Abstract
With the exponential growth of research efforts on non-coding microRNAs (miRNAs) in the past decade, miRNAs have been demonstrated to be important in many major human diseases, including diabetes, heart disease, and cancer. Due to the broad regulatory function of miRNAs, alterations of their expression can have profound consequences on multiple critical genes and pathways. One of the major issues related to the success of treating advanced colorectal cancer is chemoresistance. In this review, we will present some of the recent advancements in miRNA research related to chemoresistance mechanisms to 5-FU based chemotherapy in colorectal cancer and cancer stem cells. We believe that this miRNA-mediated resistance mechanism will offer novel strategies to develop future anti-cancer therapies.
Introduction
Colorectal cancer is the second leading cause of death from cancer in the Western World, responsible annually for an estimated 50,000 deaths in the United States and 500,000 worldwide (1). Recent studies have found that in addition to the genetic changes traditionally associated with carcinogenesis (MLH1, MLH2, MSH6, PMS2), epigenetic alterations (e.g. promoter methylation, miRNAs) also play key roles in colorectal cancer development (2, 3) and metastasis (4, 5). Additionally, there is mounting evidence indicating that post-transcriptional and translational controls mediated by various regulatory molecules, such as RNA binding proteins and non-coding miRNAs, are critically important in the development of various diseases, including cancer (6–8).
In 1993, Ambros et al. made a landmark discovery of a small non-coding RNA, lin-4, which impacted the development of Caenorhabditis elegans (9). This RNA turned out the be the first identified miRNA, a class of non-coding, single-stranded RNAs of ~22 nucleotides that modulate protein expression by promoting RNA degradation, inhibiting mRNA translation, and in some cases affecting transcription. Nearly a decade after the discovery of lin-4, the potential roles of miRNAs in cancer were first reported (8), profoundly changing our understanding of cellular networks and opening the door for new discoveries in cancer research.
To date, over 1000 mammalian miRNAs have been identifiedby cloning and sequencing approaches (4), including several hundred of human miRNAs (10). Although miRNA-mediated mRNA degradation occurs in mammals, most miRNAs are thought to use a secondary mechanism of gene regulation mainly via imperfect base-pairing to the 3′-untranslated regions (3′-UTRs) of their target mRNAs. This leads to the repression of target gene expression post-transcriptionally, likely at the translational level (9, 11, 12). Such regulation can induce rapid changes in protein synthesis without the need for transcriptional activation and subsequent steps in mRNA processing, in effect acting as a new layer of controlling expression of a given protein that is more precise, immediate and energy-efficient than transcriptional regulation alone (13). Furthermore, miRNA-mediated translational regulation has the advantage of being readily reversible, providing the cell with great flexibility in responding to various genotoxic stresses and rapid changes in growth conditions. It has been estimated that over 30% of the protein coding genes were regulated by miRNAs. Clearly, it is essential to know not only the levels of individual mRNAs, but also the extent to which mRNAs are being translated into their corresponding proteins and the miRNAs that regulate these processes. Thus, post-transcriptional and translational control mediated by miRNAs has become a new frontier for cancer research.
miRNA and Colorectal Cancer
It has been well documented that a number of miRNAs play important roles in colorectal cancer (14). Michael et al. first reported that miR-143 and miR-145 was reduced in colorectal specimens in 2003 (15). The first large scale miRNA expression profiling was reported in 2006 by Cummins et al (10). Furthermore, studies from our group have demonstrated that miR-192 and miR-215 directly contribute to one mechanism of chemotherapy resistance to fluoropyrimidines and antifolates in colorectal cancer (16–20). In this review, we will cover some of the recent advancements of miRNA research in colorectal cancer with a focus on chemoresistance.
