Abstract
A bovine interferon α (BoIFNα) gene that included signal sequence was amplified from bovine liver genomic DNA. The gene was named BoIFN-α1 according to the position at which the encoded gene of the bovine IFN was located in the bovine genome. The sequence included a 23-amino-acid signal peptide and a 166-amino-acid mature peptide. The structural characteristics and phylogenetic relationships of the BoIFN-α1 gene were analyzed. A recombinant mature BoIFN-α1 (rBoIFN-α1) was expressed in the yeast Pichia pastoris. Physicochemical characteristics and antiviral activity were determined in vitro. Recombinant BoIFN-α1 was found to be highly sensitive to trypsin and stable at pH 2.0 or 65°C. It also exhibited antiviral activity, which was neutralized by a rabbit anti-rBoIFNα polyclonal antibody. This study revealed that rBoIFN-α1 has the typical characteristics of IFNα and can be used for both research and industrial application.
Introduction
Interferon alphas (IFNαs) are encoded by a family of closely related intronless genes in all mammalian species. Multiple genes that encode IFNα subtypes have been identified in several mammalian species, such as human, equine, bovine, canine, and mouse species (Roberts and others 1998). The bovine IFNα gene cluster is distributed over ∼570 kb across the short arm of chromosome 8, resulting in at least 10 to 12 distinct bovine IFNα proteins. Thus, the bovine IFNα subtypes show more than 92% nucleotide sequence identity and at least 90% amino sequence identity. Bovine interferon α (BoIFNα) contains 162 to 168 aa, ∼65% homology relative to the human IFNα, and ∼57% homology relative to mouse IFNα (Oritani and others 2001).
Despite the high degree of identity among the IFNα subtypes from the same species, differences in function and magnitude of biologic activities are often observed. Different subtypes of human IFNα (huIFNα) exhibit various antiviral activities. Among all the subtypes, HuIFNα8 is reportedly the most potent, whereas huIFNα1 provides the lowest antiviral activity (Fostern and others 1996). Several authors have reported that murine IFNα (muIFNα4) was 5 to 10 times more active than muIFNα1 (Van and others 1998). Thus, the biological activities among these proteins vary even though they are from the same species, and each member of the IFNα family should be characterized.
In this study, a bovine IFNα gene, with a sequence that has 100% identity with the sequence The National Center for Biotechnology Information Identity XP_001251758.1 interferon alpha-H [Bos taurus], was amplified. The gene was named BoIFN-α1 according to the position of the encoded gene of the bovine IFN at the bovine genome (Zhao and others 2009). Sequence secondary structural analysis and phylogenetic relationships of the BoIFN-α1 gene were conducted. The gene that encodes the mature peptide of BoIFN-α1 was cloned and expressed by Pichia pastoris/pPICZaA. Physicochemical characteristics and antiviral effects of purified recombinant mature BoIFN-α1 (rBoIFN-α1) were tested using Vesicular stomatitis virus (VSV) as a model. The results clearly showed that rBoIFN-α1 possesses the typical characteristics of IFNα, and that it exhibits significant antiviral activity against VSV in Madin–Darby bovine kidney (MDBK) cells. Thus, BoIFN-α1 is a potential antiviral agent against infectious bovine diseases.
Materials and Methods
Escherichia coli and yeast strains
Escherichia coli DH5α (New England Biolabs) was used to construct the recombinant plasmid. Yeast P. pastoris GS115 was used as a host strain to express the recombinant plasmid.
Cell and virus strain
MDBK cell was maintained in Dulbecco's modified Eagle's medium supplemented with 10% heat-inactivated fetal bovine serum, penicillin (100 U/mL), and streptomycin (100 μg/mL). VSV (Indiana strain) was stored at −70°C. The cell and virus were purchased from the China Institute of Veterinary Drug Control. The 50% tissue culture infectious dose (TCID50) was determined through the serial titration of virus in MDBK cells.
Rabbit polyclonal antibody anti-bovine IFNα
The polyclonal antibody was prepared in our laboratory, and recombinant bovine IFNα-A was the antigen used to obtain the polyclonal antibody.
