Abstract
Current catalogs of brain expression quantitative trait loci (eQTL) are incomplete and the findings do not replicate well across studies. All existing cortical eQTL studies are small and emphasize the need for a meta-analysis. We performed a meta-analysis of 424 brain samples across five studies to identify regulatory variants influencing gene expression in human cortex. We identified 3584 genes in autosomes and chromosome X with false discovery rate q<0.05 whose expression was significantly associated with DNA sequence variation. Consistent with previous eQTL studies, local regulatory variants tended to occur symmetrically around transcription start sites and the effect was more evident in studies with large sample sizes. In contrast to random SNPs, we observed that significant eQTLs were more likely to be near 5'-untranslated regions and intersect with regulatory features. Permutation-based enrichment analysis revealed that SNPs associated with schizophrenia and bipolar disorder were enriched among brain eQTLs. Genes with significant eQTL evidence were also strongly associated with diseases from OMIM (Online Mendelian Inheritance in Man) and the NHGRI (National Human Genome Research Institute) genome-wide association study catalog. Surprisingly, we found that a large proportion (28%) of ~1000 autosomal genes encoding proteins needed for mitochondrial structure or function were eQTLs (enrichment P-value=1.3 × 10−9), suggesting a potential role for common genetic variation influencing the robustness of energy supply in brain and a possible role in the etiology of some psychiatric disorders. These systematically generated eQTL information should be a valuable resource in determining the functional mechanisms of brain gene expression and the underlying biology of associations with psychiatric disorders.
Introduction
Psychiatric disorders like schizophrenia, bipolar disorder, major depressive disorder, autism and substance use disorders account for a significant proportion of disability world-wide1 and cause enormous personal and societal burdens.2 The lifetime prevalence estimates range from 0.1% (autism spectrum disorder) to 24% (nicotine dependence).3 These disorders have a significant genetic component, with estimates of heritability ranging from 37% (major depressive disorder) to 81% (schizophrenia).3
Recent genome-wide association studies (GWAS) investigating the genetic architecture of psychiatric disorders have identified many common variants that meet consensus criteria for significance and replication.4, 5, 6 Understanding the biological mechanisms by which these common variants contribute to complex traits is challenging. The main reason is that the majority (>90%) of disease-associated variants from many GWAS lie in noncoding regions,7 making evaluation of their function difficult. However, accumulating evidence suggests that these noncoding common variants are involved in transcriptional regulatory mechanisms such as promoter and enhancer elements8 and enriched within expression quantitative trait loci (eQTL).8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20 In addition, about 77% of SNPs implicated in GWAS were within or in high linkage disequilibrium (LD) with DNase I hypersensitivity sites, a marker for open chromatin subject to transcriptional regulation.7,21,22
eQTL studies measure genetic variation and gene expression in the same individuals, and thus link DNA variation to mRNA variation.8 These studies have received particular attention due to their inherent relevance to the control of gene expression and because they provide a way to generate hypotheses about the functional meaning of GWAS findings via relatively simple data base queries.11,23,24
There are relatively few eQTL studies of human brain tissue25, 26, 27, 28 or brain disease.23,29 Current catalogs of brain eQTLs are incomplete and the findings do not replicate well across studies—all existing brain eQTL studies are small and highlight the need for a meta-analysis.30 Thus, we performed an eQTL ‘meta-analysis' of gene expression and GWAS data for 424 normal brain samples from five studies—Gibbs et al.,27 Colantuoni et al.,31 Myers et al.,26 Stanley Medical Research Institute (SMRI),32,33 and the NIH Genotype-Tissue Expression (GTEx) project.34 We identified more than 3000 genes whose expression was significantly associated with DNA sequence variation. Many of these genes have been implicated in psychiatric disorders. Surprisingly, we found that a large proportion (28%) of ~1000 autosomal genes encoding proteins needed for mitochondrial structure or function were eQTLs. This suggests a potential role for common genetic variation influencing the robustness of energy supply in brain and a possible role in the etiology of some psychiatric disorders.
Materials and methods
Gene expression data
The quality control procedures for each study are described in the Supplementary Methods. Briefly, we included brain cortical samples from neuropathologically normal subjects of European ancestry and aged ⩾20 years at death. We downloaded raw intensity files from the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) and conducted extensive quality control procedures. As in our prior work, we processed gene expression data as consistently as possible across all studies.30,35 First, we mapped the probe sequences for all expression arrays to the genome (UCSC hg19) using Bowtie36 and removed probes that did not map, mapped to multiple locations or intersected a polymorphic SNP (HapMap3 (ref. 37) and 1000 Genomes Project data38) because such probes can result in inaccurate expression.39 Second, we excluded outlier samples on the basis of inter-sample correlations30 or if phenotypic sex did not match mean expression of probesets on chrX and chrY. Third, we also used hierarchical clustering (R function hclust) and principal component analysis (PCA) to evaluate overall performance of all arrays. Samples were clustered using the R function hclust with the average link function. The distance function used in clustering was defined as 1-|corr(s1, s2)|, where s1 and s2 refer to the expression profiles of two samples. PCA was conducted using the approach of Price et al.40 To combine data from different microarrays, probes (Illumina data) or probesets (Affymetrix data) were linked to the corresponding Entrez gene identifiers from annotation files. We used mean expression if there were multiple probes/probesets for an Entrez gene identifier.
Genotype data
For each study, we excluded samples that were related (π>>0.04), moderate relatedness with many subjects (indicating contamination or poor quality), low call rate (<0.9), inconsistent phenotype-genotype sex and ancestry outliers detected by principal component analysis. We removed SNPs with low call rate (<0.9), low minor allele frequency (MAF <0.01), deviation from Hardy–Weinberg equilibrium (PHWE <1 × 10−8), and allele frequencies inconsistent with the 1000 Genomes Project reference panel (MAF difference <0.07). We imputed genotype dosages using the 1000 Genomes Project reference panel using MaCH-admix.41 MaCH-admix does not require phasing before imputation, which is suitable for small studies. ChrX was imputed in males using the option in MaCH-admix and females were imputed in the same way as autosomes. We retained SNPs with imputation R2⩾0.3 and MAF greater than max (0.05, 5/2N).
Evaluation of covariates
Detailed information about covariate evaluation is in the Supplementary Methods. Briefly, we selected covariates for gene expression in two steps. First, we assessed all covariates by computing the type III sum of squares (SAS, v9.2) that compares a full model containing all covariates to a reduced model excluding the covariate under consideration. The impact of a covariate was quantified by determining the number of genes with a false discovery rate (FDR)42 <0.05. We included an ‘impactful' covariate when ⩾1% of genes met this criterion. Second, we regressed out the selected covariates and performed PCA on the residuals. The final covariate list included impactful covariates from the first step and the significant principal components from the second step. For the genotype data from each study, we excluded outliers that fall apart from HapMap3 CEU subjects. Supplementary Figure S1 depicts genotype PC1 versus genotype PC2 of samples in each study together with HapMap3 samples. To determine the genomic PCs that need to be included for adjustment of population stratification, we employed several methods. First, we examined a scree-plot (Supplementary Figure S2) per study, which shows the proportions of total variance in genotype data explained by various PCs. Second, we tested genome-wide association between each PC as a dependent variable and SNPs as independent variables and calculated the genomic inflation factor43 from each association test. Supplementary Figure S3 shows the contribution of each PC to genomic inflation factor. In all studies, PC1 is clearly the major contributor to λGC. There are far lesser contributions from PC2-5. Third, we evaluated whether inclusion of PC1 is enough or several more PCs may be needed to control for population substratification. For the latter approach, we included PC1-PC5 in eQTL analysis for each study and meta-analyzed eQTLs explained below. The meta-analysis results from the two approaches were very similar. A scatter plot of t-test statistics from the two approaches showed that the magnitude and direction were almost identical (Supplementary Figure S4, Pearson's correlation=0.98). The clustering pattern of significant local SNP–gene eQTLs near transcription start sites remained the same (Supplementary Figure S11). Number of significant eQTLs at varying FDR thresholds reduced marginally as expected (Supplementary Table S3). We present all tables and figures in the manuscript on the basis of eQTL analysis with genomic PC1.
eQTL analysis
We used MatrixEQTL44 to conduct the gene expression linear regression GWAS for each study while controlling for study-specific covariates. We evaluated all ‘local' eQTLs (SNP–gene distance <1 Mb). We did not evaluate ‘distant' eQTLs given the large number of statistical comparisons and consequent low power.30 The local eQTLs for each study were then meta-analyzed using a fixed effects model.45 ChrX eQTL analysis was done separately for males and females in each study and then combined using fixed effects meta- analysis, and male and female results combined using Fisher's test.46 FDR was used to control for multiple comparisons. FDR was computed separately for chr1-22 and chrX as the P-values were based on different models (t-statistics for autosomes and χ2 for chrX). We used sign tests to compare the signs of t-statistics between the five studies. Under the null hypothesis, half of the signs will be the same between two studies. The significance of the observed proportion was evaluated using the binomial distribution.