5-Fluorouracil (5-FU), miRNA, chemoresistance, and colon cancer stem cells
5-fluorouracil (5-FU) was one of the rationally-designed drugs that target thymidylate sythase (TYMS, TS), an enzyme that is responsible for the de novo synthesis of thymidylate, a necessary precursor for DNA biosynthesis. Nearly half a century after its introduction, 5-FU-based chemotherapy is one of the main treatment options for advanced metastatic colorectal cancer and other solid tumors. However, despite the steady improvement in response rate with various modulation strategies such as leucovorin (LV), methotrexate (MTX), FOLFOX4 (oxaliplatin at a dose of 85 mg/m2 as a 2-hour infusion on day 1, every two weeks, plus LV/5-FU), many patients still go through 5-FU based chemotherapy without any benefit, underscoring the need for a set of reliable predictive biomarkers that can guide personalized chemotherapy. To this end, tremendous effort has been put into searching for predictive and prognostic biomarkers in colorectal cancer. Despite extensive investigations of 5-FU and its target TS, the predictive power of TS for 5-FU-based chemotherapy is still under debate (21, 22), and studies evaluating TS, thymidine phosphorylase (TP), and dihydropyrimidine dehydrogenase (DPD); a catabolic enzyme of 5-FU) as predictive biomarkers were unable to confirm the high response rates reported by several retrospective studies (23). From these inconsistent results, it is clear that the search continues for biomarkers that can be used, either alone or in combination with existing biomarkers such as TS and DPD, to predict the likely response to 5-FU-based treatment. Chemoresistance is one of the major factors for the failure of chemotherapy. One effect behind chemoresistance is that chemotherapeutic agents, which primarily affect fast-proliferating cells, are mostly ineffective against the more slowly-proliferating and/or quiescent cancer stem cells, allowing a tumor to reconstitute itself once therapy has ceased. Because of the broad influence and reversible nature of miRNA on gene expression, we believe that miRNAs may offer new insights to this resistance mechanism, in particular with 5-FU-based colorectal cancer chemotherapy. The information we gained may help to develop new strategies to overcome chemoresistance and better predict clinical outcomes.
miRNA, p53 and colorectal cancer p53 is one of the most frequently mutated and/or deleted tumor suppressor genes in colorectal cancer and other tumor types: nearly half of all colorectal cancer patients carry p53 mutations or deletions (24). Until recently, it has been thought that the effects of p53 loss were primarily related to its function in transcriptional regulation, but it is now appreciated that post-transcriptional and translational controls are equally important. Our own research efforts began by systematically investigating which miRNAs are influenced by the p53 tumor suppressor and, by extension, p53 loss. In 2006 we first reported a regulatory relationship between p53 and a number of miRNAs (16), and discovered that nearly half of the 328 miRNA putative promoter regions, including the promoters for miR-34s, miR-192/215, and miR-26a, contain one or more p53 binding sites (16). Subsequently, several groups have demonstrated that miR-34a, which acts as a tumor-suppressor by targeting several cell-cycle genes, is regulated directly by p53 (25–27). In addition, we and two other groups provided further direct evidence that miR-192 is another miRNA that is both regulated by p53 and capable of inducing cell-cycle arrest (17, 28, 29).
Our recent efforts have focused on miRNAs that suppress the expression of thymidylate synthase (TS) and dihydrofolate reductase (DHFR), two important chemotherapeutic targets. We have found that two miRNAs we have previously identified as being regulated by p53, miR-192 and miR-215, target both TS and DHFR mRNA (17, 30). Additionally, Mishra et al reported that miR-24 also regulates DHFR expression, and demonstrated that 829C→T, a naturally occurring SNP near the miR-24 binding site in the 3′ UTR of DHFR, interferes with miR-24 function and, resulting in DHFR overexpression and methotrexate resistance in colorectal tumors (31). Further studies revealed that miR-24 is a potential tumor suppressor capable of reducing tumor cell proliferation in a p53-independent fashion and mediating several key cell cycle related genes such as p21, E2F, Myc and other cell cycle control genes (32, 33). It is also tempting to reason that nature build such redundancy to adope various growth stress conditions by utilizing several different miRNAs to quickly modulate protein expression.
Another potential p53-regulated miRNA is miR-26a. We have demonstrated that miR-26a is down regulated after loss of p53, and that miR-26a may be directly regulated by tumor suppressor p53 in colon cancer (16). A recent report showed that miR-26a not only impact colorectal cancer, it also play key role in liver cancer both as potential therapeutic agent and prognosis biomarker (34, 35).
