Abstract
Cloud computing, where scalable, on-demand compute cycles and storage are available as a service, has the potential to accelerate mass spectrometry-based proteomics research by providing simple, expandable, and affordable large-scale computing to all laboratories regardless of location or information technology expertise. We present new cloud computing functionality for the Trans-Proteomic Pipeline, a free and open-source suite of tools for the processing and analysis of tandem mass spectrometry datasets. Enabled with Amazon Web Services cloud computing, the Trans-Proteomic Pipeline now accesses large scale computing resources, limited only by the available Amazon Web Services infrastructure, for all users. The Trans-Proteomic Pipeline runs in an environment fully hosted on Amazon Web Services, where all software and data reside on cloud resources to tackle large search studies. In addition, it can also be run on a local computer with computationally intensive tasks launched onto the Amazon Elastic Compute Cloud service to greatly decrease analysis times. We describe the new Trans-Proteomic Pipeline cloud service components, compare the relative performance and costs of various Elastic Compute Cloud service instance types, and present on-line tutorials that enable users to learn how to deploy cloud computing technology rapidly with the Trans-Proteomic Pipeline. We provide tools for estimating the necessary computing resources and costs given the scale of a job and demonstrate the use of cloud enabled Trans-Proteomic Pipeline by performing over 1100 tandem mass spectrometry files through four proteomic search engines in 9 h and at a very low cost.
Tandem mass spectrometry (MS/MS) of a complex mixture of digested proteins, often termed “shotgun” proteomics, is an important proteomics technique that has enabled researchers to identify and quantify proteins in complex biological samples in a high throughput manner. Mass spectrometers continue their incremental increases in sensitivity, mass accuracy, and speed of data collection, thereby generating comprehensive highly accurate data on smaller and smaller sample sizes. Software tools have likewise become more sophisticated and have enabled improved interpretation of the mass spectra that are generated all at the cost of greater computational resources (1). Simply applying cutoffs to native search scores has been replaced with algorithms that model the output scores and other attributes of the peptide-spectrum matches (PSMs) to yield improved probabilistic metrics for peptide and protein identifications (2–4).
The typical bioinformatics workflow for analyzing such shotgun data (5) relies on an algorithm that matches the set of spectra generated by the instrument against a set of candidate matches. These candidates can be either theoretical spectra generated from a set of plausible candidate peptides selected from a set of protein sequences, termed sequence searching, or a set of previously identified mass spectra, termed spectral library searching. There are a large number of both commercial and open-source sequence search engines available for use (see (5) for references to many of these). They perform comparably over a wide range of data sets, although the output scores and formats vary significantly, thereby making comparison and integration of results challenging. However, it has been shown that combining the results of several search engines does provide a significant benefit in improved identification rates and confidence (2, 6).
The Trans-Proteomic Pipeline (TPP)1 (2), developed and maintained at the Institute for Systems Biology, is an open source suite of software tools that applies sophisticated modeling to the search results of datasets with one or more search engines. The TPP includes software tools for MS data representation, MS data visualization, peptide identification and validation, protein inference, quantification, spectral library building and searching, and biological inference. An important component of the TPP is the standardized or otherwise open data formats it supports, thereby enabling the application to many different search engines, as well as the merging of results from each.
Depending on the number of input spectra, number of sequences to be searched, cleavage constraints, and the number of potential modifications to be considered, sequence searching can be a very computationally intensive task, often performed on dedicated in-house computing clusters. However, such clusters are expensive to acquire, require expert assistance to maintain, and have limited life spans adding significantly to the cost of analysis that is often overlooked. Therefore, many labs work without access to compute clusters and resort to single desktop workstations. As a result, many datasets are processed to an extent that will fit the computational resources available, rather than to the extent possible with the most advanced techniques often undervaluing the data and limiting the interpretation of the available date. Lastly, there is also emerging interest in applying proteogenomic techniques to aid in genome annotation (7), and such approaches typically require computationally expensive searches against large search spaces of possible sequences. Clearly, a mechanism that can provide greater access to easily usable computational resources would improve the state of the proteomics research environment.
