Abstract
We demonstrate that interferon (IFN)-β-1b induces an alternative-start transcript containing the C-terminal TLDc domain of nuclear receptor coactivator protein 7 (NCOA7), a member of the OXR family of oxidation resistance proteins. IFN-β-1b induces NCOA7-AS (alternative start) expression in peripheral blood mononuclear cells (PBMCs) obtained from healthy individuals and multiple sclerosis patients and human fetal brain cells, astrocytoma, neuroblastoma, and fibrosarcoma cells. NCOA7-AS is a previously undocumented IFN-β-inducible gene that contains only the last 5 exons of full-length NCOA7 plus a unique first exon (exon 10a) that is not found in longer forms of NCOA7. This exon encodes a domain closely related to an important class of bacterial aldo-keto oxido-reductase proteins that play a critical role in regulating redox activity. We demonstrate that NCOA7-AS is induced by IFN and LPS, but not by oxidative stress and exhibits, independently, oxidation resistance activity. We further demonstrate that induction of NCOA7-AS by IFN is dependent on IFN-receptor activation, the Janus kinase-signal transducers and activators of transcription (JAK-STAT) signaling pathway, and a canonical IFN-stimulated response element regulatory sequence upstream of exon 10a. We describe a new role for IFN-βs involving a mechanism of action that leads to an increase in resistance to inflammation-mediated oxidative stress.
Introduction
Type I interferons (IFN) play a major role in organizing antiviral responses, including innate and adaptive immune functions involving host-pathogen responses and cell growth (Mogensen and others 1999). IFNs are defined structurally around a tightly compact helical protein structure that interacts with a heteromeric cell surface receptor present on all mammalian cells. Evolutionary pressures have resulted in the appearance of a family of IFNs including IFN-α and IFN-β, whose expression is restricted to particular cell types (Stark and others 1998). Gene activation by IFNs requires their interaction with 2 cell surface receptor chains, IFN-alpha receptor 1 (IFNAR1) and IFN-alpha receptor 2 (IFNAR2), which become phosphorylated, upon IFN binding, on specific tyrosine amino acid residues located in the cytoplasmic domain of each receptor chain (Wagner and others 2002). Phosphorylation and subsequent gene activation are mediated by the Janus kinase (JAK) and signal transducers and activators of transcription (STAT) pathway (Stark and Darnell 2012).
IFN-stimulated response elements (ISRE) have been identified in the promoter region of a large number of genes. ISRE activation, leading to gene expression, is dependent on components of the JAK-STAT signaling pathway (Stark and Darnell 2012). IFNs appear to have coevolved over 450 million years with the ever-increasing complexity associated with innate responses to viral infection, adaptive immune responses, and cell proliferation (Pestka and others 1987). This broad biological response may explain why IFNs induce such a large number (>250) of ISRE-dependent genes (Croze 2010). Continued documentation of IFN-induced genes is warranted to further unravel the mechanism of action of IFN in support of its use in treating human disease (eg, multiple sclerosis [MS]) and its evolution as an essential protein required for mediating innate and adaptive immune responses.
The mode of action of IFN in MS includes shifting of the Th1/Th2 inflammatory response, stabilizing the blood–brain barrier, and influencing oligodendrocyte and microglia cell differentiation (Arnason and others 1996). To further explore the mode of action of IFN-βs with a focus on exon usage, we used multi-exon microarrays to examine gene induction following IFN treatment. The genome-wide picture of genetic changes has been published elsewhere (Croze and others 2013). In this study we report for the first time, IFN-β-1b induced expression of a unique variant of the oxidation resistance gene nuclear receptor coactivator protein 7 (NCOA7), a member of the OXR family of oxidation resistance genes (Durand and others 2007). The full-length form of NCOA7 functions as a nuclear coactivator that is recruited to the nucleus when estrogen binds its nuclear receptor. The NCOA7-receptor-estrogen complex then binds to promoters of estrogen-regulated genes, causing their induction. Since estrogen causes oxidative DNA damage (Liehr and others 1986; Rajapakse and others 2005; Roy and others 2007; Perillo and others 2008; Fussell and others 2011), the antioxidant activity of NCOA7 is predicted to limit this damage (Durand and others 2007). The estrogen-binding domain of full-length NCOA7 lies in exon 7 (Shao and others 2002). We show that this exon is not present in NCOA7-AS, and suggest that the AS form of NCOA7 has a function that is distinct from the proposed role of the full-length form.
Oxidative stress has been strongly implicated as a contributor to neurodegeneration in several diseases such as Alzheimer's, Parkinson's, amyotrophic lateral sclerosis (ALS), and MS (Butterfield 2006; Melo and others 2011; Butterfield and others 2012). Association of oxidative free radicals with neuronal damage occurs in the central nervous system (CNS) during neurological diseases (Gonsette 2008; Kaur and Ling 2008). A recent biological activity associated with IFNs is the ability to reduce tissue damage caused by oxidative stress (Croze and others 2009). Tissue damage caused by inflammation could partly result from an increase in local reactive oxygen species. The observation that IFN-β-1b induces an antioxidant response is therefore particularly interesting. In this study, in addition to demonstrating that NCOA7-AS is a newly discovered IFN-β-inducible gene, we determined its regulatory mechanism, its molecular structure, and demonstrate the ability of NCOA7-AS to function as an antioxidant protein.
Materials and Methods
Cell lines and reagents
Cell lines were obtained from the American Type Culture Collection, and all cell culture reagents are from Invitrogen unless otherwise indicated. HT1080 cells and its mutant derivatives, U4A, U5A, U6A, and neuroblastoma cells were cultured using Dulbecco's minimum essential medium (MEM) supplemented with 10% (v/v) heat-inactivated bovine calf serum, 2 mM l-glutamine, and 5% penicillin and streptomycin, at 37°C in 5% carbon dioxide (CO2). U4A (-JAK1), U5A (-IFNAR2c), and U6A (-STAT2) cells are a generous gift from Dr. George Stark, Dr. Richard Ransohoff, and Elise Hovey-Bates (Cleveland Clinic, Cleveland, OH) (Stark and Darnell 2012). Peripheral blood mononuclear cells (PBMCs) derived from healthy individuals were obtained from AllCells. PBMCs (1×106cells/mL) and cultured cells were incubated at 37°C and 5.0% CO2 and grown in Eagle's MEM supplemented with 10% v/v heat-inactivated fetal bovine serum and 2.0 mM l-glutamine. Antibiotics were not included in the cell culture media. The astrocytoma cell line CH235 and human fetal brain cells were cultured in MEM supplemented with 20% (v/v) heat-inactivated bovine calf serum, 2 mM l-glutamine, 5% penicillin and streptomycin, 1 μg/mL insulin, 10 mM HEPES, and 0.1 mM nonessential amino acids at 37°C in 5% CO2. Betaseron™ and IFN-β-1b were prepared as previously described (Russell-Harde and others 1995). Betaseron is IFN-β-1b formulated in human serum albumin (specific activity=3.2×107 U/mg), while IFN-β-1b represents purified, unformulated material (specific activity=2.6×107 U/mg).
Study population
MS patients undergoing Betaseron treatment were studied. Their Expanded Disability Status Scale scores had a mean of 4.4 and ranged from 2.5 to 5.5 (Kurtzke 1983; McDonald and others 2001). None received corticosteroids or immunosuppressants for 1 year prior to or during the study. Betaseron treatment, patient sampling, and purification of PBMCs were performed as previously described (Croze and others 2012). Human studies were conducted with the approval of the Institutional Review Board of the University of Chicago.
RNA isolation and GeneChip analysis from in vivo samples
Heparinized blood (35 mL) was collected from MS patients by venipuncture and PBMCs were immediately isolated using Ficoll-Paque Plus density gradients (Amersham Biosciences). RNA was isolated and processed as previously described (Reder and others 2008). RNA was hybridized to the GeneChip® Human Exon 1.0 ST array (HuEx1.0ST) (Affymetrix, Inc.) containing ∼1.4 million probe sets recognizing over 1 million exon clusters by Expression Analysis, Inc. Expression summarization was performed using the Robust multichip average (RMA) (Gentleman and others 2004) implementation in Affymetrix Power Tools (APT) version 1.12.0 (www.Affymetrix.com) (Croze and others 2012). Exon data were analyzed using Excel Array Analysis software XRAY (XRAY v2.633; Biotique Systems, Inc.).
NCOA7/NCOA7-AS mRNA expression in human cells
Cells were incubated with or without IFN-β-1b (1,000 International Units [IU]/mL, 1×107 cells), for the times indicated. Cells were collected, washed twice in cold phosphate-buffered saline (PBS), and RNA was isolated. RNA from PBMCs and other cell types was isolated using Qiagen RNeasy as previously described (Reder and others 2008). To determine the specificity of NCOA7-AS induction by IFN-β-1b, cells were stimulated with or without IFN-β-1b (1,000 IU/1×107 cells) for either 4 or 18 h. RNA isolated at each time point was analyzed by reverse transcription–polymerase chain reaction (RT-PCR; TaqMan) using specific NCOA7-AS primer sets (Applied Biosystems). Primer sets for NCOA7-AS were AS1: forward 3′ GGAGACTGGCCTATAGCACGTT 5′, reverse 3′ CGATTTCCGGTAGAGCGTCTT 5′, probe 6FAMAGCACGGGACCAGCMGBNFQ. Negative control primer set (NCOA7 N-terminal region of message) forward, 3′ GAAAATCCTTTGCCACTCACACT 5′, reverse 3′ GGCTGCTTTCTCCGTTTGC 5′, Probe, 6FAMCAGCCATGGTCCAGCMGBNFQ.
