Abstract
The superior frontal gyrus (SFG), an area of the brain frequently found to have reduced gray matter in patients with schizophrenia, is involved in self-awareness and emotion, which are impaired in schizophrenia. However, no genome-wide association studies of SFG volume have investigated in patients with schizophrenia. To identify single-nucleotide polymorphisms (SNPs) associated with SFG volumes, we demonstrated a genome-wide association study (GWAS) of gray matter volumes in the right or left SFG of 158 patients with schizophrenia and 378 healthy subjects. We attempted to bioinformatically ascertain the potential effects of the top hit polymorphism on the expression levels of genes at the genome-wide region. We found associations between five variants on 1p36.12 and the right SFG volume at a widely used benchmark for genome-wide significance (P<5.0 × 10−8). The strongest association was observed at rs4654899, an intronic SNP in the eukaryotic translation initiation factor 4 gamma, 3 (EIF4G3) gene on 1p36.12 (P=7.5 × 10−9). No SNP with genome-wide significance was found in the volume of the left SFG (P>5.0 × 10−8); however, the rs4654899 polymorphism was identified as the locus with the second strongest association with the volume of the left SFG (P=1.5 × 10−6). In silico analyses revealed a proxy SNP of rs4654899 had effect on gene expression of two genes, HP1BP3 lying 3′ to EIF4G3 (P=7.8 × 10−6) and CAPN14 at 2p (P=6.3 × 10−6), which are expressed in moderate-to-high levels throughout the adult human SFG. These results contribute to understand genetic architecture of a brain structure possibly linked to the pathophysiology of schizophrenia.
Introduction
Schizophrenia is a common and complex psychiatric disorder with a lifetime risk of approximately 1%. This disorder has a strong genetic component; the estimated heritability is 81%.1 Multiple genetic variants that have a small effect have been implicated in the pathogenesis of schizophrenia.2 A genome-wide association study (GWAS) of single-nucleotide polymorphisms (SNPs) that accesses tens of thousands of DNA samples from patients and controls can be a powerful tool for identifying common risk factors for complex diseases, such as schizophrenia. GWASs on schizophrenia have identified several genome-wide significant associated variants.3,4 Subsequently, GWASs on neurobiological quantitative traits as intermediate phenotypes that possibly reflect the underlying genetic vulnerability better than diagnostic categorization, such as schizophrenia,5,6 have been performed to minimize the clinical and genetic heterogeneity in studies of schizophrenia.7
The superior frontal gyrus (SFG) of the brain is frequently found to have reduced gray matter in individuals with first-episode schizophrenia and neuroleptic naive schizophrenia, as well as chronic patients with schizophrenia.8,9 The SFG is involved in self-awareness and emotion.10,11 Self-awareness is the cognitive ability to differentiate between self and non-self cues and is necessary to understand the behavior of other humans. Disturbance in self-awareness linked to social cognition is a core feature of schizophrenia.12 Emotional disturbances, including meaningless laughter, are often observed in patients with schizophrenia. Meaningless laughter was also observed in unaffected siblings of schizophrenia, thus indicating its heritability.13 In addition, laughter can be elicited by electrical stimulation of the SFG. Gray matter volumes of bilateral SFG have a strong genetic component, with an estimated heritability of 76–80%.14 As there is considerable inter-individual variation in the degree of reduced volume of the SFG, it appears that genetic influences have a role in determining the degree of volume reduction of the SFG in schizophrenia. Although GWASs of bilateral hippocampal volume have recently been reported,15,16 no study has investigated other brain areas in patients with schizophrenia. To identify an SNP related to SFG volumes, we conducted a GWAS of gray matter volumes in the right or left SFG of patients with schizophrenia and healthy subjects.
Materials and methods
Subjects
We selected 281 patients with schizophrenia (52.0% males, 146 males and 135 females; mean age 36.0±12.4 years) and 413 healthy controls (49.6% males, 205 males and 208 females; mean age 36.4±12.8 years) for a GWAS of schizophrenia-related phenotypes, such as structural brain morphology, neurocognitive function and neurophysiological assessments.17, 18, 19 All of the subjects were biologically unrelated, there were no first- or second-degree relatives, and all were of Japanese descent.20,21 The subjects were excluded if they had neurological or medical conditions that could potentially affect the central nervous system, such as atypical headaches, head trauma with loss of consciousness, chronic lung disease, kidney disease, chronic hepatic disease, thyroid disease, active cancer, cerebrovascular disease, epilepsy, seizures, substance-related disorders or mental retardation. Patients with schizophrenia were recruited from the Osaka University Hospital. Each patient had been diagnosed by at least two trained psychiatrists according to the criteria from the DSM-IV (Diagnostic and Statistical Manual of Mental Disorders, Fourth Edition) based on the Structured Clinical Interview for DSM-IV. Current symptoms of schizophrenia were evaluated using the positive and negative syndrome scale. Controls were recruited through local advertisements at Osaka University. The healthy subjects were evaluated using the non-patient version of the Structured Clinical Interview for DSM-IV to exclude individuals who had current or past contact with psychiatric services or who had received psychiatric medications.
