Abstract
Background:
Assessment of resistance genes is imperative, as they become disseminated to bacterial flora in plants and to the indigenous bacterial community, and thus ultimately contributes to the clinical problems of antibiotic resistant pathogens.
Objectives:
The research was to assess the antibiotic characteristics and incidence of sul3 genes of Stenotrophomonas maltophilia isolates recovered from rhizospheres plant in Nkonkobe Municipality.
Materials and Methods:
Identification and assessment of resistance genes (sul2 and sul3 genes) were carried out using polymerase chain reaction (PCR). Analytical profile index (API) was used for biochemical characterization for identification before the PCR. Antibiotic susceptibility test was carried out using the approved guidelines and standards of Clinical Laboratory Standard Institute (CLSI).
Results:
A total of 125 isolates were identified, composed of 120 (96%) from grass root rhizosphere and 5 (4%) from soil butternut root rhizosphere. In vitro antibiotic susceptibility tests showed varying resistances to meropenem (8.9%), cefuroxime (95.6 %), ampicillin-sulbactam (53.9%), ceftazidime (10.7%), cefepime (29.3 %), minocycline (2.2%), kanamycin (56.9%), ofloxacin (2.9%), levofloxacin (1.3%), moxifloxacin (2.8%), ciprofloxacin (24.3%), gatifloxacin (1.3%), polymyxin B (2.9 %), cotrimoxazole (26.1%), trimethoprim (98.6%) and aztreonam (58%). The isolates were susceptible to the fluoroquinolones (74.3-94.7%), polymycin (97.1%) and meropenem (88.1%). The newest sulphonamide resistance gene, sul3, was detected among the trimethoprim-sulfamethoxazole (cotrimoxazole)-resistant isolates, while the most frequent sulphonamide-resistant gene in animal source isolates, sul2, was not.
Conclusions:
The commensal S. maltophilia isolates in the Nkonkobe Municipality environment harbored the resistant gene sul3 as clinical counterparts, especially from the perspective of reservoirs of antibiotic resistance determinants.
Keywords: Stenotrophomonas maltophilia, Rhizosphere, Sulphonamide, Fluoroquinolones
1. Background
Stenotrophomonas maltophilia is a readily available commensal of importance (1), found in water, soil, sewage and frequently on plant or within plant rhizosphere (2). They are commensals known for multitudinous applications in biotechnology (3). The bacteria explore the depression of immune systems to cause infection (4-6), though they have also been implicated in infection of immunocompetent subjects (7-9). They are therefore important considering their infectivity and the morbidity they initiate (10, 11), which range from nosocomial to community acquired infections. They cause a wide range of human systemic infections (12, 13) after entering through the respiratory pathway (4, 14).
Falagas et al. (15) reported a high mortality rate of 37.5% from S. maltophilia infections. Multidrug resistance by S. maltophilia has been well documented (16-19), raising the mortality rate in some areas to as high as 44.4% (20). Although the drug of choice for S. maltophilia infections is the sulfonamides (21), especially the synergistic form (cotrimoxazole or trimethoprim-sulfamethoxazole), resistance to these antibiotics is rampant around the world among human and nonhuman animals (22-24) and is mediated by the sulphonamide resistance (sul) gene. Sul3, being the newest sulphonamide gene, has been fingered as the possible reason for new rise in sulphonamide resistance world-wide (25). Sul2 has also been the most widely reported gene in animals (26-30) and can be used to trace the sulphonamide resistance genes in other sources originated from animal farms.
2. Objectives
Grasses and butternuts are eaten raw by animals and human, respectively. The latter is eaten as squash salad by human (31). Therefore, the safety of consumers is hinged on the type of bacterial flora associated with the plants and their susceptibility to antibiotics when they infect consumers. This in turn depends on the pool of genes in the rhizosphere of plants, as resistance gene(s) may become disseminated to the indigenous bacterial community form one organism, and ultimately contribute to the clinical problems of antibiotic resistant pathogens. Therefore, the aim of this study was to assess the S. maltophilia isolates from plants rhizosphere in the Nkonkobe Municipality, Eastern Cape Province, South Africa, for their antibiogram characteristics and the presence of antibiotic resistance genes, sul2 and sul3, in their genomes.
3. Materials and Methods
3.1. Study Location and Samples Collection
This study was conducted within the Nkonkobe Municipality of the Eastern Cape Province, South Africa. The Municipality is situated in the Amathole District Municipality, bordering the Nxuba Municipality to the west and the Amahlathi Municipality to the east. The municipality has a predominantly rural population and has a total of 21 wards. About 80% of the population of Nkonkobe resides in rural settlements. Soil butternut and grass roots in Alice Town environment were carefully uprooted and aseptically cut with sterile scissors into sterile containers and transported in ice to the laboratory for bacteria isolation.