The positive feedback mechanism between p53 and some of these miRNAs proved to be an important part of the regulatory function and networks of p53 mediated through miRNAs. Due to the broad impact of miRNA on regulating translation rate, miRNAs may be responsible for the fine tuning of the tumor suppressor function of p53 under acute growth environmental changes including genotoxic stress. We have reasons to believe that modulation of miRNA will have a broad impact in colorectal and other cancer types.
miRNAs mediated chemoresistance
To discover miRNAs that may play a role in chemoresistance in colorectal cancer, we focused our efforts on miRNAs that may modulate the expression of key anti-cancer therapeutic targets such as TS, DHFR and histone deacetylases (HDACs). As mentioned briefly above, recent studies from our group have found that miR-215 directly targets the expression of both TS and DHFR (30). Strikingly, our studies found that that despite the reduction of TS and DHFR levels with elevated expression of miR-215, colon cancer cells became more resistant to the TS Tomudex (TDX) and DHFR inhibitor methotrexate (MTX). Further investigation found that this effect was mainly due to the enhanced G2/M cell cycle check point control and reduced cell proliferation. The enhanced G2 check point control was mediated by the suppression of denticleless (DTL), one of the components of CUL4A-DDB1 E3 ubiquitin ligase complex (36), and essential for the early G2/M checkpoint. We found that the induction of p53 and p21 was a result of DTL suppression by miR-215, and that siRNA knock-down of DTL conferred chemoresistance to TDX and also triggered G2/M arrest in colon cancer cells. Taken together, this suggests the presence of a positive feedback loop between miR-215 and p53, mediated through the suppression of DTL (Figure 1).
Figure 1.
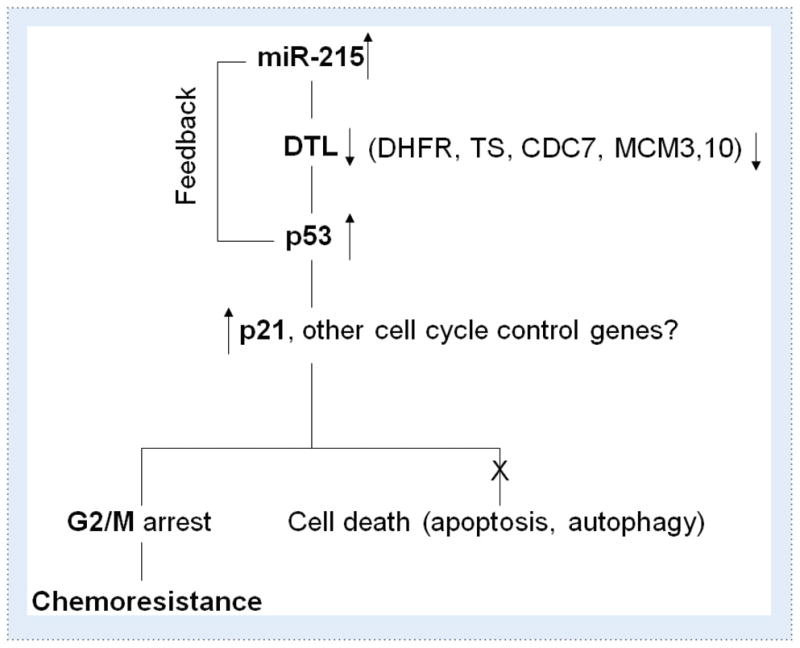
Feedback regulatory mechanism of miR-215 and p53 network in cell cycle control and chemoresistance. The expression of p53 was induced by the suppression of DTL, a critical component of th e CUL4A-DDB1 E3 ubiquitin ligase complex, along with the p53 down stream gene p21. The elevated p53 in turn up-regulates the expression of miR-215. In addition, to DTL, miR-215 also suppresses the expression of TS, DHFR, CDC7, MCM3, MCM10. miR-215 triggers cell cycle arrest at the G2/M phase without triggering cell death, which contribute to chemoresistance to TS and DHFR inhibitors.
