Abstract
Lambda-interferons (IFN-λs) have been demonstrated as having the ability to inhibit HIV replication in macrophages. However, specific differences in signaling transduction and anti-HIV activity in macrophages between different IFN-λs are unclear. Here, we showed that although all 3 members of (IFN-λ1, λ2, and λ3) IFN-λ family induced the expression of a number of genes of janus kinase/signal transducers and activators of transcription (JAK/STAT) signaling pathway in monocyte-derived macrophages, IFN-λ1 or IFN-λ3 induced higher levels of antiviral IFN-stimulated genes (ISGs) expression than did IFN-λ2. In addition, IFN-λ1 or IFN-λ3 induced higher levels of several pattern recognition receptors (PPRs) than did IFN-λ2. Incubation of IFN-λs with HIV-infected macrophages showed that IFN-λ1 or IFN-λ3 is more potent in anti-HIV activity than IFN-λ2. We also showed that IFN-λ treatment before HIV infection was more potent in HIV inhibition than that after HIV infection. Further investigations showed that the inductions of ISGs and PPRs expression by IFN-λs were largely compromised by HIV infection. These findings provide further experimental evidence that IFN-λs have therapeutic potential in treatment of HIV infection.
Introduction
Lambda-interferons (IFN-λs) are a class of recently identified members of IFN family. Including the new identified member, IFN-λ4 (Prokunina-Olsson and others 2013), the IFN-λ subfamily now has 4 members, IFN-λ1, IFN-λ2, IFN-λ3, and IFN-λ4 (Kotenko and others 2003; Prokunina-Olsson and others 2013). IFN-λs share features with both type I IFN and the interleukin (IL)-10 family cytokines (Uze and Monneron 2007). IFN-λs are structurally and genetically close to the members of IL-10 family but display type I IFN-like antiviral activity and induction of typical IFN-inducible genes (Ank and others 2006; Uze and Monneron 2007; Gad and others 2010). Similar to type I IFNs, the expression of IFN-λs could be induced by viral infections or activation of pattern recognition receptors (PPRs), including several toll-like receptors (TLRs) and retinoic acid-inducible gene I (RIG-I) (Dumoutier and others 2004; Yang and others 2005; Onoguchi and others 2007; Wang and others 2013). IFN-λs bind to their own distinctive receptor complex, IL-10Rβ and IL-28Rα, which activates janus kinase/signal transducers and activators of the transcription (JAK/STAT) signaling pathway, resulting in the phosphorylation of STAT proteins and forming of interferon-stimulated gene factor 3 (ISGF3) complex (Kotenko and others 2003; Sheppard and others 2003; Li and others 2009). The formed ISGF3 complex binds to the IFN-stimulated response element and induces ISGs that play important roles in IFN-mediated antiviral activity (Kotenko and others 2003; Zhang and others 2011). Although IFN-λs exert biological activities similar to type I IFNs, they appear to have a more specialized role in innate antiviral defense (Ank and Paludan 2009). Recently, studies have demonstrated that IFN-λs had the ability to inhibit the replication of a number of viruses, including hepatitis C virus (HCV) and hepatitis B virus (HBV) (Robek and others 2005; Marcello and others 2006), cytomegalovirus (CMV) (Brand and others 2005), apeu virus (Almeida and others 2008), herpes simplex virus type 2 (HSV 2) (Ank and others 2006), encephalomyocarditis virus (EMCV) (Kotenko and others 2003), and vesicular stomatitis virus (VSV) (Brand and others 2005).
Although it has been reported that IFN-λs had the ability to inhibit HIV replication in macrophages (Hou and others 2009; Liu and others 2012), specific differences in signaling transduction and antiviral activity against HIV replication between different members of IFN-λ family are unclear. In this study, we examined the IFN-λ-mediated gene induction profile of JAK/STAT signaling pathway and compared the antiviral activity against HIV replication of IFN-λ1, IFN-λ2, and IFN-λ3 in macrophages.
Materials and Methods
Reagents
Recombinant human IFN-λ1, IFN-λ2, and IFN-λ3 proteins were purchased from R&D Systems, Inc. RT2 First-Strand Kit and RT2 Profiler PCR Array Kit for Human JAK/STAT signaling pathway were purchased from SABiosciences, QIAGEN.
Cell culture
Peripheral blood was purchased from the Center for AIDS Research at the University of Pennsylvania. The protocol used to isolate blood from donors, purify the blood components, and distribute this material to the investigators was approved by the IRB of the Center for AIDS Research. These blood samples were screened for all normal blood-borne pathogens and certified to be pathogen free. Monocytes were purified from peripheral blood of 3 healthy adult donors according to our previously described technique (Hassan and others 1986). Freshly isolated monocytes were cultured in 48-well culture plates at a density of 2.5×105 cells/well in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal calf serum. Macrophages refer to 7-day-cultured monocytes in vitro.
IFN-λ treatment and HIV infection
Monocyte-derived macrophages were incubated either with or without IFN-λ (10, 100 ng/mL) for 24 h before infection or at day 3 postinfection with HIV Bal strain. The cells were infected with equal quantities of cell-free HIV Bal strain based on p24 protein content (30 ng/106 cells) for 2 h at 37°C in the presence or absence of IFN-λ. The cells were then washed thrice with DMEM to remove unabsorbed virus, and fresh medium was added to cell cultures. The final wash was tested for viral reverse transcription (RT) activity and shown to be free of residual inoculum. Untreated cells served as controls. The cell cultures were replaced with fresh medium with or without IFN-λ every 3 days after HIV infection. Supernatant (SN) was collected every 3 days for HIV RT activity assay. Cells were collected for HIV gag gene expression at day 12 postinfection.
PCR-based gene expression array
The pathway-focused JAK/STAT signaling pathway gene expression array was carried out according to the protocol provided by SABiosciences, QIAGEN. Briefly, total cellular RNA was extracted from monocyte-derived macrophages treated with or without IFN-λ (10 ng/mL) for 24 h using SABiosciences RT2 quantitative polymerase chain reaction-Grade RNA Isolation Kit. The elimination of genomic DNA contamination and first-strand complementary DNA synthesis were carried out using RT2 First-Strand Kit. Pathway-focused (JAK/STAT) gene expression array was performed using RT2 Profiler PCR Array System (96-Well Format). Each PCR expression array was performed twice on independent replicates. Statistical analysis of data was performed with the Data Analysis Template provided by the SABiosciences, QIAGEN.