3.2. Isolation of Test Bacteria
Isolation of the bacteria from root rhizospheres was performed following the methods of Bollet et al. (32) with slight modifications. About 1 g of the plant root sections were collected and inoculated into 10 mL of nutrient broth (bio-Merieux, Marcy-l’Etoile, France), supplemented with 0.5 mg of DL-methionine (Sigma Chemicals, South Africa) per mL. After 24 hours of incubation at 37°C, 0.1 mL was inoculated unto Mueller Hinton agar, spread to dry using a glass spreader, and allowed to stand for 15 minutes. Thereafter, four discs of 10 µg imipenem (MAST Diagnostics, Merseyside, UK) were aseptically placed on the surface of the inoculated agar. After 18 hours of incubations at 37°C, colonies that grew around the disc were subcultured for purity and subjected to preliminary identification.
3.3. Preliminary Identification of the Presumptive Stenotrophomonas Isolates
The purified isolates were Gram stained and observed under a light microscope. The Gram negative isolates were subjected to oxidase test and the oxidase negative isolates were subjected to preliminary speciation using analytic profile index 20E (API 20 E, BioMerieux, South Africa). In addition, carbon assimilation tests and other biochemical tests were carried out in the identification process. Stenotrophomonas genus positive isolates were then selected for specie confirmation.
3.4. Polymerase Chain Reaction confirmation of Stenotrophomonas maltophilia Isolates
Differentiation of S. maltophilia isolates amongst the genus isolates identified above was carried out using specie-specific polymerase chain reaction (PCR), using the primer sets SM1 (5'-CAGCCTGCGAAAAGTA-3') and SM2 (5'-TTAAGCTTGCCACGAACAG-3') (Inqaba Biotech., South Africa) (32). The PCR condition was as follows: an initial denaturation of 95°C for 5 minutes, a subsequent 30-cycle amplification including annealing at 58°C for 10 seconds, extension at 72°C for 60 seconds, and denaturation at 95°C for 10 seconds. For the last cycle, the extension step was two minutes (33). S. maltophilia DSM 50170 (ATCC 13637, type strain t20, Berlin, Germany) was used as the control.
3.5. Phenotypic Antibiotic Susceptibility Test
The disc diffusion technique was employed to determine the antibiotic susceptibility pattern of the isolates. The test antibiotics included meropenem, cefuroxime, ampicillin, ceftazidime, cefepime, minocycline, kanamycin, ofloxacin, levofloxacin, moxifloxacin, ciprofloxacin, gatifloxacin, polymyxin B, cotrimoxazole, trimethoprim, aztreonam and polymyxin B. S. maltophilia 50170 was used as the positive control, and the antibiogram was performed in accordance with standards described by the National Committee for Clinical Laboratory Standards (34) and Cheesebrough (35).
3.6. Multiple Antibiotic Resistance Index
The multiple antibiotic resistance index (MARI) was calculated as the ratio of the number of antibiotics to which resistance occurred by the isolates (a) to the total number of antibiotics to which the isolates were exposed (b), ie, MARI = a/b (36).
3.7. Assessment of Trimethoprim-Sulfamethoxazole Resistance Genes
Trimethoprim-sulfamethoxazole is the drug of choice in the treatment of infections caused by S. maltophilia. This, along with our initial observation of resistance to this antibiotic, informed the need for assessment of the presence of sul2 and sul3 genes in the resistant isolates, which were performed in accordance with the descriptions of Blahna et al. (26) using the primers (Inqaba Biotech., South Africa) listed in Table 1. The PCR condition for sul2 detection began with an enzyme activation (denaturation) stage at 94°C for five minutes, followed by 30 cycles of denaturation at 94°C for 40 seconds, annealing at 55°C for 40 seconds and extension at 72°C for 1 minute. A final extension at 72°C was run for seven minutes. For sul3 detection, the PCR condition was as follows: heating at 94°C for five minutes, 30 cycles at 94°C for 60 seconds, 55°C for 60 seconds and 72°C for 60 seconds, followed by one cycle at 72°C for seven minutes (26).
Table 1. Primers for the Assessment of Trimethoprim/Sulfamethoxazole Genes.
