Abstract
Background:
Toxin-antitoxin (TA) systems are found on the chromosomes and plasmids of many Bacteria such as Escherichia coli. The roles of TA systems in bacteria are enigmatic. Multiple biological functions of TA systems are proposed including growth modulation, persistence, and biofilm formation. Biofilms of E. coli are cause of urinary tract infections, as well as bacteraemia.
Objectives:
The current study aimed to find the association between biofilm formation and toxin-antitoxin systems in clinical isolates of E. coli.
Materials and Methods:
A total of 150 E. coli isolates were evaluated for biofilm formation by Congo red agar medium (CRA) and microtiter plate assay and the presence of different TA systems including MazEF, RelBE, hipBA, ccdAB and MqsRA.
Results:
The results of the analysis revealed that 107 E. coli isolates were potent for biofilm formation by CRA. The findings by microtiter plates showed that 102 E. coli isolates were biofilm producers. The results indicated that 80%, 85%, 70%, 91% and 82% of the isolates possessed MazEF, RelBE, hipBA, ccdAB and MqsRA TA loci, respectively.
Conclusions:
The analysis recommended that TA genes are prevalent in clinical isolates of E. coli strains. The analysis revealed that hipBA TA system is associated with biofilm formation.
Keywords: Biofilm, Escherichia coli, Toxin-Antitoxin Systems
1. Background
Toxin-antitoxin (TA) systems consist of a pair of genes in an operon which encodes a stable toxin and its labile antitoxin that inhibits the action of the toxin. TA systems depending on the antitoxin nature and manner of action are categorized in three types. Toxins are always protein, but antitoxins are either RNAs (type I and III) or proteins (type II) (1). Genes of Toxin-antitoxin systems are found on both the plasmids and chromosomes of many bacteria (2, 3). Several TA systems are identified in the chromosome of Escherichia coli including mazEF (4, 5), hipBA (6), chpBIK (7), relBE (8), yefM-yoeB (9), dinJ-yafQ (10) and mqsRA (11). Chromosomal TA gene modules mazEF and relBE are the most well studied TA systems in E. coli. mazEF TA system is known as the mediator for programmed cell death under stressful conditions (12), and regulator and responsible for inhibiting translation by cleaving mRNAs at specific sites to induce a reversible state of bacteriostasis (13). The relBE module acts as a stress response element, is activated by amino acid starvation (14), causes global translation inhibition, and leads to bacteriostasis (13).
Experimental evidences suggested that TA systems may be involved in a diverse range of behaviors, such as antiphage defense (15), persister formation (16), antibiotic-mediated programmed cell death, cellular stasis, and biofilm formation (17). Recent studies suggested the growth of bacteria in a biofilm might be a population-based strategy for bacterial survival (18). Persister cells are metabolically quiescent cells and a phenotypic subpopulation of bacteria are viable when exposed to different concentrations of antibiotics (19). Therefore, persister cells contribute to the multidrug tolerance in the bacteria of biofilms (20). Additional findings showed that there was a genetic relationship between the frequencies of persister cells and hipBA and relBE encoded by the chromosomal elements in E. coli populations (6).
Among the different kinds of TA systems studied in E. coli, the mqsRA was the first which had an obvious role in motivation of the cells to go toward the persistence phase and increased mqsR gene expression and causd persister cells, while deletion of the mqsR or mqsRA decreased the persister cell formation (21). The mqsRA is linked to motility and biofilm formation via the autoinducer-2 quorum sensing system and mqsR was first identified as an inducer in biofilm formation (22).
2. Objectives
Some of the TA systems are very important in biofilm formation and pathogenesis of bacteria. Therefore, the present study aimed to determine the association between TA genes and biofilm formation on the clinical isolates of E. coli.
3. Materials and Methods
3.1. Bacterial Strains
From 2011 to 2012, 150 E. coli clinical isolates were obtained from Milad Hospital (78 samples), Tehran, and Emmam and Mostafa hospitals (39 and 33 samples), Ilam, Iran. All of the isolates were obtained from urine samples during one year; isolated bacteria were identified by biochemical methods.
3.2. Total DNA Isolation
DNA extraction was performed by boiling method (23).
