Abstract
Background
Cholera, an infectious disease caused by Vibrio cholerae, is a major public health problem and is a particularly burden in developing countries including Nepal. Although the recent worldwide outbreaks of cholera have been due to V. cholerae El Tor, the classical biotypes are still predominant in Nepal. Serogroup O1 of the V. cholerae classical biotype was the primary cause of a cholera outbreak in Kathmandu in 2012. Thus, this study was designed to know serotypes and biotypes of V. cholerae strains causing recent outbreak with reference to drug resistant patterns. Moreover, we also report the toxigenic strains of V. cholerae from both environmental and clinical specimens by detecting the ctx gene.
Methods
Twenty four V. cholerae (n = 22 from stool samples and n = 2 from water samples) isolated in this study were subjected to Serotyping and biotyping following the standard protocols as described previously. All of the isolates were tested for antimicrobial susceptibility patterns using the modified Kirby-Bauer disk diffusion method as recommended by CLSI guidelines. The screening of the ctx genes (ctxA2-B gene) were performed by PCR method using a pair of primers; C2F (5′-AGGTGTAAAATTCCTTGACGA-3′) and C2R (5′-TCCTCAGGGTATCCTTCATC-3′) to identify the toxigenic strains of V. cholerae.
Results
Among twenty four V. cholerae isolates, 91.7% were clinical and 8.3% were from water samples. Higher rate of V. cholerae infection was found among adults of aged group 20–30 years. All isolates were serogroups O1 of the V. cholerae classical biotype and sub serotype, Ogawa. All isolates were resistant to ampicillin, nalidixic acid and cotrimoxazole. 90.9% were resistant to erythromycin however, tetracycline was found to be the most effective drug for the isolates. All isolates were multidrug resistant (MDR) and possessed a ctx gene of approximately 400 base pairs indicating the toxigenic strains.
Conclusion
Hundred percent strains of V. cholerae were MDR possessing a ctx gene. It suggests that toxigenic strains be identified and proper antibiotic susceptibility testing be conducted. This will allow effective empirical therapy to be used to treat and control cholera.
Keywords: Vibrio cholerae O1 Classical, Resistant profile, Multidrug resistant, Cholera toxin (ctx) gene
Background
Cholera is the second leading cause of mortality worldwide among children under 5 years, and is one of the main causes of morbidity in adults [1,2]. The causative agent of cholera, Vibrio cholerae is a genetically versatile bacterial species [3]. More than two hundred serogroups were identified on the basis of the somatic O antigens [4] among which O1 and O139 are two major virulent strains. Two biotypes of V. cholerae O1; classical and El Tor are the causative agents of the sixth and the seventh pandemics respectively [5]. Organisms of both biotypes of serogroup V. cholerae O1 are further subdivided into Serotypes; Inaba, Ogawa and Hikojima [6]. V. cholerae O1 is still frequently isolated from many outbreak regions of Asian countries [7]. Nepal is still a cholera endemic country where cholera outbreaks occur every year in the major cities including Kathmandu Valley causing significant morbidity and mortality [8-10].
The major virulence factors of cholera are mainly associated with the CTX genetic element which corresponds to CTX Φ (prophage), a lysogenic filamentous bacteriophage. The genetic element comprises of two gene clusters, the core and the RS2 regions. The core region contains ctx genes encoding the cholera toxin (CT) and five more genes encoding the required components for phage morphogenesis [7]. The toxin produced is transported extracellularly by type II secretion system disrupting the ion transport of intestinal epithelial cells [11]. The subsequent loss of water and electrolytes leads to severe secretory diarrhoea, a characteristic of cholera [12]. The presence of such genes confirms the toxigenic strains of V. cholerae.
The poor socio-economic status, inadequate sanitation and poor access to safe drinking water are the major predisposing factors of cholera outbreaks in many cities of Nepal. V. cholerae usually spreads by the faecal-oral route by ingesting faecally contaminated water or food, person to person transmission and direct contact with infected faeces as described in previous studies. Antimicrobial therapy is commonly recommended for shortening the duration or reducing the severity of symptoms as well as lessening bacterial excretion. However the problem of antimicrobial resistance among the agent continues to be alarming. Despite few studies on diarrhoeal diseases in Nepal, there is lack of adequate information on bacterial enteric pathogens and their antimicrobial resistance trend has been changed over a longer time period globally and in Kathmandu valley as well [9,13,14]. In addition, emergence of multidrug resistant V. cholerae isolates is a major problem in developing countries today [15]. Since most diarrhoeal diseases are treated empirically, it is important to know the susceptibility pattern of the prevalent pathogens. Hence, this study aimed to identify the toxigenic strains of V. cholerae isolates from clinical and environmental samples in Kathmandu city and to know their changing resistant profiles.