The term cloud computing has recently become popularly known to apply to the technique of augmenting (or even replacing) computation and storage to a network of computers with dynamically allocated resources to fit the need of the user. The user can benefit from nearly unlimited resources at a modest cost because of the large economies of scale afforded by the service providers. Commercial cloud computing providers include Amazon Elastic Compute Cloud (EC2) under a greater umbrella of services termed Amazon Web Services (AWS), Google's App Engine, Microsoft's Azure platform, and IBM's Softlayer. Open-source toolkits like Eucalyptus, Nimbus, and Hadoop (8) enable users to set up their own cloud computing infrastructure. As cloud computing becomes more pervasive in day to day computational tasks, it is expected that more providers will also enter the field in the coming years.
AWS provides an especially flexible platform for enabling cloud computing applications for bioinformatics (9) by providing virtualized servers that can execute custom machine images, virtually unlimited and secure file storage, as well as a messaging services that can provide job communication across AWS. Such a setup is generally called Infrastructure as a Service (IaaS). Anyone can create a custom virtual image that contains the exact operating system (e.g. a specific version of Linux or Microsoft Windows) of choice and a customized set of software applications and configuration. Then any number of virtual machines can be started using this virtual image as its boot disk. These images can be published and used by others for free or for a small fee. This infrastructure enables a broad range of applications to be deployed on AWS. One such example is the development of OneOmics by a partnership between AB SCIEX and Illumina in collaboration with ISB and ETH within Illumina's BaseSpace environment, providing fully integrated cloud computing raw data storage and analysis of both proteomic and genomic data using an App store model similar to the smartphone industry.
There have been several previous efforts to bring the power of cloud computing to proteomics data analysis. ViPDAC (10) provides an Amazon image that can be started on EC2 and enables analysis of MS/MS data on the virtual machine. However, it is a manual process to start many searches and ViPDAC is not streamlined to easily enable the processing of hundreds of MS runs across many EC2 nodes. It also does not provide the advanced post-search modeling capabilities of the TPP. MR-Tandem (11) is an adaption of the X!Tandem (12) search engine to the Hadoop architecture, with components that allow it to be run on Amazon's Elastic Map-Reduce infrastructure. Although it utilizes cloud capabilities efficiently, it is limited to a single search algorithm and therefore lacks the capability of combined analysis available using the post-processing found in TPP. If a job consists of a small number of files that would each take a long time to search, there is a benefit to splitting the files into pieces, distributing the searches across multiple machines, and then reassembling the results (13). However, if the number of files to process significantly outnumbers the available compute nodes, there is probably little benefit. The Hydra search engine (14) was specifically designed for the Hadoop architecture and automatically distributes the job over any number of nodes eliminating the need to split up files prior to processing. Hydra search results can be processed with the TPP. The Central Proteomics Facilities Pipeline (CPFP) (15) enables AWS-based cloud computing functionality within its data management system. ProteoCloud (16) also provides a mechanism to launch search jobs on AWS and combine the results.
Here we introduce new functionality distributed with the TPP to extend its use to cloud computing platforms, and specifically on AWS, either in a fully hosted form or as an extremely flexible high capacity computing resource for processing of data stored in a local lab. We focus on making TPP components and interface easy to use so that any laboratory can capitalize on the resources available via cloud computing with limited computing expertise. This development in the TPP enables easy mass spectrometry data processing and data results sharing and storage. After introducing the new functionality, we explore some cost and performance issues. We then demonstrate running an example large canine proteome dataset with 1110 MS runs through four search engines on an EC2 virtual cluster. Finally we introduce several on-line tutorials that guide users through installing the TPP and processing sample data on the Amazon EC2.
New Functionality
The TPP is a mature yet continually updated, free and open-source software suite installable on Microsoft Windows, Linux, and MacOS. It can operate interchangeably in a large group Linux environment as well as a single-user desktop machine. We have incorporated major new functionality in support of cloud computing on Amazon's Web Services platform into the TPP. Below we describe the four major facets of new functionality: (1) prebuilt Amazon machine images, (2) fully hosted TPP, (3) the amztpp command line program for remote processing on EC2 resources, and (4) GUI enhancements to make these features easy to use via the web interface of the TPP.
Amazon Machine Images
The first new resource for the TPP is a set of official Amazon machine images (AMIs) for the most recent versions of the TPP. An AMI is a template that is made from a snapshot of a system disk of a virtual computer that can be loaded onto almost any of the virtual computing resources offered by EC2. Launching new virtual computers on the EC2 is easily accomplished using such an AMI as the starting point within the Amazon EC2 console, the EC2 command line tools, or by third party tools using the EC2 application programming interface (API).
