Abstract
Expression of HOX transcript antisense intergenic RNA (HOTAIR), a large intergenic noncoding RNA (lincRNA), has been described as a metastases-associated lincRNA in various cancers including breast, liver and colon cancer cancers. We sought to determine if expression of HOTAIR could be used as a surrogate for assessing nodal metastases and evaluated RNA in situ hybridization (RNA-ISH) assay in a tissue microarray constructed from 133 breast cancer patients. The prognostic value of HOTAIR was further validated in large cohorts using The Cancer Genome Atlas (TCGA) breast cancer subjects. RNA-ISH analysis was successful in 94 cases (17% cases scored 0, 32.9% scored 1, 30.8% scored 2, and 19.1% scored 3). The expression of HOTAIR did not correlate with nodal metastasis regardless of the scoring intensity or with other study parameters (age, tumor size and grade, expression status). Further analysis of TCGA dataset showed that HOTAIR expression was lower in ductal carcinomas but higher in ER-negative tumors. Overexpression of HOTAIR was not associated with nodal metastases or prognosis in ER-positive patients. Its function as a poor prognostic indicator in ER-negative patients was restricted to node-positive patients. HOTAIR appears to be a marker for lymphatic metastases rather than hematogenous metastases in ER-negative patients.
Recent studies have shown that the idea of DNA transcription resulting in synthesis of corresponding protein is rather simplistic1. A number of additional factors are involved in the control of the transcription process. RNAs do not appear to be simple messengers but are complex species with many distinct functional subtypes. Next generation sequencing studies have estimated that 98% of the DNA is noncoding; these RNAs derived from noncoding regions have a variety of functions that include gene regulation2,3. Among the different classes of noncoding (ncRNAs), microRNAs have been the most extensively investigated4. In addition to microRNAs, long noncoding RNAs (lncRNAs) constitute another species of RNAs. LncRNAs are defined as transcribed RNA molecules that are longer than 200 nucleotides and have no obvious protein coding capacity5. They are more tissue specific than protein coding genes6. The abundance of lncRNAs in the genome, their association with the myriad of different disease phenotypes, and their diverse cellular roles are drawing the attention of the scientific community7,8,9,10,11,12,13,14,15. However, knowledge regarding the actual number of functional lncRNAs and the mechanism(s) by which they carry out their functions is still limited. Recent advances in RNA sequencing (RNA-seq) and computational methods have revealed that large intergenic noncoding RNAs (lincRNAs) are the largest class of lncRNA molecules in the human genome5. At least 8000 human lincRNAs have been identified5. Biological characterization studies suggest that many lincRNAs act as scaffolds that regulate molecular (protein, RNA, and DNA) interactions required for various signaling networks. It has been estimated that at least 30% of lncRNAs are bound to the polycomb repressive complexes and target these chromatin-modifying complexes to the target genes16.
Gupta et al hybridized RNA from normal breast epithelia, primary tumors, and distant metastases to ultrahigh density HOX tiling array17. They found 170 ncRNAs and 63 HOX exons that were differentially expressed. HOTAIR (HOX antisense intergenic RNA) was one of the metastases associated lincRNAs. Rinn et al and Woo and Kingston et al had previously identified its role in regulating HOX genes18,19. HOTAIR is a large noncoding RNA which is 2158-nucleotides long, and expressed from the HOXC locus on chromosome 12q13.1318. It contains 5′ and 3′ domains. The 5′ domain binds to PRC2, while the 3′ domain interacts with the LSD1/Co-REST/REST complex to coordinately regulate histone H3 lysine 27 methylation and lysine 4 demethylation, and epigenetically modify chromatin structure thereby regulating gene expression20. Gupta et al further showed by qPCR that HOTAIR is overexpressed 100–2000 fold in breast cancer metastases17. Its expression was sometimes high but heterogeneous in primary tumors. The expression was a significant predictor of metastases and death in a series of 132 breast cancer patients with extensive followup.