HIV RT assay
HIV RT activity was determined based on the technique of (Willey and others 1988) with modifications (Ho and others 1992). In brief, 10 μL of culture SN from macrophages infected with or without HIV was added to a cocktail containing poly(A), oligo(dT) (Amersham Biosciences, Inc.), MgCl2, and [32P]dTTP (Amersham Biosciences, Inc.) and incubated for 20 h at 37°C. Then, 30 μL of the cocktail was spotted onto DE81 paper (Whatman International Ltd.), dried, and washed 5 times with 2× saline-sodium citrate buffer and once with 95% ethanol. The filter paper was then air dried. Radioactivity was counted in a liquid scintillation counter (PerkinElmer Life Sciences).
RNA extraction and real-time RT-PCR
Total RNA from macrophages was extracted with Tri-Reagent (Molecular Research Center) as previously described (Li and others 2003). Total RNA (1 μg) was subjected to RT using the RT system (Promega) with random primers for 1 h at 42°C. The reaction was terminated by incubating the reaction mixture at 99°C for 5 min, and the mixture was then maintained at 4°C. The resulting cDNA was then used as a template for real-time PCR quantification. Real-time PCR was performed with 1/10 of the cDNA with the iQ SYBR Green Supermix (Bio-Rad Laboratories, Inc.) as previously described (Zhang and others 2005). The amplified products were visualized and analyzed using the software MyiQ provided with the thermocycler (iCycler iQ real-time PCR detection system; Bio-Rad Laboratories, Inc.). The oligonucleotide primers were synthesized by Integrated DNA Technologies, Inc. and sequences were shown in Table 1. The cDNA was amplified by PCR, and the products were measured using SYBR green I (Bio-Rad Laboratories, Inc.). The data were normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and presented as the change in induction relative to that of untreated control cells.
Table 1.
Primers for Real-Time Reverse Transcription-Polymerase Chain Reaction
| Primer | Orientation | Sequence (5′-3′) |
|---|---|---|
| GAPDH | Sense | GGTGGTCTCCTCTGACTTCAACA |
| Antisense | GTTGCTGTAGCCAAATTCGTTGT | |
| RIG-I | Sense | CTTGGCATGTTACACAGCTGAC |
| Antisense | GCTTGGGATGTGGTCTACTCA | |
| MDA-5 | Sense | ACATAACAGCAACATGGGCAGTG |
| Antisense | TTTGGTAAGGCCTGAGCTGGAG | |
| TLR-3 | Sense | AGCCACCTGAAGTTGACTCAGG |
| Antisense | CAGTCAAATTCGTGCAGAAGGC | |
| TLR-7 | Sense | AAAATGGTGTTTCCAATGTGG |
| Antisense | GGCAGAGTTTTAGGAAACCATC | |
| TLR-8 | Sense | TTATGTGTTCCAGGAACTCAGAGAA |
| Antisense | TAATACCCAAGTTGATAGTCGATAAGTTTG | |
| ISG15 | Sense | GGCTGGGAGCTGACGGTGAAG |
| Antisense | GCTCCGCCCGCCAGGCTCTGT | |
| ISG56 | Sense | TTCGGAGAAAGGCATTAGA |
| Antisense | TCCAGGGCTTCATTCATAT | |
| OAS1 | Sense | AGAAGGCAGCTCACGAAACC |
| Antisense | CCACCACCCAAGTTTCCTGTA | |
| OAS2 | Sense | CAGTCCTGGTGAGTTTGCAGT |
| Antisense | ACAGCGAGGGTAAATCCTTGA | |
| MxA | Sense | GCCGGCTGTGGATATGCTA |
| Antisense | TTTATCGAAACATCTGTGAAAGCAA | |
| Viperin | Sense | TGGGTGCTTACACCTGCTG |
| Antisense | TGAAGTGATAGTTGACGCTGGT |
Statistical analysis
Student's t-test was used to evaluate the significance of difference between groups, and multiple comparisons were performed by regression analysis and one-way analysis of variance. P values of less than 0.05 were considered significant. All data are presented as mean±SD. Statistical analyses were performed with SPSS 11.5 for Windows. Statistical significance was defined as P<0.05.
Results
IFN-λs regulate the expression of JAK/STAT pathway genes
To evaluate the IFN-λ-mediated modulation of JAK/STAT pathway gene expression in macrophages, we treated macrophages with the same concentration (10 ng/mL) of IFN-λ1, λ2, or λ3 for 24 h. Total cellular RNA was collected for gene expression array to examine the profile of IFN-λ-mediated regulation of genes in JAK/STAT signaling pathway. We showed that all 3 members of IFN-λ family were able to upregulate the expression of 23 genes more than 1.5-fold in macrophages (Table 2). We showed that most of the gene regulation levels were comparable in 3 IFN-λ members (Table 2), but some antiviral ISGs were regulated differently. In contrast to IFN-λ2, IFN-λ1 or IFN-λ3 induced higher expression levels of ISG15, and IFN-λ3 induced the highest expression level of 2′-5′-oligoadenylate synthetase 1 (OAS1) (Table 2). Chemokine (C-X-C motif) ligand 9 (CXCL9) was also induced by IFN-λ in macrophages (Table 2). In addition, the expression of the key elements in JAK/STAT signaling pathway, including JAK-2, JAK-3, STAT-1, STAT-2, STAT-4, and STAT-6, were enhanced by IFN-λs treatment (Table 2). Furthermore, the key regulators in JAK/STAT signaling pathway, such as interferon regulatory factor-1 (IRF-1) and ISGF3G (IRF-9), were also induced by IFN-λs treatment (Table 2). IFN-λs also induced suppressors of cytokine signaling (SOCS) 1 and SOCS2 expression (Table 2). In addition to the upregulated genes, IFN-λs were also able to downregulate expression of several genes in macrophages (Table 3).
Table 2.