Methods
Informed consent and ethical approval
Written consent was obtained from the participants’ involved in the research study. The research ethics was approved by Nepal Health Research Council (NHRC), Kathmandu, Nepal.
Isolation, identification and typing of V. cholerae
During cholera outbreak in Kathmandu city in 2012, a total of 450 stool samples from patients with diarrhoea and 30 drinking water samples from the cholera outbreak regions in the Kathmandu city were collected. The samples were enriched in alkaline peptone water (pH-8.4) at 37°C for 4–6 hours, followed by overnight culture on selective media; thiosulphate citrate bile sucrose agar (TCBS-HiMedia). The sucrose fermenting yellow colonies were subjected to biochemical tests [16] and Serotyping using kit (Mast Group and Denka Seiken, Japan) as per the kit’s instructions. The biotyping of the strains were assayed using the Polymyxin B (50 U) sensitivity test, Voges Proskauer reaction in methyl red Voges Proskauer (MRVP-HiMedia) broth medium and chicken RBC agglutination tests [6].
Antibiotic susceptibility test
The antimicrobial susceptibility testing of the isolates to various antimicrobial disks (HiMedia: Ampicillin-10mcg, Nalidixic acid-30mcg, Ciprofloxacin-5mcg, Cotrimoxazole1.25/23.75mcg, Cefotaxime-30mcg, Chloramphenicol-30mcg, Tetracycline-30mcg and Erythromcycin-15mcg) was performed using the modified Kirby-Bauer disk diffusion method as recommended by Clinical and Laboratory Standards Institute guidelines [17]. Escherichia coli (ATCC, 25922) was used for the standardization of the Kirby-Bauer test for correct interpretation of the zone diameters.
Molecular assay
PCR assay was selected as a molecular assay in this study. The genomic DNA of all isolates were extracted and purified from the aerobically grown culture in Luria Bertani (LB) broth and used for the specific PCR for the detection of ctx genes [18]. A pair of primers (Macrogen, Republic of Korea); C2F (5′-AGGTGTAAAATTCCTTGACGA-3′) and C2R (5′-TCCTCAGGGTATCCTTCATC-3′) were used for the gene amplifications as described by Patrick et al. [19]. The reaction mixture for the gene amplification was prepared in 25 μl consisting of 12.5 μl QIAGEN multiplex PCR master mix, 1 μl 10 μM forward primer, 1 μl 10 μM reverse primer, 9.5 μl distilled water and 1.0 μl of template DNA. The amplifications were performed as follows: an initial pre-denaturation at 94°C for 15 minutes followed by 35 cycles at 94°C for 30 seconds (denaturation), 60°C for 60 seconds (primer annealing), 72°C for 60 seconds (DNA extension) and a final elongation was performed at 72°C for 10 minutes on a thermocycler (Thermal cycler Perkin Elmer cetus P11966). The amplified products were fractionated by electrophoresis through 1.5% agarose gel with NEB 100 bp marker DNA which was visualized by staining the gel with ethidium bromide [19].
Data analysis
Data were entered and analyzed using SPSS software for Windows (version 16). Chi square test was used as a statistical tool to correlate between different age groups and V. cholerae infection rate.
Results
Altogether twenty four V. cholerae were isolated of which 91.7% (n = 22) were from patients with diarrhoea and 8.3% (n = 2) were from drinking water samples. Among the clinical isolates, 50% were isolated from adult patients of aged 20–30 years. There was significant difference in V. cholerae infection rate among 20–30 years aged patients as compared to other age groups (p = 0.018).
Serotyping and biotyping of V. cholerae
All strains were found to be serogroup O1, serotype Ogawa and the Classical biotypes (Table 1).
Table 1.
Serotyping and Biotyping of V. cholerae
| Typing methods | Tests performed | Serotypes | No of positive strains (%) | Types |
|---|---|---|---|---|
| Serotyping | Agglutination (Mast Group and Denka Seiken Kit, Japan) | Ogawa | 24 (100) | Serotypes Ogawa |
| Inaba | 0 | |||
| Hikojima | 0 | |||
| Biotyping | Voges Proskauer | 0 | Classical biotypes | |
| Polymyxin B sensitivity | 24 Sensitive (100) | |||
| Chicken cell agglutination | 0 |
Antibiotic resistance patterns
All clinical V. cholerae strains were susceptible to tetracycline. However, 90.9% was susceptibility to both ciprofloxacin and chloramphenicol. The sensitivity to cefotaxime was 81.8%. All isolates were found to be resistance to ampicillin, nalidixic acid and cotrimoxazole and 90.9% isolates were resistance to erythromycin.