Since the original publication, HOTAIR has been shown to be a poor prognostic factor in a number of cancers including breast, lung, colon, liver and gastrointestinal stromal tumors17,21,22,23,24. Its expression has been linked to increased cancer cell invasiveness and epithelial-to-mesenchymal transformation25. Gain-of-function studies show that the activities of hundreds of genes (such as the HOXD locus, progesterone receptor, cell adhesion molecules) are inhibited, while dozens of other genes (such as ABL2, SNAIL, and LAMB3) are activated17. An in vitro functional study showed that HOTAIR overexpression in four different breast cancer cell lines could promote colony growth and invasion17. Animal experiments demonstrated that the breast cancer cell line, MDA-MB-231, which expressed HOTAIR, grew quickly in primary tumor foci with increased metastatic potential to the lung17.
Given the purported importance of HOTAIR in breast cancer, we sought to address the question of whether the expression of HOTAIR could be used as a surrogate for assessing nodal metastases. We additionally analyzed the impact of high levels of HOTAIR on nodal metastases and overall survival in The Cancer Genome Atlas (TCGA) dataset.
Results
In situ HOTAIR Expression in Breast Cancer TMA Cohort
The expression of HOTAIR was analyzed in 94 of the 133 cases by RNA-ISH based on the eligibility criteria. These criteria included the presence of at least 100 tumor cell nuclei. Thirty-nine cases were excluded due to lack of adequate tumor cells, folds in tissue sections, or loss of tissue during processing.
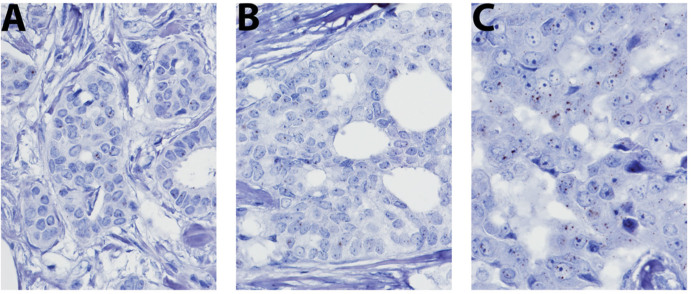
Consistent with prior descriptions, the signals were scattered in the cell and were not restricted to the nucleus26. Similarly, very few, if any, signals were identified in the nontumor stromal cells—highlighting the tissue-specific distribution of this lincRNA. The signals were absent or rare (less than 1 per 100 tumor nuclei) in 16 cases (17%); one to 10 signals per 100 nuclei in 31 cases (32.9%), 10 to 100 signals in 29 (30.8%), and innumerable in 18 cases (19.1%). Representative images of HOTAIR microRNA signals are shown in Figure 1A–C for each scoring category. The expression of HOTAIR did not correlate with nodal metastasis regardless of the scoring intensity used as a cutoff point (Table 1). It also did not correlate with other parameters such as age (categorical 50 versus > 50), tumor size and grade, and ER and HER2 status.
Figure 1. Expression of HOTAIR in a series of breast cancer samples.
Expression levels were depicted as (A) 1–10 signals, (B) 10–100 signals, and (C) multiple signals per cell.
Table 1. Clinicopathological characteristics of the 94 cases that were analyzable for the expression of HOTAIR.
| Negative (0 or 1) | Positive (2–3) | P value | |
|---|---|---|---|
| Premenopausal | 16 | 15 | 1 |
| Postmenopausal | 31 | 32 | |
| Tumor size ≤ 2 cm | 27 | 34 | 0.194 |
| Tumor size > 2 cm | 20 | 13 | |
| Grade 1 | 12 | 14 | 0.902 |
| Grade 2 | 19 | 17 | |
| Grade 3 | 16 | 16 | |
| IDC | 37 | 41 | 0.557 |
| ILC | 5 | 3 | |
| Others | 5 | 3 | |
| Node-negative | 28 | 34 | 0.276 |
| Node-positive | 19 | 13 | |
| ER-negative | 10 | 13 | 0.632 |
| ER-positive | 37 | 34 | |
| HER2-negative | 40 | 7 | 1 |
| HER2-positive | 39 | 8 |
IDC, invasive ductal carcinoma; ILC, invasive lobular carcinoma.