Upregulated Genes in JAK/STAT Pathway by IFN-λs
| Fold of control | ||||
|---|---|---|---|---|
| Gene | Description | λ1 | λ2 | λ3 |
| CDKN1A | Cyclin-dependent kinase inhibitor 1A (p21, Cip1) | 1.51 | 1.50 | 2.12 |
| CXCL9 | Chemokine (C-X-C motif) ligand 9 | 6.90 | 2.55 | 5.21 |
| IL20 | Interleukin-20 | 3.47 | 4.38 | 6.91 |
| IL2RA | Interleukin 2 receptor, alpha | 1.00 | 2.29 | 1.66 |
| IRF1 | Interferon regulatory factor 1 | 3.14 | 2.22 | 3.31 |
| IRF9 | Interferon regulatory factor 9 | 2.45 | 2.23 | 2.59 |
| ISG15 | ISG 15 ubiquitin-like modifier | 48.78 | 31.47 | 78.84 |
| JAK2 | Janus kinase 2 | 1.58 | 1.53 | 2.03 |
| JAK3 | Janus kinase 3 | 2.17 | 1.73 | 1.88 |
| JUNB | Jun B proto-oncogene | 1.84 | 1.56 | 1.55 |
| LRG1 | Leucine-rich alpha-2-glycoprotein 1 | 2.42 | 2.05 | 3.49 |
| MCL1 | Myeloid cell leukemia sequence 1 (BCL2-related) | 1.83 | 1.58 | 2.02 |
| OAS1 | 2′-5′-oligoadenylate synthetase 1, 40/46 kDa | 4.88 | 4.15 | 6.94 |
| PRLR | Prolactin receptor | 2.47 | 1.76 | 2.46 |
| SOCS1 | Suppressor of cytokine signaling 1 | 2.95 | 1.92 | 4.94 |
| SOCS2 | Suppressor of cytokine signaling 2 | 2.03 | 1.65 | 2.06 |
| SP1 | Sp1 transcription factor | 1.79 | 1.55 | 1.87 |
| STAT1 | Signal transducer and activator of transcription 1 | 6.97 | 5.297 | 6.98 |
| STAT2 | Signal transducer and activator of transcription 2 | 4.08 | 3.15 | 3.70 |
| STAT4 | Signal transducer and activator of transcription 4 | 2.08 | 2.28 | 2.71 |
| STAT6 | Signal transducer and activator of transcription 6 | 1.67 | 1.50 | 1.51 |
| TYK2 | Tyrosine kinase 2 | 1.60 | 1.37 | 1.18 |
| USF1 | Upstream transcription factor 1 | 1.87 | 1.42 | 1.65 |
Primary human monocyte-derived macrophages were treated with IFN-λ1, λ2, or λ3 (10 ng/mL) for 24 h. Total RNA was harvested and subjected to the gene array using RT2 Profiler PCR Array System (SABiosciences, QIAGEN). Data analysis was performed using Data Analysis Template from SABiosciences. The genes that displayed less than 1.5-fold upregulation relative to the control were omitted. The IFN-λs upregulated about 23 genes more than 1.5-fold in the JAK/STAT signaling pathway.
Table 3.
Downregulated Genes in JAK/STAT Pathway by IFN-λs
| Fold of control | ||||
|---|---|---|---|---|
| Gene | Description | λ1 | λ2 | λ3 |
| F2R | Coagulation factor II (thrombin) receptor | −1.54 | −1.96 | −1.73 |
| MYC | V-myc myelocytomatosis viral oncogene homolog (avian) | −2.93 | −2.42 | −3.32 |
| NOS2 | Nitric oxide synthase 2, inducible | −1.80 | −1.78 | −2.06 |
| PDGFRA | Platelet-derived growth factor receptor, alpha polypeptide | −2.86 | −2.12 | −4.30 |
| PRL | Prolactin | −2.41 | −0.77 | −1.39 |
| PTPN11 | Protein tyrosine phosphatase, nonreceptor type 11 | −1.45 | −1.64 | −1.67 |
| SMAD3 | SMAD family member 3 | −1.35 | −2.39 | −1.88 |
Primary human monocyte-derived macrophages were treated with IFN-λ1, λ2, or λ3 (10 ng/mL) for 24 h. Total RNA was harvested and subjected to the gene array using RT2 Profiler PCR Array System (SABiosciences, QIAGEN). Data analysis was performed using Data Analysis Template from SABiosciences. The genes that displayed less than 1.5-fold downregulation relative to the control were omitted. The IFN-λs downregulated about 7 genes more than 1.5-fold in the JAK/STAT signaling pathway.
IFN-λs induce antiviral ISGs expression
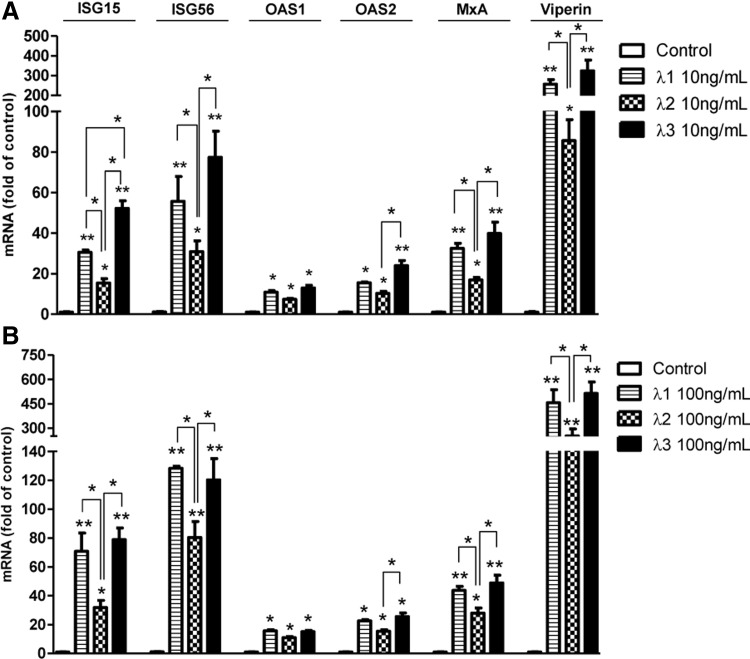
To confirm the different regulation of antiviral ISGs expression generated in PCR gene array experiment, we examined more antiviral ISGs expression in different donors with IFN-λ concentration effect. As shown in Fig. 1, all 3 IFN-λ members could induce ISG15, ISG56, OAS1, OAS2, myxovirus resistance A (MxA), and Viperin expression in macrophages. IFN-λ3 induced the highest ISGs expression in a low concentration treatment group (10 ng/mL) (Fig. 1A). IFN-λ1 and IFN-λ3 upregulated similar levels of ISGs expression in a high concentration treatment group (100 ng/mL) (Fig. 1B), and both the induction levels were higher than IFN-λ2 (Fig. 1A, B).
