Abstract
The Arabidopsis thalianaT-DNA insertion mutant rh57-1 exhibited hypersensitivity to glucose (Glc) and abscisic acid (ABA). The other two rh57 mutants also showed Glc hypersensitivity similar to rh57-1, strongly suggesting that the Glc-hypersensitive feature of these mutants results from mutation of AtRH57. rh57-1 and rh57-3 displayed severely impaired seedling growth when grown in Glc concentrations higher than 3%. The gene, AtRH57 (At3g09720), was expressed in all Arabidopsis organs and its transcript was significantly induced by ABA, high Glc and salt. The new AtRH57 belongs to class II DEAD-box RNA helicase gene family. Transient expression of AtRH57-EGFP (enhanced green fluorescent protein) in onion cells indicated that AtRH57 was localized in the nucleus and nucleolus. Purified AtRH57-His protein was shown to unwind double-stranded RNA independent of ATP in vitro. The ABA biosynthesis inhibitor fluridone profoundly redeemed seedling growth arrest mediated by sugar. rh57-1 showed increased ABA levels when exposed to high Glc. Quantitative real time polymerase chain reaction analysis showed that AtRH57 acts in a signaling network downstream of HXK1. A feedback inhibition of ABA accumulation mediated by AtRH57 exists within the sugar-mediated ABA signaling. AtRH57 mutation and high Glc conditions additively caused a severe defect in small ribosomal subunit formation. The accumulation of abnormal pre-rRNA and resistance to protein synthesis-related antibiotics were observed in rh57 mutants and in the wild-type Col-0 under high Glc conditions. These results suggested that AtRH57 plays an important role in rRNA biogenesis in Arabidopsis and participates in response to sugar involving Glc- and ABA signaling during germination and seedling growth.
Keywords: RNA helicase, glucose-hypersensitive, abscisic acid, rRNA biogenesis, seedling growth, Arabidopsis thaliana seeds
Introduction
RNA helicases contain a large gene family found in all kingdoms (Linder, 2006). They are involved in many different cellular processes including ribosome biogenesis, RNA splicing, maturation, transport, editing, RNA interference, transcription, and mRNA stabilization and degradation (Cordin et al., 2006). The DEAD-box RNA helicases, by far the largest family of RNA helicases, has the sequence of Asp-Glu-Ala-Asp (D-E-A-D) in motif II (Linder and Jankowsky, 2011). In spite of the sequence similarity of DEAD-box RNA helicases within the core helicase regions, each DEAD-box protein is believed to play various key roles in plant development (Linder and Jankowsky, 2011). The proteins have been intensively studied in animals and yeasts (Kemp and ImLer, 2009; Sahni et al., 2010) but only a few DEAD-box members have been identified in plants. In Arabidopsis, at least 120 members of the RNA helicases can be predicted using the TAIR database (http://www.arabidopsis.org/) and 58 DEAD-box RNA helicases have been identified thus far (Boudet et al., 2001). For instance, loss of the Arabidopsis DEAD-box protein ISE1 leads to abnormal mitochondria and enhanced cell-to-cell transport through plasmodesmata (Stonebloom et al., 2009). Arabidopsis DEAD-box proteins are required for the development of female gametophytes and play a role in rRNA biogenesis (Huang et al., 2010a; Liu et al., 2010). Recently, both AtRH3 and AtRH22 have been reported to affect chloroplast ribosome biogenesis (Asakura et al., 2012; Chi et al., 2012).
Aside from participating in different housekeeping processes, DEAD-box RNA helicase expression responds to various stresses (Owttrim, 2006; Vashisht and Tuteja, 2006). For example, ectopic expressions of the DEAD-box proteins PDH45 and PDH47 in peas have been respectively shown to confer salt resistance in tobacco (Sanan-Mishra et al., 2005) and promote responses to cold and salinity stresses in shoots and roots (Vashisht et al., 2005). In Arabidopsis, DEAD-box protein LOS4 has been demonstrated to be essential in exporting mRNA and regulating the expression of CBF factor under the condition of chilling stress (Gong et al., 2005). Three DEAD-box RNA helicases AtRH5, AtRH9, and AtRH25 also respond to multiple abiotic stresses in Arabidopsis (Kant et al., 2007; Kim et al., 2008). All these reports suggest a crucial role of plant helicases in stress tolerance; however, the response mechanism of RNA helicases to glucose (Glc), an important signaling factor affecting plant development has yet to be studied except that the UPF1 RNA helicase was found to alter sugar signaling in Arabidopsis (Yoine et al., 2006). DEAD-box proteins are diverse in structure and modulate various biological processes. Thus, the exact role of most plant DEAD-box proteins largely remains unclear and requires further studies.
Previous studies have demonstrated that high Glc causes abscisic acid (ABA) accumulation, resulting in a delay in germination and inhibition of the early stages of seedling growth (Rolland et al., 2002; Gibson, 2005). Under various stress conditions, the arrest of seedling growth is also mediated by ABA (Lopez-Molina et al., 2001). For gene regulation, high Glc may promote ABA biosynthesis and signaling gene expression and increase ABA content in the cell (Cheng et al., 2002). Most of the ABA biosynthesis and ABA-insensitive mutants described thus far are insensitive to high Glc. A number of negative and positive regulators controlling ABA signaling pathways have recently been identified (Stone et al., 2006; Bu et al., 2009; Carvalho et al., 2010; Huang et al., 2010b). Identification of these regulators emphasizes the complexity of the plant signaling transduction network.
The present study reports the phenotypic and molecular analyses of rh57 mutants showing defects in the Glc control of seedling growth, and gene expression. A feedback inhibition of ABA accumulation mediated by AtRH57 exists within the sugar-mediated ABA signaling. AtRH57 mutation and high amounts of Glc additively impair small ribosomal subunit formation. AtRH57 plays an important role in pre-rRNA processing in Arabidopsis.
Results
rh57 mutants are hypersensitive to Glc-dependent inhibition of germination and early seedling growth
The seedlings of T-DNA insertion mutants, rh57-1,rh57-2, and rh57-3, grew smaller and slightly pale in color when compared with wild-type (WT) of Arabidopsis thaliana under normal growth conditions (Figure1). Further, full germination of rh57-1 and rh57-3 seeds, measured as radicle protrusion from seed coat, occurred slightly later than WT seeds; approximately 2 more days were needed to reach full seed germination (Figure S1).
Figure 1.
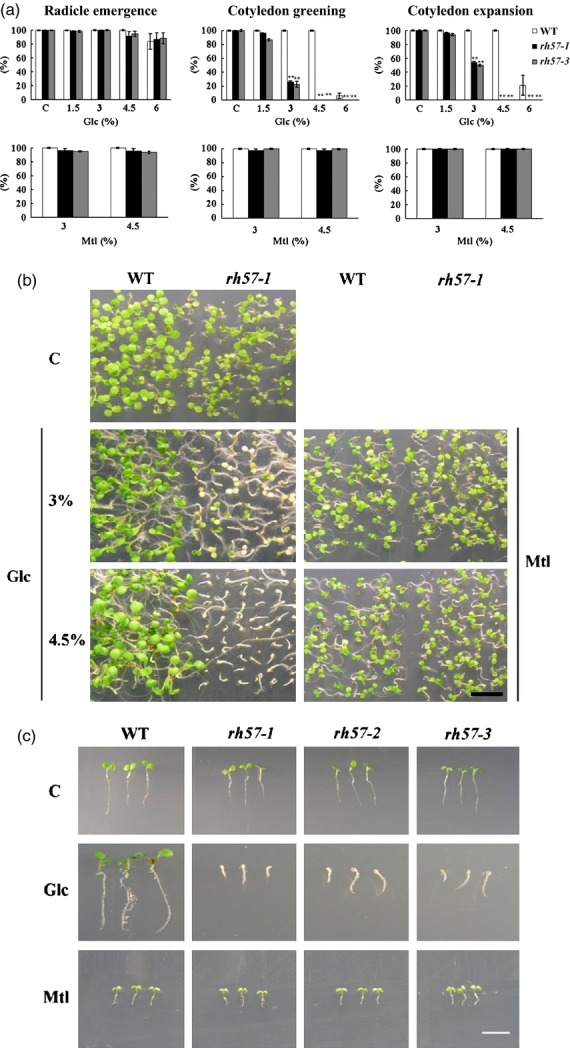
Effects of glucose (Glc) on seed germination and early seedling development of rh57 mutants. (a) Wild-type (WT, Col-0 white bars) of Arabidopsis thaliana,rh57-1 (SALK_008887, black bars), and rh57-3 (SALK_019721, gray bars) seedlings were grown in sugar-free MS medium with or without the indicated concentrations of Glc and mannitol (Mtl) as a control. The percentages of seed germination, cotyledon greening and expansion were scored 9 days after stratification. Error bars represent standard deviation (SD; t-test: ** P < 0.01). (b) Representative images of WT and rh57-1 seedlings. (c) Glc-hypersensitive phenotypes of rh57 mutants in the presence of 4.5% Glc. Data were obtained from three biologically independent experiments. Bar = 5 mm.
Addition of 6% Glc to the Murashige and Skoog (MS) medium (Murashige and Skoog, 1962) decreased germination percentage and induced post-germinative growth arrest with a concomitant block in cotyledon greening and expansion in WT Arabidopsis seedlings (Zhou et al., 1998). rh57-1 seedlings displayed a similar phenotype when grown in the presence of indicated Glc conditions (Figure1b). Further, rh57-2 and rh57-3 also displayed strong post-germinative growth arrest similar to rh57-1 when seeds were grown in MS medium that contained 4.5% Glc (Figure1c). Their cotyledons remained white to fairly pale green compared with WT seedlings. In addition, rh57 roots were markedly shorter than WT roots 9 days after germination, indicating significant effects of the mutation. Moreover, true leaves were not observed in all three rh57 backgrounds. In contrast, the three mutants showed no visible phenotypic differences compared with WT seedlings in the presence of 4.5% mannitol (Mtl). These observations suggested that the Glc-hypersensitive feature of these rh57 mutants may indeed result from mutation of AtRH57 (At3g09720). Quantitative analysis was taken to examine seed germination and cotyledon development of these mutants. When grown in MS medium for 9 days, rh57 mutant cotyledon greening and expansion were significantly affected by the addition of Glc concentrations more than 3% (Figure1a). In contrast with Glc, Mtl induced similar effects in WT and mutant seedlings under the three parameters studied. Therefore, rh57 mutants exhibit arrest in seedling development, suggesting that rh57 mutation increases plant sensitivity to relatively high concentrations of Glc.
rh57 mutation also increased the sensitivity of dark-grown Arabidopsis seedlings to the inhibitory effect of Glc on hypocotyl length (Figure S2). Both rh57-1 and rh57-3 seedlings displayed similar hypocotyl length when compared with WT in the absence of Glc addition. In contrast, the length of mutant hypocotyls was markedly decreased with Glc higher than 3%. Nevertheless, both mutants exhibited hypocotyls similar to those of WT seeds in the presence of either 3 or 4.5% Mtl (Figure S2), suggesting that phenotypic alteration specifically responds to Glc.
Molecular characterization of rh57 mutants
In addition to rh57-1, the T-DNA insertion mutants rh57-2 and rh57-3 were examined (Figure S3a). Polymerase chain reaction (PCR) analyses with the AtRH57-specific primer pair (LP + RP) flanking the T-DNA insertion site amplified the expected size of the AtRH57 gene product from WT seeds but not from mutants, indicating that AtRH57 is disrupted in these mutants (Figure S3b). In contrast, PCR with the T-DNA-specific primer (BP) and AtRH57-specific primer (RP) amplified a product from mutants but not from WT seeds (Figure S3b), confirming that insertion occurs in AtRH57.