Confirmation of the Prognostic Value of HOTAIR in Breast Cancer Using TCGA Breast Invasive Carcinoma Dataset
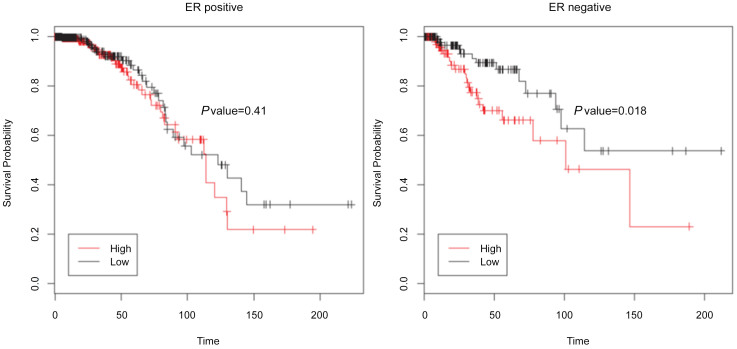
To assess the prognostic value of HOTAIR expression in breast tumors, we next analyzed its correlation with overall survival using TCGA data of breast cancer subjects. The expression of HOTAIR in these subjects was categorized based on the low (n = 476; black line) and high (n = 476; red line) expressions using the median expression as a cutoff (Figure 2A–B). The higher HOTAIR expression was associated with shorter overall survival in ER-negative breast cancer patients (P = 0.018) (Figure 2B). In the cohort with high HOTAIR levels, the overall survival probability was 60% and 46.4% at 50 and 100 months, respectively, and those with low HOTAIR levels showed a survival probability of 86% and 62.8% at 50 and 100 months, respectively. On the other hand, HOTAIR levels were not associated with overall survival in ER-positive breast cancer (P = 0.41) (Figure 2A).
Figure 2. Impact of HOTAIR expression on survival in The Cancer Genome Atlas dataset (n = 952).
KM plots show that HOTAIR expression above median is associated with poor outcome in ER-negative patients (A) but not in ER-positive patients (B).
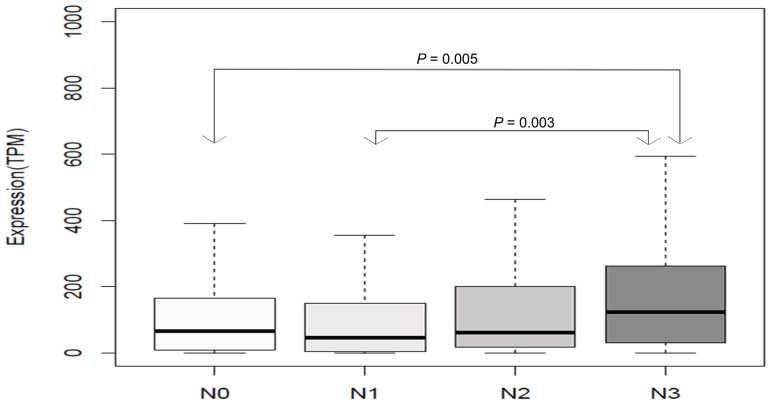
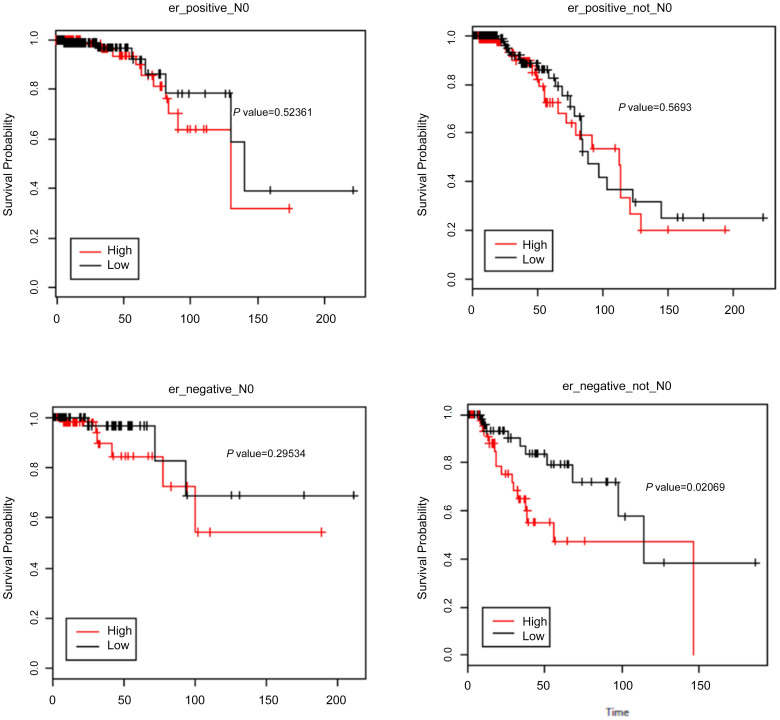
We further assessed the correlation of HOTAIR expression with overall survival and lymph node status using TCGA dataset. High expression of HOTAIR was not associated with a greater likelihood of nodal metastases. Further subset analysis of the nodal involvement showed significance only in extensive node positivity (N3) when compared with N0 (P = 0.0049) and N1 (P = 0.00035) (Figure 3). In TCGA dataset, HOTAIR was not associated with prognosis in ER-positive patients (node positive and negative). High levels were found to be associated with worse prognosis in ER-negative/ node–positive patients (P = 0.02) but not in node-negative patients (P = 0.2) (Figure 4A–D).