3.3. Polymerase Chain Reaction
Polymerase Chain Reaction (PCR) was performed by purified total DNA. Specific primers were used to amplify the mazF, relE, hipA, mqsR, and ccdB genes. The oligonucleotide sequences of the primers are listed in Table 1. PCR amplification was performed in a final volume of 25 μL containing 3 μL of purified total DNA, 10μL PCR buffer, 1.5 mM MgC12, 10mM of dNTP, 2.5 pM of each primer (Gen Fanavaran Company, Iran) and one unit of Taq polymerase (Super Taq Company, England). PCR was carried out in a thermal cycler apparatus (C1000TM; BIO RAD) with an initial denaturation step (at 95°C for five minute), following 35 cycles including denaturation at 94°C for one minute, annealing (at 58.7°C for mqsR, hipA and ccdB, at 58.5°C for mazF and at 58.6°C for relE) for 45 seconds and extension at 72°C for one minute and a final extension step at 72°C for 10 minutes. The PCR amplification products were analyzed by 1% gel agarose (Merck, Germany).
Table 1. Gene-specific Primers Used in Polymerase Chain Reaction a.
| Primer Sequence (5′ to 3′) | Length, nt | Accession Number |
|---|---|---|
| mazF | 288 | EG11249 |
| (F) ATGGTAAGCCGATACGTACCC | ||
| (R) TGGGGCAACTGTTCCTTT | ||
| relE | 267 | EG11131 |
| (F) GACGAGCGGGCACTAAAGGAAT | ||
| (R) TCAGAGAATGCGTTTGACCG | ||
| hipA | 1314 | EG10443 |
| (F) CTTGTCACTTGGATGAACAACCAG | ||
| (R) TCACTTACTACCGTATTCTCGGCT | ||
| ccdB | 272 | P62554 |
| (F) GAGAGAGCCGTTATCGTCTGTT | ||
| (R) TCCCCAGAACATCAGGTTAATG | ||
| MqsR | 194 | EG13023 |
| (F) ACGCACACCACATACACGTT | ||
| (R) GCCTGGGTCTGTAAACATCCT |
a in Toxin-antitoxin system.
3.4. Biofilm Formation Assay
3.4.1. Congo-red Agar Method (CRA)
Biofilm production in bacterial cultures was determined by Congo-red Agar method (CRA) as described previously by Subramanian et al. (24) with slight modifications. CRA medium was prepared using brain heart infusion (BHA) (Merck, Germany) supplemented with 5% sucrose (Merck, Germany) and Congo red (Merck, Germany). The medium was composed of BHA (52 g/L), sucrose (50 g/L), and Congo-red stain (0.8 g/L). Congo-red was prepared as the aqueous solution, autoclaved, and then added when the agar cooled to 55°C. Plates were inoculated and incubated for 24 hours at 37°C. The positive isolate was indicated by black and dry crystalline colonies. Weak biofilm producers usually remained pink with the darkness at the center of colonies. Intermediate results were exhibited by the darkness of the colonies with the absence of a dry crystalline.
3.5. Microtiter Plate Assay (Quantitative Assays)
The ability to form biofilms was assayed in microtiter plates as described by O’Toole and Kolter (25) and Vieira et al. (26) with slight modifications. Briefly, all isolates were incubated overnight in nutrient agar (Merck, Germany) at 37ºC; then cells were grown for 24 hours in 4 mL of LB medium (Merck, Germany) at 37ºC. Before the experiments, all the culture suspensions were vortexed and OD was adjusted to 0.45 to 0.65 at 600 nm in a spectrophotometer (Bausch and Lomb, USA). Ten microliter of Bacterial suspension was transferred to 96-well polystyrene microtiter plates and for each strain assay, it was done in triplicate. Then 190 µL of LB was added into each well and later incubated without shaking for 24 hours at 37ºC. To one of the wells for each strain just 200 µL of LB medium without bacteria was added as negative control. After incubation for 24 hours, the medium was removed and the plates were carefully washed three times with saline buffer to remove loosely attached bacteria. As the final step, the excess of buffer was removed and the biofilms were air dried for 10 minutes. Wells were stained with 200 µL of 0.5% Crystal Violet (CV) in H2O at room temperature for 15 minutes. The CV solution was removed and the wells were washed three times with sterile distilled water. To elute bound CV, 200 µL of a mixture containing 80% ethanol and 20% acetone was added to each well and the plate was incubated at room temperature for 15 minutes. Finally, the optical density was determined using a microtiter plate reader at a wavelength of 492 nm (A 492 nm).