Portions of the 1626 bp open reading frame of AtRH57 were amplified by RT-PCR to help define the physiological function of AtRH57 and describe accumulation of the AtRH57 transcript. As shown in Figure S3(c), AtRH57 transcript levels were absent or markedly reduced in the insertion lines. Low levels of the product were detected with a primer pair that amplified a region upstream of the insertion site (primer pair AtRH57-A) in rh57-1 and rh57-2 seedling mRNAs but no product was detected in rh57-3. Conversely, no product was detected in rh57-2 mRNA but low levels of transcripts were detected in rh57-1 and rh57-3 using primer pair AtRH57-B. RT-PCR analysis indicated that the gene is expressed in all plant organs (Figure S3d). GeneChip data analysis of each stage and period of seed development in Arabidopsis revealed that AtRH57 is expressed in different parts of seed tissues in various developmental stages (Figure S4).
AtRH57 is a class II DEAD-box RNA helicase gene protein and localized in the nucleus and nucleolus
In addition to the consensus phenylalanine (F) at position 139, AtRH57 possesses all nine highly conserved motifs that represent the characteristic feature of DEAD-box proteins (Figure S5). AtRH57 contains a DESD sequence in motif II and is thus classified into the DExD group. All DEAD-box RNA helicases lack long amino (N)- and carboxyl (C)-terminal extensions (Cordin et al., 2006). However, AtRH57 contains a relatively long N-terminal extension of 138 amino acids upstream from the conserved F, similar to AtRH2 (Figure S5).
All AtRH genes have been classified into four classes based on a comparison of the structures of regions coding for the catalytic domain (Boudet et al., 2001). However, this new AtRH57 gene was not included in the list at the time of classification. According to the criteria set by Boudet et al. (2001), AtRH57 can be categorized into Class II for two reasons: First, similar to other AtRH genes marked with asterisks in Figure S6(a), AtRH57 does not exhibit completely identical or partially identical structures. Second, similar to other Class II helicases, AtRH57 shares at least one intron at an identical position with other Class II genes (Figure S6b). For example, the consensus xLxLDExD (indicated by an open arrow), where x represents variable amino acids, resides at the beginning of the exon that contained DExD box (motif II). Conserved introns at positions 172, 175, and 176 immediately preceding the consensus sequence have been found in AtRH32,AtRH35, and AtRH36, respectively, and these three genes have been classified as Class II members (Boudet et al., 2001). Similarly, an intron X (indicated by a closed arrow) has also been observed in AtRH57 at a position preceding the consensus sequence, indicating that AtRH57 is a Class II helicase gene.
NucPred (http://www.sbc.su.se/~maccallr/nucpred/cgi-bin/single.cgi) analysis of the deduced amino acid sequence of AtRH57 revealed the presence of a bipartite nuclear localization sequence (NLS, 62–91 amino acid residues), suggesting that the protein is localized in the nucleus with a 93% probability. To examine the subcellular localization of AtRH57, we generated a construct of AtRH57 fused with the sequence of enhanced green fluorescent protein (EGFP) under the control of a 35S promoter. The construct was then introduced into onion epidermal cells by particle bombardment. Fluorescence microscopy revealed that AtRH57-EGFP was localized in the nucleus, with a number of dense spots co-localized with the nucleolus in expressing onion cells (Figure2a–d) whereas EGFP protein, a control, was detected in the cytoplasm and nucleus (Figure2e,f).
Figure 2.
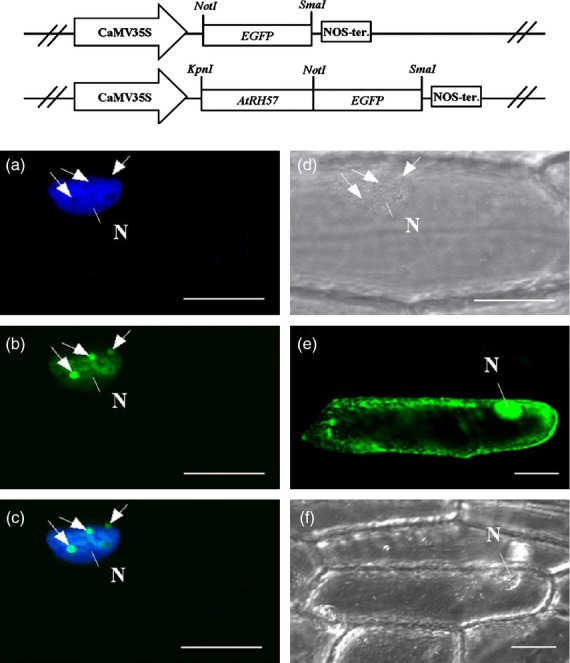
Subcellular localization of AtRH57. Top panel indicates the constructs of EGFP and AtRH57–EGFP. Transient expression of the AtRH57–EGFP (a–d) and EGFP only (e, f) in the onion epidermal cells. (a) An image of a 4′,6-diamidino-2-phenylindole (DAPI) fluorescent field. (b, e) Images of green fluorescent fields. (c) A merged image of DAPI stain (a) and green fluorescence (b). (d, f) Bright field images. Nucleoli are indicated by arrows. N, nucleus; Bar = 40 μm.
RNA unwinding activity of AtRH57
Earlier studies have shown that RNA helicases can unwind double-stranded RNA (dsRNA) in the presence of NTP in vitro (Yu and Owttrim, 2000; Chung et al., 2009). To determine the function of AtRH57, first, the AtRH57-His protein was expressed in Escherichia coli and affinity-purified (Figure3a). Next, the partial dsRNA as a substrate (Figure3b) was incubated with AtRH57-His in the presence of ATP. As shown in Figure3(c), the single-stranded RNA (ssRNA) was generated, indicating that AtRH57-His can unwind dsRNA. The level of unwinding of ssRNAs from dsRNAs was nicely correlated with the increasing concentrations of the AtRH57-His protein (lanes 3–5). The presence of ssRNA induced by AtRH57-His was also detected in the absence of ATP (Figure3c, lanes 6–8), suggesting that AtRH57 is an ATP-independent RNA helicase. Addition of proteinase K to the reaction did not cause dsRNA to unwind at all (Figure3c, lane 9).
Figure 3.
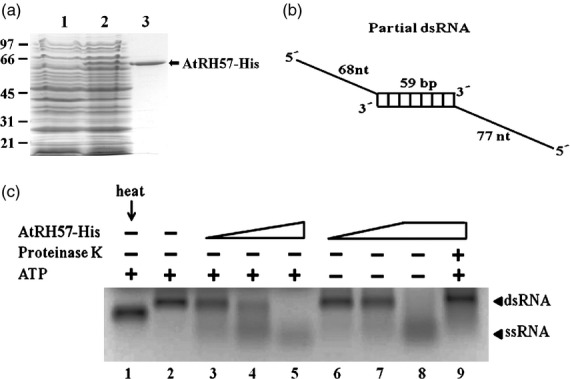
Over-expression and activity assay of AtRH57-His protein. (a) Over-expression and purification of AtRH57-His protein in Escherichia coli. The expressed vector only (lane 1) and AtRH57-His fusion protein (lane 2) were extracted, affinity-purified (lane 3) using Ni2+-NTA-agarose, fractionated, and then stained with Coomassie blue. Marker protein sizes are indicated at the left. (b) Schematic representation of the partial dsRNA substrate. The RNA substrate contains a 59-bp duplex region, with 68 and 77 nucleotide long single-stranded regions at the 5′ ends. (c) In vitroRNA helicase activity assay of AtRH57-His protein. Various amounts of AtRH57-His protein: 40 ng (lanes 3 and 6), 90 ng (lanes 4 and 7), and 376 ng (lanes 5, 8, and 9) were incubated with 1.0 μg partial dsRNA in 20 μl of helicase buffer at 30°C for 1 h and separated by 1.2% formaldehyde gel electrophoresis. Reactions were either added with (lanes 1–5 and 9) or without (lanes 6–8) 1 mmATP. The reaction mixture in lane 9 was pre-treated with proteinase K prior to incubation with partial dsRNA. Partial dsRNA was treated either with (lane 1) or without (lane 2) heat as controls. dsRNA and ssRNA bands were subsequently visualized by ethidium bromide (EtBr) staining.
Transcripts of Glc-responsive genes are altered in rh57 mutants
To investigate whether or not rh57 display a phenotype of hypersensitivity to high Glc at the molecular level, 9-day-old seedlings of WT and mutants were incubated with shaking in sugar-free MS that contained 4.5% Glc for indicated time intervals. Several sugar-responsive genes were analyzed using quantitative real-time PCR (qRT-PCR). As shown in Figure4, APL3, which encodes a large subunit of ADP-Glc pyrophosphorylase, a key enzyme for starch synthesis, and CHS, which encodes a chalcone synthase, a key enzyme for anthocyanin biosynthesis, were clearly induced by 4.5% Glc in rh57-1 and rh57-3 when compared with the WT seedlings. On the other hand, when compared with the WT seedlings, 4.5% Glc significantly repressed photosynthetic genes RBCS, which encodes ribulose-1,5-bisP carboxylase and PC2, which encodes a plastocyanin protein in both rh57 mutants either during a short period of induction (Figure4) or when seedlings were grown in the MS medium-containing plates for 9 days (Figure S7a). It is worthy of noting that APL3 and CHS were strongly induced by 4.5% Glc after a long period (9 days) of stimulation. In this experiment, Mtl was used as a control. A reasonable explanation for these results is that mutation in AtRH57 could alter gene sensitivity to Glc repression/induction in high concentrations of Glc.
Figure 4.
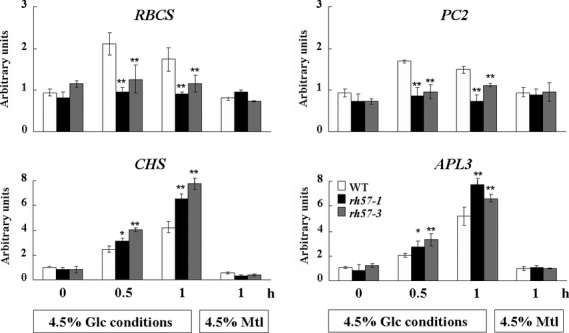
Alteration of glucose (Glc)-responsive gene expression in rh57 mutants. RNA levels of Glc-responsive genes were determined by qRT-PCR using total RNA isolated from 9-day-old seedlings of WT,rh57-1, and rh57-3 that were incubated with shaking in sugar-free MS that contained 4.5% Glc for indicated time intervals before harvest. Mannitol (Mtl) was used as a control. The analyzed Glc-responsive genes included RBCS, ribulose-1,5-bisphosphate carboxylase small subunit; PC2, plastocyanin 2; CHS, chalcone synthase; APL3, ADP-Glc pyrophosphorylase large subunit. ACTIN1 was used as an internal control. Data were obtained from three biologically independent experiments. Error bars represent standard deviation (SD;t-test: *P < 0.05; **P < 0.01).
rh57 mutants exhibit ABA hypersensitivity and enhanced ABA accumulation by Glc
Exogenous application of ABA affected germination to a higher extent in rh57-1 and rh57-3 mutants than in WT seeds (Figure5a). Addition of 0.75 μm ABA resulted in a marked decrease in seed germination (<36%) in rh57 mutants compared with WT seedlings (about 60%). It indicates that rh57 mutants exhibit hypersensitive reactions to ABA. Previous studies have shown that ABA accumulation is induced by Glc during seedling development (Cheng et al., 2002; Carvalho et al., 2010). This indication is reinforced by the fact that WT seedlings fail to arrest growth in 6% Glc supplemented with fluridone (Ullah et al., 2002). Fluridone, an inhibitor of ABA synthesis, blocks carotenoid synthesis leading to an albino-like seedling and, thus, we only measured cotyledon expansion. As shown in Figure5(b), without fluridone addition, the presence of Glc did not significantly affect early development of WT seedlings, but adverse effects on both mutant seedlings were obvious. In contrast, severe developmental arrest observed in both rh57 mutant seedlings was markedly redeemed when the medium that contained 3 or 4.5% Glc was supplemented with 1 μm fluridone (Figure5b). Therefore, this result suggests that seedling growth arrest in the mutant mainly functions through Glc-mediated ABA biosynthesis.