Cloning of bovine IFN-α1 gene and structure characteristics analysis
A bovine IFNα that included signal sequences was amplified from bovine liver genomic DNA by a polymerase chain reaction (PCR). The PCR program was as follows: 94°C for 5 min, 30 cycles of 94°C for 50 s, 63°C for 30 s, 72°C for 1 min, and 72°C for 10 min. The product was inserted into a pMD18-T vector (TaKaRa) and sent for sequencing. The putative N-glycosylation sites of the bovine IFNα gene were found using the NetNGlyc Web site (www.cbs.dtu.dk/services/NetNGlyc), and the YinOYang (www.cbs.dtu.dk/services/YinOYang/) was used to predict the putative O-glycosylation sites. Secondary structure elements were predicted using the algorithms available from Network Protein Sequence (www.npsa-pbil.ibcp.fr.).
Phylogeny reconstruction
The multiple sequence alignment of bovine IFNα was compared with its counterparts in other animals using the program ClustalX. The sequences used in the comparison were retrieved from the GenBank. A phylogenetic tree was constructed using MEGA5.0 and the Neighbor-Joining method with a bootstrap of 1,000 repetitions (Tamura and others 2011). The IFNβ of horse and sheep were chosen as outgroups.
Construction of the vector for secreted expression of BoIFN-α1
The E. coli/P. pastoris shuttle vector pPICZαA was used to obtain the secretive expression of the target proteins. A pair of primers was designed to amplify the gene encoding the mature peptide and express mature rBoIFN-α1 in yeast P. pastoris. The forward primer was 5′AGCTCTCGAGAAAAGATGCCACCTGCCTC ACTCC3′, and the reverse primer was 5′CCGGAATTCAACCAGGTGTGTGTCAG TCC3′. The underlined sequence in the forward primer indicated an Xho I restriction site, which was in-framed with the start codon of α-factor secretion signal on pPICZαA. The reverse primer indicated an EcoR I restriction site denoted as the underscored part. The PCR amplified the BoIFN-α1 gene under the following conditions: The first step was initiated at 94°C for 5 min, followed by 30 cycles of 94°C for 30 s, 58°C for 30 s, and 72°C for 1 min; the final extension was conducted at 72°C for 10 min. A 521 bp PCR product was recovered from the agarose gel, digested with Xho I and EcoR I, and cloned into the yeast expression vector pPICZαA (Invitrogen), which was predigested with the same enzymes. The construct was designated as pPICZαA-BoIFN-α1.
Transformation and screening of P. pastoris
The constructed recombinant vector pPICZαA-BoIFN-α1 was linearized by Pme I, and 10 μg of the linearized plasmid was electroporated into 100 μL of the competent cells of P. pastoris GS115 according to the user's manual of Invitrogen with minor modification (Sambrook and others 1992). The transformants were selected at 30°C on the Yeast Extract Peptone Dextrose Medium (YPDS) agar plates (10 g/L yeast extract, 20 g/L peptone, 20 g/L dextrose, 20 g/L agar, and 1 M sorbitol) containing 100 μg/mL Zeocin for 3 to 4 days. Single colonies of the transformants were randomly selected from the plates and were shaken in YPD medium at 30°C and 250 rpm. After 48 h, the recombinant Pichia genomic DNA was prepared through the boiling–freezing–boiling method (Hai and others 2003). The recombinant gene integration was verified by PCR, using yeast genomic DNA as a template. The PCR primer sets included the universal primers 5′AOX1/3′AOX1 and the specifically forward/reverse primer. The parameters of the PCR program were used in the PCR amplification. The detected positive transformants were used in the succeeding expression. The sequences of the above primers are shown in Table 1.
Table 1.
Sequences of the Primers
| Primers | Sequences |
|---|---|
| Forward | 5′ AGCTCTCGAGAAAAGATGCCACCTGCCTCACTCC3′ |
| Reverse | 5′ CCGGAATTCAACCAGGTGTGTGTCAGTCC3′ |
| 5′AOXI | 5′GACTGGTTCCAATTGACAAGC3′ |
| 3′AOXI | 5′GCAAATGGCATTCTGACATCC3′ |
The sequences underlined with a single line, double lines denoted the cleavage sites of Xho I and EcoR I, respectively; the shadowed sequences denoted the cleavage sites of Kex2.
