Abstract
Purpose:
The American College of Medical Genetics and Genomics released practice guidelines recommending reporting of incidental findings from exome and whole-genome sequencing by massively parallel (next-generation) sequencing for multiple conditions. Policy statements from other agencies are still being developed, and many attempt to take into consideration the predicted increase in workload caused by reporting incidental findings. We describe the effects of changing the sensitivity and the specificity, as well as the implications of varying diagnostic criteria and a priori prevalence, and those of increasing the number of included conditions, on rates of incidental findings.
Methods:
We developed a simple mathematical model based on binomial probability for predicting rates of incidental findings. We primed and validated the model using published variant frequencies.
Results:
The model correctly calculates observed rates of incidental findings. Changing the model's parameters shows that even minor changes in diagnostic criteria or sequencing accuracy cause large variation in rates of incidental findings.
Conclusion:
Our model correctly explains observed rates of incidental findings. Key drivers of rates include diagnostic criteria, variant frequency, disease penetrance, and sequencing and bioinformatics accuracy. Rates of incidental findings are relatively insensitive to even large increases in the number of conditions included.
Keywords: incidental findings, massively parallel sequencing, result reporting, whole-genome sequencing
Introduction
Massively parallel sequencing, also called next-generation sequencing, has dramatically lowered the cost of genomic sequencing for multiple genetic conditions. Best ethical and clinical practice for clinical diagnostic testing using traditional targeted sequencing technologies requires obtaining informed consent and ensuring the availability of genetic counseling. Testing for specific genetic conditions using massively parallel sequencing poses new challenges in terms of ensuring that informed consent is obtained because testing may discover off-target genetic conditions, known as incidental findings (IFs).1,2
The American College of Medical Genetics and Genomics (ACMG) recently published recommendations proposing the mandated clinical reporting of IFs for 24 autosomal dominant (AD) (including one semidominant) conditions.3,4,5 These conditions were selected because they are highly penetrant, asymptomatic for long periods of time, and amenable to preventive measures and/or treatments. Controversially, the ACMG recommended that genetic identification of the 24 conditions be sought and reported “without reference to patient preferences” both because of the high potential benefit to patients and because individual informed consent seemed logistically unfeasible. In its policy statement the ACMG predicted “1% of sequencing reports will include an incidental variant” from the list of 24. Other authors applied the ACMG recommendations to various data sets and found rates of IFs varying from 1.2 to 11%.6,7,8,9
We describe a simple mathematical model that calculates the rate at which AD IFs would occur in a data set or a population. Our model is based on binomial distribution, and input requires only an estimate (or range of estimates) of gene frequencies of variants (including mutations). We primed the model with variant frequencies drawn from the literature. We then validated the model by comparing its predicted rates of IFs with those found from other data sets.
Assuming that exome and whole-genome sequencing becomes increasingly routine, it is likely that the lists of recommended conditions to be reported will expand. The ACMG has recommended that only variants highly likely to be pathogenic be included among the genes for which reporting is mandatory. As the establishment of reference databases of variation continues to expand, new variants are being found and the classification of variants into pathogenic versus nonpathogenic is continuing to change.10 We used our model to study the effects of increasing the number of AD conditions included. We also used our model to predict how changes in the frequencies of variants, or of variants of unknown significance (VUS), might affect the rates of reporting of IFs; finally, we define the theoretical limits of rates of reporting of IFs.
Materials and Methods
In silico model
A simple mathematical model was developed based on binomial distribution.11
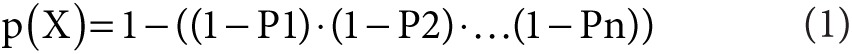 |
p(X) represents the probability of reporting at least one IF, and P1 to Pn are the pathogenic variant frequencies of the various genetic conditions.
The model assumes all conditions are inherited independently. We used gene variant prevalence data when available. For those conditions for which variant prevalence data were not available, we estimated them by assuming they were the same as the observed disease prevalence with 100% penetrance. Variations in penetrance were addressed by sensitivity analysis over a range of variant rates (described below).
Validation of model
A diagnostic panel was constructed in silico based on the 24 ACMG-recommended minimum list of genes to be reported.3 Prevalence data were obtained from the literature (Table 1). When a range of prevalence data was available, the lowest and highest values were selected, and the most likely estimate was calculated as the geometric mean. Alternatively, when only a single datum was available, half and twice this prevalence were selected as the low and high estimates, respectively.
Table 1. Prevalence and frequencies of gene variants for the 24 ACMG conditions.