General methods for performing quantitative RT-PCR (qRT-PCR) have been previously described (Reder and others 2008). RNA samples were isolated using RNeasy Midi endotoxin-free Kit (Qiagen, Inc.) and run in triplicate or quadruplets for each sample. Confirmation of exon array results by RT-PCR made use of the same RNA used for exon GeneChip® analysis. RT-PCRs were performed using a 96-well Opticon II DNA Engine (BioRad) or an ABI Prism 7900HT Sequence Detection System (Applied Biosystems). Results are presented as linearized C(t) (2−ΔC(t), log 10 scale) values normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH).
Fluorescence-activated cell sorting
1×106 cells were labeled with rabbit polyclonal antibodies recognizing either IFNAR2c (IFNAR2c-PC) or IFNAR1 (IFNAR1-PC) (Wagner and others 2004) for 1 h at 4°C, washed with ice-cold PBS (50 mM sodium phosphate pH 7.3 containing 150 mM sodium chloride), and then incubated with a fluorescein isothiocyanate-conjugated anti-rabbit antibody (DAKO Corp.) for 1 hr at 4°C. Fluorescence-activated cell sorting (FACS) analysis was performed as previously described using a FACS Vantage™ SE (Becton Dickinson Bioscience) (Wagner and others 2002).
Northern analysis
RNA is quantified by measuring the UV absorbance at 260 nm on a Nanodrop 2000 spectrophotometer (Thermo Scientific). The procedure of northern analysis has been described elsewhere (He and others 2003). Briefly, 30 μg of RNA from each sample is loaded onto 1% agarose gels with 6.7% formaldehyde, separated by electrophoresis, transferred onto nylon membranes, and detected with a 32P-labeled DNA probe specific to NCOA7-AS or as indicated in the text.
NCOA7-AS mRNA sequence analysis and complimentary DNA cloning
To determine the 5′ end sequence of the IFN-induced NCOA7-AS mRNA, total RNA was extracted from IFN-β-1b-treated HT1080 cells, and reverse transcribed using the Superscript III reverse transcriptase (Invitrogen) with primer NCOA7-2476 (ATGCCGATTTCCGGTAGAG) to produce complimentary DNA (cDNA). The cDNA was treated with RNase H to remove the RNA template and a poly-adenine sequence was added to 3′ of the cDNA using the terminal transferase (New England Biolabs). The polyadenylated cDNA was subjected to 2 rounds of PCRs using primer sets GATACCCTTGCACCCTTGC/GCTCGATGTGCACTGCTTTTTTTTTTTTTTTT and GCTCGATGTGCACTGC/CGATGTGCATATTCTCCAGG. The PCR product was gel purified and sequenced using primer NCOA7-2365r (CGATGTGCATATTCTCCAGG). The sequencing result was aligned to the human genome sequence to determine the transcription start site and the sequence of the 5′ end of the NCOA7 mRNA.
To determine the coding sequence (CDS) of the IFN-induced NCOA7-AS mRNA, total RNA was extracted from IFN-β-1b treated cells. The mRNA was purified using the Oligotex miniprep kit (Qiagen) and reverse transcribed by primer NCOA7-1218r (TGCTGTGCACTTTCGATTTT). The cDNA was then amplified by 2 rounds of PCR using primer sets AGCTCAGCGTGGCTACAAGT/GATGGCAGAAAGGAAGCATT and AACTGTGGTGTGGAAGCAGA/TTTTCCAGCCAGCTTTAGGA, and finally the PCR product was gel purified and sequenced using primer NCOA7-89f (GCCACACTTCTCACTGCTCA). Sequencing results confirmed the predicted CDS of NCOA7-AS. To determine the 3′ untranslated region of the IFN-induced NCOA7-AS mRNA, similar procedures are used except that primer NCOA7-3232r (GGCAGCAGTGAAAACATGAG) was used in reverse transcription and primer sets AGCTCAGCGTGGCTACAAGT/GGGAAAGAAAATCACTGAATGC and AACTGTGGTGTGGAAGCAGA/TCCATAAATGAAGGGTCACACA were used in the nested PCRs, and then primers NCOA7-3045r (TCCATAAATGAAGGGTCACACA) and NCOA7-739f (TTCAGGATCTGGAGGTGTGG) were used in sequencing.
To determine the 3′ end sequence of the IFN-induced NCOA7-AS, total mRNA was reversed transcribed using primer RACE1 (GCTCGATGTGCACTGCTTTTTTTTTTTTTTTT) and RNase H treated to remove the RNA templates. The cDNA was then subjected to 2 rounds of PCRs using primer pairs TGTAGCCTGGGTGTACGTTTC/GCTCGATGTGCACTGC and CCATTTACACCATCTGCAACC/GCTCGATGTGCACTGC. The PCR product was gel purified and sequenced using primer NCOA7-2970f (GAGCATATCCAGATGCTGTGTT) and a single transcriptional polyadenylation site was found.
To test oxidation resistance activity in the bacterial oxidative antimutator assay, NCOA7-AS was cloned into the bacterial expression vector pTrc99A by first amplifying its cDNA using primers Kpn-NCOA7-AS-f (GCGC-GGTACC-ATGAGAGGCCAAAGATTACCCT) and Bam-NCOA7-r (CGGC-GGATCC-TCAATCAAATGCCCACACCTC), digesting the product and vector with KpnI and BamHI, then purifying and ligating the digested products to produce the plasmid pMV1360. To use the start codon of the cDNA, the vector was first modified by digesting with NcoI, removing the 5′ ssDNA ends with mung bean nuclease, then ligating the blunt ends to remove the start codon of the vector.
Oxidative antimutator assay
Different variants of NCOA7 were inserted into the pTrc99a bacterial expression vector and tested as previously described with the exception that the instability of the C-terminal deletion plasmid and selection against its maintenance at 37°C required incubating this cell at 30°C for 5 days, rather than 37°C for 3 days to attain similar colony sizes (3–5 mm in diameter). Instability at 30°C and function of the lacZ reversion assay at this temperature was confirmed by prolonged incubation beyond 5 days at 30°C, which resulted in the production of many sectored colonies in which a sector or sectors exhibited high levels lacZ reversion within colonies lacking mutations in other regions (not shown).
Chromatin immunoprecipitation DNA sequencing (ChIP-seq) analysis
The ChIP-seq data were obtained from the Gene Expression Omnibus under the accession No. GSE54414. The data were aligned to the mouse genome (NCBI37/mm9; July 2007) using the ELAND alignment software (Illumina). Regions where the ChIP signals were enriched relative to the normal rabbit serum control were determined as described (Ramsey and others 2010; Litvak and others 2012). We used a false discovery rate of less than 1%. Integrated Genome Browser (IGB) (Nicol and others 2009) was used to display the ChIP-seq data.
Data handling and statistics
The mean values±standard deviations were determined for a given experiment. Statistical tests comparing differences between groups were performed using the Student's t-test. A P value of <0.05 was considered to be significant.
Results
NCOA7-AS induction by IFN-β-1b
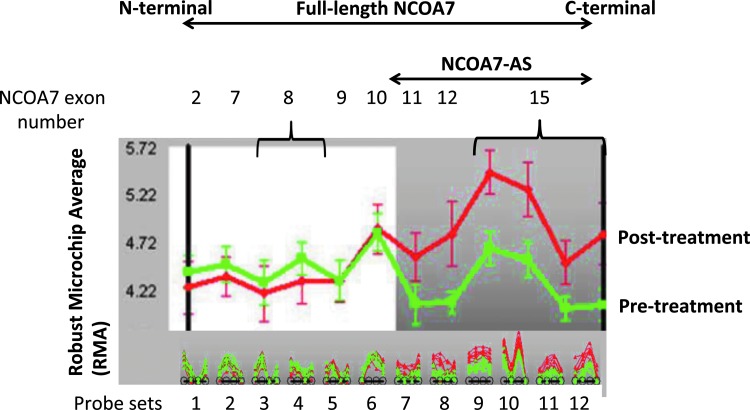
MS patients with the relapsing-remitting form of MS, previously recruited for a different study (Reder and others 2008), were initially used to first identify NCOA7-AS. The genome-wide study of total gene expression after Betaseron IFN-β-1b treatment has been published elsewhere (Croze and others 2013). These patients had all received Betaseron for a mean period of 6.9 (0.5–10.5) years and discontinued therapy for at least 64 h before the start of study. Patient blood samples were taken at 0, 4, and 18 h post injection of Betaseron and PBMCs were isolated. mRNA purified from MS patient PBMCs was analyzed using the GeneChip Human Exon 1.0 ST Exon array (Affymetrix, Inc.). NCOA7 exon expression measurements were performed using probes that identified exons 2, 7, 8, 9, 10, 11, 12, and 15. Expression levels of each of these exons of NCOA7 are shown in Fig. 1. Exon numbering is shown in Fig. 2. Figure 1 indicates that, among the exons identified in full-length NCOA7, exons 2, 7, 8, 9, and 10 are not expressed at elevated levels after Betaseron treatment, while exons 11, 12, and 15 are strongly induced. Sequences of all NCOA7 probe sets are listed in Supplementary Table S1 (Supplementary Data are available online at www.liebertpub.com/jir).
FIG. 1.
