Abstract
Although Th1 and Th2 cytokines can inhibit interleukin (IL)-17-secreting T cells, how these cells are regulated under different infectious conditions is still debated. Our previous studies have shown that vaccination of IL-4 and IL-13 gene knockout (KO) mice can induce high-avidity HIV KdGag197-205-specific CD8 T cells with better protective efficacy. In this study, when IL-13, IL-4, STAT6 KO, and wild-type BALB/c mice were prime-boost immunized with an HIV poxviral modality, elevated numbers of IL-17A+ splenic KdGag197-205-specific CD8 T cells were observed in all the KO mice compared with the wt BALB/c control. Similarly, when wt BALB/c mice were immunized with IL-13Rα2-adjuvanted HIV vaccines (that transiently inhibited IL-13 activity and induced high-avidity CD8 T cells with enhanced protective efficacy), elevated IL-17A+ KdGag197-205-specific CD8 T cells were detected both in the lung and the spleen. However, at the transcriptional level, elevated TGF-β, IL-6, ROR-γt, and IL-17A mRNA copy numbers were mainly detected in IL-4 KO, but not the IL-13 KO mice. These data suggested that TGF-β, IL-6, ROR-γt, but not IL-23a, played a role in IL-17A regulation in KdGag197-205-specific CD8 T cells. Collectively, our findings suggest that IL-4 and IL-13 differentially regulate the expression of IL-17A in KdGag197-205-specific CD8 T cells at the transcriptional and translational level, respectively, implicating IL-17A as an indirect modulator of CD8 T cell avidity and protective immunity.
Introduction
The interleukin-17 (IL-17) family of cytokines (IL-17A to F) has been shown to play a role in host defense and control of inflammatory diseases (Harrington and others 2005; Korn and others 2009). Various studies have shown that CD4 subsets that express IL-17A are associated with autoimmune and/or inflammatory conditions (Park and others 2005; Smiley and others 2007; Riol-Blanco and others 2010). Recent studies have shown that CD4 T cells (Th17), NKT, γδ T cells (Campillo-Gimenez and others 2010; Cooney and others 2011), innate lymphoid cells (ILCs) (Sutton and others 2012), and CD8 T cells under certain conditions can express IL-17A (Hamada and others 2009; Huber and others 2009). Several studies have shown that Th17 cells modulate host defense by mobilization and activation of neutrophils and upregulation of proinflammatory cytokines/chemokines by various cell types (Ye and others 2001; Wu and others 2007; Zhou and others 2008; Zhang and others 2009).
The differentiation of CD4 Th17 cells in mice requires TGF-β and IL-6, while IL-6 and IL-1β have shown to play a more important role in humans (Korn and others 2009). The CD4 Th17 cell induction requires the combined activities of IL-6 and TGF-β to activate the Janus kinase pathway, thereby initiating STAT3 signaling and ROR-γt transcription (Hwang 2010). Following differentiation, IL-21 is required for amplification of Th17 responses, while IL-23 is associated with the maintenance of Th17 expression. Studies have shown that IFN-γ, IL-4, and/or IL-13 through their transcription factors T-bet and GATA3, respectively, can suppress ROR-γt activity resulting in the inhibition of IL-17A production by CD4 T cells (Harrington and others 2005; Park and others 2005). For example, studies by Cruz and others (2006) have demonstrated that IFN-γ regulates the expression of IL-17A production in CD4+ T cells. Similarly, Newcomb and others (2009, 2011) have shown that a functional IL-13 receptor (IL-13Rα1) is expressed on CD4+ Th17 cells, and IL-13 negatively regulates IL-17A production by downregulating ROR-γt while increasing STAT6 and GATA3 expression. Similarly in asthma studies, IL-4 has been associated with the inhibition of IL-17A activity in the lung (Campillo-Gimenez and others 2010). In the context of HIV/SIV infection, the loss of IL-17A production in the intestinal mucosae has been associated with SIV/HIV disease progression in macaques (Cecchinato and others 2008; Xu and others 2012). Indeed, an inverse correlation between HIV-1 plasma RNA and the IL-17RA/Th17 cell number has also been reported (Salgado and others 2011).
Studies in our laboratory have shown that IL-4/IL-13 play an important role in HIV-specific CD8 T cell avidity (Ranasinghe and others 2007; Ranasinghe and Ramshaw 2009). Specifically, our recently developed recombinant IL-13Rα2-adjuvanted vaccines [fowlpox virus (FPV)-HIV IL-13Rα2/vaccinia virus (VV)-HIV IL-13Rα2] have shown to induce elevated high-avidity mucosal and systemic HIV-gag specific CD8 T cells with better protective efficacy (Ranasinghe and others 2013). However, whether IL-13, IL-4, and/or IFN-γ are involved in modulating IL-17A activity in KdGag197-205-specific CD8 T cells and resulting protective efficacy is not established. Therefore, in this study, we have investigated IL-17A expression by KdGag197-205-specific CD8 T cells at the transcriptional and translational level in the spleen and the lung of knockout (KO) mice given the FPV-HIV/VV-HIV prime-boost vaccination and wild-type BALB/c mice given the novel IL-13Rα2-adjuvanted vaccination and a surrogate mucosal HIV challenge.
Materials and Methods
Mice and vaccines
Pathogen-free, 6- to 8-week-old female wild-type BALB/c (H-2d), IL-13, IL-4, and STAT6 KO mice on a BALB/c background were obtained from the Australian National University (ANU) animal breeding facility. These animals were used and maintained in accordance with the approved ANU animal experimentation ethics guidelines. The parent recombinant FPV-HIV gag/pol (mut) and VV-HIV gag/pol (mut) original vaccine stocks were kindly provided by Dr David Boyle at the CSIRO Australian Animal Health Laboratory and were grown in the laboratory. The HIV vaccines coexpressing IL-13Rα2 were constructed as described previously. (Ranasinghe and others 2013; Jackson and others 2014).
Immunization
Mice (n=5–10) were primed intranasally (i.n.) with either 107 plaque-forming units (pfu) rFPV-HIV gag/pol or rFPV-HIV gag/pol IL-13Rα2 and boosted intramuscularly (i.m.) with 107 pfu rVV-HIV gag/pol or rVV-HIV gag/pol IL-13Rα2, 2 weeks apart. Before each immunization, viruses were diluted in phosphate-buffered saline and sonicated to obtain a homogeneous viral suspension as described previously (Ranasinghe and others 2013). During i.n. immunization, 10 μL of rFPV was delivered to each nostril (total 20 μL) and during i.m. immunization, 50 μL of rVV per quadriceps muscle. To evaluate protective immunity at 6 weeks after the final vaccine booster, mice were challenged intranasally with 75 pfu of the influenza virus PR8 expressing the KdGag197-205 epitope of HIV in the neuraminidase stalk, and body weight was monitored for 9–10 days after the challenge as described previously (Ranasinghe and others 2013).
Preparation of mucosal and systemic lymphocytes
To measure the mucosal and systemic immune response, mice were sacrificed at different time intervals (3 days, 14 days, and 8 weeks, post challenge), spleen and lung samples were harvested, and single cell suspensions were prepared as described previously (Xi and others 2012; Ranasinghe and others 2013). Briefly, spleen samples were prepared by passing spleen cell suspensions through a Falcon™ nylon cell strainer into the Roswell Park-Memorial Institute (RPMI) medium, washed, and treated with red cell lysis buffer, and cells were suspended in complete RPMI. Lung samples were first cut into small pieces and digested in 2 mL of complete RPMI (with 5% fetal bovine serum), containing 2 mg/mL collagenase (Sigma Aldrich), 2.4 mg/mL Dispase (Gibco), and 5 U/mL DNAse (Calbiochem), and incubated at 37°C in a water bath for 1 h with gentle vortexing every 10–15 min. The cells were next passed through a Falcon nylon cell strainer, washed, and the samples were treated with red cell lysis buffer, washed again twice and passed though a sterile gauze, and finally resuspended in complete RPMI. To evaluate IL-17 expression following surrogate mucosal challenge, the spleen and lung cell suspensions were obtained from the experiments described in Ranasinghe and others (2013).
