Abstract
The serotonin transporter gene (5-HTT/SLC6A4)-linked polymorphic region has been suggested to have a modulatory role in mediating effects of early-life stress exposure on psychopathology rendering carriers of the low-expression short (s)-variant more vulnerable to environmental adversity in later life. The underlying molecular mechanisms of this gene-by-environment interaction are not well understood, but epigenetic regulation including differential DNA methylation has been postulated to have a critical role. Recently, we used a maternal restraint stress paradigm of prenatal stress (PS) in 5-HTT-deficient mice and showed that the effects on behavior and gene expression were particularly marked in the hippocampus of female 5-Htt+/− offspring. Here, we examined to which extent these effects are mediated by differential methylation of DNA. For this purpose, we performed a genome-wide hippocampal DNA methylation screening using methylated-DNA immunoprecipitation (MeDIP) on Affymetrix GeneChip Mouse Promoter 1.0 R arrays. Using hippocampal DNA from the same mice as assessed before enabled us to correlate gene-specific DNA methylation, mRNA expression and behavior. We found that 5-Htt genotype, PS and their interaction differentially affected the DNA methylation signature of numerous genes, a subset of which showed overlap with the expression profiles of the corresponding transcripts. For example, a differentially methylated region in the gene encoding myelin basic protein (Mbp) was associated with its expression in a 5-Htt-, PS- and 5-Htt × PS-dependent manner. Subsequent fine-mapping of this Mbp locus linked the methylation status of two specific CpG sites to Mbp expression and anxiety-related behavior. In conclusion, hippocampal DNA methylation patterns and expression profiles of female prenatally stressed 5-Htt+/− mice suggest that distinct molecular mechanisms, some of which are promoter methylation-dependent, contribute to the behavioral effects of the 5-Htt genotype, PS exposure and their interaction.
Introduction
Prenatal stress (PS) exposure has been shown to affect fetal brain development and to increase the risk for later-life psychopathology.1 Furthermore, the serotonin transporter gene (5-HTT/SLC6A4)-linked polymorphic region (5-HTTLPR) is assumed to modulate the interaction between developmental stress exposure and emotional dysregulation later in life in both nonhuman primates and humans.2,3 The exact molecular mechanisms underlying this gene-by-environment (G × E) interaction, however, remain to be elucidated.
Recently, using a maternal restraint stress paradigm of PS in wild-type (WT, +/+) and heterozygous (+/−) 5-Htt deficient mice, we have shown that the long-term behavioral effects of PS are partly dependent on the 5-Htt genotype.4 Although mice carrying the 5-Htt+/− genotype showed well-defined adaptive capacity, for example, in terms of cognitive performance, this advantage came at the expense of an increased vulnerability to PS exposure especially in female offspring. Moreover, gene expression profiles of the hippocampus indicated that distinct molecular mechanisms were associated with effects of 5-Htt+/− genotype, PS exposure and their interaction. More specifically, mitogen-activated protein kinase and neurotrophin signaling were regulated by both the 5-Htt+/− genotype and PS exposure, whereas cytokine and wingless-type MMTV integration site family (Wnt) signaling were affected in a 5-Htt genotype × PS manner, indicating a G × E interaction at the molecular level.
Epigenetic processes such as DNA methylation and histone modifications translate external stimuli into changes of gene expression and, as such, have a pivotal role in the interaction of genetic and environmental factors in determining an individual's phenotype. Thus, epigenetic programming during development may contribute to the etiology and influence the course of neuropsychiatric disorders.5 Recent work in rodents has shown that epigenetic mechanisms regulate various signaling pathways previously implicated in anxiety- and depression-like behavior.6,7 In light of these findings, the present study aimed to examine the role of DNA methylation in mediating differential gene expression observed in the 5-Htt × PS paradigm. Genome-wide promoter methylation was assessed using methylated-DNA immunoprecipitation (MeDIP) followed by high-resolution DNA promoter array analysis (MeDIP-on-Chip). Resulting DNA methylation signatures were subsequently linked to gene expression profiles and behavioral observations.
Materials and methods
Experimental animals
Hippocampal tissue derived from female C57BL6/J mice of four groups of mice4 were used, that is, 5-Htt+/− mice (B6.129[Cg]-Slc6a4tm1Kpl/J) exposed to PS (5-Htt+/− PS) or left undisturbed during pregnancy (5-Htt+/− C) and the corresponding 5-Htt+/+ groups (that is, 5-Htt+/+ PS and 5-Htt+/+ C, respectively). Around P95, following behavioral analysis and 1 week after measuring stress-induced plasma corticosterone secretion,4 the mice were killed. Brains were removed, immediately placed on dry ice and stored at −80 °C for future experiments. No more than two female pups per litter were used to prevent litter effects.8 All experiments were approved by the Animals Ethics Board of Maastricht University (Permit number: OE 2007-109) and all efforts were made to minimize suffering. Animal group sizes for pyrosequencing and quantitative PCR with reverse transcription (RT-qPCR) were: 5-Htt+/+ C=7, 5-Htt+/+ PS=9, 5-Htt+/− C=10, 5-Htt+/− PS=10. For the DNA methylation promoter array, the same group sizes apply, except for 5-Htt+/+ PS=6.
DNA methylation analysis
DNA methylation was assessed by MeDIP (Diagenode, Liège, Belgium) using an antibody against 5-methylcytosine followed by GeneChip Mouse Promoter 1.0 R tiling array (Affymetrix, Santa Clara, CA, USA) using the right part of the hippocampus (see Supplementary Methods for more details). The MeDIP array data were subjected to within-sample pairwise LOESS normalization and calculation of MeDIP-input signal log2 ratios. Between-sample quantile normalization was applied to the signal log2 ratios, followed by a 300 bp sliding-window median smoothing. Autosomal regions enriched by the MeDIP procedure were detected by the CMARRT algorithm.9 With the present (=1) and absent (=0) calls for MeDIP enrichment, effect directions were determined as previously described.4 Briefly, genotype (G) effect directions (d) were calculated by
Gd=((5-Htt+/− C+5-Htt+/− PS)−(5-Htt+/+ C+5-Htt+/+ PS)) × 0.5,
environment (E) effect directions by
Ed=((5-Htt+/+ PS+5-Htt+/− PS)−(5-Htt+/+ C+5-Htt+/− C)) × 0.5
and interaction (G × E) effect directions by
G × Ed=((5-Htt+/− PS−5-Htt+/− C)−(5-Htt+/+ PS−5-Htt+/+ C)) × 0.5.
For a detailed description of the bioinformatics, see Supplementary Methods. No ‘cutoff' value for the fold change was applied. For optimizing discrimination of differentially methylated regions (DMRs) associated with the same gene, a unique DMR identification number was assigned to each DMR. Sometimes, more than one gene was annotated to one DMR. The validity of the methylation data obtained from the promoter array was tested by pyrosequencing and MeDIP quantitative real-time PCR (MeDIP-qPCR). For more details on procedures and methods, as well as PCR and sequencing primers used, see Supplementary Material. Microarray data has been deposited in MIAME-compliant form in Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) with the accession number GSE51634.
Overlap of DNA methylation with gene expression
In addition to focusing solely on differential DNA methylation, DMRs were also assessed in the context of gene expression changes, as investigated previously in the same mice using GeneChip Mouse Genome 430 2.0 Arrays (Affymetrix).4 For this purpose, we identified genes that both displayed a modified DNA methylation signature and were differentially expressed in our 5-Htt × PS paradigm. Since their expression data were not available, miRNA were not included in the analyses. Gene expression microarray results were validated and analyzed with regard to specific splice variants using RT-qPCR (for details and primer sequences, see Supplementary Methods and Material).4
Results
Genome-wide DNA methylation
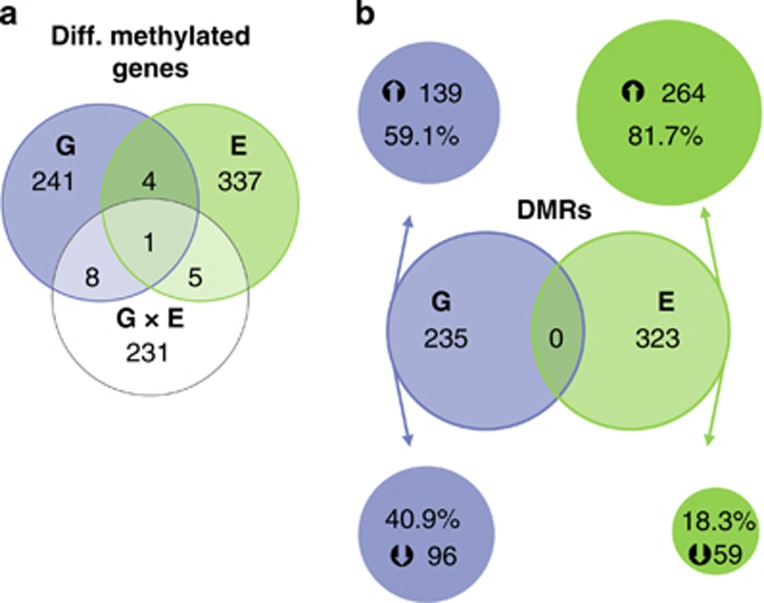
The MeDIP promoter array analysis revealed that the methylation status of two to four hundred genomic regions was significantly modified by either the 5-Htt+/− genotype (G), PS exposure (E) or their interaction (G × E) (Figure 1; Supplementary Tables S1–S3 for a complete overview of all genes significantly affected by G, E and G × E, respectively). More specifically, 235 DMRs, involving 254 genes, were differentially methylated in 5-Htt+/− when compared with 5-Htt+/+ mice, with 40.9% of the DMRs showing a decreased and 59.1% displaying an increased degree of methylation in 5-Htt+/− mice (Figure 1a). Furthermore, 323 DMRs corresponding to 347 genes were affected by PS exposure. As depicted in Figure 1b, 81.7% DMRs showed an increase in methylation in PS mice compared with controls, whereas in merely 18.3% DMRs methylation was decreased. Finally, the DNA methylation status of 218 genomic regions involving 245 genes was affected by a 5-Htt × PS interaction. Supplementary Table 2 depicts enriched annotation term clusters determined by Functional Annotation Clustering using DAVID10 (enrichment score >1.3) for DMRs. Two clusters have been found enriched due to genotype, that is, ribosome-associated terms and ion-binding terms. For DMRs affected by PS, five clusters were identified, for example, WD repeat terms containing PAK1 interacting protein 1 (Pak1ip1) and striatin, calmodulin binding protein 3 (Strn3, also known as Sg2na). Another five clusters were enriched due to an interaction of genotype and PS, among them two clusters with cytoskeleton terms and one with EGF-associated terms. The latter comprised genes such as Notch3 and FAT tumor suppressor homolog 1 (Drosophila, Fat1) encoding a gene of the cadherin superfamily.