TPP's AMIs are built from a recent version of the official Ubuntu Linux AMIs with TPP installed, along with several other popular free and open-source proteomics software packages, including ProteoWizard's (17) msconvert, Comet (18), X!Tandem (12), OMSSA (19), MyriMatch (20), and InsPecT (21). As of TPP version 4.7, the AMIs also include popular RNASeq software packages such as TopHat, Cufflinks, and samtools (22–24), to enable sample specific sequence FASTA protein database construction for use in mass spectrometry data searching algorithms included in the TPP release. These TPP AMI's are updated and tested to ensure functionality as intended by the developers. See http://tools.proteomecenter.org/wiki/index.php?title = Amazon_EC2_AMI for access to all available images, the configuration of the AMIs, getting-started guides, and other developer and automation information.
There are several significant advantages to providing prebuilt official AMIs to the community. For new users, they provide a very easy mechanism for trying out TPP without having to install and configure the TPP on their own systems. Also, because the developers of TPP maintain the AMIs, end users need not concern themselves with the effort of updating images as new features, releases, and patches become available. Users who require more compute resources than they may have on hand can easily launch TPP-ready EC2 instances with high memory and compute capabilities with ease. They also provide a historical reference to allow comparisons of results processed with deprecated versions of TPP. Further, anyone is free to modify and use a TPP AMI as a basis for creating their own AMIs containing additional applications or data.
Gaining access to Amazon Web Services is very simple, requiring only some basic user information and a credit card for payment of service charges. No up-front cost or subscription fee is applied; charges only accrue for actual use of resources. EC2 instances based on this AMI are most easily launched via the TPP as described below, but can also be managed through the AWS management console.
Fully Hosted TPP
Because TPP provides a web interface, utilizing a purely AWS-hosted TPP solution is as simple as creating an AWS account, launching a virtual instance of a TPP image using the AWS management console and directing your web browser to the virtual server. However, for novice users, the AWS console can be daunting to use. Therefore, we also provide a simple web application, the TPP Web Launcher for Amazon (TWA), which takes the complexity out of choosing the correct TPP image and EC2 options.
The virtual server will remain up and running for as long as the user wishes, accruing a charge in 1 h increments that amounts to $1–$20 (all cost figures are expressed in USD as of May 2014) per day, depending on the instance type. The virtual server can be shut down at any time when it is not needed such as nights or weekends so charges due not accrue. Also built into the TPP image is a “dead man's switch” that can be enabled and will guarantee that an instance is shut down after a chosen period of time. Another advantage of such a system is that the TPP instance is available to the user from anywhere and available to any collaborators with whom the machine URL and login information are shared allowing the sharing of usage and results regardless of geographical location. This is the easiest method to test the TPP because no local software installation is required beyond a web browser.
In such a scenario, all data to be processed and explored can be stored either on Amazon's Simple Storage Solution (S3), in an Elastic Block Store (EBS), or on the local instance file system. Local instance storage can range from 160 GB to 1690 GB depending on the EC2 type chosen and it is not persistent, as it only lasts the life of the instance. Alternatively, one or more EBS stores which can store 1 GB to 1 TB can be mounted as devices and persist beyond the life of the instance allowing one to stop and start instances as needed. EBS charges costs start at $0.10 per 1 million I/O requests and $0.10 per GB-month for provisioned storage.
