Abstract
Background:
Pseudomonas aeruginosa is the most common pathogen causing nosocomial infections. Resistance of P. aeruginosa strains to broad-spectrum cephalosporins may be mediated by extended-spectrum β-lactamases (ESBLs).
Objectives:
We intended to investigate the prevalence of ESBLs and antimicrobial susceptibilities of P. aeruginosa isolated from patients in Zahedan, Iran.
Materials and Methods:
In this cross-sectional study, during 2012–2013, 116 P. aeruginosa isolates were collected from a teaching hospital in Zahedan, Iran. Susceptibility to eight antimicrobial agents was carried out by disk diffusion method. The ESBL producing strains were detected by combination disk test (CDT). ESBL positive isolates as well as other isolates showing minimum inhibitory concentrations (MICs) ≥ 4 μg/mL for ceftazidime, cefotaxime, ceftriaxone and aztreonam, were screened for the presence of the genes encoding blaTEM, blaSHV, blaPER-1 and blaVEB-1, by polymerase chain reaction (PCR).
Results:
Ciprofloxacin and piperacillin were the most efficient antipseudomonal agents. The results disclosed that 19 (16.37%) of the isolates were multidrug resistant and 8 (6.89%) were ESBL-positive. Of the 116 isolates, 30 (25.86%) were resistant to at least one of the antibiotics ceftazidime, ceftriaxone, cefotaxime or aztreonam and among these 30 (100%), 4 (13.3%), 2 (6.6%) and 2 (6.6%), amplified blaTEM, blaVEB-1, blaPER-1 and blaSHV, respectively. From the 30 TEM-positive isolates, 22 were ESBL-negative. Sequencing of the ESBL genes verified the accuracy of the PCR products.
Conclusions:
According to our results, blaTEM-116 was the most frequent isolated ESBL gene among the P. aeruginosa strains isolated from patients.
Keywords: Pseudomonas aeruginosa, Extended-Spectrum beta-Lactamase
1. Background
Pseudomonas aeruginosa is an obligate aerobic Gram-negative bacillus, the natural flora of the skin and gastrointestinal tract of humans which can be found in water and soil (1). This bacterium is an opportunistic pathogen and a major cause of nosocomial infections in immunocompromised patients, including patients with a wide range of malignancies, cystic fibrosis, burns, etc. This bacterium comprises a variety of pathogens and has a high resistance to most common antibiotics (2, 3).
One of the most prominent attributes of these strains is their resistance to multiple clinically important antibiotics like third generation of cephalosporins, imipenem and aztreonam. A great number of P. aeruginosa strains generate various classes of extended spectrum β-lactamases (ESBLs) which allow the bacterium to tolerate against extended–spectrum cephalosporins, such as cefotaxime, ceftriaxone and ceftazidime and they have been reported with a developing frequency (4). Conventionally, ESBLs have been derivatives of TEM and SHV parent enzymes. Although, an explosion of developments in ESBLs of non-TEM, non-SHV lineage was seen in Europe last year. Notably, the CTX-M type ESBLs have become widespread (5).
ESBLs are undergoing continuum mutations, causing the progression of new enzymes, showing expanded substrate profiles. So far, there have been more than 300 different ESBL variants which have been clustered into nine different structural and evolutionary families based on their amino acid sequences. TEM and SHV were the major types. Nevertheless, CTX-M type is more frequent in some countries (6). PER-1 that can hydrolyze penicillins, oxyimino-cephalosporins and aztreonam, but not oxacillins, cephamycins and carbapenems, was first detected on a plasmid of P. aeruginosa RNL-1 in France in 1991 (7).
In 1996, VEB-1 enzyme was first identified in an Escherichia coli isolate from Vietnam with four further sequence variants since detected in widely scattered countries, in members of the Enterobacteriaceae or in the nonfermenting genera, particularly Pseudomonas spp. (8). Detection of TEM and SHV genes by molecular methods in ESBL-producing bacteria and their pattern of antimicrobial resistance can provide beneficial data about their epidemiology and risk factors associated with these infections (9).
2. Objectives
The purpose of this study was to isolate and identify the types of ESBLs produced by P. aeruginosa isolated from various specimens (urine, blood, tracheal tube, wound, ear discharge) of patients hospitalized in different units of educational hospitals of Zahedan, Iran.