Figure 1.
(a) Venn diagram illustrating the number of genes of which the DNA methylation status was altered by genotype (G, 5-Htt+/− versus 5-Htt+/+), the environment (E; that is, prenatal stressed (PS) versus control animals) or in an interactive manner (G × E, indicating those genes of which the effect of the environment depends on the genotype) in the hippocampus of female mice. DNA methylation analysis was performed by applying hippocampal MeDIP DNA on Affymetrix Mouse Promoter 1.0R arrays. (b) Venn diagram showing the number of differentially (diff.) methylated regions (DMRs) regulated by G or E and the corresponding direction in which the 5-Htt+/− genotype and PS regulated the corresponding DMRs. The direction of the change in methylation does not necessarily implicate a change of the gene's expression to the opposite direction (see Discussion for further details). MeDIP, methylated-DNA immunoprecipitation.
Differential DNA methylation associated with gene expression changes
Next, we examined to which extent these differentially methylated genes also displayed changes in their expression. We compared DMR data with the hippocampal gene expression profiles from the same mice as previously reported.4 Table 1a lists the 25 genes that were both differentially methylated and expressed (P<0.05) in 5-Htt+/− compared with 5-Htt+/+ offspring. These 25 genes corresponded to 26 DMRs, half of which showed downregulation and the other half upregulation of methylation. Among the differentially methylated/expressed genes were E2F transcription factor 3 (E2f3), kinesin family member 13 A (Kif13a), low density lipoprotein receptor class A domain containing 3 (Ldlrad3), fibroblast growth factor receptor 4 (Fgfr4) and bone morphogenic protein receptor 1b (Bmpr1b).
Table 1. Differentially methylated and expressed genes.
| Symbol | Gene name |
Methylation |
Expression |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Chr | Start | End | FC | in/de | FC | Change | Affy ID | ||
| A) Genotype (G) | |||||||||
| Bbx | Bobby sox homolog (Drosophila) | 16 | 50435836 | 50436363 | 1.58 | ↑ | 1.26 | ↑ | 1430820_a_at |
| Ldlrad3 | Low density lipoprotein receptor class A domain containing 3 | 2 | 102029692 | 102029893 | 1.28 | ↑ | 1.12 | ↓ | 1438666_at |
| Bcl2l11 // Acoxl | BCL2-like 11 (apoptosis facilitator) // acyl-Coenzyme A oxidase-like | 2 | 127946767 | 127947010 | 1.26 | ↑ | 1.11 | ↓ | 1456005_a_at |
| Bcl2l11 // Acoxl | BCL2-like 11 (apoptosis facilitator) // acyl-Coenzyme A oxidase-like | 2 | 127946767 | 127947010 | 1.26 | ↑ | 1.35 | ↑ | 1456006_at |
| Kif13a | Kinesin family member 13A | 13 | 47028519 | 47029667 | 1.23 | ↓ | 1.12 | ↓ | 1451890_at |
| Spsb1 | splA/ryaNodine receptor domain and SOCS box containing 1 | 4 | 149331144 | 149332126 | 1.23 | ↑ | 1.19 | ↓ | 1428472_at |
| Fank1 // Dhx32 | Fibronectin type 3 and ankyrin repeat domains 1 // DEAH(Asp-Glu-Ala-His) box polypeptide 32 | 7 | 140975216 | 140978199 | 1.19 | ↓ | 1.17 | ↓ | 1447495_at |
| Dhx32 // Fank | DEAH (Asp-Glu-Ala-His) box polypeptide 32 // fibronectin type 3 and ankyrin repeat domains 1 | 7 | 140975216 | 140978199 | 1.19 | ↓ | 1.19 | ↓ | 1420427_a_at |
| Fgfr4 | Fibroblast growth factor receptor 4 | 13 | 55249814 | 55250054 | 1.17 | ↑ | 1.15 | ↓ | 1427845_at |
| Fgfr4 | Fibroblast growth factor receptor 4 | 13 | 55249814 | 55250054 | 1.17 | ↑ | 1.26 | ↓ | 1427776_a_at |
| E2f3 | E2F transcription factor 3 | 13 | 30080878 | 30081078 | 1.17 | ↓ | 1.14 | ↑ | 1427462_at |
| Ttn | Titin | 2 | 76823368 | 76823897 | 1.17 | ↓ | 1.21 | ↑ | 1427446_s_at |
| Nr5a1 | Nuclear receptor subfamily 5, group A, member 1 | 2 | 38568797 | 38569145 | 1.15 | ↓ | 1.17 | ↓ | 1421730_at |
| BC030307 | cDNA sequence BC030307 | 10 | 86175833 | 86176109 | 1.14 | ↑ | 1.15 | ↑ | 1441409_at |
| Bmpr1b | Bone morphogenetic protein receptor, type 1B | 3 | 141593558 | 141593900 | 1.14 | ↑ | 1.19 | ↓ | 1422872_at |
| C030046E11Rik | RIKEN cDNA C030046E11 gene | 19 | 29654132 | 29654407 | 1.14 | ↓ | 1.14 | ↑ | 1431023_at |
| Cybrd1 | Cytochrome b reductase 1 | 2 | 70957388 | 70957814 | 1.12 | ↓ | 1.13 | ↑ | 1425040_at |
| Fabp6 | Fatty acid binding protein 6, ileal (gastrotropin) | 11 | 43418691 | 43419752 | 1.10 | ↑ | 1.27 | ↓ | 1450682_at |
| Akap6 // n-R5s58 | A kinase (PRKA) anchor protein 6 // nuclearencoded rRNA 5S 58 | 12 | 53980545 | 53980725 | 1.09 | ↓ | 1.14 | ↑ | 1440859_at |
| Krt23 | Keratin 23 | 11 | 99352405 | 99352894 | 1.07 | ↓ | 1.21 | ↓ | 1418213_at |
| Nr5a1 | Nuclear receptor subfamily 5, group A, member 1 | 2 | 38569234 | 38569481 | 1.07 | ↓ | 1.17 | ↓ | 1421730_at |
| Lgr5 | Leucine rich repeat containing G protein coupled receptor 5 | 10 | 115028742 | 115029312 | 1.05 | ↑ | 1.13 | ↑ | 1444519_at |
| Mttp | Microsomal triglyceride transfer protein | 3 | 137796736 | 137797014 | 1.04 | ↑ | 1.25 | ↑ | 1419399_at |
| Tmem100 | Transmembrane protein 100 | 11 | 89896819 | 89897137 | 1.03 | ↓ | 1.22 | ↑ | 1446625_at |
| Pdlim4 | PDZ and LIM domain 4 | 11 | 53883752 | 53883994 | 1.02 | ↑ | 1.21 | ↓ | 1417928_at |
| Pla2g5 // Pla2g2a | Phospholipase A2, group V // phospholipase A2, group IIA (platelets, synovial fluid) | 4 | 138390529 | 138390886 | 1.02 | ↑ | 1.59 | ↓ | 1417814_at |
| Kcnj5 | Potassium inwardly-rectifying channel, subfamily J, member 5 | 9 | 32130003 | 32130278 | 1.01 | ↑ | 1.18 | ↑ | 1421762_at |
| 0610040J01Rik | RIKEN cDNA 0610040J01 gene | 5 | 64268747 | 64268986 | 1.01 | ↓ | 1.12 | ↓ | 1424404_at |
| B) Environment (E) | |||||||||
| Ddx46 | Dead (Asp-Glu-Ala-Asp) box polypeptide 46 | 13 | 55749228 | 55749440 | 1.33 | ↑ | 1.16 | ↓ | 1424569_at |
| Kcnh3 | Potassium voltage-gated channel, subfamily H (eag-related), member 3 | 15 | 99050982 | 99051487 | 1.31 | ↑ | 1.11 | ↓ | 1459107_at |
| Tlr12 | Toll-like receptor 12 | 4 | 128291909 | 128292687 | 1.31 | ↑ | 1.10 | ↓ | 1437931_at |
| Ropn1l // Gm6361 | Ropporin 1-like // predictedgene 6361 | 15 | 31378527 | 31378689 | 1.26 | ↑ | 1.15 | ↓ | 1423959_at |
| Tmtc1 | Transmembrane and tetratricopeptide repeat containing 2 // Smallnucleolar RNA SNORA17 | 10 | 104626225 | 104626657 | 1.25 | ↓ | 1.14 | ↑ | 1441033_at |
| Fjx1 | Four jointed box 1 (Drosophila | 2 | 102295770 | 102296667 | 1.23 | ↑ | 1.20 | ↓ | 1450728_at |
| Fjx1 | Four jointed box 1 (Drosophila) | 2 | 102295770 | 102296667 | 1.23 | ↑ | 1.17 | ↓ | 1422733_at |
| Cdc73 | Cell division cycle 73, Paf1/RNA polymerase II complex component, homolog (S. cerevisiae) | 1 | 144990530 | 144990925 | 1.23 | ↓ | 1.15 | ↑ | 1427972_at |
| Sox6 | SRY-box containing gene 6 | 7 | 123171580 | 123171989 | 1.23 | ↓ | 1.14 | ↑ | 1427677_a_at |
| Bcl10 // Ddah1 | B cell leukemia/lymphoma 10 // dimethylarginine dimethylamiNohydrolase 1 | 3 | 145538088 | 145538612 | 1.21 | ↓ | 1.18 | ↑ | 1443524_x_at |
| Mettl7a1 | Methyltransferase like 7A1 | 15 | 100131133 | 100131965 | 1.21 | ↑ | 1.16 | ↑ | 1434151_at |
| Mettl7a1 | Methyltransferase like 7A1 | 15 | 100131133 | 100131965 | 1.21 | ↑ | 1.13 | ↑ | 1454858_x_at |
| Prkab1 // Cit | Protein kinase, AMP-activated, beta 1 Non-catalytic subunit // citron | 5 | 116457782 | 116458147 | 1.20 | ↑ | 1.13 | ↓ | 1424119_at |
| BC051628 // BC006779 | cDNA sequence BC051628 // cDNA sequence BC006779 | 2 | 180964456 | 180964941 | 1.19 | ↓ | 1.11 | ↑ | 1431758_at |
| Pnlip | Pancreatic lipase | 19 | 58744425 | 58744760 | 1.17 | ↑ | 1.25 | ↑ | 1433431_at |
| Zfp64 | Zinc finger protein 64 | 2 | 168808365 | 168808848 | 1.16 | ↑ | 1.21 | ↑ | 1456431_at |
| Zfp64 | Zinc finger protein 64 | 2 | 168808365 | 168808848 | 1.16 | ↑ | 1.12 | ↓ | 1451696_at |
| Xpot | Exportin, tRNA (nuclear export receptor for tRNAs) | 10 | 121044448 | 121044882 | 1.16 | ↑ | 1.13 | ↑ | 1441681_at |