Intracellular cytokine staining of IFN-γ and IL-17A
First, 4×106 cells per well were aliquoted into 96-well U-bottom plates and stimulated with 5 μg/mL immunodominant H-2Kd-binding 197AMQMLKETI205 9 mer Gag peptide (prepared at the Biomolecular Resource facility at the JCSMR/ANU) for 16 h at 37°C with 5% CO2 and 1× Brefeldin-A (e-Biosciences) was added to each well and further incubated for 4–5 h. (expression kinetics studies in our laboratory have indicated that the peak IL-17A expression is detected 16–20 h post peptide stimulation). The intracellular cytokine staining (ICS) was performed as described previously (Ranasinghe and others 2006, 2007, 2013). Briefly, the cells were first surface stained with anti-CD8 PE (BD Pharmingen), and then fixed and permeabilized before staining with conjugated anticytokine antibodies (IFN-.γ FITC and IL-17A APC). The samples were analyzed on a 4 color FACS Calibur Flow cytometer (Becton and Dickinson) and 500,000 events were acquired, and the data were analyzed using Cell Quest Pro software. When plotting the graphs, the percentages of the KdGag197-205-specific CD8 T cells producing cytokines were calculated after subtracting the stimulated values from the unstimulated background values.
IL-17A ELISpot
CD8+ T cells were first isolated by negative selection (from pooled spleens) using the EasySep mouse CD8+ T cell enrichment kit according to the manufacturer's instructions (Stem Cell Technologies), and IL-17A expression by KdGag197-205-specific CD8+ T cells was measured by IL-17A ELISpot (e-Biosciences). Briefly, enriched spleen cells and nonenriched lung cells at a density of 4×106 cells/mL in 100 μL were added to Millipore polyvinylidene difluoride plates (Millipore) in duplicates and coated with 4 μg/mL concentration of anti-mouse IL-17A capture antibody. Cells were then stimulated with (5 μg/mL) immunodominant H-2Kd-binding 197AMQMLKETI205 9 mer Gag or with (5 μg/mL) Con-A (Sigma) as a positive control, and the unstimulated cells were included as background control. The cells were then incubated at 37°C with 5% CO2 for 20–22 h, washed, and incubated with 2 μg/mL of biotinylated anti-mouse IL-17A antibody (e-Biosciences) for 2 h at room temperature. The plates were washed and streptavidin–alkaline phosphatase 1:1,000 (GE Healthcare) was added and incubated for 1.5 h at room temperature before developing with NBT/BCIP (Moss.). The spot-forming units (SFU) were counted using ELISpot Bioreader-4000 PRO-X (BioSys). Other than where stated, unstimulated cell counts were subtracted from stimulated counts before plotting the data.
mRNA preparation and quantitative real-time polymerase chain reaction analysis
Briefly, enriched CD8+ T cells were stimulated with 5 μg/mL immunodominant H-2Kd-binding 197AMQMLKETI205, 9 mer Gag peptide for 16 h. Following stimulation, mRNA was prepared and cDNA was synthesized as described previously (Ranasinghe and others 2007). As mRNA copy numbers of the cytokines (IL-6, IL-17, and IL-23a) and transcription factors (TGF-β and ROR-γt) of interests were low, the samples were first preamplified with gene-specific primers (Table 1) using Hot Star Taq Master mix (Qiagen) (cycling as 1×95°C 2 min for Taq polymerase activation, followed by 20×95°C 15 s and 60°C 4 min amplification cycle), and the preamplified cDNA samples were diluted 1:5 and 2 μL of the diluted sample was used in each qRT-PCR reaction using primers indicated in Table 1. qRT-PCR was performed as 1×50°C, 2 min, 95°C, 10 min, followed by 40 cycles of (95°C, 15s, 60°C, 1 min) using an ABI Prism™ 7700 Sequence Detection System (Perkin Elmer/PE Applied Biosystems). All reactions were performed in duplicate and to ensure that single products were obtained after each reaction, the melting curves of the primers were also tested by dissociation runs as described previously (Ranasinghe and others 2007). The arbitrary copy numbers were calculated as follows: Arbitrary copy number=[105/2(Ct-17)]×104, where 1 Ct value=105 mRNA copies (de Mestre and others 2007).
Table 1.
The Primers Used in This Study
| Primer | Sense primer sequence 5′-3′ | Antisense primer sequence 5′-3′ | References |
|---|---|---|---|
| IL-6 | GAGGATACCACTCCCAACAGACC | AAGTGCATCATCGTTGTTCATACA | Ranasinghe and others (2007) |
| IL-17-A | TCCAGAAGGCCCTCAGACTA | AGCATCTTCTCGACCCTGAA | Lexberg and others (2008) |
| IL-23a | TGGCTGTGCCTAGGAGTAG | AGATGCCCAGCCTGAGTTCT | Wang and Seed (2003) |
| ROR-γt | TGCAAGACTCATCGACAAGG | AGGGGATTCAACATCAGTGC | Lexberg and others (2008) |
| TGF-β | CGTGGAAATCAACGGG | CAGAAGTTGGCATGGT | Lexberg and others (2008) |
| L32 | GCTGGAGGTGCTGCTGATGTG | CGTTGGGATTGGTGACTCTGATGG | Ranasinghe and others (2007) |
Statistical analysis
The P values were calculated by performing a two-tailed unpaired Student's t-test. Other than where stated, the experiments were repeated at least thrice. For lung samples, statistics were calculated from pooled samples (n=5–10 mice per group) using minimum 3 separate repeats.
Results
Evaluation of IL-17A expression by KdGag197-205-specific CD8 T cells in the spleen and the lung
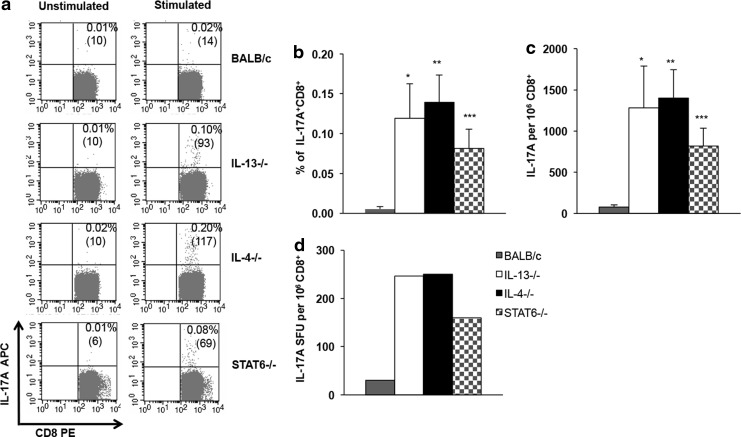
To determine whether IL-4, IL-13, and STAT6 can dampen IL-17A expression by antigen-specific CD8 T cells, IL-4, IL-13, and STAT6 KO and wild-type BALB/c mice were prime-boost immunized with i.n. FPV-HIV gag/pol/i.m. VV-HIVgag/pol as indicated in the Materials and Methods section and IL-17A expression was evaluated at 14 days postbooster vaccination. The ICS results indicated that the IL-17A+ KdGag197-205-specific CD8 T cell numbers in spleens obtained from IL-4, IL-13, and STAT6 KO mice were significantly higher compared with the wild-type BALB/c control (*P=0.017 BALB/c vs. IL-13−/−; **P=0.001 BALB/c vs. IL-4−/−; and ***P=0.005 BALB/c vs. STAT6−/−) (Fig. 1). However, no statistical differences were observed between the 3 KO groups tested. Next, when splenic CD8 T cells were negatively enriched and the total number of KdGag197-205-specific CD8 T cells producing IL-17A were further analyzed using ELISpot, data confirmed that the expression of IL-17A spot-forming cell numbers was elevated in IL-4, IL-13, and STAT6 KO animals compared with the wild-type BALB/c control, eliciting an expression hierarchy of IL-4 KO/IL-13 KO>STAT6 KO>wt BALB/c (Fig. 1).