In another study, we identifiedthat one of the key targets of miR-140 is HDAC4 (37). Ectopic expression of miR-140 reduced colon cancer cell proliferation and triggers cell cycle arrest. miR-140 over-expression also rendered colon cancer cells more resistant to TDX and MTX treatment. Further studies from our laboratory revealed that colon cancer stem-like cell populations, known to be highly resistant to 5-FU treatment, contained high levels of miR-215 and miR-140 (37). By blocking the activity of miR-140 using anti-miR-140 LNA oligonucleotides, we were able to sensitize colon cancer stem cells to 5-FU treatment (37).
These findings raise an interesting point: previously, miRNAs have been classified as either pro- or anti-oncogenic. However, given the breadth of targets and pathways impacted by a given miRNA, it can target both oncogenes and tumor suppressor genes resulting in a net phenotypic outcome is rather complex. For example, miR-215 and miR-140 appear at first glance to act as tumor suppressors because they can slow tumor growth and reduce tumor size in vivo. However, despite their anti-proliferative effect, both miRNAs produced cells resistant to chemotherapy. This may be a reason that we can cure mice by shrink human tumor xenografts with various approaches, but most of these approaches will not work in human as we were unable to monitor tumor relapse from the small population of chemoresistant tumor cells due to the short life span of the mice. Innovative strategies have to be developed to overcome such difficulties.
miRNA as diagnostic, predictive, and prognostic biomarkers in colorectal cancer
The effort of discovering colorectal cancer related miRNAs begin with expression profiling of miRNAs using colorectal cancer cell lines and tumor specimens. Previously, our lab demonstrated that miRNAs are adequately stable in archival formalin fixed paraffin embedded (FFPE) tissue samples for expression analysis (38), allowing our own and numerous other labs to use large numbers of archival clinical specimens for miRNA-based cancer biomarker discovery. In addition to FFPE samples, circulating miRNAs in body fluids (e.g. blood) have been discovered to be potential cancer biomarker candidates for cancer detection (39). Several recent reports have demonstrated the potential of miRNAs as biomarkers in the diagnosis colorectal cancer (40, 41). Ng et al. revealed that the expression of miR-92 is significantly elevated in plasma of patients with colorectal cancer and can be a potential non-invasive molecular marker for colorectal cancer screening (41). Huang et al. also reported that miR-29a and miR-92a also have value for the diagnosis of advanced colorectal cancer (40). Table 1 summarizes some of the important miRNAs as potential biomarkers in colorectal cancer.
TABLE 1.
| miRNAs | References |
|---|---|
|
| |
| Diagonstic marker
| |
| miR-29a | 40 |
| miR-92a | 41 |
|
| |
| Key targets
| |
| TYMS, TS (miR-192, miR-215 | 17,30 |
| DHFR (miR-192, miR-215, miR-24) | 17,30,31 |
| Bcl-2 (miR-34) | 25–27 |
| HDAC4 (miR-140) | 18 |
| ZEB1, ZEB2 (miR-200c) | 43,44 |
|
| |
| Predictive Biomarker
| |
| miR-181b | 19 |
| let-7g | 19 |
|
| |
| Prognosis
| |
| miR-145 | 42 |
| miR-21 | 45,46 |
| miR-200c | 20 |
| miR-320 | 42 |
| miR-498 | 42 |
Biomarker candidates (diagnostic, predictive and prognostic) of miRNAs in colorectal cancer.
In addition to its use as a diagnostic marker, it has also been shown that miRNA may useful in predicting the possible outcome of treatment. Our laboratory has investigated the potential of miRNAs as a predictive biomarker in colorectal cancer patients treated with 5-FU based chemotherapy, and also investigated the biomarker potential of miRNAs in colorectal cancer patients treated with S-1, the next generation oral 5-FU-based compound for treating colorectal cancer and other solid tumors (19), and have demonstrated that the expression of miR-181b and let-7g was significantly associated with chemoresponse to S-1.
A number of studies have also demonstrated the potential of miRNAs markers as prognostic markers in colorectal cancer. A recent microarray-based miRNA expression profiling study of 10 samples of normal mucosa and 49 stage II colon cancers revealed several differentially expressed miRNAs between normal tissue and tumor microsatellite subtypes, with miR-145 showing the lowest expression in cancer relative to normal tissue. They further demonstrated that a biomarker based on these miRNA expression profiles could not only correctly predict the microsatellite status for the majority of cancers, but also predict recurrence of disease with an overall performance accuracy of 81%, indicating a potential role of miRNAs in determining tumor aggressiveness (42). The expression levels of miR-320 and miR-498, both included in the predictive biomarker, correlated with the probability of recurrence-free survival by multivariate analysis (42).