Expression and purification of pPICZαA-BoIFN-α1 in P. pastoris
The expression of pPICZαA-BoIFN-α1 in P. pastoris was conducted in a flask as described (Hou and others 2011). After harvesting the culture supernatant, the protein was purified by ammonium sulfate precipitation from the supernatant, dialyzed for 2 days against phosphate-buffered saline (PBS), filtered through a 0.22 μm filter, and stored at −70°C for further use. The final protein concentration was quantified by the BCA Protein Assay kit.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis and western blot analysis of rBoIFN-α1
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis was performed with 5% stacking gel and 15% (v/v) resolving gel and stained by Coomassie Brilliant blue G-250 (Cao 2004). For western blot analysis, after SDS-PAGE, proteins were transferred onto the nitrocellulose and the membrane was blocked for 2 h at 37°C with PBST [PBS and 0.5% Tween-20 (pH 7.4)] containing 5% skimmed milk. The membrane was incubated for 2 h at 37°C with a rabbit anti-bovine IFNα polyclonal diluted 1:250 in PBST containing 5% skimmed milk. The membrane was then incubated for 1 h at 37°C with a goat anti-rabbit IgG conjugated to horseradish peroxidase and diluted 1:5,000. Finally, the membrane was visualized by 4-chloro-1-naphthol. The rBoIFNαA, which was also expressed in P. pastoris, was used as a control.
TCID50 assay for VSV
VSV titers were determined through an endpoint dilution assay, and the titers were expressed as the TCID50 per milliliter using the Reed–Muench method (Reed and Muench 1938). The virus titers were calculated by determining the dilution, giving 50% of wells containing cells that displayed a cytopathic effect.
Antiviral assay of rBoIFN-α1 in vitro
The antiviral activity of rBoIFN-α1 was assayed through the ability of rBoIFN-α1 to inhibit the cytopathic effect of the VSV on MDBK cells (Gresser and others 1974; Meager 2002). The specific activity was determined with reference to rBoIFNαA. Briefly, monolayers of MDBK cells cultured in 96-well plates were treated with 100 μL of 4-fold serial dilutions of rBoIFN-α1 or rBoIFNαA for 24 h and challenged by VSV (100 TCID50/well) after extensive washing with PBS. The wells without viruses were used as the cell controls, and the wells without any IFN were used as the virus controls. The plate was then reincubated at 37°C under a humidified 5% CO2 atmosphere for 18 to 24 h. One unit of IFN activity was defined as the amount required to inhibit the cytopathic effect by 50%.
To neutralize the rBoIFN-α1 antiviral activity, the rBoIFN-α1 or rBoIFNαA samples were preincubated with a serial 2-fold diluted rabbit anti-BoIFNα polyclonal antibody for 1 h at 37°C, after which the antiviral activity was determined by MDBK/VSV system. The preimmune rabbit serum served as a negative control.
The detection of important physicochemical characteristics of rBoIFN-α1
Trypsin sensitivity assay of rBoIFN-α1
The rBoIFN-α1 samples were combined with 1% trypsin to a final concentration of 0.25% trypsin and placed in a water bath for 1 h at 37°C, after which the antiviral activity was determined by the MDBK/VSV system. The antiviral activities of the treated and untreated samples were compared.
pH sensitivity assay of rBoIFN-α1
The rBoIFN-α1 samples were combined with hydrogen chloride or sodium hydroxide to adjusted pH levels of 2.0, 4.0, 10.0, and 12.0 for 24 h at 4°C, after which they were adjusted back to original pH (7.0). The antiviral activity was determined by the MDBK/VSV system.
Temperature sensitivity assay of rBoIFN-α1
The rBoIFN-α1 samples were placed in a 42°C, 56°C, and 63°C water bath for 4 h, after which they were rapidly placed in an icebox for cooling. The antiviral activity was determined by the MDBK/VSV system.
Statistical analysis
All statistical analyses were performed by one-way analysis of variance using an SPSS 16.0 software package (version 16.0; SPSS Inc.). The data were expressed as the mean±standard deviation. A value of P<0.05 was considered statistically significant.
Results
Structural characteristics analysis of bovine IFN-α1
The predicted secondary structure and possible residues involved in the biological activity of bovine IFN-α1 were identified (Fig. 1). The sequence of bovine IFN-α1 contained an N-terminal secretory signal peptide from residues 1–23 and a mature peptide that contained 166 amino-acid residues and 3 putative O-glycosylation sites at positions 25, 27, and 153 of the mature peptide. The amino-acid residues Arg-33 and Tyr-123 are conserved residues, and two domains (residues 29–35 and 123–140) are highly conserved. There are four cysteine residues at positions 1, 29, 99, and 139 of the mature peptide-formed two disulfide bonds. The investigation of secondary structure indicated that bovine IFN-α1 had 5 putative α helices, labeled A to E, from residues 11–21, 54–66, 81–101, 112–133, and 139–158, respectively.