Local TPP with AWS as Compute Resource Solution
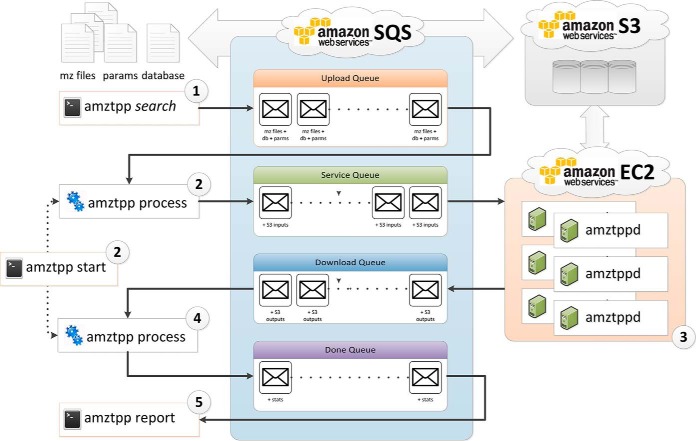
Many users already have a local TPP installation and are comfortable with their disk management practices but wish for more computing power to perform more searches with greater search spaces to enable a more complete analysis of their datasets. For such a use case, the amztpp command-line program for queuing and executing computationally expensive MS searches and other programs were developed and are now available to use with the TPP. This program manages all aspects of running cloud computing based MS/MS searches for the X!Tandem, Comet, OMMSA, MyriMatch, and InsPecT search engines. It utilizes three Amazon Web Services, the Simple Queue Messaging service (SQS) for scheduling work and interprocess communication, the Simple Storage Service (S3) for cloud file storage, and the Elastic Compute Cloud (EC2) for virtual computing. Conceptually, a set of MS/MS run files (e.g. in mzML (25) or mzXML (26) format) are submitted to the cloud in a message to SQS along with a search parameter file and sequence database via a command “amztpp <search engine> *.mzML,” either via the command line or via the Petunia graphical user interface. All these data components are uploaded to S3, one or more EC2 instances are started, the data are searched, and the results are downloaded back to the client and the remote instances are terminated. The above tasks occur somewhat in parallel in a time-efficient manner. The general workflow is summarized in Fig. 1. See supplemental Material S1 for additional information on provisioning AWS instances.
Fig. 1.
Overview of the amztpp workflow. 1. The client program amztpp queues a message containing the input files for each search to run to an upload queue. 2. The amztpp background process is started and begins polling the upload queue. For each message dequeued it uploads the input files to S3 and enqueues a message with the S3 locations to the service queue. Depending on current available instances the client may also initiate a new EC2 instance. 3. The amztppd daemon running on each EC2 instance polls the service queue. For each service message received, the input files are downloaded from S3 to the local file system and the search is run. When the search completes, all results (pep.xml, .tandem, output_sequences, captured stderr, and stdout) are uploaded to S3 and a message is enqueued in the download queue. 4. The client program polls the download queue and for each received message downloads the output files to the local client's file system and then queues a final message in the done queue containing statistics and any error messages. 5. General performance statistics and status can be obtained using the amztpp report command to query all messages found in the done queue.
Control of amztpp via the TPP Graphical User Interface
The amztpp components are command-line programs that are therefore amenable for automated pipelines and processing of a large number of datasets. However, to make these tools more accessible to more users, we have also integrated the use of the programs into the easy-to-use graphical user interface (GUI) of the TPP. This allows users who prefer a GUI to leverage the full capability of this new functionality. Users can set up searches in the same way as is normally done to run on the local machine, but now there is an additional selection component that enables a choice of where to submit the jobs: on the local machine as before, or to a cluster of EC2 computers (See supplemental Fig. S5). The option of an EC2 cluster target is enabled after a user enters AWS account configuration into the TPP's Petunia interface (27).
After a search is submitted, it may be monitored via the new job monitoring page, which lists all active and historical jobs, along with their current status and hyperlinks to more information (supplemental Fig. S4). There is also a new AWS status page that provides the current status of AWS resource usage including: how many EC2 computers are running, the number of messages in the SQS queues, details on files in S3, how many jobs are running and pending, and links to additional information about the user's AWS account and server logs (supplemental Fig. S3).
Below we discuss some considerations and implications for processing data using AWS, and then provide an example of speed and costs when using different back-end processing solutions.
Amazon's EC2 provides a wide variety of different instance types from which to choose with different memory, disk storage, CPU, and networking capacity. The TPP instances will run on most instance types except for the micro instances, which are suitable only for minimal processing. We compared the relative performance per dollar spent for various instance types, and find that the c1.medium instance can perform the task with the lowest cost, and the c1.xlarge is the most efficient for sequence searching with X!Tandem when considering a cost versus processing time tradeoff. See supplemental Material S3 for a full discussion of instance type considerations.