Superior frontal volumes obtained from the magnetic resonance imaging data were assessed in 158 patients with schizophrenia and 378 healthy subjects. Detailed demographic information is shown in Supplementary Table S1. Mean age and handedness did not differ significantly between the cases and controls (P>0.50); however, the gender ratio, years of education and estimated premorbid intelligence quotient differed significantly between the cases and controls (P<0.05). The ratio of male was higher in patients with schizophrenia compared with the controls. The years of education and estimated premorbid intelligence quotient were significantly lower in patients with schizophrenia compared with the controls. When the genotype groups in the top five SNPs with genome-wide significance of the right SFG volume were compared within the patient and control groups, we found no differences across the demographic variables, except for the gender ratio in the controls (rs6700718, rs1354792, rs10218584, and rs6702110; P<0.05). Written informed consent was obtained from all the subjects after the procedures had been fully explained. This study was performed in accordance with the World Medical Association's Declaration of Helsinki and was approved by the Research Ethical Committee of Osaka University.
Magnetic resonance imaging procedure and extraction of SFG volumes
All magnetic resonance imaging data were obtained using a 1.5-T GE Signa EXCITE system (Tokyo, Japan). A three-dimensional volumetric acquisition of a T1-weighted gradient echo sequence produced a gapless series of 124 sagittal sections using a spoiled gradient-recalled acquisition in the steady state (SPGR) sequence (TE/TR, 4.2/12.6 ms; flip angle, 15° acquisition matrix, 256 × 256; 1NEX, FOV, 24 × 24 cm; slice thickness, 1.4 mm). We screened all scans and found no gross abnormalities, such as infarcts, hemorrhages or brain tumors, in any of the subjects. Each image was visually examined to eliminate any images with motion or metal artifacts, and the anterior commissure–posterior commissure line was adjusted.22 MR images were processed with the VBM8 toolbox (http://dbm.neuro.uni-jena.de/vbm/download/) implemented for SPM8 (Wellcome Department of Imaging Neuroscience, University College London, UK, http://www.fil.ion.ucl.ac.uk/spm) running in MATLAB (The Mathworks, Natick, MA, USA) for tissue segmentation and anatomical normalization, as described elsewhere.23, 24, 25 The voxel values of the normalized gray matter images were modulated according to the nonlinear component of the transformation, which resulted in approximating brain-size-adjusted gray matter volumes while preserving local volume changes.26 Gray matter volumes of the bilateral SFG were then calculated by using the maximum probabilistic atlas using 20 hand-labeled images (Supplementary Figure S1).27,28
SNP selection and SNP genotyping
Genotyping was performed using the Affymetrix Genome-Wide Human SNP Array 6.0 (Affymetrix, Santa Clara, CA, USA), according to the manufacturer's protocol. The genotypes were called from the CEL files using Birdseed v2 for the 6.0 chip implemented in the Genotyping Console software (Affymetrix). We then applied the following quality control (QC) criteria to exclude samples: (i) arrays with low QC (<0.4) according to Birdseed v2 (N=0), (ii) samples for which <95% of the genotypes were called (N=0) and (iii) samples in the same family according to π̂ (>0.4, N=0). Next, we excluded SNPs that: (i) had low call rates (<0.95), (ii) were duplicated, (iii) were localized to sex chromosomes, (iv) deviated from Hardy–Weinberg equilibrium in the controls (P<0.0001) or (v) had low minor allele frequencies <0.05. After all of these exclusions, 517 946 SNPs that underwent QC remained for experimental analysis.
To test for the existence of a genetic structure in the data, we performed a principal component analysis using EIGENSTRAT 3.0 software.29 Ten eigenvectors were calculated. Genotype information from the JPT (Japanese in Tokyo, Japan), CHB (Han Chinese in Beijing, China), CEU (Utah residents with ancestors from northern and western Europe) and YRI (Yoruba in Ibadan, Nigeria) in HapMap phase III was compared with our data set to check for population stratification (Supplementary Figure S2).