Our group has discovered that miR-200c also has potential as a prognostic biomarker by providing experimental evidence that high levels of miR-200c were associated with poor prognosis in colorectal cancer (20). The close association of let-7g and miR-200c with clinical outcomes of colorectal cancer is no accident: it is now known that both let-7g and miR-200c plays roles in regulating epithelial-mesenchymal transition (EMT), a key event in cancer progression (43). In the case of miR-200c, this is accomplished by regulating the expression of ZEB1 and ZEB2 (44).
The predictive and prognostic potential of another miRNA, miR-21, has also been demonstrated to be closely associated with clinical outcome in colorectal cancer (45, 46). Shetter et al. found several miRNAs, including highly elevated miR-21, to be associated with low sensitivity and poor response to chemotherapy and poor survival.
Conclusion and Future Perspectives
Post-transcriptional control, miRNAs in particular, are now been recognized as important cellular processes, alteration of which can contribute to a number of human diseases. miRNA-mediated regulation allows cells to fine-tune the expression of multiple genes and pathways via an energy-efficient mechanism that can be both quickly effected and readily reversed, providing an incredible degree of flexibility constantly changing conditions. This highly-adaptive system, however, is also inherited by these cells that become cancerous, contributing to chemoresistance and other survival mechanisms. In the present review, we summarized the role of miRNAs in colorectal cancer and chemoresistance mechanism. The incredible complexity of the miRNA layer of regulation will certainly provide a host of future discoveries of new regulatory mechanisms ranging from the level of the individual genes to vast regulatory networks. With the continual advancement of knowledge in the domains of miRNA, mRNA, protein expression, and computational biology, we come ever closer to fully understanding the complex miRNA-mediated regulatory pathways and networks in both cancer and normal cells. This knowledge, in turn, will no doubt continually offer new therapeutic strategies and diagnostic/prognostic biomarkers for the treatment of colorectal cancer and other human diseases.
Acknowledgments
The author thanks Matthew A. Titimus for his critical reading and comments on the manuscript. This work was supported by Stony-Brook Translational Research Laboratory Start-up fund (J. Ju) and NIH CA114043 (J. Ju)
Biography
Jingfang Ju, Ph.D. is currently the Co-Director of the Translational Research Laboratory at Stony Brook University Medical Center. He was the founding Director of the Cancer Genomics Program at the Mitchell Cancer Institute before joining Stony Brook University. His research is currently focusin on elucidating translational control mediated by non-coding miRNAs and their impact on chemoresistance and cancer stem cells. He earned his Ph.D. in Molecular Biology and Biochemistry from the University of Southern California, Los Angeles, in which he investigated the chemoresistance mechanism of fluoropyrimidines in colon cancer.