FIG. 1.
Evaluation of IL-17A+ KdGag197-205-specific splenic CD8 T cells in KO mice compared with wild-type BALB/c. IL-4 KO (black), IL-13 KO (white), STAT6 KO (stripes) on BALB/c background, and wt BALB/c (gray) mice (n=4 per group) were immunized i.n./i.m. with AE FPV-HIV/AE VV-HIV. At 14 days booster vaccination, KdGag197-205-specific T cell responses were measured by (a–c) ICS and (d) IL-17A ELISpot using negatively isolated pooled CD8+ T cells as described in the Materials and Methods section. In all, FACS plot numbers indicate IL-17A-producing CD8+ T cells as a percentage (top) and as the gated number of events (bottom within brackets). The graph (c) indicates the total number of IL-17A SFU per 106 CD8+ T cells measured by ELISpot compared to the number of IL-17A cells per 106 CD8+ T cells as measured by ICS (b). ICS data represent mean of 3 independent experiments (n=12 mice) and ELISpot data are representative of the same 3 independent experiments. Unstimulated cells were used as background controls and values were subtracted from each sample before plotting the data (In ELISpot, the average background counts were less than 10; and in the ICS, the unstimulated average sample values were in the rage of 0.01%). The error bars represent standard error of the mean (SEM). In ICS data, the P values were calculated using Student's two-tailed two-sample unpaired t-test *P=0.017 (BALB/c vs. IL-13−/−), **P=0.001 (BALB/c vs. IL-4−/−), and ***P=0.005 (BALB/c vs. STAT6−/−). FPV, fowlpox virus; ICS, intracellular cytokine staining; IL, interleukin; i.m., intramuscularly; i.n., intranasally; KO, knockout; SFU, spot-forming units; VV, vaccinia virus.
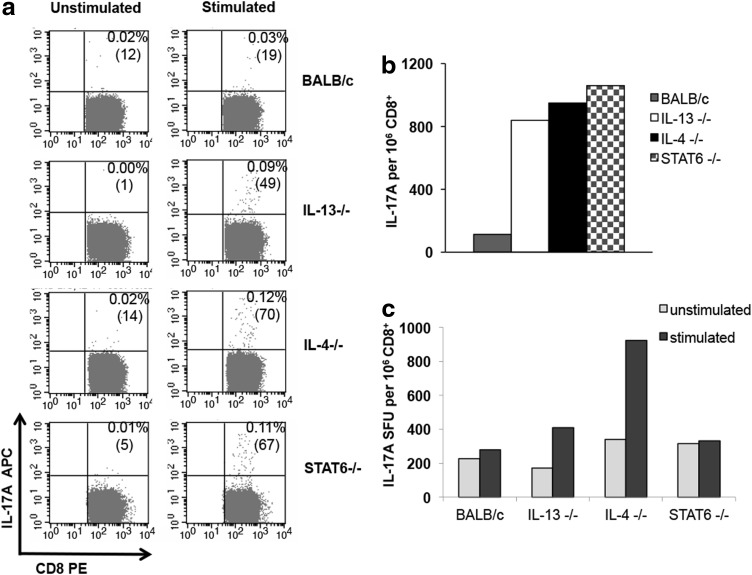
Several studies have shown that following viral infections, CD4 and CD8 T cells can express elevated IL-17A in the lung. As the rFPV-HIV gag/pol priming vaccine was delivered to the lung mucosae, the number of IL-17A+ KdGag197-205-specific lung CD8+ T cells was also evaluated. The ICS data indicated that IL-4, IL-13, and STAT6 KO mice induced elevated numbers of KdGag197-205-specific lung CD8 T cells expressing IL-17A compared with the wild-type BALB/c control (Fig. 2a, b). Due to high endogenous IL-17A expression in the lung, the background ELISpot counts were higher making it difficult to accurately estimate the number of KdGag197-205-specific IL-17A SFU by ELISpot at the lung mucosae, unlike the ICS analysis (Fig. 2c). Note that unlike the spleen samples, lung cells were not negatively enriched for CD8+ T cells and other IL-17A+ cell populations such as ILC3 (Maloy and Uhlig 2013) could be contributing to this high background.
FIG. 2.
Evaluation of IL-17A+ KdGag197-205-specific lung CD8 T cells in KO mice compared with wild-type BALB/c. H-2d background IL-4 KO (black), IL-13 KO (white), STAT6 KO (stripes), and wild-type BALB/c (gray) mice (n=4 per group) were immunized i.n./i.m. with AE FPV-HIV/AE VV-HIV. At 14 days booster vaccination, KdGag197-205-specific T cell responses were measured by (a, b) IL-17A ICS and (c) IL-17A ELISpot (unlike the spleen, the lung samples were not negatively enriched for CD8 T cells due to small sample size) as described in the Materials and Methods section. The graphs represent IL-17A-expressing HIV-specific CD8 T cells per 1×106 CD8+ T cells as a pooled value. In ICS data, unstimulated cells were used as background controls and these values (ranging 0.01–0.02%) were subtracted from each sample before plotting the graphs. The data are representative of 3 independent experiments.
Evaluation of IFN-γ+IL-17A+ KdGag197-205-specific CD8 T cells
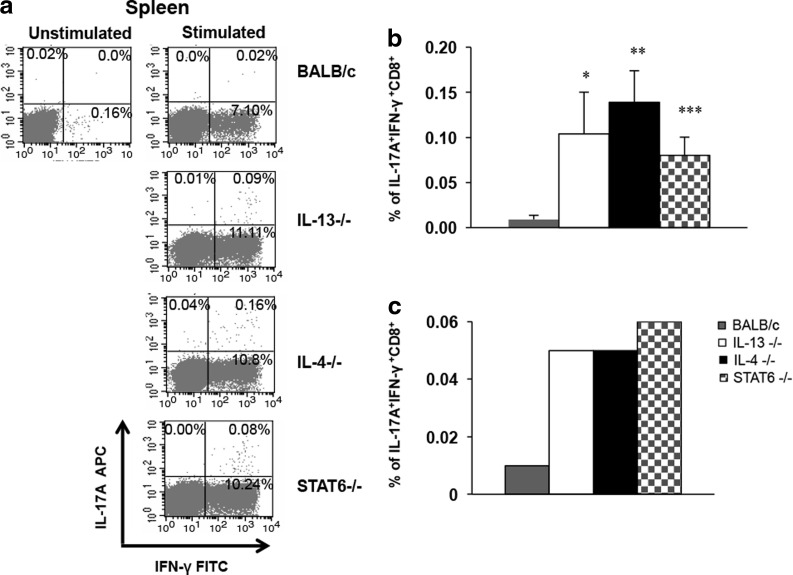
To determine whether these IL-17A-producing KdGag197-205-specific CD8+ T cells also expressed IFN-γ, the KO and wild-type BALB/c mice were prime-boost immunized with i.n. FPV-HIV gag/pol/i.m. VV-HIV gag/pol as indicated previously and IL-17A and IFN-.γ double-positive CD8 T cells were evaluated by ICS. Results indicated that majority of the IL-17A-producing KdGag197-205-specific CD8 T cells were also positive for IFN-γ (Fig. 3a). The IL-17A+IFN-γ+ KdGag197-205-specific CD8 T cell numbers in the spleen were significantly elevated in KO mice compared with wt control; *P=0.05 (BALB/c vs. IL-13−/−), **P=0.001(BALB/c vs. IL-4−/−), and ***P=0.004 (BALB/c vs. STAT6−/−) (Fig. 3b). The IL-17A+IFN-γ+ CD8 cell numbers were much lower in the lung compared to the spleen (Fig. 3b, c). The double-positive expression hierarchy in the spleen was similar to what was observed in Fig. 1.