Statistical analyses
Statistical analyses of the demographic variables were performed using PASW Statistics 18.0 software (SPSS Japan, Tokyo, Japan). Differences in the clinical characteristics between patients and controls were analyzed using χ2 tests for the categorical variables and the Mann–Whitney U-test for the continuous variables. Multiple linear regression analysis was performed to compare the gray matter volumes in the right and left SFG regions among genotypes (the number of major alleles; 0, 1 or 2) using PLINK 1.07 software. Diagnosis, age and gender were included as covariates. Quantile–Quantile is listed in Supplementary Figure S3.
Results
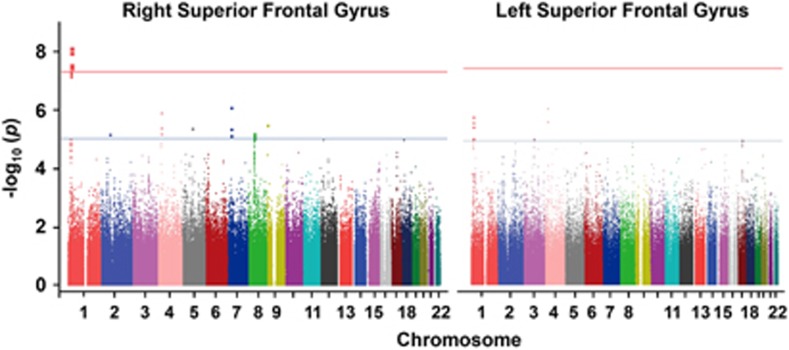
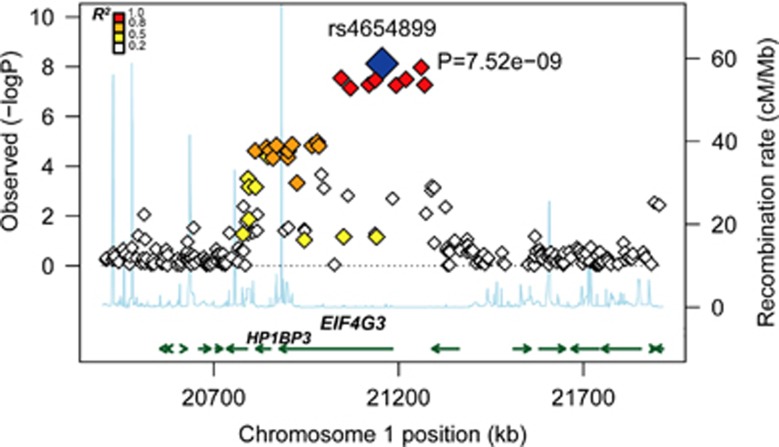
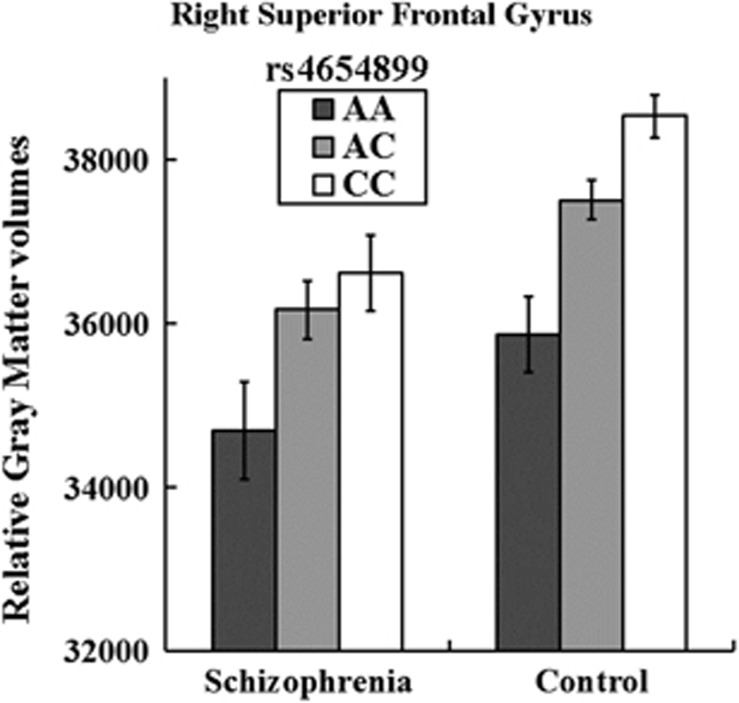
We observed associations between five variants (rs4654899, rs6702110, rs6700718, rs10218584 and rs1354792) on 1p36.12 and the right SFG volume at a widely used benchmark for genome-wide significance (P<5.0 × 10−8, r2 among SNPs >0.8; Figure 1). The strongest association was observed at rs4654899, an intronic SNP in the eukaryotic translation initiation factor 4 gamma, 3 (EIF4G3) gene on 1p36.12 (P=7.5 × 10−9; Figure 2). No SNP with genome-wide significance was found in the volume of the left SFG; however, the rs4654899 polymorphism was identified as the locus with the second strongest association with the volume of the left SFG (P=1.5 × 10−6; Figure 1). The top 10 and top 200 markers on each SFG are shown in Tables 1 and 2 and Supplementary Tables S2 and S3. Post hoc analyses separately assessed in patients and controls also revealed reduced but significant associations (Tables 1 and 2 and Supplementary Tables S2 and S3). Genotype effects of rs4654899 on gray matter volume of right superior frontal gyrus were found in patients with schizophrenia and controls (Figure 3). We attempted to bioinformatically ascertain the potential effects of the rs4654899 polymorphism on the expression levels of genes at the genome-wide region by using the mRNA by SNP Browser 1.0.1 database (http://www.sph.umich.edu/csg/liang/asthma/). Significant effects of the rs3767248 proxy SNP for rs4654899 (r2=1.0) were identified in the expressions of the heterochromatin protein 1, binding protein 3 (HP1BP3) gene (P=7.8 × 10−6), which lies 3′ to EIF4G3, as a cis-acting effect (<200 kb), and the calpain 14 (CAPN14) gene (P=6.3 × 10−6), as a trans-acting effect (>200 kb; Supplementary Table S4). Both HP1BP3 and CAPN14 are expressed in moderate-to-high levels throughout the adult human SFG (Supplementary Figures S4 and Supplementary Figure S5), as visualized in the Allen Institute Human Brain Atlas Explorer 2 software (http://human.brain-map.org/static/brainexplorer).