| Nr2c1 // Ndufa12 | Nuclear receptor subfamily 2, group C, member 1 // NADHdehydrogenase (ubiquinone) 1 alpha subcomplex, 12 | 10 | 93657585 | 93657974 | 1.14 | ↑ | 1.19 | ↓ | 1449157_at |
| Mast4 // Cd179 | Microtubule-associated serine/threonine kinase family member 4 // CD180antigen | 13 | 103564348 | 103564628 | 1.14 | ↓ | 1.14 | ↑ | 1459387_at |
| Clstn2 | Calsyntenin 2 | 9 | 97936180 | 97936420 | 1.12 | ↑ | 1.19 | ↓ | 1422158_at |
| Cldn18 | Claudin 18 | 9 | 99617339 | 99617897 | 1.12 | ↑ | 1.19 | ↑ | 1425445_a_at |
| Map3k1 | Mitogen-activated protein kinase kinase kinase 1 | 13 | 112601065 | 112601546 | 1.12 | ↑ | 1.24 | ↑ | 1443540_at |
| Akap13 | A kinase (PRKA) anchor protein 13 | 7 | 82870878 | 82871179 | 1.11 | ↑ | 1.10 | ↓ | 1433722_at |
| Kdm6b // Dnahc2 | KDM1 lysine (K)-specific demethylase 6B // dynein, axonemal, heavy chain 2 | 11 | 69290421 | 69290898 | 1.11 | ↑ | 1.26 | ↑ | 1440346_at |
| Kdm6b // Dnahc2 | KDM1 lysine (K)-specific demethylase 6B // dynein, axonemal, heavy chain 2 | 11 | 69290421 | 69290898 | 1.11 | ↑ | 1.14 | ↑ | 1456610_at |
| Kat2a // Dhx58 | K(lysine) acetyltransferase 2A //DEXH(Asp-Glu-X-His) box polypeptide 58 | 11 | 100566819 | 100567318 | 1.10 | ↑ | 1.15 | ↓ | 1422972_s_at |
| Prok2 | Prokineticin 2 | 6 | 99674161 | 99674602 | 1.10 | ↑ | 1.15 | ↑ | 1451952_at |
| Ptpn21 | Protein tyrosine phosphatase, Non-receptor type 21 | 12 | 99923550 | 99923778 | 1.09 | ↑ | 1.15 | ↑ | 1419055_a_at |
| Rad9b | RAD9 homolog B (S. cerevisiae) | 5 | 122803144 | 122803311 | 1.09 | ↑ | 1.18 | ↓ | 1425800_at |
| Mcm3 | Minichromosome maintenance deficient 3 (S. cerevisiae) | 1 | 20807895 | 20808068 | 1.09 | ↑ | 1.17 | ↑ | 1420029_at |
| Msi1 | Musashi homolog 1(Drosophila) | 5 | 115873837 | 115874905 | 1.08 | ↑ | 1.24 | ↑ | 1444306_at |
| Stt3b | STT3, subunit of the oligosaccharyltransferase complex, homolog B (S. cerevisiae) | 9 | 115162948 | 115163232 | 1.08 | ↑ | 1.22 | ↑ | 1431541_at |
| Dna2 // Slc25a16 | DNA replication helicase 2 homolog (yeast) // solute carrier family 25 (mitochondrial carrier, Graves disease autoantigen), member 16 | 10 | 62407000 | 62407211 | 1.08 | ↑ | 1.25 | ↑ | 1457909_at |
| Slc30a1 // 1700034H15Rik | Solute carrier family 30 (zinc transporter), member 1 // RIKEN cDNA 1700034H15 gene | 1 | 193732789 | 193733167 | 1.08 | ↑ | 1.15 | ↓ | 1436164_at |
| Nos1 | Nitric oxide synthase 1, neuronal | 5 | 118312626 | 118312968 | 1.07 | ↑ | 1.20 | ↓ | 1438483_at |
| Prss23 // 1700019G06Rik | Protease, serine, 23 // RIKEN cDNA 1700019G06 gene | 7 | 96658688 | 96659035 | 1.07 | ↓ | 1.17 | ↓ | 1446560_at |
| Chaf1b // Morc3 | Chromatin assembly factor 1, subunit B (p60) // microrchidia 3 | 16 | 93865780 | 93866055 | 1.07 | ↑ | 1.24 | ↑ | 1431275_at |
| Gja1 | Gap junction protein, alpha 1 | 10 | 56216896 | 56217111 | 1.04 | ↑ | 1.42 | ↓ | 1415801_at |
| C) G × E | |||||||||
| 2010001K21Rik | RIKEN cDNA 2010001K21 gene | 13 | 47102005 | 47102256 | — | — | — | — | 1447576_at |
| 5430421N21Rik | RIKEN cDNA 5430421N21 gene | 15 | 101325243 | 101325528 | — | — | — | — | 1427118_at |
| Ank3 | Ankyrin 3, epithelial | 10 | 68991227 | 68992000 | — | — | — | — | 1447259_at |
| Atp10b | ATPase, class V, type 10B | 11 | 42964973 | 42965240 | — | — | — | — | 1444911_at |
| Cabin1 | Calcineurin binding protein 1 | 10 | 75229253 | 75229611 | — | — | — | — | 1437794_at |
| Cage1 | Cancer antigen 1 | 13 | 38127687 | 38128016 | — | — | — | — | 1434810_a_at |
| Chrna9 | Cholinergic receptor, nicotinic, alpha polypeptide 9 | 5 | 66361850 | 66362047 | — | — | — | — | 1447214_at |
| Chrna9 | Cholinergic receptor, nicotinic, alpha polypeptide 9 | 5 | 66362494 | 66362774 | — | — | — | — | 1447214_at |
| Clpb // Phox2a | ClpB caseinolytic peptidase B homolog (E. coli) // paired-likehomeobox 2a | 7 | 108938653 | 108939395 | — | — | — | — | 1454168_a_at |
| Dppa3 | Developmental pluripotency-associated 3 | 6 | 122594236 | 122594653 | — | — | — | — | 1424295_at |
| F10 // Proz | Coagulation factor X // proteinZ, vitamin K-dependent plasma glycoprotein | 8 | 13055218 | 13055381 | — | — | — | — | 1449305_at |
| Fam135a // 1110058L19Rik | Family with sequence similarity 135, member A // RIKEN cDNA 1110058L19 gene | 1 | 24034551 | 24034832 | — | — | — | — | 1453122_at |
| Fam170a | Family with sequence similarity 170, member A | 18 | 50438458 | 50438693 | — | — | — | — | 1456560_at |
| Foxj1 // Rnf157 | Forkhead box J1 // ring finger protein 157 | 11 | 116218086 | 116218410 | — | — | — | — | 1425291_at |
| Glra1 | Glycine receptor, alpha 1 subunit | 11 | 55418779 | 55418960 | — | — | — | — | 1422277_at |
| Heatr1 // Lgals8 | Heat repeat containing 1 // lectin,galactose binding, soluble 8 | 13 | 12520880 | 12521218 | — | — | — | — | 1452419_at |
| Mbp // Rpl21-ps8 | Myelin basic protein // ribosomalprotein L21, pseudogene 8 | 18 | 82693616 | 82694069 | — | — | — | — | 1451961_a_at |
| Parp14 | Poly (ADP-ribose) polymerase family, member 14 | 16 | 35858088 | 35858439 | — | — | — | — | 1451564_at |
| Pim1 | Proviral integration site 1 | 17 | 29656335 | 29656796 | — | — | — | — | 1423006_at |
| Pla2g5 // Pla2g2a | Phospholipase A2, group V // phospholipase A2, group IIA (platelets, synovial fluid) | 4 | 138382101 | 138382425 | — | — | — | — | 1417814_at |
| Psd3 | Pleckstrin and Sec7 domain containing 3 | 8 | 70220885 | 70221160 | — | — | — | — | 1418749_at |
| Tcof1 | Treacher Collins Franceschetti syndrome 1, homolog | 18 | 61013037 | 61013345 | — | — | — | — | 1423600_a_at |
| Uevld | UEV and lactate/malate dehyrogenase domains | 7 | 54224905 | 54225077 | — | — | — | — | 1421785_at |
| Wdr66 | WD repeat domain 66 | 5 | 123745990 | 123746262 | — | — | — | — | 1447215_at |
Abbreviations: FC, fold change; in/de, increased or decreased expression.
Genes of which both DNA methylation status and gene expression level were significantly altered by genotype (G; that is, 5-Htt+/− versus 5-Htt+/+), the environment (E; that is, prenatal stressed versus control mice) or in an interactive manner (G × E; that is, indicating those genes of which the effect of the environment depends upon the genotype) in the hippocampus of female mice. DNA methylation analysis was performed by applying hippocampal DNA enriched by methylated-DNA immunoprecipitation (MeDIP) on Affymetrix (Affy) Mouse Promoter 1.0R arrays. Gene expression data as described in van den Hove et al.4
Among the 35 genes of which DNA methylation and expression were differentially affected by PS (Table 1b), the majority, that is, 28 genes, showed decreased methylation. Among those differentially methylated and expressed genes were the histone acetyltransferase K[lysine] acetyltransferase 2 A (Kat2a), nitric oxide synthase 1 (Nos1), calsyntenin 2 (Clstn2), Musashi homolog 1 (Msi1) and four jointed box 1 (Fjx1). We furthermore detected a DMR about 10 kb upstream of Mir124-2.
Moreover, we found 23 differentially methylated/expressed genes that were affected in a 5-Htt × PS fashion (Table 1c). Among them were ankyrin 3, epithelial (Ank3), calcineurin binding protein 1 (Cabin1), myelin basic protein (Mbp), phospholipase A2, group V (Pla2g5) and the guanine nucleotide exchange factor pleckstrin and Sec7 domain containing 3 (Psd3).
Of note, 60% and 63% of the differentially methylated/expressed genes that were affected by a G and E effect, respectively, followed the canonical anticorrelation of promoter methylation and gene expression, that is, increased promoter methylation and decreased gene expression or vice versa. The remaining differentially methylated/expressed genes showed an ‘atypical' pattern.