Analysis of Human Exon 1.0 ST GeneChip® data using mRNA collected from multiple sclerosis (MS) patients treated with Betaseron (interferon [IFN]-β-1b). XRAY v2.63 screen shot. Untreated (0 h, green) and IFN-β-1b treated (4 h, red) MS patients (n=7). The bottom shows individual probe levels for the 12 nuclear receptor coactivator protein 7 (NCOA7) probe sets per Affymetrix core annotations. The top of the screen shot shows probe set levels for the comparison; the dark background highlights the region of differential exon expression. The y-axis displays Robust multichip average (RMA) expression value. NCOA7-AS exons (exons 10–16) induced by administration of Betaseron are shown. Probes and probe sets are shown along the x-axis. Sequences of probe sets are listed in Supplementary Table S1.
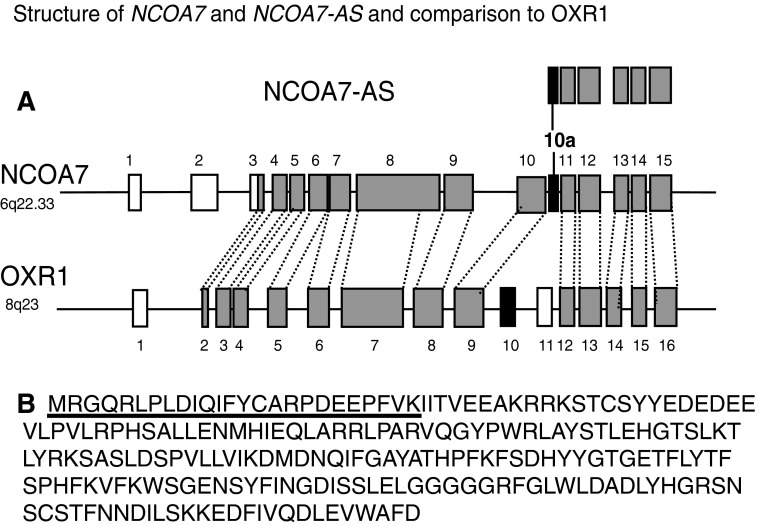
FIG. 2.
Structure of NCOA7 and NCOA7-AS compared to the OXR1 family of genes. (A) Alignment of NCOA7, NCOA7-AS, and OXR1. Gray boxes represent highly similar exons of NCOA7 and OXR1. White and black boxes are unique to either OXR1 or NCOA7. Black boxes are used as first exons in alternative start forms of OXR1 and NCOA7. Alignment of individual exons of NCOA7 and OXR1 indicates that the amino acid sequences of exons 3, 4, 7, 9, 12, 13, 14, and 15 of NCOA7 are more than 50% identical to their OXR homologs; exon 5, 6, 10, and 11 are 25%–50% identical to their OXR1 homologs; exon 8 is about 25% identical to its OXR1 homolog. NCOA7 exon numbering is used. (B) Amino acid sequence predicted from the complimentary DNA sequence. The sequence of the unique exon 10a is underlined.
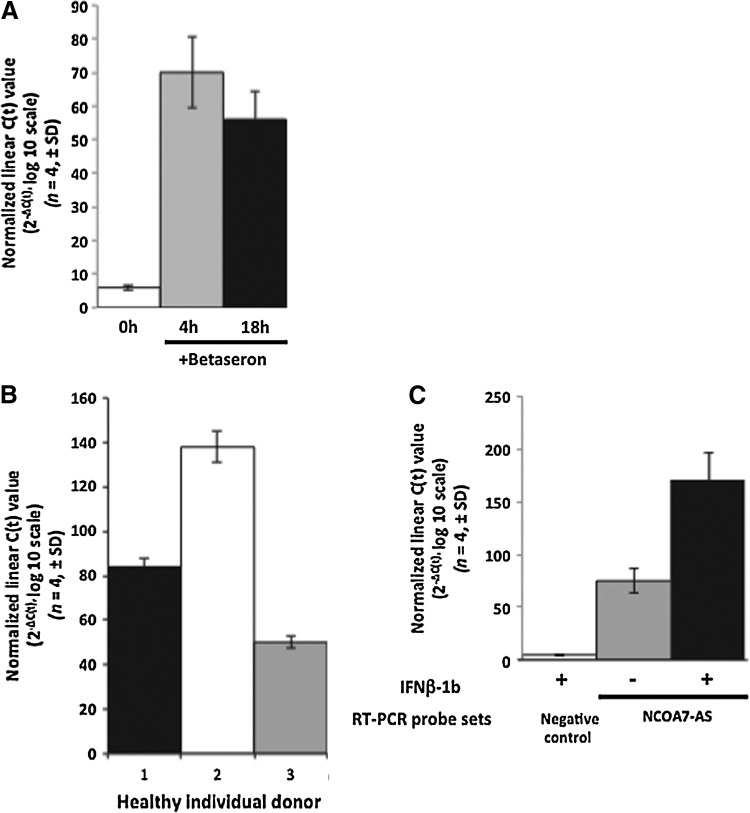
The induction of NCOA7-AS observed in MS patients using the all-exon GeneChip was confirmed by RT-PCR using NCOA7-AS-specific primers (see Materials and Methods section) and the same mRNA used to hybridize to the GeneChip. Induction is seen both at 4 and 18 h post-treatment (Fig. 3A). NCOA7-AS mRNA levels increase rapidly after treatment, reaching maximum expression at 4 h, then decline at 18 h. Figure 3B shows that NCOA7-AS mRNA induction also occurred in vitro in PBMCs collected from 3 healthy volunteers 4 h post-stimulation with IFN-β-1b. Induction of NCOA7-AS mRNA by IFN-β-1b was normalized to unstimulated PMBCs. Comparison of all 3 healthy individuals did reveal some variability in the degree of the NCOA7-AS response in vitro to IFN-β-1b treatment. Induction of NCOA7-AS (C-Terminal region of NCOA7) was then evaluated in primary human fetal brain cells relative to a primer set recognizing the N-terminal region of NCOA7, which is not expected to be induced by IFN-β-1b (Fig. 3C). Collectively, these results demonstrate that NCOA7-AS is induced by IFN-β-1b in PBMCs of healthy individuals, MS patients, and in cells derived from the CNS.
FIG. 3.
Confirmation in vivo of NCOA7-AS microarray results using reverse transcription–polymerase chain reaction (RT-PCR) in primary cells and 3 different cell lines. (A) TaqMan (RT-PCR) detection of NCOA7-AS mRNA obtained from peripheral blood mononuclear cells (PBMCs) isolated from MS patients treated with Betaseron. The same RNA used for the microarray was used to independently validate expression of NCOA7-AS using RT-PCR at 0, 4, and 18 h post-Betaseron administration. (B) Induction of NCOA7-AS in PBMCs of healthy individuals stimulated 4 h with IFN-β-1b in vitro. Three healthy individual donors were evaluated (1–3). Data are presented relative to unstimulated cells. (C) Induction of NCOA7-AS in human fetal brain cells stimulated 4 h with IFN-β-1b. A primer set for amplification of the N-terminal region of NCOA7 preceding the start of NCOA7-AS was used as a negative control primer. All results were normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and expressed as normalized linearized C(t) values (2−ΔC(t), log 10 scale, n=4, ±standard deviation [SD]).
IFN-β-1b induction of NCOA7-AS in human cell lines
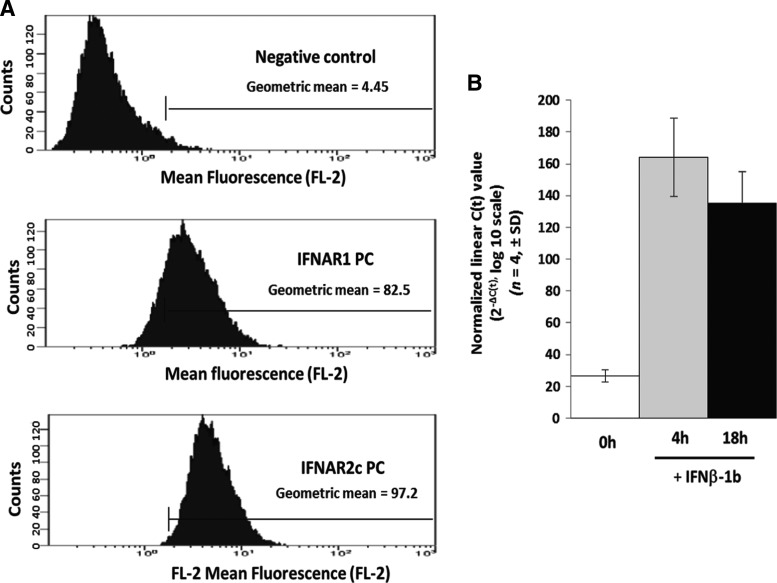
FACS, using IFN receptor specific antibodies, showed that the IFNAR1 and IFNAR2 are expressed on the surface of both peripheral immune cells and cells originating from the CNS, such as CH235 astrocytoma cells, demonstrating the presence of the human IFN receptor in cells derived from the CNS (Fig. 4A). CH235 cells, when stimulated in vitro with IFN-β-1b, express NCOA7-AS mRNA 4 and 18 h poststimulation (Fig. 4B). IFN-β-1b stimulation resulted in a 5 to 6-fold induction of NCOA7-AS over endogenously expressed NCOA7-AS.
FIG. 4.
Expression of the type I IFN-receptor and NCOA7-AS induction in cultured human astrocytes. (A) Fluorescence-activated cell sorting analysis of IFN-alpha receptor 1 (IFNAR1) and IFN-alpha receptor 2 (IFNAR2) on the surface of CH235 astrocytic cells. Negative control rabbit preimmune sera (Negative control; geometric mean=4.45) is compared to IFNAR1 polyclonal antisera (IFNAR1PC; geometric mean=82.5) and IFNAR2c polyclonal antisera (IFNAR2cPC; geometric mean=97.2). The x-axis represents the number of cell counts and the y-axis the mean fluorescence (FL-2; log 10). (B) NCOA7-AS mRNA induction in CH235 cells at t=0 h (0h), 4 h (4h), and 18 h (18h) after stimulation with IFN-β-1b. All results were normalized to GAPDH and expressed as normalized linearized C(t) values (2−ΔC(t), log 10 scale, n=4, ±SD).