3. Materials and Methods
The project was approved by the Zahedan University of Medical Sciences Ethical Committee (No. 2496).
3.1. Bacterial Isolates and Detection of Extended Spectrum β-Lactamases
This descriptive study was performed in Ali-Ebne-Abitaleb and Khatam-e-Anbia Hosbitals in Zahedan, Iran, during one year (2012-2013). P. aeruginosa was isolated from patients enrolled in this study. The isolates were cultured on triple sugar iron agar (Merck, Germany) and incubated at 37°C in air. The isolates with positive oxidase test, prepared from a pigment on Mueller–Hinton agar (Merck, Germany) and grown aerobically in OF (Merck, Germany) medium and oxidation of glucose, were identified as P. aeruginosa. The organisms were isolated from urine (n = 39), tracheal tube (n = 62), wound (n = 3), blood (n = 10), and ear discharge (n = 2).
Among the 116 collected isolates, 24 (20.9%) were isolated from children. There was 37 (31.9%) female and 79 (68.1%) male. The susceptibilities of the bacterial isolates to eight antimicrobial agents including imipenem (IMI; 10 µg), ciprofloxacin (CIP; 5 µg), ceftazidime (CAZ; 30 µg), ceftriaxone (CRO; 30 µg), cefotaxime (CTX; 30 µg), aztreonam (ATM; 30 µg), piperacillin (PRL; 100 µg) and gentamicin (GM; 10 µg) (Mast, UK) were determined by the disk diffusion method depending on the criteria published by the Clinical and Laboratory Standards Institute (CLSI). E. coli ATCC 25922 and P. aeruginosa ATCC 27853 were used as controls (10). Multidrug resistant (MDR) isolates were defined as those resistant to three or more classes of antipseudomonal agents (carbapenems, penicillins, cephalosporins, monobactams, fluoroquinolones and aminoglycosides) (11).
Minimum inhibitory concentration (MIC) E-test (Liofilchem, Italy) method for cefotaxime (0.002-32 µg), ceftriaxone (0.016-256 µg), ceftazidime (0.016-256 µg) and aztreonam (0.016-256 µg), the recommendations (CLSI) for resistant isolates, was performed and interpreted (10). ESBL production in all of the isolates was identified by combination disk test (CDT). For each test, disks containing cephalosporin alone (CTX and CAZ) and in combination with clavulanic acid are applied. The inhibition zone around the cephalosporin disk/tablet combined with clavulanic acid is compared with the zone around the disk/tablet with the cephalosporin alone. The test is positive if the inhibition zone diameter is ≥ 5 mm larger with clavulanic acid than without (12).
3.1.2. DNA Extraction and Polymerase Chain Reaction
ESBL-positive isolates and others showing MICs ≥ 4 μg/mL for CAZ, CTX, CRO and ATM were cultured in LB (Luria Bertani) broth at 37°C overnight and the plasmid DNA was extracted by plasmid isolation kit (MBST, Iran). Specific primers and the annealing temperature for amplifying the blaSHV, blaTEM, blaPER-1, blaCTX-M1 and blaVEB-1 genes by PCR are shown in Table 1 (13, 14). PCR was carried out by PCR PreMix 20 µL (Bioneer, Korea). K. pneumoniae 7881 containing the blaSHV, blaCTX-M and blaTEM genes, P. aeruginosa KOAS strain containing the blaPER gene, and P. aeruginosa 10.2 containing the blaVEB gene (Pasteur Institute of Iran) were used as controls.
Table 1. Primers Used in This Study.