FIG. 3.
IFN-γ+ IL-17A+ KdGag197-205-specific CD8+ T cell responses in KO mice compared with responses in wild-type BALB/c. H-2d background IL-4 KO (black), IL-13 KO (white), STAT6 KO (stripes), and wild-type BALB/c (gray) mice (n=4 per group) were immunized i.n./i.m. with AE FPV-HIV/AE VV-HIV. At 14 days booster immunization, KdGag197-205-specific T cells expressing IFN-γ+ IL-17A responses were measured by ICS (a) representative FACs plots from each group, (b) spleen data, and (c) lung data. In all graphs, unstimulated cells were used as background controls and were subtracted from each sample before plotting the data. In spleen samples, data are representative of 3 independent experiments and error bars represent standard error of the mean (SEM). P values were calculated using student's two-tailed unpaired t-test *P=0.05 (BALB/c vs. IL-13−/−), **P=0.001(BALB/c vs. IL-4−/−), and ***P=0.004 (BALB/c vs. STAT6−/−). In lung samples, each group represents the pooled value from n=4 mice/group. Data are representative of 3 experiments.
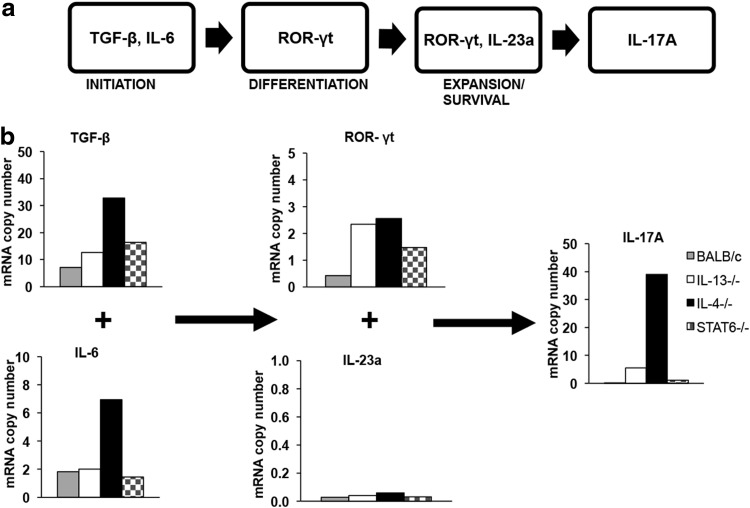
Transcriptional regulation profile of IL-17A in KdGag197-205-specific CD8 T cells in wt BALB/c compared with IL-4, IL-13, and STAT6 KO mice
Next, following FPV-HIV gag/pol/VV-HIV gag/pol vaccination, the IL-17A post-transcription regulatory factors (TGF-β, IL-6, ROR-γt, and IL-23a) in KdGag197-205-specific CD8+ T cells were evaluated in IL-4, IL-13, and STAT6 KO and wild-type BALB/c mice at the mRNA level. Interestingly, unlike the protein expression, the relative IL-17A mRNA level and also TGF-β and IL-6 mRNA expression levels in KdGag197-205-specific CD8 T cells were enhanced in the vaccinated IL-4 KO mice compared with the IL-13 or STAT6 KO mice tested (Fig. 4b). The ROR-γt mRNA copy numbers in KdGag197-205-specific CD8 T cells were elevated in all the KO mice compared with the BALB/c wild-type control and the expression of IL-23a mRNA was extremely low in all mice groups tested (Table 2).
FIG. 4.
Transcriptional regulation profile of IL-17A in KdGag197-205-specific CD8+ T cells following i.n./i.m. prime-boost immunization. (a) Schematic diagram of IL-17A regulation in TH17 cells (Hwang 2010). (b) H-2d Background mice (n=4) from IL-4−/− (black), IL-13−/− (white), STAT6−/− (stripes), and BALB/c (gray) were prime-boost immunized with FPV-HIV/VV-HIV (control vaccine). At 14 days postbooster immunization, splenic CD8+ T cells were negatively enriched and stimulated with KdGag197-205 peptide for 16 h. RNA was extracted, cDNA was synthesized, and quantitative real-time polymerase chain reaction was performed to assess the changes in the mRNA levels. Data show the relative mRNA copy numbers of TGF-β, IL-6, ROR-γt, IL-23a, and IL-17A of stimulated cells after normalizing against L32 expression calculated as indicated in the Materials and Methods section. Data are representative of 3 experiments.
Table 2.
Table Summarizing Transcriptional Regulation Profile of IL-17A in KO and wt BALB/c Mice Following Vaccination
| IL-6 | TGF-β | RORγt | IL-23a | IL-17A | |
|---|---|---|---|---|---|
| IL-4−/− | +++ | +++ | ++± | − | ++++ |
| IL-13−/− | + | ++ | ++± | − | ++ |
| STAT6−/− | ± | ++ | +± | − | + |
| BALB/c | + | ± | ± | − | − |
“+” indicates level of gene expression; “−” indicates no gene expression.