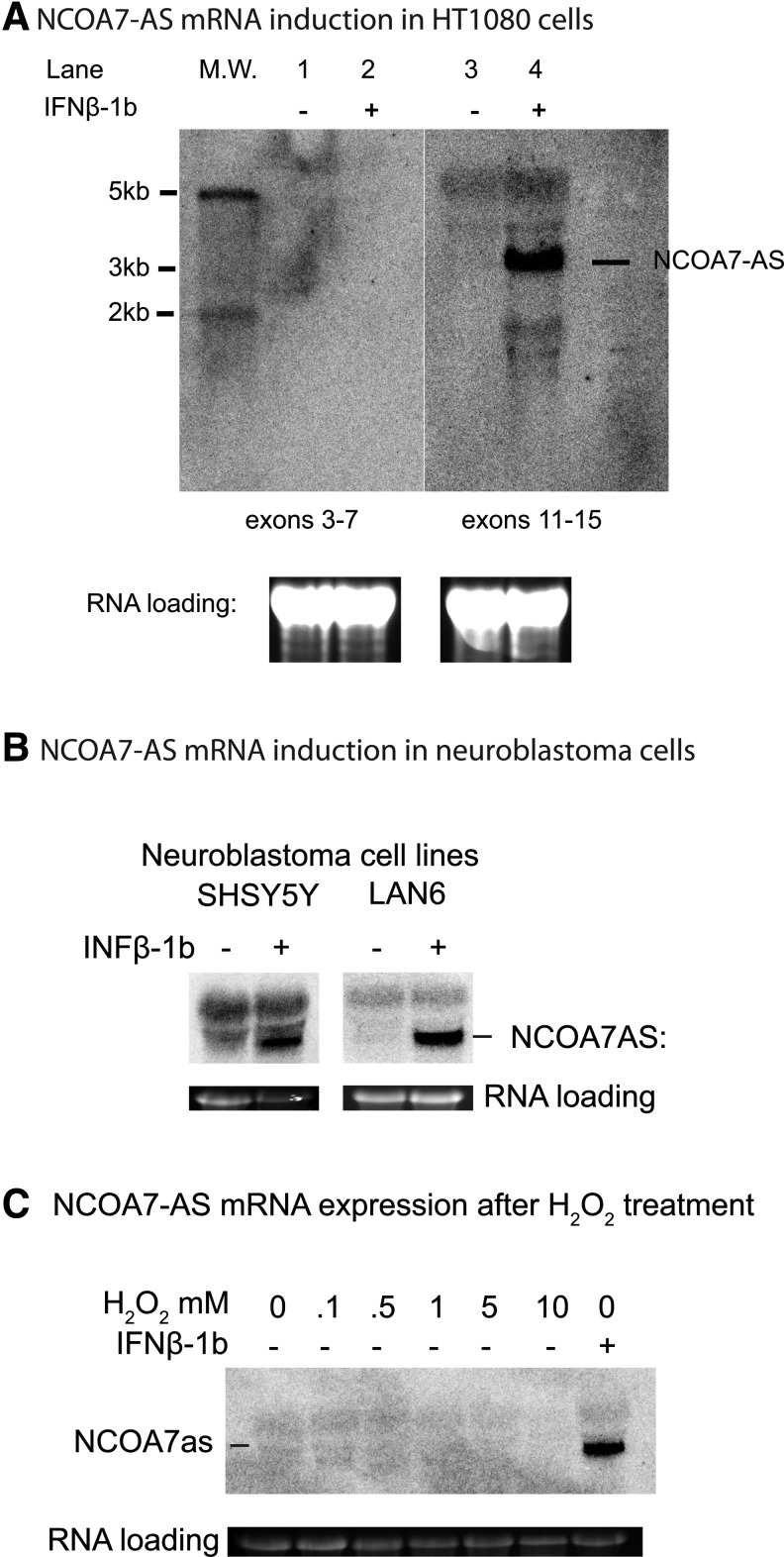
Human HT1080 fibrosarcoma cells express IFNAR1 and IFNAR2 and respond to IFN-β-1b (Der and others 1998). As is the case with PBMCs isolated from MS patients or healthy individuals, NCOA7-AS is strongly induced in the HT1080 cell line and in 2 neuroblastoma cell lines, SHSY5Y and LAN6 (Fig. 5A, B). A larger species of mRNA that hybridizes to the exon 11-15probe is seen in HT1080 cells and several larger species are seen in neuronal cells. The larger mRNAs do not change in intensity when compared to IFN treated and untreated cells and are therefore not inducible. The presence of additional NCOA7 mRNAs in neuronal cells is expected since NCOA7 is expressed at high levels in neurons (Shao and others 2002). However, since larger forms of NCOA7 do not include exon 10a, the proteins encoded by the larger mRNAs are likely to be functionally different from the IFN-inducible NCOA7-AS (UCSC genome database [http://genome.ucsc.edu]; also see Results and Discussion of Fig. 6). Together, these results indicate that the IFN-induced expression of NCOA7-AS in PBMCs derived from MS patients is recapitulated in various cells originating from connective tissue, the CNS, and peripheral neurons.
FIG. 5.
Induction of NCOA7-AS by IFN-β-1b treatment but not H2O2 treatment. (A) Induction of NCOA7-AS in HT1080 fibrosarcoma cells, assayed with hybridization probes to exons 3–7 (left panel) and hybridization probes to exons 11–15 (right panel). (B) A probe to exons 11–15 was used to detect NCOA7-AS in SHSY5Y and LAN6 neuroblastoma cells. (C) Cells were treated with H2O2 at the indicated concentrations, or IFN-β-1b (1000 U) for 4 h before mRNA isolation. Ethidium bromide staining of 28S rRNA is shown as a gel loading control in all panels.
FIG. 6.
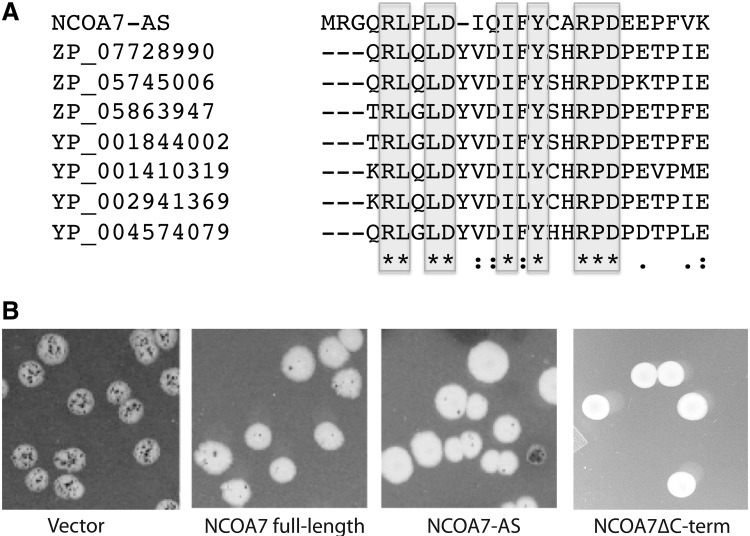
NCOA7-AS structure and function. (A) Alignment of exon 10a with bacterial aldo-keto oxidoreductases. Identical amino acids are indicated by a star (*), highly conservative substitutions are indicated by a colon (:), conservative substitutions are indicated by a dot (.). (B) Oxidation resistance activity of NCOA7-AS, full-length NCOA7, and a C-terminal deletion mutant of the full-length NCOA7 gene truncated at the PstI site within exon 9. Reduction in the number of oxidation induced lacZ mutants (dark microcolonies within the parent colony) resulting from expression of different forms of NCOA7 relative to the number seen in the vector control (left panel), indicates the level of suppression of oxidative damage resulting from expression of NCOA7. All colonies were grown until the parent colony reached the same diameter (3–5 mm) (see Materials and Methods section for assay details.)
Previous experiments demonstrated that the full-length NCOA7 gene is not induced in response to oxidative stress (Durand and others 2007). To test whether the response to oxidative stress of the NCOA7-AS variant is similar to its full-length counterpart or not, we treated HT1080 cells with different doses of H2O2 and measured the levels of NCOA7-AS-specific mRNA after 4 h. Figure 5C shows that the NCOA7-AS variant, like full-length NCOA7 (Durand and others 2007), is not induced in response to H2O2 treatment, but shows strong induction after IFN-β-1b treatment. It therefore differs from a closely related ortholog, OXR1, which is strongly induced in response to oxidative stress (Elliott and Volkert 2004).
Sequence of NCOA7-AS
We determined the precise structure of NCOA7-AS by first performing 5′ and 3′ RACE to sequence the ends of NCOA7-AS cDNA from IFN-β-1b-induced HT1080 cells. After sequencing each end, the cDNA was amplified using primers Kpn-NCOA7-AS-f and Bam-NCOA7-AS-r (see Materials and Methods section) and cloned into the KpnI and BamHI restriction sites of the pTrc99A bacterial expression vector, and the remainder of the gene was sequenced (Accession No. KC238672). Analysis of the sequence indicates that NCOA7-AS includes exons 11–15 of the full-length NCOA7 gene and a new exon (10a) that is encoded by genomic sequences that lie between exons 10 and 11. Database analysis of all currently known NCOA7 cDNA sequences reveals that exon 10a appears to be found only in variants that begin with exon 10a and this exon is not incorporated into any known forms of NCOA7 that include exons upstream of exon 10a. Figure 2A shows the structure of NCOA7-AS in comparison with full-length NCOA7 and its close ortholog OXR1. Figure 2B shows the amino acid sequence of NCOA7-AS determined from the translated mRNA.
