Callejón M. (2005) Simultaneous determination of pharmaceutically active compounds in wastewater samples by solid phase extraction and high-performance liquid chromatography with diode array and fluorescence detectors. Anal. Chim. Acta 550, 116–122 [Google Scholar]
- 34. Mashimoto M., Ushijima I., Suetsugi I., Akimoto T., Watanabe K., Yamada M. (1998) Stress-dependent anti-conceptive effects of carbamazepine: a study in stressed and nonstressed. Prog. Neuro-Psychoph. 22, 159–168 [DOI] [PubMed] [Google Scholar]
- 35. Tateishi T., Asoh M., Nakura H., Watanabe M., Tanaka M., Kumai T., Kobayashi S. (1999) Carbamazepine induces multiple cytochrome P450 subfamilies in rats. Chem-Biol. Interact. 117, 257–268 [DOI] [PubMed] [Google Scholar]
- 36. Biswas N. M., Gupta R. S., Chattopadhyay A., Choudhury G. R., Sarkar M. (1962) Effect of atenolol on cadmium-induced testicular toxicity in male rats. Reprod. Toxicol. 15, 699–704 [DOI] [PubMed] [Google Scholar]
- 37. Trumper L., Coux G., Monasterolo L. A., Molinas S., García V. M. C., Elías M. M. (2005) Effect of acetaminophen on the membrane anchoring of Na+, K+ATPase of rat renal cortical cells. BBA-Mol. Basis Dis. 1740, 332–339 [DOI] [PubMed] [Google Scholar]
- 38. van der Ven K., De Wit M., Keil D., Moens L., van Leemput K., Naudts B., De Coen. W. (2005) Development and application of a brain-specific cDNA microarray for effect evaluation of neuro-active pharmaceuticals in zebrafish (Danio rerio). Comp. Biochem. Phys. B 141, 408–417 [DOI] [PubMed] [Google Scholar]
- 39. Nunes B., Gaio A. R., Carvalho F., Guilhermino L. (2008) Behavior and biomarkers of oxidative stress in Gambusia holbrooki after acute exposure to widely used pharmaceuticals and a detergent. Ecotoxicol. Environ. Saf. 71, 341–354 [DOI] [PubMed] [Google Scholar]
- 40. Crane M., Watts C., Boucard T. (2006) Chronic aquatic environmental risks from exposure to human pharmaceuticals. Sci. Total Environ. 367, 23–41 [DOI] [PubMed] [Google Scholar]
- 41. Hampel M., Alonso E., Aparicio I., Santos J. L., Leaver M. (2011) Differential liver protein expression in the Atlantic salmon after exposure to environmentally relevant concentrations of three representative pharmaceutical compounds. 16th International Symposium on Pollutant Responses in Marine Organisms PRIMO16, Long Beach, CA [Google Scholar]
- 42. Yamashita H., Kazawa T., Minatogawa Y., Ebisawa T., Yamauchi T. (2002) Time-course of hepatic cytochrome P450 subfamily induction by chronic carbamazepine treatment in rats. Int. J. Neuropsychopharmacol. 5, 47–52 [DOI] [PubMed] [Google Scholar]
- 43. Tateishi T., Asoh M., Nakura H., Watanabe M., Tanaka M., Kumai T., Kobayashi S. (1999) Carbamazepine induces multiple cytochrome P450 subfamilies in rats. Chem. Biol. Interact. 117, 257–268 [DOI] [PubMed] [Google Scholar]
- 44. Spina E., Pisani F., Perucca E. (1996) Clinically significant pharmacokinetic drug interactions with carbamazepine. An update. Clin. Pharmacokinet. 31, 198–214 [DOI] [PubMed] [Google Scholar]
- 45. Mondal S., Das S., Bandyopadhyay S., Datta A., Sardar S., Ghosal P. K., Tripathi S. K. (2013) Hepatotoxicity of atenolol therapy–A report of two cases. J. Acute Dis. 2, 246–249 [Google Scholar]
- 46. Moyle P. B., Cech J. J. (1996) Fishes: an introduction to ichthyology, 3rd edn Prentice Hall, Upper Saddle River, New Jersey. [Google Scholar]
- 47. Kerner J., Hoppel C. (2000) Fatty acid import into mitochondria. Biochim. Biophys. Acta 1486, 1–17 [DOI] [PubMed] [Google Scholar]
- 48. Masubuchi Y., Suda C., Horie T. (2005) Involvement of mitocondrial permeability transition in acetaminophen-induced injury in mice. J. Hepatol. 42, 110–116 [DOI] [PubMed] [Google Scholar]
- 49. Bernardi P. (1996) The permeability transition pore control points of a cyclosporin A-sensitive mitochondrial channel involved in cell death. Biochim. Biophys. Acta 1275, 5–9 [DOI] [PubMed] [Google Scholar]