| Primers | Sequence (5´ to 3´) | PCR | Cycles | Gene and Its Size, bp | ||
|---|---|---|---|---|---|---|
| Denaturation | Annealing | Extension | ||||
| VEB-1 F | CGACTTCCATTTCCCGATGC | 95°C, 1 min | 60°C, 1 min | 72°C, 1 min | 30 | blaVEB643 |
| VEB-1 R | GGACTCTGCAACAAATACGC | |||||
| PER-1 F | ATGAATGTCATTATAAAAGC | 94°C, 30 s | 51.5°C, 1 min | 72°C, 1 min | 25 | blaPER925 |
| PER-1 R | AATTTGGGCTTAGGGCAGAA | |||||
| TEM F | GAGTATTCAACATTTCCGTGTC | 94°C, 30 s | 57°C, 1 min | 72°C, 1 min | 30 | blaTEM861 |
| TEM R | TAATCAGTGAGGCACCTATCTC | |||||
| SHV F | AAGATCCACTATCGCCAGCAG | 94°C, 30 s | 64°C, 1 min | 72°C, 1 min | 25 | blaSHV231 |
| SHV R | ATTCAGTTCCGTTTCCCAGCGG | |||||
| CTX-m1 F | GACGATGTCACTGGCTGAGC | 94°C, 30 s | 56°C, 1 min | 72°C, 1 min | 25 | blaCTX-M1 499 |
| CTX-m1 R | AGCCGCCGACGCTAATACA | |||||
3.1.3. Sequencing of Polymerase Chain Reaction Products
DNA sequencing for the whole 30 strains was performed with a DNA sequencing kit and analyzed in an automatic DNA sequencer (Bioneer Company, Korea) to determine the detected bla genes, using primers shown in Table 1. Homological analyses were performed by BLAST software in GenBank.
3.1.4. Statistical Analysis
Descriptive statistical analyses were performed using SPSS (PASW) version 18 software.
4. Results
The findings of antimicrobial tests (Kirby Bauer) showed that CIP, PRL and GM were the most efficient antipseudomonal agents. P. aeruginosa revealed resistance to CRO (23.3%), CTX (22.4%), IMI (17.2%), CAZ (14.7%), ATM (14.7%), GM (12.1%), PRL (7.8%) and CIP (3.4%), respectively (Table 2). Overall, 19 (16.37%) MDR isolates and 8 (6.89%) ESBL-positive isolates were extracted.
Table 2. Antimicrobial Susceptibility Tests on 116 P. aeruginosa Isolated From Patients of Two Hospitals in Zahedan, Iran a.
| Antimicrobial Agents, Concentration (µg) | Resistant | Intermediate | Susceptible |
|---|---|---|---|
| Piperacillin, 100 | 9 (7.8) | 0 (0) | 107 (92.2) |
| Ceftazidime, 30 | 17 (14.7) | 0 (0) | 99 (85.3) |
| Cefotaxime, 30 | 26 (22.4) | 70 (60.3) | 20 (17.2) |
| Ceftriaxone, 30 | 27 (23.3) | 31 (26.7) | 58 (50) |
| Aztreonam, 30 | 17 (14.7) | 3 (2.6) | 96 (82.8) |
| Imipenem, 10 | 20 (17.2) | 3 (2.6) | 93 (80.2) |
| Gentamicin, 10 | 14 (12.1) | 6 (5.2) | 96 (82.8) |
| Ciprofloxacin, 5 | 4 (3.4) | 1 (0.9) | 111 (95.7) |
a Data are presented as No. (%).
Among the MDR isolates, 2 (10.5%) were resistant to 8 antimicrobial agents simultaneously (Table 3). The maximum ESBL production was found in blood samples. All the eight ESBL-positive isolates were MDR. Of the 116 isolates, 30 (25.86%) were resistant at least to one of the following antibiotics: CAZ, CRO, CTX and ATM. Among these isolates, 30 (100%), 4 (13.3%), 2 (6.6%), 2 (6.6%), and 0 (0%), amplified the blaTEM, blaVEB-1, blaPER-1, blaSHV, and blaCTX-M1 genes, respectively. From 30 isolates of TEM-positive, 22 isolates were ESBL-negative phenotype.
Table 3. Resistance to Two or More Antimicrobials Among Multidrug Resistant Isolated From Patients of the Two Hospitals in Zahedan, Iran a.
| Resistance to Two or More Antimicrobials | MDR |
|---|---|
| CRO, CTX | 19 (100) |
| CRO, CTX, CAZ | 17 (89.5) |
| CRO, CTX, CAZ, ATM | 16 (84.2) |
| CRO, CTX, CAZ, ATM, GM | 13 (68.4) |
| CRO, CTX, CAZ, ATM, GM, PRL | 8 (42.1) |
| CRO, CTX, CAZ, ATM, GM, PRL, IMI | 4 (21) |
| CRO, CTX, CAZ, ATM, GM, PRL, IMI, CIP | 2 (10.5) |
a Data are presented as No. (%).