Figure 5.
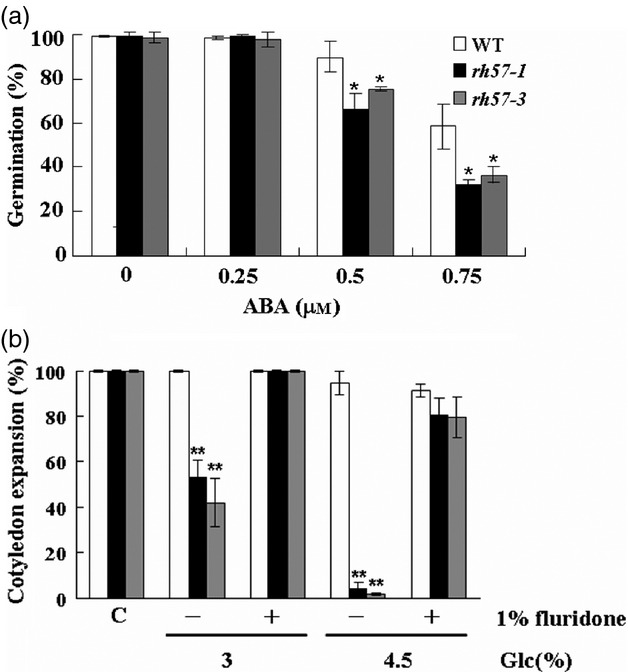
Abscisic acid (ABA) phenotypes of rh57 mutants. (a) rh57-1 and rh57-3 seedlings are hypersensitive to ABA during germination. WT (white bars), rh57-1 (black bars), and rh57-3 (gray bars) seedlings were grown either in sugar-free medium with or without the indicated concentrations of ABA. The percentage of seed germination was scored 9 days after stratification. (b) The addition of fluridone reduces the Glc-induced inhibition of cotyledon expansion. WT and rh57 seedlings were grown in sugar-free MS medium that contained 3 or 4.5% Glc and supplemented with or without 1 μm fluridone. WT and rh57 seedlings with cotyledon expansion were scored 9 days after stratification. Data were obtained from three biologically independent experiments. Error bars represent the standard deviation (SD;t-test: *P < 0.05; **P < 0.01).
This finding prompted us to determine ABA content in rh57-1 and WT seedlings. As rh57-1 seedlings grown in the presence of 4.5% Glc are too small to collect enough sample for quantitation, 14-day-old seedlings of rh57-1 grown in the MS medium solution with or without 4.5% Glc was used and incubated with shaking for 4 h. The ABA-deficient aba2 mutant (Cheng et al., 2002) was included as a control. As shown in Table1, no significant difference in ABA content between WT and mutant seedlings was observed in the absence of sugar. The presence of 4.5% Glc induced a fivefold increase in endogenous levels of ABA in WT seedlings, while no significant increase in ABA levels was observed in aba2. In contrast, in the presence of 4.5% Glc, ABA content in rh57-1 seedlings increased more than ninefold than that in WT. The hypersensitive feature of rh57-1 to high Glc could thus be attributed to the increased ABA levels.
Table 1.
Abscisic acid (ABA) content is significantly higher in rh57-1 than in Col-0 seedlings in the presence of high Glc
| Treatment | ABA content (ng g−1 fresh weight) | ||
|---|---|---|---|
| WT | rh57-1 | aba2 | |
| Control | 4.324 ± 0.004 | 4.232 ± 0.646 | 3.796 ± 0.016 |
| 4.5% Glc | 23.926 ± 0.701 | 39.141 ± 4.356** | 4.124 ± 0.204 |
Quantification of ABA levels in 14-day-old seedlings of Col-0 and mutant seedlings in MS medium supplemented with or without 4.5% Glc, and incubated with shaking for 4 h before harvest. Data are expressed as the mean ± standard deviation (SD) of three experiments. aba2, an ABA-deficient mutant plant, is shown as a control. The difference in ABA levels between rh57-1 and Col-0 seedlings is significant as evaluated by t-test (**P < 0.01).
Transcripts of ABA biosynthesis and signaling genes are induced by high Glc in rh57 mutants
As rh57 accumulates ABA in response to Glc, we examined the expression of three ABA biosynthesis genes in mutant seedlings and WT using qRT-PCR. Exposure of 9-day-old seedlings of WT and rh57 mutants to 4.5% Glc for a short period of induction resulted in higher expressions of ABA2,ABA3, and AAO3 in rh57 than those in WT (Figure6). This result is consistent with a marked rise in ABA content in rh57-1 when compared with WT in the presence of 4.5% Glc (Table1). However, the induced expressions of ABA2,ABA3, and AAO3 in rh57 were indiscernible when plants were grown in the MS medium that contained 4.5% Glc for 9 days before harvest (Figure S7b).
Figure 6.
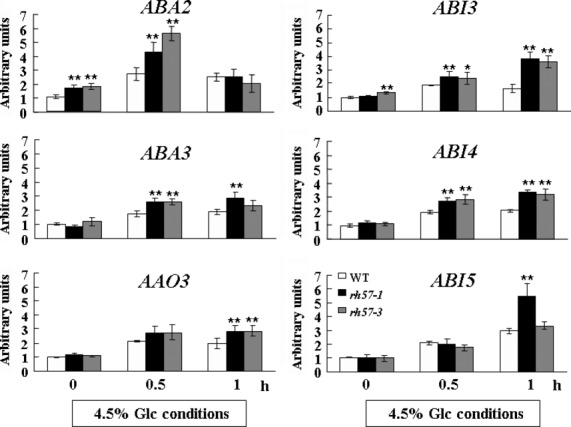
Alteration of abscisic acid (ABA) biosynthesis and signaling gene expression in rh57 mutants. RNA levels of ABA biosynthesis and signaling genes were determined by qRT-PCR using total RNA isolated from 9-day-old seedlings of WT,rh57-1, and rh57-3 that were incubated with shaking in sugar-free MS that contained 4.5% Glc for indicated time intervals before harvest. ACTIN1 was used as an internal control. ABA2,ABA3, and AAO3 are ABA biosynthesis genes, while ABI3,ABI4, and ABI5 are ABA signaling genes. Data were obtained from three biologically independent experiments. Error bars represent standard deviation (SD;t-test: *P < 0.05; **P < 0.01).
ABI5 acts downstream of ABI3 to cause seedling growth arrest during germination (Lopez-Molina et al., 2002). ABI4 also enhances ABI5 gene expression in response to sugar (Arroyo et al., 2003); the involvement of the ABI4 gene in sugar responses reveals the important player of ABA in sugar-induced process (Arenas-Huertero et al., 2000). Thus, transcription of the three key signaling genes in rh57 mutant seedlings was examined in this study by qRT-PCR. It revealed that the ABA signaling genes, ABI3,ABI4, and ABI5 whose transcripts were significantly increased by 4.5% Glc in rh57-1 and rh57-3 after a short period of induction (Figure6); however, the induction was much more pronounced after a long period (9 days) of stimulation (Figure S7b). The results were consistent with the ABA-hypersensitive feature of rh57 mutants, indicating that AtRH57 may be involved in transcriptional down-regulation of these ABA signaling genes under high Glc conditions.
AtRH57 transcripts are significantly induced by ABA, high Glc and salt
Expression of AtRH57 under various conditions was examined by qRT-PCR analysis. The AtRH57 transcript was strongly induced when 14-day-old WT Col-0 seedlings were incubated in medium that contained 4.5% Glc for 3 and 6 h (Figure S8). Significant induction was also observed when 14-day-old Col-0 seedlings were incubated in medium that contained either 100 μm ABA or 100 mm NaCl. The application of 100 μm ABA markedly induced ABI5, the control gene. ABI5 was also induced in the presence of either high Glc or salt (Figure S8).
AtRH57 acts downstream of HXK1 and mediates a unique feedback inhibition of ABA accumulation
To further elucidate the role of AtRH57 in sugar signaling, we performed qRT-PCR using total RNA obtained from 9-day-old seedlings of WT, rh57-1,hxk1 and abi4 that were incubated with shaking in sugar-free MS that contained 4.5% Glc for indicated time intervals. Given that HXK1 and ABI4 are essential for sugar and ABA signaling (Dekkers et al., 2008; Lee et al., 2012), we examined the relationship of HXK1 and ABI4 with AtRH57. As shown in Figure7, no changes in HXK1 expression were observed in rh57-1 mutants compared with that in WT, whereas AtRH57 expression was significantly reduced in hxk1 mutants. Parallel to the observation that the exogenous addition of 100 μm ABA or 100 mm NaCl significantly induced AtRH57 expression (Figure S8), we suggest that AtRH57 acts in a signaling network downstream of ABA. Similarly, no changes in HXK1 expression were observed in abi4 compared with that in WT, whereas ABI4 expression was significantly reduced in hxk1 mutants. Therefore, similar to AtRH57,ABI4 acts downstream of HXK1. The results are consistent with an earlier report showing that ABI4 acts downstream of HXK1 Glc sensors (Arenas-Huertero et al., 2000; Leόn and Sheen, 2003).
Figure 7.
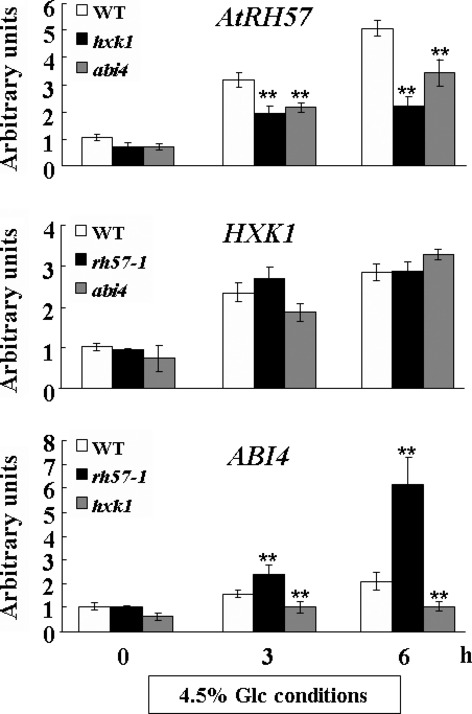
AtRH57 acts in a signaling network downstream of HXK1. RNA levels of AtRH57,HXK1 and ABI4 were determined by qRT-PCR using total RNA isolated from 9-day-old seedlings of WT,rh57-1,hxk1 (SALK_070739) and abi4 (CS8104) that were incubated with shaking in sugar-free MS medium that contained 4.5% Glc for indicated time intervals before harvest. ACTIN1 was used as an internal control. ABI4 is an ABA signaling gene. HXK1, hexokinase1. Data were obtained from three biologically independent experiments. Error bars represent standard deviation (SD;t-test: **P < 0.01).
qRT-PCR analysis was used to examine the relationship between ABI4 and AtRH57. The significant reduction of AtRH57 expression in abi4 compared with that in WT suggests that ABI4 promotes AtRH57 expression (Figure7). Conversely, the significantly higher ABI4 expression levels in rh57-1 and rh57-3 mutants compared those in WT suggest that AtRH57 exerts an inhibitory effect on ABI4 expression (Figures6 and 7). The inhibitory effect of AtRH57 on ABI4 expression is consistent with the observation that the induced expressions of ABA2,ABA3, and AAO3 in rh57 mutants were greater than those in WT (Figure6). This result is also reflected by the observation that a marked rise in ABA content occurred in rh57-1 mutants in the presence of 4.5% Glc (Table1). Therefore, we conclude that AtRH57 down-regulates ABA biosynthesis genes under high Glc conditions, which subsequently resulted in a decrease in ABA accumulation in Arabidopsis plants. As a consequence, the expression of ABA signaling genes such as ABI4 decreased. Altogether, a unique feedback inhibition of ABA biosynthesis genes and ABA accumulation by AtRH57 exists in the sugar-mediated ABA signaling pathway (Figure8).