Methylation of an intronic sequence in the Mbp gene
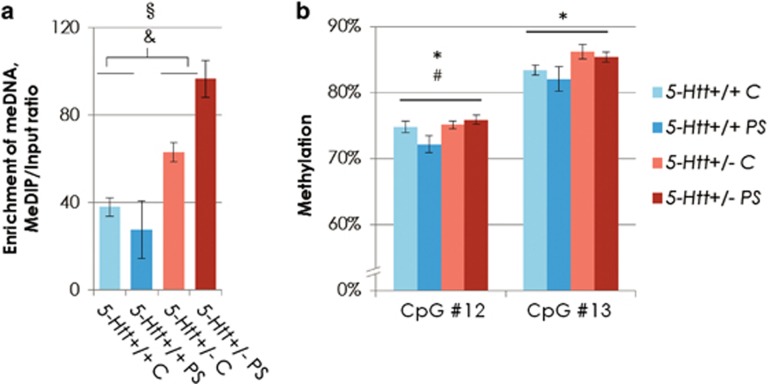
Mbp, which was both differentially methylated and expressed in a G × E interaction dependent manner, was first validated in terms of methylation, using MeDIP-qPCR. As presented in Table 1c, Mbp methylation, as assessed by MeDIP-chip, was decreased in 5-Htt+/+ mice exposed to PS compared with 5-Htt+/+ controls, whereas almost no PS-dependent change in methylation was observed in 5-Htt+/− mice. RT-qPCR with the same MeDIP DNA as used for the promoter array detected high enrichment of methylated DNA (meDNA) at the described locus (Figure 2a) and a similar methylation pattern as obtained from the array, that is, slight decrease in methylation in 5-Htt+/+ animals and an increase in methylation in 5-Htt+/− animal after PS. Moreover, we found an increase in meDNA-enrichment in 5-Htt+/− animals compared with 5-Htt+/+ (Kruskal–Wallis P=0.046, Mann–Whitney P=0.004). As a negative control, we analyzed part of the 5′-flanking region of Mir137 that did not harbor a DMR in our analysis (data not shown) and confirmed that there were no changes in methylation at this gene locus. We then assessed whether the observed signal was based on a methylation change covering the whole genomic region, or whether it originated from one or more specific CpG sites. For this purpose, we analyzed DNA methylation of 13 CpG sites in the Mbp DMR using pyrosequencing on bisulfite-treated DNA. Supplementary Figure 1 shows the percentage of unconverted cytosines, representing methylated cytosines, at CpG sites 1 through 13. As indicated by MeDIP RT-qPCR, pyrosequencing showed a high degree of methylation, that is, 70–90%. The pyrosequencing results revealed that CpG sites 12 and 13 showed a modest but significant increase in methylation in 5-Htt+/- mice when compared with 5-Htt+/+ animals (two-way analysis of variance (ANOVA) P=0.031 and P=0.019, respectively, Figure 2b). Interestingly, the methylation pattern at CpG site 12 matched the observed methylation pattern of the array and, in addition, showed a trend toward a 5-Htt × PS interaction effect (two-way ANOVA P=0.066). Moreover, there was a significant negative correlation of methylation status at CpG site 12 and 13 with Mbp expression (Spearman correlation, r=−0.412, P=0.012 and r=−0426, P=0.010, respectively). Noteworthy, the methylation status at CpG site 12 further correlated with the time spent in the open arms of and distance moved in the elevated zero maze (EZM; Spearman correlation, r=0.396, P=0.020 and r=0.345, P=0.046, respectively). Similar effects were observed when correlating individual Mbp expression levels obtained by RT-qPCR with time spent in the open arms of and distance moved in the EZM (Spearman correlation, r=−0.452, P=0.007 and r=−0.358, P=0.037, respectively).
Figure 2.
(a) Enrichment of methylated DNA at the Mbp locus (chr18:82 694 078–82 694 196) in the hippocampus of female 5-Htt+/− or 5-Htt+/+ mice exposed to PS or not (controls, C). Enrichment obtained by RT-qPCR using MeDIP and input DNA. §P=0.047 Kruskal–Wallis, &P=0.004 Mann–Whitney test when comparing all 5-Htt+/− with all 5-Htt+/+ mice (G effect). (b) Mbp methylation rate obtained by pyrosequencing of bisulfite-treated DNA at the Mbp locus (chr18: 82 693,582–82,694,175) in the same animals. Bars representing means, error bars representing s.e.m. *P<0.05 G effect, #P=0.066 trend of a G × E effect, two-way analysis of variance. E, environment; G, genotype; PS, prenatal stress; RT-qPCR, quantitative real-time PCR.
Moreover, since different splice variants of Mbp have different functions, we proceeded to elucidate which exact Mbp splice variants were differentially expressed. In our previous work,4 we found a G, E and G × E effect on total Mbp expression. As Supplementary Figure 1 illustrates, the Golli/Mbp gene locus harbors a variety of splice variants. The primers were located in the 3′ UTR of most of the Mbp transcripts, thus the signal could not be attributed to a specific splice form. By using primer pairs either recognizing exon I and exon III (Mbp without exII) or exon II and exon III (Mbp with exII) of Mbp for RT-qPCR, we found a G × E interaction and a PS effect for Mbp lacking exII (two-way ANOVA, P=0.017 and P=0.021, Supplementary Figure 2) and an increase in expression of Mbp transcripts containing exon II in PS mice (Mann–Whitney U P=0.009). We furthermore found that the expression of two Golli transcripts was changed in a G × E manner (two-way ANOVA P=0.017, data not shown). When looking at other myelin protein encoding genes, they all showed the same expression pattern as Mbp. As depicted in Supplementary Figure 2, we found a G × E interaction and a G effect for myelin oligodendrocyte glycoprotein (Mog; two-way ANOVA, P=0.035 and P=0.049). Expression of myelin-associated glycoprotein (Mag) showed a trend for an increase in 5-Htt+/+ mice exposed to PS (Kruskal–Wallis P=0.027, Mann–Whitney P=0.081), but not in 5-Htt+/−, hinting toward a G × E interaction. Moreover, the expression of Mbp w/o exII, Mag, Mog and Sox10 was highly correlated with each other within the hippocampus (Spearman correlation, 0.673<r<0.952, P<0.0001, Bonferroni-corrected). The expression changes of proteolipid protein (myelin) 1 (Plp1) did not reach statistical significance, although clearly displaying the same expression pattern. We furthermore found a PS effect on Sox10 expression (two-way ANOVA, P=0.011). A Modulated Modularity Clustering11 analysis using our array expression data of myelin-associated genes revealed several clusters of correlating myelin-associated genes (Supplementary Figure 3 and Supplementary Table 4). The first large cluster, module 5, comprises 15 myelin-associated genes, among them Mbp, Mobp, Mag, Tspan2, Pten, Mal, Ugt8a and Plp1 that correlate highly in expression (r=0.73).
Discussion
In the present study, a genome-wide DNA methylation screening was performed in offspring of 5-HTT-deficient mice subjected to PS. 5-Htt genotype, PS and their interaction differentially affected the DNA methylation signature of more than 800 genes to which the expression of a subset of these genes was related in a genotype-, PS- or G × E-specific manner.
We had previously reported various behavioral alterations using the same maternal restraint stress paradigm of PS in 5-Htt+/− mice (overview summarized in Supplementary Table 3).4 Exposure of 5-Htt+/− mice to PS was associated with increased depression-like behavior in the forced-swim test, an effect that was particularly pronounced in female offspring. Similar preliminary results were obtained by Markova et al.12 in the modified forced-swim test. On the other hand, control 5-Htt+/− showed relatively lower levels of depression-like behavior, hence, a 5-HTT deficit may also be beneficial when facing an acute challenge in adulthood. This indicates a complex interaction of variation in 5-Htt genotype and early-life environment in different sexes where a deficit in 5-HTT is not necessarily a disadvantage.13 We furthermore found that the 5-Htt genotype, PS and their interaction differentially affected the expression of numerous genes and related pathways in the hippocampus of female mice. In the present study, we analyzed hippocampal genome-wide promoter DNA methylation signatures in the same mice by applying MeDIP DNA to a genome-wide promoter tiling array. Several hundred DMRs that were influenced by 5-Htt genotype, PS or their interaction were identified. Functional Annotation Clustering using DAVID revealed enrichment of ribosome-associated annotations and ion-binding terms due to genotype and enrichment of WD repeat and SH3 domain terms due to PS. The latter term comprises Pak1ip1, which is involved in the p53-dependent regulation of cell growth and proliferation in response to stressors,14 and Strn3, a gene encoding a Ca2+-calmodulin binding protein mainly expressed in the brain, cerebellum, muscle and lung.15 In the hippocampus, it is expressed in pyramidal and granular cells.15 Pacsin2 is a member of the protein kinase C and casein kinase substrate-in-neurons family and involved in linking the actin cytoskeleton with vesicle formation by regulating tubulin polymerization. Terms enriched for a G × E effect comprised two clusters of cytoskeleton-associated terms and one EGF-associated cluster. The EGF cluster contained Notch3, a gene expressed in the ventricular zones and suggested to have a role in CNS development.16 Of note, in zebrafish, Notch3 was found to be essential for oligodendrocyte development and a Notch3 mutation led to a decrease in oligodendrocyte numbers accompanied by decreased Mbp expression.17 Fat1, a gene encoding a protein of the cadherin family that is highly expressed in the proliferating ventricular zones during prenatal mouse development.18 Variation in FAT1 has been associated with bipolar disorder.19,20
In addition, the DMR-linked genes showed partial overlap with differential gene expression profiles. For example, considering differentially expressed genes, we found increased methylation in response to PS for Clstn2 and Kat2a, whereas methylation levels of Ank3, Cabin1, Mbp, Pla2g5 and Psd3 were influenced in a G × E manner.
We found an increase in methylation in ~60% and a decrease in ~40% of the DMRs when comparing 5-Htt+/− to 5-Htt+/+ mice. Generally, human s-allele carriers as well as 5-Htt-deficient mice are more sensitive to early-life programming under the influence of (both adverse and positive) environmental changes.13 Although mostly speculative, considering the important role of 5-HT during brain development, altered central levels of 5-HT during early life may affect the extent to which environmental variation is perceived by and translated or epigenetically programmed into long-term changes in brain function and associated behavioral phenotypes. Notably, ~80% of the DMRs showed an increase in methylation when comparing PS with C offspring, whereas only 20% showed a decrease in methylation. This effect might be the consequence of a temporary increase in DNA methyl-transferase activity in PS mice during development. Although this notion awaits further exploration, Mychasiuk et al.21,22 found an increase in global DNA methylation in the hippocampus of young female rat offspring in a mild PS as well as in a bystander PS paradigm, in which the cage mate of the pregnant dam is stressed and not the dam directly.