FIG. 1.
Interferon (IFN)-λs induce IFN-stimulated genes (ISGs) expression. Seven-day-cultured macrophages were treated with 10 ng/mL (A) or 100 ng/mL (B) IFN-λs for 24 h. Total RNA extracted from cells was subjected to the real-time reverse transcriptase (RT)-PCR for the mRNA levels of ISG15, ISG56, MxA, OAS-1, OAS-2, and Viperin. The data are expressed as mRNA levels for relative (fold) to the control (without treatment, which is defined as 1). The results shown are mean±SD of triplicate cultures, representative of 3 experiments (IFN-λ vs. control, *P<0.05, **P<0.01).
IFN-λs induce PPRs expression
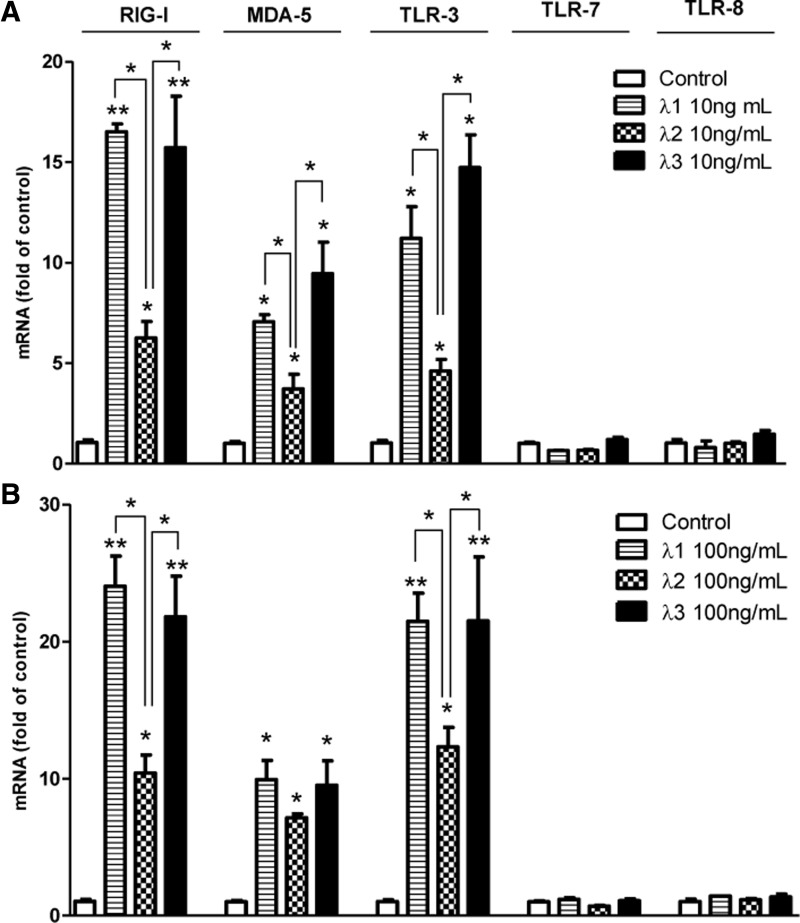
We next examined whether IFN-λs regulated different expression of several PPRs that play important roles during RNA virus infection (Thompson and Locarnini 2007; Jensen and Thomsen 2012). We showed that IFN-λs could induce RIG-I, melanoma differentiation-associated protein 5 (MDA-5), and TLR-3 expression in macrophages, but had little effect on TLR-7 and TLR-8 expression in macrophages (Fig. 2). IFN-λ1- and IFN-λ3-induced RIG-I, MDA-5, and TLR-3 expression was comparable in macrophages, and both were higher than IFN-λ2 (Fig. 2).
FIG. 2.
IFN-λs induce pattern recognition receptors (PPRs) expression. Seven-day-cultured macrophages were treated with 10 ng/mL (A) or 100 ng/mL (B) IFN-λs for 24 h. Total RNA extracted from cells was subjected to the real-time RT-PCR for the mRNA levels of RIG-I, MDA-5, TLR-3, TLR-7, and TLR-8. The data are expressed as mRNA levels for relative (fold) to the control (without treatment, which is defined as 1). The results shown are mean±SD of triplicate cultures, representative of 3 experiments (IFN-λ vs. control, *P<0.05, **P<0.01).
IFN-λs inhibit HIV replication
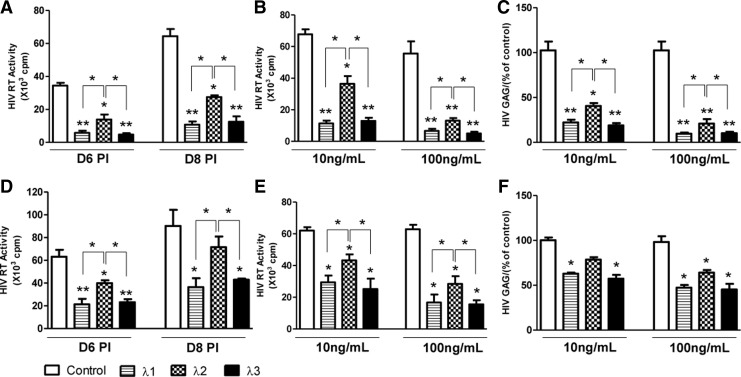
In order to compare the antiviral activity of IFN-λ1, λ2, and λ3 against HIV replication, macrophages were treated with different concentrations (10,100 ng/mL) of IFN-λ1, λ2, or λ3, respectively. As shown in Fig. 3A and B, cells that were treated with IFN-λs for 24 h before HIV Bal strain infection showed a significant decrease in RT activity in a concentration-dependent manner. IFN-λ-mediated inhibition of HIV replication was also confirmed by decreased HIV gag gene expression in macrophages treated with IFN-λs (Fig. 3C). The data showed that pretreatment of cells with 10 ng/mL of either IFN-λ1 or λ3 decreased almost 85% of HIV RT activity, while pretreatment with IFN-λ2 only decreased about 50% of HIV RT activity (Fig. 3A, B). Thus, the anti-HIV activity of IFN-λ1 and IFN-λ3 was comparable, and both were more efficient as compared with IFN-λ2. These results were also confirmed by IFN-λ-mediated inhibition of HIV gag gene expression (Fig. 3C). In addition, IFN-λ1 or IFN-λ3 also had better antiviral activity against HIV as compared with IFN-λ2 in treatment groups of day 3 postinfection (Fig. 3D–F). However, IFN-λs treatment before HIV infection (Fig. 3A–C) was more potent in HIV inhibition than that after HIV infection (Fig. 3D–F).