Evaluation of IL-17A expression by KdGag197-205-specific CD8+ T cells following IL-13Rα2-adjuvanted vaccination
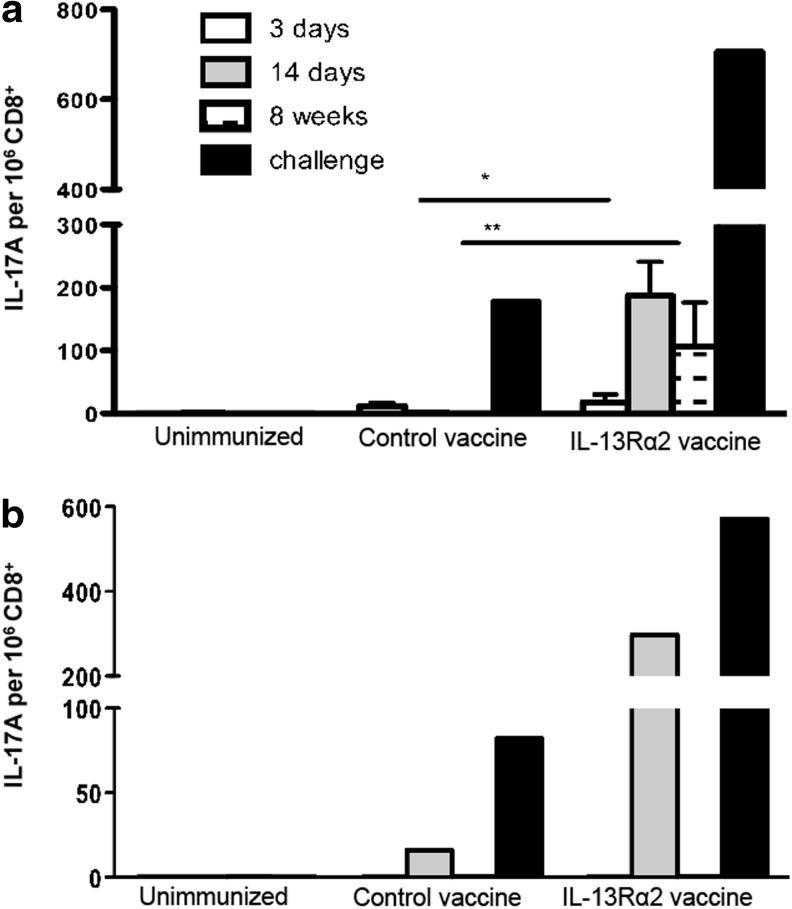
We next evaluated the IL-17A expression following the HIV IL-13Rα2-adjuvanted vaccine strategy (FPV-HIV gag/pol IL-13Rα2/VV-HIV gag/pol IL-13Rα2) that has shown to enhance both the magnitude and avidity of KdGag197-205-specific CD8 T cells (Ranasinghe and others 2013). Specifically, the responses in the spleen and the lung were evaluated at acute (3 days), effector (14 days), memory (8 weeks) stages postbooster, and following influenza-HIV challenge. ICS data indicated that in the spleen, both the IL-13Rα2-adjuvanted and the control HIV vaccines induced lower numbers of IL-17A-expressing KdGag197-205-specific CD8 T cells at the acute stage (3 days) compared to the effector stage (14 days) of immunity (Fig. 5a). However, during the memory phase (8 weeks), the IL-17A+ splenic KdGag197-205-specific CD8+ T cell frequency was significantly elevated in the IL-13Rα2-adjuvanted vaccine group compared with the mice that received the control vaccine (P=0.038). Similar to previous experiments, when the CD8 T cells were negatively enriched and the total numbers of KdGag197-205-specific CD8 T cells producing IL-17A were further evaluated by IL-17 ELISpot, results obtained were highly consistent to the ICS data (data not shown). Results also indicated that following influenza-HIV challenge, the number of IL-17A-expressing cells were much greater compared to the other time points tested (Fig. 5a). In summary, the IL-17A expression hierarchy in the spleen was found to be as follows: postchallenge >14 days >8 weeks >3 days.
FIG. 5.
Expression of CD8 T cells producing IL-17A following HIV IL-13Rα2-adjuvanted vaccine compared to HIV control vaccination. BALB/c (H-2d background) mice (n=5–8) were prime-boost immunized with FPV-HIV/VV-HIV (control vaccine) or FPV-HIV IL-13Rα2/VV-HIV IL-13Rα2 (IL-13 inhibitor vaccine). At 3 days (acute), 14 days (effector), 8 weeks (memory), and following influenza-HIV challenge, spleen and lung samples were harvested and 4×106 cells were stimulated with KdGag197-205 peptide for 16–20 h, and the expression of IL-17A by KdGag197-205-specific (a) splenic and (b) lung CD8+ T cells were evaluated by ICS. The spleen data represent mean of 3 independent experiments and error bars indicate standard error of the mean (SEM). The P values were calculated using student's two-tailed unpaired t-test; 14 days *P=0.025 and 8 weeks **P=0.038, L-13Rα2-adjuvanted vaccinated group compared with control. The lung data and the challenge data represent pooled values. The challenge data are representative of 2 independent experiments.
Similarly, when IL-17A+ KdGag197-205-specific lung CD8 T cell numbers were evaluated, at 14 days and 10 days following influenza-HIV challenge (exactly as described in Ranasinghe and others 2013), the numbers of IL-17A+ cells were found to be greatly enhanced in the IL-13Rα2-adjuvanted vaccine group compared with the control or the unimmunized mice. In contrast, no IL-17A+ CD8+ T cells were detected at 3 days or 8 weeks postbooster immunization (Fig. 5b), which was different to the profile observed in the spleen (Fig. 5a).
Discussion
Several studies have shown that IL-17A plays an important role in the mucosae and protective immunity. However, majority of these studies have mainly focused on the role of IL-17A+ CD4, not CD8 T cells (Khader and others 2007; Smiley and others 2007; Pietrella and others 2011). We have previously shown that inhibition of IL-4 and IL-13 can induce KdGag197-205-specific CD8 T cells of higher avidity and this was vaccine route dependent (Ranasinghe and others 2007). Our current studies indicate that the IL-17A+ KdGag197-205-specific CD8 T cell numbers were significantly enhanced in IL-4, IL-13, and also STAT6 KO mice following i.n. FPV-HIV/i.m. VV-HIV prime-boost vaccination compared with wild-type BALB/c. As IL-4 and IL-13 signal using the common IL-4Rα receptor complex (Tabata and others 2006; Tabata and Khurana Hershey 2007), the current results suggest that both these cytokines play an important role in IL-17A regulation in KdGag197-205-specific CD8 T cells in a STAT6-dependent manner. Interestingly, IL-4/IL-13 double KO mice subjected to epicutaneous sensitization with ovalbumin have also shown exaggerated IL-17 mRNA production in lungs after antigen challenge. These studies have demonstrated that of these 2 cytokines, IL-4 was the key regulator of IL-17A activity at the systemic level and the lung mucosae (He and others 2009). Conney and others (2011) have also shown that downregulation of IL-17A by IL-4 is STAT6 dependent and mediated by inhibition of STAT3 binding to the IL-17A promoter, which is highly consistent with our transcriptional analysis data where IL-4KO mice showed the highest IL-17A expression by KdGag197-205-specific CD8 T cells compared with the IL-13 KO mice. These data also suggest that IL-4 may act directly on the T cells, while the effect of IL-13 could be indirect through other cell types such as ILCs required for stimulation.
ILCs are a recently identified lymphocyte population lacking specific antigen receptors that serve to orchestrate protective immune responses toward infecting microorganisms, lymphoid tissue formation, and tissue homeostasis (Spits and others 2013). The 3 recognized ILC groups ILC1/NK (IFN-γ, TNF-α, T-bet), ILC2 (IL-5, IL-9, IL-13, ROR-α, GATA3), and ILC3 (IL-17, IL-22, ROR-γt) are differentiated based on their effector cytokine and transcription factor profiles. With plasticity between the ILC3 to ILC1 subsets dependent upon IL-12 and IL-18 activation (Maloy and Uhlig 2013). The transient inhibition of IL-13 in our IL13Rα2-adjuvanted vaccine regime resulted in enhanced numbers of IL-17A+IFN-γ+ HIV-Gag-specific CD8+ T cells. We have shown that the inhibition of IL-13 activity occurs within the first 6–48 h post FPV-HIV-IL13Rα2 prime immunization with significant effects upon antigen-presenting cell populations (Trivedi and others 2014), suggesting that a major source of IL-13 could be ILC2 cells. It has been shown that genetic ablation of ILC2 cells, and thus IL-13 expression, causes enhanced levels of inflammatory cytokine expression, including TNF-α and increased activation of IL-17A-producing γδ T cells upon allergen challenge, implicating ILC2 cells in the suppression of inflammatory cytokine signals (Van Dyken and others 2014). These results suggest that inhibition of ILC2 cells or IL-13 activity results in enhanced activation of ILC3/1 cells and IL-17A and TNF-α/IFN-γ expression, which subsequently influence the development of other lymphocyte subsets. We also have found that the transient inhibition of IL-13 does not influence the IL-4 activity at the vaccination site (Ranasinghe Jackson unpublished observations).