Figure 8.
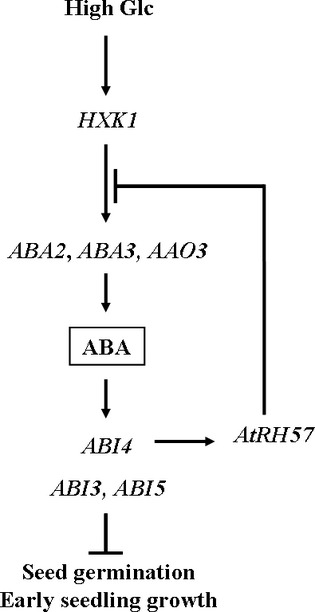
A schematic representation of glucose (Glc) signaling wherein AtRH57 acts as a crucial player in the feedback inhibition of ABA accumulation within the sugar-mediated ABA signaling pathway during Arabidopsis seed germination and early seedling growth. HXK1, hexokinase1; ABA, abscisic acid; ABA biosynthesis genes: ABA2,ABA3, and AAO3; ABA signaling genes: ABI3,ABI4, and ABI5.
Decreased processing of pre-rRNA products in rh57 mutants
The 45S/35S pre-rRNA is composed of various parts of sequences: a 5′ external transcribed sequence (ETS), internal transcribed sequences (ITS) 1 and 2, and a 3′ ETS that underwent a series of rRNA processing steps (Figure9a). To test the effect of AtRH57 mutation on the processing of 45S/35S pre-rRNA, primers specific to different regions of the 45S/35S pre-rRNA were used to measure the corresponding RNA levels in 9-day-old rh57 and WT seedlings. The relative levels of various sequences were expressed as rh57/Col-0. qRT-PCR analysis revealed that the levels of the 5′ ETS, ITS1, ITS2 and 3′ ETS sequences were approximately 2.5-fold higher in rh57-1 than in WT (Figure9b). Similarly, the relative levels of pre-rRNA were also higher in rh57-3, in particular the ITS1, ITS2 and 3′ ETS sequences. The results suggested that accumulation of pre-rRNA is relatively higher in both rh57 mutants when compared with WT plants. Therefore, the normal processing of 45S/35S pre-rRNA was indeed affected when AtRH57 was down-regulated.
Figure 9.
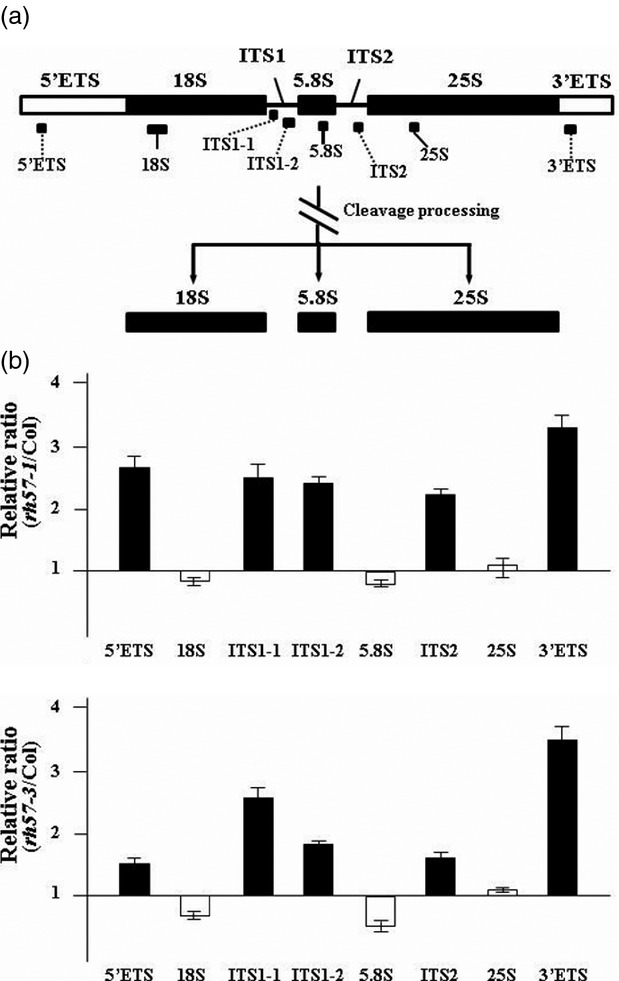
Reduced pre-rRNA processing in rh57 mutants. Comparison of the relative levels of the various regions of pre-rRNA in 9-day-old WT Col-0 and rh57 mutant plants. (a) Primer sets specific to various regions of 45S/35S pre-rRNA were used for synthesis of the first-stranded cDNA and determination of the levels of corresponding RNA sequences by qRT-PCR analysis. (b) Signal values for each PCR product were compared between rh57 and Col-0 plants. Black bars represent ETS and ITS sequences, and white bars represent 18S, 5.8S and 25S rRNAs. The values represent the means ± standard deviation (SD) for two independent isolations of RNAs.
To further characterize the ribosomal defect in rh57 mutants, we examined RNA blot analyses to visualize rRNA precursor species. Total RNA was extracted from WT, rh57-1, and rh57-3 plants, electrophoresed, and blotted on the membranes. The membranes were then probed with specific oligonucleotides to identify pre-rRNA precursors (Figure10a) and the pre-rRNA precursors were scanned for quantification (Table S2). A decrease in 18S rRNA steady-state levels (indicated by an arrow) was observed in rh57 mutants compared with that in WT (Figure10a and Table S2). In comparison, 25S rRNA levels in the mutants remained similar to the levels in WT. Probing with oligonucleotides specific to either the 5′ ETS or the ITS1 region evidenced a considerable accumulation of the 45S/35S pre-rRNA (indicated by an arrowhead) and the abnormal pre-rRNA precursors (indicated by asterisks) in the rh57 mutants (Figure10a and Table S2).
Figure 10.
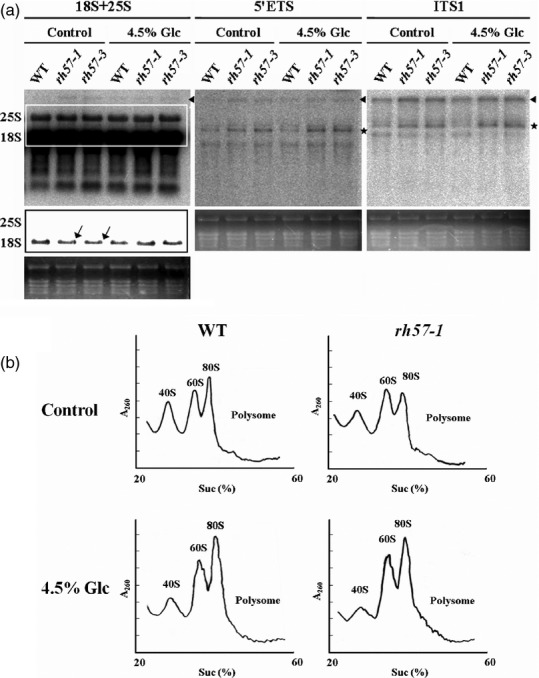
Pre-rRNA processing defects and polysome profile of the rh57 mutants. (a) Total RNA was isolated from 9-day-old seedlings of WT,rh57-1 and rh57-3 mutant plants that were incubated with shaking in sugar-free MS with or without 4.5% Glc for 30 min before harvest. The isolated total RNA (5 μg) was fractionated and blotted onto hybridization membranes for analysis. The 45S/35S pre-rRNA (indicated by an arrowhead) is represented in Figure9 with the position of selected oligonucleotides used as probes: 18S + 25S, 5′ETS, and ITS1. The abnormal pre-rRNA precursors are indicated by an asterisk. The box in the left panel indicates the18S image of reduced intensity for easier comparison. Data were obtained from two biologically independent experiments. (b). Polysome profile of the WT and rh57-1 plants. The 9-day-old seedlings of WT Col-0 and rh57-1 mutants were transferred to medium solutions supplemented with or without 4.5% Glc, and incubated for 30 min before harvest. Equivalent amounts of polysome extracts were analyzed by sucrose gradients in each case. The positions of 40S and 60S subunits are indicated as well as the monosome (80S) and polysome peaks. Data were obtained from three biologically independent experiments.
Similar results were obtained from 9-day-old seedlings of WT, rh57-1 and rh57-3 mutants incubated with shaking in sugar-free MS that contained 4.5% Glc for 30 min. The rh57 mutants showed accumulation of the aberrant pre-rRNA precursors (indicated by asterisks). However, a slight increase in 18S rRNA steady-state levels was observed in rh57 mutants compared with WT in the presence of high Glc (Figure10a). Taken together, these results demonstrated that AtRH57 mutation may affect the normal processing of 45S/35S pre-rRNA in plants grown in the presence/absence of high Glc.
AtRH57 mutation and high Glc conditions additively cause a defect in small ribosomal subunit formation
To determine whether AtRH57 mutation affects the formation of ribosomal subunits, the cell ribosome population of rh57-1 was analyzed on sucrose gradients and compared with that of WT. As shown in Figure10(b), the free 40S and 80S subunits were decreased in rh57-1. The total amount of free 60S and 40S subunits was further determined in WT and rh57-1 mutant plants. The 60S/40S ratio was 0.92 in WT and 1.38 in rh57-1 (Figure S9), thereby confirming that AtRH57 mutation reduces the steady-state level of 40S subunits.
Polysome profile analysis of WT and rh57-1 mutant plants in the presence of 4.5% Glc was also performed. Results clearly show that free 40S subunits were less detectable in the presence of high Glc, whereas the 60S and 80S subunits increased in WT (Figure10b). The 60S/40S ratio was 1.44 in WT, similar to that in rh57-1 without Glc treatment (1.38; Figure S9). Overall polysome levels in all cases, however, were indiscernible. Notably, the addition of high Glc increased the 60S/40S ratio in rh57-1 (2.50), which was approximately twofold higher than in WT and in the mutant without the addition of Glc. The results suggest that AtRH57 mutation and high amounts of Glc additively impair small ribosomal subunit formation, which may account for the observed hypersensitivity of rh57 mutants.
rh57 mutants show the reduced function of protein synthesis
To further examine whether rh57 mutants have reduced function of protein synthesis, cycloheximide (CHX), a eukaryotic protein synthesis inhibitor, was used for the test. CHX binds the eukaryotic ribosomal complexes in a stoichiometric manner (Oleinick, 1977). In addition, CHX profoundly inhibits germination in Arabidopsis because de novo protein synthesis for radicle protrusion is impeded (Rajjou et al., 2004). As shown in Figure11(a), the rh57-1 mutant showed lower percentage radicle protrusion, compared with that of WT in the presence of 1 μg ml−1 CHX. A similar result was also obtained in the medium that contained 2 μg ml−1 CHX (Figure11a). Therefore, the rh57-1 mutant showed increased sensitivity to CHX, indicating of a reduced activity of protein synthesis, possibly caused by an aberrant function in ribosomes. Without the addition of CHX, both WT and rh57-1 mutant seeds grown in MS medium 9 days after sowing developed into normal seedlings (Figure11a).
Figure 11.