Figure 1.
Manhattan plots derived from the multiple linear regression analysis of the bilateral superior frontal volumes. The blue line indicates a P-value of 1.0E−05. The red line indicates a P-value of 5.0E−08.
Figure 2.
The strongest association with the right superior frontal gyrus was found for rs4654899. P-values (−log10) are shown in regions peripheral to rs4654899 (±750 kb).
Table 1. TOP 10 SNPs for the right superior frontal gyrus.
| Rank | SNP | Chr | Bp | m | M | MAF |
Combined subjects |
Schizophrenia |
Controls |
Closest gene | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | β | P | N | β | P | N | β | P | ||||||||
| 1 | rs4654899 | 1 | 21410231 | A | C | 0.33 | 509 | −969.5 | 7.52E−09 | 153 | −1164 | 3.71E−04 | 356 | −935.9 | 1.86E−06 | EIF4G3 |
| 2 | rs6702110 | 1 | 21515906 | G | A | 0.32 | 526 | −965.3 | 1.07E−08 | 155 | −1255 | 1.79E−04 | 371 | −902.3 | 4.34E−06 | EIF4G3 |
| 3 | rs6700718 | 1 | 21299363 | A | C | 0.33 | 537 | −895.1 | 2.89E−08 | 159 | −1110 | 4.68E−04 | 378 | −847.7 | 6.81E−06 | EIF4G3 |
| 4 | rs10218584 | 1 | 21474480 | G | C | 0.33 | 537 | −891.8 | 3.21E−08 | 159 | −1110 | 4.68E−04 | 378 | −842.7 | 7.68E−06 | EIF4G3 |
| 5 | rs1354792 | 1 | 21391875 | C | T | 0.33 | 535 | −892.7 | 3.46E−08 | 157 | −1114 | 5.35E−04 | 378 | −847.7 | 6.81E−06 | EIF4G3 |
| 6 | rs6703227 | 1 | 21374810 | C | T | 0.33 | 537 | −879.3 | 5.28E−08 | 159 | −1056 | 9.55E−04 | 378 | −847.7 | 6.81E−06 | EIF4G3 |
| 7 | rs1609558 | 1 | 21525228 | C | T | 0.30 | 533 | −899.8 | 5.36E−08 | 158 | −1031 | 1.18E−03 | 375 | −870.9 | 7.67E−06 | EIF4G3 |
| 8 | rs12402486 | 1 | 21447935 | A | G | 0.34 | 527 | −880.9 | 5.58E−08 | 159 | −1110 | 4.68E−04 | 368 | −830 | 1.20E−05 | EIF4G3 |
| 9 | rs2874367 | 1 | 21324491 | A | C | 0.33 | 531 | −874.5 | 7.19E−08 | 157 | −1092 | 6.24E−04 | 374 | −829 | 1.25E−05 | EIF4G3 |
| 10 | rs6945071 | 7 | 26122423 | G | A | 0.16 | 537 | −1035 | 8.46E−07 | 159 | −1120 | 3.25E−03 | 378 | −983.2 | 1.02E−04 | NFE2L3 |
Abbreviations: Chr, chromosome; Bp, nucleotide location; m, minor allele; M, major allele; MAF, minor allele frequency; SNP, single-nucleotide polymorphism. Genome-wide significant P-values are shown as bold font and are underlined.