| Primers | Primer Sequence | Size |
|---|---|---|
| Sul2 F | 5′-GCGCTCAAGGCAGATGGCATT-3′ | 285 |
| Sul2 R | 5′-GCGTTTGATACCGGCACCCGT-3′ | 285 |
| Sul3 F | 5′- GAGCAAGATTTTTGGAATCG -3′ | 799 |
| Sul3 R | 5′- CATCTGCAGCTAACCTAGGGCTTTGGA -3′ | 799 |
4. Results
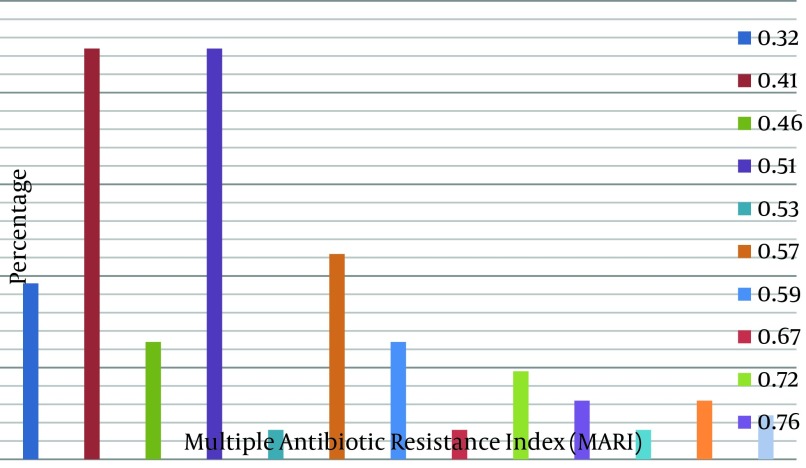
One hundred and twenty (96%) S. maltophilia isolates were recovered from grass root rhizosphere, while 5 (4%) were recovered from soil butternut rhizosphere (Table 2). About 8.9% of the isolates were resistant to meropenem, while resistance to the other antibiotics was as follows: cefuroxime 95.6%, ampicillin-sulbactam 53.9%, ceftazidime 10.7%, cefepime 29.3 %, minocycline 2.2%, kanamycin 56.9%, ofloxacin 2.9%, levofloxacin 3%, moxifloxacin (2.8%), ciprofloxacin 24.3%, gatifloxacin 1.3%, polymyxin B 2.9% and aztreonam 58% (Table 3). Variable susceptibilities to the cephalosporins (with carbapenem) were observed. About 88% of the isolates were susceptible to meropenem and ceftazidime, while 58.7% were susceptible to cefepime. In addition, 97.8% and 97.1% of the isolates were susceptible to minocycline and polymycin B, respectively. With regards to the fluoroquinolones, about 94.7% of the isolates were susceptible to both gatifloxacin and levofloxacin, while 90% and 87.1% were susceptible to moxifloxacin and ofloxacin, respectively (Table 3). A lower resistance (26.1%) to cotrimoxazole was observed in comparison with 98.6% resistance to trimethoprim (Table 3), and the MARI ranged 0.32-0.9 (Figure 1). Furthermore, four isolates were positive for sul3 genes while none were for sul2 gene (Table 4).
Table 2. Total Number and Percentage of Stenotrophomonas maltophilia Recovered Per Sourcea.
| Source | Recovered |
|---|---|
| Grass Root Rhizosphere | 120 (96.0) |
| Soil Butternut Rhizosphere | 5 (4.0) |
| Total | 125 (100) |
aData are presented as No. (%).
Table 3. Antibiotic Susceptibility Profile of Stenotrophomonas maltophilia Isolates.
| Antibiotics | Responses, % | ||
|---|---|---|---|
| Susceptible | Intermediate | Resistant | |
| Meropenem | 88.1 | 3.0 | 8.9 |
| Cefuroxime | 1.5 | 2.9 | 95.6 |
| Ampicillin-sulbactam | 44.6 | 1.5 | 53.9 |
| Ceftazidime | 88.0 | 1.3 | 10.7 |
| Cefepime | 58.7 | 12.0 | 29.3 |
| Minocycline | 97.8 | 0.0 | 2.2 |
| Kanamycin | 38.5 | 4.6 | 56.9 |
| Ofloxacin | 87.1 | 10.0 | 2.9 |
| Levofloxacin | 94.7 | 4.0 | 1.3 |
| Moxifloxacin | 90.0 | 7.2 | 2.8 |
| Ciprofloxacin | 74.3 | 1.4 | 24.3 |
| Gatifloxacin | 94.7 | 8.0 | 1.3 |
| Polymyxin B | 97.1 | 0.0 | 2.9 |
| Aztreonam | 14.5 | 27.5 | 58.0 |
| Cotrimoxazole | 63.8 | 10.1 | 26.1 |
| Trimethoprim | 0 | 11.4 | 98.6 |
Figure 1. Percentage of Isolates Versus Specific Multiple Antibiotic Resistance Index.
Horizontal axis, multiple antibiotic resistance; vertical axis, percentage of the isolates.
Table 4. Sulphonamide Resistance Genes Detected From Resistant Isolates of Stenotrophomonas maltophilia .
| Resistance Genes | Percentage Detected/phenotypic resistance, % |
|---|---|
| Sul3 | 12.1 |
| Sul2 | 0 |
5. Discussion
Commensal S. maltophilia may end up as an opportunistic pathogen (37). As revealed in this study, these bacteria are easily culturable, and appear ubiquitous, probably due to their resilience in the face of environmental stress (38). Our experience in this study suggests that the recovery of the organisms varies from place to place. As some studies have reported the isolation of this bacteria from soil butternut and walnut rhizosphere (39, 40), only 5 (4%) were isolated from the soil butternut rhizosphere compared to 120 (96%) from grass rhizosphere. The intrinsic resistance of this organism to imipenem was exploited for their isolation and API, supported by molecular identification, allowed convenient discrimination between the Stenotrophomonas species and other imipenem-resistant bacteria only (32).