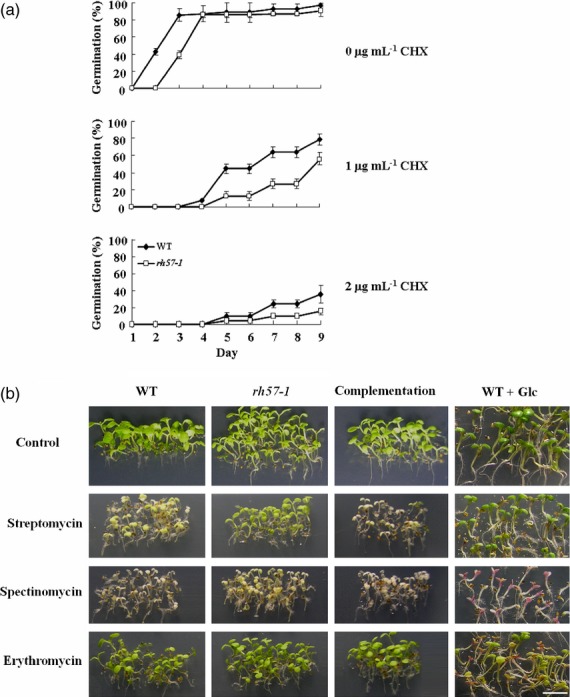
The rh57-1 mutant shows defects in protein synthesis and ribosome biogenesis. (a) The rh57-1 mutant shows increased sensitivity to CHX. Seeds were sown in sugar-free MS medium that contained the indicated CHX. Germination was counted until 9 days after seed sowing. Data were obtained from three biologically independent experiments. (b) The rh57-1 mutant and high Glc show resistance to streptomycin and spectinomycin. The rh57-1 mutant, complementation of rh57-1, WT with 4.5% Glc and WT seeds alone were directly germinated on sugar-free MS media with or without various antibiotics including streptomycin, spectinomycin and erythromycin, each with the same concentration of 50 μg ml−1. Seedlings were photographed after seeds were sown for 9 days. Data were obtained from three biologically independent experiments. Bar = 5 mm.
rh57-1 mutants show resistance to antibiotics
Given that antibiotics affect protein synthesis, its effect on WT, rh57-1, and a complementation line in the rh57-1 mutant background was tested. As shown in Figure11(b), the rh57-1 mutants showed similar sensitivity to erythromycin as the WT, but exhibited significant resistance to streptomycin and a mild resistance to spectinomycin. In the presence of streptomycin, the cotyledons and leaves of the rh57-1 mutants exhibited better growth than those of WT (Figure11b). Similar results were also obtained in rh57-1 in the presence of spectinomycin. The rh57-1 seedlings grew slightly better than the WT, although their growth was not as good as that of rh57-1 seedlings in the presence of streptomycin (Figure11b). The phenotype of the complementation line reversed its sensitivity to streptomycin or spectinomycin similar to the WT (Figure11b). Streptomycin and spectinomycin are protein synthesis inhibitors that bind to the 16S rRNA of the small (30S) subunit of the bacterial ribosome (Honoré and Cole, 1994; Galimand et al., 2000). Taken together, these findings suggest that the ribosomes may be affected in the rh57 mutants and such an alteration conferring changes in antibiotic binding sites of ribosomal proteins. However, the effects of antibiotics on eukaryotic ribosomes remain largely unknown.
In addition, the WT seedlings in the presence of 4.5% Glc showed similar sensitivity to erythromycin as the WT without Glc addition, but exhibited significant resistance to streptomycin; their cotyledons exhibited better growth than those of WT without 4.5% Glc addition (Figure11b). The WT seedlings in the presence of 4.5% Glc also grew differently in the presence of spectinomycin; the WT cotyledons turned into red instead of green color that might be reflected by a marked increase in CHS gene expression in high Glc conditions (Figure4). Therefore, the markedly reduced sensitivity to streptomycin or spectinomycin in WT seedlings in the presence of 4.5% Glc is consistent with the reduction in steady-state level of the small ribosomal subunit (40S) (Figure10b and Figure S9) and a decrease in steady-state levels of 18S rRNA (Figure10a) under the same conditions of high Glc.
Discussion
DEAD-box proteins play a crucial role in the development and stress responses of various organisms, thus they remain a subject of increasing importance (Gong et al., 2002; Sanan-Mishra et al., 2005; Kant et al., 2007; Huang et al., 2010a). The response mechanism of RNA helicases to Glc, an important signaling pathway affecting plant development, has yet to be fully elucidated. In this report, we characterized rh57 mutants that exhibit enhanced Glc and ABA hypersensitivity during germination and early seedling development in Arabidopsis. The AtRH57 gene encodes a Class II DEAD-box RNA helicase localized in the nucleus and nucleolus. AtRH57 may be involved in rRNA processing and plays a key role in rRNA biogenesis in Arabidopsis.
rh57 mutants exhibit enhanced sensitivity to Glc during germination and seedling development (Figure1 and Figure S2). The changes in Glc sensitivity are attributed to the loss of function of RH57, suggesting that AtRH57 plays a crucial role during early seedling development. A 2-day delay in seed germination may also be attributed to the fact that AtRH57 gene defects were observed at early seed development. Furthermore, AtRH57 is expressed in all organs (Figure S3d) and in different parts of seed tissues (Figure S4), also suggesting of the importance of this gene in seed development. rh57 mutants also exhibit a Glc phenotype at the molecular level. About 4.5% Glc is sufficient to profoundly repress the photosynthetic RBCS and PC2 genes in rh57 mutants. It correlates with hypersensitive phenotype of rh57. In addition, anthocyanin biosynthesis (CHS) and starch synthesis (APL3) genes are markedly induced in both mutants (Figure4). Therefore, the AtRH57 protein is involved in sugar-responsive gene expression.
Similar to the Glc response, rh57 seeds showed greatly enhanced sensitivity to ABA during germination (Figure5a). rh57-1 indeed increases ABA levels under high Glc conditions (Table1). It is consistent with the enhanced levels of mRNA of ABA biosynthesis genes in rh57 when compared with those in WT after a short time of induction. When the collected plants were grown for 9 days instead of a short time of stimulation, it is possible that the mRNA levels of ABA biosynthesis genes in rh57 become decreased. The result is correlated with the observation of decreased levels of mRNA in salt and drought stress for a prolonged period of time (Qiu and Yu, 2009; Dong et al., 2011). In contrast with ABA biosynthesis genes, the expression of ABA signaling genes drastically induced in rh57 mutants when the two mutants were subjected to a long period of stimulation (Figure S7b). Furthermore, ABA inhibitory analysis indicates that the seedling growth arrest is resulted from increased ABA levels in rh57 mutants under high Glc conditions (Figure5b). It clearly suggests that the AtRH57-mediated Glc-signaling pathway is mainly controlled by Glc-induced ABA accumulation.
The ABA signaling genes are significantly induced by Glc in rh57 mutants (Figure6). Thus, the AtRH57 protein involved in the repression of these ABA signaling genes is likely via a decrease in ABA levels in the cell. Further, given that AtRH57 exerts an inhibitory effect on ABI4 expression and ABI4 promotes AtRH57 expression (Figure7), a feedback inhibition of ABA biosynthesis genes and ABA accumulation thus exists within the sugar-mediated ABA signaling. As proposed in Figure8, high Glc levels up-regulate the HXK1-mediated expression of ABA biosynthesis genes. However, the product of AtRH57 plays a crucial role in feedback inhibition involving a decrease in the expression of ABA biosynthesis genes and subsequently, ABA decreases its level of accumulation in plants. As a consequence, the induction of ABA signaling genes is reduced, resulting in counteracting of sugar-mediated growth arrest. Conversely, AtRH57 mutation relieves inhibition of ABA biosynthesis and signaling genes and thus, the level of ABA increases in rh57, resulting in hypersensitivity phenotype of the mutants.
ABI4 is crucial for ABA signaling transduction pathways during seed development and germination (Finkelstein et al., 1998). This transcription factor is also important for chloroplast retrograde signaling (Koussevitzky et al., 2007), bidirectional communication between plastid and nucleus (Lee et al., 2012), lipid mobilization from the embryo (Penfield et al., 2006), and various Glc responses (Arenas-Huertero et al., 2000; Laby et al., 2000). In addition, ABI4 is a required activator of its own expression during development (Bossi et al., 2009) and a negative regulator of proline accumulation at low water potential, as well as serves to connect proline accumulation to sugar sensing (Verslues and Bray, 2006). Aside from the diverse roles in ABA/sugar signaling, we also found a feedback inhibition of ABA accumulation mediated by AtRH57 (Figure8). This feedback inhibition can be induced by ABI4. An alternative feedback inhibition of ABA accumulation was previously reported by Verslues and Bray (2006), who suggested that ABI1/2 plays a role in controlling feedback regulation of ABA accumulation by inhibiting ABA catabolism or conjugation.
Our result reveals that AtRH57 unwinds dsRNAs into ssRNAs without ATP in vitro (Figure3). The varied positions of ssRNA between heat treatment and AtRH57 additions may be possibly due to the presence of secondary structure despite that the sample was heated. It is similar to the observation of GST-GmRH protein where the mobility of ssRNA with GmRH addition is faster than ssRNA treated with heat (Chung et al., 2009). Limited reports on RNA helicases independent of ATP are available. AvDH1 and GmRH induced by salt stress exhibit ATP-independent RNA helicase activity (Liu et al., 2008; Chung et al., 2009). Although the mechanism for the ATP-independent RNA helicase activity of AtRH57 remains unclear, its activity independent of ATP may be related to substrate specificity.
rh57 mutants show an increased accumulation of pre-rRNA compared with Col-0 (Figures9 and 10a). The reduced pre-rRNA processing or the accumulation of abnormal pre-rRNA can be visualized by RNA blot analysis. Further, we have demonstrated that AtRH57 mutation and high Glc conditions additively cause a defect in small ribosomal subunit formation, causing the reduced ribosome biogenesis in the mutants. The less free 40S resulting in reduced ribosomes and function of protein synthesis in rh57 may thus retard germination and seedling growth. It correlates with the fact that 2 additional days are needed for rh57-1 and rh57-3 to reach full seed germination (Figure S1). Furthermore, rh57 cotyledon grew smaller and slightly pale in color when compared with WT under normal growth conditions (Figure1). Despite the fact that high Glc conditions may impair ribosome biogenesis, enough ribosomes are produced in WT plants to sustain their growth. When the rh57 mutants were grown in the presence of high Glc conditions, AtRH57 mutation and high amounts of Glc additively impair small ribosomal subunit formation, which may account for the observed hypersensitivity of rh57 mutants. Recently, Guo et al. (2011) revealed that the ribosome profile of ABA-treated Arabidopsis seedlings display reduced polysome levels and concomitant accumulation of 60S subunit and 80 monosome, demonstrating that ABA inhibits global protein translation. In this study, despite that high Glc conditions accumulate the 60S ribosomal subunits and 80S monosome, the 40S subunit is significantly reduced and overall polysome levels are indiscernible in WT (Figure10b). Therefore, the changes in ribosome profile under high Glc conditions differ from that by ABA treatment although Glc signaling is partly mediated by ABA.
The defect in small ribosomal subunit is also supported by the facts that rh57-1 seedlings showed significant resistance to streptomycin and a mild resistance to spectinomycin (Figure1b). Both streptomycin and spectinomycin are protein synthesis inhibitors that bind to the 16S rRNA of the small subunit of the bacterial ribosome. However, the effect of antibiotics on eukaryotic ribosomes remains largely unknown. It was reported that mutations in eukaryotic 18S ribosomal RNA affect translation and resistance to aminoglycoside antibiotics including streptomycin (Chernoff et al., 1994). Recently, Rosado et al. (2010) demonstrated that the absence of RPL4 confers resistance to erythromycin and chloramphenicol in Arabidopsis. Mutation in RPL4, a component of the ribosomal 60S subunits, induces the conformational change of the tunnel entrance on which the RPL4 protein covers. As a result, the antibiotics cannot bind to the ribosomal subunits (Gabashvili et al., 2001). In addition, Abbasi et al. (2010) have suggested that mutation of APUM23 confers resistance to streptomycin, an antibiotic that targets the decoding center of the ribosome. APUM23 mutation displayed a phenotype similar to those mutants that have a defect in ribosomal protein gene. Thus, ribosomes are probably affected in the apum23-1 mutant and such an influence conferring changes in antibiotic binding sites of ribosomal proteins. It was proposed by Abbasi et al. (2010) that the APUM23 mutation causes the production of abnormal rRNA species incorporated into the ribosomes and accordingly, generates abnormal ribosomes that can no longer bind streptomycin. The observation that rh57 mutants show abnormal pre-rRNA precursors and reduced level of 40S subunits argues in favor of this concept.