Enrichment analyses
Common variants identified from GWAS may influence susceptibility to diseases via regulation of gene expression.8,9 We evaluated this broad hypothesis using enrichment analyses.
First, we assessed whether SNPs associated with psychiatric disorders were enriched among genetic variants that were part of a cortical eQTL (that is, SNP–gene pair) using permutation tests.10 Specifically, we evaluated the overlap between eQTLs in human cortex with five psychiatric disorders studied by the Psychiatric Genomics Consortium (PGC): attention-deficit hyperactivity disorder, autism, bipolar disorder, major depressive disorder and schizophrenia (SCZ).5 We obtained results files from the PGC website (https://pgc.unc.edu/Sharing.php#SharingOpp) from a GWAS meta-analysis of these disorders in independent cases and controls.47 There were 1 065 656 GWAS SNPs common to the five PGC results files and our brain eQTL SNPs. We excluded the extended major histocompatibility locus (eMHC, chr6:25–34 Mb) given its high gene density, LD and functionally clustered genes. We compared LD-pruned sets of GWAS SNPs generated via PLINK (—indep-pairwise 100 25 0.8).48 For each disorder, we generated 10 000 randomized SNP sets, each the same size as the original list of associated GWAS SNPs at a given P-value threshold matched on MAF distribution of the original list and sampled without replacement from the null set. For each set, we determined the number of significant eQTL SNPs at FDR threshold of 0.05. These permutations yielded an empirical enrichment P-value, calculated as the proportion of 10 000 randomized sets in which the number of eQTL SNPs exceeds the originally observed number of eQTL SNPs at the FDR threshold. We repeated this analysis for a recent larger SCZ GWAS.49
Second, we evaluated whether genes that were part of a SNP–gene eQTL in brain were enriched for functional roles in biological pathways or similar cellular functions. We evaluated the following gene sets previously associated with SCZ: expert-curated lists of synaptic genes,50 genes encoding postsynaptic density proteins,51 genes encoding the NMDA (N-methyl-D-aspartate) receptor52 and activity-regulated cytoskeleton-associated protein complex,52 genes whose mRNAs interact with FMRP,53 genes encoding components of voltage-gated calcium channels (all CACN* RefSeq genes)49 and genes whose proteins interact with a calcium channel subunit.54 We also evaluated OMIM disease genes,55 genes with an eQTL in peripheral blood from the largest human eQTL study,30 and the human orthologs of genes with local eQTLs in mouse brain.56 We tested for enrichment using a right-sided Fisher's exact test compared with all 17 537 autosomal genes tested in our meta-analysis and for 9855 brain-expressed genes (defined as mean expression greater than the 25% quantile in three of the five studies in this meta-analysis).
Bioinformatic evaluation
We evaluated whether brain eQTLs were enriched for variants implicated in complex diseases using the NHGRI GWAS catalog (http://www.genome.gov/26525384, downloaded September 2013).57 We selected GWAS SNP associations with reported P-values <1 × 10−9 and keeping the SNP-trait association with the smallest P-value. Many local eQTLs span an extended region containing multiple eQTL SNPs with significant correlation. These are not independent associations but result from high LD between associated eQTL SNPs. We summarized these regions using ‘clumping' (that is, the index eQTL SNP with the strongest association plus the genomic range defined by other eQTL SNPs in high LD with the index SNP) for each significant gene.49 We focused on genes with q<0.05 and performed clumping using PLINK to retain eQTL SNPs with r2<0.6 within 500-kb windows (—clump-P1 0.05—clump-P2 0.05—clump-r2 0.6—clump-kb 500). To guard against a falsely inflated intersection rate with the GWAS catalog SNPs, we used q-values rather than P-values as input for clumping and identified GWAS catalog SNPs with reported P-values <1 × 10−9 that were intersecting the clumped regions.
Results
Meta-analysis of eQTL
We first conducted eQTL analyses for each of the five cortical studies. After quality control, sample sizes ranged from 24 to 189 and the numbers of transcripts ranged from 10 038 to 15 857 per study (17 537 genes evaluated at least one study). Supplementary Figures S5–S9 show plots of gene location versus eQTL location. We defined a local eQTL as an SNP–gene eQTL±1 Mb of the transcription start or end sites for a gene and distant otherwise. As expected, local eQTLs tended to have stronger effects than distant eQTLs. Studies with small sample sizes showed much weaker local eQTLs.
We next conducted a meta-analysis of 424 brain samples across five studies to identify regulatory variants influencing gene expression in human cortex (Table 1). As previous eQTL studies14,30,35,58 reported that distant eQTLs are usually weaker than local eQTLs, replicate poorly and require considerably larger sample sizes for reliable detection, we restricted our meta-analysis to the detection of local eQTLs. To evaluate the consistency of effects between studies, we performed sign tests between all pairs of studies (Supplementary Table S1). Sign tests were done in two ways: selecting SNP–gene pairs from the first study with P<1 × 10−5 and evaluating the signs for the same SNP–gene pairs in the second study, and by reversing the procedure. Sign tests for the results of the three studies with N>50 were always highly significant (concordance rates=69–80%, P=1.5 × 10−7 or smaller). More stringent P-value cutoffs gave similar results. The sign tests indicate the general comparability of the five data sets and also show the dramatic effect of increased sample size.
Table 1. Sample summary.
| Study | Subjects | Cortical region | Genes | SNPs | RNA array | DNA array |
|---|---|---|---|---|---|---|
| Colantuoni et al.31 | 56 | Frontal | 14 308 | 7 887 134 | IL Human 49 K Oligo array | IL 650 K |
| Gibbs et al.27 | 124 | Frontal | 12 010 | 7 874 224 | IL HumanRef-8 | IL 550 K |
| GTEx | 24 | Frontal | 13 524 | 7 780 852 | RNA-seq (IL HiSeq 2000) | IL Omni 5 M |
| Myers et al.26 | 189 | Frontal, temporal | 15 857 | 6 391 052 | IL HumanRef-8 | AF 500 K |
| SMRI | 31 | Frontal | 10 038 | 6 779 110 | AF HG-U133a | AF 5.0 |
Abbreviations: AF, Affymetrix; GTEx, Genotype-Tissue Expression project; IL, Illumina; SMRI, Stanley Medical Research Institute; SNP, single-nucleotide polymorphism.
Number of European-ancestry subjects aged ⩾20 after quality control.
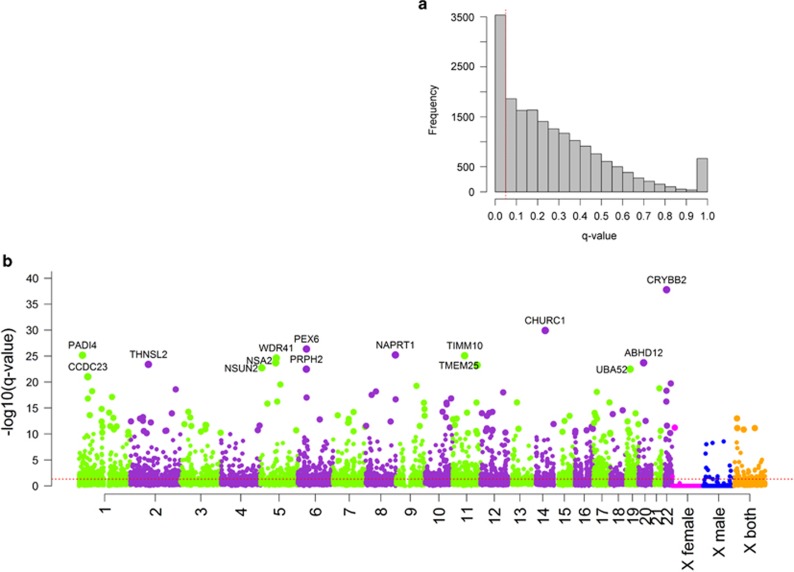
The number of eQTLs at varying FDR thresholds is summarized in Supplementary Table S2. We used a standard FDR threshold <0.05 throughout the paper. At this threshold, there were 176 794 significant autosomal SNP–gene pairs arising from 159 151 SNPs and 3520 genes. This includes more SNP–gene pairs than that found in individual studies (Table 2). As expected, we detected more eQTLs from studies with larger sample sizes. Figure 1a shows distribution of q-values for the most significant association for each gene in autosomes and chromosome X. Figure 1b depicts a Manhattan plot for the primary results, showing the q-value of the most significant SNP for each of 17 546 autosomal genes and 650 genes tested on chrX. The most significant association was in CRYBB2 (rs997872, q=1.75 × 10−38). CRYBB2 encodes βB2-crystallin, which is a structural protein in the ocular lens,59 but recent studies demonstrated that this gene is expressed in several regions of the mammalian brain60 and βB2-crystallin has a role in hippocampal function and behavioral phenotypes.59 The second most significant signal was for CHURC1 (churchill domain containing protein 1; rs10131002, q=1.18 × 10−30). A prior study61 identified CHURC1 as a potential candidate gene for autism. Another top association signal was for NSUN2 (rs567289, q=1.94 × 10−23) which encodes a tRNA methyltransferase and causes an autosomal form of mental retardation (OMIM 611091).62,63
Table 2. Summary of meta-analysis results in autosomes (q<0.05).