The recovery rate of this bacterium appears to be increasing with time compared to when the bacteria was initially discovered. This scenario is buttressed by our findings as well as those of Gulmez et al. (41) which showed a higher frequency of occurrence of this specie than previously observed. S. maltophilia has been reported to be resistant to myriads of antibiotics (42, 43). This high resistance characteristic which was peculiar to clinical isolates has now been observed among environmental strains (44, 45). The resistance observed to kanamycin and trimethoprim in this study was in agreement with the report of Musa et al. (46) on commensal S. maltophilia from Osphronemus goramy. Similarly, S. maltophilia resistance to cephalosporin was higher in this study compared to that reported previously (47).
Berg et al. (48) and Crossman et al. (49) also noted that resistance to conventional antibiotics would have helped S. maltophilia to compete with other rhizospheric bacteria and made them survive in their habitat. This assertion is pertinent as all the isolates here showed MARI > 0.2, which implies that they have arisen from high-risk sources where antibiotics is in constant arbitrary use resulting in high selective pressure, as reported by Suresh et al. (50). Fluoroquinolone and polymycin B, both of which showed good activities against the S. maltophilia isolates, are usually the antibiotics of choice in the treatment of infections by the bacteria. The activities of these antibiotics against the bacteria have been similarly reported by Gales et al. (10) and Tripodi et al. (51). Valdezate et al. (52) observed that > 95% (94.7% in this study) of the bacterial isolates in their study were susceptible to a fluoroquinolone. However, it is known that trimethoprim-sulfamethoxazole is the drug of therapeutic choice against S. maltophilia infections (10, 53-55); but several reports have shown that the prevalence of S. maltophilia strains that are resistant to trimethoprim-sulfamethoxazole are increasing (56-58).
In this study, about 26% of the S. maltophilia isolates were resistant to this antibiotic compared with 2% reported elsewhere (10). The trend continues to threaten the public health of individuals, especially in an HIV/AIDS infested populations where the immune system is weakened. Resistance to trimethoprim-sulfamethoxazole is mediated by the sulphonamide resistance sul genes among other determinants (59). A study in Portugal by Antunes et al. (60) detected sul1, sul2, or sul3 genes in some Gram-negative isolates. The sul3 gene was observed to meditate trimethoprim-sulfamethoxazole resistance (61). This gene was earlier detected in some Gram-negative isolates recovered from animals and foods in Switzerland and Germany (22-24), suggesting commensal S. maltophilia to be as important as its clinical counterpart.
The presence of sul3 genes in this study may imply that the endophytic and clinical strains possess a similar level of antibiotic resistance, which may be more extensive among some endophytic strains of S. maltophilia (2). This probably explains the resistance against cotrimoxazole observed in this study. The rise in this sulphonamide resistance worldwide has been attributed to the newest sulphonamide resistance gene, sul3, especially in nonclinical (human) specimens like fresh water and soil (used in this study), sewage loving animals and animal farm (25, 62); but the isolates harboring these genes can still infect human. The potential threat that such resistant isolates could be to public health, informed the call for a surveillance study of the sul gene and phenotypic sulfamethoxazole by Toleman et al. (59).
Commensal S. maltophilia appears to be an important commensal with comparable antibiogram characteristics to its clinical strains. It also appears to be abundant in grass and soil butternut rhizosphere in the Eastern Cape Province of South Africa. The MARIs of the bacterial isolates suggest that their sources have been under antibiotics selective pressure, which could be related to abuse of antibiotics. Their antibiogram characteristics also suggest that the bacterium is an important reservoir of antibiotic resistant determinants (especially sulphonamide resistance (sul3) genes) in the environment.
Acknowledgments
The authors acknowledge the Govan Mbeki Research Development Council (GMRDC) for support, including the payment for handling the fees, as well as Dr. A.O. Aiyegoro and the University of Uyo for their supports.
Footnotes
Funding/Support:Funding was provided by the Govan Mbeki Research Development Council (GMRDC).