Figure 3. Correlation of HOTAIR expression with nodal status using The Cancer Genome Atlas dataset.
Comparison of the cases by nodal status revealed only significant association between N3 versus N0 (P = 0.005) or N1 (P = 0.003). Other comparisons were not significant.
Figure 4. Kaplan–Meier overall survival in regards to HOTAIR expression and nodal status.
Expression above median is not associated with poor outcome in ER-positive (regardless of lymph node status) (A & B). In ER-negative cases (C & D), high HOTAIR was only associated with poor outcome in lymph node–positive tumors.
Discussion
Metastasis is the leading cause of breast cancer mortality. The prevention and treatment of metastasis, however, remains a significant clinical challenge. The spread of breast tumors to local and regional lymph nodes is an important means of tumor dissemination. The presence and the number of involved lymph nodes remains the single best indicator of whether or not the cancer has become widely metastatic. Identification of the underlying molecular mechanisms of lymph node metastasis and a better understanding how to modulate these will be a significant step in the goal of prevention of metastases.
Recent studies have shown that lincRNA is a novel class of molecules that regulates cancer progression and metastasis27. LincRNAs can serve as scaffolds to control chromatin states and epigenetic changes17,20. LincRNA HOTAIR was shown to regulate metastatic progression by reprogramming the chromatin state17,20. The high expression of HOTAIR was a significant predictor with poor prognosis and metastasis in breast carcinomas17. Further analysis performed by the same group showed that HOTAIR was also increased in the metastatic carcinomas when compared in matched primary and metastatic cancers26. A separate study found HOTAIR expression to be an important independent indicator for predicting metastasis, especially in ER-positive breast cancer patients28.
The current study investigated the role of HOTAIR expression in relation to nodal metastases in a breast cancer TMA cohort. Using RNA-ISH, in situ overexpression of HOTAIR was not associated with nodal metastases. We also did not observe any association with age, tumor grade, ER, and HER2. The difference in observed results could be due to a number of parameters including assay methods and size of cohort including the number of ER-positive cases. In the original study, Gupta et al identified increased HOTAIR expression in primary and metastatic breast cancers using RT-PCR based methods17. The same group (Chisholm et al) developed an in situ hybridization assay using well-established cell line controls26. In this followup study, they observed a trend of higher HOTAIR expression in the metastatic than in the primary breast cancers. Importantly, they could not confirm the survival data reported in the original study. However, based on the analysis of only 6 patients of the original 243 cases, they reported a trend for tumors overexpressing EZH2 and HOTAIR to have a poor prognosis. Sorensen et al analyzed the expression of HOTAIR in a case control design study using an Agilent-based microarray platform28. They found high HOTAIR expression in primary tumors from patients who developed metastases as opposed to patients who did not. The association was significantly in patients with ER-positive tumors, but not in ER-negative tumors28. Lu et al analyzed the expression of HOTAIR in a series of 348 patients using quantitative RT-PCR22. They did not find a significant association of HOTAIR expression with prognosis in univariate analysis. These studies together indicate that the differences observed cannot be entirely explained by assay method variability.