Table 2. TOP 10 SNPs for the left superior frontal gyrus.
| Rank | SNP | Chr | Bp | m | M | MAF |
Combined subjects |
Schizophrenia |
Controls |
Closest gene | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | β | P | N | β | P | N | β | P | ||||||||
| 1 | rs4574391 | 4 | 27212223 | C | T | 0.25 | 533 | −880 | 7.63E−07 | 158 | −693 | 4.27E−02 | 375 | −965 | 3.51E−06 | STIM2 |
| 2 | rs4654899 | 1 | 21410231 | A | C | 0.33 | 509 | −787 | 1.51E−06 | 153 | −691 | 3.81E−02 | 356 | −863 | 4.22E−06 | EIF4G3 |
| 3 | rs2046701 | 4 | 27211040 | C | A | 0.25 | 532 | −826 | 2.22E−06 | 158 | −697 | 3.87E−02 | 374 | −887 | 1.28E−05 | STIM2 |
| 4 | rs1609558 | 1 | 21525228 | C | T | 0.3 | 533 | −763 | 2.33E−06 | 158 | −524 | 1.11E−01 | 375 | −885 | 1.67E−06 | EIF4G3 |
| 5 | rs6702110 | 1 | 21515906 | G | A | 0.32 | 526 | −769 | 3.37E−06 | 155 | −714 | 3.90E−02 | 371 | −834 | 9.21E−06 | EIF4G3 |
| 6 | rs10218584 | 1 | 21474480 | G | C | 0.33 | 537 | −704 | 8.32E−06 | 159 | −566 | 8.48E−02 | 378 | −791 | 1.05E−05 | EIF4G3 |
| 7 | rs1354792 | 1 | 21391875 | C | T | 0.33 | 535 | −704 | 8.69E−06 | 157 | −625 | 5.92E−02 | 378 | −776 | 1.59E−05 | EIF4G3 |
| 8 | rs2623384 | 3 | 99064220 | G | A | 0.39 | 525 | −724 | 9.15E−06 | 157 | −894 | 6.79E−03 | 368 | −642 | 5.92E−04 | COL8A1 |
| 9 | rs2292343 | 17 | 45455670 | C | G | 0.34 | 524 | −721 | 9.74E−06 | 155 | −836 | 1.22E−02 | 369 | −666 | 3.45E−04 | EFCAB13 |
| 10 | rs3883317 | 17 | 45484111 | A | G | 0.34 | 534 | −703 | 1.03E−05 | 158 | −743 | 2.70E−02 | 376 | −675 | 1.72E−04 | EFCAB13 |
Abbreviations: Chr, chromosome; Bp, nucleotide location; m, minor allele; M, major allele; MAF, minor allele frequency; SNP, single-nucleotide polymorphism.
Figure 3.
Impact of the rs4654899 genotype of the EIF4G3 gene on the right superior frontal gyrus. Each column shows relative gray matter volumes of the right superior frontal gyrus. Error bars represent the standard error.