| Study | SNP–gene eQTL pairs | Unique SNPs | Unique genes | Genes overlapping with meta-analysis |
|---|---|---|---|---|
| Colantuoni et al.31 | 10 792 | 10 767 | 364 | 250 (7%) |
| Gibbs et al.27 | 110 541 | 103 099 | 2101 | 1279 (36%) |
| GTEx | 0 | 0 | 0 | 0 (0%) |
| Myers et al.26 | 67 534 | 63 768 | 1308 | 864 (25%) |
| SMRI | 1018 | 1018 | 42 | 22 (1%) |
| Meta-analysis | 176 794 | 159 151 | 3520 | 3520 (100%) |
Abbreviations: GTEx, Genotype-Tissue Expression project; SMRI, Stanley Medical Research Institute; SNP, single-nucleotide polymorphism.
Overlapping genes compare an individual study to the 3520 genes with q<0.05 in the meta-analysis.
Figure 1.
Distribution of q-values and Manhattan plot. (a) Distribution of q-values for the most significant association for each gene in autosomes and chromosome X. Red dotted line denotes q-value of 0.05. (b) Manhattan plot of −log10(q) for the most significant association for each of 17 546 autosomal genes and 650 genes in chromosome X. Genes with −log10(q) >20 are highlighted. Red dotted line denotes q=0.05.
The X chromosome is enriched for genes important in brain development, mental retardation and intellectual disability.64, 65, 66, 67, 68 Of 650 chrX genes, there were 1486 significant SNP–gene pairs arising from 1321 SNPs and 64 genes (Figure 1b). Comparison of significant eQTL genes with OMIM revealed that several genes previously implicated in mental retardation showed association signals from sex-combined analysis: OPHN1 (OMIM 300127, q=0.004 with rs618306 ), ARHGEF6 (OMIM 300267, q=0.005 with rs150810369), ACSL4 (OMIM 300157, q=0.01 with X:108198176), SRPX2 (OMIM 300642, q=0.03 with rs35728723), IGBP1 (OMIM 300139, q=0.03 with rs12846068), IL1RAPL1 (OMIM 300206, q=0.04 with rs7471803).
We next asked whether there are genomic regions with unusually higher or lower numbers of eQTLs. We evaluated the relationship between the numbers of genes and eQTLs within each 1 Mb interval of the genome. The MHC region (chr6:31–33 Mb) showed more significant genes than expected (41 of 93 genes were significant eQTLs). On the other hand, none of the 32 genes in chr1:152–153 Mb was an eQTL. This region contains a cluster of late cornified envelope genes, which are specifically expressed in skin.
Characteristics of local SNPs
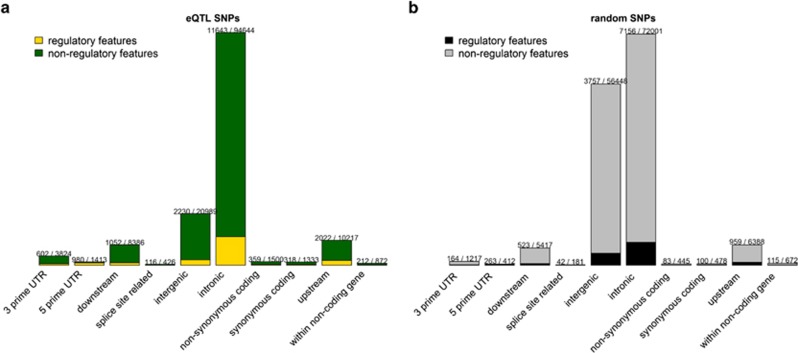
We examined functional consequences of significant SNPs with q<0.05 (143 679 unique SNPs) using the Ensembl Variant Effect Predictor (VEP) tool.69 VEP reports a set of consequence terms defined by the Sequence Ontology (http://www.sequenceontology.org). Detailed information can be found at http://useast.ensembl.org/info/genome/variation/predicted_data.html#consequences. We used options (--most_severe, --per_gene) to output only the most severe consequence per gene. If a gene has multiple transcripts, an SNP was assigned to the transcript with the most severe predicted consequence. If more than one transcript has the same predicted consequence, VEP tool selected the transcript at random. Most significant SNPs were intronic (65.9%) followed by intergenic (14.6%), or upstream (7.1%) or downstream (5.8%) of protein coding sequence. Notably, only a small number of SNPs were exonic (2.0%). As shown in Figure 2a, SNPs in the 5'-untranslated region had the highest overlap with regulatory features (69.4%), and intergenic SNPs had the lowest overlap (10.6%). The overall proportion of variants in a regulatory feature was 13.6%. Next, we asked whether similar functional consequences could be observed from nonsignificant SNPs. We randomly selected the same number (143 679) of MAF-matched SNPs from SNP–gene pairs with q>0.5 and repeated the procedure (Figure 2b). Similar to the significant eQTL SNPs, most of the nonsignificant SNPs were intronic (50.1%), followed by intergenic (39.3%), upstream (4.4%), downstream (3.8%) and exonic regions (0.6%). Notably, the percentage of SNPs in intergenic regions increased by 25%. SNPs in the 5'-untranslated region again had the highest rate (63.8%) of overlap with regulatory features and SNPs in intergenic regions had the lowest rate (6.7%) of overlapping with regulatory features. The overall proportion of variants that fall in a regulatory feature was lower (9.2%) than the set of significant SNPs.
Figure 2.
Predicted functional consequences of local eQTL SNPs. (a) Functional consequences of significant eQTLs (q<0.05, 143 679 unique SNPs) using Ensembl Variant Effect Predictor tool. Each SNP was assigned to the most severe predicted consequence. The ratio on each bar represents number of SNPs with regulatory features divided by number of SNPs in each functional category. (b) Functional consequences of randomly selected, MAF-matched, insignificant eQTLs (q>0.5, 143 679 unique SNPs). eQTL, expression quantitative trait loci; MAF, minor allele frequency; SNP, single-nucleotide polymorphism.
We evaluated whether there were significant differences between the classifications of significant and randomly selected nonsignificant eQTL SNPs. The overall distributions were significantly different between the two sets of SNPs (χ2 P<1 × 10−4). Each functional consequence relative to intergenic also revealed significant difference between the two sets of SNPs (Supplementary Table S4). Odds ratios ranged from 3.5 to 9.2 and all P-values were <1 × 10−4. SNPs in 5'-untranslated region showed the largest difference and were 9.2 times more likely to be significant eQTLs.
Prior studies observed clustering of significant local SNP–gene eQTLs near transcription start sites.11,26,30,70,71 We replicated this pattern (Supplementary Figure S10) which was more evident in the studies with larger samples.
Brain eQTLs and psychiatric disorders
We evaluated enrichment of brain eQTLs in regard to SNPs associated with five psychiatric disorders studied by the PGC and a recent larger SCZ GWAS (Table 3). As shown in prior studies,47 SNPs associated with SCZ and bipolar disorder showed highly significant enrichments for eQTLs. This enrichment in brain eQTLs remained significant regardless of different statistical cutoffs used to generate the SNPs of interest. SNPs associated with attention-deficit hyperactivity disorder, autism and major depressive disorder showed much less enrichment in eQTLs, possibly owing to smaller sample sizes and fewer significant SNPs in the GWAS results.