Oxidation resistance activity of NCOA7-AS
To test whether NCOA7-AS is active as an antioxidant protein, we used the bacterial oxidative antimutator assay (Durand and others 2007; Volkert and others 2008). NCOA7-AS was expressed in an E. coli strain that carries mutations in mutM and mutY, which function to prevent GC→TA transversions by repairing 8-oxoG and its 8-oxoG:A mispaired premutagenic intermediate (Michaels and others 1992a, 1992b; Wang and others 2004; Volkert and others 2008). This E. coli strain also carries the lacZcc104 allele, which reverts to Lac+ only by GC→TA transversion and serves as the reporter of oxidative DNA damage (Michaels and Miller 1992; Michaels and others 1992a; Wang and others 2004; Volkert and others 2008).
Figure 6B, left panel shows the elevated level of spontaneous GC→TA transversion mutations when only the pTrc99A vector plasmid is present in the cell. The frequency of GC→TA transversion mutations can be approximated by counting the number of dark blue, Lac+ microcolonies within the white Lac− parent colony. Figure 6B, center panels show that introduction of the pTrc99a bacterial expression vector carrying either full-length NCOA7 or NCOA7-AS suppresses oxidative mutagenesis, indicating that both proteins have a strong protective activity and therefore have an antioxidant function that reduces the number of 8-oxoG lesions produced. The active site for antioxidant function of the closely related protein, OXR1, is localized to a domain that resides within exon 8 (Murphy and Volkert 2012). This exon corresponds to exon 9 of NCOA7. Deletion studies of full-length NCOA7 indicate that exon 9 contains a region required for its oxidative damage resistance activity, because N-terminal deletions lacking exons 1–8 (Durand and others 2007) and C-terminal deletions lacking exons 10–15 (Fig. 6B, right panel) exhibit oxidation resistance activity and because exons 11–15, which are present in both NCOA7 and NCOA7-AS, are dispensable for this activity. Since exon 9 is not present in NCOA7-AS, it suggests exon 10a may be required for the activity of the AS form. Direct tests of deletions resulted in proteins too unstable for analysis, precluding a direct test of the requirement of exon 10a for activity. Since there is no detectable homology between exons 9 and 10a, the 2 NCOA7 proteins are likely to result in oxidation resistance via different biochemical mechanisms.
Expression of NCOA7-AS requires the JAK-STAT pathway
Genes regulated by IFN-β-1b are induced when IFN interacts with the IFNAR1 and IFNAR2 chains on the surface of IFN-responsive cells. IFN-β-1b binding to these receptor chains results in the formation of a heteromeric receptor complex and results in the phosphorylation of specific tyrosine residues located in the cytoplasmic domain of each receptor chain (Stark and others 1998; Mogensen and others 1999; Stark and Darnell 2012). Phosphorylation of the IFN receptor chains is mediated by JAK1 and Tyrosine Kinase 2 (Tyk2) and results in the binding and subsequent activation of STATs. Gene induction ensues when activated STATs (STAT1 and STAT2) together with IFN response factor 9 (IRF9) translocate from the cytoplasm into the nucleus and bind to the ISRE activating IFN-dependent gene transcription (Stark and Darnell 2012).
Examination of genomic sequences upstream of the transcription initiating exon of NCOA7-AS (exon 10a in Fig. 2) reveals the presence of 3 overlapping regions that lie between exon 10 and 10a. The sequences of the overlapping regions are similar in sequence to the STAT, ISRE, and IRF sites found in the IFN-β-1b-responsive oligoanenylate synthetase (OAS1a) and OAS1b genes (Fig. 7A) and include a match to the canonical minimal ISRE sequence A/GNGAAANNGAAACT (Kerr and Stark 1991; Tenoever and others 2007; Pulit-Penaloza and others 2012), suggesting NCOA7-AS transcription initiates from this region and requires the JAK-STAT signaling pathway.
FIG. 7.
Identification of the NCOA7-AS promoter region, which contains elements similar to signal transducers and activators of transcription (STAT), IFN response factor (IRF), and IFN-stimulated response element (ISRE) regulatory elements. (A) Boxes indicate the conserved STAT, ISRE, and IRF elements of OAS1a, OAS1b, and NCOA7-AS of mouse and humans (Pulit-Penaloza and others 2012). The canonical ISRE sequence is shown in bold. The sequence shown lies immediately upstream of the human mRNA start site of exon 10a, which is indicated by the upper arrow. The mRNA start sites of OAS1a and OAS1b are aligned and are indicated by the lower arrow. (B) Induction of NCOA7-AS is dependent on the Janus kinase (JAK)-STAT signaling pathway. NCOA7-AS induction is blocked by mutations in several key genes of the JAK-STAT regulatory pathway. Reintroduction of the wild-type alleles of IFNAR2 and JAK with expression plasmids restores induction in the appropriate mutant. The somewhat weaker response in the complemented strains is presumably due to poor growth of complemented mutant cell lines or plasmid instability. All cell lines are derivatives of HT1080 and have been described elsewhere (Pellegrini and others 1989; Watling and others 1993; Lutfalla and others 1995). U4A (JAK1 mutant), U5A (IFNAR2c mutant), U6A (STAT2 mutant), U4A JAK1 (U4A transfected with a JAK1 expression plasmid) U5AR2c (U5A transfected with a IFNAR2c expression plasmid), and the wild-type HT1080 control. Ethidium bromide staining of 28 S rRNA is shown as a gel loading control.
To test the JAK-STAT requirement directly, we obtained mutant derivatives of HT1080 cells in which the JAK1, STAT2, or the IFNAR2 genes are inactive. Figure 7B shows that unlike wild-type cells, IFN-β-1b induction of NCOA7-AS is completely abolished in cell lines carrying each of these 3 mutations, demonstrating that induction requires all 3 of these key components of the JAK-STAT signaling pathway and that induction presumably results from the interaction of an activated STAT transcription complex with the sequence elements shown in Fig. 7A. To confirm the requirement for JAK-STAT signaling, we tested whether reintroduction of the appropriate gene (JAK1 or IFNAR2c) restored induction. Restoration of induction is observed when either the JAK1 or IFNAR2c gene is reintroduced into the corresponding mutant cell line, although NCOA7-AS induction by IFN-β-1b stimulation of the “rescued” U6A or U5A mutant cell line did not reach wild-type levels. Poor induction in strains carrying both mutant and wild-type alleles is not unexpected, since point mutations can often result in competition between mutant and wild-type proteins.
NCOA7-AS induction by IFN-β and LPS in macrophages
The observation that the oxidation resistance NCOA7-AS gene is induced by IFN-β-1b raises the possibility that the natural function of this protein may be to protect the host from reactive oxygen species (ROS) generated by the immune system to counteract bacterial and viral infections. This hypothesis predicts that NCOA7-AS will be induced when cells mount an immune response.
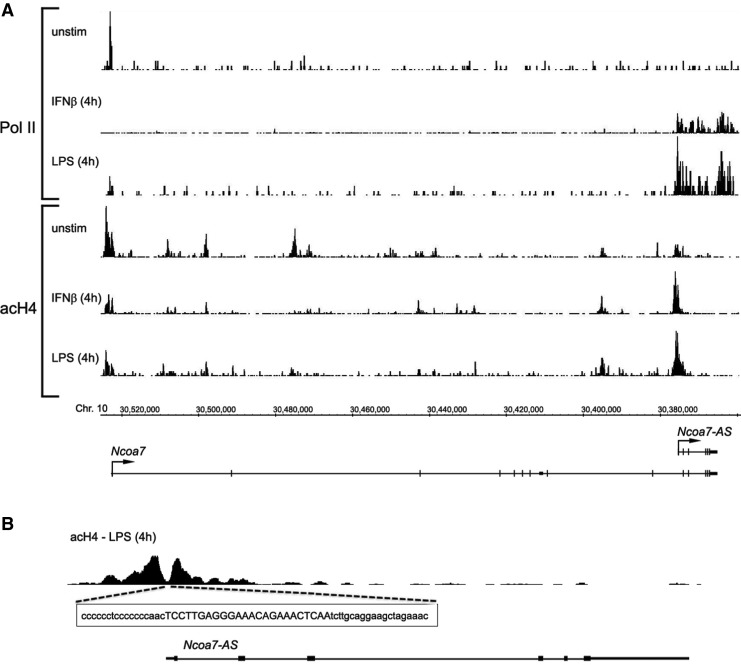
To test this possibility, we first confirmed that IFN-β induces NCOA7-AS in mouse macrophages by measuring Pol II chromatin immunoprecipitation and sequencing of the bound DNA fragments (ChIP-seq). This serves as an independent measurement of transcription levels by determining the number of RNA Pol II molecules bound to specific regions of DNA. Figure 8a (unstim) shows that Pol II molecules are bound to the promoter of the full-length NCOA7 gene prior to IFN-β treatment. However, little or no Pol II is seen downstream of the promoter, indicating little active transcription, suggesting that Pol II is stalled at the promoter. This is consistent with the lack of a signal for either full-length NCOA7 or NCOA7-AS in northern blots using probes to the upstream exons present only in the full-length form, or in uninduced cells (Fig. 5). Four hours after IFN-β treatment, Pol II levels at the upstream promoter for full-length NCOA7 is reduced and high levels of Pol II are detected throughout the NCOA7-AS coding region (Fig. 8a, IFNβ [4h]), indicating transcription is induced and the gene is actively being transcribed. No increase in Pol II occupancy is seen in regions encoding exons upstream of exon 10a, indicating this region is not being transcribed. Thus, transcription must initiate at a promoter just upstream of exon 10a. These results support the conclusions reached using micro arrays, RT-PCR, and northern blots.