A significant advantage of using EC2 instances is that many instances can be started to make a large volume of computing occur quickly. And in general, as long as the computing required can be partitioned into segments of just under one hour, then it is almost the same cost to run N instances for one hour as it is to run one instance for N hours, meaning that large jobs can be completed very quickly for nearly the same cost as doing it more slowly on a single machine. We show the results from a real example of this in supplemental Fig. S9 along with discussion of this topic in supplemental Material S4. In order to help users estimate what resources would be optimal for a particular search job, we have developed a simulator called amzsim. The simulator considers numerous parameters including the number of mzML files, the average upload/download speeds, average file sizes, and average search times. The simulated results include costs for EC2, SQS, and S3 services, a timeline of the simulated jobs, and a table containing all of the simulated data as shown in supplemental Fig. S8. Further discussion of the simulator is found in supplemental Material S4.
An additional element that can save cost is to use Amazon's spot pricing, which is an attempt to provide attractive lower pricing for surplus available computing power. The spot pricing mechanism allows users to bid a price that they are willing to pay, and if the spot price of instances is less than the bid amount, then instances are started for the task. But if the spot price exceeds the bid price because of increased demand from others, instances may be terminated to meet higher paying or bidding demand. Use of this system is supported by the TPP, including sophisticated logic to restart processing work that was terminated when the spot price exceeded the bid price. For additional details see supplemental Material S5.
As a demonstration of the capabilities of the amztpp system on a large set of data, we have processed 1110 mzML files through four different search engines in order to build the Canine PeptideAtlas. Using spot pricing all processing was completed with 654 machine-hours over an elapsed time of 9.2 h for a total cost of $88.12. A complete description of this demonstration project is provided in supplemental Material S6.
A final important consideration is the network connection speed between the data source and the AWS processing power. As long as the time it takes to process an MS run significantly exceeds the time that it takes to upload the data and download the results, then the work can be spread among multiple nodes. However, if the upload component is slower than the processing, then it will be difficult to keep more than one instance busy and capitalize on the potential of elastic cloud computing. For a more thorough discussion of this, see supplemental Material S7.
In has been noted that the community would benefit from more standard operating procedures (SOPs) or case studies of informatics processing of proteomics data to provide a resource for researchers trying to improve the search results from their data (28). In order to guide new users through an example of processing proteomics shotgun data through the TPP using AWS cloud computing infrastructure, we have created a set of step-by-step tutorials. Similar to our previously published tutorial on installing and using the TPP on a local Windows machine (and searching locally) (27), we have prepared one tutorial that starts up an AWS instance with the TPP running via TWA and guides the user through processing and exploration of the dataset in easy steps, requiring no resources except a web browser and a credit card to which ∼$2 can be charged. This is the same dataset used in a previous tutorial (27) as well as the biannual proteomics informatics course taught by the developers at ISB (see http://www.proteomecenter.org/course.php).
A second tutorial guides the user through a local installation of the TPP on a Windows desktop, configuration of AWS components, and processing of the same dataset using AWS only as a remote compute resource. Exploration of the results is performed on the local desktop installation and the only charge is for the actual processing of data, well under $1. Both these tutorials are available on the TPP web site at http://tools.proteomecenter.org/wiki/index.php?title=TPP_Tutorials along with other tutorials that are unrelated to this article. The on-line tutorials are maintained and adjusted as the TPP evolves through continued maintenance and development. Although the local TPP with remote AMZ tutorial is tuned for Windows users, the same tutorial can be used under Linux or MacOS, except that the installation step requires additional expert input and not part of the tutorial itself (but the procedure is available elsewhere on the web site). The TWA tutorial is based on an EC2 instance natively running on Linux, but the operating system is largely invisible to the user. Complete information on downloads, tutorials, and other links is available at http://tools.proteomecenter.org/wiki/index.php?title=TPP:Cloud.
Supplementary Material
Footnotes
Author contributions: J.S. and E.W.D. designed research; J.S., L.M., and D.S. performed research; J.S. analyzed data; J.S., L.M., D.S., E.W.D., and R.L.M. wrote the paper.
* This work was supported in part with Federal funds from the National Institute of General Medical Sciences under grant No. R01 GM087221, 2P50 GM076547/Center for Systems Biology and S10RR027584, the National Science Foundation MRI grant No. 0923536, the American Recovery and Reinvestment Act (ARRA) funds through National Institutes of Health, the National Human Genome Research Institute grant No. RC2 HG005805, and the National Institute of Biomedical Imaging and Bioengineering grant No. U54EB020406. AWS costs were funded by compute time credits provided through an educational grant from Amazon Web Services Inc.