References
- 1.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 2.Weisenberger DJ, Siegmund KD, Campan M, Young J, Long TI, Faasse MA, Kang GH, Widschwendter M, Weener D, Buchanan D, Koh H, Simms L, Barker M, Leggett B, Levine J, Kim M, French AJ, Thibodeau SN, Jass J, Haile R, Laird PW. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet. 2006;38:787–793. doi: 10.1038/ng1834. [DOI] [PubMed] [Google Scholar]
- 3.Widschwendter M, Fiegl H, Egle D, Mueller-Holzner E, Spizzo G, Marth C, Weisenberger DJ, Campan M, Young J, Jacobs I, Laird PW. Epigenetic stem cell signature in cancer. Nat Genet. 2007;39:157–158. doi: 10.1038/ng1941. [DOI] [PubMed] [Google Scholar]
- 4.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 5.Calin GACC. MicroRNA-cancer connection: the beginning of a new tale. Cancer Res. 2006;66:7390–7394. doi: 10.1158/0008-5472.CAN-06-0800. [DOI] [PubMed] [Google Scholar]
- 6.Ju J, Pedersen-Lane J, Maley F, Chu E. Regulation of p53 expression by thymidylate synthase. Proc Natl Acad Sci U S A. 1999;96:3769–3774. doi: 10.1073/pnas.96.7.3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fu L, Minden MD, Benchimol S. Translational regulation of human p53 gene expression. Embo J. 1996;15:4392–4401. [PMC free article] [PubMed] [Google Scholar]
- 8.Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, Rassenti L, Kipps T, Negrini M, Bullrich F, Croce CM. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 10.Cummins JM, He Y, Leary RJ, Pagliarini R, Diaz LA, Jr, Sjoblom T, Barad O, Bentwich Z, Szafranska AE, Labourier E, Raymond CK, Roberts BS, Juhl H, Kinzler KW, Vogelstein B, Velculescu VE. The colorectal microRNAome. Proc Natl Acad Sci U S A. 2006;103:3687–3692. doi: 10.1073/pnas.0511155103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pillai RS, Bhattacharyya SN, Artus CG, Zoller T, Cougot N, Basyuk E, Bertrand E, Filipowicz W. Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science. 2005;309:1573–1576. doi: 10.1126/science.1115079. [DOI] [PubMed] [Google Scholar]
- 12.Ruvkun G. Clarifications on miRNA and cancer. Science. 2006;311:36–37. doi: 10.1126/science.311.5757.36d. [DOI] [PubMed] [Google Scholar]
- 13.Dony C, Kessel M, Gruss P. Post-transcriptional control of myc and p53 expression during differentiation of the embryonal carcinoma cell line F9. Nature. 1985;317:636–639. doi: 10.1038/317636a0. [DOI] [PubMed] [Google Scholar]
- 14.Slaby O, Svoboda M, Michalek J, Vyzula R. MicroRNAs in colorectal cancer: translation of molecular biology into clinical application. Mol Cancer. 2009;8:102. doi: 10.1186/1476-4598-8-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Michael MZ, van Holst Pellekaan OCSMNG, Young GP, James RJ. Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol Cancer Res. 2003;1:882–891. [PubMed] [Google Scholar]
- 16.Xi Y, Shalgi R, Fodstad O, Pilpel Y, Ju J. Differentially regulated micro-RNAs and actively translated messenger RNA transcripts by tumor suppressor p53 in colon cancer. Clin Cancer Res. 2006;12:2014–2024. doi: 10.1158/1078-0432.CCR-05-1853. [DOI] [PubMed] [Google Scholar]
- 17.Song B, Wang Y, Kudo K, Gavin EJ, Xi Y, Ju J. miR-192 Regulates dihydrofolate reductase and cellular proliferation through the p53-microRNA circuit. Clin Cancer Res. 2008;14:8080–8086. doi: 10.1158/1078-0432.CCR-08-1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Song B, Wang Y, Xi Y, Kudo K, Bruheim S, Botchkina GI, Gavin E, Wan Y, Formentini A, Kornmann M, Fodstad O, Ju J. Mechanism of chemoresistance mediated by miR-140 in human osteosarcoma and colon cancer cells. Oncogene. 2009;28:4065–4074. doi: 10.1038/onc.2009.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakajima G, Hayashi K, Xi Y, Kudo K, Uchida K, Takasaki K, Yamamoto M, Ju J. Non-coding MicroRNAs hsa-let-7g and hsa-miR-181b are Associated with Chemoresponse to S-1 in Colon Cancer. Cancer Genomics Proteomics. 2006;3:317–324. [PMC free article] [PubMed] [Google Scholar]