FIG. 3.
IFN-λs inhibit HIV replication in macrophages. Seven-day-cultured macrophages were treated with IFN-λs (10, 100 ng/mL) for 24 h before HIV Bal infection (A–C) or day 3 postinfection (D–F). Culture supernatant (SN) was subjected to HIV RT assay (A, B, D, and E); total RNA from cells was subjected to HIV gag gene expression by real-time PCR (C, F). The results shown are mean±SD of triplicate cultures, representative of 3 experiments (IFN-λ vs. control, *P<0.05, **P<0.01).
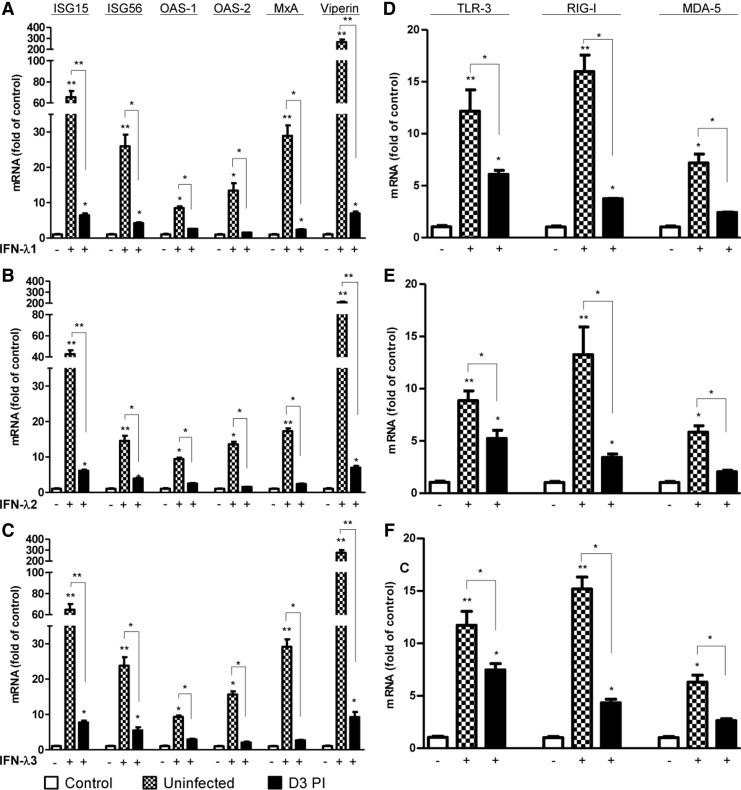
HIV infection compromises IFN-λ-induced ISGs and PPRs expression
As indicated, IFN-λ treatment before HIV infection was more potent in HIV inhibition than that after HIV infection. We next examined whether HIV had the ability to compromise the IFN-λ-induced antiviral ISGs expression. We showed that HIV infection could significantly compromise the IFN-λ-induced antiviral ISGs expression in macrophages (Fig. 4A–C). Furthermore, HIV infection also suppressed the RIG-I, MDA-5, and TLR-3 expression induced by IFN-λs treatment (Fig. 4D–F).
FIG. 4.
HIV infection compromises IFN-λ-induced IFN-stimulated genes (ISGs) and PPRs expression. Seven-day-cultured macrophages infected with (day 3 postinfection) or without HIV Bal were treated with IFN-λs (100 ng/mL) for 24 h. Total RNA extracted from cells was subjected to the real-time RT-PCR for the mRNA levels of ISG15, ISG56, MxA, OAS-1, OAS-2, Viperin, RIG-I, MDA-5, TLR-3, TLR-7, and TLR-8. The data are expressed as mRNA levels for relative (fold) to the control (without treatment, which is defined as 1). The results shown are mean±SD of triplicate cultures, representative of 3 experiments (IFN-λ vs. control, *P<0.05, **P<0.01).
Discussion
IFNs are important players in host innate immunity, as they possess innate antiviral activity against viral infections. In addition to type I IFNs and type II IFN that have been known for decades as antiviral cytokines, type III IFNs (IFN-λs) are also shown to have strong antiviral function (Ank and Paludan 2009). Although IFN-λs exert biological activities similar to type I IFNs, they appear to have a more specialized role in innate antiviral defense (Ank and Paludan 2009). The members (IFN-λ1, IFN-λ2, and IFN-λ3) of the IFN-λ family are composed of 3 structure-related cytokines, encoded by IL-29, IL-28A, and IL-28B gene, respectively. IFN-λ1 protein shares about 80% amino-acid identity with IFN-λ2 or IFN-λ3, while IFN-λ2 and IFN-λ3 proteins are 95% identical (Donnelly and others 2004). IFN-λs exert their bioactivity through binding to their own distinctive receptor complex, IL-10Rβ and IL-28Rα (Kotenko and others 2003; Sheppard and others 2003; Li and others 2009). Although studies showed that monocytes do not express functional IFN-λ receptors (Witte and others 2009; Diegelmann and others 2010; Dickensheets and others 2013), we demonstrated that macrophages express both units of IFN-λ receptors. Our previous study (Hou and others 2009) showed that macrophages express IL-10Rβ at both mRNA and protein levels, and express IL-28Rα at mRNA level. In addition, here we observed that macrophages also express IL-28Rα at protein level detected by Flow Cytometry using a specific antibody of IL-28Rα (PE-conjugated anti-IL-28Ra antibody, R&D, Supplementary Fig. S1; Supplementary Data are available online at www.liebertpub.com/jir). IFN-λs have been reported that had the ability to inhibit the replication of a number of viruses (Kotenko and others 2003; Brand and others 2005; Robek and others 2005; Ank and others 2006; Marcello and others 2006; Almeida and others 2008), including HIV (Hou and others 2009; Liu and others 2012); however, specific differences in signaling transduction and antiviral activities against HIV replication in macrophages between different IFN-λs are unclear. In this study, we performed PCR-based gene array to examine the profile of genes in JAK/STAT signaling pathway in IFN-λ-treated macrophages. We showed that all 3 members of the IFN-λ family were able to upregulate expression of 23 genes in macrophages (Table 2). Interestingly, we found that most of the gene regulation levels were comparable in these 3 IFN-λ members, but some antiviral genes were regulated differently (Table 2). IFN-λ1 or IFN-λ3 induced a higher expression level of ISG15 than IFN-λ2, and IFN-λ3 induced the highest expression level of OAS1. CXCL9, an IFN-γ-inducible chemokine (Kouroumalis and others 2005), was also induced by IFN-λs treatment in macrophages (Table 2). In addition, the expression of the key elements in the JAK/STAT signaling pathway, including JAK-2, JAK-3, STAT-1, STAT-2, STAT-4, and STAT-6 (Rawlings and others 2004), was enhanced by IFN-λs treatment (Table 2). Furthermore, the key regulators in the JAK/STAT signaling pathway, such as IRF-1 and ISGF3G (IRF-9) (Taniguchi and others 2001), were also induced by IFN-λs treatment (Table 2). To our surprise, IFN-λs also induced SOCS1 and SOCS2 expression, which are negative regulators