3.6. Quantitative Analysis
The mean absorbance from the control wells (Ac) was computed in A492 nm. The microtiter plate assay was performed in triplicate and the average of the three wells for each strain was calculated. Strains were classified as follows: A ≤ Ac = no biofilm producer, Ac < A ≤ (2 × Ac) = weak biofilm producer, (2 × Ac) < A ≤ (4 × Ac) = moderate biofilm producer and (4 × Ac) < A = strong biofilm producer (27).
4. Results
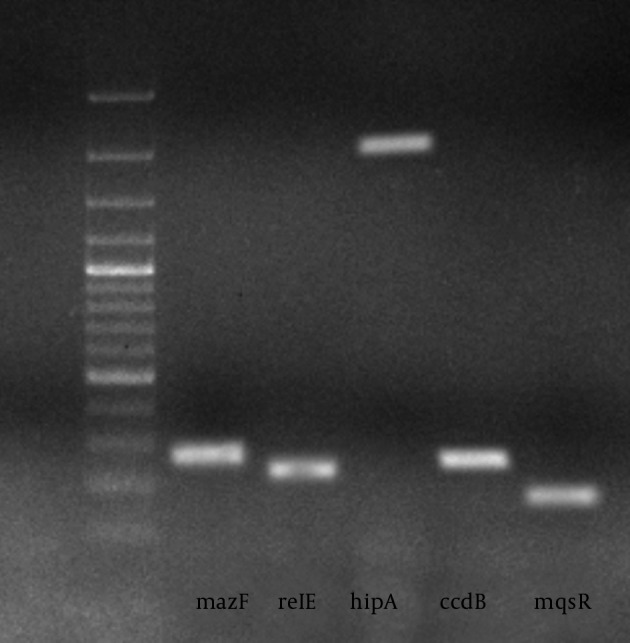
Chromosomal DNA of all E. coli clinical isolates were subjected for PCR, the results were positive if PCR products with the expected size were observed on the agarose gel. The PCR results revealed that the mazEF, relBE, hipBA, ccdAB, and mqsRA were present in 121 (80%), 128 (85%), 106 (70%), 137 (91%), and 123 (82%) of the isolates, respectively. The Prevalence of different TA genes is displayed in Tables 2 and 3. The PCR results are shown in Figure 1.
Table 2. Frequency of Toxin-Antitoxin Genes in Biofilm Positive and Negative Isolates by Congo-red Assay a.
| Toxin-Antitoxin Genes | Biofilm Positive Isolates | Biofilm Negative Isolates | Total |
|---|---|---|---|
| mazF | |||
| Negative | 21 (72.4) | 8 (27.6) | 29 (100) |
| Positive | 86 (71.1) | 35 (28.9) | 121 (100) |
| relE | |||
| Negative | 13 (59.1) | 9 (40.9) | 22 (100) |
| Positive | 94 (73.4) | 34 (26.6) | 128 (100) |
| ccdB | |||
| Negative | 8 (61.5) | 5 (38.5) | 13 (100) |
| Positive | 99 (72.3) | 38 (27.7) | 137 (100) |
| hipA | |||
| Negative | 26 (59.1) | 18 (40.9) | 44 (100) |
| Positive | 81 (76.4) | 25 (23.6) | 106 (100) |
| MqsR | |||
| Negative | 20 (74.1) | 7 (25.9) | 27 (100) |
| Positive | 87 (70.7) | 36 (29.3) | 123 (100) |
a Data are presented as No. (%).
Table 3. Frequency of Toxin-Antitoxin Genes in Biofilm Positive and Negative Isolates by Microtiter Plate Assay a.
| Toxin-Antitoxin Genes | Biofilm positive isolates | Biofilm negative isolates | Total |
|---|---|---|---|
| mazF | |||
| Negative | 22 (75.9) | 7 (24.1) | 29 (100) |
| Positive | 80 (66.1) | 41 (33.9) | 121 (100) |
| relE | |||
| Negative | 15 (68.2) | 7 (31.8) | 22 (100) |
| Positive | 87 (68) | 41 (32) | 128 (100) |
| ccdB | |||
| Negative | 6 (46.2) | 7 (53.8) | 13 (100) |
| Positive | 96 (70.1) | 41 (29.9) | 137 (100) |
| hipA | |||
| Negative | 27 (61.4) | 17 (38.6) | 44 (100) |
| Positive | 75 (70.8) | 31 (29.2) | 106 (100) |
| MqsR | |||
| Negative | 21 (77.8) | 6 (22.2) | 27 (100) |
| Positive | 81 (65.9) | 42 (34.1) | 123 (100) |
a Data are presented as No. (%).