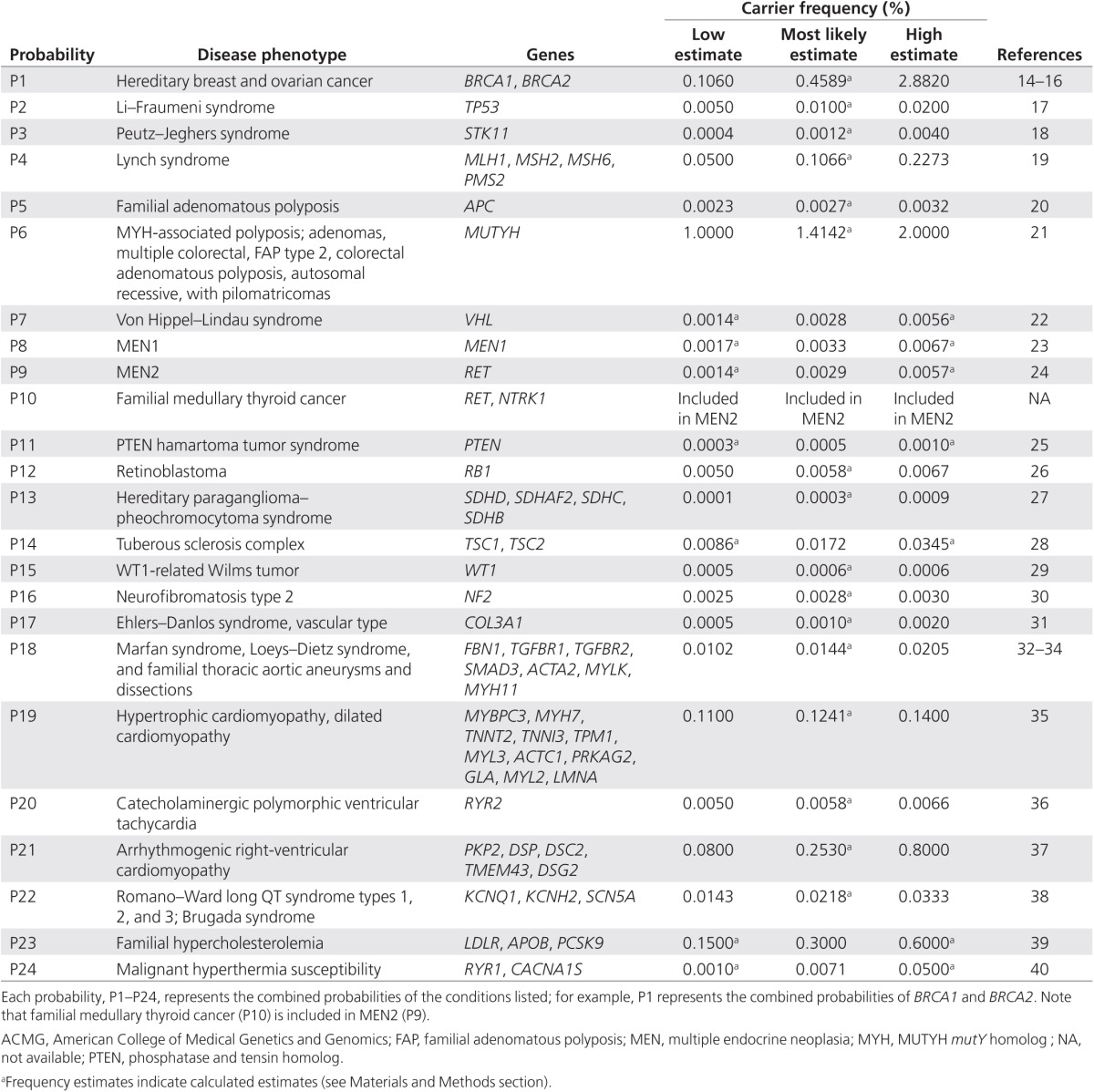
The predicted rate of IFs was calculated by applying these gene frequencies (Table 1) to our model (Eq. 1). Calculations were repeated separately for the lower and higher limits.
Assessing the impact of deviations from reported variant prevalence rates
To simulate the effect of altering variant prevalence rates (because of changes in variant classification, variations in disease penetrance, inaccuracies in the literature data, or differences in populations), we performed sensitivity analysis by repeating our calculations over a range of three log2 orders of magnitude of variant frequencies, using one-quarter or one-half the lower or twice the upper reported estimates of prevalence for all conditions in Table 1. This simulation would also account for errors in sequencing or incorrect annotations in variant databases resulting in incorrectly calling variants pathogenic, nonpathogenic, or VUS. The range of values was chosen to cover the range of described inaccuracies of current variant databases.10,12
Simulating the effects of increasing the number of tested conditions
To calculate the increase in reporting frequency that occurs with the inclusion of additional conditions, we first ordered the list of variant frequencies from most to least frequent. We then successively considered including only the most common condition, the two most common conditions, the three most common conditions, and so forth, until all conditions were included for the calculation of the cumulative frequency of predicted significant findings. To determine the incremental contribution of each additional condition tested to the overall rate of findings, we calculated the percentage difference between the predicted frequency of findings for the first n most common conditions and the (n + 1) next most common condition. We identified the value of n at which there was less than a 1% relative increase in findings, as well as the value of n at which the relative increase in findings was less than 0.1%.
To simulate the effect of further increasing the number of included conditions in some future panel of recommended reporting conditions, we extended our model. We allocated each of the conditions examined into one of 10 “bins” based on orders of magnitude of variant frequency; these bins ranged from 2, 1, 0.1, 0.01, and 0.001 down to 10–8%. This allocation was done in two separate experiments. Each condition was initially assigned to the bin with the closest frequency that it did not exceed (to produce a maximum estimate), and then each was assigned to the bin with closest frequency that it did exceed (to produce a minimum estimate). We repeated the in silico simulation of all conditions in Table 1 to validate the modification of the model. We then observed the effects of changing the number of conditions in each variant frequency bin.
Results
IFs and AD inheritance
Applying our model to the proposed ACMG-recommended screening panel of 24 conditions (Table 1), we calculated that ~2.7% (range: 1.5–6.5%) of screened individuals would have an IF (Tables 2 and 3).
Table 2. Modeling how the overall rates of IFs are influenced by prevalence rates for the 24 ACMG conditions.