Furthermore, four out of eight ESBL-positive isolates had the blaTEM and blaVEB genes, two had blaTEM and blaSHV genes, and two carried blaTEM and blaPER genes simultaneously. In addition, the sequencing results manifested that blaTEM, blaVEB-1 and blaPER-1 of the isolates were 100% homogenized with the blaTEM-116, blaVEB-1 and blaPER-1 genes, respectively. However, the blaSHV gene was compatible with blaSHV-12 about 98%.
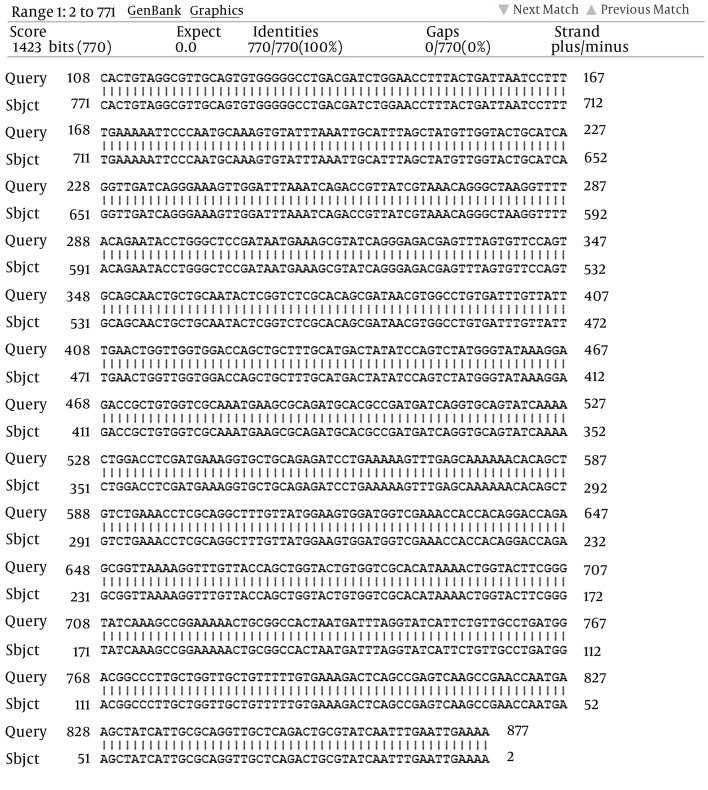
Figure 1. The Analysis of P18 DNA Partial Sequence Identity on blaPER-1.
Subject: blaPER-1 (accession number: AY953376.2 in GenBank); Query: P18 sample. Its partial sequence shared 100% identity with AY953376.2.
5. Discussion
P. aeruginosa is the most frequent pathogen causing nosocomial infections. Plasmid-mediated antibiotic resistance is commonplace in P. aeruginosa isolated from burn infected patients in different hospitals in Iran. Resistance of P. aeruginosa strains to the broad-spectrum cephalosporins may be mediated by ESBLs. The occurrence of MDR P. aeruginosa strains is growing in the world and limiting the therapeutic options (11). Our investigation showed the resistance rate in the following order: CRO, CTX, IMI, ATM, CAZ, GM, PRL and CIP. The results showed that CIP (95.7%) and PRL (92.2%) were the most effective antipseudomonal agents.
In this study, the prevalence of ESBL-producing strains of P. aeruginosa was determined about 6.8%. While, in a study by Tavajjohi et al. in Kashan, Iran, it was 9.2%. Moreover, Woodford et al. in the UK and Lim et al. in Malaysia reported it as 3.7% and 4.2%, respectively (8, 11, 15). However, the results of other investigators indicate high prevalence of ESBLs in P. aeruginosa strains, observed as about 28% in Thailand, 20.3% in India, 39.4% in the burn unit of Tehran, Iran, and 35.9% in the burn unit of northwest Pakistan patients. In the study by Shacheraghi et al. in Tehran, the frequency of the blaVEB-1 gene was 100% (4). Mirsalehian et al. in Tehran observed that the frequency of blaPER-1 and blaVEB-1 were 49.25% and 31.34%, respectively (16). However, in this study, the blaTEM gene frequency was 100% (16-19). The blaTEM gene in this study had 100% identity with blaTEM-116, which has been reported for the first time in Iran. blaTEM-116 is derived from blaTEM-1, which are different in two amino acids. Amino acid 84 in TEM-116 is Ile but in TEM-1 it is Val and amino acid 184 in TEM-116 is Val but in TEM-1 it is Ala. blaTEM-1 was the most frequent β-lactamase and conferred resistance to AMP, PRL, and cephalothin. TEM-116 had a spectrum that was expanded to CAZ, CTX, and ATM (20), which are in agreement with our study.