Among the environmental V. cholerae isolates, all were resistance to ampicillin, nalidixic acid, cotrimoxazole and erythromycin. Fifty percent (n = 1) isolates were resistance to chloramphenicol as well (Table 2).
Table 2.
General antibiotic susceptibility pattern of V. cholerae (total no = 24)
| Antibiotics used (HiMedia) | Clinical isolates (n = 22) | Environmental isolates (n = 2) | ||
|---|---|---|---|---|
| No. of Resistant (%) | No. of Sensitive (%) | No. of Resistant (%) | No. of Sensitive (%) | |
| Ampicillin | 22 (100) | 0 | 2 (100) | 0 |
| Nalidixic acid | 22 (100) | 0 | 2 (100) | 0 |
| Ciprofloxacin | 2 (9.1) | 20 (90.9) | 0 | 2 (100) |
| Cotrimoxazole | 22 (100) | 0 (81.8) | 2 (100) | 0 |
| Cefotaxime | 4 (18.2) | 18 | 0 | 2 (100) |
| Chloramphenicol | 2 (9.1) | 20 (90.9) | 1 (50) | 1 (50) |
| Tetracycline | 0 | 22 (100) | 0 | 2 (100) |
| Erythromcycin | 20 (90.9) | 2 (9.1) | 2 (100) | 0 |
Antibiotic resistance profiles
Five different types of resistant profiles were observed among the clinical isolates which were named as the clinical resistant type 1 to type 5 profiles. Two isolates were resistance to only three antibiotics; ampicillin, nalidixic acid, cotrimoxazole named as clinical resistant type 1 (CR1) profile. Higher no. of isolates; 68.2% (n = 15) were of the CR2 type. Similarly, the resistant types, CR3, CR4 and CR5 were seen in 13.6%, 4.5% and 4.5% isolates respectively (Table 3).
Table 3.
Antibiotic resistant profile of V. cholerae isolates
| Resistant types | Resistant profiles | No. of isolates (%) |
|---|---|---|
| Clinical isolates | ||
| CR1 | Ampicillin, Nalidixic acid, Cotrimoxazole | 2 (9.1) |
| CR2 | Ampicillin, Nalidixic acid, Cotrimoxazole, Erythromycin | 15 (68.2) |
| CR3 | Ampicillin, Nalidixic acid, Cotrimoxazole, Erythromycin, Cefotaxime | 3 (13.6) |
| CR4 | Ampicillin, Nalidixic acid, Cotrimoxazole, Erythromycin, Chloramphenicol, Ciprofloxacin | 1 (4.5) |
| CR5 | Ampicillin, Nalidixic acid, Cotrimoxazole, Erythromycin, Chloramphenicol, Ciprofloxacin, Cefotaxime | 1 (4.5) |
| Environmental isolates | ||
| ER1 | Ampicillin, Nalidixic acid, Cotrimoxazole, Erythromycin | 1 (50) |
| ER2 | Ampicillin, Nalidixic acid, Cotrimoxazole, Erythromycin, Chloramphenicol | 1 (50) |
Note: CR-Clinical resistant type, ER- Environmental resistant type.
Similarly two different resistant types were observed among V. cholerae isolates from environmental specimen. Fifty percent (n = 1) was of the environmental resistant type 1 (ER1) profile which was found to resist antibiotics such as ampicillin, nalidixic acid, cotrimoxazole and erythromycin. The remaining 50% showed an ER2 type (Table 3).
Multidrug resistant V. cholerae and detection of ctx gene
All of the isolates were found to be multidrug resistant (Table 2) and highly pathogenic strains possessing the ctx gene of approximately 400 base pairs (Figures 1 and 2).
Figure 1.
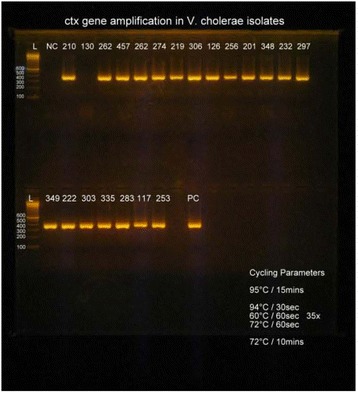
Amplification of ctx gene in V. cholerae isolates (L-Ladder, PC-Positive control, NC-Negative control, clinical V. cholerae isolate; 210, 130, 262, 457 202, 274, 219, 306, 126, 256, 201, 248, 232, 297, 349, 222, 303, 335, 283, 117, 253 showing positive ctx band).