FIG. 8.
RNA Pol II occupancy and histone H4 acetylation pattern at the NCOA7 gene locus. (A) The binding of RNA Pol II (Pol II) and histone H4 acetylation (acH4) profile at the NCOA7 gene locus are determined by ChIP-seq analysis of IFN-β and LPS-treated murine bone marrow-derived macrophages. All ChIP-seq data tracks (vertical bars at 10 bp intervals) are in tags per million and are on the same vertical scale. Unstimulated cells (unstim), 4 h after IFN-β (IFN-β [4h]) or LPS (LPS [4h]) addition. Histone acetylation pattern at the cis-regulatory region of NCOA7-AS gene. (B) Expansion of the ChIP-seq analysis of histone H4 acetylation (acH4) at the promoter region of the NCOA7-AS gene in LPS-stimulated macrophages is shown, as is the relevant portion of the DNA sequence that lies within the valley between the 2 histone acetylation peaks. The conserved ISRE element is indicated by the capital letters.
To test whether macrophages show a similar transcription profile during infection, they were treated with the bacterial antigen LPS and RNA Pol II occupancy was measured. As is the case with IFN-β treatment, LPS triggers a strong induction of NCOA7-AS transcription, as indicated by the high level of Pol II occupancy 4 h after treatment (Fig. 8a, LPS [4h]). LPS also causes a similar reduction in Pol II occupancy at the upstream full-length NCOA7 promoter as is seen after IFN-β treatment.
To further examine the promoter activity, we performed ChIP-seq of acetylated histone H4 in the NCOA7 coding region of the chromosome (Fig. 8a, acH4). In unstimulated cells, histone H4 acetylation is high in region of the upstream promoter for full-length NCOA7 and some histone acetylation is detected in the predicted downstream NCOA7-AS promoter region. Four hours after LPS treatment, histone H4 acetylation is reduced at the upstream promoter for full-length NCOA7 and is enhanced in the region of the predicted downstream NCOA7-AS promoter; the same region that marks the upstream border of NCOA7-AS where Pol II occupancy region begins. The valley between the 2 peaks of acetylation is a typical pattern that delineates the histone-free region required for transcription factor binding and transcription initiation (Ramsey and others 2010). Examination of the sequence within the valley (Fig. 8b) indicates it contains the predicted ISRE, STAT, and IRF binding sequences (Figs. 7 and 8b), thus demonstrating that the ISRE sequence encodes an internal promoter that drives IFN induced, JAK-STAT-dependent transcription of the NCOA7-AS gene. These results also demonstrate that the canonical ISRE element is completely conserved and identical in mouse and human DNA (Fig. 7A, bold letters) and that induction does indeed occur in macrophages shortly upon IFN-β or LPS treatment.
Discussion
Results described in this study demonstrate induction of a previously unidentified IFN-inducible gene, NCOA7-AS. Its identification was made possible by the use of multi-exon microarrays. Previous efforts to characterize IFN-inducible genes (Waddell and others 2010) (Ottoboni and others 2012) failed to identify NCOA7-AS, presumably because only 1 of 3 NCOA7-specific probe sets used in standard (3′-biased) microarrays hybridizes to the IFN-β-1b-inducible NCOA7-AS mRNA.
We demonstrate NCOA7-AS expression in fibroblasts, CNS-derived human astrocytes, fetal brain cells, and 2 peripheral nerve cell neuroblastomas. The distribution of NCOA7-AS in cells located in the CNS and the periphery suggests an important function for NCOA7-AS in maintaining ROS resistance in disparate types of cells.
NCOA7-AS contains the unique exon 10a that is homologous to bacterial aldo-keto oxido-reductases (Fig. 6A), which are known to protect cells from lipid peroxidation products and reduce cellular levels of ROS (Jin and Penning 2007; Li and others 2012). An increase in ROS has been observed in areas of neurodegeneration occuring within the CNS in association with a variety of diseases. Furthermore, human aldo-keto oxido-reductases have been directly linked to Alzheimer's disease (Picklo and others 2001). Alignment of the active site domain of bacterial oxido-reductases with NCOA7 reveals a high degree of homology with exon 10a of NCOA7-AS and that this exon contains the conserved tyrosine and aspartic acid residues required for activity (Fig. 6A). While the exact nature of the antioxidant activity of NCOA7-AS remains to be determined, the lack of similarity between exon 10a and exon 9 of the full-length form suggests NCOA7-AS and NCOA7 proteins may vary mechanistically in their mode of action, and that NCOA7-AS likely shares biological activity with the aldo-keto class of redox active proteins.
Alternative start sites are a common feature of eukaryotic genes that contribute to mRNA and protein diversity (Rojas-Duran and Gilbert 2012). At least 25% of all expressed genes appear to initiate from multiple promoters and therefore produce mRNAs with different 5′ ends (Carninci and others 2006; Kimura and others 2006; Pal and others 2011; Yamashita and others 2011). In addition to incorporation of exon 10a, NCOA7-AS lacks the estrogen-binding domain found in exon 8 of NCOA7 (Shao and others 2002) suggesting that interaction with the estrogen bound estrogen receptor complex is a unique function of longer forms of NCOA7 that is not shared by NCOA7-AS.
Free radical formation, oxidative stress, and inability to control ROS levels can result in neuronal and tissue damage, DNA mutagenesis, lipid bilayer disruption, modification of proteins, and ATP-related energy failure (Gonsette 2008). Maintaining a correct balance between oxygen usage and free radical formation is necessary for maintaining cellular homeostasis especially in the CNS (Kaur and Ling 2008).
Type I IFN-regulated genes are typically induced in response to viral infection or inflammation (Pestka and others 1987). The presence of an ISRE sequence upstream of the transcriptional start site of the NCOA7-AS gene, the requirement for the JAK-STAT signaling pathway for its expression, and the observation that increased expression is accompanied by histone acetylation in regions flanking the histone-free ISRE element (Fig. 8b) indicates that NCOA7-AS is an alternative start-site variant that initiates transcription from the ISRE site in a JAK-STAT-dependent manner. The lack of Pol II binding to regions upstream of the NCOA7-AS CDSs indicates that regions upstream of exon 10a are not transcribed, further supporting the conclusion that NCOA7-AS is a newly discovered IFN-responsive gene product.
IFNs function as a first line of defense against various pathogens by activating innate and adaptive immune responses. This raises the possibility that NCOA7-AS may have evolved to limit immune-mediated tissue damage resulting from an increase in reactive oxygen species produced in response to infection (Akaike and others 1998). Increases in ROS associated with tissue damage have been described previously and involve immune activation of macrophages, leukocytes, astrocytes, and microglia cells (Columbano and others 1981; Weitzman and Gordon 1990; Rehermann and Nascimbeni 2005); hepatitis virus infection of the liver leading to development of hepatocellular carcinoma (Hagen and others 1994); and constitutive nitric oxide (NO) expression in mouse activated peritoneal macrophages (PM) (Guillemard and others 1999). Furthermore, inhibition of NO-producing enzymes in PMs by l-arginine-NG–amine (AA) is accompanied by expression of IFNs and IFN regulatory factor 1 and a correlated decrease in encephalomyocarditis virus replication. Mechanisms of action associated with IFNs that reduce cellular damage caused by oxidative stress is an important new function ascribed to IFNs and warrants further study. It should be noted that NO production itself can have both detrimental and beneficial immunomodulatory effects, resulting in a need to maintain a tight balance in the levels of reactive oxygen and nitrogen species (Wang and others 2001).
Reactive oxygen species are associated with formation of MS lesions and neuronal degeneration (Gonsette 2008; Kaur and Ling 2008). Neurodegeneration in MS has been linked to astrocyte activation, gliosis, lack of remyelination in early lesions, and reduced oligodendrocyte viability and maturation (Zhang and others 2011). Inflammation and oxidative stress in the CNS have been linked to all these factors (Amor and others 2010). Oxidative stress is likely to play a role in other neurological diseases (Dauer and Przedborski 2003). However, antioxidant treatments in neurological diseases have met with little or no success, possibly due to the importance of maintaining a strict balance between the negative and positive effects of oxygen generated free radicals.
The observation that IFN-β-1b treatment induces a previously unrecognized antioxidant protein suggests IFN-β-1b plays an important role in regulating potentially damaging levels of oxidative stress brought on by inflammation. Since NCOA7-AS, like NCOA7, has antioxidant activity, its induction may well assist other cellular antioxidant activities to protect neurons under immunological attack in MS. The observation that NCOA7-AS is not induced by ROS suggests its induction by IFN-β-1b may provide event- and tissue-specific resistance to oxidative stress beyond general cellular resistance mechanisms that are normally induced directly in response to oxidative stress.