- 20.Xi Y, Formentini A, Chien M, Weir DB, Russo JJ, Ju J, Kornmann M. Prognostic Values of microRNAs in Colorectal Cancer. Biomark Insights. 2006;2:113–121. [PMC free article] [PubMed] [Google Scholar]
- 21.Salonga D, Danenberg KD, Johnson M, Metzger R, Groshen S, Tsao-Wei DD, Lenz HJ, Leichman CG, Leichman L, Diasio RB, Danenberg PV. Colorectal tumors responding to 5-fluorouracil have low gene expression levels of dihydropyrimidine dehydrogenase, thymidylate synthase, and thymidine phosphorylase. Clin Cancer Res. 2000;6:1322–1327. [PubMed] [Google Scholar]
- 22.Showalter SL, Showalter TN, Witkiewicz A, Havens R, Kennedy EP, Hucl T, Kern SE, Yeo CJ, Brody JR. Evaluating the drug-target relationship between thymidylate synthase expression and tumor response to 5-fluorouracil. Is it time to move forward? Cancer Biol Ther. 2008;7:986–994. doi: 10.4161/cbt.7.7.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smorenburg CH, Peters GJ, van Groeningen CJ, Noordhuis P, Smid K, van Riel AM, Dercksen W, Pinedo HM, Giaccone G. Phase II study of tailored chemotherapy for advanced colorectal cancer with either 5-fluouracil and leucovorin or oxaliplatin and irinotecan based on the expression of thymidylate synthase and dihydropyrimidine dehydrogenase. Ann Oncol. 2006;17:35–42. doi: 10.1093/annonc/mdj046. [DOI] [PubMed] [Google Scholar]
- 24.Hollstein M, Sidransky D, Vogelstein B, Harris CC. p53 mutations in human cancers. Science. 1991;253:49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- 25.He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, Jackson AL, Linsley PS, Chen C, Lowe SW, Cleary MA, Hannon GJ. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chang TC, Wentzel EA, Kent OA, Ramachandran K, Mullendore M, Lee KH, Feldmann G, Yamakuchi M, Ferlito M, Lowenstein CJ, Arking DE, Beer MA, Maitra A, Mendell JT. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell. 2007;26:745–752. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raver-Shapira NME, Meiri E, Spector Y, Rosenfeld N, Moskovits N, Bentwich Z, Oren M. Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol Cell. 2007;26:731–743. doi: 10.1016/j.molcel.2007.05.017. [DOI] [PubMed] [Google Scholar]
- 28.Georges Sara A, MCB, Kim Soo-yeon, Schelter Janell M, Guo Jane, Chang Aaron N, Jackson Aimee L, Carleton Michael O, Linsley Peter S, Cleary Michele A, Nelson Chau B. Coordinated Regulation of Cell Cycle Transcripts by p53-Inducible microRNAs, miR-192 and miR-215. Cancer Res. 2008;68:10105–10112. doi: 10.1158/0008-5472.CAN-08-1846. [DOI] [PubMed] [Google Scholar]
- 29.Braun Christian J, XZ, Savelyeva Irina, Wolff Sonja, Moll Ute M, Schepeler Troels, Ørntoft Torben F, Andersen Claus L, Dobbelstein Matthias. p53-Responsive MicroRNAs 192 and 215 Are Capable of Inducing Cell Cycle Arrest. Cancer Res. 2008;68:10094–10104. doi: 10.1158/0008-5472.CAN-08-1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Song B, Wang T, Titmus M, Botchkina G, Formentini A, Kornmann M, Ju J. Molecular mechanism of chemoresistance by miR-215 in osteosarcoma and colon cancer cells Mol Cancer 9:96. 2010 doi: 10.1186/1476-4598-9-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mishra PJ, HR, Mishra PJ, Longo-Sorbello GS, Banerjee D, Bertino JR. A miR-24 microRNA binding-site polymorphism in dihydrofolate reductase gene leads to methotrexate resistance. Proc Natl Acad Sci U S A. 2007;104:13513–13518. doi: 10.1073/pnas.0706217104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mishra PJ, Song B, Wang Y, Humeniuk R, Banerjee D, Merlino G, Ju J, Bertino JR. MiR-24 tumor suppressor activity is regulated independent of p53 and through a target site polymorphism. PLoS One. 2009;4:e8445. doi: 10.1371/journal.pone.0008445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lal A, Navarro F, Maher CA, Maliszewski LE, Yan N, O’Day E, Chowdhury D, Dykxhoorn DM, Tsai P, Hofmann O, Becker KG, Gorospe M, Hide W, Lieberman J. miR-24 Inhibits cell proliferation by targeting E2F2, MYC, and other cell-cycle genes via binding to “seedless” 3′UTR microRNA recognition elements. Mol Cell. 2009;35:610–625. doi: 10.1016/j.molcel.2009.