Figure 2.
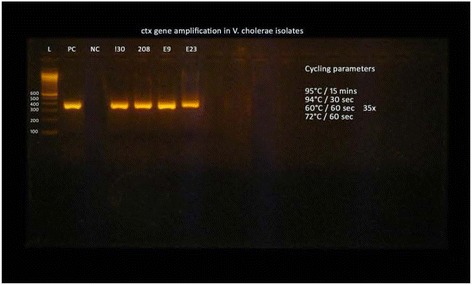
Amplification of ctx gene in V. cholerae isolates (L-Ladder, PC-Positive control, NC-Negative control, clinical V. cholerae isolate; 130, 208 and environmental isolates; E9 E23 showing positive ctx band).
Discussion
Cholera is one of the most predominant diarrhoeal diseases in Nepal even these days. In this study, we found evidence of V. cholerae in 4.9% of cholera cases among patients with diarrhoea and in 6.67% of drinking water samples. A study by Karki and Tiwari in Kathmandu reported 25.1% cholera cases in 2004 [20] and study by Tamang et al. in Kavre reported 31% of positive cases for V. cholerae in the same year [21]. The frequencies of V. cholerae among patients with diarrhoea were found to be still higher in the studies carried out by Kansakar et al. (11.17%), Karki et al. (27.1%) and Shah et al. (8.21%) [22-24]. The higher rate of the pathogens in the previous studies might be due to the hospital based analysis, however the prevalence may be lower among community based studies. The diarrhoeal cases were not only due to V. cholerae in our study. This study also reported 4% of diarrhoea caused by Shigella spp (Shigella flexneri; n = 14 and Shigella sonnei; n = 4) and 1.33% was due to intestinal parasites (Entamoeba histolytica; n = 3, Cyclospora cayetanesis; n = 2 and Blasocystis hominis; n = 1). The detail of results was not shown here. The predisposing factors such as poor sanitation, lack of safe drinking water and unhygienic foods preparations were found to be responsible for the repeated occurrence of the pathogens in Kathmandu and other districts of Nepal.
Vibrio cholerae El Tor O1 Ogawa was responsible for the endemics in Nepal before 2012 [21,24] and previous outbreaks of cholera in Kathmandu valley in 2004 [25]. In contrast, all of the isolates in this study were the V. cholerae O1 serogroups Ogawa and the classical biotype. Infections with classical strains are generally more severe than those with El Tor strains [6]. Three strains; V. cholerae O1 biotype El Tor, V. cholerae O1 biotype Classical and V. cholerae O139 have been frequently isolated in cholera outbreaks in Asian countries [26]. Although classical V. cholerae O1 caused the fifth and sixth pandemics, and presumably the earlier pandemics, the seventh pandemic was attributed to the El Tor biotype, which has been replaced by the classical biotype in this study. The Inaba and Hikojima sero subtypes were not found in this study. Other research conducted in Nepal had reported the occurrence of both Ogawa and Inaba serotypes with an interval of several years [21,27-29]. Children and the elderly people are mostly affected by cholera [2,30,31]. Contrary to this, adult populations of age group 20–30 years were highly infected accounting for 8.7% as compared to all other aged groups (3.4%) in our context (p = 0.018). The studies by Kansakar et al. [22] and Yadav et al. [32] found similar results in which most of the infected patients were adults aged 20 to 29 years and 15 to 29 years respectively. The greater incidence of infections in these groups was found because of their food habits outside the home including consumption of street food.