Supplementary Material
Acknowledgments
Dr. Peter Bringmann (Bayer Healthcare) and Dr. Hugh Salamon (Bayer Healthcare, Knowledge Synthesis Inc.) for help in generating exon array data and bioinformatics. Dr. Volker Knappertz (Bayer Healthcare, Teva Pharmaceuticals) and Dr. Dirk Pleimes (Bayer Healthcare) for helpful discussions and financial support and Dr. Pedro Paz (Bayer Healthcare) and Dr. Sharlene Velichko (Bayer Healthcare, Veracyte) for assistance in developing the primer sets for NCOA7-AS; Drs. Alonzo Ross and Daniel Kilpatrick (University of Massachusetts Medical School) for neuroblastoma cell lines and Drs. George Stark, Richard Ransohoff, and Elise Hovey-Bates (Cleveland Clinic) for wild-type and mutant HT1080 cell lines. This work was supported in part by a research grant from Bayer Healthcare to MRV and by a grant (RG No. 4509A) from the U.S. National MS Society to ATR.
Authors' Contributions
L.Y. and M.R.V. designed and performed experiments, cellular and molecular studies of gene expression and function, and conducted data analysis. M.R.V. drafted the article, with assistance by all participants in the study. E.C. initiated the patient analysis and validation study and with K.D.Y. designed and coordinated the microarray experiments and with T.T. designed and performed the validation experiments. K.D.Y. conducted the genomic data analysis. A.T.R. was responsible for patient analysis, conduct, and collection of patient samples. V.L. conducted the ChIP-seq studies of transcription.
Author Disclosure Statement
Ed Croze, Ken D. Yamaguchi, and Tiffany Tran were employees of Bayer Healthcare while conducting several of the experiments described in this article. Anthony T. Reder and Michael R. Volkert received funding from Bayer Healthcare. Bayer Healthcare was not involved in the decision to publish or in determining the content of the article.
References
- Akaike T, Suga M, Maeda H. 1998. Free radicals in viral pathogenesis: molecular mechanisms involving superoxide and NO. Proc Soc Exp Biol Med 217(1):64–73 [DOI] [PubMed] [Google Scholar]
- Amor S, Puentes F, Baker D, van der Valk P. 2010. Inflammation in neurodegenerative diseases. Immunology 129(2):154–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnason BG, Dayal A, Qu ZX, Jensen MA, Genc K, Reder AT. 1996. Mechanisms of action of interferon-beta in multiple sclerosis. Springer Semin Immunopathol 18(1):125–148 [DOI] [PubMed] [Google Scholar]
- Butterfield DA. 2006. Oxidative stress in neurodegenerative disorders. Antioxid Redox Signal 8(11–12):1971–1973 [DOI] [PubMed] [Google Scholar]
- Butterfield DA, Perluigi M, Reed T, Muharib T, Hughes CP, Robinson RA, Sultana R. 2012. Redox proteomics in selected neurodegenerative disorders: from its infancy to future applications. Antioxid Redox Signal 17(11):1610–1655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carninci P, Sandelin A, Lenhard B, Katayama S, Shimokawa K, Ponjavic J, Semple CA, Taylor MS, Engstrom PG, Frith MC, Forrest AR, Alkema WB, Tan SL, Plessy C, Kodzius R, Ravasi T, Kasukawa T, Fukuda S, Kanamori-Katayama M, Kitazume Y, Kawaji H, Kai C, Nakamura M, Konno H, Nakano K, Mottagui-Tabar S, Arner P, Chesi A, Gustincich S, Persichetti F, Suzuki H, Grimmond SM, Wells CA, Orlando V, Wahlestedt C, Liu ET, Harbers M, Kawai J, Bajic VB, Hume DA, Hayashizaki Y. 2006. Genome-wide analysis of mammalian promoter architecture and evolution. Nat Genet 38(6):626–635 [DOI] [PubMed] [Google Scholar]
- Columbano A, Rajalakshmi S, Sarma DS. 1981. Requirement of cell proliferation for the initiation of liver carcinogenesis as assayed by three different procedures. Cancer Res 41(6):2079–2083 [PubMed] [Google Scholar]
- Croze E. 2010. Differential gene expression and translational approaches to identify biomarkers of interferon beta activity in multiple sclerosis. J Interferon Cytokine Res 30(10):743–749 [DOI] [PubMed] [Google Scholar]
- Croze E, Veichko S, Tran T, Yamaguchi KD, Salamon H, Reder AT, Kanppertz V, Paz P. 2009. Betaseron induces a novel alternate start transcript in cells obtained from relapsing-remitting multiple sclerosis patients and human brain that is associated with control of oxidative resistance. Multiple sclerosis 15(S138):475 [Google Scholar]
- Croze E, Yamaguchi KD, Knappertz V, Reder AT, Salamon H. 2012. Interferon-beta-1b-induced short- and long-term signatures of treatment activity in multiple sclerosis. Pharmacogenomics J 15 (S138):475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croze E, Yamaguchi KD, Knappertz V, Reder AT, Salamon H. 2013. Interferon-beta-1b-induced short- and long-term signatures of treatment activity in multiple sclerosis. Pharmacogenomics J 13(5):443–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dauer W, Przedborski S. 2003. Parkinson's disease: mechanisms and models. Neuron 39(6):889–909 [DOI] [PubMed] [Google Scholar]
- Der SD, Zhou A, Williams BR, Silverman RH. 1998. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc Natl Acad Sci U S A 95(26):15623–15628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand M, Kolpak A, Farrell T, Elliott NA, Shao W, Brown M, Volkert MR. 2007. The OXR domain defines a conserved family of eukaryotic oxidation resistance proteins. BMC Cell Biol 8:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott NA, Volkert MR. 2004. Stress induction and mitochondrial localization of Oxr1 proteins in yeast and humans. Mol Cell Biol 24(8):3180–3187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fussell KC, Udasin RG, Smith PJ, Gallo MA, Laskin JD. 2011. Catechol metabolites of endogenous estrogens induce redox cycling and generate reactive oxygen species in breast epithelial cells. Carcinogenesis 32(8):1285–1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JY, Zhang J. 2004. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5(10):R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonsette RE. 2008. Neurodegeneration in multiple sclerosis: the role of oxidative stress and excitotoxicity. J Neurol Sci 274(1–2):48–53 [DOI] [PubMed] [Google Scholar]
- Guillemard E, Varano B, Belardelli F, Quero AM, Gessani S. 1999. Inhibitory activity of constitutive nitric oxide on the expression of alpha/beta interferon genes in murine peritoneal macrophages. J Virol 73(9):7328–7333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen TM, Huang S, Curnutte J, Fowler P, Martinez V, Wehr CM, Ames BN, Chisari FV. 1994. Extensive oxidative DNA damage in hepatocytes of transgenic mice with chronic active hepatitis destined to develop hepatocellular carcinoma. Proc Natl Acad Sci U S A 91(26):12808–12812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F, Li X, Spatrick P, Casillo R, Dong S, Jacobson A. 2003. Genome-wide analysis of mRNAs regulated by the nonsense-mediated and 5′ to 3′ mRNA decay pathways in yeast. Mol Cell 12(6):1439–1452 [DOI] [PubMed] [Google Scholar]
- Jin Y, Penning TM. 2007. Aldo-keto reductases and bioactivation/detoxication. Annu Rev Pharmacol Toxicol 47:263–292 [DOI] [PubMed] [Google Scholar]
- Kaur C, Ling EA. 2008. Antioxidants and neuroprotection in the adult and developing central nervous system. Curr Med Chem 15(29):3068–3080 [DOI] [PubMed] [Google Scholar]
- Kerr IM, Stark GR. 1991. The control of interferon-inducible gene expression. FEBS Lett 285(2):194–198 [DOI] [PubMed] [Google Scholar]
- Kimura K, Wakamatsu A, Suzuki Y, Ota T, Nishikawa T, Yamashita R, Yamamoto J, Sekine M, Tsuritani K, Wakaguri H, Ishii S, Sugiyama T, Saito K, Isono Y, Irie R, Kushida N, Yoneyama T, Otsuka R, Kanda K, Yokoi T, Kondo H, Wagatsuma M, Murakawa K, Ishida S, Ishibashi T, Takahashi-Fujii A, Tanase T, Nagai K, Kikuchi H, Nakai K, Isogai T, Sugano S. 2006. Diversification of transcriptional modulation: large-scale identification and characterization of putative alternative promoters of human genes. Genome Res 16(1):55–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtzke JF. 1983. Rating neurologic impairment in multiple sclerosis: an expanded disability status scale (EDSS). Neurology 33(11):1444–1452 [DOI] [PubMed] [Google Scholar]
- Li D, Ferrari M, Ellis EM. 2012. Human aldo-keto reductase AKR7A2 protects against the cytotoxicity and mutagenicity of reactive aldehydes and lowers intracellular reactive oxygen species in hamster V79-4 cells. Chem Biol Interact 195(1):25–34 [DOI] [PubMed] [Google Scholar]
- Liehr JG, Ulubelen AA, Strobel HW. 1986. Cytochrome P-450-mediated redox cycling of estrogens. J Biol Chem 261(36):16865–16870 [PubMed] [Google Scholar]