Mbp emerged from our previous study4 as a gene of interest that was regulated at the mRNA expression level by 5-Htt genotype, PS and the interaction of both. In the present study, the tiling array revealed a DMR at the Mbp locus that was modified by a 5-Htt × PS interaction. Subsequent pyrosequencing indicated differential effects on methylation of the 5-Htt genotype at two out of 13 analyzed CpG sites and a tendency for a G × E interaction effect at CpG site 12. Moreover, methylation at CpG site 12 and 13 correlated negatively with Mbp expression and anxiety-like behavior in the EZM, suggesting functional methylation of Mbp. Noteworthy, similar to our Mbp mRNA expression, findings assessed by RT-qPCR (see van den Hove et al.4 for more details), Föcking et al.23 found an increase in hippocampal MBP protein levels in adult 5-Htt+/+ mice after exposure to the exact same PS paradigm. The Golli-Mbp locus comprises three transcription start sites, which result in two different forms of transcripts, the ‘classical' Mbp transcripts and the later discovered Golli-Mbp transcripts that contain additional exons at the 5′ end. The MBP protein family is expressed in oligodendrocytes and Schwann cells, whereas the Golli-specific proteins are expressed in myelin-producing and neuronal cells.24, 25, 26 Expression at the Golli-Mbp locus is subject to complex developmental regulation, involving various splice variants and subsequent posttranslational modifications resulting in MBP proteins ranging from 14 to 21.5 kD in size.27 The 18.5 kD classic MBP that is essential for CNS myelin formation is the main structural component of myelin in the mature brain. It is forming the major dense line of myelin and functions as a ‘molecular sieve' by preventing proteins of the paranodal loops to diffuse into compact myelin.27, 28, 29, 30 The shiverer mouse model, which is characterized by early-onset generalized tremors and seizures—both becoming more prominent with aging—as well as a shortened lifespan, has sparse and relatively unstructured brain myelin due to a deletion in Mbp.31,32 MBPs are intrinsically disordered proteins33,34 and due to their structural flexibility they have also been implicated in other functions, for example, as regulators of the cytoskeleton35, 36, 37 and voltage-gated Ca2+ channels.38, 39, 40, 41 Intrinsically disordered proteins have moreover been suggested to function as interaction hubs, which might also be a possible function of MBP, taking into account its cellular abundance.27,42, 43, 44 We detected an increase in both Mbp transcripts lacking and containing exon II in PS mice when compared with C mice. Of note, exon II encodes a nuclear-trafficking signal. The nuclear-localized 21.5 kD MBP, which arises from a Mbp transcript containing exon II, is important for early myelinisation and has also been suggested to contribute to the differentiation of oligodendrocytes. Smith et al.45 showed that 21.5 kD MBP facilitates neurite outgrowth and branching of neuronal cells in a coculture of N2a neuronal cells and N19-oligodendrocytes. Furthermore, Kikusui et al.46 reported that early weaning induced increased anxiety-like behavior in male-outbred IRC mice, paralleled by a decrease in two out of the four investigated MBP isoforms in brain homogenates at 5 weeks of age. These changes in MBP levels were, however, not found in Wistar rats using an early-weaning paradigm.47 Furthermore, mice deficient for Fyn, a gene encoding a protein-tyrosine kinase involved in posttranscriptional regulation of MBP, show attenuated MBP expression in the forebrain48 and were reported to show increased anxiety-like behavior when compared with 5-Htt+/+ controls.49 On the other hand, intracranial injection of apotransferrin into 3-day-old male rats led to increased deposition of abnormally composed myelin and anxiolytic behavior in the elevated plus maze in adult rats when compared with controls.50 It is assumed that myelinisation influences GABAergic transmission in the hippocampus thereby impacting anxiety-like behavior,46,50 since hippocampal GABAergic fibers are myelinated and the majority of cholinergic fibers are not.51 In line with this notion, our data show a modest but significant negative correlation between Mbp expression and anxiety-like behavior. Various studies in rodents have shown a functional role of the ventral hippocampus in innate anxiety response and particularly its regulatory function for the exploration time of the open arms of the elevated plus maze.52, 53, 54 For example, injection of the GABA a2 agonist TPA023 into the ventral hippocampus has anxiolytic effects in rats during elevated plus maze exploration.52 Moreover, MBP is being discussed as a risk gene for schizophrenic disorders and alterations in myelination and oligodendrocyte numbers were found in patients with schizophrenic disorder, major depressive disorder and bipolar disorder.55, 56, 57, 58, 59, 60, 61, 62, 63 Analysis of the Mbp sequence assessed for differential methylation using JASPAR64 revealed a putative SOX10 binding site a few base pairs upstream of CpG site 12. SOX10 is a transcription factor essential for terminal differentiation of oligodendroglia and directly impacts Mbp expression.65 Intriguingly, Mbp expression was reported to be induced by SOX10 in cooperative action with Krox20 and the chromatin remodeling enzymes SWI/SNF.66 As NG2-positive oligodendrocyte progenitor cells express the glucocorticoid receptor, as well as the glucocorticoid receptor-cofactors SRC-1 and p300, it might be possible that PS exposure affects oligodendrocyte development.67 Interestingly, Xu et al.68 found a negative effect of PS on myelination in the hippocampus of 22-day-old Sprague–Dawley rats. Furthermore, Miyata et al.69 showed that chronic water immersion and restraints stress leads to morphological alterations in oligodendrocytes of the corpus callosum. They furthermore found that dexamethasone exposure of oligodendrocytes in vitro triggers a 1.5-fold increase in MBP-positive oligodendrocyte cell diameter.
Bone morphogenetic protein receptor, type 1B (Bmpr1b) belongs to the class I BMP receptor genes, which are, while functionally redundant, involved in BMP signaling. We detected an increase in DNA methylation in the first intron of Bmpr1b and a decrease in Bmpr1b mRNA expression in 5-Htt+/− mice compared with 5-Htt+/+ controls. BMP signaling has a pivotal role in dendate gyrus (DG) development and double null mutant Bmpr1b::Bmpr1a mice have smaller DG than controls, which might reflect impaired granule cell production during DG development, and a reduced DG neuronal progenitor pool.70 Finally, these mice also showed less anxiety-like behavior in the elevated plus maze when compared with WT controls.70
Our promoter array analysis moreover revealed a PS effect on the methylation status of Calsyntenin 2 (Clstn2) and K(lysine) acetyltransferase 2A (Kat2a). More specifically, PS animals showed an increase in methylation at a DMR 5′ upstream of the Clstn2 gene when compared with C animals. Clstn2 has been identified as one of three calsyntenin genes coding for a postsynaptic protein exclusively expressed in the brain. Hintsch et al.71 found that, in the hippocampus, Clnst2 was primarily expressed in pyramidal cells of the CA2 and CA3 region, in some scattered interneurons in the pyramidal cell layer of CA1 and in the granular cell layer of the DG. However, little is known about the role of Clnst2 regarding stress exposure. Furthermore, we found an increase in methylation at a DMR (Chr. 9, 97936180 to 97936420) covering several exons and introns in a gene involved in chromatin remodeling, that is, Kat2a, as well as a decrease in expression in this gene in PS mice compared with controls. KAT2A is a histone acetyltransferase linked to transcriptional activation72 that targets K9, K14 and K18 of histone H3, as well as all four amino-terminal K residues of histone H4.73 This acetyltransferase can be recruited by several DNA binding factors such as myc,74,75 E2F76 and p5377 to induce changes in chromatin structure and activate transcription. Kat2a is critical for both normal embryonal development78 and normal brain growth, and loss of function of Kat2a results in decreased stem cell proliferation in the murine cortex.79
Remarkably, we detected both a G and G × E effect on both expression and methylation of the secretory phospholipase A2, group V (Pla2g5) gene. Proteins of the phospholipase A2 family release unesterified arachidonic acid (AA) from membrane phospholipids and are hence key enzymes in the activation of the arachidonic acid cascade.80,81 The ω-6 polyunsaturated fatty acid, arachidonic acid, and its metabolites are involved in neurotransmitter release, cerebral blood flow regulation and inflammatory processes, which also have a role in multiple sclerosis and Alzheimer's disease.82,83 Other genes that were regulated in a G × E fashion in our study comprise ankyrin 3, epithelial (Ank3, also known as Ankyrin-G), calcineurin binding protein 1 (Cabin1) and pleckstrin and Sec7 domain containing 3 (Psd3, also known as EFA6D). Ank3 encodes a protein that in the brain is localized at the nodes of Ranvier and axonal initial segments84 where it is involved in the localization of transmembrane adhesion molecules, voltage-gated sodium channels, the spectrin membrane skeleton and potassium channels.85,86 Several studies suggested ANK3 as a candidate gene for bipolar disorder.87, 88, 89 Psd3 is expressed throughout the brain, with highest expression levels found in olfactory bulb, cerebral cortex, hippocampal pyramidal cell layer and cerebellar granule cell layer.90 Psd3 encodes a guanine nucleotide exchange factor for the small GTPase ARF6,90 which regulates membrane trafficking and the actin cytoskeleton and is involved in neuronal functions including neurite formation,91,92 spine density and maintenance,93,94 endo- and exocytosis of synaptic vesicles95,96 and receptor internalization.97
While comparing DNA methylation and gene expression profiles, we noted that only a modest part of the genes for which we found a DMR were also differentially expressed and a considerable proportion of genes did not show an inverse relationship between DNA methylation and expression. This is not surprising as the relation of DNA methylation and gene expression is highly complex.98, 99, 100, 101 Genome-wide studies show that the influence of DNA methylation on gene expression is very much context dependent.98 Guo et al.,100 for example, have found only a modest correlation between CpG methylation near transcription start sites and gene expression when analyzing neural activity-induced changes in methylation in murine dentate granule neurons. Depending on CpG density, DMRs have previously been categorized in low-, intermediate- and high-CpG promoters. Methylation of intermediate-CpG promoters and high-CpG promoters was associated with inactivity of the promoter in human primary somatic and germline cells, however, only a part of the inactive high-CpG promoters were methylated at all, indicating that methylation of a HPC is not compatible with expression, but lacking methylation does not necessarily indicate promoter activity. In contrast, Weber et al.98 detected no correlation between the methylation status of low-CpG promoters and promoter activity. Furthermore, both chromatin accessibility, which is also strongly regulated by histone modifications, and the availability of transcription factors are needed for a gene to be expressed, adding another level of complexity to its regulation. In addition, technical and analytical limitations may have contributed to this observation in our study. The tiling array used in this study comprised only promoter regions and thus did not cover all possible gene-related regulatory elements that could have had an influence on gene expression, such as enhancers, silencers or remote control regions. Individual DMRs were also occasionally assigned to two flanking or embedded genes, not necessarily implicating a functional role for both genes. In most cases, expression profiles did not convey splice form-specific information thereby possibly mitigating and concealing the effects of 5-Htt genotype and PS on expression. Comparing methylation levels to gene expression was further complicated by the presence of different cell types with different gene expression and DNA methylation profiles in the hippocampal homogenates that were used in this study.