It has been known that rRNA biogenesis mainly proceeds in the nucleolus. The function of AtRH57 involved in the process of pre-rRNA in rh57 mutants is in agreement with the nucleolar localization of the protein (Figure2). It was reported that AtRH36 also plays an key role in pre-rRNA processing in Arabidopsis; however, atrh36-1 heterozygous plants generated short siliques that contained defective seeds (Huang et al., 2010a). In contrast, rh57 mutants are homozygous and produce siliques and seeds as normal as Col-0. Recently, both AtRH3 and AtRH22 have been reported to affect chloroplast ribosome biogenesis (Asakura et al., 2012; Chi et al., 2012).
In conclusion, rh57 mutants exhibit enhanced Glc and ABA hypersensitivity during seed germination and seedling development in Arabidopsis. The hypersensitivity feature of rh57 is suggestively due to a severe defect in small ribosomal subunit formation in the presence of high Glc. The AtRH57 gene encodes a Class II DEAD-box protein localized in the nucleus and nucleolus. AtRH57 acts in a signaling network downstream of HXK1. A unique feedback inhibition of ABA accumulation mediated by ABI4-induced AtRH57 exists within the sugar-mediated ABA signaling. AtRH57 mutation and high Glc additively cause a severe defect in small ribosomal subunit formation. AtRH57 is involved in rRNA processing and plays a crucial role in rRNA biogenesis. These results show that the AtRH57 protein participates in response to sugar by repressing Glc-mediated ABA accumulation and signaling gene expression during early seedling development.
Experimental procedures
Plant materials, growth conditions and stress treatments
All Arabidopsis (A. thaliana) plants used in this study were on the Col-0 ecotype background. Seeds of rh57-1,rh57-2 and rh57-3 are T-DNA insertion lines (SALK_008887, CS917749, and SALK_019721, respectively), and were obtained from the Arabidopsis Biological Resource Center (Ohio State University, Columbus, OH, USA). hxk1 (SALK_070739), abi4-1 (CS8104), and aba2 (also known as glucose-insensitive1,gin1; Cheng et al., 2002) were also used in this study. The hxk1 mutant has a T-DNA insertion in the first intron and showed no HXK1 product when tested by HXK1 polyclonal antibodies. When grown in soil, seeds were sown on a 1:1:8 mixture of vermiculite, perlite, and peat moss and watered every other day.
Seed germination and post-germination assays
WT and mutant seeds (approximately 100 seeds each) were aseptically treated and rinsed in accordance with Yang et al. (2005). To measure Glc and ABA sensitivity, seeds were sown in MS medium (Murashige and Skoog, 1962) plates and placed at 22°C with a 16-h light photoperiod. Different concentrations of d-Glc, Mtl, ABA, and 1 μm fluridone (Wako, http://www.wako-chem.co.jp/english/) were added where indicated. Triplicates were performed for each treatment. Germination, cotyledon greening and expansion were measured at indicated days. The number of cotyledon greening and expansion was expressed as a percentage of the total number of germinated seeds. For the measurement of hypocotyl length, germinated seeds were grown vertically on solid MS with Glc and Mtl in complete darkness at 22°C.
Genomic DNA isolation and genotyping of mutant plants
Arabidopsis DNA was isolated from seedling according to Maliga et al. (1995). Mutant plants with the insertion of T-DNA were identified by PCR analysis. For genotyping, PCR analysis was carried out using AtRH57-specific primers for the WT and in combination with T-DNA-specific primers (Table S1) for the mutants. The DNA fragments were fractionated on 1.5% agarose gel and stained with ethidium bromide (EtBr).
RNA isolation, RT-PCR and qRT-PCR
For different abiotic stresses, 14-day-old seedlings of WT Col-0 were transferred to MS medium solutions that contained either 4.5% Glc or 100 mm NaCl or 100 μm ABA, and incubated with shaking for indicated time intervals before harvest. Total RNA were then extracted from treated seedlings. For RT-PCR, the first-stranded cDNA was synthesized with 3 μg total RNA using oligo (dT) primers in accordance with the manufacturer's protocol (Invitrogen, https://www.invitrogen.com). RT-PCR analyses were conducted using the gene-specific primer pairs such as AtRH57-A, -B, and -partial listed in Table S1. qRT-PCR analysis was performed in accordance with the procedures described by Hsu et al. (2011). WT Col-0, rh57-1, and rh57-3 seedlings were grown in MS plates supplemented with or without 4.5% Glc or Mtl and collected 9 days after stratification. For a short period of induction, the 9-day-old WT, rh57-1,rh57-3,abi4, and hxk1 seedlings were incubated with shaking in sugar-free MS medium with or without 4.5% Glc for indicated time intervals before harvest. The processing analysis of pre-rRNA was carried out in accordance with Kojima et al. (2007). qRT-PCR was performed using the primer sets listed in Table S1. The melting temperatures to generate specific product were obtained as such: ACTIN (85°C), RBCS (85°C), PC2 (86°C), CHS (86°C), APL3 (84°C), ABA2 (85°C), ABA3 (85°C), AAO3 (84°C), ABI3 (83°C), ABI4 (89°C), ABI5 (87°C), HXK1 (86°C), AtRH57 (80°C), 5.8S rRNA (82°C), 18S rRNA (82°C), 25S rRNA (84°C), ITS1-1 (87°C), ITS1-2 (89°C), ITS2 (84°C), 5′ ETS (85°C), 3′ ETS (80°C), and ACTIN (85°C).
RNA blot
RNA blot analysis was in accordance with the procedures described by Yang et al. (2005) with some modifications. The WT, rh57-1 and rh57-3 9-day-old mutant seedlings were incubated with shaking in sugar-free MS medium with or without 4.5% Glc for 30 min before harvest. Total RNA were isolated and an appropriate amount of each sample (5 μg) was loaded on the agarose gel. Primers used for amplifying specific probes: 25S, 18S, 5′ ETS, and ITS1 are listed in Table S1. The probes were labeled by random priming with (α-32P)-dCTP by Random prime DNA labeling system kit (Amersham International, http://www.kaker.com/mvd/data/Amersham_International.html). The membranes were exposed to phosphoimaging film and analyzed using a phosphoimage analyzer (Bioimage-Analyzer System-1000; Fuji, http://www.labmerchant.com/).
Measurement of ABA
For quantification of ABA content, 14-day-old seedlings of WT Col-0 and rh57-1 and aba2 mutants were transferred to MS medium solutions supplemented with or without 4.5% Glc, and incubated with shaking for 4 h before harvest. ABA measurement was in accordance with the method of Xiong et al. (2001). ABA in the solution was measured using the Phytodetek ABA immunoassay kit (Idetek, http://www.idetek.com/).
Construction, over-expression and purification of AtRH57-His fusion protein
An entire coding region of AtRH57 gene was amplified by PCR with KpnI-AtRH57o and NotI-AtRH57o primers (Table S1). The amplified AtRH57 DNA fragment was then excised with KpnI and NotI and inserted into the bacterial expression vector pET-29a (Novagen, http://www.emdmillipore.com/) to create a construct of pET-AtRH57. The nucleotide sequence of the insert was confirmed by DNA sequencing. The confirmed pET-AtRH57 plasmid was transformed into E. coli competent BL21 cells for expression. The lysate of transformed cells was centrifuged at 12 000 g for 20 min at 4°C after which the supernatant was decanted, mixed with 1.5 ml of nickel-chelating resin (Qiagen, http://www.qiagen.com/) for 3 h before the mixture was packed into a column. The AtRH57-His fusion protein was eluted with 6 ml of elution buffer (250 mm imidazole, 0.1 m NaCl, and 20 mm Tris-HCl, pH 7.5) and dialyzed by the dialysis buffer (20 mm Tris–HCl, pH 7.5, 0.1 m NaCl) overnight. The concentrated protein was fractionated and stained with Coomassie blue.
Preparation of dsRNA substrates and activity assay of RNA helicases
RNA substrates were synthesized by in vitro transcription of multiple cloning sites from the pGEM-T Easy vector (Promega, www.promega.com/). The vector was linearized after XhoI digested and transcribed with SP6 polymerase to generate a transcript of 136 nucleotides. Similarly, the vector was linearized with SpeI and transcribed with T7 polymerase to generate a transcript of 127 nucleotides. After treated with DNase (Promega) and extracted with phenol/chloroform, the generated transcripts were precipitated and resuspended in RNase free water.
The in vitro RNA helicase activity assay was carried out as described in Tai et al. (1996) with some modifications. Partially double-stranded RNA (1 μg) was incubated with various concentration recombinant AtRH57 proteins (40–376 ng) in 20 μl of helicase buffer [20 mm HEPES-KOH (pH 8.0), 1 mm ATP, 0.1 mg ml−1 BSA, 20 U ml−1 RNasin (New England Biolabs, https://www.neb.com/)]. After incubation at 30°C for 1 h, reactions were terminated by the addition of 5 μl of 5× RNA loading dye (50% glycerol, 1 mm EDTA, pH 8.0, 0.4% bromophenol blue, 0.4% xylene cyanol). An aliquot (10 μl) from the reaction mixture was electrophoresed and EtBr-stained on a 1.2% formaldehyde gel.
Construction and subcellular localization of AtRH57-EGFP fusion protein
To make the pUC18 (Thermo Scientific, http://www.thermoscientificbio.com/) construct that contained the 35S promoter, the 35S-GUS (β-glucuronidase) fragment was released from the pBI121 vector (BD Biosciences Clontech, http://www.clontech.com/) with HindIII and EcoRI and ligated into pUC18. To create the EGFP construct in the modified pUC18, the complete open reading frame (ORF) of EGFP was amplified by PCR with a primer pair (Table S1) of which the 5′-primer of EGFP contained three consecutive restriction sites XbaI, KpnI, and NotI and the 3′-primer of EGFP contained three consecutive restriction sites SmaI, BamHI, and SacI. To further generate the AtRH57-EGFP construct in the modified pUC18, the complete ORF of AtRH57 was amplified by PCR with AtRH57-5′ and AtRH57-3′ primer pairs (Table S1). The PCR-amplified fragment of AtRH57 was restricted with KpnI and NotI, then was ligated into pUC-EGFP that was also cut with the same restriction sites. The sequence of pAtRH57-EGFP was confirmed by DNA sequencing.
The method of particle bombardment was in accordance with Sanford et al. (1993) with some modifications. Both the AtRH57-EGFP and EGFP constructs were coated onto gold particles (1 μm) and introduced into onion cells with the Biolistic PDS-1000-He apparatus (Bio-Rad, www.bio-rad.com/) under a vacuum of 27 inches of Hg. Onion epidermal cells were placed into 0.5× MS medium with 1% (w/v) sucrose before bombardment and incubated in the dark at 24°C for 16–24 h after particle delivery. The nuclei were stained with 0.2 μg ml−1 DAPI for 30 min at room temperature. EGFP and DAPI fluorescence were observed with confocal laser scanning biological microscope FV 1000 (Olympus, www.olympus-global.com/). Excitation wavelengths and emission filters were 488 nm/bandpass 505–530 nm for EGFP and 358 nm/461 nm for DAPI.