FIG. 1.
The results of secondary structure prediction. The sequence underlined with a single line is a signal sequence, the arrow denotes the signal sequence cleavage site, and underlined with wavy lines are the two highly conserved domains. Cysteine residues forming disulfide bonds are marked with gray circles. The conserved amino-acid residues Arg-33 and Tyr-123 are marked with a gray box and a white circle. Letters A to E refer to the α-helices in bovine IFN-α1. IFN-α1, interferon α1.
Reconstruction of phylogeny
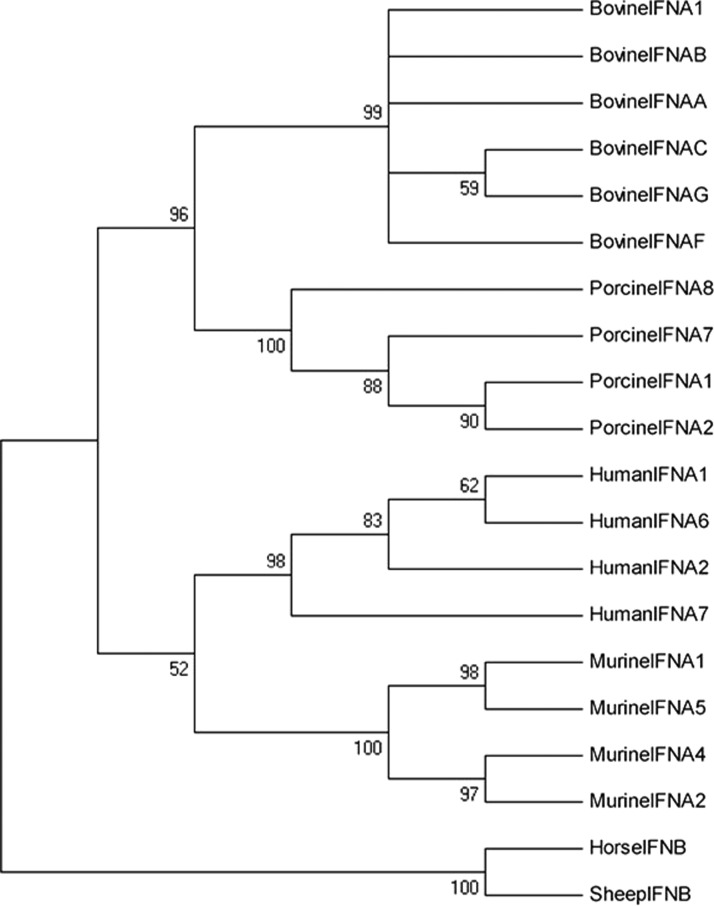
A phylogenetic tree was constructed to elucidate the evolutionary position of bovine IFN-α1. The phylogenetic relationships among the bovine, human, porcine, and mouse IFNα types were inferred through the neighbor-joining method. All bovine IFNα genes clustered in the same group (Fig. 2). Bovine IFNα clustered with IFNs from other mammals, such as those from pig, human, and mouse, resulting in an in-group with regard to other mammalian species.
FIG. 2.
Phylogenetic tree based on nucleotide sequences of bovine IFN-α1 and type I IFN from different species by the neighbor-joining method. GenBank accession numbers are as follows: human IFNα1 (NM_024013), human IFNα2 (NM_000605), human IFNα6 (NM_021002), human IFNα7 (BC074992), murine IFNα1 (NM_010502), murine IFNα2 (NM_010503), murine IFNα4 (NM_010504), murine IFNα5 (NM_010505), porcine IFNα1 (DQ249000), porcine IFNα2 (DQ249002), porcine IFNα7 (DQ872660), porcine IFNα8 (DQ248999), bovine IFN-α1 (XP_001251758.1), bovine IFNαA (EU276064), bovine IFNαB (M10953), bovine IFNαC (NM_174085), bovine IFNαF (NM_001172042), bovine IFNαG (EU276064), horse IFNβ (NM_001172040), and sheep IFNβ (JX458084). The IFNβ of horse and sheep were selected as outgroups.