Assessing the relation of DNA methylation and gene expression is however more complex than initially anticipated. It should be noted that pyrosequencing, which was applied to determine the methylation rate of single CpG sites, is a bisulfite-treated DNA-based technique and thus cannot distinguish between 5-methyl-cytosine (DNA methylation) and 5-hydroxymethyl-cytosine (DNA hydroxymethylation). This adds another level of complexity when comparing pyrosequencing data with MeDIP data, which are based on a 5-methyl-cytosine-specific antibody. Furthermore, P-values represent nominal P-values and n-numbers for promoter array analysis reflect two to three arrays per group. These limitations prompted us to interpret the data with caution and further research is required to reveal to which extent the molecular players identified here may provide useful targets in the development of intervention strategies for stress-related disorders of emotion regulation. The analysis of a single brain region, such as the hippocampus, poses an additional constraint, as other structures, such as the amygdala and the prefrontal cortex, are also known to be involved in stress and emotion regulation. Furthermore, as the left hippocampus was used for the gene expression study and the right hippocampus for the DNA methylation analysis, we cannot exclude that left–right asymmetries might have influenced the results.102 Finally, we cannot exclude that behavioral testing might have affected hippocampal DNA methylation/gene expression.
In conclusion, our data show that the effects of PS on DNA methylation in the hippocampus of female offspring are partially dependent on the 5-Htt genotype and that various genes which displayed changes of their methylation signature were also differentially expressed. Specifically, a differentially methylated genomic region in Mbp was associated with the gene's expression in a genotype-, PS- and G × E-dependent manner. The methylation rate of two CpG sites at the Mbp locus was related to Mbp expression and anxiety-related behavior, suggesting functional DNA methylation regulation of the Mbp gene. Our findings are the basis for further research that is expected to further explore the role of developmental epigenetic programming by PS, 5-Htt and their interaction.
Acknowledgments
This work was supported by the Deutsche Forschungsgemeinschaft (DFG) Sonderforschungsbereich Transregio (SFB TRR) 58/A1 and A5 to K-PL and AGS, as well as C2 to KD, B6/Z2 to AR; and Interdisziplinäres Zentrum für Klinische Forschung (IZKF) grant Z-6 to CJS, N-221 to AR and TH and the European Community; EC: AGGRESSOTYPE FP7/No. 602805 to K-PL, TS and DLvdH. K-GS was supported by a PhD fellowship of the Graduate School of Life Sciences (GSLS), University of Würzburg. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on the Translational Psychiatry website (http://www.nature.com/tp)
Supplementary Material
References
- Darnaudery M, Maccari S. Epigenetic programming of the stress response in male and female rats by prenatal restraint stress. Brain Res Rev. 2008;57:571–585. doi: 10.1016/j.brainresrev.2007.11.004. [DOI] [PubMed] [Google Scholar]
- Canli T, Lesch KP. Long story short: the serotonin transporter in emotion regulation and social cognition. Nat Neurosci. 2007;10:1103–1109. doi: 10.1038/nn1964. [DOI] [PubMed] [Google Scholar]
- Caspi A, Hariri AR, Holmes A, Uher R, Moffitt TE. Genetic sensitivity to the environment: the case of the serotonin transporter gene and its implications for studying complex diseases and traits. Am J Psychiatry. 2010;167:509–527. doi: 10.1176/appi.ajp.2010.09101452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Hove DL, Jakob SB, Schraut KG, Kenis G, Schmitt AG, Kneitz S, et al. Differential effects of prenatal stress in 5-Htt deficient mice: towards molecular mechanisms of gene x environment interactions. PLoS One. 2011;6:e22715. doi: 10.1371/journal.pone.0022715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bredy TW, Sun YE, Kobor MS. How the epigenome contributes to the development of psychiatric disorders. Dev Psychobiol. 2010;52:331–342. doi: 10.1002/dev.20424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsankova N, Renthal W, Kumar A, Nestler EJ. Epigenetic regulation in psychiatric disorders. Nat Rev Neurosci. 2007;8:355–367. doi: 10.1038/nrn2132. [DOI] [PubMed] [Google Scholar]
- Schroeder M, Krebs MO, Bleich S, Frieling H. Epigenetics and depression: current challenges and new therapeutic options. Curr Opin Psychiatry. 2010;23:588–592. doi: 10.1097/YCO.0b013e32833d16c1. [DOI] [PubMed] [Google Scholar]
- Chapman RH, Stern JM. Failure of severe maternal stress or ACTH during pregnancy to affect emotionality of male rat offspring: implications of litter effects for prenatal studies. Dev Psychobiol. 1979;12:255–267. doi: 10.1002/dev.420120309. [DOI] [PubMed] [Google Scholar]
- Kuan PF, Chun H, Keles S. CMARRT: a tool for the analysis of ChIP-chip data from tiling arrays by incorporating the correlation structure. Pac Symp Biocomput. 2008. pp. 515–526. [PMC free article] [PubMed]
- Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone EA, Ayroles JF. Modulated modularity clustering as an exploratory tool for functional genomic inference. PLoS Genet. 2009;5:e1000479. doi: 10.1371/journal.pgen.1000479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markova N, Shevtsova E, Bachurin S, Steinbusch HM, Strekalova T.Individual susceptibility to depressive-like traits in two mouse models of depression and hippocampal levels of GSK3 beta FEBS J 2013280((Suppl 1219–220. [Google Scholar]
- Homberg JR, van den Hove DL. The serotonin transporter gene and functional and pathological adaptation to environmental variation across the life span. Prog Neurobiol. 2012;99:117–127. doi: 10.1016/j.pneurobio.2012.08.003. [DOI] [PubMed] [Google Scholar]
- Yu W, Qiu Z, Gao N, Wang L, Cui H, Qian Y, et al. PAK1IP1, a ribosomal stress-induced nucleolar protein, regulates cell proliferation via the p53-MDM2 loop. Nucleic Acids Res. 2011;39:2234–2248. doi: 10.1093/nar/gkq1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castets F, Rakitina T, Gaillard S, Moqrich A, Mattei MG, Monneron A. Zinedin, SG2NA, and striatin are calmodulin-binding, WD repeat proteins principally expressed in the brain. J Biol Chem. 2000;275:19970–19977. doi: 10.1074/jbc.M909782199. [DOI] [PubMed] [Google Scholar]
- Irvin DK, Zurcher SD, Nguyen T, Weinmaster G, Kornblum HI. Expression patterns of Notch1, Notch2, and Notch3 suggest multiple functional roles for the Notch-DSL signaling system during brain development. J Comp Neurol. 2001;436:167–181. [PubMed] [Google Scholar]
- Zaucker A, Mercurio S, Sternheim N, Talbot WS, Marlow FL. notch3 is essential for oligodendrocyte development and vascular integrity in zebrafish. Dis Model Mech. 2013;6:1246–1259. doi: 10.1242/dmm.012005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox B, Hadjantonakis AK, Collins JE, Magee AI. Cloning and expression throughout mouse development of mfat1, a homologue of the Drosophila tumour suppressor gene fat. Dev Dyn. 2000;217:233–240. doi: 10.1002/(SICI)1097-0177(200003)217:3<233::AID-DVDY1>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- Light KJ, Miller AL, Doughty CJ, Joyce PR, Olds RJ, Kennedy MA. FAT and bipolar affective disorder. Mol Psychiatry. 2007;12:899–900. doi: 10.1038/sj.mp.4002040. [DOI] [PubMed] [Google Scholar]
- Abou Jamra R, Becker T, Georgi A, Feulner T, Schumacher J, Stromaier J, et al. Genetic variation of the FAT gene at 4q35 is associated with bipolar affective disorder. Mol Psychiatry. 2008;13:277–284. doi: 10.1038/sj.mp.4002111. [DOI] [PubMed] [Google Scholar]
- Mychasiuk R, Schmold N, Ilnytskyy S, Kovalchuk O, Kolb B, Gibb R. Prenatal bystander stress alters brain, behavior, and the epigenome of developing rat offspring. Dev Neurosci. 2011;33:159–169. doi: 10.1159/000330034. [DOI] [PubMed] [Google Scholar]
- Mychasiuk R, Ilnytskyy S, Kovalchuk O, Kolb B, Gibb R. Intensity matters: brain, behaviour and the epigenome of prenatally stressed rats. Neuroscience. 2011;180:105–110. doi: 10.1016/j.neuroscience.2011.02.026. [DOI] [PubMed] [Google Scholar]
- Föcking MOR, Prickaerts J, Steinbusch HWM, Dunn MJ, van den Hove DLA, Cotter DR. Proteomic investigation of the hippocampus in prenatally stressed mice implicates changes in membrane trafficking, cytoskeletal and metabolic function. Dev Neurosci. 2014;36:432–442. doi: 10.1159/000365327. [DOI] [PubMed] [Google Scholar]
- Landry CF, Ellison J, Skinner E, Campagnoni AT. Golli-MBP proteins mark the earliest stages of fiber extension and terminal arboration in the mouse peripheral nervous system. J Neurosci Res. 1997;50:265–271. doi: 10.1002/(SICI)1097-4547(19971015)50:2<265::AID-JNR15>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Landry CF, Ellison JA, Pribyl TM, Campagnoni C, Kampf K, Campagnoni AT. Myelin basic protein gene expression in neurons: developmental and regional changes in protein targeting within neuronal nuclei, cell bodies, and processes. J Neurosci. 1996;16:2452–2462. doi: 10.1523/JNEUROSCI.16-08-02452.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry CF, Pribyl TM, Ellison JA, Givogri MI, Kampf K, Campagnoni CW, et al. Embryonic expression of the myelin basic protein gene: identification of a promoter region that targets transgene expression to pioneer neurons. J Neurosci. 1998;18:7315–7327. doi: 10.1523/JNEUROSCI.18-18-07315.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harauz G, Boggs JM. Myelin management by the 18.5-kDa and 21.5-kDa classic myelin basic protein isoforms. J Neurochem. 2013;125:334–361. doi: 10.1111/jnc.12195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Readhead C, Hood L. The dysmyelinating mouse mutations shiverer (shi) and myelin deficient (shimld) Behav Genet. 1990;20:213–234. doi: 10.1007/BF01067791. [DOI] [PubMed] [Google Scholar]