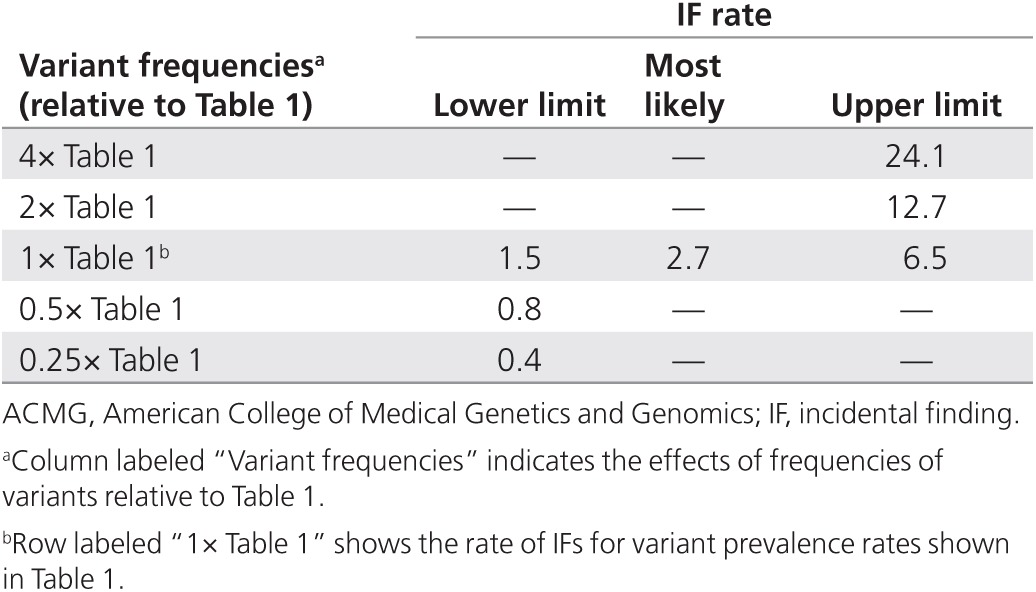
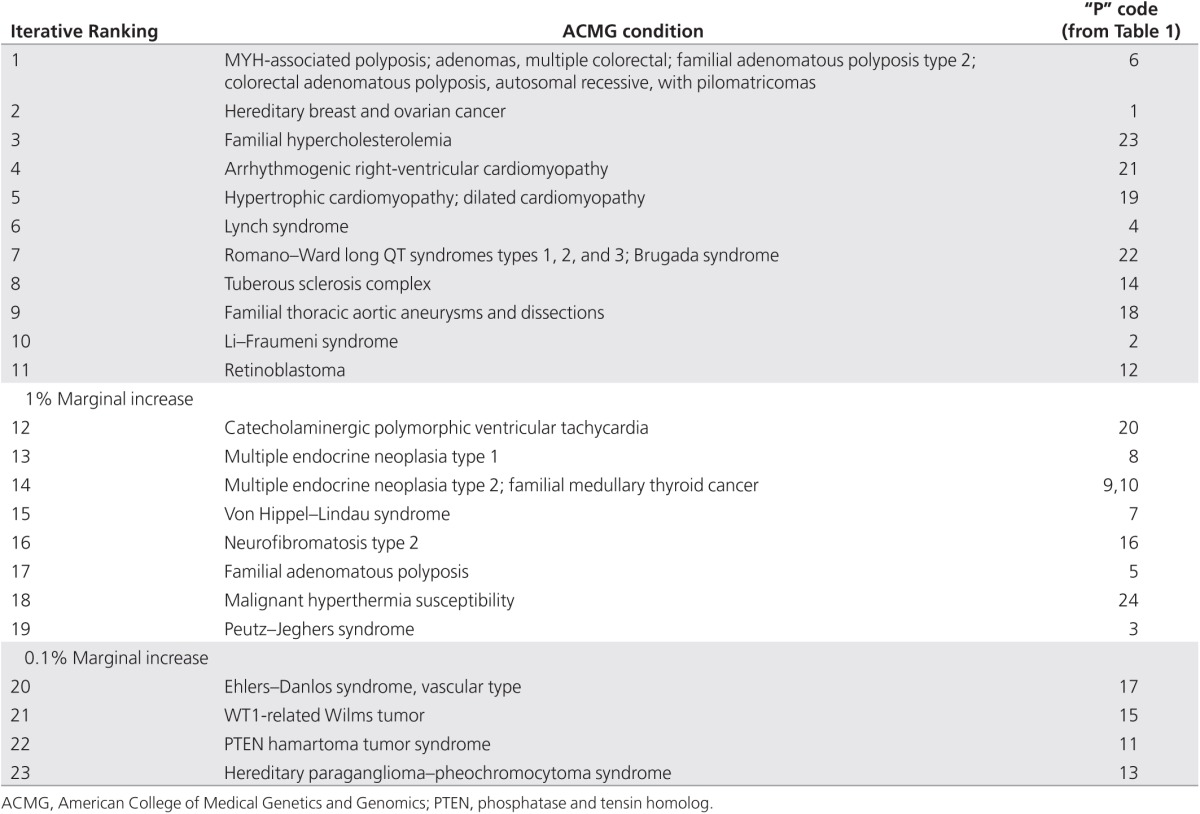
Table 3. ACMG condition and identifier code from Table 1.