of the JAK/STAT signaling pathway (Okugawa and others 2013). The different regulated antiviral ISGs expression by IFN-λs was also confirmed in different donors with IFN-λ concentration effect. In addition to upregulated gene expression, IFN-λs could also downregulate expression of several genes in macrophages, such as V-myc avian myelocytomatosis viral oncogene homolog (MYC) and platelet-derived growth factor receptor, alpha polypeptide (PDGFRA). MYC is the viral homolog of c-myc transduced by several acute transforming retroviruses (Lee and Reddy 1999). MYC expression is driven by the retroviral long terminal repeat in case of retroviral transduction (Lee and Reddy 1999). Studies (Crouch and others 2005) showed that the activity of MYC can be potentiated by virally derived mutations. MYC may also play a role in the processes of HIV infection and replication. PDGFRA belongs to a family of proteins called receptor tyrosine kinases (RTKs) (Rubin and others 2007). Binding of its ligand (PDGF) leads to activation of several downstream signaling molecules, including RAS/MAPK and PI3K (Rubin and others 2007). The signaling pathways stimulated by the PDGFRA protein control many important cellular processes such as cell growth, proliferation, and other oncogenic processes (Rubin and others 2007). It is reported that IFN-γ could decrease the expression of PDGFRA in rat oligodendrocyte progenitor cells (Tanner and others 2011). In addition, a study showed that PDGFRA activation is required for human CMV infection (Soroceanu and others 2008). Overall, further studies are needed to investigate the roles of these downregulated genes in IFN-λ-mediated anti-HIV activity.
Although all 3 members of IFN-λ had the ability to inhibit virus replication, it was reported that IFN-λ3 is more potent than IFN-λ1 and λ2 (Dellgren and others 2009). We compared the anti-HIV activity of IFN-λ1, IFN-λ2, and IFN-λ3 in macrophages. We showed that IFN-λ1 or IFN-λ3 had a better antiviral activity against HIV as compared with IFN-λ2 in treatment groups both before and after virus infection. The different antiviral properties may be due to the different ISGs expression regulated by different members of the IFN-λ family, as these ISGs have been reported to play important roles in IFN-mediated antiviral activity (Lenschow and others 2005; Raychoudhuri and others 2011; Schoggins and Rice 2011).
Induction of the IFN-based antiviral innate immunity depends on several TLRs and RIG-I-like receptors (RLR). TLR-3, TLR-7, and TLR-8 are reported that can recognize viral RNA to trigger the IFN signaling pathway (Thompson and Locarnini 2007; Jensen and Thomsen 2012). In addition to TLRs, RIG-I and MDA-5, which belong to the RLR family, also can recognize viral RNA during viral infections (Thompson and Locarnini 2007; Jensen and Thomsen 2012). Here, we showed that IFN-λs could induce TLR-3, RIG-I, and MDA-5 expression in macrophages, and the induction levels were higher in the IFN-λ1 or IFN-λ3 treatment group than in the IFN-λ2 treatment group. Furthermore, we found that IFN-λ1 or IFN-λ3 also induced higher expression levels of some members of APOBEC3 family than IFN-λ2 (data not shown). These members are cellular cytidine deaminases that have the ability to inhibit the mobility of HIV (Yu and others 2004; Holmes and others 2007). Tetherin, which has been identified as an IFN-α inducible cellular factor (Neil and others 2008), was also induced in macrophages treated with IFN-λs (data not shown). Tetherin is a transmembrane protein that specifically restricts HIV by preventing its release from infected cells (Neil and others 2008). These findings provide sound mechanisms for an IFN-λ-mediated anti-HIV effect in macrophages.
In order to establish a persistent infection in the target cells, virus evolves strategies to evade host immune response, including inhibition of intracellular innate immunity. We showed that the IFN-λ-mediated inhibition of HIV replication was less potent after HIV infection compared with pretreatment, suggesting that HIV may impair IFN-λ-mediated antiviral activity. When we treated cells infected with HIV at day 3 postinfection with IFN-λs, the IFN-λ-mediated antiviral ISGs expression was largely compromised by HIV infection compared with uninfected cells, as well as the TLR-3, RIG-I, and MDA-5 expression. Further studies are needed to investigate the mechanism(s) of the action of HIV-mediated disruption of IFN-λ signaling pathway.
Finally, the new member of type III IFNs, IFN-λ4, had been reported that was associated with impaired clearance of HCV and had anti-HCV activity (Prokunina-Olsson and others 2013); its potential anti-HIV properties need to be investigated in the near future.
Supplementary Material
Acknowledgment
This work was supported by the grants (DA12815, DA22177, DA27550, DA36413, and DA36163) from the National Institutes of Health.
Author Disclosure Statement
The authors declare that there are no conflicts of interest.