- Aggarwal S, Yurlova L, Snaidero N, Reetz C, Frey S, Zimmermann J, et al. A size barrier limits protein diffusion at the cell surface to generate lipid-rich myelin-membrane sheets. Dev Cell. 2011;21:445–456. doi: 10.1016/j.devcel.2011.08.001. [DOI] [PubMed] [Google Scholar]
- Pedraza L, Huang JK, Colman DR. Organizing principles of the axoglial apparatus. Neuron. 2001;30:335–344. doi: 10.1016/s0896-6273(01)00306-3. [DOI] [PubMed] [Google Scholar]
- Dupouey P, Jacque C, Bourre JM, Cesselin F, Privat A, Baumann N. Immunochemical studies of myelin basic protein in shiverer mouse devoid of major dense line of myelin. Neurosci Lett. 1979;12:113–118. doi: 10.1016/0304-3940(79)91490-3. [DOI] [PubMed] [Google Scholar]
- Privat A, Jacque C, Bourre JM, Dupouey P, Baumann N. Absence of the major dense line in myelin of the mutant mouse "shiverer". Neurosci Lett. 1979;12:107–112. doi: 10.1016/0304-3940(79)91489-7. [DOI] [PubMed] [Google Scholar]
- Hill CM, Bates IR, White GF, Hallett FR, Harauz G. Effects of the osmolyte trimethylamine-N-oxide on conformation, self-association, and two-dimensional crystallization of myelin basic protein. J Struct Biol. 2002;139:13–26. doi: 10.1016/s1047-8477(02)00513-0. [DOI] [PubMed] [Google Scholar]
- Harauz G, Ishiyama N, Hill CM, Bates IR, Libich DS, Fares C. Myelin basic protein-diverse conformational states of an intrinsically unstructured protein and its roles in myelin assembly and multiple sclerosis. Micron. 2004;35:503–542. doi: 10.1016/j.micron.2004.04.005. [DOI] [PubMed] [Google Scholar]
- Smith GS, Homchaudhuri L, Boggs JM, Harauz G. Classic 18.5- and 21.5-kDa myelin basic protein isoforms associate with cytoskeletal and SH3-domain proteins in the immortalized N19-oligodendroglial cell line stimulated by phorbol ester and IGF-1. Neurochem Res. 2012;37:1277–1295. doi: 10.1007/s11064-011-0700-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamm VV, De Avila M, Smith GS, Ahmed MA, Harauz G. Structured functional domains of myelin basic protein: cross talk between actin polymerization and Ca(2+)-dependent calmodulin interaction. Biophys J. 2011;101:1248–1256. doi: 10.1016/j.bpj.2011.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boggs JM, Rangaraj G, Hill CM, Bates IR, Heng YM, Harauz G. Effect of arginine loss in myelin basic protein, as occurs in its deiminated charge isoform, on mediation of actin polymerization and actin binding to a lipid membrane in vitro. Biochemistry. 2005;44:3524–3534. doi: 10.1021/bi0473760. [DOI] [PubMed] [Google Scholar]
- Paez PM, Fulton D, Spreuer V, Handley V, Campagnoni AT. Modulation of canonical transient receptor potential channel 1 in the proliferation of oligodendrocyte precursor cells by the golli products of the myelin basic protein gene. J Neurosci. 2011;31:3625–3637. doi: 10.1523/JNEUROSCI.4424-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paez PM, Fulton DJ, Spreuer V, Handley V, Campagnoni CW, Campagnoni AT. Regulation of store-operated and voltage-operated Ca2+ channels in the proliferation and death of oligodendrocyte precursor cells by golli proteins. ASN Neuro. 2009;1:1. doi: 10.1042/AN20090003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paez PM, Fulton DJ, Spreuer V, Handley V, Campagnoni CW, Macklin WB, et al. Golli myelin basic proteins regulate oligodendroglial progenitor cell migration through voltage-gated Ca2+ influx. J Neurosci. 2009;29:6663–6676. doi: 10.1523/JNEUROSCI.5806-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith GS, Paez PM, Spreuer V, Campagnoni CW, Boggs JM, Campagnoni AT, et al. Classical 18.5-and 21.5-kDa isoforms of myelin basic protein inhibit calcium influx into oligodendroglial cells, in contrast to golli isoforms. J Neurosci Res. 2011;89:467–480. doi: 10.1002/jnr.22570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagliyan O, Proctor EA, D'Auria KM, Ding F, Dokholyan NV. Structural and dynamic determinants of protein-peptide recognition. Structure. 2011;19:1837–1845. doi: 10.1016/j.str.2011.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dogan J, Schmidt T, Mu X, Engstrom A, Jemth P. Fast association and slow transitions in the interaction between two intrinsically disordered protein domains. J Biol Chem. 2012;287:34316–34324. doi: 10.1074/jbc.M112.399436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganguly D, Otieno S, Waddell B, Iconaru L, Kriwacki RW, Chen J. Electrostatically accelerated coupled binding and folding of intrinsically disordered proteins. J Mol Biol. 2012;422:674–684. doi: 10.1016/j.jmb.2012.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith GS, Samborska B, Hawley SP, Klaiman JM, Gillis TE, Jones N, et al. Nucleus-localized 21.5-kDa myelin basic protein promotes oligodendrocyte proliferation and enhances neurite outgrowth in coculture, unlike the plasma membrane-associated 18.5-kDa isoform. J Neurosci Res. 2013;91:349–362. doi: 10.1002/jnr.23166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikusui T, Kiyokawa Y, Mori Y. Deprivation of mother-pup interaction by early weaning alters myelin formation in male, but not female, ICR mice. Brain Res. 2007;1133:115–122. doi: 10.1016/j.brainres.2006.11.031. [DOI] [PubMed] [Google Scholar]
- Kodama Y, Kikusui T, Takeuchi Y, Mori Y. Effects of early weaning on anxiety and prefrontal cortical and hippocampal myelination in male and female Wistar rats. Dev Psychobiol. 2008;50:332–342. doi: 10.1002/dev.20289. [DOI] [PubMed] [Google Scholar]
- Lu Z, Ku L, Chen Y, Feng Y. Developmental abnormalities of myelin basic protein expression in fyn knock-out brain reveal a role of Fyn in posttranscriptional regulation. J Biol Chem. 2005;280:389–395. doi: 10.1074/jbc.M405973200. [DOI] [PubMed] [Google Scholar]
- Miyakawa T, Yagi T, Kagiyama A, Niki H. Radial maze performance, open-field and elevated plus-maze behaviors in Fyn-kinase deficient mice: further evidence for increased fearfulness. Brain Res Mol Brain Res. 1996;37:145–150. doi: 10.1016/0169-328x(95)00300-h. [DOI] [PubMed] [Google Scholar]
- Viola H, Marta CB, Medina JH, Soto EF, Pasquini JM. Anxiolytic-like behavior in rats is induced by the neonatal intracranial injection of apotransferrin. J Neurosci Res. 2001;63:196–199. doi: 10.1002/1097-4547(20010115)63:2<196::AID-JNR1011>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Gartner U, Hartig W, Brauer K, Bruckner G, Arendt T. Immunofluorescence and immunoelectron microscopic evidence for differences in myelination of GABAergic and cholinergic septohippocampal fibres. Int J Dev Neurosci. 2001;19:347–352. doi: 10.1016/s0736-5748(01)00019-3. [DOI] [PubMed] [Google Scholar]
- McEown K, Treit D. Alpha2 GABAA receptor sub-units in the ventral hippocampus and alpha5 GABAA receptor sub-units in the dorsal hippocampus mediate anxiety and fear memory. Neuroscience. 2013;252:169–177. doi: 10.1016/j.neuroscience.2013.08.012. [DOI] [PubMed] [Google Scholar]
- Trent NL, Menard JL. The ventral hippocampus and the lateral septum work in tandem to regulate rats' open-arm exploration in the elevated plus-maze. Physiol Behav. 2010;101:141–152. doi: 10.1016/j.physbeh.2010.04.035. [DOI] [PubMed] [Google Scholar]
- Zhang WN, Bast T, Xu Y, Feldon J. Temporary inhibition of dorsal or ventral hippocampus by muscimol: distinct effects on measures of innate anxiety on the elevated plus maze, but similar disruption of contextual fear conditioning. Behav Brain Res. 2014;262:47–56. doi: 10.1016/j.bbr.2013.10.044. [DOI] [PubMed] [Google Scholar]
- Sibille E, Wang Y, Joeyen-Waldorf J, Gaiteri C, Surget A, Oh S, et al. A molecular signature of depression in the amygdala. Am J Psychiatry. 2009;166:1011–1024. doi: 10.1176/appi.ajp.2009.08121760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parlapani E, Schmitt A, Erdmann A, Bernstein HG, Breunig B, Gruber O, et al. Association between myelin basic protein expression and left entorhinal cortex pre-alpha cell layer disorganization in schizophrenia. Brain Res. 2009;1301:126–134. doi: 10.1016/j.brainres.2009.09.007. [DOI] [PubMed] [Google Scholar]
- Le-Niculescu H, Balaraman Y, Patel S, Tan J, Sidhu K, Jerome RE, et al. Towards understanding the schizophrenia code: an expanded convergent functional genomics approach. Am J Med Genet B Neuropsychiatr Genet. 2007;144B:129–158. doi: 10.1002/ajmg.b.30481. [DOI] [PubMed] [Google Scholar]
- Chambers JS, Perrone-Bizzozero NI. Altered myelination of the hippocampal formation in subjects with schizophrenia and bipolar disorder. Neurochem Res. 2004;29:2293–2302. doi: 10.1007/s11064-004-7039-x. [DOI] [PubMed] [Google Scholar]
- Ayalew M, Le-Niculescu H, Levey DF, Jain N, Changala B, Patel SD, et al. Convergent functional genomics of schizophrenia: from comprehensive understanding to genetic risk prediction. Mol Psychiatry. 2012;17:887–905. doi: 10.1038/mp.2012.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews PR, Eastwood SL, Harrison PJ. Reduced myelin basic protein and actin-related gene expression in visual cortex in schizophrenia. PLoS One. 2012;7:e38211. doi: 10.1371/journal.pone.0038211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le-Niculescu H, Kurian SM, Yehyawi N, Dike C, Patel SD, Edenberg HJ, et al. Identifying blood biomarkers for mood disorders using convergent functional genomics. Mol Psychiatry. 2009;14:156–174. doi: 10.1038/mp.2008.11. [DOI] [PubMed] [Google Scholar]
- Mosebach J, Keilhoff G, Gos T, Schiltz K, Schoeneck L, Dobrowolny H, et al. Increased nuclear Olig1-expression in the pregenual anterior cingulate white matter of patients with major depression: a regenerative attempt to compensate oligodendrocyte loss. J Psychiatr Res. 2013;47:1069–1079. doi: 10.1016/j.jpsychires.2013.03.018. [DOI] [PubMed] [Google Scholar]
- Hayashi Y, Nihonmatsu-Kikuchi N, Yu X, Ishimoto K, Hisanaga SI, Tatebayashi Y. A novel, rapid, quantitative cell-counting method reveals oligodendroglial reduction in the frontopolar cortex in major depressive disorder. Mol Psychiatry. 2011;16:1155–1158. doi: 10.1038/mp.2011.84. [DOI] [PubMed] [Google Scholar]