Construction of vector for secreted expression of BoIFN-α1
The gene that encoded the mature peptide of BoIFN-α1 was cloned by the forward/reverse primer pairs. The target gene was then cleaved by Xho I and EcoR I, and recombined into vector pPICZαA, forming pPICZaA-BoIFN-α1. The result of the enzyme cleavage by Xho I and EcoR I showed two fragments at 3.6 and 521 bp (data not shown). These recombinant plasmids were sent for sequencing. All these results indicated the successful construction of the recombinant plasmids.
PCR determination of transformants
The linearized plasmid pPICZαA-BoIFN-α1 was transformed into P. pastoris GS115, and PCR was employed to precisely detect the positive transformants with the target gene (Fig. 3A, B). We used the specific primers pairs obtained 521-bp-long brand, and the specific reverse primer and 5′AOXI primers pairs obtained 853-bp product, respectively. These results suggested that the recombinant plasmid pPICZαA-BoIFN-α1 was successfully transformed into the host yeast and was accurately integrated into the yeast genome. A selected positive transformant was induced for expression.
FIG. 3.
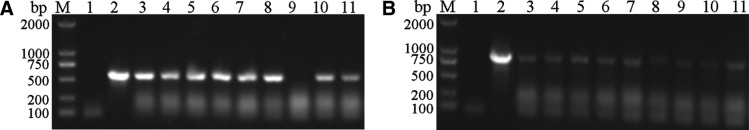
Polymerase chain reaction determination of the transformants of Pichia pastoris containing target gene. (A) The transformed cells of yeast containing pPICαA-BoIFN-α1 with the specific forward/reverse primers. (B) The transformed cells of yeast containing pPICαA-BoIFN-α1 with the specific reverse/5′AOXI primers. BoIFN-α1, bovine interferon α1.
Expression of target genes and SDS-PAGE and western blot analysis of rBoIFN-α1
Methanol was used to induce the secretive expression of the target genes in yeast. After being induced by methanol, the BoIFN-α1 was expressed in yeast, while the number reduced was not expressed. The concentration of purified protein of approximately 0.2 mg/mL was quantified by the BCA Protein Assay kit. The purified rBoIFN-α1 was identified by SDS-PAGE and western blot analysis. Two bands were found, that is, one was at 18 kDa, which was consistent with the predicted molecular weight from the sequence, and the other was at ∼20 kDa (Fig. 4A, B). The 20 kDa band was still observed after treatment with O-glycanases (New England Biolabs). These results indicated that the 20 kDa band could not have been caused by the glycosylation at the three putative O-glycosylation sites at residues 25, 27, and 153 in the mature peptide. Instead, the band might have resulted from the incomplete cleavage of the α-factor signal peptide sequence in the expression vector pPICZαA or some other post-translational modifications. The rBoIFNαA also showed two bands.
FIG. 4.
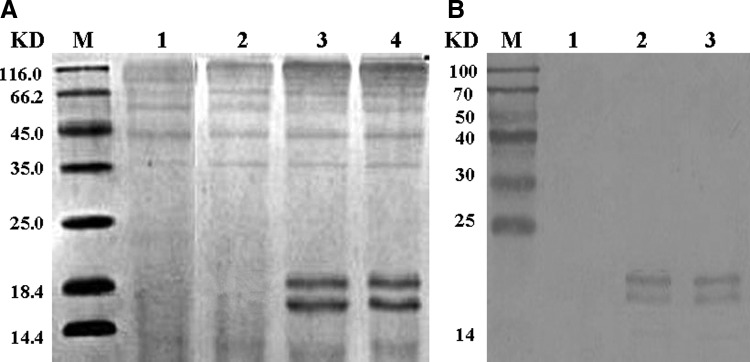
SDS-PAGE and western blotting analysis on purified rBoIFN-α1 and rBoIFNαA. (A) Purified rBoIFN-α1 and rBoIFNαA separated on 15% SDS-PAGE and stained with Coomassie Brilliant blue G-250. M, sizes (KD) of molecular-weight markers; Lane 1: supernatant of Pichia pastoris culture transformed with pPICZaA plasmid (control); Lane 2: supernatant of P. pastoris culture transformed with pPICαA-BoIFN-α1 plasmid number induced; Lane 3: rBoIFN-α1; Lane 4: rBoIFNαA. (B) Western blot analysis of rBoIFN-α1 and rBoIFNαA with a goat anti-rabbit IgG conjugated to horseradish peroxidase antibody against rabbit anti-bovine IFNα polyclonal antibody. M, sizes (KD) of molecular-weight markers; Lane 1: supernatant of P. pastoris culture transformed with pPICZaA plasmid (control); Lane 2: rBoIFN-α1; Lane 3: rBoIFNαA. SDS-PAGE, sodium dodecyl sulfate-polyacrylamide gel electrophoresis; rBoIFN-α1, recombinant mature BoIFN-α1.