Phenotypic detection of ESBLs by clavulanate in P. aeruginosa due to various reasons such as the presence of chromosomal AmpC, broad-spectrum oxacillinase, resistant to clavulanate or other resistance mechanisms such as low permeability of the membrane and efflux pumps, was more difficult than the Enterobacteriaceae and leads to false negative results (13). In our study, this topic clearly indicated that despite of eight ESBL-positive phenotype isolates, 22 ESBL-negative phenotype isolates were the blaTEM genes. Therefore, molecular techniques such as PCR for detection of ESBLs seem to be necessary.
Acknowledgments
We gratefully acknowledge Dr. Davood Esmaeili for his guidance in Analysis of the sequencing results.
Footnotes
Authors’ Contributions:Study concept and design: Morteza Soltanian Bajgiran, Mohmmad Bokaeian, Shahram Shahraki Zahedani. Search in literature: Morteza Soltanian Bajgiran, Mohmmad Bokaeian, Shahram Shahraki Zahedani. Drafting of the manuscript: Morteza Soltanian Bajgiran. Critical revision of the manuscript for important intellectual content: Morteza Soltanian Bajgiran, Mohmmad Bokaeian, Shahram Shahraki Zahedani. Statistical analysis: Morteza Soltanian Bajgiran, Alireza Ansari Moghaddam.
Funding/Support:This study was resulted from the MSc. thesis under the code 30/K, sponsored by the Research Deputy of Zahedan University of Medical Sciences.
References
- 1.Palmer GC, Palmer KL, Jorth PA, Whiteley M. Characterization of the Pseudomonas aeruginosa transcriptional response to phenylalanine and tyrosine. J Bacteriol. 2010;192(11):2722–8. doi: 10.1128/JB.00112-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bonomo RA, Szabo D. Mechanisms of multidrug resistance in Acinetobacter species and Pseudomonas aeruginosa. Clin Infect Dis. 2006;43 Suppl 2:S49–56. doi: 10.1086/504477. [DOI] [PubMed] [Google Scholar]
- 3.Lister PD, Wolter DJ, Hanson ND. Antibacterial-resistant Pseudomonas aeruginosa: clinical impact and complex regulation of chromosomally encoded resistance mechanisms. Clin Microbiol Rev. 2009;22(4):582–610. doi: 10.1128/CMR.00040-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shacheraghi F, Shakibaie MR, Noveiri H. Molecular Identification of ESBL genes blaGES-1, blaVEB-1, blaCTX-M blaOXA-1, blaOXA-4, blaOXA-10 and blaPER-1 in Pseudomonas aeruginosa strains isolated from burn patients by PCR, RFLP and sequencing techniques. Int J Biol life Sci. 2010;3(6):138–42. [Google Scholar]
- 5.Paterson DL. Extended-spectrum beta-lactamases: the European experience. Curr Opin Infect Dis. 2001;14(6):697–701. doi: 10.1097/00001432-200112000-00006. [DOI] [PubMed] [Google Scholar]
- 6.Bali E. B., Acik L., Sultan N. Phenotypic and molecular characterization of SHV,TEM, CTX-M and extended-spectrum b-lactamase produced by Escherichia coli, Acinobacter baumannii and Klebsiella isolates in a Turkish hospital. Afr J Microbiol Res. 2010;4(8):650–4. [Google Scholar]
- 7.Bae IK, Jang SJ, Kim J, Jeong SH, Cho B, Lee K. Interspecies dissemination of the bla gene encoding PER-1 extended-spectrum beta-lactamase. Antimicrob Agents Chemother. 2011;55(3):1305–7. doi: 10.1128/AAC.00994-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Woodford N, Zhang J, Kaufmann ME, Yarde S, Tomas Mdel M, Faris C, et al. Detection of Pseudomonas aeruginosa isolates producing VEB-type extended-spectrum beta-lactamases in the United Kingdom. J Antimicrob Chemother. 2008;62(6):1265–8. doi: 10.1093/jac/dkn400. [DOI] [PubMed] [Google Scholar]