References
- Almeida GM, de Oliveira DB, Magalhaes CL, Bonjardim CA, Ferreira PC, Kroon EG. 2008. Antiviral activity of type I interferons and interleukins 29 and 28a (type III interferons) against Apeu virus. Antiviral Res 80(3):302–308 [DOI] [PubMed] [Google Scholar]
- Ank N, Paludan SR. 2009. Type III IFNs: new layers of complexity in innate antiviral immunity. Biofactors 35(1):82–87 [DOI] [PubMed] [Google Scholar]
- Ank N, West H, Bartholdy C, Eriksson K, Thomsen AR, Paludan SR. 2006. Lambda interferon (IFN-lambda), a type III IFN, is induced by viruses and IFNs and displays potent antiviral activity against select virus infections in vivo. J Virol 80(9):4501–4509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand S, Beigel F, Olszak T, Zitzmann K, Eichhorst ST, Otte JM, Diebold J, Diepolder H, Adler B, Auernhammer CJ, Goke B, Dambacher J. 2005. IL-28A and IL-29 mediate antiproliferative and antiviral signals in intestinal epithelial cells and murine CMV infection increases colonic IL-28A expression. Am J Physiol Gastrointest Liver Physiol 289(5):G960– G968 [DOI] [PubMed] [Google Scholar]
- Crouch DH, Fisher F, La Rocca SA, Goding CR, Gillespie DA. 2005. Viral mutations enhance the Max binding properties of the vMyc b-HLH-LZ domain. Nucleic Acids Res 33(16):5235–5242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellgren C, Gad HH, Hamming OJ, Melchjorsen J, Hartmann R. 2009. Human interferon-lambda3 is a potent member of the type III interferon family. Genes Immun 10(2):125–131 [DOI] [PubMed] [Google Scholar]
- Dickensheets H, Sheikh F, Park O, Gao B, Donnelly RP. 2013. Interferon-lambda (IFN-lambda) induces signal transduction and gene expression in human hepatocytes, but not in lymphocytes or monocytes. J Leukoc Biol 93(3):377–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diegelmann J, Beigel F, Zitzmann K, Kaul A, Goke B, Auernhammer CJ, Bartenschlager R, Diepolder HM, Brand S. 2010. Comparative analysis of the lambda-interferons IL-28A and IL-29 regarding their transcriptome and their antiviral properties against hepatitis C virus. PLoS One 5(12):e15200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly RP, Sheikh F, Kotenko SV, Dickensheets H. 2004. The expanded family of class II cytokines that share the IL-10 receptor-2 (IL-10R2) chain. J Leukoc Biol 76(2):314–321 [DOI] [PubMed] [Google Scholar]
- Dumoutier L, Tounsi A, Michiels T, Sommereyns C, Kotenko SV, Renauld JC. 2004. Role of the interleukin (IL)-28 receptor tyrosine residues for antiviral and antiproliferative activity of IL-29/interferon-lambda 1: similarities with type I interferon signaling. J Biol Chem 279(31):32269–32274 [DOI] [PubMed] [Google Scholar]
- Gad HH, Hamming OJ, Hartmann R. 2010. The structure of human interferon lambda and what it has taught us. J Interferon Cytokine Res 30(8):565–571 [DOI] [PubMed] [Google Scholar]
- Hassan NF, Campbell DE, Douglas SD. 1986. Purification of human monocytes on gelatin-coated surfaces. J Immunol Methods 95(2):273–276 [DOI] [PubMed] [Google Scholar]
- Ho WZ, Lioy J, Song L, Cutilli JR, Polin RA, Douglas SD. 1992. Infection of cord blood monocyte-derived macrophages with human immunodeficiency virus type 1. J Virol 66(1):573–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes RK, Malim MH, Bishop KN. 2007. APOBEC-mediated viral restriction: not simply editing? Trends Biochem Sci 32(3):118–128 [DOI] [PubMed] [Google Scholar]
- Hou W, Wang X, Ye L, Zhou L, Yang ZQ, Riedel E, Ho WZ. 2009. Lambda interferon inhibits human immunodeficiency virus type 1 infection of macrophages. J Virol 83(8):3834–3842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen S, Thomsen AR. 2012. Sensing of RNA viruses: a review of innate immune receptors involved in recognizing RNA virus invasion. J Virol 86(6):2900–2910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotenko SV, Gallagher G, Baurin VV, Lewis-Antes A, Shen M, Shah NK, Langer JA, Sheikh F, Dickensheets H, Donnelly RP. 2003. IFN-lambdas mediate antiviral protection through a distinct class II cytokine receptor complex. Nat Immunol 4(1):69–77 [DOI] [PubMed] [Google Scholar]
- Kouroumalis A, Nibbs RJ, Aptel H, Wright KL, Kolios G, Ward SG. 2005. The chemokines CXCL9, CXCL10, and CXCL11 differentially stimulate G alpha i-independent signaling and actin responses in human intestinal myofibroblasts. J Immunol 175(8):5403–5411 [DOI] [PubMed] [Google Scholar]
- Lee CM, Reddy EP. 1999. The v-myc oncogene. Oncogene 18(19):2997–3003 [DOI] [PubMed] [Google Scholar]
- Lenschow DJ, Giannakopoulos NV, Gunn LJ, Johnston C, O'Guin AK, Schmidt RE, Levine B, Virgin HWt. 2005. Identification of interferon-stimulated gene 15 as an antiviral molecule during Sindbis virus infection in vivo. J Virol 79(22):13974–13983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Liu X, Zhou Y, Su SB. 2009. Interferon-lambdas: the modulators of antivirus, antitumor, and immune responses. J Leukoc Biol 86(1):23–32 [DOI] [PubMed] [Google Scholar]
- Li Y, Zhang T, Douglas SD, Lai JP, Xiao WD, Pleasure DE, Ho WZ. 2003. Morphine enhances hepatitis C virus (HCV) replicon expression. Am J Pathol 163(3):1167–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu MQ, Zhou DJ, Wang X, Zhou W, Ye L, Li JL, Wang YZ, Ho WZ. 2012. IFN-lambda3 inhibits HIV infection of macrophages through the JAK-STAT pathway. PLoS One 7(4):e35902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcello T, Grakoui A, Barba-Spaeth G, Machlin ES, Kotenko SV, MacDonald MR, Rice CM. 2006. Interferons alpha and lambda inhibit hepatitis C virus replication with distinct signal transduction and gene regulation kinetics. Gastroenterology 131(6):1887–1898 [DOI] [PubMed] [Google Scholar]