Figure 1. PCR Analysis of Toxin-Antitoxin Genes in E. coli Strains.
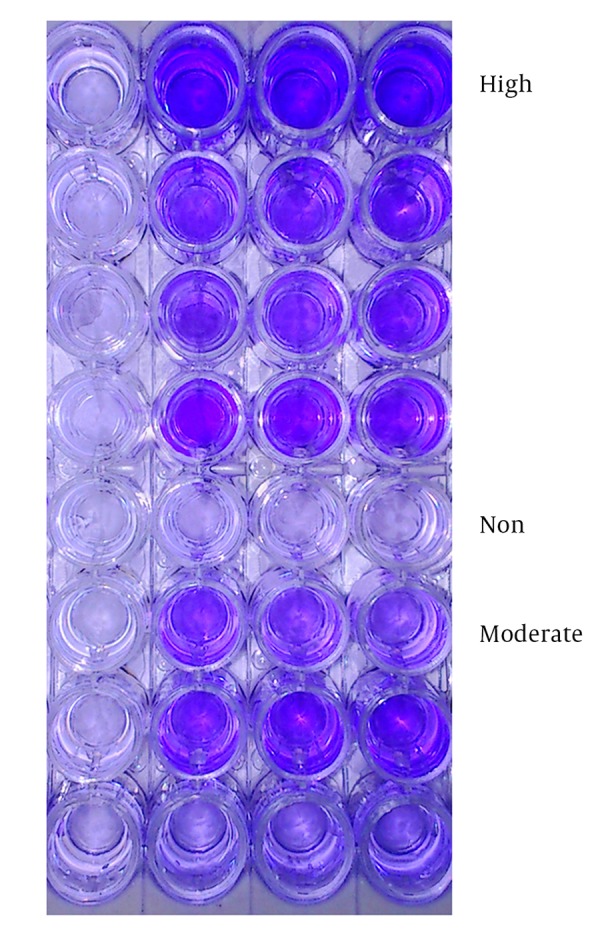
The Congo-red Agar method (CRA) for biofilm screening showed that 107 isolates (71.3%) were positive for biofilm production. Among them, 61 isolates (40.6%) showed dry crystalline and black colonies at the Congo-red Agar culture, which were considered as strong biofilm producers; 46 isolates (30.6%) did not show dry crystalline black colonies and were identified as moderately biofilm producers. Non-biofilm producers were 43 isolates (28.6%) that showed pink (15.3%) or yellow (13.3%) colonies. In the quantitative assay for the biofilm formation by the microtiter plate method, the isolates were also classified as strongly biofilm producer, moderate and weakly adherent isolates, and non-biofilm producers. Quantitative microtiter assay for biofilm formation showed positive results in 102 isolates (68%), among which 44 (29.3%), 58 (38.6%), and 35 (23.3%) of the isolates were strongly, moderately, and weakly biofilm producers, while the remaining 13 (8.6%) isolates were non-biofilm producers. Thirty-five weakly adherent isolates were considered as negative or non-biofilm producers. Isolates screened for biofilm formation by microtiter plate assay and CRA methods are shown in Figures 2 and 3.
Figure 2. Biofilm Producing Strain (Black Colonies) and Non-Biofilm-Producing Strain (Red Colonies) of E. coli Grown on Congo-red Medium.

Figure 3. Screening the Biofilm Producers by Microtiter Plate Method: High, Moderate and Non-Biofilm Producers Differentiated With Crystal Violet Staining in Microtiter Plates.
Among mazEF, relBE, ccdAB, hipBA and mqsRA positive isolates, 71.1%, 73.4%, 72.3%, 76.4% and 70.7% isolates showed biofilm formation by CRA method, respectively, and 66.1%, 68%, 70.1%, 70.8% and 65.9% of E. coli clinical isolates were positive by microtiter plate assay respectively; while 72.4%, 59.1%, 61.5%, 59.1%, 74.1% of mazEF, relBE, ccdAB, hipBA and mqsRA TA loci negative isolates were biofilm producers by CRA method and 75.9%, 68.2%, 46.2%, 61.4%, 77.8% by microtiter plate assay, respectively. These results showed no significant relationship between TA positive isolates and biofilm formation (P > 0.05) except between hipBA positive isolates and biofilm producing by CRA assay (P < 0.05), since biofilm formation among hipBA positive isolates is more frequent than those of its negative isolates. The differences in biofilm formation and presence of TA systems were analyzed by SPSS software and crosstabs test for independent samples. Chi square test was used to find the correlation between biofilm formation and TA systems.