Table 3. eQTL enrichment analysis of LD-pruned SNPs from PGC GWAS meta-analyses.
| GWAS threshold | PGC autism | PGC ADHD | PGC bipolar | PGC MDD | PGC SCZ | Sweden SCZ |
|---|---|---|---|---|---|---|
| P<0.0001 | 6 (6e−4) | 0 (1.0) | 9 (2e−4) | 0 (1.0) | 9 (0.0047) | 86 (<1e−4) |
| P<0.01 | 91 (0.11) | 61 (0.89) | 153 (<1e−4) | 78 (0.40) | 165 (<1e−4) | 816 (<1e−4) |
| P<0.1 | 719 (0.141) | 630 (0.88) | 580 (<1e−4) | 756 (0.0075) | 982 (<1e−4) | 3639 (<1e−4) |
Abbreviations: ADHD, attention-deficit hyperactivity disorder; eQTL, expression quantitative trait loci; GWAS, genome-wide association study; LD, linkage disequilibrium; MDD, major depressive disorder; PGC, Psychiatric Genomics Consortium; SCZ, schizophrenia; SNP, single-nucleotide polymorphism.
eQTL SNPs with q<0.05 were used for enrichment tests. Each cell gives the number of overlapping SNPs between eQTL SNPs and SNPs at the given threshold for a GWAS data set. The parentheses are empirical P-values obtained after 10 000 permutations with ‘<1e−4' meaning that none of 10 000 simulations yielded an eQTL count greater than the observed count. Due to high LD, the extended MHC region (chr6: 25–34 Mb) was removed. The first five data sets were imputed to HapMap3, and the rightmost column to 1000 Genomes and so the numbers of overlapping SNPs are greater.
To complement the permutation-based enrichment tests, we also applied the Bayesian framework implemented in SHERLOCK72 to the Sweden SCZ GWAS results.49 Briefly, SHERLOCK aligns the genetic architecture of SCZ against eQTL results to evaluate overlap and to summarize the evidence that a given SNP supports a functional role for the gene. A gene showing a high positive Bayes factor supports the evidence that it is more likely to be associated with SCZ via transcriptional regulation. Supplementary Table S5 lists the top predicted genes and their supporting SNPs of SCZ that meet logarithm Bayes factor >4.0 (that is, the posterior probability of the gene being associated with SCZ is >e4 or >55 times more likely). Among 27 predicted genes with logarithm Bayes factor >4.0 (Supplementary Table S5), many are associated with SCZ as expected and also in regions known to be associated with bipolar disorder (ITIH4, GLT8D1, GNL3, NEK4), five major psychiatric disorders (FTSJ2, PCGEM1, C10orf32) and Parkinson's disease (C10orf32). Some of these genes were in high LD regions (for example, chr10:104.5–105.2 Mb and chr3:52.2–53.2 Mb, Supplementary Figures S12 and S13).
Gene set enrichment
Genomic studies of SCZ have implicated biological pathways using multiple types of genomic data (common variation, rare CNVs and rare exonic variation).49,50,52,73,74 We asked whether genes in significant SNP–gene eQTLs in human cortex were enriched for pathways previously implicated in SCZ. Table 4 summarizes the results of enrichment tests using the gene sets selected from the literature. In either all genes or brain-expressed genes, we did not observe enrichment of eQTL genes among any gene sets hypothesized to be associated with SCZ.
Table 4. Enrichment test of eQTL genes (q<0.05) in gene sets.
| Gene set | Genes | Overlap | OR1 (P) | OR2 (P) |
|---|---|---|---|---|
| Implicated in schizophrenia | ||||
| FMRP interactor53 | 780 | 125 | 0.75 (0.99) | 0.72 (0.99) |
| Synaptic functional gene group50 | 947 | 170 | 0.86 (0.96) | 0.82 (0.98) |
| Synaptic signaling pathways50 | 121 | 29 | 1.26 (0.17) | 1.28 (0.18) |
| Calcium channel subunit interactor54 | 189 | 28 | 0.69 (0.98) | 0.68 (0.97) |
| Calcium subunit49 | 24 | 4 | 0.80 (0.74) | 0.62 (0.83) |
| Postsynaptic genes51 | 687 | 113 | 0.78 (0.99) | 0.68 (0.99) |
| NMDA receptor52 | 58 | 9 | 0.73 (0.85) | 0.65 (0.91) |
| ARC52 | 24 | 2 | 0.36 (0.97) | 0.31 (0.98) |
| Other gene sets | ||||
| Nuclear-encoded mitochondrial genes75 | 914 | 257 | 1.60 (1.3 × 10−9) | 1.39 (1.0 × 10−4) |
| Oxidative phosphorylation genes76 | 88 | 24 | 1.50 (0.06) | 1.41 (0.11) |
| Nuclear regulators of mitochondria genes76 | 14 | 2 | 0.66 (0.80) | 0.31 (0.95) |
| OMIM55 | 2914 | 609 | 1.06 (0.12) | 1.15 (0.009) |
| ‘godot' eQTL study in blood (q<0.001)30 | 6662 | 1578 | 1.43 (9.4 × 10−21) | 1.36 (6.9 × 10−11) |
| Significant strain effect in mouse brain diallel56 | 9534 | 1986 | 1.11 (0.003) | 0.90 (0.98) |
Abbreviations: ARC, activity-regulated cytoskeleton; eQTL, expression quantitative trait loci; NMDA, N-methyl-D-aspartate; OMIM, Online Mendelian Inheritance in Man.
For enrichment tests, we used 3520 eQTL genes in autosomes with false discovery rate (FDR) q<0.05. OR1 is odds ratio using all tested autosomal genes (17 537), and OR2 is odds ratio using brain-expressed, autosomal genes (9855). P-values are from right-sided Fisher's exact tests.
We conducted a series of enrichment analyses with the list of genes with significant eQTL evidence (Table 4). First, using functional annotation clustering in DAVID,77 genes with eQTL evidence were very significantly enriched (Fisher's exact test P-value=3.24 × 10−9) for GO gene sets related to multiple mitochondrial gene sets (Supplementary Table S6). To adjust for common biases due to gene size, LD within and between genes, and pathway sizes, we generated a set of independent, nominally associated genomic intervals using clumping implemented in PLINK and then tested it for enrichment in GO gene sets using InRich.78 Analysis was restricted to 4074 GO categories containing genes between 5 and 3000 to account for pathway sizes. We used permutation to get empirical P-values per pathway and to correct for multiple-testing. Multiple GO pathways related to mitochondrial structure and function were ranked as top pathways (Supplementary Table S7). This result is consistent with the DAVID results, indicating that mitochondrial pathways are robust findings regardless of different gene-set enrichment methods. We tested for enrichment in mitochondrial pathways by further analyses using nuclear-encoded mitochondrial genes from MitoCarta (http://www.broadinstitute.org/pubs/MitoCarta),75 autosomal oxidative phosphorylation genes,76 and nuclear-encoded transcriptional regulators of mitochondrial genes.76,79 Of 914 nuclear-encoded mitochondrial genes, 257 genes (28%) overlapped with genes showing significant eQTL evidence. We observed strong enrichment of significant eQTL genes in autosomal mitochondrial genes (odds ratio=1.60, P=1.3 × 10−9 using all genes; odds ratio=1.39, P=1.0 × 10−4 using brain-expressed genes). However, no enrichment was observed for nuclear regulators of mitochondrial genes (P=0.80 using all genes, P=0.95 using brain-expressed genes) and oxidative phosphorylation genes (P=0.06 using all genes, P=0.11 using brain-expressed genes).
Second, genes expressed in multiple tissues tend to have local regulatory elements.80 To evaluate the hypothesis, we compared eQTL genes in peripheral blood30 with genes having local eQTLs in our meta-analysis. We found strong overlap between eQTLs in these two tissues. Of 6662 genes with significant local eQTL evidence in peripheral blood (q<0.001), 1578 genes (24%) overlapped with genes with local eQTL evidence in brain (odds ratio=1.43, P=9.4 × 10−21 using all genes). Restricting to 5261 genes expressed in both tissues, 70.7% of 1193 eQTL genes in brain were eQTL genes in blood (Supplementary Figure S14).
Third, gene regulation might be conserved across species. We compared our brain eQTL genes with their mouse orthologs with local eQTL evidence in a carefully conducted brain RNA-seq study. The enrichment of human brain eQTLs in genes from mouse brain samples was significant (odds ratio=1.11, P=0.003).56
Brain eQTLs, GWAS and OMIM
To assess the biological relevance of brain eQTLs for annotation of variants implicated in human diseases, we compared unique SNPs in autosomes and chrX (after filtering at q<0.05 and keeping the SNP with the smallest q-value if there were multiple SNP–gene pairs) to the NHGRI GWAS catalog.57 We restricted our search to GWAS SNPs with P<1 × 10−9, yielding 2946 SNPs for 471 traits from 869 papers. Of the 2946 SNPs implicated by GWAS, 528 (17.9%) were part of a local eQTL (178 directly associated with 94 traits and 350 SNPs indirectly via a proxy SNP with r2>0.2). The 10 most frequent traits were height (12 SNPs), inflammatory bowel disease (11), Crohn's disease (9), plasma phospholipid levels (6), total cholesterol (5), chronic kidney disease (4), coronary heart disease (4), HDL cholesterol (4), metabolite levels (4) and red blood cell traits (4). We evaluated brain eQTLs that overlap with the SNPs associated with central nervous system-related phenotypes (Table 5), and identified overlap with bipolar disorder, Parkinson's disease and nicotine dependence.