References
- 1.Alfieri N, Ramotar K, Armstrong P, Spornitz ME, Ross G, Winnick J, et al. Two consecutive outbreaks of Stenotrophomonas maltophilia (Xanthomonas maltophilia) in an intensive-care unit defined by restriction fragment-length polymorphism typing. Infect Control Hosp Epidemiol. 1999;20(8):553–6. doi: 10.1086/501668. [DOI] [PubMed] [Google Scholar]
- 2.Ryan RP, Monchy S, Cardinale M, Taghavi S, Crossman L, Avison MB, et al. The versatility and adaptation of bacteria from the genus Stenotrophomonas. Nat Rev Microbiol. 2009;7(7):514–25. doi: 10.1038/nrmicro2163. [DOI] [PubMed] [Google Scholar]
- 3.Adegoke AA, Tom M, Okoh AI. Sten. maltophilia as important commensal Biotechnology. J Pure Appl Microbiol. 2012;6(2):111–8. [Google Scholar]
- 4.Denton M, Keer V, Hawkey PM. Correlation between genotype and beta-lactamases of clinical and environmental strains of Stenotrophomonas maltophilia. J Antimicrob Chemother. 1999;43(4):555–8. doi: 10.1093/jac/43.4.555. [DOI] [PubMed] [Google Scholar]
- 5.Mendoza DL, Darin M, Waterer GW, Wunderink RG. Update on Stenotrophomonas maltophilia Infection in the ICU. Clin Pulm Med. 2007;14(1):17–22. doi: 10.1097/01.cpm.0000252835.11240.42. [DOI] [Google Scholar]
- 6.Gnanasekaran I, Bajaj R. Stenotrophomonas maltophiliabacteremia in end-stage renal disease patients receiving maintenance hemodialysis. Dial Transplant. 2009;38(1):30–2. doi: 10.1002/dat.20276. [DOI] [Google Scholar]
- 7.Kim JH, Kim SW, Kang HR, Bae GB, Park JH, Nam EJ, et al. Two episodes of Stenotrophomonas maltophilia endocarditis of prosthetic mitral valve: report of a case and review of the literature. J Korean Med Sci. 2002;17(2):263–5. doi: 10.3346/jkms.2002.17.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pruvost C, May L, Davous N, Petit A. [Plantar pyoderma due to Stenotrophomonas maltophilia]. Ann Dermatol Venereol. 2002;129(6-7):886–7. [PubMed] [Google Scholar]
- 9.Thomas J, Prabhu VN, Varaprasad IR, Agrawal S, Narsimulu G. Stenotrophomonas maltophilia: a very rare cause of tropical pyomyositis. Int J Rheum Dis. 2010;13(1):89–90. doi: 10.1111/j.1756-185X.2009.01447.x. [DOI] [PubMed] [Google Scholar]
- 10.Gales AC, Jones RN, Forward KR, Linares J, Sader HS, Verhoef J. Emerging importance of multidrug-resistant Acinetobacter species and Stenotrophomonas maltophilia as pathogens in seriously ill patients: geographic patterns, epidemiological features, and trends in the SENTRY Antimicrobial Surveillance Program (1997-1999). Clin Infect Dis. 2001;32 Suppl 2:S104–13. doi: 10.1086/320183. [DOI] [PubMed] [Google Scholar]
- 11.Pathmanathan A, Waterer GW. Significance of positive Stenotrophomonas maltophilia culture in acute respiratory tract infection. Eur Respir J. 2005;25(5):911–4. doi: 10.1183/09031936.05.00096704. [DOI] [PubMed] [Google Scholar]
- 12.Munter RG, Yinnon AM, Schlesinger Y, Hershko C. Infective endocarditis due to Stenotrophomonas (Xanthomonas) maltophilia. Eur J Clin Microbiol Infect Dis. 1998;17(5):353–6. doi: 10.1007/BF01709460. [DOI] [PubMed] [Google Scholar]
- 13.Labarca JA, Leber AL, Kern VL, Territo MC, Brankovic LE, Bruckner DA, et al. Outbreak of Stenotrophomonas maltophilia bacteremia in allogenic bone marrow transplant patients: role of severe neutropenia and mucositis. Clin Infect Dis. 2000;30(1):195–7. doi: 10.1086/313591. [DOI] [PubMed] [Google Scholar]
- 14.Fujita J, Yamadori I, Xu G, Hojo S, Negayama K, Miyawaki H, et al. Clinical features of Stenotrophomonas maltophilia pneumonia in immunocompromised patients. Respiratory Medicine. 1996;90(1):35–8. doi: 10.1016/s0954-6111(96)90242-5. [DOI] [PubMed] [Google Scholar]
- 15.Falagas ME, Kastoris AC, Vouloumanou EK, Rafailidis PI, Kapaskelis AM, Dimopoulos G. Attributable mortality of Stenotrophomonas maltophilia infections: a systematic review of the literature. Future Microbiol. 2009;4(9):1103–9. doi: 10.2217/fmb.09.84. [DOI] [PubMed] [Google Scholar]
- 16.Denton M, Todd NJ, Littlewood JM. Role of anti-pseudomonal antibiotics in the emergence of Stenotrophomonas maltophilia in cystic fibrosis patients. Eur J Clin Microbiol Infect Dis. 1996;15(5):402–5. doi: 10.1007/BF01690098. [DOI] [PubMed] [Google Scholar]