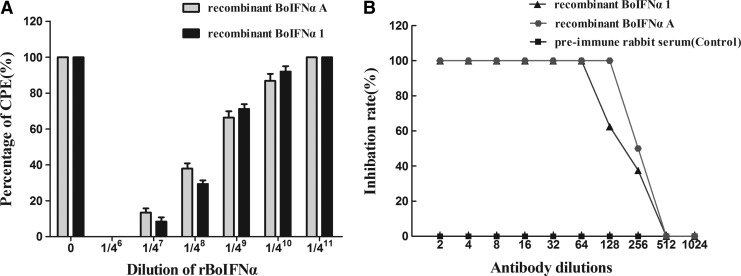
Antiviral activities of rBoIFN-α1 in vitro
The antiviral activity of the rBoIFNα proteins against VSV was tested in MDBK cells, which were pretreated for 24 h with purified rBoIFN-α1/rBoIFNαA and challenged with VSV. Both recombinant proteins contained between 65,536 and 262,144 U/mL of antiviral activity against VSV, and the antiviral activity of rBoIFN-α1 was lower than that of rBoIFNαA (Fig. 5A). The specificity of the response was determined by incubating the proteins with rabbit anti-bovine IFNα polyclonal antibody before the assay. The antiviral activity of rBoIFN-α1 was completely neutralized by the rabbit anti-BoIFNα polyclonal antibody at a dilution of 1:64, whereas that of rBoIFNαA was neutralized at a dilution of 1:128. No neutralization of antiviral activity was observed on the addition of preimmune rabbit serum (Fig. 5B).
FIG. 5.
Characterization of rBoIFN-α1 biological activity. (A) Antiviral activity of rBoIFN-α1 in MDBK/VSV system. (B) Neutralization of antiviral activity with anti-BoIFNα polyclonal antibody in MDBK/VSV system. MDBK, Madin–Darby bovine kidney; VSV, vesicular stomatitis virus.
Important physicochemical characteristics of rBoIFN-α1
After treatment with 0.25% trypsin, the rBoIFN-α1 samples completely lost their antiviral activity. This finding demonstrated that rBoIFN-α1 was highly sensitive to trypsin (data not shown).
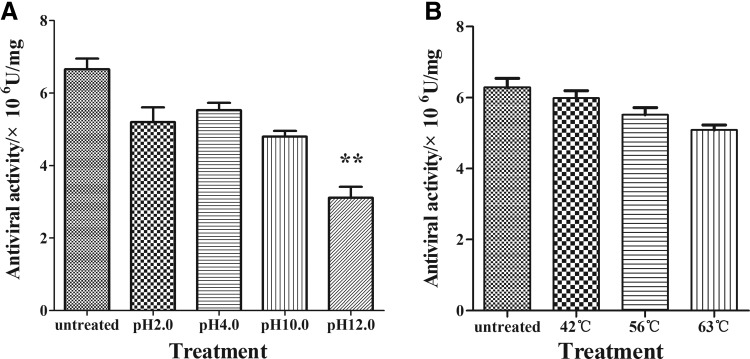
To determine the pH sensitivity, the rBoIFN-α1 samples were treated at pH 2.0, 4.0, 10.0, and 12.0 for 24 h at 4°C, after which the pH levels were adjusted back to their original pH level, and the antiviral activity against VSV was measured. The untreated sample was taken as the control. The samples treated with different pH levels retained their antiviral activity. The loss in the antiviral activities of the samples treated at pH 2.0, 4.0, 10.0, and 12.0 were 0.24-, 0.19-, 0.45-, and 0.78-fold lower than that of the untreated sample, respectively (Fig. 6A). The antiviral activity of the sample treated at pH 12.0 was significantly lower than that of the control (P<0.01). The difference between the antiviral activity of the samples treated at pH 2.0, 4.0, or 10.0 and that of the control was not significant (P>0.05).
FIG. 6.