Antibiotics and CHX assays of seedlings
Genomic DNA was isolated from 14-day-old WT seedlings. The regulatory region 1500 bp upstream of ATG plus the entire coding region of AtRH57 was amplified from genomic DNA by PCR with KpnI–AtRH57c and NcoI–AtRH57c primers (Table S1). The sequence was then subcloned into the binary pCAMBIA1301 vector, confirmed by DNA sequencing and introduced into Agrobacterium tumefaciens for floral dip transformation (Clough and Bent, 1998) of homozygous rh57-1 plants. The medium that contained hygromycin was used to select plants and T1 plants were grown to maturity and selfed. More than two independent lines in the rh57-1 mutant background were selected from each transformation.
For various antibiotics assays, 50–100 seeds of WT, rh57-1 and a T3 complementation line in the rh57-1 mutant background were aseptically treated, rinsed and water-imbibed as described above. Seeds were then sown in solid MS medium that contained 50 μg ml−1 of streptomycin, spectinomycin or erythromycin, respectively and vertically placed in the growth chamber at 22°C with a 16-h light photoperiod. Seedlings were photographed after seeds were sown for 9 days. For CHX assay, seeds were sown in solid MS medium that contained 1 or 2 μg ml−1 of CHX, respectively and germination was counted every day in the growth chamber for 9 days.
Polysome and ribosomal subunit analyses
The 9-day-old WT Col-0 and rh57-1 seedlings grown in sugar-free MS medium were transferred to medium solutions supplemented with or without 4.5% Glc, and incubated for 30 min before harvest. For fractionation of total polysomes, ribosomes were isolated from Arabidopsis seedlings according to Zanetti et al. (2005). Briefly, 0.3 g of the pulverized frozen tissue was suspended in 0.7 ml of PEB buffer [200 mm Tris–HCl, pH 9.0, 200 mm KCl, 25 mm EGTA, 36 mm MgCl2, 1% (v/v) β-mercaptoethanol, 50 μg ml−1 cycloheximide, 50 μg ml−1 chloramphenicol, 0.5 mg ml−1 heparin, 1% (v/v) Triton X-100, 1% (v/v) Tween 20, 1% (w/v) Brij-35, 1% (v/v) NP-40, 2% (v/v) polyoxyethylene, and 1% (w/v) deoxycholic acid]. The crude extract was centrifuged at 12 000 g for 10 min at 4°C, after which the supernatant was loaded onto an 8-ml continuous sucrose gradient (20–60%) and spun for 3.5 h at 170 000 g at 4°C. Fractions were obtained using a gradient fractionator that was connected to an ultra-violet (UV) light detector (ISCO, Lincoln, NE, USA, http://www.isco.com/). The distribution profile of ribosomes was examined by a UV260 absorbance.
Analysis of GeneChip hybridization data
The analysis of microarray experiments was performed using the ATH1 Arabidopsis GeneChip microarray that contained 22 810 cDNAs (Le et al., 2010). All samples were represented by two biological replicates that were harvested independently. Signal intensities (relative mRNA prevalences) were generated using mas 5.0 and imported into Microsoft Excel and Microsoft Access for further analysis.
Acknowledgments
This work was supported by the National Science Council of Republic of China (Taiwan) under grants NSC101-2911-I-005-301, NSC-102-2911-I-005-301, and the Ministry of Education, Taiwan, China under the ATU plan.
Conflict of interest
The authors do not have any conflict of interest to declare.
Supporting Information
Figure S1.rh57 mutants exhibit hypersensitivity to Glc during germination.
Figure S2. Inhibitory effects of Glc on hypocotyl length of rh57 in the dark.
Figure S3.Characterization of Arabidopsis rh57 mutants.
Figure S4.AtRH57 transcript expression during seed development.
Figure S5. Alignment of AtRH57 with related DEAD-box RNA helicases of other species.
Figure S6. Identification of AtRH57 as a member of Class II DEAD-box RNA helicases.
Figure S7. Alteration of glucose (Glc)-responsive, ABA biosynthesis and signaling gene expression in rh57 mutants in the presence and absence of 4.5% Glc for 9 days.
Figure S8. Induction of AtRH57 transcript expression in Arabidopsis thaliana under ABA, high Glc and NaCl conditions.
Figure S9. Relative abundance of the 40S and 60S subunits.
Table S1.Sequences of the gene-specific primers used in the experiments.
Table S2. Density quantitation of ribosomes and pre-rRNA precursors on RNA blots.
References
- Abbasi N, Kim HB, Park NI, Kim HS, Kim YK, Park YI, Choi SB. APUM23, a nucleolar Puf domain protein, is involved in pre-ribosomal RNA processing and normal growth patterning in Arabidopsis. Plant J. 2010;64:960–976. doi: 10.1111/j.1365-313X.2010.04393.x. [DOI] [PubMed] [Google Scholar]
- Arenas-Huertero F, Arroyo A, Zhou L, Sheen J, León P. Analysis of Arabidopsis glucose-insensitive mutants, gin5 and gin6, reveals a central role of the plant hormone ABA in the regulation of plant vegetative development by sugar. Genes Dev. 2000;14:2085–2096. [PMC free article] [PubMed] [Google Scholar]
- Arroyo A, Bossi F, Finkelstein RR, León P. Three genes that affect sugar sensing (abscisic acid insensitive 4, abscisic acid insensitive 5, and constitutive triple response 1) are differentially regulated by glucose in Arabidopsis. Plant Physiol. 2003;133:231–242. doi: 10.1104/pp.103.021089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asakura Y, Galarneau E, Watkins KP, Barkan A, van Wijk KJ. Chloroplast RH3 DEAD-box RNA helicases in maize and Arabidopsis function in splicing of specific group II introns and affect chloroplast ribosome biogenesis. Plant Physiol. 2012;159:961–974. doi: 10.1104/pp.112.197525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossi F, Cordoba E, Dupré P, Mendoza MS, Román CS, León P. The Arabidopsis ABA-INSENSITIVE (ABI) 4 factor acts as a central transcription activator of the expression of its own gene, and for the induction of ABI5 and SBE2.2 genes during sugar signaling. Plant J. 2009;59:359–374. doi: 10.1111/j.1365-313X.2009.03877.x. [DOI] [PubMed] [Google Scholar]
- Boudet N, Aubourg S, Toffano-Nioche C, Kreis M, Lecharny A. Evolution of intron/exon structure of DEAD helicase family genes in ArabidopsisCaenorhabditis, and Drosophila. Genome Res. 2001;11:2101–2114. doi: 10.1101/gr.200801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bu Q, Li H, Zhao Q, et al. The Arabidopsis RING finger E3 ligase RHA2a is a novel positive regulator of abscisic acid signaling during seed germination and early seedling development. Plant Physiol. 2009;150:463–481. doi: 10.1104/pp.109.135269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho RF, Carvalho SD, Duque P. The plant-specific SR45 protein negatively regulates glucose and ABA signaling during early seedling development in Arabidopsis. Plant Physiol. 2010;154:772–783. doi: 10.1104/pp.110.155523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng WH, Endo A, Zhou L, et al. A unique short-chain dehydrogenase/reductase in Arabidopsis glucose signaling and abscisic acid biosynthesis and functions. Plant Cell. 2002;14:2723–2743. doi: 10.1105/tpc.006494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernoff YO, Vincent A, Liebman SW. Mutations in eukaryotic 18S ribosomal RNA affect translational fidelity and resistance to aminoglycoside antibiotics. EMBO J. 1994;13:906–913. doi: 10.1002/j.1460-2075.1994.tb06334.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi W, He B, Mao J, Li Q, Ma J, Ji D, Zou M, Zhang L. The function of RH22, a DEAD RNA helicase, in the biogenesis of the 50S ribosomal subunits of Arabidopsis chloroplasts. Plant Physiol. 2012;158:693–707. doi: 10.1104/pp.111.186775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung E, Cho CW, Yun BH, Choi HK, So HA, Lee SW, Lee JH. Molecular cloning and characterization of the soybean DEAD-box RNA helicase gene induced by low temperature and high salinity stress. Gene. 2009;443:91–99. doi: 10.1016/j.gene.2009.05.005. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Cordin O, Banroques J, Tanner NK, Linder P. The DEAD-box protein family of RNA helicases. Gene. 2006;367:17–37. doi: 10.1016/j.gene.2005.10.019. [DOI] [PubMed] [Google Scholar]
- Dekkers BJ, Schuurmans JA, Smeekens SC. Interaction between sugar and abscisic acid signalling during early seedling development in Arabidopsis. Plant Mol. Biol. 2008;67:151–167. doi: 10.1007/s11103-008-9308-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong H, Zhen Z, Peng J, Chang L, Gong Q, Wang NN. Loss of ACS7 confers abiotic stress tolerance by modulating ABA sensitivity and accumulation in Arabidopsis. J. Exp. Bot. 2011;62:4875–4887. doi: 10.1093/jxb/err143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR, Wang ML, Lynch TJ, Rao S, Goodman HM. The Arabidopsis abscisic acid response locus ABI4 encodes an APETALA 2 domain protein. Plant Cell. 1998;10:1043–1054. doi: 10.1105/tpc.10.6.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabashvili IS, Gregory ST, Valle M, Grassucci R, Worbs M, Wahl MC, Dahlberg AE, Frank J. The polypeptide tunnel system in the ribosome and its gating in erythromycin resistance mutants of L4 and L22. Mol. Cell. 2001;8:181–188. doi: 10.1016/s1097-2765(01)00293-3. [DOI] [PubMed] [Google Scholar]
- Galimand M, Gerbaud G, Courvalin P. Spectinomycin resistance in Neisseria spp. due to mutations in 16S rRNA. Antimicrob. Agents Chemother. 2000;44:1365–1366. doi: 10.1128/aac.44.5.1365-1366.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson SI. Control of plant development and gene expression by sugar signaling. Curr. Opin. Plant Biol. 2005;8:93–102. doi: 10.1016/j.pbi.2004.11.003. [DOI] [PubMed] [Google Scholar]
- Gong Z, Lee H, Xiong L, Jagendorf A, Stevenson B, Zhu JK. RNA helicase-like protein as an early regulator of transcription factors for plant chilling and freezing tolerance. Proc. Natl Acad. Sci. USA. 2002;99:11507–11512. doi: 10.1073/pnas.172399299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Z, Dong CH, Lee H, Zhu J, Xiong L, Gong D, Stevenson B, Zhu JK. A DEAD-box RNA helicase is essential for mRNA export and important for development and stress responses in Arabidopsis. Plant Cell. 2005;17:256–267. doi: 10.1105/tpc.104.027557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Wang S, Valerius O, Hall H, Zeng Q, Li JF, Weston DJ, Ellis BE, Chen JG. Involvement of Arabidopsis RACK1 in protein translation and its regulation by abscisic acid. Plant Physiol. 2011;155:370–383. doi: 10.1104/pp.110.160663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honoré N, Cole ST. Streptomycin resistance in mycobacteria. Antimicrob. Agents Chemother. 1994;38:238–242. doi: 10.1128/aac.38.2.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu YF, Yu SC, Yang CY, Wang CS. Lily ASR protein-conferred cold and freezing resistance in Arabidopsis. Plant Physiol. Biochem. 2011;49:937–945. doi: 10.1016/j.plaphy.2011.07.002. [DOI] [PubMed] [Google Scholar]
- Huang CK, Huang LF, Huang JJ, Wu SJ, Yeh CH, Lu CA. A DEAD-box protein, AtRH36, is essential for female gametophyte development and is involved in rRNA biogenesis in Arabidopsis. Plant Cell Physiol. 2010a;51:694–706. doi: 10.1093/pcp/pcq045. [DOI] [PubMed] [Google Scholar]