Discussion
To date, it remained unclear whether there were genetic variants strongly related to SFG volume in patients with schizophrenia and healthy subjects. This study is the first GWAS to identify the SNPs associated with the SFG, which have an important role in schizophrenia-related social functions and is reduced in patients with schizophrenia. We revealed that there were associations at the genome-wide significant level between SFG and genetic variants of the EIF4G3 gene on 1p36.12. Individuals with minor A-allele of the most significant variant rs4654899 had smaller right SFG volumes compared with those with major C-allele in both patients and controls. Bioinformatical data indicate that the rs3767248 proxy SNP for rs4654899 has important roles in the expression of the HP1BP3 and CAPN14 genes, which are expressed in human adult SFG. The HP1BP3 and CAPN14 gene expressions of the minor G-allele of the rs3767248 polymorphism were significantly lower than those of the major A-allele. However, whether the expression levels of these genes in the brains or serums of patients with schizophrenia are lower or higher than those in healthy subjects is unknown. Further study is needed to investigate the difference of the expressions between patients and controls.
To our knowledge, no study has reported associations between these genes and schizophrenia, although the chromosomal region (1p36.12) related to the risk of schizophrenia has been reported.30 The exact functions of these two genes are unknown; however, HP1BP3 is predicted to bind to DNA and have a role in nucleosome assembly. CAPN14, which belongs to the calpain large subunit family, is a cytosolic calcium-activated cysteine protease involved in a variety of cellular processes, including apoptosis, cell division, modulation of integrin–cytoskeletal interactions and synaptic plasticity.
In this study, we examined the effects of genotypes on SFG volumes in a combined sample of patients and controls, and found similar effects of genotypes in patients and controls. Susceptibility genes for schizophrenia do not directly encode for their clinical syndrome/behaviors. The syndrome/behaviors observed in schizophrenia are produced by intermediate steps that occur between genes and syndrome/behaviors; and intermediate steps, such as changes of brain volumes, underlie the syndrome/behavior of schizophrenia. The intermediate phenotypes are located on the pathogenesis path, and are likely associated with a more basic and proximal etiological process rather than pathogenesis of disease itself.5,6 Therefore, each genetic variant is related to controls as well as patients, and accumulations of each genetic variant could contribute to pathogenesis of schizophrenia through intermediate steps.
To date, although abnormal brain lateralization in schizophrenia causing a failure of left hemisphere dominance has been reported,31 there is no evidence of SFG lateralization in schizophrenia. In addition, there is no report for developmental/functional differences between the right and left SFG. We found genome-wide significant variants related to right SFG volumes, whereas these variants were not related to left SFG volumes at genome-wide significant level. The difference of significance between right and left SFG was due to a difference of genotype effects in patients (for example, rs4654899, right: P=3.71 × 10−4, left: P=3.81 × 10−2) but not in controls (right: P=1.86 × 10−6, left: P=4.22 × 10−6). As it has been reported that gray matter volume deficits were more extensive in individuals with first-episode schizophrenia and neuroleptic naive than that of their neuroleptic-treated counterparts in left SFG,9 confounding factors, such as duration of antipsychotic treatment or dose of antipsychotics, might affect our results.
In this study, we provide new insights into the genetic architecture of a brain structure closely linked to schizophrenia. It is still unclear whether and to what extent the effects of the genetic variant on SFG volumes observed here might be associated with an increased risk for schizophrenia. We suggest that the variant may have a role in the impairments of self-awareness and emotion noted in patients with schizophrenia through volumetric vulnerability of the SFG.
There were several limitations to this study. We recruited a relatively large sample with an only Japanese ethnicity to avoid population stratification. However, the existence of a false-positive association cannot be excluded as an explanation for our results. Further investigations of other samples with much larger sample sizes and/or with different ethnicities are needed to confirm our findings. It is unclear whether our results are directly/indirectly linked to the rs4654899 SNP, to other SNPs in high linkage disequilibrium with this SNP or to interactions between this SNP and other SNPs. To determine whether rs4654899 is the most strongly associated variant for SFG volume in the chromosomal region, an extensive search such as sequencing for other functional variants at this locus could provide further information underlying the genomic mechanism for this variant.
In conclusion, we found that genetic variants of the EIF4G3 gene could be associated with structural vulnerability of the SFG. Further replication studies are necessary to confirm our findings. Identification of causal variants and the functional effects of these genes may help to reveal additional genetic variables involved in the neurodevelopment and pathogenesis of schizophrenia.