 This article contains supplemental Figs. S1 to S11, Material S1 to S7, and Tables S1 to S3.
This article contains supplemental Figs. S1 to S11, Material S1 to S7, and Tables S1 to S3.
1 The abbreviations used are:
- TPP
- trans-proteomic pipeline
- AWS
- Amazon web services
- AMI
- Amazon machine image.
REFERENCES
- 1. Nesvizhskii A. I., Vitek O., Aebersold R. (2007) Analysis and validation of proteomic data generated by tandem mass spectrometry. Nat. Methods 4, 787–797 [DOI] [PubMed] [Google Scholar]
- 2. Keller A., Eng J., Zhang N., Li X. J., Aebersold R. (2005) A uniform proteomics MS/MS analysis platform utilizing open XML file formats. Mol. Syst. Biol. 1, 2005.0017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kall L., Canterbury J. D., Weston J., Noble W. S., MacCoss M. J. (2007) Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat. Methods 4, 923–925 [DOI] [PubMed] [Google Scholar]
- 4. Ma Z. Q., Dasari S., Chambers M. C., Litton M. D., Sobecki S. M., Zimmerman L. J., Halvey P. J., Schilling B., Drake P. M., Gibson B. W., Tabb D. L. (2009) IDPicker 2.0: improved protein assembly with high discrimination peptide identification filtering. J. Proteome Res. 8, 3872–3881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Deutsch E. W., Lam H., Aebersold R. (2008) Data analysis and bioinformatics tools for tandem mass spectrometry in proteomics. Physiol. Genomics 33, 18–25 [DOI] [PubMed] [Google Scholar]
- 6. Shteynberg D., Deutsch E. W., Lam H., Eng J. K., Sun Z., Tasman N., Mendoza L., Moritz R. L., Aebersold R., Nesvizhskii A. I. (2011) iProphet: multi-level integrative analysis of shotgun proteomic data improves peptide and protein identification rates and error estimates. Mol. Cell Proteomics 10, M111 007690. 10.1074/mcp.M111.007690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jaffe J. D., Berg H. C., Church G. M. (2004) Proteogenomic mapping as a complementary method to perform genome annotation. Proteomics 4, 59–77 10.1002/pmic.200300511 [DOI] [PubMed] [Google Scholar]
- 8. White T. (2009) Hadoop: The Definitive Guide. O'Reilly Media; Sebastopol, CA, USA [Google Scholar]
- 9. Fusaro V. A., Patil P., Gafni E., Wall D. P., Tonellato P. J. (2011) Biomedical cloud computing with Amazon Web Services. PLoS Comput. Biol. 7, e1002147. 10.1371/journal.pcbi.1002147 PCOMPBIOL-D-10-00322 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Halligan B. D., Geiger J. F., Vallejos A. K., Greene A. S., Twigger S. N. (2009) Low cost, scalable proteomics data analysis using Amazon's cloud computing services and open source search algorithms. J. Proteome Res. 8, 3148–3153 10.1021/pr800970z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pratt B., Howbert J. J., Tasman N. I., Nilsson E. J. (2012) MR-Tandem: parallel X!Tandem using Hadoop MapReduce on Amazon Web Services. Bioinformatics 28, 136–137 doi: btr615 [pii] 10.1093/bioinformatics/btr615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Craig R., Beavis R. C. (2004) TANDEM: matching proteins with tandem mass spectra. Bioinformatics 20, 1466–1467 [DOI] [PubMed] [Google Scholar]
- 13. Mohammed Y., Mostovenko E., Henneman A. A., Marissen R. J., Deelder A. M., Palmblad M. (2012) Cloud parallel processing of tandem mass spectrometry based proteomics data. J. Proteome Res. 10.1021/pr300561q [DOI] [PubMed] [Google Scholar]
- 14. Lewis S., Csordas A., Killcoyne S., Hermjakob H., Hoopmann M. R., Moritz R. L., Deutsch E. W., Boyle J. (2012) Hydra: a scalable proteomic search engine which utilizes the Hadoop distributed computing framework. BMC Bioinformatics 13, 324 10.1186/1471-2105-13-324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Trudgian D. C., Mirzaei H. (2012) Cloud CPFP: a shotgun proteomics data analysis pipeline using cloud and high performance computing. J. Proteome Res. 11, 6282–6290 10.1021/pr300694b [DOI] [PubMed] [Google Scholar]