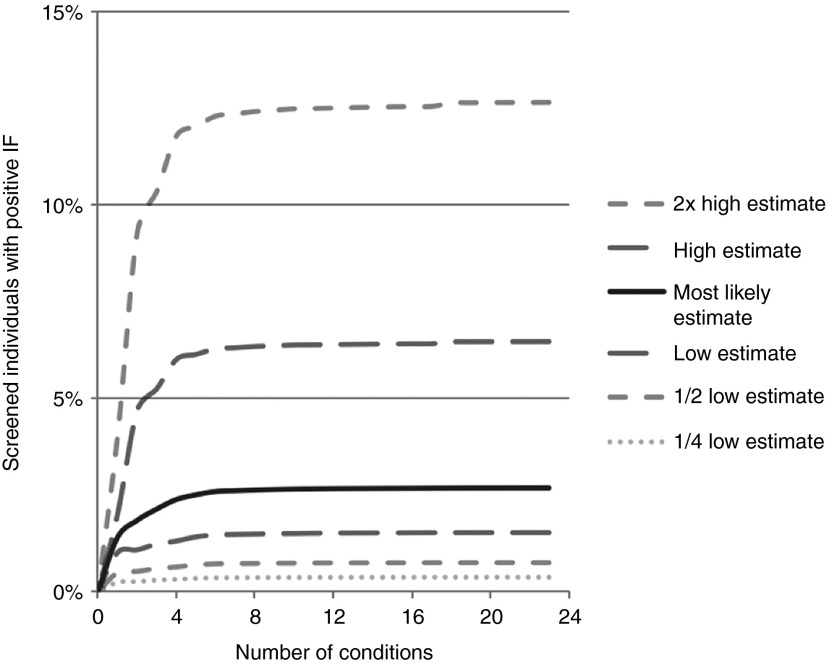
We considered the impact of IFs on the rate of reporting if variant frequencies were to change or if there were sequencing errors or errors in the subsequent bioinformatics analyses and database annotations. For each condition in Table 1, we repeated our calculations by assuming the highest and lowest variant prevalence rates were incorrect by a factor of two or four (Figure 1 and Tables 2 and 3).
Figure 1.
Effects on rate of incidental findings (IFs) over a range of variant frequencies and prevalence. For each of the 24 American College of Medical Genetics and Genomics conditions, we determined the proportion of screened individuals who would have a reported IF for varying prevalence rates of each condition. Table 1 lists the lowest, most likely, and highest rates as determined from literature review, and these are graphically displayed here. We then repeated our calculations over a three log2 order of magnitude range of variant frequencies, using one-quarter or one-half the lower or twice the upper reported estimates of variant prevalence.
Increasing the list of mandatory reporting conditions
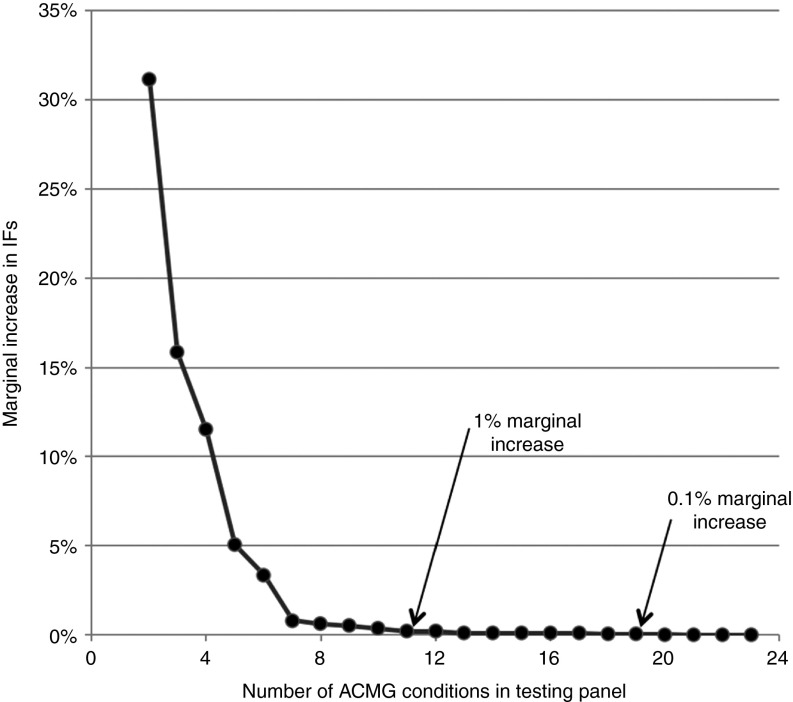
We studied the effects of increasing the number of tested AD conditions on the predicted number of IFs reported. Commencing with the one condition in the ACMG list with the highest variant prevalence, we simulated the effects of testing for only that one condition, or for that condition plus the next most prevalent condition, or for those two conditions plus the next most prevalent condition, and so on, until all 24 conditions were included. The resulting rates of IFs and the marginal increase in IFs with each addition to the number of conditions tested are shown in Figure 2. Using only the seven genes with the highest variant prevalence of the 24 genes recommended for mandatory reporting contributed 97% toward the total number of predicted IFs; including the 11 most prevalent of the 24 genes contributed 99%, and by the time 19 of the 24 genes had been considered, more than 99.9% of all IFs discoverable with the full ACMG list would have been reported.
Figure 2.
Marginal increases in the number of reported incidental findings (IFs). As described in the Materials and Methods section, we calculated the marginal increase in IFs by iteratively considering the effect of adding to the first n most common conditions, one additional condition, until all 24 conditions were included for the calculation of the cumulative frequency of predicted IFs. Each data point in the series represents the relative marginal increase in IFs beyond the IFs identified by the previous point in the series. We identified the value of n at which there was less than a 1% relative increase in IFs (occurring at n = 11) and the value of n at which the relative increase in IFs was less than 0.1% (occurring at n = 19). Table 3 indicates the sorted order of analyses of the 24 conditions from Table 1.