Acknowledgments
We thank all the individuals who participated in this study. This work was supported by research grants from the Japanese Ministry of Health, Labor and Welfare (H22-seishin-ippan-001); KAKENHI, 22390225-Grant-in-Aid for Scientific Research (B), 23659565-Grant-in-Aid for Challenging Exploratory Research and Grant-in-Aid for Scientific Research on Innovative Areas (Comprehensive Brain Science Network) from the Japanese Ministry of Education, Culture, Sports, Science and Technology (MEXT) and the Japan Foundation for Neuroscience and Mental Health.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on the Translational Psychiatry website (http://www.nature.com/tp)
Supplementary Material
References
- Sullivan PF, Kendler KS, Neale MC. Schizophrenia as a complex trait: evidence from a meta-analysis of twin studies. Arch Gen Psychiatry. 2003;60:1187–1192. doi: 10.1001/archpsyc.60.12.1187. [DOI] [PubMed] [Google Scholar]
- Sun J, Kuo PH, Riley BP, Kendler KS, Zhao Z. Candidate genes for schizophrenia: a survey of association studies and gene ranking. Am J Med Genet B Neuropsychiatr Genet. 2008;147B:1173–1181. doi: 10.1002/ajmg.b.30743. [DOI] [PubMed] [Google Scholar]
- Stefansson H, Ophoff RA, Steinberg S, Andreassen OA, Cichon S, Rujescu D, et al. Common variants conferring risk of schizophrenia. Nature. 2009;460:744–747. doi: 10.1038/nature08186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donovan MC, Craddock N, Norton N, Williams H, Peirce T, Moskvina V, et al. Identification of loci associated with schizophrenia by genome-wide association and follow-up. Nat Genet. 2008;40:1053–1055. doi: 10.1038/ng.201. [DOI] [PubMed] [Google Scholar]
- Meyer-Lindenberg A, Weinberger DR. Intermediate phenotypes and genetic mechanisms of psychiatric disorders. Nat Rev Neurosci. 2006;7:818–827. doi: 10.1038/nrn1993. [DOI] [PubMed] [Google Scholar]
- Tan HY, Callicott JH, Weinberger DR. Intermediate phenotypes in schizophrenia genetics redux: is it a no brainer. Mol Psychiatry. 2008;13:233–238. doi: 10.1038/sj.mp.4002145. [DOI] [PubMed] [Google Scholar]
- Hashimoto R, Ikeda M, Ohi K, Yasuda Y, Yamamori H, Fukumoto M, et al. Genome-wide association study of cognitive decline in schizophrenia. Am J Psychiatry. 2013;170:683–684. doi: 10.1176/appi.ajp.2013.12091228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan RC, Di X, McAlonan GM, Gong QY. Brain anatomical abnormalities in high-risk individuals, first-episode, and chronic schizophrenia: an activation likelihood estimation meta-analysis of illness progression. Schizophr Bull. 2011;37:177–188. doi: 10.1093/schbul/sbp073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung M, Cheung C, Yu K, Yip B, Sham P, Li Q, et al. Gray matter in first-episode schizophrenia before and after antipsychotic drug treatment. Anatomical likelihood estimation meta-analyses with sample size weighting. Schizophr Bull. 2011;37:199–211. doi: 10.1093/schbul/sbp099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg II, Harel M, Malach R. When the brain loses its self: prefrontal inactivation during sensorimotor processing. Neuron. 2006;50:329–339. doi: 10.1016/j.neuron.2006.03.015. [DOI] [PubMed] [Google Scholar]
- Fried I, Wilson CL, MacDonald KA, Behnke EJ. Electric current stimulates laughter. Nature. 1998;391:650. doi: 10.1038/35536. [DOI] [PubMed] [Google Scholar]
- Sass LA, Parnas J. Schizophrenia, consciousness, and the self. Schizophr Bull. 2003;29:427–444. doi: 10.1093/oxfordjournals.schbul.a007017. [DOI] [PubMed] [Google Scholar]
- Bearden CE, Rosso IM, Hollister JM, Sanchez LE, Hadley T, Cannon TD. A prospective cohort study of childhood behavioral deviance and language abnormalities as predictors of adult schizophrenia. Schizophr Bull. 2000;26:395–410. doi: 10.1093/oxfordjournals.schbul.a033461. [DOI] [PubMed] [Google Scholar]