Previous studies have indicated that IL-6 and TGF-β directs the initiation of IL-17A expression in CD4 Th17 cells (Korn and others 2008; Hou and others 2009). TGF-β has also shown to enhance the IL-6 receptor activity, by downregulating IL-4, IFN-γ, and suppressor of cytokine signaling 3, and helping the differentiation of IL-17-expressing cells (Veldhoen and others 2006; Gao and others 2012). In our studies, IL-4 KO mice showed the highest TGF-β and IL-6 mRNA expression resulting in higher IL-17A mRNA expression compared with the other KO groups tested, suggesting that IL-4 may be directly involved in negatively regulating IL-6, TGF-β, and IL-17A. Interestingly, reduced IL-17-producing cells have been associated with mice deficient of ROR-γt (Ivanov and others 2006). ROR-γt is regarded as the master transcriptional regulator of IL-17A activity in CD4 T cells (Korn and others 2007). In this study, ROR-γt mRNA copy numbers were found to be higher in all the KO mice tested compared with BALB/c control. Collectively, our mRNA data suggest that at the transcriptional level, TGF-β, IL-6, and ROR-γt, but not IL-23a, appear to play a role in IL-17A regulation in KdGag197-205-specific CD8 T cells and that is IL-4 dependent.
Whether IFN-γ activity can dampen IL-17A expression has been highly debated. Several studies have indicated that Th17-like CD8 T cells have reduced cytotoxicity (Hou and others 2009); Huber and others (2009) have associated this to lower T-box transcription factor eomesodermin, granzyme B, and IFN-γ expression. Similarly, IFN-γ-deficient mice infected with Mycobacterium bovis bacille Calmette Guérin (BCG), showed elevated expression of IL-17A, and addition of exogenous IFN-γ has been shown to increase IL-12 and decreased IL-23-mediated CD4 IL-17A expression (Cruz and others 2006). While many other studies have shown that IL-17A plays a role in protective immunity, for example, following Helicobacter pylori, Bordetella pertussis, and Pseudomonas aerogenosa infections, IL-17A expression by CD4 T cells has been associated with rapid bacterial clearance (Higgins and others 2006; Priebe and others 2008; DeLyria and others 2009). Similarly, following vaccination and challenge, IL-23-mediated IL-17+IFN-γ+ CD4 T cells in the lung have been associated with reduced Mycobacterium tuberculosis growth (Khader and others 2007). In a recent study, it was shown that early, but not late, neutrophil recruitment is essential for IL-17A-mediated long-term control of M. tuberculosis infection and that IFN-γ was not sufficient to control M. tuberculosis growth when the IL-17RA pathway was blocked (Freches and others 2013). In the context of viral infections, vaccination against rotavirus generated elevated IL-17+IFN-γ+ expression associated with reducing fecal viral shedding (VanCott and others 2006; McNeal and others 2007). Interestingly, Wang and others (2011) have shown that IL-17−/− mice infected with H5N1 influenza exhibited increased weight loss, reduced survival rate, and high lung pathology. Elevated IL-17A+ CD8 T cells and protective efficacy have been associated with recruitment of large numbers of neutrophils to the lung mucosae following influenza virus challenge (Hamada and others 2009). Other viral infection studies using IL-17−/− mice have shown that IL-17A-secreting cells were not critical for viral clearance, but instead were involved in regulating antiviral immunity through an additional mechanism such as neutrophil recruitment (Yeh and others 2010).
In our studies, the majority of the KdGag197-205-specific IL-17A-expressing CD8 T cells also expressed IFN-γ. The most abundant IL-17A+IFN-γ+CD8+ subset was detected in IL-4 KO mice, suggesting that IL-4 strongly regulated IL-17A. We have previously shown that the IL-13Rα2-adjuvanted vaccines given to wt BALB/c mice can induce enhanced effector and memory CD8+IFN-γ+ T cells of higher avidity with better protective efficacy compared to the control vaccination (Ranasinghe and others 2013). In the current study, elevated IL-17A+ KdGag197-205-specific splenic CD8 T cells were observed following IL-13Rα2-adjuvanted vaccine at effector, memory, and following influenza-HIV challenge compared to the control vaccine and unimmunized controls. However, at 14 days, the IL-17A+ KdGag197-205-specific CD8 T cell numbers observed in BALB/c mice immunized with the IL-13Rα2-adjuvanted vaccine were much lower compared with the IL-13KO mice given the control vaccine. This is not entirely surprising, as IL-13Rα2-adjuvanted vaccines transiently inhibited IL-13 activity at the vaccination site, unlike a total absence of IL-13 in IL-13 KO mice. Interestingly, in the lung the elevated IL-17A+ KdGag197-205-specific CD8 T cells were detected only at 14 days post booster vaccination and following influenza-HIV challenge, suggesting that IL-17A expression in the lung is most likely associated with the activation status of CD8 T cells. Studies by Yen and others (2009) have shown that the transcription factor STAT3 could be involved in the polarization of IL-17-expressing cells and plays a functional role in inducing IL-17A+IFN-γ+ CD8 T cells, suggesting that the cell phenotypes that express IL-17A have the ability to be functionally plastic.
Collectively, our observations, together with published literature, suggest that according to the pathogen and the environment they encounter, the induction and regulation of IL-17A in CD4 or CD8 T cell subsets could be vastly different. In summary, current data indicate the following: (i) although at the transcriptional level, IL-4 appears to play a predominant role in IL-17A regulation in antigen-specific CD8+ T cells, at the protein level both IL-4 and IL-13 appear to play an important role in IL-17A regulation; and (ii) as IL-4 and IL-13 are directly associated with regulating CD8+ CTL avidity, in a vaccine context, IL-17A may also be indirectly involved in modulating CD8 T cell avidity and protective immunity.
Acknowledgments
The authors would like to thank the ACRF BRF/JCSMR for synthesizing the HIV KdGag197-205-specific peptide, Dr David Boyle at the CSIRO Australian Animal Health Laboratories for providing the rFPV and rVV parent vaccine stocks, and Lisa Pavlinovic and Annette Buchanan for help in the laboratory. This work was supported by the NHMRC project grant 525431(CR), the development grant award APP1000703, the Bill and Melinda Gates Foundation GCE Phase I grant OPP10115149 (C.R.), and the ACH2 EOI grant (C.R.).
Author Disclosure Statement
The authors have no conflicts of interest.
Author Contributions
J.R. conducted all the experiments, data analysis, and preparation of the initial manuscript. R.J.J. designed and constructed the rFPV, rVV, and IL-13Ra2-adjuvanted vaccines. S.T. helped with some initial studies and manuscript revisions. C.R. helped design the experiments, data analysis, and preparation of the manuscript.