The size of the cohort and the distribution of cases based on molecular classification and nodal positivity could explain some of the observed differences. To circumvent these issues, we analyzed the expression of HOTAIR in TCGA dataset (n = 952) and correlated it with nodal metastases and overall survival. Consistent with our in situ hybridization data, high HOTAIR expression in the entire dataset was not associated with nodal involvement (P = 0.33). High HOTAIR was associated with worse outcome in patients with ER-negative breast tumors (P = 0.018), but not in ER-positive patients (P = 0.41). This observation is in contrast to the results reported by Sorensen et al but are similar to those reported by Lu et al28,29. Some of the differences could be explained by the size of the cohorts, relative proportions of ER-positive and ER-negative patients, and the endpoints used. The endpoint in the current study is overall survival while many of the studies used distant metastases-free survival.
Association of high HOTAIR expression was observed with poor overall survival in ER-negative tumors; interestingly, this was observed only with patients having nodal metastases. It has been well documented that nodal involvement in triple-negative tumors is less prognostic in ER-negative tumors as opposed to ER-positive tumors. Several studies have reported that triple-negative cancers have less nodal metastasis, although the tumors are categorized as high grade30,31,32,33.
It is thus possible that the high expression of HOTAIR in ER-negative tumors could be an indicator of activation of pathways associated with lymphatic metastases rather than vascular metastases.
A strong association was seen between HOTAIR expression and histology. Ductal carcinomas (not otherwise specified) were less likely to have high levels of HOTAIR compared to special histological types such as lobular carcinomas (P < 0.00001). This finding is somewhat in variance with the high expression seen in ER-negative tumors.
In summary, our study shows that the prognostic role of HOTAIR expression is more or less restricted to ER-negative, lymph node–positive tumors, where its expression could possibly be used as a potential prognostic marker identifying patients at greater risk for poor overall survival. It does not seem to be useful in prognostication of ER-positive breast cancer or identification of patients likely to have nodal metastases. Given the well-recognized followup limitations of TCGA cohort, further studies are necessary to clarify the role of HOTAIR in metastases in cohorts well annotated for tumor histology, nodal status, and survival information.
Methods
Patient Cohort and Tissue Microarray (TMA)
Appropriate Institutional Review Board (IRB) approval from the Indiana University Research Ethics Committee was obtained. The methods were carried out in "accordance" with the approved IRB guidelines; informed consent was obtained from all subjects. A TMA was constructed from 133 consecutive patients with breast cancer. The procedure involved extracting 1 mm punches from tumors and implanting them in a new paraffin block. The array information was recorded in an Excel sheet. The cohort consists of 133 women with diagnosed with invasive carcinoma in the same year. The TMA was constructed using duplicate 1 mm cores from tumors. Clinical data with respect to age, tumor size, and ER and HER2 expression status were collected from the clinical charts. The ER and HER2 analyses were performed in a CLIA-certified lab using cutoffs recommended by the current ASCO-CAP guidelines34,35. All patients had undergone some form of nodal assessment (either sentinel node biopsy or axillary nodal sampling); none had received neoadjuvant chemotherapy.
RNA In Situ Hybridization Assay (RNA ISH)
In situ detection for HOTAIR was performed using the RNAscope (Brown) FFPE kit (Advanced Cell Diagnostics, Hayward, CA) according to the manufacturer's instructions. Briefly, TMAs were cut in 4 μm thick sections. The tissue was baked for 1 hour at 60°C. The samples were then placed in Cytosol and brought down to water. Solution 1 was applied for 15 minutes at room temperature. The tissue sections were boiled at 95°C for 15 minutes in solution 2. Solution 3 (protease treatment) was then applied at 40°C. The provided probe and probe solution were applied. The slides were covered with agarose gel and placed in a rack at 40°C for hybridization for 2 hours. The tissue samples were then washed in the provided washing buffer. The 3,3′-diaminobenzidine solutions A and B were mixed in equal volume and left on for 10 minutes at room temperature. Hematoxylin counterstaining was performed.