All the strains in this study were resistant to nalidixic acid, cotrimoxazole and ampicillin suggesting these drugs should not be used in the treatment of cholera. Das et al. also reported 100% resistance to the above three antibiotics [33]. A high incidence of cotrimoxazole resistant V. cholerae O1 strains has been reported in the studies in Africa, Asia and South America [34,35]. The study by Karki and Tiwari [20] found that all the V. cholerae strains were resistance to ampicillin while 97.8% isolates were susceptible to ciprofloxacin. Generally, fluoroquinolones have excellent activity against cholera [20] however; fluoroquinolone resistant strains of V. cholerae have recently been reported from India [33,36,37]. The majority of V. cholerae strains in our study were susceptible to tetracycline (100%), ciprofloxacin (90.9%), cefotaxime (81.8%) and chloramphenicol (90.9%) which may be effective alternative drugs for the treatment of cholera. However the development of resistance needs to be monitored. A similar result was also found by Shah et al. [24] showing sensitivity of 90% and 77.3% to cefotaxime and chloramphenicol respectively. However they showed that 81.8% of strains were resistant to tetracycline. Garg et al., reported high-level resistance to chloramphenicol in India. This result contrasted to our findings [13]. Macrolide resistance was rarely reported in the studies by Harris et al., 2012 and Kanskar et al., 2011 [2,22], yet a high level erythromycin resistance (90.9%) was found in our study. Resistance to erythromycin and other antimicrobial agents among V. cholerae can be acquired through selected mutations over the time, or due to widespread use of antibiotics for prophylaxis in asymptomatic individuals [38].
All V. cholerae were found to be multidrug resistance in the study. MDR cholera epidemics have been reported from Bangladesh [39], Pakistan [40] and Nepal [9,23]. Indiscriminate use of antibiotics in the treatment of cholera and other enteric diseases has led to the emergence of antibiotic resistance among V. cholerae. Epidemics of MDR cholera (both classical and El Tor biotypes) have been reported worldwide [41]. MDR in V. cholerae can be attributed to either a spontaneous mutation or to the horizontal transfer of resistance genes between members of gut coliform or other co-existing microflora and Vibrio spp [42].
All V. cholerae strains tested in our study possessed the ctx gene. Toxigenic strains of V. cholerae contained the essential genetic element, CTX [43,44]. The isolates in the study were thus confirmed as toxigenic strains. The ctx genes are located in the CTX element and encode the cholera toxin CT. This toxin is primarily responsible for the severe secretory diarrhoea in infected person. Thus we screened all the isolates for the presence of ctx gene. Our results showed the presence of ctx gene of approximately 400 bp (~385 bp) in all the tested strains similar as described by Patrick et al. [19]. Similar genes were also detected in the environmental isolates. Chakraborty et al. [45] also found the critical virulence genes in the environmental strains of V. cholerae.
Conclusions
V. cholerae is one of the major agents associated with diarrhoea outbreaks in Nepal with highest propensity during the rainy seasons. The burden of diarrhoea depends upon the strain and not all V. cholerae are toxigenic and epidemic. So, it is suggested that V. cholerae regularly be examined for the presence of the ctx gene from clinical and non-clinical samples to ensure identification of the toxigenic strain. Proper antibiotic susceptibility testing of V. cholerae is important to guide appropriate antimicrobial therapy.
Acknowledgment
The authors are thankful to University Grants Commission (UGC), Sanothimi, Bhaktapur, Nepal for financial support to this work as well as all the participants in this research. The authors are also grateful to related individuals from different households for providing drinking water samples and NHRC for ethically approving this research work. Finally, we appreciate Dr. Hannah Brindle for copy edit of this manuscript.
Abbreviations
- AST
Antibiotic Susceptibility test
- ATCC
American Type Culture Collection
- CLSI
Clinical Laboratory Standard Institutes
- CR
Resistant profile of Clinical V. cholerae isolates
- CT
Cholera Toxin
- Ctx
Cholera toxin producing gene
- DNA
Deoxyribonucleic Acid
- ER
Resistant profile of Environmental V. cholerae isolates
- KCMS
Kantipur College of Medical Sciences
- KTM
Kathmandu
- LB
Luria Bertani
- MDR
Multi Drug Resistance
- NHRC
Nepal Health Research Council
- RLABB
Research Laboratory for Biotechnology and Biochemistry
- TCBS
Thiosulphate Citrate Bile Sucrose Agar
- UGC
University Grants Commission
- PCR
Polymerase Chain Reaction
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
UTS developed this research proposal with help of NA which was supported financially from University Grants Commission, Nepal, performed the molecular work and drafted the initial version of this manuscript. NA contributed to the manuscript writing. RM processed stool samples for isolating and identifying V. cholerae and worked on obtaining ethical approval of this research from Nepal Health Research Council (NHRC), Nepal. MRB analyzed data and edited the manuscript. KRR collected water samples from Kathmandu city and processed on the isolation and identification of V. cholerae. SRB supervised microbiological works. VPA helped and guided molecular biology works. All authors reviewed and approved the final version of manuscript.
Contributor Information
Upendra Thapa Shrestha, Email: upendrats@gmail.com.