References
- Campillo-Gimenez L, Cumont MC, Fay M, Kared H, Monceaux V, Diop O, Muller-Trutwin M, Hurtrel B, Levy Y, Zaunders J, Dy M, Leite-de-Moraes MC, Elbim C, Estaquier J. 2010. AIDS progression is associated with the emergence of IL-17-producing cells early after simian immunodeficiency virus infection. J Immunol 184(2):984–992 [DOI] [PubMed] [Google Scholar]
- Cecchinato V, Trindade CJ, Laurence A, Heraud JM, Brenchley JM, Ferrari MG, Zaffiri L, Tryniszewska E, Tsai WP, Vaccari M, Parks RW, Venzon D, Douek DC, O'Shea JJ, Franchini G. 2008. Altered balance between Th17 and Th1 cells at mucosal sites predicts AIDS progression in simian immunodeficiency virus-infected macaques. Mucosal Immunol 1(4):279–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooney LA, Towery K, Endres J, Fox DA. 2011. Sensitivity and resistance to regulation by IL-4 during Th17 maturation. J Immunol 187(9):4440–4450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz A, Khader SA, Torrado E, Fraga A, Pearl JE, Pedrosa J, Cooper AM, Castro AG. 2006. Cutting edge: IFN-gamma regulates the induction and expansion of IL-17-producing CD4 T cells during mycobacterial infection. J Immunol 177(3):1416–1420 [DOI] [PubMed] [Google Scholar]
- de Mestre AM, Soe-Htwe T, Sutcliffe EL, Rao S, Pagler EB, Hornby JR, Hulett MD. 2007. Regulation of mouse Heparanase gene expression in T lymphocytes and tumor cells. Immunol Cell Biol 85(3):205–214 [DOI] [PubMed] [Google Scholar]
- DeLyria ES, Redline RW, Blanchard TG. 2009. Vaccination of mice against H pylori induces a strong Th-17 response and immunity that is neutrophil dependent. Gastroenterology 136(1):247–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freches D, Korf H, Denis O, Havaux X, Huygen K, Romano M. 2013. Mice genetically inactivated in interleukin-17A receptor are defective in long-term control of Mycobacterium tuberculosis infection. Immunology 140(2):220–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z, Gao Y, Li Z, Chen Z, Lu D, Tsun A, Li B. 2012. Synergy between IL-6 and TGF-beta signaling promotes FOXP3 degradation. Int J Clin Exp Pathol 5(7):626–633 [PMC free article] [PubMed] [Google Scholar]
- Hamada H, Garcia-Hernandez Mde L, Reome JB, Misra SK, Strutt TM, McKinstry KK, Cooper AM, Swain SL, Dutton RW. 2009. Tc17, a unique subset of CD8 T cells that can protect against lethal influenza challenge. J Immunol 182(6):3469–3481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington LE, Hatton RD, Mangan PR, Turner H, Murphy TL, Murphy KM, Weaver CT. 2005. Interleukin 17-producing CD4+ effector T cells develop via a lineage distinct from the T helper type 1 and 2 lineages. Nat Immunol 6(11):1123–1132 [DOI] [PubMed] [Google Scholar]
- He R, Kim HY, Yoon J, Oyoshi MK, MacGinnitie A, Goya S, Freyschmidt EJ, Bryce P, McKenzie AN, Umetsu DT, Oettgen HC, Geha RS. 2009. Exaggerated IL-17 response to epicutaneous sensitization mediates airway inflammation in the absence of IL-4 and IL-13. J Allergy Clin Immunol 124(4):761–770 e1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins SC, Jarnicki AG, Lavelle EC, Mills KH. 2006. TLR4 mediates vaccine-induced protective cellular immunity to Bordetella pertussis: role of IL-17-producing T cells. J Immunol 177(11):7980–7989 [DOI] [PubMed] [Google Scholar]
- Hou W, Kang HS, Kim BS. 2009. Th17 cells enhance viral persistence and inhibit T cell cytotoxicity in a model of chronic virus infection. J Exp Med 206(2):313–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber M, Heink S, Grothe H, Guralnik A, Reinhard K, Elflein K, Hunig T, Mittrucker HW, Brustle A, Kamradt T, Lohoff M. 2009. A Th17-like developmental process leads to CD8(+) Tc17 cells with reduced cytotoxic activity. Eur J Immunol 39(7):1716–1725 [DOI] [PubMed] [Google Scholar]
- Hwang ES. 2010. Transcriptional regulation of T helper 17 cell differentiation. Yonsei Med J 51(4):484–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov II, McKenzie BS, Zhou L, Tadokoro CE, Lepelley A, Lafaille JJ, Cua DJ, Littman DR. 2006. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell 126(6):1121–1133 [DOI] [PubMed] [Google Scholar]
- Jackson RJ, Boyle DB, Ranasinghe C. 2014. Heterologous prime-boost regimens in DNA vaccination. In: Rinaldi M, Fioretti D, Iurescia S, eds. Methods in Molecular Biology; Springer: [DOI] [PubMed] [Google Scholar]
- Khader SA, Bell GK, Pearl JE, Fountain JJ, Rangel-Moreno J, Cilley GE, Shen F, Eaton SM, Gaffen SL, Swain SL, Locksley RM, Haynes L, Randall TD, Cooper AM. 2007. IL-23 and IL-17 in the establishment of protective pulmonary CD4+ T cell responses after vaccination and during Mycobacterium tuberculosis challenge. Nat Immunol 8(4):369–377 [DOI] [PubMed] [Google Scholar]
- Korn T, Bettelli E, Oukka M, Kuchroo VK. 2009. IL-17 and Th17 Cells. Annu Rev Immunol 27:485–517 [DOI] [PubMed] [Google Scholar]
- Korn T, Mitsdoerffer M, Croxford AL, Awasthi A, Dardalhon VA, Galileos G, Vollmar P, Stritesky GL, Kaplan MH, Waisman A, Kuchroo VK, Oukka M. 2008. IL-6 controls Th17 immunity in vivo by inhibiting the conversion of conventional T cells into Foxp3+ regulatory T cells. Proc Natl Acad Sci U S A 105(47):18460–18465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korn T, Oukka M, Kuchroo V, Bettelli E. 2007. Th17 cells: effector T cells with inflammatory properties. Semin Immunol 19(6):362–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lexberg MH, Taubner A, Förster A, Albrecht I, Richter A, Kamradt T, Radbruch A, Chang HD. 2008. Th memory for interleukin-17 expression is stable in vivo. Eur J Immunol 38(10):2654–2664 [DOI] [PubMed] [Google Scholar]
- Maloy KJ, Uhlig HH. 2013. ILC1 populations join the border patrol. Immunity 38(4):630–632 [DOI] [PubMed] [Google Scholar]
- McNeal MM, Basu M, Bean JA, Clements JD, Lycke NY, Ramne A, Lowenadler B, Choi AH, Ward RL. 2007. Intrarectal immunization of mice with VP6 and either LT(R192G) or CTA1-DD as adjuvant protects against fecal rotavirus shedding after EDIM challenge. Vaccine 25(33):6224–6231 [DOI] [PubMed] [Google Scholar]
- Newcomb DC, Boswell MG, Zhou W, Huckabee MM, Goleniewska K, Sevin CM, Hershey GK, Kolls JK, Peebles RS., Jr.2011. Human TH17 cells express a functional IL-13 receptor and IL-13 attenuates IL-17A production. J Allergy Clin Immunol 127(4):1006–1013.e1–e4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newcomb DC, Zhou W, Moore ML, Goleniewska K, Hershey GK, Kolls JK, Peebles RS., Jr.2009. A functional IL-13 receptor is expressed on polarized murine CD4+ Th17 cells and IL-13 signaling attenuates Th17 cytokine production. J Immunol 182(9):5317–5321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H, Li Z, Yang XO, Chang SH, Nurieva R, Wang YH, Wang Y, Hood L, Zhu Z, Tian Q, Dong C. 2005. A distinct lineage of CD4 T cells regulates tissue inflammation by producing interleukin 17. Nat Immunol 6(11):1133–1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietrella D, Rachini A, Pines M, Pandey N, Mosci P, Bistoni F, d'Enfert C, Vecchiarelli A. 2011. Th17 cells and IL-17 in protective immunity to vaginal candidiasis. PLoS One 6(7):e22770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priebe GP, Walsh RL, Cederroth TA, Kamei A, Coutinho-Sledge YS, Goldberg JB, Pier GB. 2008. IL-17 is a critical component of vaccine-induced protection against lung infection by lipopolysaccharide-heterologous strains of Pseudomonas aeruginosa. J Immunol 181(7):4965–4975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranasinghe C, Medveczky JC, Woltring D, Gao K, Thomson S, Coupar BE, Boyle DB, Ramsay AJ, Ramshaw IA. 2006. Evaluation of fowlpox-vaccinia virus prime-boost vaccine strategies for high-level mucosal and systemic immunity against HIV-1. Vaccine 24(31–32):5881–5895 [DOI] [PubMed] [Google Scholar]