Table 5. eQTL SNP clumping regions and brain diseases from the NHGRI GWAS catalog.
| Region | eQTL SNPs | Index eQTL SNP | eQTL SNP q | Gene | Disease | PMID | GWAS SNP | GWAS P | Other genes |
|---|---|---|---|---|---|---|---|---|---|
| chr2:97351561–97476219 | 84 | rs62152787 | 0.002058 | CNNM4 | BIP | 22182935 | rs2271893 | 2E−10 | hsa-mir-3127, MIR3127, LMAN2L, FER1L5, CNNM4 |
| chr3:52214610–52529899 | 106 | rs9863753 | 1.9E−6 | STAB1 | BIP | 22182935 | rs7618915 | 3E−10 | hsa-mir-135a-1, hsa-let7g, WDR82, TWF2, TNNC1, TLR9, STAB1, SEMA3G, PPM1M, PHF7, NISCH, MIRLET7G, MIR135A1, GLYCTK, DNAH1, BAP1, ALAS1 |
| chr3:52537969–52898427 | 42 | rs11712872 | 2.92E−12 | ITIH3 | AUT, ADHD, BIP, MDD, SCZ | 23453885 | rs2535629 | 3E−12 | TMEM110-MUSTN1, TMEM110, STAB1, SPCS1, SNORD69, SNORD19B, SNORD19, PBRM1, NT5DC2, NEK4, MUSTN1, ITIH3, ITIH4, ITIH1, HBII-210, HBII108B, HBII108, GNL3, GLT8D1, C3orf78 |
| chr6:28162780–28239681 | 12 | rs1531681 | 0.008303 | NKAPL | SCZ | 22037552 | rs1635 | 7E−12 | chr6.tRNA62- SerGCT, ZNF193, ZNF187, ZKSCAN4, NKAPL, LOC222699 |
| chr15:78808235–78926018 | 121 | rs12902493 | 0.000168 | AGPHD1 | ND | 18385739 | rs1051730 | 6E−20 | PSMA4, CHRNB4, CHRNA5, CHRNA3, AGPHD1 |
| chr17:43543075–43848638 | 2 | 17_43543075 | 4.64E−9 | PLEKHM1 | PD | 21292315 | rs2942168 | 1E−28 | hsa-mir-4315-1, PLEKHM1, MIR4315-1, MIR4315-2, MGC57346, LRRC37A4, LOC644172, CRHR1, C17orf69 |
| chr17:43848412–43931907 | 277 | rs116853553 | 1.03E−8 | CRHR1 | PD | 21738487 | rs12185268 | 3E−14 | LOC100128977, IMP5, CRHR1 |
| chr17:44762589–44865603 | 74 | rs199448 | 8.23E−9 | NSF | PD | 19915575 | rs199533 | 1E−14 | WNT3, NSF |
Abbreviations: ADHD, attention-deficit hyperactivity disorder; AUT, autism; BIP, bipolar disorder; eQTL, expression quantitative trait loci; GWAS, genome-wide association study; MDD, major depressive disorder; ND, nicotine dependence; NHGRI, National Human Genome Research Institute; PD, Parkinson's disease; SCZ, schizophrenia; SNP, single-nucleotide polymorphism.
The ‘Gene' column refers to the gene in which an index eQTL SNP was located. ‘Other genes' column shows other named genes in the genomic interval. This table includes GWAS SNP associations with reported P-values <1 × 10−9 from the NHGRI GWAS catalog and keeping the GWAS SNP-trait association with the smallest P-value. The extended MHC region (chr6:25–34 Mb) was excluded.
We compared our eQTL genes to OMIM55 which catalogs genes often containing rare variation with strong effects. We observed significant enrichment of genes with significant brain eQTL evidence in OMIM disease genes (odds ratio=1.15, P=0.009). Mitochondrial complex I deficiency and Leigh syndrome were the second most frequent diseases in our data (FOXRED1, NDUFA2, NDUFA10, NDUFAF1, NDUFAF2, NDUFAF4 and NDUFS2).
Protein–protein interaction
We used DAPPLE81 to evaluate whether genes with strong evidence of local eQTLs connected via protein–protein interactions. Genes with evidence of local eQTLs showed somewhat higher network connectivity (direct P=0.04 and indirect P=0.01). However, many of these genes were in small networks rather than a single network, suggesting that there is no a dominant functional network related to all these genes (Supplementary Figure S15).
Discussion
We performed a meta-analysis of local regulatory variation of 424 postmortem brain samples from five human brain eQTL studies. Our analysis of local eQTLs in this relatively large sample size allowed us to identify more eQTLs than those from individual studies.
Consistent with prior findings, we observed that local regulatory variants tend to occur symmetrically around transcription start sites, and effect was more evident in studies with large sample sizes. Significant eQTLs tended to be near 5'-untranslated regions and intersect with regulatory features. In accordance with previous eQTL studies showing that eQTLs are more likely to overlap with SNPs implicated in GWAS,10 we observed that SNPs associated with SCZ and bipolar disorder were enriched among brain eQTLs. Many brain eQTLs are also associated with central nervous system-related diseases (Table 5).
We compared our results with previous findings from the literature. Myers et al.26 and Liu et al.28 reported significant associations between RPS26 and rs11171739 in prefrontal cortex. Cheung et al.82 reported a significant association between RPS26 and rs2271194 (in high LD with rs11171739) in lymphoblastoid cells. We observed strong associations for both RPS26–rs11171739 (q=7.7 × 10−11) and RPS26–rs2271194 (q=1.3 × 10−10). Another eQTL study of human liver identified a significant RPS26 and rs2292239 relationship and suggested RPS26 as a candidate susceptibility gene for type 1 diabetes.83 We observed a strong correlation for the RPS26–rs2292239 pair (q=5 × 10−5) as well.
There are expression variants that are specific to tissues, cells, anatomical regions and diseases.14,34 However, the substantial overlap (24%) between eQTLs from the largest eQTL study in peripheral blood30 and eQTLs in brain implies that there are many local regulatory variants showing more ubiquitous effects independent of tissue types. Indeed, sample size is a key factor in replicating findings within and between tissues30 may be the most important factor of successful eQTL mapping. Another study that examined eQTLs in blood, adipose tissue and liver reported that about 30% of local eQTLs overlap in those three tissues, suggesting that there are shared expression control mechanisms between tissues.83
We identified a significant number of brain eQTLs that influence the expression of nuclear-encoded genes involved in mitochondrial function and strong evidence of functional clusters related to mitochondrial function (for example, nuclear-encoded mitochondrial genes, P=1.3 × 10−9). Moreover, mitochondrial complex I deficiency genes involved in local eQTL were a frequent overlap. This raises an intriguing possibility, that common genetic variation influences the expression of sets of autosomal genes that influence the number and/or function of mitochondria. Nuclear-encoded autosomal genes (~1000 based on MitoCarta75) and mitochondrial-encoded genes (13 genes in human84) are involved in ATP synthesis, cellular energy metabolism and oxidative phosphorylation, as well as regulation of cellular calcium levels, steroid synthesis, production of free radicals and regulation of apoptosis.85 The central nervous system has a very high metabolic rate because neurons require large amounts of ATP for maintenance of ionic gradients across the cell membranes and for neurotransmission. Neuronal function and survival depend critically on mitochondrial function and oxygen supply.86 Thus, it is conceivable that minor deviations from normal mitochondria functioning can have devastating consequences on the integrity of cells and influence a variety of diseases, including aging,87 cancer,88 metabolic traits,89 neurodegenerative diseases85 and psychiatric disorders.90,91
Although most patients with psychiatric disorders do not have classical mitochondrial diseases caused by mutations of nuclear or mitochondrial DNA, multiple lines of evidences support that impairment in any processes related to normal mitochondria function may be critical in neurobiology of psychiatric disorders.85,91 A study of large, rare CNVs in SCZ observed significant enrichment in gene products localized to mitochondria.92 Impaired neuronal differentiation in hair follicle-derived induced pluripotent stem cells from SCZ cases is associated with mitochondrial dysfunction.93 A recent meta-analysis of autism spectrum disorders suggests an association with mitochondrial dysfunction.94 Mutations and deletions in mitochondrial DNA have been reported to be associated with mood disorders and bipolar disorder.95,96 Postmortem brain samples of bipolar disorder cases showed a pronounced decrease in the expression of nuclear genes regulating oxidative phosphorylation.97 Taken together, gene pathways or networks involving mitochondria function may have an etiological role for some psychiatric disorders.