Nabaraj Adhikari, Email: adhinarinaba2004@yahoo.com.
Rojina Maharjan, Email: maharjan.rojina@gmail.com.
Megha R Banjara, Email: banjaramr@gmail.com.
Komal R Rijal, Email: rijalkomal@gmail.com.
Shital R Basnyat, Email: basnyatshitalraj@yahoo.com.
Vishwanath P Agrawal, Email: vpa@wlink.com.np.
References
- 1.Bryce J, Boschi-Pinto C, Shibuya K, Black RE, the WHO Child Health Epidemiology Reference Group WHO estimates of the causes of death in children. Lancet. 2005;365:1147–52. doi: 10.1016/S0140-6736(05)71877-8. [DOI] [PubMed] [Google Scholar]
- 2.Harris JB, LaRocque RC, Qadri F, Ryan ET, Calderwood SB. Cholera. Lancet. 2012;379:2466–76. doi: 10.1016/S0140-6736(12)60436-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alam M, Rashed SM, Mannan SB, Islan T, Lizarraga-Partida ML, Delgado G, et al. Occurrence in Mexico, 1998–2008, of Vibro cholerae CTX+ El Tor carrying an additional truncated CTX Prophage. Proc Natl Acad Sci U S A. 2014;111(27):9917–22. doi: 10.1073/pnas.1323408111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chatterjee SN, Chaudhuli K. Lipopolysaccharides of Vibrio cholerae. I. Physical and Chemical Characterization. Biochim Biophys Acta. 2003;1639:65–79. doi: 10.1016/j.bbadis.2003.08.004. [DOI] [PubMed] [Google Scholar]
- 5.Ramamurthy T, Nair GB. Evolving identity of epidemic Vibrio cholerae past and present. Sci Cult. 2010;76:153–9. [Google Scholar]
- 6.Kaper JB, Morris JG, Levine MM. Cholera. Clin Microbiol Rev. 1995;8:48–86. doi: 10.1128/cmr.8.1.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yi Y, Lu N, Liu F, Li J, Zhang R, Jia L, et al. Genome sequence and comparative analysis of a Vibrio cholerae O139 strain E306 isolated from a cholera case in China. Gut Pathogens. 2014;6:3. doi: 10.1186/1757-4749-6-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dixit SM, Johura FT, Manandhar S, Sadique A, Rajbhandari RM, Mannan SB, Rashid M, Islam S, Karmacharya D, Watanabe H, Sack RB, Cravioto A, Alam M: Cholera outbreaks (2012) in three districts of Nepal reveal clonal transmission of multi-drug resistant Vibrio cholerae O1. BMC Infectious Diseases, 2014, 14:392, http://www.biomedcentral.com/1471-2334/14/392. [DOI] [PMC free article] [PubMed]
- 9.Pun SR, Maharjan R, Shrestha D, Pokharel D, Shah Y, Bastola A, et al. An outbreak of Vibrio cholerae in 2012, Kathmandu. Nepal Trop Med Surg. 2013;1:115. [Google Scholar]
- 10.Shrestha UT, Sujakhu H. Coliform and Vibrio cholerae analysis of drinking water collected from cholera outbreak region of Bhaktapur Municipality. International Journal of Environment. 2014;3(3):139–45. doi: 10.3126/ije.v3i3.11073. [DOI] [Google Scholar]
- 11.Sanchez J, Holmgren J. Cholera toxin- afoe & a friend. Indian J Med Res. 2011;133:153–63. [PMC free article] [PubMed] [Google Scholar]
- 12.Maheshari M, Nelapati K, Kiranmayi B. Vibrio cholerae-A Review. Veterinary world. 2011;4(9):423–8. doi: 10.5455/vetworld.2011.423-428. [DOI] [Google Scholar]
- 13.Garg P, Chakraborty S, Basu I, Datta S, Rajendran K, Bhattacharya T, et al. Expanding multiple antibiotic resistance among clinical strains of Vibrio cholerae isolated from 1992–7 in Calcutta, India. Epidemiol Infect. 2000;124:393–9. doi: 10.1017/S0950268899003957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.World Health Organization, WHO Bulletin, Drug-resistant Salmonella, Available http://www.who.int/ mediacentre/factsheets/fs139/en. Accessed March 20, 2009.