5. Discussion
Early reports showed that TA systems did not play a role in biofilm formation. For example, a study by Lemos et al. (28) demonstrated that mutants lacking homologues of the mazF and relE genes in Streptococcus mutans had no effect on biofilm formation compared to parental strains (28). The first TA system linked to biofilm formation was mqsRA in E. coli (22). The importance of this TA system in biofilm formation was linked to mqsRA in motility, biofilm formation, and the autoinducer-2 quorum sensing system (29). These initial results of Kasari et al. (30) confirmed that the relationship between mqsRA gene, biofilm formation, and deletion of mqsRA caused a significant reduction in biofilm formation (30).
Further evidence was obtained by Tsilibaris et al. (31) regarding the role of TA systems in biofilm formation. They used a strain, named Δ5 that lacked five of the most-studied TA pairs: mazF/mazE, relE/relB, yoeB/yefM, yafQ/dinJ, and chpB. They reported that these five deletions had no impact on the stress response of the bacterial cells (31); however, Ren et al. (22) reported that the TA systems were important for biofilm formation based on their microarray results (22). According to their findings, upon deletion of these five TA systems, biofilm formation decreased after eight hours and increased after 24 hours in rich medium at 37°C. Therefore, their work provided additional evidence that TA pairs have important role in biofilm formation (32).
Kolodkin-Gal et al. (33) wanted to find out which of the TA systems plays a greater role in biofilm formation. They studied the effect TA systems on biofilm formation. Using the E. coli deletion mutants, they observed a significant decrease in biofilm formation in both the DmazEF and the DdinJ-yafQ mutants at the early 8 hours and after 24 hours , compared to their parental strain, however they indicated only a partial decrease in biofilm formation in the DyefM-yoeB, DchpBIK and DrelBE mutants (33).
In general, the current study demonstrated a high tendency among the clinical isolates of E. coli to form biofilm. Biofilm production in E. coli might promote the increased colonization and persistence responsible for the device-related infections. In addition, there was a significant association of biofilms with the hipBA system. The hipBA locus was related to persistence since once toxin HipA levels reached a threshold, persistence occurred. Persister cells arise mainly in biofilms and in stationary-phase cultures. It was demonstrated that deletion of the hipBA genes caused a 10 to 100-fold reduction in persister production under the stationary and biofilm cultures. This phenotype contributes to the tolerance of biofilm bacteria to antibiotics which is the cause of severe human infections. However the current study showed that there is no difference in the presence of other TA systems in the biofilm producer and non-biofilm producer isolates but reverse-transcripting studies are needed to define the amount of TA gene expression in all isolates. On the other hand, the current survey showed that TA systems were highly prevalent in clinical isolates of E. coli. Additionally studies determined that if TA systems are prevalent and functional in clinical isolates of the bacteria, they could be used as a novel antimicrobial strategy, since a small molecule capable of neutralizing effect of the antitoxin and free the toxin can kill the cell.
Acknowledgments
Authors would like to thank Ilam University of Medical Sciences, Clinical Microbiology Research Center to have financed this research, and Milad Hospital in Tehran and Emmam and Mostafa hospitals in Ilam for supplying the bacterial isolates.
Footnotes
Funding/Support:This research has been supported by the grant of Ilam University of Medical Sciences, Clinical Microbiology Research Center.