There are several limitations of this study. First, more data are required. Our sample size was less than that required for confident local eQTL identification.30 Second, this investigation included only normal adult brain samples. Inclusion of data from cases with psychiatric disorders or from earlier developmental stages would likely be informative. Third, although consistent quality control steps were applied, different DNA and RNA platforms across studies may have impacted our findings. To evaluate the impact of between-study heterogeneity, we performed a random-effect meta-analysis using ‘REML' method in metaphor R package (Supplementary File, http://cran.r-project.org/web/packages/metafor/index.html). We observed that the P-values from random-effect model tend to be larger than fixed-effect model. This is not surprising since fixed-effect models are known to produce tighter confidence intervals and more significant P-values than random-effect models in the presence of between-study heterogeneity.98, 99, 100 The genomic control inflation factors for the fixed-effect and random-effect analyses were 1.08 and 0.87, respectively. Top signals from random-effect model and fixed-effect model were quite different. Many significant SNP–gene pairs from fixed-effect model became nonsignificant via random-effect model.101 Small sample sizes, different expression platforms and unknown differences across our studies could possibly introduce such a large variation in effect sizes and thus inflated between-study heterogeneity. We need to be more cautious about interpretation of the fixed-effect results. On the other hand, there can be a large uncertainty in meta-analysis about the presence and the extent of between-study heterogeneity with limited number of studies. It was pointed out that strong inferences about heterogeneity or lack thereof should be avoided.98 Finally, analysis of postmortem human brain tissues face many challenges as we cannot fully control for all potential confounders that might have impacted the integrity of brain expression assessment (for example, antemortem history, medication use, licit or illicit substance use disorders, cause of death or postmortem delay).
Despite these limitations, the eQTLs and pathways identified in this investigation warrant further exploration as potential candidates involved in pathogenesis of psychiatric disorders. Annotating SNPs identified from GWAS of psychiatric disorders with brain eQTL information will be a valuable resource to characterize the functions of causal variants and generate testable hypotheses for the mechanism underlying GWAS findings.
Acknowledgments
We thank Duan Qing and Yun Li for the help and support for genotype imputation using MaCH-Admix. We also thank Wei Sun for the help in eQTL analysis. Funding for this project was from the US National Institutes of Health who had no role in the design, execution, analysis and manuscript preparation. This work was funded by grant #08R-1959 (VV) from the Stanley Medical Research Institute and by grant #R01MH097283 (EvdO) from the National Institute of Mental Health.
Dr Sullivan was on the SAB of Expression Analysis (Durham, NC, USA). The remaining authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on the Translational Psychiatry website (http://www.nature.com/tp)
Supplementary Material
References
- World Health Organization . The Global Burden of Disease: 2004 Update. WHO Press: Geneva, Switzerland; 2008. [Google Scholar]
- Collins PY, Patel V, Joestl SS, March D, Insel TR, Daar AS, et al. Grand challenges in global mental health. Nature. 2011;475:27–30. doi: 10.1038/475027a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan PF, Daly MJ, O'Donovan M. Genetic architectures of psychiatric disorders: the emerging picture and its implications. Nat Rev Genet. 2012;13:537–551. doi: 10.1038/nrg3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Psychiatric GWAS Consortium Bipolar Disorder Working Group Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nat Genet. 2011;43:977–983. doi: 10.1038/ng.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Psychiatric GWAS Consortium Steering Committee A framework for interpreting genomewide association studies of psychiatric disorders. Mol Psychiatry. 2009;14:10–17. doi: 10.1038/mp.2008.126. [DOI] [PubMed] [Google Scholar]
- Psychiatric GWAS Consortium Coordinating Committee, Cichon S, Craddock N, Daly M, Faraone SV, Gejman PV, et al. Genome-wide association studies: history, rationale, and prospects for psychiatric disorders. Am J Psychiatry. 2009;166:540–546. doi: 10.1176/appi.ajp.2008.08091354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurano MT, Humbert R, Rynes E, Thurman RE, Haugen E, Wang H, et al. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;237:1190–1195. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cookson W, Liang L, Abecasis G, Moffatt M, Lathrop M. Mapping complex disease traits with global gene expression. Nat Rev Genet. 2009;10:184–194. doi: 10.1038/nrg2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majewski J, Pastinen T. The study of eQTL variations by RNA-seq: from SNPs to phenotypes. Trends Genet. 2011;27:72–79. doi: 10.1016/j.tig.2010.10.006. [DOI] [PubMed] [Google Scholar]
- Nicolae DL, Gamazon E, Zhang W, Duan S, Dolan ME, Cox NJ. Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLoS Genet. 2010;6:e1000888. doi: 10.1371/journal.pgen.1000888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stranger BE, Montgomery SB, Dimas AS, Parts L, Stegle O, Ingle CE, et al. Patterns of cis regulatory variation in diverse human populations. PLoS Genet. 2012;8:e1002639. doi: 10.1371/journal.pgen.1002639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundberg E, Small KS, Hedman AK, Nica AC, Buil A, Keildson S, et al. Mapping cis- and trans-regulatory effects across multiple tissues in twins. Nat Genet. 2012;44:1084–1089. doi: 10.1038/ng.2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lango Allen H, Estrada K, Lettre G, Berndt SI, Weedon MN, Rivadeneira F, et al. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature. 2010;467:832–838. doi: 10.1038/nature09410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emilsson V, Thorleifsson G, Zhang B, Leonardson AS, Zink F, Zhu J, et al. Genetics of gene expression and its effect on disease. Nature. 2008;452:423–428. doi: 10.1038/nature06758. [DOI] [PubMed] [Google Scholar]
- de Jong S, van Eijk KR, Zeegers DW, Strengman E, Janson E, Veldink JH, et al. Expression QTL analysis of top loci from GWAS meta-analysis highlights additional schizophrenia candidate genes. Eur J Hum Genet. 2012;20:1004–1008. doi: 10.1038/ejhg.2012.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fransen K, Visschedijk MC, van Sommeren S, Fu JY, Franke L, Festen EA, et al. Analysis of SNPs with an effect on gene expression identifies UBE2L3 and BCL3 as potential new risk genes for Crohn's disease. Hum Mol Genet. 2010;19:3482–3488. doi: 10.1093/hmg/ddq264. [DOI] [PubMed] [Google Scholar]
- Luo R, Sanders SJ, Tian Y, Voineagu I, Huang N, Chu SH, et al. Genome-wide Transcriptome Profiling Reveals the Functional Impact of Rare De Novo and Recurrent CNVs in Autism Spectrum Disorders. Am J Hum Genet. 2012;91:38–55. doi: 10.1016/j.ajhg.2012.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speliotes EK, Willer CJ, Berndt SI, Monda KL, Thorleifsson G, Jackson AU, et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet. 2010;42:937–948. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeller T, Wild P, Szymczak S, Rotival M, Schillert A, Castagne R, et al. Genetics and beyond—the transcriptome of human monocytes and disease susceptibility. PLoS One. 2010;5:e10693. doi: 10.1371/journal.pone.0010693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamazon ER, Huang RS, Cox NJ, Dolan ME. Chemotherapeutic drug susceptibility associated SNPs are enriched in expression quantitative trait loci. Proc Natl Acad Sci USA. 2010;107:9287–9292. doi: 10.1073/pnas.1001827107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurman RE, Rynes E, Humbert R, Vierstra J, Maurano MT, Haugen E, et al. The accessible chromatin landscape of the human genome. Nature. 2012;489:75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degner JF, Pai AA, Pique-Regi R, Veyrieras JB, Gaffney DJ, Pickrell JK, et al. DNase I sensitivity QTLs are a major determinant of human expression variation. Nature. 2012;482:390–394. doi: 10.1038/nature10808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou F, Chai HS, Younkin CS, Allen M, Crook J, Pankratz VS, et al. Brain expression genome-wide association study (eGWAS) identifies human disease-associated variants. PLoS Genet. 2012;8:e1002707. doi: 10.1371/journal.pgen.1002707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton PR, Clayton DG, Cardon LR, Craddock N, Deloukas P, Duncanson A, et al. Association scan of 14,500 nonsynonymous SNPs in four diseases identifies autoimmunity variants. Nat Genet. 2007;39:1329–1337. doi: 10.1038/ng.2007.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinzen EL, Ge D, Cronin KD, Maia JM, Shianna KV, Gabriel WN, et al. Tissue-specific genetic control of splicing: implications for the study of complex traits. PLoS Biol. 2008;6:e1. doi: 10.1371/journal.pbio.1000001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers AJ, Gibbs JR, Webster JA, Rohrer K, Zhao A, Marlowe L, et al. A survey of genetic human cortical gene expression. Nat Genet. 2007;39:1494–1499. doi: 10.1038/ng.2007.16. [DOI] [PubMed] [Google Scholar]