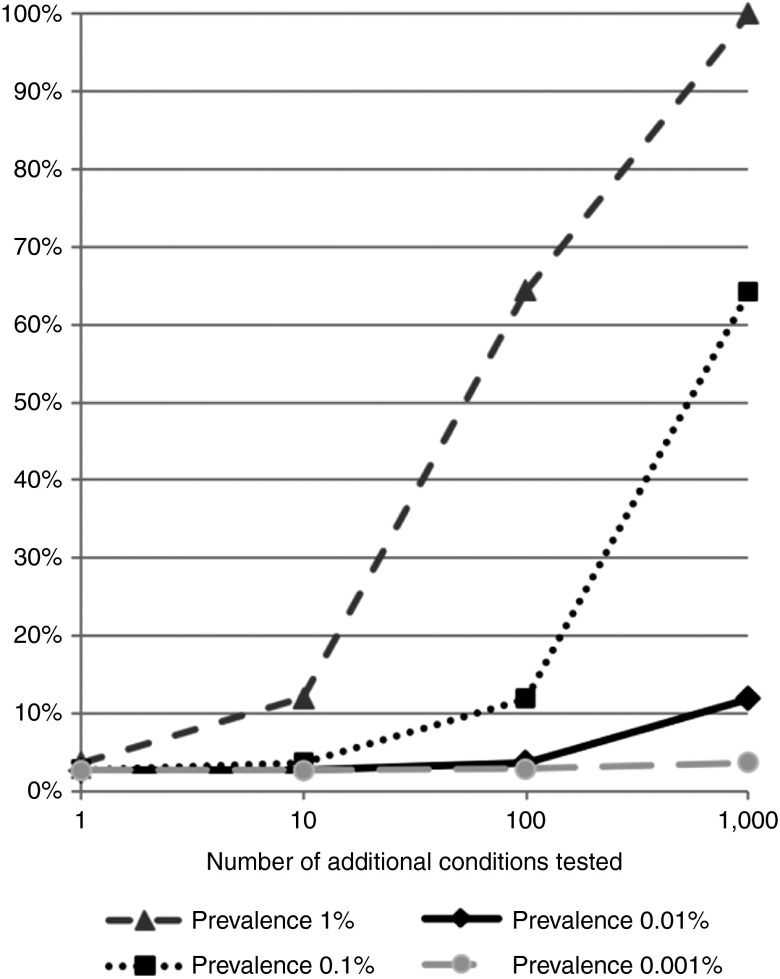
To generalize our understanding of the rate at which significant findings will be reported as the number of conditions being considered is increased, we modified our model slightly by introducing “bins” of logarithmic variant frequencies. This modification did not significantly change our calculation of the rate of IFs for the ACMG panel of AD conditions: the range of IFs predicted by our original model (1.5–6.5%; Table 1) became 1.5–6.3% using the binned model. However, using the binned model, we were able to simulate the effect of introducing additional conditions. We found that increasing the number of conditions being considered had very different effects, depending on the variant frequencies of those conditions. Adding only a small number of conditions with high variant frequencies (>0.1%) has a large effect on the number of IFs. By contrast, the addition of 100 conditions with variant frequency of ~10–2, or even 1,000 conditions with variant frequency of ~10–3, did not have as large an effect on the number of IFs (Figure 3).
Figure 3.
Numbers of significant diagnoses with increasing numbers of tested conditions. We calculated the predicted number of additional significant findings that would be reported if we were to add to the American College of Medical Genetics and Genomics (ACMG)–recommended testing panel additional conditions of the indicated prevalence. Note that the 24 current ACMG recommendations have prevalences ranging from 1.4 to 0.0003% (see Table 1).
Discussion
Massively parallel sequencing technologies will greatly increase the number of genetic diagnoses made primarily through laboratory testing. High rates of reporting of significant genetic findings will result in downstream costs to and impact on the health system because of the need for genetic counseling, confirmatory testing, medical consultation, and potential intervention. However, the individuals identified may derive significant benefit from identification of these findings, and the high rates of reporting may ultimately provide cost benefits through improved screening and early medical intervention. The costs to implement the recommendations need to be compared with the potential benefits, cost offsets, and utility of reporting and acting on these findings.
We have developed a model to simulate the impact of changes in variant frequencies and classification and of expanding the list of conditions recommended for mandatory reporting. The impact of expanding the list of conditions for reporting can also be considered a model to demonstrate the impact of reporting VUS in addition to pathogenic mutations. We used this model to explore the likely impact of expanded testing on the rates of diagnoses of AD conditions (and especially IFs).
The rate of IFs
The implementation of the ACMG Recommendations for Clinical Reporting of Incidental Findings suggested only a modest increase in the use of health system resources, with an estimated IF frequency of ~1%.3 Recent studies by others have begun to explore these implications, initially by applying the ACMG Recommendations to various large data sets.6,8,9 These studies suggest that the rate of IFs may be higher than originally anticipated but that some of this increase may be artifactual because of measurement uncertainty in the sequencing or bioinformatics phases, or even the incorrect classification of pathogenicity.
Our modeling of the ACMG Recommendations for Clinical Reporting of Incidental Findings confirms that a nontrivial percentage (1.5–6.5%) of screened individuals will have a significant reportable finding (Tables 2 and 3). Although slightly higher than the value of ~1% IFs originally forecast by the ACMG,3 it is in broad agreement with experimental values of 1.2% (Americans with African ancestry) and 3.4% (Americans with European ancestry).8
This value is lower than those found by Xue et al.,6 who reported a rate of IFs of 11% among 179 individuals from the 1000 Genomes Project, and by Cassa et al.,9 who reported a rate of 8.5% from a set of 1,092 individuals drawn from various studies in the literature. However, both these studies included in their data sets a wider range of inheritance modes than those considered in our model.
The ACMG recommendations were based on an AD mode of inheritance, whereas Xue et al.6 included both AD and homozygous autosomal recessive conditions in their calculations. If one corrects the findings of Xue et al.6 to include only AD inherited conditions (as listed in their Table 2), a revised rate of IFs of 6.9% is obtained; further reduction in their rate of IFs might be appropriately made by noting that Xue et al.6 included yet more conditions, some of which (e.g., loose anagen hair syndrome) were pathogenic but had significantly less serious consequences to patients and some of which were disease-causing variants from databases that were incompletely validated. Noting these qualifications of Xue et al6., their adjusted rate of IFs of 6.9% is consistent with the upper limit of 6.5% predicted by our model.