References
- 1.Van Melderen L. Toxin-antitoxin systems: why so many, what for? Curr Opin Microbiol. 2010;13(6):781–5. doi: 10.1016/j.mib.2010.10.006. [DOI] [PubMed] [Google Scholar]
- 2.Kolodkin-Gal I, Engelberg-Kulka H. Induction of Escherichia coli chromosomal mazEF by stressful conditions causes an irreversible loss of viability. J Bacteriol. 2006;188(9):3420–3. doi: 10.1128/JB.188.9.3420-3423.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Erental A, Sharon I, Engelberg-Kulka H. Two programmed cell death systems in Escherichia coli: an apoptotic-like death is inhibited by the mazEF-mediated death pathway. PLoS Biol. 2012;10(3) doi: 10.1371/journal.pbio.1001281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Engelberg-Kulka H, Sat B, Reches M, Amitai S, Hazan R. Bacterial programmed cell death systems as targets for antibiotics. Trends Microbiol. 2004;12(2):66–71. doi: 10.1016/j.tim.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 5.Amitai S, Kolodkin-Gal I, Hananya-Meltabashi M, Sacher A, Engelberg-Kulka H. Escherichia coli MazF leads to the simultaneous selective synthesis of both "death proteins" and "survival proteins". PLoS Genet. 2009;5(3) doi: 10.1371/journal.pgen.1000390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schumacher MA, Piro KM, Xu W, Hansen S, Lewis K, Brennan RG. Molecular mechanisms of HipA-mediated multidrug tolerance and its neutralization by HipB. Science. 2009;323(5912):396–401. doi: 10.1126/science.1163806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Masuda Y, Ohtsubo E. Mapping and disruption of the chpB locus in Escherichia coli. J Bacteriol. 1994;176(18):5861–3. doi: 10.1128/jb.176.18.5861-5863.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Overgaard M, Borch J, Gerdes K. RelB and RelE of Escherichia coli form a tight complex that represses transcription via the ribbon-helix-helix motif in RelB. J Mol Biol. 2009;394(2):183–96. doi: 10.1016/j.jmb.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grady R, Hayes F. Axe-Txe, a broad-spectrum proteic toxin-antitoxin system specified by a multidrug-resistant, clinical isolate of Enterococcus faecium. Mol Microbiol. 2003;47(5):1419–32. doi: 10.1046/j.1365-2958.2003.03387.x. [DOI] [PubMed] [Google Scholar]
- 10.Christensen SK, Maenhaut-Michel G, Mine N, Gottesman S, Gerdes K, Van Melderen L. Overproduction of the Lon protease triggers inhibition of translation in Escherichia coli: involvement of the yefM-yoeB toxin-antitoxin system. Mol Microbiol. 2004;51(6):1705–17. doi: 10.1046/j.1365-2958.2003.03941.x. [DOI] [PubMed] [Google Scholar]
- 11.Wang X, Kim Y, Hong SH, Ma Q, Brown BL, Pu M, et al. Antitoxin MqsA helps mediate the bacterial general stress response. Nat Chem Biol. 2011;7(6):359–66. doi: 10.1038/nchembio.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Williams JJ, Hergenrother PJ. Exposing plasmids as the Achilles' heel of drug-resistant bacteria. Curr Opin Chem Biol. 2008;12(4):389–99. doi: 10.1016/j.cbpa.2008.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Williams JJ, Halvorsen EM, Dwyer EM, DiFazio RM, Hergenrother PJ. Toxin-antitoxin (TA) systems are prevalent and transcribed in clinical isolates of Pseudomonas aeruginosa and methicillin-resistant Staphylococcus aureus. FEMS Microbiol Lett. 2011;322(1):41–50. doi: 10.1111/j.1574-6968.2011.02330.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Christensen SK, Mikkelsen M, Pedersen K, Gerdes K. RelE, a global inhibitor of translation, is activated during nutritional stress. Proc Natl Acad Sci U S A. 2001;98(25):14328–33. doi: 10.1073/pnas.251327898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pecota DC, Wood TK. Exclusion of T4 phage by the hok/sok killer locus from plasmid R1. J Bacteriol. 1996;178(7):2044–50. doi: 10.1128/jb.178.7.2044-2050.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Magnuson RD. Hypothetical functions of toxin-antitoxin systems. J Bacteriol. 2007;189(17):6089–92. doi: 10.1128/JB.00958-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Engelberg-Kulka H, Amitai S, Kolodkin-Gal I, Hazan R. Bacterial programmed cell death and multicellular behavior in bacteria. PLoS Genet. 2006;2(10) doi: 10.1371/journal.pgen.0020135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harrison JJ, Ceri H, Turner RJ. Multimetal resistance and tolerance in microbial biofilms. Nat Rev Microbiol. 2007;5(12):928–38. doi: 10.1038/nrmicro1774. [DOI] [PubMed] [Google Scholar]