- Huang Y, Li CY, Pattison DL, Gray WM, Park S, Gibson SI. SUGAR-INSENSITIVE3, a RING E3 ligase, is a new player in plant sugar response. Plant Physiol. 2010b;152:1889–1900. doi: 10.1104/pp.109.150573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kant P, Kant S, Gordon M, Shaked R, Barak S. STRESS RESPONSE SUPPRESSOR1 and STRESS RESPONSE SUPPRESSOR2, two DEAD-box RNA helicases that attenuate Arabidopsis responses to multiple abiotic stresses. Plant Physiol. 2007;145:814–830. doi: 10.1104/pp.107.099895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp C, ImLer JL. Antiviral immunity in drosophila. Curr. Opin. Immunol. 2009;21:3–9. doi: 10.1016/j.coi.2009.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JS, Kim KA, Oh TR, Park CM, Kang H. Functional characterization of DEAD-box RNA helicases in Arabidopsis thaliana under abiotic stress conditions. Plant Cell Physiol. 2008;49:1563–1571. doi: 10.1093/pcp/pcn125. [DOI] [PubMed] [Google Scholar]
- Kojima H, Suzuki T, Kato T, et al. Sugar-inducible expression of the nucleolin-1 gene of Arabidopsis thaliana and its role in ribosome synthesis, growth and development. Plant J. 2007;49:1053–1063. doi: 10.1111/j.1365-313X.2006.03016.x. [DOI] [PubMed] [Google Scholar]
- Koussevitzky S, Nott A, Mockler TC, Hong F, Sachetto-Martins G, Surpin M, Lim J, Mittler R, Chory J. Signals from chloroplasts converge to regulate nuclear gene expression. Science. 2007;316:715–719. [PubMed] [Google Scholar]
- Laby RJ, Kincaid MS, Kim D, Gibson SI. The Arabidopsis sugar-insensitive mutants sis4 and sis5 are defective in abscisic acid synthesis and response. Plant J. 2000;23:587–596. doi: 10.1046/j.1365-313x.2000.00833.x. [DOI] [PubMed] [Google Scholar]
- Le BH, Cheng C, Bui AQ, et al. Global analysis of gene activity during Arabidopsis seed development and identification of seed-specific transcription factors. Proc. Natl Acad. Sci. USA. 2010;107:8063–8070. doi: 10.1073/pnas.1003530107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SA, Yoon EK, Heo JO, Lee MH, Hwang I, Cheong H, Lee WS, Hwang YS, Lim J. Analysis of Arabidopsis glucose-insensitive growth mutants reveals the involvement of the plastidial copper transporter PAA1 in glucose-induced intracellular signaling. Plant Physiol. 2012;159:1001–1012. doi: 10.1104/pp.111.191726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leόn P, Sheen J. Sugar and hormone connections. Trends Plant Sci. 2003;8:110–116. doi: 10.1016/S1360-1385(03)00011-6. [DOI] [PubMed] [Google Scholar]
- Linder P. Dead-box proteins: a family affair—active and passive players in RNP-remodeling. Nucleic Acids Res. 2006;34:4168–4180. doi: 10.1093/nar/gkl468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder P, Jankowsky E. From unwinding to clamping - the DEAD-box RNA helicase family. Nat. Rev. Mol. Cell Biol. 2011;12:505–516. doi: 10.1038/nrm3154. [DOI] [PubMed] [Google Scholar]
- Liu HH, Liu J, Fan SL, Song MZ, Han XL, Liu F, Shen FF. Molecular cloning and characterization of a salinity stress-induced gene encoding DEAD-box helicase from the halophyte Apocynum venetum. J. Exp. Bot. 2008;59:633–644. doi: 10.1093/jxb/erm355. [DOI] [PubMed] [Google Scholar]
- Liu M, Shi DQ, Yuan L, Liu J, Yang WC. SLOW WALKER3, encoding a putative DEAD-box RNA helicase, is essential for female gametogenesis in Arabidopsis. J. Integr. Plant Biol. 2010;52:817–828. doi: 10.1111/j.1744-7909.2010.00972.x. [DOI] [PubMed] [Google Scholar]
- Lopez-Molina L, Mongrand S, Chua NH. A post-germination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proc. Natl Acad. Sci. USA. 2001;98:4782–4787. doi: 10.1073/pnas.081594298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Molina L, Mongrand S, McLachlin DT, Chait BT, Chua NH. ABI5 acts downstream of ABI3 to execute an ABA-dependent growth arrest during germination. Plant J. 2002;32:317–328. doi: 10.1046/j.1365-313x.2002.01430.x. [DOI] [PubMed] [Google Scholar]
- Maliga P, Klessig DF, Cashmore AR, Gruissem W, Varner JE. Methods in Plant Molecular Biology: A Laboratory Course Manual. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 1995. [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco. Physiol. Plant. 1962;15:473–497. [Google Scholar]
- Oleinick NL. Initiation and elongation of protein synthesis in growing cells: differential inhibition by cycloheximide and emetine. Arch. Biochem. Biophys. 1977;182:171–180. doi: 10.1016/0003-9861(77)90296-x. [DOI] [PubMed] [Google Scholar]
- Owttrim GW. RNA helicases and abiotic stress. Nucleic Acids Res. 2006;34:3220–3230. doi: 10.1093/nar/gkl408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penfield S, Li Y, Gilday AD, Graham S, Graham IA. Arabidopsis ABA INSENSITIVE4 regulates lipid mobilization in the embryo and reveals repression of seed germination by the endosperm. Plant Cell. 2006;18:1887–1899. doi: 10.1105/tpc.106.041277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Y, Yu D. Over-expression of the stress-induced OsWRKY45 enhances disease resistance and drought tolerance in Arabidopsis. Environ. Exp. Bot. 2009;65:35–47. [Google Scholar]
- Rajjou L, Gallardo K, Debeaujon I, Vandekerckhove J, Job C, Job D. The effect of α-amanitin on the Arabidopsis seed proteome highlights the distinct roles of stored and neosynthesized mRNAs during germination. Plant Physiol. 2004;134:1598–1613. doi: 10.1104/pp.103.036293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolland F, Moore B, Sheen J. Sugar sensing and signaling in plants. Plant Cell. 2002;14(Suppl):S185–S205. doi: 10.1105/tpc.010455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosado A, Sohn EJ, Drakakaki G, Pan S, Swidergal A, Xiong Y, Kang BH, Bressan RA, Raikhel NV. Auxin-mediated ribosomal biogenesis regulates vacuolar trafficking in Arabidopsis. Plant Cell. 2010;22:143–158. doi: 10.1105/tpc.109.068320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahni A, Wang N, Alexis JD. UAP56 is an important regulator of protein synthesis and growth in cardiomyocytes. Biochem. Biophys. Res. Commun. 2010;393:106–110. doi: 10.1016/j.bbrc.2010.01.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanan-Mishra N, Pham XH, Sopory SK, Tuteja N. Pea DNA helicase 45 over-expression in tobacco confers high salinity tolerance without affecting yield. Proc. Natl Acad. Sci. USA. 2005;102:509–514. doi: 10.1073/pnas.0406485102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanford JC, Smith FD, Russell JA. Optimizing the biolistic process for different biological applications. Methods Enzymol. 1993;217:483–509. doi: 10.1016/0076-6879(93)17086-k. [DOI] [PubMed] [Google Scholar]
- Stone SL, Williams LA, Farmer LM, Vierstra RD, Callis J. KEEP ON GOING, a RING E3 ligase essential for Arabidopsis growth and development, is involved in abscisic acid signaling. Plant Cell. 2006;18:3415–3428. doi: 10.1105/tpc.106.046532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stonebloom S, Burch-Smith T, Kim I, Meinke D, Mindrinos M, Zambryski P. Loss of the plant DEAD-box protein ISE1 leads to defective mitochondria and increased cell-to-cell transport via plasmodesmata. Proc. Natl Acad. Sci. USA. 2009;106:17229–17234. doi: 10.1073/pnas.0909229106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai CL, Chi WK, Chen DS, Hwang LH. The helicase activity associated with hepatitis C virus nonstructural protein 3 (NS3) J. Virol. 1996;70:8477–8484. doi: 10.1128/jvi.70.12.8477-8484.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullah H, Chen JG, Wang S, Jones AM. Role of a heterotrimeric G protein in regulation of Arabidopsis seed germination. Plant Physiol. 2002;129:897–907. doi: 10.1104/pp.005017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vashisht AA, Tuteja N. Stress-responsive DEAD-box helicases, a new pathway to engineer plant stress tolerance. J. Photochem. Photobiol. B. 2006;84:150–160. doi: 10.1016/j.jphotobiol.2006.02.010. [DOI] [PubMed] [Google Scholar]
- Vashisht AA, Pradhan A, Tuteja R, Tuteja N. Cold- and salinity stress-induced bipolar pea DNA helicase 47 is involved in protein synthesis and stimulated by phosphorylation with protein kinase C. Plant J. 2005;44:76–87. doi: 10.1111/j.1365-313X.2005.02511.x. [DOI] [PubMed] [Google Scholar]
- Verslues PE, Bray EA. Role of abscisic acid (ABA) and Arabidopsis thaliana ABA-insensitive loci in low water potential-induced ABA and proline accumulation. J. Exp. Bot. 2006;57:201–212. doi: 10.1093/jxb/erj026. [DOI] [PubMed] [Google Scholar]
- Xiong L, Ishitani M, Lee H, Zhu JK. The Arabidopsis LOS5/ABA3 locus encodes a molybdenum cofactor sulfurase and modulates cold stress- and osmotic stress-responsive gene expression. Plant Cell. 2001;13:2063–2083. doi: 10.1105/TPC.010101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CY, Chen YC, Jauh GY, Wang CS. A lily ASR protein involves abscisic acid signaling and confers drought and salt resistance in Arabidopsis. Plant Physiol. 2005;139:836–846. doi: 10.1104/pp.105.065458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoine M, Ohto MA, Onai K, Mita S, Nakamura K. The lba1 mutation of UPF1 RNA helicase involved in nonsense-mediated mRNA decay causes pleiotropic phenotypic changes and altered sugar signalling in Arabidopsis. Plant J. 2006;47:49–62. doi: 10.1111/j.1365-313X.2006.02771.x. [DOI] [PubMed] [Google Scholar]
- Yu E, Owttrim GW. Characterization of the cold stress-induced cyanobacterial DEAD-box protein CrhC as an RNA helicase. Nucleic Acids Res. 2000;28:3926–3934. doi: 10.1093/nar/28.20.3926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanetti ME, Chang I-F, Gong F, Galbraith DW, Bailey-Serres J. Immunopurification of polyribosomal complexes of Arabidopsis for global analysis of gene expression. Plant Physiol. 2005;138:624–635. doi: 10.1104/pp.105.059477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Jang JC, Jones TL, Sheen J. Glucose and ethylene signal transduction crosstalk revealed by an Arabidopsis glucose-insensitive mutant. Proc. Natl Acad. Sci. USA. 1998;95:10294–10299. doi: 10.1073/pnas.95.17.10294. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1.rh57 mutants exhibit hypersensitivity to Glc during germination.
Figure S2. Inhibitory effects of Glc on hypocotyl length of rh57 in the dark.
Figure S3.Characterization of Arabidopsis rh57 mutants.
Figure S4.AtRH57 transcript expression during seed development.
Figure S5. Alignment of AtRH57 with related DEAD-box RNA helicases of other species.
Figure S6. Identification of AtRH57 as a member of Class II DEAD-box RNA helicases.
Figure S7. Alteration of glucose (Glc)-responsive, ABA biosynthesis and signaling gene expression in rh57 mutants in the presence and absence of 4.5% Glc for 9 days.
Figure S8. Induction of AtRH57 transcript expression in Arabidopsis thaliana under ABA, high Glc and NaCl conditions.
Figure S9. Relative abundance of the 40S and 60S subunits.
Table S1.Sequences of the gene-specific primers used in the experiments.
Table S2. Density quantitation of ribosomes and pre-rRNA precursors on RNA blots.