- 15.Vila J, Pal T. Update on antibacterial resistance in low-income countries factors favoring the emergence of resistance. The Open Infectious Diseases Journal. 2010;4:38–54. [Google Scholar]
- 16.Forbes BA, Sahm DF, Weissfeild AS, Bailey WR. Bailey and Scotts’ Diagnostic Microbiology (Eds. Forbes BA, Sahm DF, Weissfeild AS). 12th edn, Elsevier Mosby Publisher, 2007, Original from The University of Michigan, pp-105-119, 371–379, ISBN-0323030653, 9780323030656
- 17.CLSI- Clinical Laboratory Standard Institute . Performance standards for antimicrobial susceptibility testing; twenty second informational supplement, 32:M100-S22. 2012. [Google Scholar]
- 18.Ranjbar R, Pourshafie MR, Sadeghifard N, Karami A, Hamidian M, Izadi M, et al. Molecular Characterization of Epidemic Isolates of Vibrio cholerae O1 by Arbitrarily primed PCR (AP-PCR) Iranian J Publ Health. 2008;37(2):83–7. [Google Scholar]
- 19.Patrick GB, Nishibuchi M, Tunung R, Son R. Molecular characterization of clinical isolate of Vibrio cholerae isolated from outbreaks cases in Malaysia. International Food Research Journal. 2012;19(3):1267–74. [Google Scholar]
- 20.Karki A, Tiwari BR. Prevalence of acute diarrhea in Kathmandu valley. J Nepal Med Assoc. 2007;46:175–9. [PubMed] [Google Scholar]
- 21.Tamang MD, Sharma N, Makaju RK, Sarma AN, Koju R, Nepali N, et al. An outbreak of El Tor cholera in Kavre district, Nepal. Kathmandu Univ Med J. 2005;3:138–42. [PubMed] [Google Scholar]
- 22.Kansakar P, Baral P, Malla S, Ghimire GR. Antimicrobial susceptibilities of enteric bacterial pathogens isolated in Kathmandu, Nepal, during 2002–2004. J Infect Dev Ctries. 2011;5:163–8. doi: 10.3855/jidc.1016. [DOI] [PubMed] [Google Scholar]
- 23.Karki R, Bhatta DR, Malla S, Dumre SP. Cholera incidence among patients with diarrhea visiting National public health laboratory, Nepal. Jpn J Infect Dis. 2010;63:185–7. [PubMed] [Google Scholar]
- 24.Shah BK, Sharma S, Shakya G, Upadhyay BP. Multiple drug resistant Vibrio cholerae, Salmonella and Shigella from Nepalgunj Cholera outbreak and different hospitals of Nepal. Nepalese Journal of Biosciences. 2012;2:31–9. [Google Scholar]
- 25.Malla S, Kansakar P, Ghimirey G. Cholera outbreak in Kathmandu valley in 2004: A review of National Public Health Laboratory findings. J Nepal assoc Med Lab Sciences. 2005;7:20–3. [Google Scholar]
- 26.Faruque SM, Albert MJ, Mekalanos JJ. Epidemiology, Genetics, and Ecology of Toxigenic Vibrio cholerae. Microbiol Mol Biol Rev. 1998;62:1301–14. doi: 10.1128/mmbr.62.4.1301-1314.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pokharel M, Sherchand JB, Upreti HC, Katuwal A, Gauchan P. A perspective study on the etiology of diarrhea in children less than 12 years of age attending Kanti Children’s Hospital. J Nepal Paediat Soc. 2009;29:10–6. [Google Scholar]
- 28.Sherchand JB, Yokoo M, Sherchand O, Pant AR, Nakagom O. Burden of enteropathogens associated diarrheal diseases in children hospital. Nepal Scientific World. 2009;7:71–5. [Google Scholar]
- 29.Shrestha SD, Malla S, Adhikari BR, Shakya G, Basnyat SR, Sharma S. Antibiotic susceptibility patterns of Vibrio cholerae isolates. J Nepal Med Assoc. 2010;49(179):232–6. [PubMed] [Google Scholar]
- 30.Rai SK, Bhatta DR. Study of enteropathogens and its predisposing factors in gastroenteritis suspected children attending Kanti Children Hospital, Kathmandu, Nepal. J Nepal Asso Med Lab Sciences. 2004;6:48–53. [Google Scholar]
- 31.Taneja N, Mohan B, Khurana S, Sharma M. Antimicrobial resistance in selected bacterial enteropathogens in north India. Indian J Med Res. 2004;120:39–43. [PubMed] [Google Scholar]