- Bryne JC, Valen E, Tang MH, Marstrand T, Winther O, da Piedade I, et al. JASPAR, the open access database of transcription factor-binding profiles: new content and tools in the 2008 update Nucleic Acids Res 200836((Database issueD102–D106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolt CC, Rehberg S, Ader M, Lommes P, Riethmacher D, Schachner M, et al. Terminal differentiation of myelin-forming oligodendrocytes depends on the transcription factor Sox10. Genes Dev. 2002;16:165–170. doi: 10.1101/gad.215802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marathe HG, Mehta G, Zhang X, Datar I, Mehrotra A, Yeung KC, et al. SWI/SNF Enzymes Promote SOX10- Mediated Activation of Myelin Gene Expression. PLoS One. 2013;8:e69037. doi: 10.1371/journal.pone.0069037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsusue Y, Horii-Hayashi N, Kirita T, Nishi M. Distribution of corticosteroid receptors in mature oligodendrocytes and oligodendrocyte progenitors of the adult mouse brain. J Histochem Cytochem. 2014;62:211–226. doi: 10.1369/0022155413517700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Yang B, Yan C, Hu H, Cai S, Liu J, et al. Effects of duration and timing of prenatal stress on hippocampal myelination and synaptophysin expression. Brain Res. 2013;1527:57–66. doi: 10.1016/j.brainres.2013.06.025. [DOI] [PubMed] [Google Scholar]
- Miyata S, Koyama Y, Takemoto K, Yoshikawa K, Ishikawa T, Taniguchi M, et al. Plasma corticosterone activates SGK1 and induces morphological changes in oligodendrocytes in corpus callosum. PLoS One. 2011;6:e19859. doi: 10.1371/journal.pone.0019859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caronia G, Wilcoxon J, Feldman P, Grove EA. Bone morphogenetic protein signaling in the developing telencephalon controls formation of the hippocampal dentate gyrus and modifies fear-related behavior. J Neurosci. 2010;30:6291–6301. doi: 10.1523/JNEUROSCI.0550-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hintsch G, Zurlinden A, Meskenaite V, Steuble M, Fink-Widmer K, Kinter J, et al. The calsyntenins—a family of postsynaptic membrane proteins with distinct neuronal expression patterns. Mol Cell Neurosci. 2002;21:393–409. doi: 10.1006/mcne.2002.1181. [DOI] [PubMed] [Google Scholar]
- Grant PA, Duggan L, Cote J, Roberts SM, Brownell JE, Candau R, et al. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 1997;11:1640–1650. doi: 10.1101/gad.11.13.1640. [DOI] [PubMed] [Google Scholar]
- Ciurciu A, Komonyi O, Pankotai T, Boros IM. The Drosophila histone acetyltransferase Gcn5 and transcriptional adaptor Ada2a are involved in nucleosomal histone H4 acetylation. Mol Cell Biol. 2006;26:9413–9423. doi: 10.1128/MCB.01401-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon SB, Wood MA, Cole MD. The essential cofactor TRRAP recruits the histone acetyltransferase hGCN5 to c-Myc. Mol Cell Biol. 2000;20:556–562. doi: 10.1128/mcb.20.2.556-562.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenneth NS, Ramsbottom BA, Gomez-Roman N, Marshall L, Cole PA, White RJ. TRRAP and GCN5 are used by c-Myc to activate RNA polymerase III transcription. Proc Natl Acad Sci USA. 2007;104:14917–14922. doi: 10.1073/pnas.0702909104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang SE, McMahon SB, Cole MD, Hearing P. E2F transcriptional activation requires TRRAP and GCN5 cofactors. J Biol Chem. 2001;276:32627–32634. doi: 10.1074/jbc.M102067200. [DOI] [PubMed] [Google Scholar]
- Candau R, Zhou JX, Allis CD, Berger SL. Histone acetyltransferase activity and interaction with ADA2 are critical for GCN5 function in vivo. EMBO J. 1997;16:555–565. doi: 10.1093/emboj/16.3.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Edmondson DG, Evrard YA, Wakamiya M, Behringer RR, Roth SY. Loss of Gcn5l2 leads to increased apoptosis and mesodermal defects during mouse development. Nat Genet. 2000;26:229–232. doi: 10.1038/79973. [DOI] [PubMed] [Google Scholar]
- Martinez-Cerdeno V, Lemen JM, Chan V, Wey A, Lin W, Dent SR, et al. N-Myc and GCN5 regulate significantly overlapping transcriptional programs in neural stem cells. PLoS One. 2012;7:e39456. doi: 10.1371/journal.pone.0039456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balboa MA, Balsinde J, Winstead MV, Tischfield JA, Dennis EA. Novel group V phospholipase A2 involved in arachidonic acid mobilization in murine P388D1 macrophages. J Biol Chem. 1996;271:32381–32384. doi: 10.1074/jbc.271.50.32381. [DOI] [PubMed] [Google Scholar]
- Shinohara H, Balboa MA, Johnson CA, Balsinde J, Dennis EA. Regulation of delayed prostaglandin production in activated P388D1 macrophages by group IV cytosolic and group V secretory phospholipase A2s. J Biol Chem. 1999;274:12263–12268. doi: 10.1074/jbc.274.18.12263. [DOI] [PubMed] [Google Scholar]
- Cho W. Structure, function, and regulation of group V phospholipase A(2) Biochim Biophys Acta. 2000;1488:48–58. doi: 10.1016/s1388-1981(00)00109-8. [DOI] [PubMed] [Google Scholar]
- Sun GY, Shelat PB, Jensen MB, He Y, Sun AY, Simonyi A. Phospholipases A2 and inflammatory responses in the central nervous system. Neuromol Med. 2010;12:133–148. doi: 10.1007/s12017-009-8092-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kordeli E, Lambert S, Bennett V. Ankyrin G: a new ankyrin gene with neural-specific isoforms localized at the axonal initial segment and node of Ranvier. J Biol Chem. 1995;270:2352–2359. doi: 10.1074/jbc.270.5.2352. [DOI] [PubMed] [Google Scholar]
- Jenkins SM, Bennett V. Ankyrin-G coordinates assembly of the spectrin-based membrane skeleton, voltage-gated sodium channels, and L1 CAMs at Purkinje neuron initial segments. J Cell Biol. 2001;155:739–746. doi: 10.1083/jcb.200109026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Z, Kao T, Horvath Z, Lemos J, Sul JY, Cranstoun SD, et al. A common ankyrin-G-based mechanism retains KCNQ and NaV channels at electrically active domains of the axon. J Neurosci. 2006;26:2599–2613. doi: 10.1523/JNEUROSCI.4314-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze TG, Detera-Wadleigh SD, Akula N, Gupta A, Kassem L, Steele J, et al. Two variants in Ankyrin 3 (ANK3) are independent genetic risk factors for bipolar disorder. Mol Psychiatry. 2009;14:487–491. doi: 10.1038/mp.2008.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith EN, Bloss CS, Badner JA, Barrett T, Belmonte PL, Berrettini W, et al. Genome-wide association study of bipolar disorder in European American and African American individuals. Mol Psychiatry. 2009;14:755–763. doi: 10.1038/mp.2009.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rueckert EH, Barker D, Ruderfer D, Bergen SE, O'Dushlaine C, Luce CJ, et al. Cis-acting regulation of brain-specific ANK3 gene expression by a genetic variant associated with bipolar disorder. Mol Psychiatry. 2013;18:922–929. doi: 10.1038/mp.2012.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakagami H, Suzuki H, Kamata A, Owada Y, Fukunaga K, Mayanagi H, et al. Distinct spatiotemporal expression of EFA6D, a guanine nucleotide exchange factor for ARF6, among the EFA6 family in mouse brain. Brain Res. 2006;1093:1–11. doi: 10.1016/j.brainres.2006.02.058. [DOI] [PubMed] [Google Scholar]
- Hernandez-Deviez DJ, Casanova JE, Wilson JM. Regulation of dendritic development by the ARF exchange factor ARNO. Nat Neurosci. 2002;5:623–624. doi: 10.1038/nn865. [DOI] [PubMed] [Google Scholar]
- Hernandez-Deviez DJ, Roth MG, Casanova JE, Wilson JM. ARNO and ARF6 regulate axonal elongation and branching through downstream activation of phosphatidylinositol 4-phosphate 5-kinase alpha. Mol Biol Cell. 2004;15:111–120. doi: 10.1091/mbc.E03-06-0410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki H, Yamazaki M, Watanabe H, Maehama T, Yokozeki T, Kanaho Y. The small GTPase ADP-ribosylation factor 6 negatively regulates dendritic spine formation. FEBS Lett. 2005;579:6834–6838. doi: 10.1016/j.febslet.2005.11.022. [DOI] [PubMed] [Google Scholar]
- Choi S, Ko J, Lee JR, Lee HW, Kim K, Chung HS, et al. ARF6 and EFA6A regulate the development and maintenance of dendritic spines. J Neurosci. 2006;26:4811–4819. doi: 10.1523/JNEUROSCI.4182-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galas MC, Helms JB, Vitale N, Thierse D, Aunis D, Bader MF. Regulated exocytosis in chromaffin cells. A potential role for a secretory granule-associated ARF6 protein. J Biol Chem. 1997;272:2788–2793. doi: 10.1074/jbc.272.5.2788. [DOI] [PubMed] [Google Scholar]
- Krauss M, Kinuta M, Wenk MR, De Camilli P, Takei K, Haucke V. ARF6 stimulates clathrin/AP-2 recruitment to synaptic membranes by activating phosphatidylinositol phosphate kinase type Igamma. J Cell Biol. 2003;162:113–124. doi: 10.1083/jcb.200301006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaney KA, Murph MM, Brown LM, Radhakrishna H. Transfer of M2 muscarinic acetylcholine receptors to clathrin-derived early endosomes following clathrin-independent endocytosis. J Biol Chem. 2002;277:33439–33446. doi: 10.1074/jbc.M205293200. [DOI] [PubMed] [Google Scholar]
- Weber M, Hellmann I, Stadler MB, Ramos L, Paabo S, Rebhan M, et al. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet. 2007;39:457–466. doi: 10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- Deaton AM, Bird A. CpG islands and the regulation of transcription. Genes Dev. 2011;25:1010–1022. doi: 10.1101/gad.2037511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JU, Ma DK, Mo H, Ball MP, Jang MH, Bonaguidi MA, et al. Neuronal activity modifies the DNA methylation landscape in the adult brain. Nat Neurosci. 2011;14:1345–1351. doi: 10.1038/nn.2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat Rev Genet. 2008;9:465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- Moskal JR, Kroes RA, Otto NJ, Rahimi O, Claiborne BJ. Distinct patterns of gene expression in the left and right hippocampal formation of developing rats. Hippocampus. 2006;16:629–634. doi: 10.1002/hipo.20198. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.