- Ranasinghe C, Ramshaw IA. 2009. Immunisation route-dependent expression of IL-4/IL-13 can modulate HIV-specific CD8(+) CTL avidity. Eur J Immunol 39(7):1819–1830 [DOI] [PubMed] [Google Scholar]
- Ranasinghe C, Trivedi S, Stambas J, Jackson RJ. 2013. Unique IL-13Ralpha2-based HIV-1 vaccine strategy to enhance mucosal immunity, CD8(+) T-cell avidity and protective immunity. Mucosal Immunol 6(6):1068–1080 [DOI] [PubMed] [Google Scholar]
- Ranasinghe C, Turner SJ, McArthur C, Sutherland DB, Kim JH, Doherty PC, Ramshaw IA. 2007. Mucosal HIV-1 pox virus prime-boost immunization induces high-avidity CD8+ T cells with regime-dependent cytokine/granzyme B profiles. J Immunol 178(4):2370–2379 [DOI] [PubMed] [Google Scholar]
- Riol-Blanco L, Lazarevic V, Awasthi A, Mitsdoerffer M, Wilson BS, Croxford A, Waisman A, Kuchroo VK, Glimcher LH, Oukka M. 2010. IL-23 receptor regulates unconventional IL-17-producing T cells that control bacterial infections. J Immunol 184(4):1710–1720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salgado M, Rallon NI, Rodes B, Lopez M, Soriano V, Benito JM. 2011. Long-term non-progressors display a greater number of Th17 cells than HIV-infected typical progressors. Clin Immunol 139(2):110–114 [DOI] [PubMed] [Google Scholar]
- Smiley KL, McNeal MM, Basu M, Choi AH, Clements JD, Ward RL. 2007. Association of gamma interferon and interleukin-17 production in intestinal CD4+ T cells with protection against rotavirus shedding in mice intranasally immunized with VP6 and the adjuvant LT(R192G). J Virol 81(8):3740–3748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spits H, Artis D, Colonna M, Diefenbach A, Di Santo JP, Eberl G, Koyasu S, Locksley RM, McKenzie AN, Mebius RE, Powrie F, Vivier E. 2013. Innate lymphoid cells—a proposal for uniform nomenclature. Nat Rev Immunol 13(2):145–149 [DOI] [PubMed] [Google Scholar]
- Sutton CE, Mielke LA, Mills KH. 2012. IL-17-producing gammadelta T cells and innate lymphoid cells. Eur J Immunol 42(9):2221–2231 [DOI] [PubMed] [Google Scholar]
- Tabata Y, Chen W, Warrier MR, Gibson AM, Daines MO, Hershey GK. 2006. Allergy-driven alternative splicing of IL-13 receptor alpha2 yields distinct membrane and soluble forms. J Immunol 177(11):7905–7912 [DOI] [PubMed] [Google Scholar]
- Tabata Y, Khurana Hershey GK. 2007. IL-13 receptor isoforms: breaking through the complexity. Curr Allergy Asthma Rep 7(5):338–345 [DOI] [PubMed] [Google Scholar]
- Trivedi S, Jackson RJ, Ranasinghe C. 2014. Different HIV pox viral vector-based vaccines and adjuvants can induce unique antigen presenting cells that modulate CD8 T cell avidity. Virology 468–470C:479–489 [DOI] [PubMed] [Google Scholar]
- Van Dyken SJ, Mohapatra A, Nussbaum JC, Molofsky AB, Thornton EE, Ziegler SF, McKenzie AN, Krummel MF, Liang HE, Locksley RM. 2014. Chitin activates parallel immune modules that direct distinct inflammatory responses via innate lymphoid type 2 and gammadelta T cells. Immunity 40(3):414–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanCott JL, Prada AE, McNeal MM, Stone SC, Basu M, Huffer B, Jr., Smiley KL, Shao M, Bean JA, Clements JD, Choi AH, Ward RL. 2006. Mice develop effective but delayed protective immune responses when immunized as neonates either intranasally with nonliving VP6/LT(R192G) or orally with live rhesus rotavirus vaccine candidates. J Virol 80(10):4949–4961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldhoen M, Hocking RJ, Atkins CJ, Locksley RM, Stockinger B. 2006. TGFbeta in the context of an inflammatory cytokine milieu supports de novo differentiation of IL-17-producing T cells. Immunity 24(2):179–189 [DOI] [PubMed] [Google Scholar]
- Wang X, Chan CC, Yang M, Deng J, Poon VK, Leung VH, Ko KH, Zhou J, Yuen KY, Zheng BJ, Lu L. 2011. A critical role of IL-17 in modulating the B-cell response during H5N1 influenza virus infection. Cell Mol Immunol 8(6):462–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Seed B. 2003. A PCR primer bank for quantitative gene expression analysis. Nucleic Acids Res. 31(24):154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q, Martin RJ, Rino JG, Breed R, Torres RM, Chu HW. 2007. IL-23-dependent IL-17 production is essential in neutrophil recruitment and activity in mouse lung defense against respiratory Mycoplasma pneumoniae infection. Microbes Infect 9(1):78–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi Y, Day SL, Jackson RJ, Ranasinghe C. 2012. Role of novel type I interferon epsilon in viral infection and mucosal immunity. Mucosal Immunol 5(6):610–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Wang X, Liu DX, Moroney-Rasmussen T, Lackner AA, Veazey RS. 2012. IL-17-producing innate lymphoid cells are restricted to mucosal tissues and are depleted in SIV-infected macaques. Mucosal Immunol 2012(6):39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye P, Garvey PB, Zhang P, Nelson S, Bagby G, Summer WR, Schwarzenberger P, Shellito JE, Kolls JK. 2001. Interleukin-17 and lung host defense against Klebsiella pneumoniae infection. Am J Respir Cell Mol Biol 25(3):335–340 [DOI] [PubMed] [Google Scholar]
- Yeh N, Glosson NL, Wang N, Guindon L, McKinley C, Hamada H, Li Q, Dutton RW, Shrikant P, Zhou B, Brutkiewicz RR, Blum JS, Kaplan MH. 2010. Tc17 cells are capable of mediating immunity to vaccinia virus by acquisition of a cytotoxic phenotype. J Immunol 185(4):2089–2098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen HR, Harris TJ, Wada S, Grosso JF, Getnet D, Goldberg MV, Liang KL, Bruno TC, Pyle KJ, Chan SL, Anders RA, Trimble CL, Adler AJ, Lin TY, Pardoll DM, Huang CT, Drake CG. 2009. Tc17 CD8 T cells: functional plasticity and subset diversity. J Immunol 183(11):7161–7168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Clarke TB, Weiser JN. 2009. Cellular effectors mediating Th17-dependent clearance of pneumococcal colonization in mice. J Clin Invest 119(7):1899–1909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M, Yang B, Ma R, Wu C. 2008. Memory Th-17 cells specific for C. albicans are persistent in human peripheral blood. Immunol Lett 118(1):72–81 [DOI] [PubMed] [Google Scholar]