- 19.Stewart PS. Mechanisms of antibiotic resistance in bacterial biofilms. Int J Med Microbiol. 2002;292(2):107–13. doi: 10.1078/1438-4221-00196. [DOI] [PubMed] [Google Scholar]
- 20.Keren I, Shah D, Spoering A, Kaldalu N, Lewis K. Specialized persister cells and the mechanism of multidrug tolerance in Escherichia coli. J Bacteriol. 2004;186(24):8172–80. doi: 10.1128/JB.186.24.8172-8180.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim Y, Wood TK. Toxins Hha and CspD and small RNA regulator Hfq are involved in persister cell formation through MqsR in Escherichia coli. Biochem Biophys Res Commun. 2010;391(1):209–13. doi: 10.1016/j.bbrc.2009.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ren D, Bedzyk LA, Thomas SM, Ye RW, Wood TK. Gene expression in Escherichia coli biofilms. Appl Microbiol Biotechnol. 2004;64(4):515–24. doi: 10.1007/s00253-003-1517-y. [DOI] [PubMed] [Google Scholar]
- 23.Queipo-Ortuno MI, De Dios Colmenero J, Macias M, Bravo MJ, Morata P. Preparation of bacterial DNA template by boiling and effect of immunoglobulin G as an inhibitor in real-time PCR for serum samples from patients with brucellosis. Clin Vaccine Immunol. 2008;15(2):293–6. doi: 10.1128/CVI.00270-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Subramanian P, Shanmugam N, Sivaraman U, Kumar S, Selvaraj S. Antiobiotic resistance pattern of biofilm-forming uropathogens isolated from catheterised patients in Pondicherry, India. Australas Med J. 2012;5(7):344–8. doi: 10.4066/AMJ.2012.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O'Toole GA, Kolter R. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol Microbiol. 1998;28(3):449–61. doi: 10.1046/j.1365-2958.1998.00797.x. [DOI] [PubMed] [Google Scholar]
- 26.Vieira HL, Freire P, Arraiano CM. Effect of Escherichia coli morphogene bolA on biofilms. Appl Environ Microbiol. 2004;70(9):5682–4. doi: 10.1128/AEM.70.9.5682-5684.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stepanovic S, Cirkovic I, Ranin L, Svabic-Vlahovic M. Biofilm formation by Salmonella spp. and Listeria monocytogenes on plastic surface. Lett Appl Microbiol. 2004;38(5):428–32. doi: 10.1111/j.1472-765X.2004.01513.x. [DOI] [PubMed] [Google Scholar]
- 28.Lemos JA, Brown TA, Jr., Abranches J, Burne RA. Characteristics of Streptococcus mutans strains lacking the MazEF and RelBE toxin-antitoxin modules. FEMS Microbiol Lett. 2005;253(2):251–7. doi: 10.1016/j.femsle.2005.09.045. [DOI] [PubMed] [Google Scholar]
- 29.Gonzalez Barrios AF, Zuo R, Hashimoto Y, Yang L, Bentley WE, Wood TK. Autoinducer 2 controls biofilm formation in Escherichia coli through a novel motility quorum-sensing regulator (MqsR, B3022). J Bacteriol. 2006;188(1):305–16. doi: 10.1128/JB.188.1.305-316.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kasari V, Kurg K, Margus T, Tenson T, Kaldalu N. The Escherichia coli mqsR and ygiT genes encode a new toxin-antitoxin pair. J Bacteriol. 2010;192(11):2908–19. doi: 10.1128/JB.01266-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsilibaris V, Maenhaut-Michel G, Mine N, Van Melderen L. What is the benefit to Escherichia coli of having multiple toxin-antitoxin systems in its genome? J Bacteriol. 2007;189(17):6101–8. doi: 10.1128/JB.00527-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim Y, Wang X, Ma Q, Zhang XS, Wood TK. Toxin-antitoxin systems in Escherichia coli influence biofilm formation through YjgK (TabA) and fimbriae. J Bacteriol. 2009;191(4):1258–67. doi: 10.1128/JB.01465-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kolodkin-Gal I, Verdiger R, Shlosberg-Fedida A, Engelberg-Kulka H. A differential effect of E. coli toxin-antitoxin systems on cell death in liquid media and biofilm formation. PLoS One. 2009;4(8) doi: 10.1371/journal.pone.0006785. [DOI] [PMC free article] [PubMed] [Google Scholar]