- 50. Lemasters J. J., Nieminen A. L., Qian T., Trost L. C., Elmore S. P., Nishimura Y., Crowe R. A., Cascio W. E., Bradham C. A., Brenner D. A., Herman B. (1998) The mitochondrial permeability transition in cell death: a common mechanism in necrosis, apoptosis, and autophagy. Biochim. Biophys. Acta 1366, 177–196 [DOI] [PubMed] [Google Scholar]
- 51. Masubuchi Y., Nakayama S., Horie T. (2002) Role of mitochondrial permeability transition in diclofenac-induced hepatocyte injury in rats. Hepatology 35, 544–551 [DOI] [PubMed] [Google Scholar]
- 52. Jaeschke H., Knight T. R., Bajt M. L. (2003) The role of oxidant stress and reactive nitrogen species in acetaminophen hepatotoxicity. Toxicol. Lett. 144, 279–288 [DOI] [PubMed] [Google Scholar]
- 53. James L. P., McCullough S. S., Lamps L. W., Hinson J. A. (2003) Effect of Nacetylcysteine on acetaminophen toxicity in mice: relationship to reactive nitrogen and cytokine formation. Toxicol. Sci. 75, 458–467 [DOI] [PubMed] [Google Scholar]
- 54. Mongkolsuk S., Praituan W., Loprasert S., Fuangthong M., Chamnongpol S. (1998) Identification and characterization of a new organic hydroperoxide resistance (ohr) gene with a novel pattern of oxidative stress regulation from Xanthomonas campestris pv. phaseoli. J Bacteriol. 180, 2636–2643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Riis B., Rattan S. I. S., Clark B. F. C., Merrick W. C. (1990) Eukaryotic protein elongation factors. Trends Biochem. Sci. 15, 420–424 [DOI] [PubMed] [Google Scholar]
- 56. Chen G., Pan B., Hawver D. B., Wright C. B., Potter W. Z., Manji H. K. (1996) Attenuation of cyclic AMP production by carbamazepine. J. Neurochem. 67, 2079–2086 [DOI] [PubMed] [Google Scholar]
- 57. Montezinho L. P., Mork A., Duarte C. B., Penschuck S., Geraldes C. F., Castro M. M. (2007) Effects of mood stabilizers on the inhibition of adenylate cyclase via dopamine D(2)-like receptors. Bipolar Disord. 9, 290–297 [DOI] [PubMed] [Google Scholar]
- 58. Pauwels P. J., Leysen J. E., Janssen P. A. J. (1989) β-Adrenoceptor-mediated cAMP accumulation in cardiac cells: effects of nebivolol. Eur. J. Pharm-Molec Ph. 172, 471–479 [DOI] [PubMed] [Google Scholar]
- 59. Vernouillet G., Eullaffroy P., Lajeunesse A., Blaise C., Gagné F., Juneau P. (2010) Toxic effects and bioaccumulation of carbamazepine evaluated by biomarkers measured in organisms of different trophic levels. Chemosphere 80, 1062–1068 [DOI] [PubMed] [Google Scholar]
- 60. Tagawa T., Sumi K., Uno R., Itagaki Y., Fujii F., Yamaguchi H. (1997) Pure red cell aplasia during carbamazepine monotherapy. Brain Dev. 19, 300–302 [DOI] [PubMed] [Google Scholar]
- 61. Petrak J., Ivanek R., Toman O., Cmejla R., Cmejlova J., Vyoral D., Zivny J., Vulpe C. D. (2008) Déjà vu in proteomics. A hit parade of repeatedly identified differentially expressed proteins. Proteomics 8, 1744–1749 [DOI] [PubMed] [Google Scholar]
- 62. Lopez J. L. (2005) Role of proteomics in taxonomy: the Mytilus complex as a model of study. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 815, 261–274 [DOI] [PubMed] [Google Scholar]
- 63. Heijne W. H. M., Stierum R. H., Slijper M., van Bladeren P. J., van Ommen B. (2003) Toxicogenomics of bromobenzene hepatotoxicity: a combined transcriptomics and proteomics approach. Biochem. Pharmacol. 65, 857–875 [DOI] [PubMed] [Google Scholar]
- 64. Vizcaíno J. A., Deutsch E. W., Wang R., Csordas A., Reisinger F., Ríos D., Dianes J. A., Sun Z., Farrah T., Bandeira N., Binz P. A., Xenarios I., Eisenacher M., Mayer G., Gatto L., Campos A., Chalkley R. J., Kraus H. J., Albar J. P., Martinez-Bartolomé S., Apweiler R., Omenn G. S., Martens L., Jones A. R., Hermjakob H. (2014) ProteomeXchange provides globally co-ordinated proteomics data submission and dissemination. Nature Biotechnol. 30(3), 223–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.