- 32.Yadav DK, Tamrakar D, Baral R, Jha P, Gautam S, Pokharel PK. Outbreak of Cholera in Tilathi VDC Saptari Nepal. Kathmandu Univ Med J. 2012;10:36–9. doi: 10.3126/kumj.v10i4.10992. [DOI] [PubMed] [Google Scholar]
- 33.Das S, Choudhry S, Saha R, Ramachandran VG, Kaur K, Sarkar BL. Emergence of multiple drug resistance Vibrio cholerae O1 in East Delhi. J Infect Dev Ctries. 2011;5:294–8. doi: 10.3855/jidc.1153. [DOI] [PubMed] [Google Scholar]
- 34.Goel AK, Jiang SC. Genetic determinants of virulence, antibiogram and altered biotype among the Vibrio cholerae O1 isolates from different cholera outbreaks in India. Infect Genet Evol. 2010;10:814–8. doi: 10.1016/j.meegid.2009.06.022. [DOI] [PubMed] [Google Scholar]
- 35.Mandomando I, Espasa M, Valles X, Sacarlal J, Sigauque B, Ruiz J, et al. Antimicrobial resistance of Vibrio cholerae O1 serotype Ogawa isolated in Manhica District Hospital, southern Mozambique. J Antimicrob Chemother. 2007;60:662–4. doi: 10.1093/jac/dkm257. [DOI] [PubMed] [Google Scholar]
- 36.Garg P, Sinha S, Chakraborty R, Bhattacharya SK, Nair GB, Ramamurthy T, et al. Emergence of fluoroquinolone-resistant strains of Vibrio cholerae O1 biotype El Tor among hospitalized patients with cholera in Calcutta, India. Antimicrob Agents Chemother. 2001;45:1605–6. doi: 10.1128/AAC.45.5.1605-1606.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mukhopadhyay AK, Basu I, Bhattacharya SK, Bhattacharya MK, Nair GB. Emergence of fluoroquinolone resistance in strains of Vibrio cholerae isolated from hospitalized patients with acute diarrhea in Calcutta, India. Antimicrob Agents Chemother. 1998;42:206–7. doi: 10.1128/aac.42.1.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sharifi-Mood B, Metanat M. Diagnosis, Clinical Management, Prevention, and Control of Cholera. A Review Study Int J Infect. 2014;1(1):e18303. [Google Scholar]
- 39.Shah M, Faruque M, Islam J, Ahmad QS, Biswas K, Faruque ASG. An improved technique for isolation of environmental Vibrio cholerae with epidemic potential: monitoring the emergence of a multiple-antibiotic–resistant epidemic strain in Bangladesh. J Infect Dis. 2006;193:1029–36. doi: 10.1086/500953. [DOI] [PubMed] [Google Scholar]
- 40.Siddique AK, Baqui HA, Eusof A, Haider K, Hossain MA, Bashir I, et al. Survival of classic cholera in Bangladesh. Lancet. 1991;337:1125–7. doi: 10.1016/0140-6736(91)92789-5. [DOI] [PubMed] [Google Scholar]
- 41.Mandal S, Mandal MD, Kumar Pal NK. Cholera: a great global concern. Asian Pac J Trop Med. 2011;4:573–80. doi: 10.1016/S1995-7645(11)60149-1. [DOI] [PubMed] [Google Scholar]
- 42.Speer BS, Shoemaker NB, Salyers AA. Bacterial Resistance to tetracycline: mechanisms, transfer and clinical significance. Clin Microbiol Rev. 1992;25:387–99. doi: 10.1128/cmr.5.4.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Juliana MVB, Ronaldo MA, Irma NGR, Dalia PR, David KRK, Campos C. Prevalence of virulence-associated genes in clinical and environmental Vibrio cholerae strains isolated in Brazil between 1991 and 1999. FEMS Microbiol Lett. 2002;215:15–21. doi: 10.1111/j.1574-6968.2002.tb11364.x. [DOI] [PubMed] [Google Scholar]
- 44.Chen CH, Shimada T, Elhadi N, Radu S, Nishibushi M. Phenotypic and Genotypic characteristic and epidemiological significance of ctx + strains of Vibrio cholerae isolated from seafood in Malaysia. Appl Environ Microbiol. 2004;70:1964–72. doi: 10.1128/AEM.70.4.1964-1972.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chakraborty S, Mukhopadhyay AK, Bhadra RK, Ghosh AN, Mitra R, Shimada T, et al. Virulence genes in Environmental strains of Vibrio cholerae. Appl Environ Microbiol. 2000;66(9):4022–8. doi: 10.1128/AEM.66.9.4022-4028.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]