Cassa et al.9 similarly included in their study a broader range of conditions and inheritance modes (and thus diagnostic triggers for reporting an IF) than the specific set of 24 conditions defined in the ACMG recommendations3; ~24% of their reported conditions were homozygous minor variants. To make their data more comparable with the AD recommendations of the ACMG, correcting their findings by excluding these 24% homozygous variants obtained a revised rate of IFs of 6.5%, and further reduction in their rate of IFs might be appropriate by noting their conclusions that at least some of the variants detected may be erroneous findings or have lower penetrance than previously expected. Again, this revised rate of 6.5% is consistent with the upper limit predicted by our model.
We conclude that our base model is fit for the purpose of describing the expected rates of IFs. However, we note the many cautions expressed by these investigators6,8,9 regarding erroneous reports and the source of possible errors, and we used our model to explore the significance of these factors.
Effects on IFs of changing the input parameters
Information about the population frequencies of rare conditions is necessarily limited, and calculations based on such limited information must be regarded with caution. Most of the estimates of variant frequency for these rare conditions are based on the prevalence of a particular disorder in a population, rather than the prevalence of pathogenic allelic variants for that condition.
Because our model relies on estimated rates of variants of these genetic conditions rather than prevalence of the conditions themselves, we must rely on some implicit assumptions.6,8,9,12,13 To derive rates of variant prevalence from rates of condition prevalence, our calculations assume 100% penetrance, 100% identification of causative variants (sensitivity), and 100% specificity. In reality, these assumptions are unlikely to hold true. For example, the degree of penetrance and expressivity, and thus the true variant frequency, is not known for many genetic conditions13; this would lead to an incorrect estimation of the predicted number of IFs. Some conditions (such as malignant hyperthermia) may manifest themselves only after a rare environmental event, and should this event not be encountered during the life of the individual, then the true incidence of these conditions will be underestimated. Furthermore, diseases such as these, which have had their genetic basis determined by sequencing high-risk patients and looking for the common themes, are likely to overestimate the penetrance of the associated genetic cause.12 Finally, because some pathogenic phenotypes will be due to genetic variations not yet described or currently considered VUS, estimation of the number of reported IFs will contain additional imprecision. Even experts may disagree when clinical cases are reviewed: manual curation is time consuming and, even then, what is actionable may not always be agreed on.6,8,13
To attempt to account for these various uncertainties in the estimates, we performed a sensitivity analysis of our model. We studied the effect on the predicted rate of IFs by varying the estimated prevalence of all conditions over a range of values. We chose factors of two- and fourfold increase above or decrease below the most likely values; this range would be sufficient to encompass reported alterations in misclassifications of variants in major databases.10 As noted by Johnston et al.,7 when the goal is to identify only variations likely to be causative and to minimize false positives, laboratory and bioinformatics decisions need to make a trade-off between diagnostic sensitivity and specificity because small variations in the diagnostic decision matrix will greatly affect the apparent rate of IFs. We found that the rate at which IFs are reported is highly dependent on the actual clinical prevalence of the condition and the prevalence of pathogenic variants for that gene (Figure 1).
Adding to this uncertainty, the analytical processes involved in massively parallel sequencing/whole-genome sequencing are not error free. Sequencing errors,12 variations in bioinformatics assembly and pipelines,6 and limitations, inadequacies, or errors in the annotations of current databases6,10,12 all introduce significant measurement uncertainty and analytical error, which can lead to over- or underreporting of IFs.
Figure 1 shows that errors in sequencing and in bioinformatics analysis after sequencing, errors in database annotations, and difficulties in determining the true incidence of disease and the true prevalence of pathogenic variants all are major determinants of the rate of IFs. When other parameters are held constant, a 4-fold change in the value of any one of these key drivers can produce up to a 10-fold difference in the rate of IFs (Tables 2 and 3).
Effects of increasing the number of AD conditions in the ACMG recommendations
Ongoing research continues to reveal the genetic basis for more and more conditions. The ACMG list of recommendations is likely to grow over time. We therefore modeled the likely impact of this list increasing in number. Although the number of IFs reported will increase as the number of tested conditions increases, our modeling suggests this increase is not linear. This nonlinearity is partly because of the underlying mathematics of the binomial calculations used in the model and partly because (we assume) the most common monogenic, AD, highly penetrant conditions have already been identified and included. A consequence of this nonlinear behavior is that there is a diminishing marginal increase in reported IFs as more conditions are added. From the ACMG-recommended list of 24 conditions, those with the highest variant frequencies contribute between 99 and 99.9% (the top 11 and 19 conditions, respectively) to potential IFs (Figure 2).
We modeled the effects of expanding the list of conditions tested by adding more AD conditions. We found that the most significant driver of the rate of IFs is not the number of additional conditions per se (Figure 2) but rather the variant frequency of the most common of these additional conditions (Figure 3). If we assume that the current ACMG list already contains the most frequent of the AD conditions to be considered, then expanding the list to 100 or even 1,000 additional conditions only modestly increases the rate of IFs. However, adding even a single condition whose variant frequency is of the order of 1% sharply increases the number of IFs. These effects of variant frequencies on increasing numbers of IFs can also be used to model the effect of reporting VUS for these conditions.