- Neil SJ, Zang T, Bieniasz PD. 2008. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature 451(7177):425–430 [DOI] [PubMed] [Google Scholar]
- Okugawa S, Mekata T, Inada M, Kihara K, Shiki A, Kannabiran K, Kono T, Sakai M, Yoshida T, Itami T, Sudhakaran R. 2013. The SOCS and STAT from JAK/STAT signaling pathway of kuruma shrimp Marsupenaeus japonicus: molecular cloning, characterization and expression analysis. Mol Cell Probes 27(1):6–14 [DOI] [PubMed] [Google Scholar]
- Onoguchi K, Yoneyama M, Takemura A, Akira S, Taniguchi T, Namiki H, Fujita T. 2007. Viral infections activate types I and III interferon genes through a common mechanism. J Biol Chem 282(10):7576–7581 [DOI] [PubMed] [Google Scholar]
- Prokunina-Olsson L, Muchmore B, Tang W, Pfeiffer RM, Park H, Dickensheets H, Hergott D, Porter-Gill P, Mumy A, Kohaar I, Chen S, Brand N, Tarway M, Liu L, Sheikh F, Astemborski J, Bonkovsky HL, Edlin BR, Howell CD, Morgan TR, Thomas DL, Rehermann B, Donnelly RP, O'Brien TR. 2013. A variant upstream of IFNL3 (IL28B) creating a new interferon gene IFNL4 is associated with impaired clearance of hepatitis C virus. Nat Genet 45(2):164–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlings JS, Rosler KM, Harrison DA. 2004. The JAK/STAT signaling pathway. J Cell Sci 117(Pt 8):1281–1283 [DOI] [PubMed] [Google Scholar]
- Raychoudhuri A, Shrivastava S, Steele R, Kim H, Ray R, Ray RB. 2011. ISG56 and IFITM1 proteins inhibit hepatitis C virus replication. J Virol 85(24):12881–12889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robek MD, Boyd BS, Chisari FV. 2005. Lambda interferon inhibits hepatitis B and C virus replication. J Virol 79(6):3851–3854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin BP, Heinrich MC, Corless CL. 2007. Gastrointestinal stromal tumour. Lancet 369(9574):1731–1741 [DOI] [PubMed] [Google Scholar]
- Schoggins JW, Rice CM. 2011. Interferon-stimulated genes and their antiviral effector functions. Curr Opin Virol 1(6):519–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard P, Kindsvogel W, Xu W, Henderson K, Schlutsmeyer S, Whitmore TE, Kuestner R, Garrigues U, Birks C, Roraback J, Ostrander C, Dong D, Shin J, Presnell S, Fox B, Haldeman B, Cooper E, Taft D, Gilbert T, Grant FJ, Tackett M, Krivan W, McKnight G, Clegg C, Foster D, Klucher KM. 2003. IL-28, IL-29 and their class II cytokine receptor IL-28R. Nat Immunol 4(1):63–68 [DOI] [PubMed] [Google Scholar]
- Soroceanu L, Akhavan A, Cobbs CS. 2008. Platelet-derived growth factor-alpha receptor activation is required for human cytomegalovirus infection. Nature 455(7211):391–395 [DOI] [PubMed] [Google Scholar]
- Taniguchi T, Ogasawara K, Takaoka A, Tanaka N. 2001. IRF family of transcription factors as regulators of host defense. Annu Rev Immunol 19:623–655 [DOI] [PubMed] [Google Scholar]
- Tanner DC, Cherry JD, Mayer-Proschel M. 2011. Oligodendrocyte progenitors reversibly exit the cell cycle and give rise to astrocytes in response to interferon-gamma. J Neurosci 31(16):6235–6246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson AJ, Locarnini SA. 2007. Toll-like receptors, RIG-I-like RNA helicases and the antiviral innate immune response. Immunol Cell Biol 85(6):435–445 [DOI] [PubMed] [Google Scholar]
- Uze G, Monneron D. 2007. IL-28 and IL-29: newcomers to the interferon family. Biochimie 89(6–7):729–734 [DOI] [PubMed] [Google Scholar]
- Wang Y, Ye L, Wang X, Li J, Song L, Ho W. 2013. Retinoic acid inducible gene-I (RIG-I) signaling of hepatic stellate cells inhibits hepatitis C virus replication in hepatocytes. Innate Immun 19(2):193–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willey RL, Smith DH, Lasky LA, Theodore TS, Earl PL, Moss B, Capon DJ, Martin MA. 1988. In vitro mutagenesis identifies a region within the envelope gene of the human immunodeficiency virus that is critical for infectivity. J Virol 62(1):139–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witte K, Gruetz G, Volk HD, Looman AC, Asadullah K, Sterry W, Sabat R, Wolk K. 2009. Despite IFN-lambda receptor expression, blood immune cells, but not keratinocytes or melanocytes, have an impaired response to type III interferons: implications for therapeutic applications of these cytokines. Genes Immun 10(8):702–714 [DOI] [PubMed] [Google Scholar]
- Yang K, Puel A, Zhang S, Eidenschenk C, Ku CL, Casrouge A, Picard C, von Bernuth H, Senechal B, Plancoulaine S, Al-Hajjar S, Al-Ghonaium A, Marodi L, Davidson D, Speert D, Roifman C, Garty BZ, Ozinsky A, Barrat FJ, Coffman RL, Miller RL, Li X, Lebon P, Rodriguez-Gallego C, Chapel H, Geissmann F, Jouanguy E, Casanova JL. 2005. Human TLR-7-, -8-, and -9-mediated induction of IFN-alpha/beta and -lambda Is IRAK-4 dependent and redundant for protective immunity to viruses. Immunity 23(5):465–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Q, Chen D, Konig R, Mariani R, Unutmaz D, Landau NR. 2004. APOBEC3B and APOBEC3C are potent inhibitors of simian immunodeficiency virus replication. J Biol Chem 279(51):53379–53386 [DOI] [PubMed] [Google Scholar]
- Zhang L, Jilg N, Shao RX, Lin W, Fusco DN, Zhao H, Goto K, Peng LF, Chen WC, Chung RT. 2011. IL28B inhibits hepatitis C virus replication through the JAK-STAT pathway. J Hepatol 55(2):289–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Lin RT, Li Y, Douglas SD, Maxcey C, Ho C, Lai JP, Wang YJ, Wan Q, Ho WZ. 2005. Hepatitis C virus inhibits intracellular interferon alpha expression in human hepatic cell lines. Hepatology 42(4):819–827 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.