- Gibbs JR, van der Brug MP, Hernandez DG, Traynor BJ, Nalls MA, Lai SL, et al. Abundant quantitative trait loci exist for DNA methylation and gene expression in human brain. PLoS Genet. 2010;6:e1000952. doi: 10.1371/journal.pgen.1000952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Cheng L, Badner JA, Zhang D, Craig DW, Redman M, et al. Whole-genome association mapping of gene expression in the human prefrontal cortex. Mol Psychiatry. 2010;15:779–784. doi: 10.1038/mp.2009.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster JA, Gibbs JR, Clarke J, Ray M, Zhang W, Holmans P, et al. Genetic control of human brain transcript expression in Alzheimer disease. Am J Hum Genet. 2009;84:445–458. doi: 10.1016/j.ajhg.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright FA, Sullivan PF, Brooks AI, Zou F, Sun W, Xia K, et al. Heritability and genomics of gene expression in peripheral blood. Nat Genet. 2014;46:430–437. doi: 10.1038/ng.2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colantuoni C, Lipska BK, Ye T, Hyde TM, Tao R, Leek JT, et al. Temporal dynamics and genetic control of transcription in the human prefrontal cortex. Nature. 2011;478:519–523. doi: 10.1038/nature10524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrey EF, Barci BM, Webster MJ, Bartko JJ, Meador-Woodruff JH, Knable MB. Neurochemical markers for schizophrenia, bipolar disorder, and major depression in postmortem brains. Biol Psychiatry. 2005;57:252–260. doi: 10.1016/j.biopsych.2004.10.019. [DOI] [PubMed] [Google Scholar]
- Torrey EF, Webster M, Knable M, Johnston N, Yolken RH. The stanley foundation brain collection and neuropathology consortium. Schizophr Res. 2000;44:151–155. doi: 10.1016/S0920-9964(99)00192-9. [DOI] [PubMed] [Google Scholar]
- GTEx Consortium The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia K, Shabalin AA, Huang S, Madar V, Zhou Y, Wang W, et al. seeQTL: a searchable database for human eQTLs. Bioinformatics. 2011;28:451–452. doi: 10.1093/bioinformatics/btr678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altshuler DM, Gibbs RA, Peltonen L, Dermitzakis E, Schaffner SF, Yu F, et al. Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467:52–58. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durbin RM, Abecasis GR, Altshuler DL, Auton A, Brooks LD, Gibbs RA, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramasamy A, Trabzuni D, Gibbs JR, Dillman A, Hernandez DG, Arepalli S, et al. Resolving the polymorphism-in-probe problem is critical for correct interpretation of expression QTL studies. Nucleic Acids Res. 2013;41:e88. doi: 10.1093/nar/gkt069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- Liu EY, Li M, Wang W, Li Y. MaCH-admix: genotype imputation for admixed populations. Genet Epidemiol. 2013;37:25–37. doi: 10.1002/gepi.21690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57:289–300. [Google Scholar]
- Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- Shabalin AA. Matrix eQTL: ultra fast eQTL analysis via large matrix operations. Bioinformatics. 2012;28:1353–1358. doi: 10.1093/bioinformatics/bts163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft P, Zeggini E, Ioannidis JP. Replication in genome-wide association studies. Stat Sci. 2009;24:561–573. doi: 10.1214/09-STS290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher RA. On the interpretation of Chi-square from contingency tables and the calculation of P. J R Stat Soc. 1922;85:87–94. [Google Scholar]
- Cross-Disorder Group of the Psychiatric Genomics Consortium Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet. 2013;381:1371–1379. doi: 10.1016/S0140-6736(12)62129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira M, Bender D, et al. PLINK: a toolset for whole-genome association and population-based linkage analysis. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripke S, O'Dushlaine C, Chambert K, Moran JL, Kahler AK, Akterin S, et al. Genome-wide association analysis identifies 13 new risk loci for schizophrenia. Nat Genet. 2013;45:1150–1159. doi: 10.1038/ng.2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruano D, Abecasis GR, Glaser B, Lips ES, Cornelisse LN, de Jong AP, et al. Functional gene group analysis reveals a role of synaptic heterotrimeric G proteins in cognitive ability. Am J Hum Genet. 2010;86:113–125. doi: 10.1016/j.ajhg.2009.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croning MD, Marshall MC, McLaren P, Armstrong JD, Grant SG. G2Cdb: the Genes to Cognition database. Nucleic Acids Res. 2009;37:D846–D851. doi: 10.1093/nar/gkn700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirov G, Pocklington AJ, Holmans P, Ivanov D, Ikeda M, Ruderfer D, et al. De novo CNV analysis implicates specific abnormalities of postsynaptic signalling complexes in the pathogenesis of schizophrenia. Mol Psychiatry. 2012;17:142–153. doi: 10.1038/mp.2011.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JC, Van Driesche SJ, Zhang C, Hung KY, Mele A, Fraser CE, et al. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell. 2011;146:247–261. doi: 10.1016/j.cell.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller CS, Haupt A, Bildl W, Schindler J, Knaus HG, Meissner M, et al. Quantitative proteomics of the Cav2 channel nano-environments in the mammalian brain. Proc Natl Acad Sci USA. 2010;107:14950–14957. doi: 10.1073/pnas.1005940107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKusick VA. Mendelian inheritance in man and its online version, OMIM. Am J Hum Genet. 2007;80:588–604. doi: 10.1086/514346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowley JJ, Zhabotynsky V, Sun W, Huang S, Pakatci IK, Kim Y, et al. Pervasive allelic imbalance revealed by allele-specific gene expression in highly divergent mouse crosses(Submitted). [DOI] [PMC free article] [PubMed]
- Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci USA. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon AL, Liang L, Moffatt MF, Chen W, Heath S, Wong KC, et al. A genome-wide association study of global gene expression. Nat Genet. 2007;39:1202–1207. doi: 10.1038/ng2109. [DOI] [PubMed] [Google Scholar]
- Sun M, Holter SM, Stepan J, Garrett L, Genius J, Kremmer E, et al. Crybb2 coding for betaB2-crystallin affects sensorimotor gating and hippocampal function. Mamm Genome. 2013;24:333–348. doi: 10.1007/s00335-013-9478-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganguly K, Favor J, Neuhauser-Klaus A, Sandulache R, Puk O, Beckers J, et al. Novel allele of crybb2 in the mouse and its expression in the brain. Invest Ophthalmol Vis Sci. 2008;49:1533–1541. doi: 10.1167/iovs.07-0788. [DOI] [PubMed] [Google Scholar]
- Griswold AJ, Ma D, Sacharow SJ, Robinson JL, Jaworski JM, Wright HH, et al. A de novo 1.5 Mb microdeletion on chromosome 14q23.2-23.3 in a patient with autism and spherocytosis. Autism Res. 2011;4:221–227. doi: 10.1002/aur.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan MA, Rafiq MA, Noor A, Hussain S, Flores JV, Rupp V, et al. Mutation in NSUN2, which encodes an RNA methyltransferase, causes autosomal-recessive intellectual disability. Am J Hum Genet. 2012;90:856–863. doi: 10.1016/j.ajhg.2012.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abbasi-Moheb L, Mertel S, Gonsior M, Nouri-Vahid L, Kahrizi K, Cirak S, et al. Mutations in NSUN2 cause autosomal-recessive intellectual disability. Am J Hum Genet. 2012;90:847–855. doi: 10.1016/j.ajhg.2012.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Maldergem L, Hou Q, Kalscheuer VM, Rio M, Doco-Fenzy M, Medeira A, et al. Loss of function of KIAA2022 causes mild to severe intellectual disability with an autism spectrum disorder and impairs neurite outgrowth. Hum Mol Genet. 2013;22:3306–3314. doi: 10.1093/hmg/ddt187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee NR, Wallace GL, Adeyemi EI, Lopez KC, Blumenthal JD, Clasen LS, et al. Dosage effects of X and Y chromosomes on language and social functioning in children with supernumerary sex chromosome aneuploidies: implications for idiopathic language impairment and autism spectrum disorders. J Child Psychol Psychiatry. 2012;53:1072–1081. doi: 10.1111/j.1469-7610.2012.02573.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najm J, Horn D, Wimplinger I, Golden JA, Chizhikov VV, Sudi J, et al. Mutations of CASK cause an X-linked brain malformation phenotype with microcephaly and hypoplasia of the brainstem and cerebellum. Nat Genet. 2008;40:1065–1067. doi: 10.1038/ng.194. [DOI] [PubMed] [Google Scholar]