- 17.Zhang L, Li XZ, Poole K. SmeDEF multidrug efflux pump contributes to intrinsic multidrug resistance in Stenotrophomonas maltophilia. Antimicrob Agents Chemother. 2001;45(12):3497–503. doi: 10.1128/AAC.45.12.3497-3503.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brooke JS. Stenotrophomonas maltophilia: an emerging global opportunistic pathogen. Clin Microbiol Rev. 2012;25(1):2–41. doi: 10.1128/CMR.00019-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vartivarian S, Anaissie E, Bodey G, Sprigg H, Rolston K. A changing pattern of susceptibility of Xanthomonas maltophilia to antimicrobial agents: implications for therapy. Antimicrob Agents Chemother. 1994;38(3):624–7. doi: 10.1128/aac.38.3.624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maningo E, Watanakunakorn C. Xanthomonas maltophilia and Pseudomonas cepacia in lower respiratory tracts of patients in critical care units. J Infect. 1995;31(2):89–92. doi: 10.1016/s0163-4453(95)91985-6. [DOI] [PubMed] [Google Scholar]
- 21.Bin Abdulhak AA, Zimmerman V, Al Beirouti BT, Baddour LM, Tleyjeh IM. Stenotrophomonas maltophilia infections of intact skin: a systematic review of the literature. Diagn Microbiol Infect Dis. 2009;63(3):330–3. doi: 10.1016/j.diagmicrobio.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 22.Grape M, Sundstrom L, Kronvall G. Sulphonamide resistance gene sul3 found in Escherichia coli isolates from human sources. J Antimicrob Chemother. 2003;52(6):1022–4. doi: 10.1093/jac/dkg473. [DOI] [PubMed] [Google Scholar]
- 23.Riesenfeld CS, Goodman RM, Handelsman J. Uncultured soil bacteria are a reservoir of new antibiotic resistance genes. Environ Microbiol. 2004;6(9):981–9. doi: 10.1111/j.1462-2920.2004.00664.x. [DOI] [PubMed] [Google Scholar]
- 24.Schluter A, Szczepanowski R, Puhler A, Top EM. Genomics of IncP-1 antibiotic resistance plasmids isolated from wastewater treatment plants provides evidence for a widely accessible drug resistance gene pool. FEMS Microbiol Rev. 2007;31(4):449–77. doi: 10.1111/j.1574-6976.2007.00074.x. [DOI] [PubMed] [Google Scholar]
- 25.Perreten V, Boerlin P. A new sulfonamide resistance gene (sul3) in Escherichia coli is widespread in the pig population of Switzerland. Antimicrob Agents Chemother. 2003;47(3):1169–72. doi: 10.1128/AAC.47.3.1169-1172.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blahna MT, Zalewski CA, Reuer J, Kahlmeter G, Foxman B, Marrs CF. The role of horizontal gene transfer in the spread of trimethoprim-sulfamethoxazole resistance among uropathogenic Escherichia coli in Europe and Canada. J Antimicrob Chemother. 2006;57(4):666–72. doi: 10.1093/jac/dkl020. [DOI] [PubMed] [Google Scholar]
- 27.Gow SP, Waldner CL, Harel J, Boerlin P. Associations between antimicrobial resistance genes in fecal generic Escherichia coli isolates from cow-calf herds in western Canada. Appl Environ Microbiol. 2008;74(12):3658–66. doi: 10.1128/AEM.02505-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hammerum AM, Sandvang D, Andersen SR, Seyfarth AM, Porsbo LJ, Frimodt-Moller N, et al. Detection of sul1, sul2 and sul3 in sulphonamide resistant Escherichia coli isolates obtained from healthy humans, pork and pigs in Denmark. Int J Food Microbiol. 2006;106(2):235–7. doi: 10.1016/j.ijfoodmicro.2005.06.023. [DOI] [PubMed] [Google Scholar]
- 29.Sunde M, Norstrom M. The prevalence of, associations between and conjugal transfer of antibiotic resistance genes in Escherichia coli isolated from Norwegian meat and meat products. J Antimicrob Chemother. 2006;58(4):741–7. doi: 10.1093/jac/dkl294. [DOI] [PubMed] [Google Scholar]
- 30.Trobos M, Jakobsen L, Olsen KE, Frimodt-Moller N, Hammerum AM, Pedersen K, et al. Prevalence of sulphonamide resistance and class 1 integron genes in Escherichia coli isolates obtained from broilers, broiler meat, healthy humans and urinary infections in Denmark. Int J Antimicrob Agents. 2008;32(4):367–9. doi: 10.1016/j.ijantimicag.2008.04.021. [DOI] [PubMed] [Google Scholar]
- 31.Anon Butternut squash nutrition facts. 2009 Available from: http://www.nutrition-and-you.com/butternut-squash.html.