Conclusion
We developed and validated a simple model that allows the effects of various lists of AD conditions to be simulated and allows the rates of reporting of IFs and genetic carriers to be readily estimated. Our model shows that these rates are highly dependent on the apparent prevalence of included conditions, the actual prevalence of the genetic variants that cause these conditions, and the accuracy and quality of the sequencing and bioinformatics analyses.
This study has two key findings: (i) The accuracy of variant annotation of the underlying genomic databases is a significant factor in the proportion of individuals who will be flagged with a reportable finding. (ii) The proportion of individuals with IFs rapidly becomes asymptotic and self-limiting, even with the addition of many more highly penetrant AD condition/gene pairs to the list of reportable findings. There is a diminishing marginal increase in reported IFs as more mutations are added. The major benefits in identifying the most clinically significant IFs may be achieved by including as few as 11 (for 99% benefits) or 19 (for 99.9% benefits) of the currently recommended ACMG panel of 24 conditions.
However, although the proportion of individuals with IFs may become limiting and constant, different challenges will emerge as the range of potential condition/gene pairs is expanded. These will include issues such as the need for clinicians to be familiar with an ever-widening corpus of knowledge and information and the assessment of cost/utility of testing for this broadening range of conditions balanced by the availability, cost, and complexity of potential therapeutic options that may in turn be offset by a reduction in later costs through early intervention and disease prevention.
Disclosure
The authors declare no conflict of interest.
Acknowledgments
The authors thank Yemima (Mimi) Berman, Melody Caramins, Nigel Clarke, Lisa Koe, Raelia Lew, and Graeme Suthers for their critical review of the manuscript and their valuable comments.
References
- Crawford G, Foulds N, Fenwick A, Hallowell N, Lucassen A. Genetic medicine and incidental findings: it is more complicated than deciding whether to disclose or not. Genet Med. 2013;15:896–899. doi: 10.1038/gim.2013.165. [DOI] [PubMed] [Google Scholar]
- Presidential Commission for the Study of Bioethical Issues Anticipate and Communicate: Ethical Management of Incidental and Secondary Findings in the Clinical, Research, and Direct-to-Consumer Contexts . http://bioethics.gov/sites/default/files/FINALAnticipateCommunicate_PCSBI_0.pdf . 2013 [DOI] [PubMed]
- Green RC, Berg JS, Grody WW, et al. American College of Medical Genetics and Genomics ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genet Med. 2013;15:565–574. doi: 10.1038/gim.2013.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- American College of Medical Genetics and Genomics Incidental findings in clinical genomics: a clarification. Genet Med. 2013;15:664–666. doi: 10.1038/gim.2013.82. [DOI] [PubMed] [Google Scholar]
- American College of Medical Genetics and Genomics ACMG Updates Recommendation on “Opt Out” for Genome Sequencing Return of Results . http://www.acmg.net/docs/Release_ACMGUpdatesRecommendations_final.pdf . Accessed 1 May 2014.
- Xue Y, Chen Y, Ayub Q, et al. 1000 Genomes Project Consortium Deleterious- and disease-allele prevalence in healthy individuals: insights from current predictions, mutation databases, and population-scale resequencing. Am J Hum Genet. 2012;91:1022–1032. doi: 10.1016/j.ajhg.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston JJ, Rubinstein WS, Facio FM, et al. Secondary variants in individuals undergoing exome sequencing: screening of 572 individuals identifies high-penetrance mutations in cancer-susceptibility genes. Am J Hum Genet. 2012;91:97–108. doi: 10.1016/j.ajhg.2012.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorschner MO, Amendola LM, Turner EH, et al. National Heart, Lung, and Blood Institute Grand Opportunity Exome Sequencing Project Actionable, pathogenic incidental findings in 1,000 participants' exomes. Am J Hum Genet. 2013;93:631–640. doi: 10.1016/j.ajhg.2013.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassa CA, Tong MY, Jordan DM. Large numbers of genetic variants considered to be pathogenic are common in asymptomatic individuals. Hum Mutat. 2013;34:1216–1220. doi: 10.1002/humu.22375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson BA, Spurdle AB, Plazzer JP, et al. Application of a 5-tiered scheme for standardized classification of 2,360 unique mismatch repair gene variants in the InSiGHT locus-specific database. Nat Genet. 2014;46:107–115. doi: 10.1038/ng.2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiegel MR, Boxer RW. Schaum's Outline of Theory and Problems of Statistics SI (Metric) Edition. McGraw-Hill, New York; 1981. [Google Scholar]
- Kohane IS, Hsing M, Kong SW. Taxonomizing, sizing, and overcoming the incidentalome. Genet Med. 2012;14:399–404. doi: 10.1038/gim.2011.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krier JB, Green RC. Management of Incidental Findings in Clinical Genomic Sequencing. Wiley; Hoboken, NJ; 2001. [DOI] [PubMed] [Google Scholar]