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ji J, Shi J, Budhu A, Yu Z, Forgues M, Roessler S, Ambs S, Chen Y, Meltzer PS, Croce CM, Qin LX, Man K, Lo CM, Lee J, Ng IO, Fan J, Tang ZY, Sun HC, Wang XW. MicroRNA expression, survival, and response to interferon in liver cancer. N Engl J Med. 2009;361:1437–1447. doi: 10.1056/NEJMoa0901282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kota J, Chivukula RR, O’Donnell KA, Wentzel EA, Montgomery CL, Hwang HW, Chang TC, Vivekanandan P, Torbenson M, Clark KR, Mendell JR, Mendell JT. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell. 2009;137:1005–1017. doi: 10.1016/j.cell.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sansam CL, Shepard JL, Lai K, Ianari A, Danielian PS, Amsterdam A, Hopkins N, Lees JA. DTL/CDT2 is essential for both CDT1 regulation and the early G2/M checkpoint. Genes Dev. 2006;20:3117–3129. doi: 10.1101/gad.1482106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Song B, Wang Y, Xi Y, Kudo K, Bruheim S, Botchkina GI, Gavin E, Wan Y, Formentini A, Kornmann M, Fodstad O, Ju J. Mechanism of chemoresistance mediated by miR-140 in human osteosarcoma and colon cancer cells. Oncogene. 2009;28:4065–4074. doi: 10.1038/onc.2009.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xi Y, Nakajima G, Gavin E, Morris CG, Kudo K, Hayashi K, Ju J. Systematic analysis of microRNA expression of RNA extracted from fresh frozen and formalin-fixed paraffin-embedded samples. Rna. 2007;13:1668–1674. doi: 10.1261/rna.642907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O’Briant KC, Allen A, Lin DW, Urban N, Drescher CW, Knudsen BS, Stirewalt DL, Gentleman R, Vessella RL, Nelson PS, Martin DB, Tewari M. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang Z, Huang D, Ni S, Peng Z, Sheng W, Du X. Plasma microRNAs are promising novel biomarkers for early detection of colorectal cancer. Int J Cancer. 2009;27(1):118–126. doi: 10.1002/ijc.25007. [DOI] [PubMed] [Google Scholar]
- 41.Ng EK, Chong WW, Jin H, Lam EK, Shin VY, Yu J, Poon TC, Ng SS, Sung JJ. Differential expression of microRNAs in plasma of patients with colorectal cancer: a potential marker for colorectal cancer screening. Gut. 2009;58:1375–1381. doi: 10.1136/gut.2008.167817. [DOI] [PubMed] [Google Scholar]
- 42.Schepeler T, Reinert JT, Ostenfeld MS, Christensen LL, Silahtaroglu AN, Dyrskjot L, Wiuf C, Sorensen FJ, Kruhoffer M, Laurberg S, Kauppinen S, Orntoft TF, Andersen CL. Diagnostic and prognostic microRNAs in stage II colon cancer. Cancer Res. 2008;68:6416–6424. doi: 10.1158/0008-5472.CAN-07-6110. [DOI] [PubMed] [Google Scholar]
- 43.Peter ME. Let-7 and miR-200 microRNAs: guardians against pluripotency and cancer progression. Cell Cycle. 2009;8:843–852. doi: 10.4161/cc.8.6.7907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y, Goodall GJ. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 45.Schetter AJ, Leung SY, Sohn JJ, Zanetti KA, Bowman ED, Yanaihara N, Yuen ST, Chan TL, Kwong DL, Au GK, Liu CG, Calin GA, Croce CM, Harris CC. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA. 2008;299:425–436. doi: 10.1001/jama.299.4.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Slaby O, Svoboda M, Fabian P, Smerdova T, Knoflickova D, Bednarikova M, Nenutil R, Vyzula R. Altered expression of miR-21, miR-31, miR-143 and miR-145 is related to clinicopathologic features of colorectal cancer. Oncology. 2007;72:397–402. doi: 10.1159/000113489. [DOI] [PubMed] [Google Scholar]