- Litvak V, Ratushny AV, Lampano AE, Schmitz F, Huang AC, Raman A, Rust AG, Bergthaler A, Aitchison JD, Aderem A. 2012. A FOXO3-IRF7 gene regulatory circuit limits inflammatory sequelae of antiviral responses. Nature 490(7420):421–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutfalla G, Holland SJ, Cinato E, Monneron D. Reboul J, Rogers NC, Smith JM, Stark GR, Gardiner K, Mogensen KE, et al. 1995. Mutant U5A cells are complemented by an interferon-alpha beta receptor subunit generated by alternative processing of a new member of a cytokine receptor gene cluster. EMBO J 14(20):5100–5108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald WI, Compston A, Edan G, Goodkin D, Hartung HP, Lublin FD, McFarland HF, Paty DW, Polman CH, Reingold SC, Sandberg-Wollheim M, Sibley W, Thompson A, van den Noort S, Weinshenker BY, Wolinsky JS. 2001. Recommended diagnostic criteria for multiple sclerosis: guidelines from the International Panel on the diagnosis of multiple sclerosis. Ann Neurol 50(1):121–127 [DOI] [PubMed] [Google Scholar]
- Melo A, Monteiro L, Lima RM, Oliveira DM, Cerqueira MD, El-Bacha RS. 2011. Oxidative stress in neurodegenerative diseases: mechanisms and therapeutic perspectives. Oxid Med Cell Longev 2011:467180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels ML, Cruz C, Grollman AP, Miller JH. 1992a. Evidence that MutY and MutM combine to prevent mutations by an oxidatively damaged form of guanine in DNA. Proc Natl Acad Sci USA 89:7022–7025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels ML, Miller JH. 1992. The GO system protects organisms from the mutagenic effect of the spontaneous lesion 8-hydroxyguanine (7,8-dihydro-8-oxoguanine). J Bacteriol 174(20):6321–6325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels ML, Tchou J, Grollman AP, Miller JH. 1992b. A repair system for 8-oxo-7,8-dihydrodeoxyguanine. Biochemistry 31:10964–10968 [DOI] [PubMed] [Google Scholar]
- Mogensen KE, Lewerenz M, Reboul J, Lutfalla G, Uze G. 1999. The type I interferon receptor: structure, function, and evolution of a family business. J Interferon Cytokine Res 19(10):1069–1098 [DOI] [PubMed] [Google Scholar]
- Murphy KC, Volkert MR. 2012. Structural/functional analysis of the human OXR1 protein: identification of exon 8 as the anti-oxidant encoding function. BMC Mol Biol 13(1):26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicol JW, Helt GA, Blanchard SG, Jr., Raja A, Loraine AE. 2009. The Integrated Genome Browser: free software for distribution and exploration of genome-scale datasets. Bioinformatics 25(20):2730–2731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottoboni L, Keenan BT, Tamayo P, Kuchroo M, Mesirov JP, Buckle GJ, Khoury SJ, Hafler DA, Weiner HL, De Jager PL. 2012. An RNA profile identifies two subsets of multiple sclerosis patients differing in disease activity. Sci Transl Med 4(153):153ra131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal S, Gupta R, Kim H, Wickramasinghe P, Baubet V, Showe LC, Dahmane N, Davuluri RV. 2011. Alternative transcription exceeds alternative splicing in generating the transcriptome diversity of cerebellar development. Genome Res 21(8):1260–1272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrini S, John J, Shearer M, Kerr IM, Stark GR. 1989. Use of a selectable marker regulated by alpha interferon to obtain mutations in the signaling pathway. Mol Cell Biol 9(11):4605–4612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perillo B, Ombra MN, Bertoni A, Cuozzo C, Sacchetti S, Sasso A, Chiariotti L, Malorni A, Abbondanza C, Avvedimento EV. 2008. DNA oxidation as triggered by H3K9me2 demethylation drives estrogen-induced gene expression. Science 319(5860):202–206 [DOI] [PubMed] [Google Scholar]
- Pestka S, Langer JA, Zoon KC, Samuel CE. 1987. Interferons and their actions. Annu Rev Biochem 56:727–777 [DOI] [PubMed] [Google Scholar]
- Picklo MJ, Olson SJ, Markesbery WR, Montine TJ. 2001. Expression and activities of aldo-keto oxidoreductases in Alzheimer disease. J Neuropathol Exp Neurol 60(7):686–695 [DOI] [PubMed] [Google Scholar]
- Pulit-Penaloza JA, Scherbik SV, Brinton MA. 2012. Activation of Oas1a gene expression by type I IFN requires both STAT1 and STAT2 while only STAT2 is required for Oas1b activation. Virology 425(2):71–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajapakse N, Butterworth M, Kortenkamp A. 2005. Detection of DNA strand breaks and oxidized DNA bases at the single-cell level resulting from exposure to estradiol and hydroxylated metabolites. Environ Mol Mutagen 45(4):397–404 [DOI] [PubMed] [Google Scholar]
- Ramsey SA, Knijnenburg TA, Kennedy KA, Zak DE, Gilchrist M, Gold ES, Johnson CD, Lampano AE, Litvak V, Navarro G, Stolyar T, Aderem A, Shmulevich I. 2010. Genome-wide histone acetylation data improve prediction of mammalian transcription factor binding sites. Bioinformatics 26(17):2071–2075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reder AT, Velichko S, Yamaguchi KD, Hamamcioglu K, Ku K, Beekman J, Wagner TC, Perez HD, Salamon H, Croze E. 2008. IFN-beta1b induces transient and variable gene expression in relapsing-remitting multiple sclerosis patients independent of neutralizing antibodies or changes in IFN receptor RNA expression. J Interferon Cytokine Res 28(5):317–331 [DOI] [PubMed] [Google Scholar]
- Rehermann B, Nascimbeni M. 2005. Immunology of hepatitis B virus and hepatitis C virus infection. Nat Rev Immunol 5(3):215–229 [DOI] [PubMed] [Google Scholar]
- Rojas-Duran MF, Gilbert WV. 2012. Alternative transcription start site selection leads to large differences in translation activity in yeast. RNA 18(12):2299–2305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy D, Cai Q, Felty Q, Narayan S. 2007. Estrogen-induced generation of reactive oxygen and nitrogen species, gene damage, and estrogen-dependent cancers. J Toxicol Environ Health B Crit Rev 10(4):235–257 [DOI] [PubMed] [Google Scholar]
- Russell-Harde D, Knauf M, Croze E. 1995. The use of Zwittergent 3–14 in the purification of recombinant human interferon-beta Ser17 (Betaseron). J Interferon Cytokine Res 15(1):31–37 [DOI] [PubMed] [Google Scholar]
- Shao W, Halachmi S, Brown M. 2002. ERAP140, a conserved tissue-specific nuclear receptor coactivator. Mol Cell Biol 22(10):3358–3372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark GR, Darnell JE., Jr.2012. The JAK-STAT pathway at twenty. Immunity 36(4):503–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark GR, Kerr IM, Williams BR, Silverman RH, Schreiber RD. 1998. How cells respond to interferons. Annu Rev Biochem 67:227–264 [DOI] [PubMed] [Google Scholar]
- Tenoever BR, Ng SL, Chua MA, McWhirter SM, Garcia-Sastre A, Maniatis T. 2007. Multiple functions of the IKK-related kinase IKKepsilon in interferon-mediated antiviral immunity. Science 315(5816):1274–1278 [DOI] [PubMed] [Google Scholar]
- Volkert MR, Wang JY, Elliott NA. 2008. A functional genomics approach to identify and characterize oxidation resistance genes. Methods Mol Biol 477:331–342 [DOI] [PubMed] [Google Scholar]
- Waddell SJ, Popper SJ, Rubins KH, Griffiths MJ, Brown PO, Levin M, Relman DA. 2010. Dissecting interferon-induced transcriptional programs in human peripheral blood cells. PLoS One 5(3):e9753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner TC, Velichko S, Chesney SK, Biroc S, Harde D, Vogel D, Croze E. 2004. Interferon receptor expression regulates the antiproliferative effects of interferons on cancer cells and solid tumors. Int J Cancer 111(1):32–42 [DOI] [PubMed] [Google Scholar]
- Wagner TC, Velichko S, Vogel D, Rani MR, Leung S, Ransohoff RM, Stark GR, Perez HD, Croze E. 2002. Interferon signaling is dependent on specific tyrosines located within the intracellular domain of IFNAR2c. Expression of IFNAR2c tyrosine mutants in U5A cells. J Biol Chem 277(2):1493–1499 [DOI] [PubMed] [Google Scholar]
- Wang B, Xiong Q, Shi Q, Le X, Abbruzzese JL, Xie K. 2001. Intact nitric oxide synthase II gene is required for interferon-beta-mediated suppression of growth and metastasis of pancreatic adenocarcinoma. Cancer Res 61(1):71–75 [PubMed] [Google Scholar]
- Wang JY, Sarker AH, Cooper PK, Volkert MR. 2004. The single-strand DNA binding activity of human PC4 prevents mutagenesis and killing by oxidative DNA damage. Mol Cell Biol 24(13):6084–6093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watling D, Guschin D, Muller M, Silvennoinen O, Witthuhn BA, Quelle FW, Rogers NC, Schindler C, Stark GR, Ihle JN, et al. 1993. Complementation by the protein tyrosine kinase JAK2 of a mutant cell line defective in the interferon-gamma signal transduction pathway. Nature 366(6451):166–170 [DOI] [PubMed] [Google Scholar]
- Weitzman SA, Gordon LI. 1990. Inflammation and cancer: role of phagocyte-generated oxidants in carcinogenesis. Blood 76(4):655–663 [PubMed] [Google Scholar]
- Yamashita R, Sathira NP, Kanai A, Tanimoto K, Arauchi T, Tanaka Y, Hashimoto S, Sugano S, Nakai K, Suzuki Y. 2011. Genome-wide characterization of transcriptional start sites in humans by integrative transcriptome analysis. Genome Res 21(5):775–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Kramer EG, Mahase S, Dutta DJ, Bonnamain V, Argaw AT, John GR. 2011. Targeting oligodendrocyte protection and remyelination in multiple sclerosis. Mt Sinai J Med 78(2):244–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.