The important physicochemical characteristics of rBoIFN-α1. (A) The results of pH sensitivity of rBoIFN-α1. (B) The results of temperature sensitivity of the rBoIFN-α1. **P<0.01.
To determine the temperature sensitivity, the rBoIFN-α1 samples were placed in a 42°C, 56°C, and 63°C water bath for 4 h, and their antiviral activity against VSV was measured. The results in Fig. 6B show that, after treatment under different temperatures, the antiviral activity of the treated samples did not significantly differ from that of the untreated samples (P>0.05). All the samples retained relatively high antiviral activity.
Discussion
Previous studies have identified the multiple genes that encode the bovine IFNα subtypes, and the BoIFN-α1 gene was first cloned from the epithelium cells of the rotavirus-infected calf in 1996 (Chaplin and others 1996a). However, bovine IFNα subtypes has not been systematically examined, and to date, no information on the characteristics of BoIFN-α1 is available. We amplified the BoIFN-α1 from bovine liver genomic DNA and analyzed the sequence secondary structure and phylogenetic relationships; then, we predicted the three-dimensional model. The mature peptide gene of BoIFN-α1 was cloned into the vector pPICZαA; we obtained the rBoIFN-α1 and used the expression system of P. pastoris successfully. The molecular weight of rBoIFN-α1 is about 18 kDa and is secreted into culture medium, and the concentration is approximately 0.2 mg/mL. Further, we determined the biological and physicochemical characteristics of rBoIFN-α1. This study provides a basis for the industrial production of rBoIFN-α1 and its potential use in the prevention and cure of viral bovine diseases.
The bovine IFNα1 contains conserved residues that are essential for the activity of HuIFN, such as the amino-acid residues Arg-33 and Tyr-23 (Fish and others 1989). These two domains (residues 29–35 and 123–140) are highly conserved between the BoIFNα subtypes, although their level of conservation is lower across other species (Chaplin and others 1996b). Similar to HuIFN, the bovine IFN-α1 has two disulfide bonds formed by the cysteine residues at positions 1 and 99 as well as at 29 and 139. The region around the critical Cys-29 to Cys-139 bond is especially important to IFNα (Wetzel and others 1982). The secondary structure predicts that bovine IFN-α1 has 5 putative α helices. The residues in these regions mediate the IFNα to interact with the receptor. All these structures are essential to the activity of IFNα.
The phylogenetic analysis suggests that all the IFNα subtypes originated from common ancestral genes and that the duplications led to the current subtypes after the divergence of bovine, human, pig, and mouse species. The bovine IFN-α1 and other bovine IFNα genes clustered in the same group, which indicates that they share a common ancestor.
The methylotrophic yeast P. pastoris expression system has a strong AOX1 promoter, so it can highly express heterologous proteins with proper folding and considerable post-translational modifications. In addition, it can secrete in vitro heterologous proteins into the culture medium to facilitate the purification of the recombinant protein (Damasceno and others 2012). These advantages were considered in the expression of the rBoIFN-α1 in yeast.
The rBoIFN-α1 exhibited comparable antiviral activity against VSV in MDBK cells, and its antiviral activity was neutralized completely by rabbit anti-BoIFNα polyclonal antibody. This finding indicated that rBoIFN-α1 had good specificity and that its antiviral activity was relatively stable.
The rBoIFN-α1 has some important physicochemical characteristics, such as high sensitivity to trypsin, insensitivity to temperature, sensitivity to alkali, and resistance to acid. These characteristics are similar to the typical physicochemical characteristics of IFNα. These results demonstrated that bovine IFNα was successfully cloned.
In summary, the mature peptide gene of rBoIFN-α1 was cloned, and biologically active protein in P. pastoris GS115 was successfully secreted. The characteristics of rBoIFN-α1 were systematically investigated in vitro; the phylogenetic relationships and secondary structure of rBoIFN-α1 were analyzed. Further studies should be performed to evaluate the antiviral effects of BoIFN-α1, compare its antiviral effects with the other subtypes of BoIFNα, and determine its antiviral effects in vivo by animal experiments. These studies will pave the foundation for utilizing rBoIFN-α1 as a useful antiviral agent against infectious and viral bovine diseases.