- 32.Bollet C, Davin-Regli A, De Micco P. A Simple Method for Selective Isolation of Stenotrophomonas maltophilia from Environmental Samples. Appl Environ Microbiol. 1995;61(4):1653–4. doi: 10.1128/aem.61.4.1653-1654.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Whitby PW, Carter KB, Burns JL, Royall JA, LiPuma JJ, Stull TL. Identification and detection of Stenotrophomonas maltophilia by rRNA-directed PCR. J Clin Microbiol. 2000;38(12):4305–9. doi: 10.1128/jcm.38.12.4305-4309.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.National Committee for Clinical Laboratory Standards. Performance standards for antimicrobial susceptibility testing, 9th informational supplement M100-S9. Wayne Pa: National Committee for Clinical Laboratory Standards; 1999. [Google Scholar]
- 35.Cheesebrough. M. District Laboratory Practice in Tropical Countries. 2nd ed. Cambridge University Press; 2006. [Google Scholar]
- 36.Krumperman PH. Multiple antibiotic resistance indexing of Escherichia coli to identify high-risk sources of fecal contamination of foods. Appl Environ Microbiol. 1983;46(1):165–70. doi: 10.1128/aem.46.1.165-170.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nyc O, Matejkova J. Stenotrophomonas maltophilia: Significant contemporary hospital pathogen - review. Folia Microbiol (Praha). 2010;55(3):286–94. doi: 10.1007/s12223-010-0043-4. [DOI] [PubMed] [Google Scholar]
- 38.Borner D, Marsch WC, Fischer M. [Necrotizing otitis externa caused by Stenotrophomonas maltophilia]. Hautarzt. 2003;54(11):1080–2. doi: 10.1007/s00105-003-0551-0. [DOI] [PubMed] [Google Scholar]
- 39.Rettenmaier H, Lingens F. Purification and some properties of two isofunctional juglone hydroxylases from Pseudomonas putida J1. Biol Chem Hoppe Seyler. 1985;366(7):637–46. doi: 10.1515/bchm3.1985.366.2.637. [DOI] [PubMed] [Google Scholar]
- 40.Kan FL, Chen ZY, Wang ET, Tian CF, Sui XH, Chen WX. Characterization of symbiotic and endophytic bacteria isolated from root nodules of herbaceous legumes grown in Qinghai-Tibet plateau and in other zones of China. Arch Microbiol. 2007;188(2):103–15. doi: 10.1007/s00203-007-0211-3. [DOI] [PubMed] [Google Scholar]
- 41.Gulmez D, Hascelik G. Stenotrophomonas maltophilia: antimicrobial resistance and molecular typing of an emerging pathogen in a Turkish university hospital. Clin Microbiol Infect. 2005;11(11):880–6. doi: 10.1111/j.1469-0691.2005.01257.x. [DOI] [PubMed] [Google Scholar]
- 42.Alonso A, Sanchez P, Martinez JL. Stenotrophomonas maltophilia D457R contains a cluster of genes from gram-positive bacteria involved in antibiotic and heavy metal resistance. Antimicrob Agents Chemother. 2000;44(7):1778–82. doi: 10.1128/aac.44.7.1778-1782.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Song JH, Sung JY, Kwon KC, Park JW, Cho HH, Shin SY, et al. [Analysis of acquired resistance genes in Stenotrophomonas maltophilia]. Korean J Lab Med. 2010;30(3):295–300. doi: 10.3343/kjlm.2010.30.3.295. [DOI] [PubMed] [Google Scholar]
- 44.Liaw SJ, Teng LJ, Hsueh PR, Ho SW, Luh KT. In vitro activities of antimicrobial combinations against clinical isolates of Stenotrophomonas maltophilia. J Formos Med Assoc. 2002;101(7):495–501. [PubMed] [Google Scholar]
- 45.Tan CK, Liaw SJ, Yu CJ, Teng LJ, Hsueh PR. Extensively drug-resistant Stenotrophomonas maltophilia in a tertiary care hospital in Taiwan: microbiologic characteristics, clinical features, and outcomes. Diagn Microbiol Infect Dis. 2008;60(2):205–10. doi: 10.1016/j.diagmicrobio.2007.09.007. [DOI] [PubMed] [Google Scholar]
- 46.Musa N, Wei SL, Shaharom F, Wee W. Surveillance of bacteria species in diseased freshwater ornamental fish from aquarium shop. World Appl Sci J. 2008;3(6):903–5. [Google Scholar]
- 47.Jones RN, Sader HS, Beach ML. Contemporary in vitro spectrum of activity summary for antimicrobial agents tested against 18 569 strains non-fermentative Gram-negative bacilli isolated in the SENTRY Antimicrobial Surveillance Program (1997–2001). Int J Antimicrob Agents. 2003;22(6):551–6. doi: 10.1016/s0924-8579(03)00245-0. [DOI] [PubMed] [Google Scholar]