- 16. Muth T., Peters J., Blackburn J., Rapp E., Martens L. (2013) ProteoCloud: a full-featured open source proteomics cloud computing pipeline. J. Proteomics 88, 104–108 10.1016/j.jprot.2012.12.026 [DOI] [PubMed] [Google Scholar]
- 17. Kessner D., Chambers M., Burke R., Agus D., Mallick P. (2008) ProteoWizard: open source software for rapid proteomics tools development. Bioinformatics 24, 2534–2536 10.1093/bioinformatics/btn323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Eng J. K., Jahan T. A., Hoopmann M. R. (2012) Comet: an open source tandem mass spectrometry sequence database search tool. Proteomics 10.1002/pmic.201200439 [DOI] [PubMed] [Google Scholar]
- 19. Geer L. Y., Markey S. P., Kowalak J. A., Wagner L., Xu M., Maynard D. M., Yang X., Shi W., Bryant S. H. (2004) Open mass spectrometry search algorithm. J. Proteome Res. 3, 958–964 [DOI] [PubMed] [Google Scholar]
- 20. Tabb D. L., Fernando C. G., Chambers M. C. (2007) MyriMatch: highly accurate tandem mass spectral peptide identification by multivariate hypergeometric analysis. J. Proteome Res. 6, 654–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tanner S., Shu H., Frank A., Wang L. C., Zandi E., Mumby M., Pevzner P. A., Bafna V. (2005) InsPecT: identification of posttranslationally modified peptides from tandem mass spectra. Anal. Chem. 77, 4626–4639 [DOI] [PubMed] [Google Scholar]
- 22. Trapnell C., Williams B. A., Pertea G., Mortazavi A., Kwan G., van Baren M. J., Salzberg S. L., Wold B. J., Pachter L. (2010) Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515 10.1038/nbt.1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Trapnell C., Pachter L., Salzberg S. L. (2009) TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111 10.1093/bioinformatics/btp120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li H., Durbin R. (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Martens L., Chambers M., Sturm M., Kessner D., Levander F., Shofstahl J., Tang W. H., Rompp A., Neumann S., Pizarro A. D., Montecchi-Palazzi L., Tasman N., Coleman M., Reisinger F., Souda P., Hermjakob H., Binz P. A., Deutsch E. W. (2011) mzML–a community standard for mass spectrometry data. Mol. Cell Proteomics 10, R110 000133. 10.1074/mcp.R110.000133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pedrioli P. G., Eng J. K., Hubley R., Vogelzang M., Deutsch E. W., Raught B., Pratt B., Nilsson E., Angeletti R. H., Apweiler R., Cheung K., Costello C. E., Hermjakob H., Huang S., Julian R. K., Kapp E., McComb M. E., Oliver S. G., Omenn G., Paton N. W., Simpson R., Smith R., Taylor C. F., Zhu W., Aebersold R. (2004) A common open representation of mass spectrometry data and its application to proteomics research. Nat. Biotechnol. 22, 1459–1466 [DOI] [PubMed] [Google Scholar]
- 27. Deutsch E. W., Mendoza L., Shteynberg D., Farrah T., Lam H., Tasman N., Sun Z., Nilsson E., Pratt B., Prazen B., Eng J. K., Martin D. B., Nesvizhskii A. I., Aebersold R. (2010) A guided tour of the Trans-Proteomic Pipeline. Proteomics 10, 1150–1159 10.1002/pmic.200900375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kinsinger C. R., Apffel J., Baker M., Bian X., Borchers C. H., Bradshaw R., Brusniak M. Y., Chan D. W., Deutsch E. W., Domon B., Gorman J., Grimm R., Hancock W., Hermjakob H., Horn D., Hunter C., Kolar P., Kraus H. J., Langen H., Linding R., Moritz R. L., Omenn G. S., Orlando R., Pandey A., Ping P., Rahbar A., Rivers R., Seymour S. L., Simpson R. J., Slotta D., Smith R. D., Stein S. E., Tabb D. L., Tagle D., Yates J. R., 3rd, Rodriguez H. (2011) Recommendations for mass spectrometry data quality metrics for open access data (corollary to the Amsterdam Principles). Mol. Cell Proteomics 10, O111 015446 doi: O111.015446 [pii] 10.1074/mcp.O111.015446 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.