- Hulshoff Pol HE, Schnack HG, Posthuma D, Mandl RC, Baare WF, van Oel C, et al. Genetic contributions to human brain morphology and intelligence. J Neurosci. 2006;26:10235–10242. doi: 10.1523/JNEUROSCI.1312-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bis JC, DeCarli C, Smith AV, van der Lijn F, Crivello F, Fornage M, et al. Common variants at 12q14 and 12q24 are associated with hippocampal volume. Nat Genet. 2012;44:545–551. doi: 10.1038/ng.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein JL, Medland SE, Vasquez AA, Hibar DP, Senstad RE, Winkler AM, et al. Identification of common variants associated with human hippocampal and intracranial volumes. Nat Genet. 2012;44:552–561. doi: 10.1038/ng.2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto R, Ohi K, Yasuda Y, Fukumoto M, Yamamori H, Kamino K, et al. The KCNH2 gene is associated with neurocognition and the risk of schizophrenia. World J Biol Psychiatry. 2013;14:114–120. doi: 10.3109/15622975.2011.604350. [DOI] [PubMed] [Google Scholar]
- Ohi K, Hashimoto R, Yasuda Y, Nemoto K, Ohnishi T, Fukumoto M, et al. Impact of the genome wide supported NRGN gene on anterior cingulate morphology in schizophrenia. PLoS One. 2012;7:e29780. doi: 10.1371/journal.pone.0029780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto R, Ohi K, Yasuda Y, Fukumoto M, Yamamori H, Takahashi H, et al. Variants of the RELA gene are associated with schizophrenia and their startle responses. Neuropsychopharmacology. 2011;36:1921–1931. doi: 10.1038/npp.2011.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto R, Ohi K, Yasuda Y, Fukumoto M, Iwase M, Iike N, et al. The impact of a genome-wide supported psychosis variant in the ZNF804A gene on memory function in schizophrenia. Am J Med Genet B Neuropsychiatr Genet. 2010;153B:1459–1464. doi: 10.1002/ajmg.b.31123. [DOI] [PubMed] [Google Scholar]
- Ohi K, Hashimoto R, Yasuda Y, Fukumoto M, Nemoto K, Ohnishi T, et al. The AKT1 gene is associated with attention and brain morphology in schizophrenia. World J Biol Psychiatry. 2013;14:100–113. doi: 10.3109/15622975.2011.591826. [DOI] [PubMed] [Google Scholar]
- Evans W. An encephalographic ratio for estimating ventricular enlargement and cerebral atrophy. Arch Neurol Psychiatry. 1942;47:931–937. [Google Scholar]
- Wilke M, Holland SK, Altaye M, Gaser C. Template-O-Matic: a toolbox for creating customized pediatric templates. Neuroimage. 2008;41:903–913. doi: 10.1016/j.neuroimage.2008.02.056. [DOI] [PubMed] [Google Scholar]
- Ashburner J. A fast diffeomorphic image registration algorithm. Neuroimage. 2007;38:95–113. doi: 10.1016/j.neuroimage.2007.07.007. [DOI] [PubMed] [Google Scholar]
- Ohi K, Hashimoto R, Yamamori H, Yasuda Y, Fujimoto M, Umeda-Yano S, et al. The impact of the genome-wide supported variant in the cyclin M2 gene on gray matter morphology in schizophrenia. Behav Brain Funct. 2013;9:40. doi: 10.1186/1744-9081-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good CD, Johnsrude IS, Ashburner J, Henson RN, Friston KJ, Frackowiak RS. A voxel-based morphometric study of ageing in 465 normal adult human brains. Neuroimage. 2001;14:21–36. doi: 10.1006/nimg.2001.0786. [DOI] [PubMed] [Google Scholar]
- Hammers A, Allom R, Koepp MJ, Free SL, Myers R, Lemieux L, et al. Three-dimensional maximum probability atlas of the human brain, with particular reference to the temporal lobe. Hum Brain Mapp. 2003;19:224–247. doi: 10.1002/hbm.10123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckemann RA, Hajnal JV, Aljabar P, Rueckert D. Hammers A. Automatic anatomical brain MRI segmentation combining label propagation and decision fusion. Neuroimage. 2006;33:115–126. doi: 10.1016/j.neuroimage.2006.05.061. [DOI] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- Hong KS, Won HH, Cho EY, Jeun HO, Cho SS, Lee YS, et al. Genome-widely significant evidence of linkage of schizophrenia to chromosomes 2p24.3 and 6q27 in an SNP-Based analysis of Korean families. Am J Med Genet B Neuropsychiatr Genet. 2009;150B:647–652. doi: 10.1002/ajmg.b.30884. [DOI] [PubMed] [Google Scholar]
- Ribolsi M, Koch G, Magni V, Di Lorenzo G, Rubino IA, Siracusano A, et al. Abnormal brain lateralization and connectivity in schizophrenia. Rev Neurosci. 2009;20:61–70. doi: 10.1515/revneuro.2009.20.1.61. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.