A provided positive control probe ubiquitin C was used. For negative control, the enclosed negative control probe was applied. In addition, samples where the probe was omitted were included to exclude background staining.
The slides were independently evaluated by two separate observers (ITV and SB). Positive staining was indicated by signals as brown punctate dots present in the nucleus and/or cytoplasm as described in prior studies26. The number of signal staining was counted in 100 tumor cells. The study was performed on tissue microarrays (TMAs) which had 1 mm cores. This relatively small tissue core in many ways circumvented issues related to hotspots and tissue heterogeneity. The number 100 was chosen to ensure good representation of the tumor without loss of “huge” number of cases. The continuous number was categorized into four categories for statistical analysis. These categories were 0 = less than 1 signal per 100 cells; 1 = 1–10 signals; 2 = 11–100 signals, and 3 = > 101 signals in 100 cells.
The expression of the HOTAIR lincRNA was primarily correlated with nodal status. The secondary endpoints included correlations with other clinicopathological parameters such as age, tumor size, grade, and ER, PR, and HER2 expression status.
Analysis of The Cancer Genome Atlas (TCGA)
To validate the clinical relevance of HOTAIR levels in a larger cohort of breast cancer, we obtained the normalized levels of HOTAIR expression (Level 3 data) in 952 breast cancer patients enrolled in TCGA database breast invasive carcinoma study (available at https://tcga-data.nci.nih.gov/tcga/tcgaHome2.jsp). Patients with breast cancer were categorized based on the ER status (n = 924; 656 ER positive), HER2 (n = 336; 145 HER2 positive), and lymph node status. The clinical information for each patient was also obtained. To model survival, gene expression at or below median was considered low and above median was considered high. Overall survival was calculated from the date of initial diagnosis of breast cancer to disease-specific deaths (patients whose vital status is termed dead) and months to last followup (patients who are alive). Kaplan–Meier survival analysis was used to estimate association of HOTAIR expression with survival of patients and with ER, HER2, and nodal status. The “survival” package in R (R Foundation for Statistical Computing) was used for statistical analyses.
Statistical Analysis
Data were analyzed using Statistical Package for Social Sciences v.17.0 (SPSS Institute, Chicago, IL, USA) software to determine the correlations between HOTAIR and multiple clinicopathological parameters. Fisher exact test and chi-squared test were performed. All P values were two-sided, and 0.05 was taken as the significance level. Survival analyses by Kaplan–Meier method were performed for both epithelial and stromal cells as appropriate. The differences in survival were tested using the Log rank test.
Author Contributions
Y.G.-P. and S.B. designed, interpreted the analyses, and wrote the main manuscript. I.T.V. and S.B. evaluated the RNA-ISH TMA cohort data, Y.N. and S.C.J. did the TCGA analysis. All authors reviewed the manuscript.
Acknowledgments
Sunil Badve is supported by the Susan G. Komen for the Cure Scholar Award and 100 Voices of Hope.
References
- Shen X., Qi P. & Du X. Long non-coding RNAs in cancer invasion and metastasis. Mod Pathol 28, 4–13 (2015). [DOI] [PubMed] [Google Scholar]
- Harrow J. et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res 22, 1760–1774 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium E. P. et al. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentwich I. et al. Identification of hundreds of conserved and nonconserved human microRNAs. Nat Genet 37, 766–770 (2005). [DOI] [PubMed] [Google Scholar]
- Cabili M. N. et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev 25, 1915–1927 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien T. et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res 22, 1775–1789 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volders P. J. et al. LNCipedia: a database for annotated human lncRNA transcript sequences and structures. Nucleic Acids Res 41, D246–251 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carninci P. et al. The transcriptional landscape of the mammalian genome. Science 309, 1559–1563 (2005). [DOI] [PubMed] [Google Scholar]
- Wapinski O. & Chang H. Y. Long noncoding RNAs and human disease. Trends Cell Biol 21, 354–361 (2011). [DOI] [PubMed] [Google Scholar]