Acknowledgments
This study was supported by grants from the Earmarked Fund for China Agriculture Research System (No. CARS-37), the subproject of National 12th 5-year Support Key Projects (2012BAD12B03 and 2012BAD12B05), the fund of Food Safety and Nutrition Collaborative Innovation Center, and the Key Technologies Research and Development Program of Heilongjiang province (GA09B302). The authors would like to express their sincere appreciation to the reviewers for their insightful recommendations, which have greatly aided them in improving the quality of this article.
Author Disclosure Statement
No competing financial interests exist.
References
- Cao ZW. 2004. An effective method of Tricine-SDS-PAGE for separating the 1 kDa peptide. China Biotechnol 24:74–76 [Google Scholar]
- Chaplin PJ, Entrican G, Gelder KI, Collins RA. 1996a. Cloning and biologic activities of a bovine interferon-α isolated from the epithelium of rotavirus-infected calf. J Interferon Cytokine Res 16:25–30 [DOI] [PubMed] [Google Scholar]
- Chaplin PJ, Parsons KR, Collins RA. 1996b. The cloning of cattle interferon-A subtypes isolated from the gut epithelium of rotavirus-infected calves. Immunogenetics 44:143–145 [DOI] [PubMed] [Google Scholar]
- Damasceno LM, Huang CJ, Batt CA. 2012. Protein secretion in Pichia pastoris and advances in protein production. Appl Microbiol Biotechnol 93:31–39 [DOI] [PubMed] [Google Scholar]
- Fish EN, Banejee NK, Stebbing N. 1989. The role of three domains in the biological activity of human IFN-α. J Interferon Res 9:97–114 [DOI] [PubMed] [Google Scholar]
- Fostern GR, Rodrigues O, Ghouze F, Schulte-Frohlinde E, Testa D, Liao MJ, Stark GR, Lead beater L, Thomas HC. 1996. Different relative activities of human cell-derived interferon subtypes: IFN-α8 has very high antiviral potency. J Interferon Cytokine Res 16:1027–1033 [DOI] [PubMed] [Google Scholar]
- Gresser I, Bandu MT, Brouty-boye D, Tovey M. 1974. Pronounced antiviral activity of human interferon on bovine and porcine cells. Nature 251:543–545 [DOI] [PubMed] [Google Scholar]
- Hai JU, Liang D, Guo G, Zhang J. 2003. Comparison of four methods to prepare Pichia genomic DNA for PCR. Tianjin Med J 31:270–272 [Google Scholar]
- Hou F, Liu K, Chen P, et al. . 2011. Antiviral activity of rChIFN-α against vesicular stomatitis virus and Newcastle disease virus: a novel recombinant chicken interferon-α showed high antiviral activity. Res Vet Sci 91:e73–e79 [DOI] [PubMed] [Google Scholar]
- Meager A. 2002. Biological assays for interferons. J Immunol Methods 261:21–36 [DOI] [PubMed] [Google Scholar]
- Oritani K, Kincade PW, Zhang C, Tomiyama Y, Matsuzawa Y. 2001. Type I interferons and limitin: a comparison of structures, receptors, and functions. Cytokine Growth Factor Rev 12:337–347 [DOI] [PubMed] [Google Scholar]
- Reed LJ, Muench H. 1938. A simple method of estimating fifty percent endpoints. Am J Hygiene 27:493–497 [Google Scholar]
- Roberts RM, Liu L, Guo Q, Leaman D, Bixby J. 1998. The evolution of the type I interferons. J Interferon Cytokine Res 18:805–816 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Translated by Jin DY, Li MF, Hou YD. 1992. Molecular Cloning: A Laboratory Manual, 2nd ed. Beijing: Science Press, pp 16–68 [Google Scholar]
- Tamura K, Petersonn D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Heuvel M, Bosveld IJ, Klaassen P, Zwarthoff EC, Trapman J. 1998. Structure–function analysis of murine interferon-alpha: antiviral properties of novel hybrid interferons. J Interferon Res 8:5–14 [DOI] [PubMed] [Google Scholar]
- Wetzel R, Levine HL, Estell DA, Shire S, Finer-Moore J, Stroud RM, Bewley TA. 1982. Structure-function studies on human alpha interferon. In: Interferons. Academic Press: NY, pp 365–376 [Google Scholar]
- Zhao X, Cheng G, Yan W, Liu M, He Y, Zheng Z. 2009. Characterization and virus-induced expression profiles of the porcine interferon-ω multigene family. J Interferon Cytokine Res 10:687–693 [DOI] [PubMed] [Google Scholar]