- 48.Berg G, Eberl L, Hartmann A. The rhizosphere as a reservoir for opportunistic human pathogenic bacteria. Environ Microbiol. 2005;7(11):1673–85. doi: 10.1111/j.1462-2920.2005.00891.x. [DOI] [PubMed] [Google Scholar]
- 49.Crossman LC, Gould VC, Dow JM, Vernikos GS, Okazaki A, Sebaihia M, et al. The complete genome, comparative and functional analysis of Stenotrophomonas maltophilia reveals an organism heavily shielded by drug resistance determinants. Genome Biol. 2008;9(4):R74. doi: 10.1186/gb-2008-9-4-r74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Suresh T, Srinivasan D, Hatha AAM, Lakshmanaperumalsamy P. The Incidence, Antibiotic Resistance and Survival of Salmonella and Escherichia coli Isolated from Broiler Chicken Retail Outlets. Microbes Environments. 2000;15(3):173–81. doi: 10.1264/jsme2.2000.173. [DOI] [Google Scholar]
- 51.Tripodi MF, Andreana A, Sarnataro G, Ragone E, Adinolfi LE, Utili R. Comparative activities of isepamicin, amikacin, cefepime, and ciprofloxacin alone or in combination with other antibiotics against Stenotrophomonas maltophilia. Eur J Clin Microbiol Infect Dis. 2001;20(1):73–5. doi: 10.1007/pl00011239. [DOI] [PubMed] [Google Scholar]
- 52.Valdezate S, Vindel A, Loza E, Baquero F, Canton R. Antimicrobial susceptibilities of unique Stenotrophomonas maltophilia clinical strains. Antimicrob Agents Chemother. 2001;45(5):1581–4. doi: 10.1128/AAC.45.5.1581-1584.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Denton M, Kerr KG. Microbiological and clinical aspects of infection associated with Stenotrophomonas maltophilia. Clin Microbiol Rev. 1998;11(1):57–80. doi: 10.1128/cmr.11.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Betriu C, Rodriguez-Avial I, Sanchez BA, Gomez M, Picazo JJ. Comparative in vitro activities of tigecycline (GAR-936) and other antimicrobial agents against Stenotrophomonas maltophilia. J Antimicrob Chemother. 2002;50(5):758–9. doi: 10.1093/jac/dkf196. [DOI] [PubMed] [Google Scholar]
- 55.Krueger TS, Clark EA, Nix DE. In vitro susceptibility of Stenotrophomonas maltophilia to various antimicrobial combinations. Diagn Microbiol Infect Dis. 2001;41(1-2):71–8. doi: 10.1016/s0732-8893(01)00281-4. [DOI] [PubMed] [Google Scholar]
- 56.Micozzi A, Venditti M, Monaco M, Friedrich A, Taglietti F, Santilli S, et al. Bacteremia due to Stenotrophomonas maltophilia in patients with hematologic malignancies. Clin Infect Dis. 2000;31(3):705–11. doi: 10.1086/314043. [DOI] [PubMed] [Google Scholar]
- 57.Tsiodras S, Pittet D, Carmeli Y, Eliopoulos G, Boucher H, Harbarth S. Clinical implications of stenotrophomonas maltophilia resistant to trimethoprim-sulfamethoxazole: a study of 69 patients at 2 university hospitals. Scand J Infect Dis. 2000;32(6):651–6. doi: 10.1080/003655400459577. [DOI] [PubMed] [Google Scholar]
- 58.Al-Jasser AM. Stenotrophomonas maltophilia resistant to trimethoprim-sulfamethoxazole: an increasing problem. Ann Clin Microbiol Antimicrob. 2006;5:23. doi: 10.1186/1476-0711-5-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Toleman MA, Bennett PM, Bennett DM, Jones RN, Walsh TR. Global emergence of trimethoprim/sulfamethoxazole resistance in Stenotrophomonas maltophilia mediated by acquisition of sul genes. Emerg Infect Dis. 2007;13(4):559–65. doi: 10.3201/eid1304.061378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Antunes P, Machado J, Sousa JC, Peixe L. Dissemination of sulfonamide resistance genes (sul1, sul2, and sul3) in Portuguese Salmonella enterica strains and relation with integrons. Antimicrob Agents Chemother. 2005;49(2):836–9. doi: 10.1128/AAC.49.2.836-839.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Enne VI, King A, Livermore DM, Hall LM. Sulfonamide resistance in Haemophilus influenzae mediated by acquisition of sul2 or a short insertion in chromosomal folP. Antimicrob Agents Chemother. 2002;46(6):1934–9. doi: 10.1128/AAC.46.6.1934-1939.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Enne VI, Livermore DM, Stephens P, Hall LM. Persistence of sulphonamide resistance in Escherichia coli in the UK despite national prescribing restriction. Lancet. 2001;357(9265):1325–8. doi: 10.1016/S0140-6736(00)04519-0. [DOI] [PubMed] [Google Scholar]