- Gutschner T. & Diederichs S. The hallmarks of cancer: a long non-coding RNA point of view. RNA Biol 9, 703–719 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunner A. L. et al. Transcriptional profiling of lncRNAs and novel transcribed regions across a diverse panel of archived human cancers. Genome Biol 13, R75 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qureshi I. A., Mattick J. S. & Mehler M. F. Long non-coding RNAs in nervous system function and disease. Brain Res 1338, 20–35 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibb E. A. et al. Human cancer long non-coding RNA transcriptomes. PLoS One 6, e25915 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer T. R., Dinger M. E. & Mattick J. S. Long non-coding RNAs: insights into functions. Nat Rev Genet 10, 155–159 (2009). [DOI] [PubMed] [Google Scholar]
- Amaral P. P., Clark M. B., Gascoigne D. K., Dinger M. E. & Mattick J. S. lncRNAdb: a reference database for long noncoding RNAs. Nucleic Acids Res 39, D146–151 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil A. M. et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A 106, 11667–11672 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R. A. et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 464, 1071–1076 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn J. L. et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129, 1311–1323 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo C. J. & Kingston R. E. HOTAIR lifts noncoding RNAs to new levels. Cell 129, 1257–1259 (2007). [DOI] [PubMed] [Google Scholar]
- Tsai M. C. et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science 329, 689–693 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhan A. et al. Bisphenol-A and diethylstilbestrol exposure induces the expression of breast cancer associated long noncoding RNA HOTAIR in vitro and in vivo. J Steroid Biochem Mol Biol 141, 160–170 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L. et al. Association of large noncoding RNA HOTAIR expression and its downstream intergenic CpG island methylation with survival in breast cancer. Breast Cancer Res Treat 136, 875–883 (2012). [DOI] [PubMed] [Google Scholar]
- Nakagawa T. et al. Large noncoding RNA HOTAIR enhances aggressive biological behavior and is associated with short disease-free survival in human non-small cell lung cancer. Biochem Biophys Res Commun 436, 319–324 (2013). [DOI] [PubMed] [Google Scholar]
- Zhuang Y. et al. Induction of long intergenic non-coding RNA HOTAIR in lung cancer cells by type I collagen. J Hematol Oncol 6, 35 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padua Alves C. et al. Brief report: The lincRNA Hotair is required for epithelial-to-mesenchymal transition and stemness maintenance of cancer cell lines. Stem Cells 31, 2827–2832 (2013). [DOI] [PubMed] [Google Scholar]
- Chisholm K. M. et al. Detection of Long Non-Coding RNA in Archival Tissue: Correlation with Polycomb Protein Expression in Primary and Metastatic Breast Carcinoma. PLoS ONE 7, e47998 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai M. C., Spitale R. C. & Chang H. Y. Long intergenic noncoding RNAs: new links in cancer progression. Cancer Res 71, 3–7 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorensen K. P. et al. Long non-coding RNA HOTAIR is an independent prognostic marker of metastasis in estrogen receptor-positive primary breast cancer. Breast Cancer Res Treat 142, 529–536 (2013). [DOI] [PubMed] [Google Scholar]
- Liu X. H. et al. Lnc RNA HOTAIR functions as a competing endogenous RNA to regulate HER2 expression by sponging miR-331-3p in gastric cancer. Mol Cancer 13, 92 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitale A., Mazzola P., Soldini D., Mazzucchelli L. & Bordoni A. Breast cancer classification according to immunohistochemical markers: clinicopathologic features and short-term survival analysis in a population-based study from the South of Switzerland. Ann Oncol 20, 628–635 (2009). [DOI] [PubMed] [Google Scholar]
- Carey L. A. et al. Race, breast cancer subtypes, and survival in the Carolina Breast Cancer Study. JAMA 295, 2492–2502 (2006). [DOI] [PubMed] [Google Scholar]
- Da Silva L., Clarke C. & Lakhani S. R. Demystifying basal-like breast carcinomas. J Clin Pathol 60, 1328–1332 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakha E. A. et al. Prognostic markers in triple-negative breast cancer. Cancer 109, 25–32 (2007). [DOI] [PubMed] [Google Scholar]
- Hammond M. E. et al. American Society of Clinical Oncology/College Of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer. J Clin Oncol 28, 2784–2795 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff A. C. et al. Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. J Clin Oncol 31, 3997–4013 (2013). [DOI] [PubMed] [Google Scholar]