- Ford D, Easton DF, Peto J. Estimates of the gene frequency of BRCA1 and its contribution to breast and ovarian cancer incidence. Am J Hum Genet. 1995;57:1457–1462. [PMC free article] [PubMed] [Google Scholar]
- Antoniou AC, Pharoah PD, McMullan G, et al. A comprehensive model for familial breast cancer incorporating BRCA1, BRCA2 and other genes. Br J Cancer. 2002;86:76–83. doi: 10.1038/sj.bjc.6600008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittemore AS, Gong G, John EM, et al. Prevalence of BRCA1 mutation carriers among U.S. non-Hispanic Whites. Cancer Epidemiol Biomarkers Prev. 2004;13:2078–2083. [PubMed] [Google Scholar]
- Schneider K, Zelley K, Nichols KE, Garber J.Li-Fraumeni syndrome. Pagon RA, Adam MP, Ardinger HH.et al (eds.) GeneReviews University of Washington; Seattle, WA; 2013 [PubMed] [Google Scholar]
- McGarrity TJ, Amos CI, Frazier ML, Wei C.Peutz-Jeghers syndrome. Pagon RA, Adam MP, Ardinger HH.et al (eds.). GeneReviews University of Washington; Seattle, WA; 2013 [PubMed] [Google Scholar]
- Kohlmann W, Gruber SB.Lynch syndrome. Pagon RA, Adam MP, Ardinger HH.et al (eds.). GeneReviews University of Washington; Seattle, WA; 2012 [Google Scholar]
- Jasperson KW, Burt RW.APC-associated polyposis conditions. Pagon RA, Adam MP, Ardinger HH.et al (eds.). GeneReviews University of Washington; Seattle, WA; 2011 [PubMed] [Google Scholar]
- Brand R, Nielsen M, Lynch H,, Infante E.MUTYH-associated polyposis. Pagon RA, Adam MP, Ardinger HH.et al (eds.). GeneReviews University of Washington; Seattle, WA; 2012 [Google Scholar]
- Frantzen C, Links TP,, Giles RH.Von Hippel-Lindau disease. Pagon RA, Adam MP, Ardinger HH.et al (eds.). GeneReviews University of Washington; Seattle, WA; 2012 [Google Scholar]
- Giusti F, Marini F, Brandi ML.Multiple endocrine neoplasia type 1. Pagon RA, Adam MP, Ardinger HH.et al (eds.). GeneReviews University of Washington; Seattle, WA; 2012 [PubMed] [Google Scholar]
- Moline J, Eng C.Multiple endocrine neoplasia type 2. Pagon RA, Adam MP, Ardinger HH.et al (eds.). GeneReviews University of Washington; Seattle, WA; 2013 [PubMed] [Google Scholar]
- Eng C.PTEN hamartoma tumor syndrome (PHTS). Pagon RA, Adam MP, Ardinger HH.et al (eds.). GeneReviews University of Washington; Seattle, WA; 2012 [PubMed] [Google Scholar]
- Lohmann DR, Gallie BL.Retinoblastoma. Pagon RA, Adam MP, Ardinger HH.et al (eds.). GeneReviews University of Washington; Seattle, WA; 2013 [PubMed] [Google Scholar]
- Gimenez-Roqueplo P.A.-P.Hereditary pheochromocytoma-paraganglioma . http://www.orpha.net/consor/cgi-bin/OC_Exp.php?Lng=EN&Expert=29072 . Accessed 19 July 2014.
- Northrup H, Koenig MK, Au KS.Tuberous sclerosis complex. Pagon RA, Adam MP, Ardinger HH.et al (eds.). GeneReviews University of Washington; Seattle, WA; 2011 [PubMed] [Google Scholar]
- Dome JS, Huff V.Wilms tumor overview. Pagon RA, Adam MP, Ardinger HH.et al (eds.). GeneReviews University of Washington; Seattle, WA; 2013 [Google Scholar]
- Evans DG.Neurofibromatosis 2. Pagon RA, Adam MP, Ardinger HH.et al (eds.). GeneReviews University of Washington; Seattle, WA; 2011 [Google Scholar]
- Pepin MG, Byers PH.Ehlers-Danlos syndrome type IV. Pagon RA, Adam MP, Ardinger HH.et al (eds.). GeneReviews University of Washington; Seattle, WA; 2011 [Google Scholar]
- Dietz HC.Marfan syndrome. Pagon RA, Adam MP, Ardinger HH.et al (eds.). GeneReviews University of Washington; Seattle, WA; 2011 [Google Scholar]
- Milewicz DM, Regalado E.Thoracic aortic aneurysms and aortic dissections. Pagon RA, Adam MP, Ardinger HH.et al (eds.). GeneReviews University of Washington; Seattle, WA; 2012 [Google Scholar]
- Clouse WD, Hallett JW, Jr, Schaff HV, et al. Acute aortic dissection: population-based incidence compared with degenerative aortic aneurysm rupture. Mayo Clin Proc. 2004;79:176–180. doi: 10.4065/79.2.176. [DOI] [PubMed] [Google Scholar]
- Cirino AL, Ho C.Familial hypertrophic cardiomyopathy overview. Pagon RA, Adam MP, Ardinger HH.et al (eds.). GeneReviews University of Washington; Seattle, WA; 2014 [PubMed] [Google Scholar]
- Napolitano C, Priori SG, Bloise R.Catecholaminergic polymorphic ventricular tachycardia. Pagon RA, Adam MP, Ardinger HH.et al (eds.). GeneReviews University of Washington; Seattle, WA; 2013 [PubMed] [Google Scholar]
- McNally E, MacLeod H, Dellefave-Castillo L.Arrhythmogenic right ventricular dysplasia/cardiomyopathy, autosomal dominant. Pagon RA, Adam MP, Ardinger HH.et al (eds.). GeneReviews University of Washington; Seattle, WA; 2014 [Google Scholar]
- Alders M, Mannens M.Romano-Ward syndrome. Pagon RA, Adam MP, Ardinger HH.et al (eds.). GeneReviews University of Washington; Seattle, WA; 2012 [Google Scholar]
- Rader DJ, Cohen J, Hobbs HH. Monogenic hypercholesterolemia: new insights in pathogenesis and treatment. J Clin Invest. 2003;111:1795–1803. doi: 10.1172/JCI18925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg H, Sambuughin N, Riazi S, Dirksen R.Malignant hyperthermia susceptibility. Pagon RA, Adam MP, Ardinger HH.et al (eds.). GeneReviews University of Washington; Seattle, WA; 2013 [PubMed] [Google Scholar]