- 9.Jain A, Mondal R. TEM & SHV genes in extended spectrum beta-lactamase producing Klebsiella species beta their antimicrobial resistance pattern. Indian J Med Res. 2008;128(6):759–64. [PubMed] [Google Scholar]
- 10.Clinical Laboratory Standards Institute . Performance standards for antimicrobial disk susceptibility testing: approved standard. National Committee for Clinical Laboratory Standards; 2012. [Google Scholar]
- 11.Tavajjohi Z, Moniri R, Khorshidi A. Detection and characterization of multidrug resistance and extended-spectrum-beta-lactamase-producing (ESBLS) Pseudomonas aeruginosa isolates in teaching hospital. African J Microbiol Res. 2011;5(20):3223–8. [Google Scholar]
- 12.European Committee on Antimicrobial Susceptibility Testing . EUCAST guidelines for detection of resistance mechanisms and specific resistances of clinical and/or epidemiological importance. 2012. [Google Scholar]
- 13.Weldhagen GF, Poirel L, Nordmann P. Ambler class A extended-spectrum beta-lactamases in Pseudomonas aeruginosa: novel developments and clinical impact. Antimicrob Agents Chemother. 2003;47(8):2385–92. doi: 10.1128/AAC.47.8.2385-2392.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dashti AA, Jadaon MM, Gomaa HH, Noronha B, Udo EE. Transmission of a Klebsiella pneumoniae clone harbouring genes for CTX-M-15-like and SHV-112 enzymes in a neonatal intensive care unit of a Kuwaiti hospital. J Med Microbiol. 2010;59(Pt 6):687–92. doi: 10.1099/jmm.0.019208-0. [DOI] [PubMed] [Google Scholar]
- 15.Lim KT, Yasin RM, Yeo CC, Puthucheary SD, Balan G, Maning N, et al. Genetic fingerprinting and antimicrobial susceptibility profiles of Pseudomonas aeruginosa hospital isolates in Malaysia. J Microbiol Immunol Infect. 2009;42(3):197–209. [PubMed] [Google Scholar]
- 16.Mirsalehian A, Feizabadi M, Nakhjavani FA, Jabalameli F, Goli H, Kalantari N. Detection of VEB-1, OXA-10 and PER-1 genotypes in extended-spectrum beta-lactamase-producing Pseudomonas aeruginosa strains isolated from burn patients. Burns. 2010;36(1):70–4. doi: 10.1016/j.burns.2009.01.015. [DOI] [PubMed] [Google Scholar]
- 17.Ullah F, Malik SA, Ahmed J. Antimicrobial susceptibility and ESBL prevalence in Pseudomonas aeruginosa isolated from burn patients in the North West of Pakistan. Burns. 2009;35(7):1020–5. doi: 10.1016/j.burns.2009.01.005. [DOI] [PubMed] [Google Scholar]
- 18.Chanawong A, M'Zali FH, Heritage J, Lulitanond A, Hawkey PM. SHV-12, SHV-5, SHV-2a and VEB-1 extended-spectrum beta-lactamases in Gram-negative bacteria isolated in a university hospital in Thailand. J Antimicrob Chemother. 2001;48(6):839–52. doi: 10.1093/jac/48.6.839. [DOI] [PubMed] [Google Scholar]
- 19.Aggarwal R, Chaudhary U, Bala K. Detection of extended-spectrum beta-lactamase in Pseudomonas aeruginosa. Indian J Pathol Microbiol. 2008;51(2):222–4. doi: 10.4103/0377-4929.41693. [DOI] [PubMed] [Google Scholar]
- 20.Song JS, Jeon JH, Lee JH, Jeong SH, Jeong BC, Kim SJ, et al. Molecular characterization of TEM-type beta-lactamases identified in cold-seep sediments of Edison Seamount (south of Lihir Island, Papua New Guinea). J Microbiol. 2005;43(2):172–8. [PubMed] [Google Scholar]