- Ropers HH, Hamel BC. X-linked mental retardation. Nat Rev Genet. 2005;6:46–57. doi: 10.1038/nrg1501. [DOI] [PubMed] [Google Scholar]
- Bassani S, Zapata J, Gerosa L, Moretto E, Murru L, Passafaro M. The neurobiology of X-linked intellectual disability. Neuroscientist. 2013;19:541–552. doi: 10.1177/1073858413493972. [DOI] [PubMed] [Google Scholar]
- McLaren W, Pritchard B, Rios D, Chen Y, Flicek P, Cunningham F. Deriving the consequences of genomic variants with the Ensembl API and SNP effect predictor. Bioinformatics. 2010;26:2069–2070. doi: 10.1093/bioinformatics/btq330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stranger BE, Forrest MS, Dunning M, Ingle CE, Beazley C, Thorne N, et al. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science. 2007;315:848–853. doi: 10.1126/science.1136678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stranger BE, Nica AC, Forrest MS, Dimas A, Bird CP, Beazley C, et al. Population genomics of human gene expression. Nat Genet. 2007;39:1217–1224. doi: 10.1038/ng2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Fuller CK, Song Y, Meng Q, Zhang B, Yang X, et al. Sherlock: detecting gene-disease associations by matching patterns of expression QTL and GWAS. Am J Hum Genet. 2013;92:667–680. doi: 10.1016/j.ajhg.2013.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lips ES, Cornelisse LN, Toonen RF, Min JL, Hultman CM, Holmans PA, et al. Functional gene group analysis identifies synaptic gene groups as risk factor for schizophrenia. Mol Psychiatry. 2012;17:996–1006. doi: 10.1038/mp.2011.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell SM, Moran JL, Fromer M, Ruderfer D, Solovieff N, Roussos P, et al. A polygenic burden of rare disruptive mutations in schizophrenia. Nature. 2014;506:185–190. doi: 10.1038/nature12975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagliarini DJ, Calvo SE, Chang B, Sheth SA, Vafai SB, Ong SE, et al. A mitochondrial protein compendium elucidates complex I disease biology. Cell. 2008;134:112–123. doi: 10.1016/j.cell.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segre AV, Groop L, Mootha VK, Daly MJ, Altshuler D. Common inherited variation in mitochondrial genes is not enriched for associations with type 2 diabetes or related glycemic traits. PLoS Genet. 2010;6:8. doi: 10.1371/journal.pgen.1001058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Tan Q, Kir J, Liu D, Bryant D, et al. DAVID Bioinformatics Resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res. 2007;35:W169–W175. doi: 10.1093/nar/gkm415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee P, O'Dushlaine C, Thomas B, Purcell S. INRICH: interval-based enrichment analysis for genome-wide association studies. Bioinformatics. 2012;28:1797–1799. doi: 10.1093/bioinformatics/bts191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly DP, Scarpulla RC. Transcriptional regulatory circuits controlling mitochondrial biogenesis and function. Genes Dev. 2004;18:357–368. doi: 10.1101/gad.1177604. [DOI] [PubMed] [Google Scholar]
- Schadt EE, Woo S, Hao K. Bayesian method to predict individual SNP genotypes from gene expression data. Nat Genet. 2012;44:603–608. doi: 10.1038/ng.2248. [DOI] [PubMed] [Google Scholar]
- Rossin EJ, Lage K, Raychaudhuri S, Xavier RJ, Tatar D, Benita Y, et al. Proteins encoded in genomic regions associated with immune-mediated disease physically interact and suggest underlying biology. PLoS Genet. 2011;7:e1001273. doi: 10.1371/journal.pgen.1001273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung VG, Spielman RS, Ewens KG, Weber TM, Morley M, Burdick JT. Mapping determinants of human gene expression by regional and genome-wide association. Nature. 2005;437:1365–1369. doi: 10.1038/nature04244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schadt EE, Molony C, Chudin E, Hao K, Yang X, Lum PY, et al. Mapping the genetic architecture of gene expression in human liver. PLoS Biol. 2008;6:e107. doi: 10.1371/journal.pbio.0060107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnery PF. Searching for nuclear-mitochondrial genes. Trends Genet. 2003;19:60–62. doi: 10.1016/s0168-9525(02)00030-6. [DOI] [PubMed] [Google Scholar]
- Manji H, Kato T, Di Prospero NA, Ness S, Beal MF, Krams M, et al. Impaired mitochondrial function in psychiatric disorders. Nat Rev Neurosci. 2012;13:293–307. doi: 10.1038/nrn3229. [DOI] [PubMed] [Google Scholar]
- Kann O, Kovacs R. Mitochondria and neuronal activity. Am J Physiol Cell Physiol. 2007;292:C641–C657. doi: 10.1152/ajpcell.00222.2006. [DOI] [PubMed] [Google Scholar]
- Yankner BA, Lu T, Loerch P. The aging brain. Annu Rev Pathol. 2008;3:41–66. doi: 10.1146/annurev.pathmechdis.2.010506.092044. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- Knoll N, Jarick I, Volckmar AL, Klingenspor M, Illig T, Grallert H, et al. Gene set of nuclear-encoded mitochondrial regulators is enriched for common inherited variation in obesity. PLoS One. 2013;8:e55884. doi: 10.1371/journal.pone.0055884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Shachar D. Mitochondrial dysfunction in schizophrenia: a possible linkage to dopamine. J Neurochem. 2002;83:1241–1251. doi: 10.1046/j.1471-4159.2002.01263.x. [DOI] [PubMed] [Google Scholar]
- Shao L, Martin MV, Watson SJ, Schatzberg A, Akil H, Myers RM, et al. Mitochondrial involvement in psychiatric disorders. Ann Med. 2008;40:281–295. doi: 10.1080/07853890801923753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szatkiewicz JP, O'Dushlaine C, Chen G, Chambert K, Moran JL, Neale BM, et al. Copy number variation in schizophrenia in Sweden. Mol Psychiatry. 2014;19:762–773. doi: 10.1038/mp.2014.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robicsek O, Karry R, Petit I, Salman-Kesner N, Muller FJ, Klein E, et al. Abnormal neuronal differentiation and mitochondrial dysfunction in hair follicle-derived induced pluripotent stem cells of schizophrenia patients. Mol Psychiatry. 2013;18:1067–1076. doi: 10.1038/mp.2013.67. [DOI] [PubMed] [Google Scholar]
- Rossignol DA, Frye RE. Mitochondrial dysfunction in autism spectrum disorders: a systematic review and meta-analysis. Mol Psychiatry. 2012;17:290–314. doi: 10.1038/mp.2010.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siciliano G, Tessa A, Petrini S, Mancuso M, Bruno C, Grieco GS, et al. Autosomal dominant external ophthalmoplegia and bipolar affective disorder associated with a mutation in the ANT1 gene. Neuromuscul Disord. 2003;13:162–165. doi: 10.1016/s0960-8966(02)00221-3. [DOI] [PubMed] [Google Scholar]
- Bergemann ER, Boles RG. Maternal inheritance in recurrent early-onset depression. Psychiatr Genet. 2010;20:31–34. doi: 10.1097/YPG.0b013e3283351153. [DOI] [PubMed] [Google Scholar]
- Konradi C, Eaton M, MacDonald ML, Walsh J, Benes FM, Heckers S. Molecular evidence for mitochondrial dysfunction in bipolar disorder. Arch Gen Psychiatry. 2004;61:300–308. doi: 10.1001/archpsyc.61.3.300. [DOI] [PubMed] [Google Scholar]
- Kavvoura FK, Ioannidis JP. Methods for meta-analysis in genetic association studies: a review of their potential and pitfalls. Hum Genet. 2008;123:1–14. doi: 10.1007/s00439-007-0445-9. [DOI] [PubMed] [Google Scholar]
- Ioannidis JP, Patsopoulos NA, Evangelou E. Uncertainty in heterogeneity estimates in meta-analyses. BMJ. 2007;335:914–916. doi: 10.1136/bmj.39343.408449.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioannidis JP, Patsopoulos NA, Evangelou E. Heterogeneity in meta-analyses of genome-wide association investigations. PLoS One. 2007;2:e841. doi: 10.1371/journal.pone.0000841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21:1539–1558. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.