Abstract
Matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) mass spectrometry (MS) represents a revolution in routine pathogen identification in clinical microbiology laboratories. A MALDI-TOF MS was introduced to tropical Africa in the clinical microbiology laboratory of the Hôpital Principal de Dakar (Senegal) and used for routine pathogen identification. Using MS, 2,429 bacteria and fungi isolated from patients were directly assayed, leading to the identification of 2,082 bacteria (85.7%) and 206 fungi (8.5%) at the species level, 109 bacteria (4.5%) at the genus level, and 16 bacteria (0.75%) at the family level. Sixteen isolates remained unidentified (0.75%). Escherichia coli was the most prevalent species (25.8%) followed by Klebsiella pneumoniae (14.8%), Streptococcus agalactiae (6.2%), Acinetobacter baumannii (6.1%), Pseudomonas aeruginosa (5.9%), and Staphylococcus aureus (5.9%). MALDI-TOF MS has also enabled the detection of rare bacteria and fungi. MALDI-TOF MS is a powerful tool for the identification of bacterial and fungal species involved in infectious diseases in tropical Africa.
Introduction
The routine identification of bacteria and fungi by matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) mass spectrometry (MS) became prevalent 5 years ago and represents a revolution in clinical microbiology laboratories.1–3 This technique enables the identification of bacteria and fungi in less than 1 hour starting with pure culture without a priori knowledge of the types of microorganisms in the sample. MALDI-TOF MS is becoming a powerful tool for routine identification, replacing Gram staining and all fastidious biochemical identifications.4 The high cost of MALDI-TOF instruments, the limitations of existing bioinformatics tools, and the lack of convenient preparations of the required chemicals previously limited the development of this technology,5 but more recently, the reduction in the cost of the instrument has facilitated access to this technology.5 Thus, its use has become widespread in many clinical laboratories, first primarily in Europe and Asia.1,2,5–11
Currently, the specific identification of microorganisms in Africa raises several issues, including that there are no regulations governing pathogen identification in many countries. Although culturing capabilities are available in major hospitals in Africa, performance limitations of the biochemical identification methods may still be encountered. When biochemical tests are available, they can still be laborious and difficult to interpret, and they can lead to poor identifications. A series of standardized and miniaturized biochemical tests associated with a database (by numerical identification) can be used to make identification easier and more accurate; however, this approach requires many kits (for Enterobacteriaceae, non-Enterobacteriaceae Gram-negative bacilli, Staphylococcus, Streptocococcus, and others) along with other dedicated reagents. Kits and reagents should, furthermore, be stored under specific conditions and have expiration dates. Reagent supply issues are frequently associated with potential problems or backorders. Furthermore, biochemical methods are frequently time-consuming, often require knowledge about the type of microorganism being tested, and fail to accurately identify several microorganisms.1,4,12
Herein, we implemented MALDI-TOF MS in a clinical microbiology laboratory in an African hospital (Hôpital Principal de Dakar [Principal Hospital of Dakar]) in Dakar, Senegal and evaluated its potential for the reliable and rapid identification of common microorganisms.
Materials and Methods
Constraints on the acquisition of MALDI-TOF MS instrumentation in Africa.
Funding for the acquisition of MALDI-TOF MS instrumentation.
The current cost for a MALDI-TOF MS instrument is estimated to be between approximately 100,000 and 200,000 Euros.5 The acquisition of the MALDI-TOF MS instrument (VITEK MS RUO; bioMérieux, Marcy l'Etoile, France) in use in Dakar was supported by the Méditerranée Infection Hospital–University Institute (IHU Méditerranée Infection; http://www.mediterranee-infection.com/), the Research Institute for Development (IRD; http://www.ird.fr/), and the French Ministry of Foreign Affairs. Created in 2012, the aim of IHU Méditerranée Infection is to fight infectious diseases on a global scale. Since 2007, the Infectiopôle South Foundation, a department of the IHU, promotes north–south trade and the coordination of scientific projects in the field of research on infectious diseases, including financial support for foreign students from the south. IRD is a public French organization involved in research with and for southern countries. Through its research, training, and innovation in partnership, IRD is involved in work in more than 50 countries in Africa, the Mediterranean area, Asia, Latin America, and French overseas territories in other regions.
Cooperation agreement and convention.
The Principal Hospital of Dakar (HPD; http://www.hopitalprincipal.sn/) is a public health hospital with special status as a military teaching hospital in Senegal. It has 420 beds and participates in the care of Senegalese patients and other patients from the surrounding area. The Clinical Microbiology Laboratory at the hospital is open 24 hours per day throughout the year. The MALDI-TOF MS was installed in this laboratory. A convention was signed between the HPD, IRD, and IHU Méditerranée Infection. The cooperation agreement stated that the instrument belongs to the HPD. In return, the hospital must ensure the recruitment of staff responsible for operating the MALDI-TOF MS instrument, provide suitable premises for its installation, provide open access to research programs of the IRD, and help other hospitals in the area to identify microorganisms.
Constraints on the installation of the MALDI-TOF MS instrument in Africa.
Technical constraints.
The chemical matrix, which is a unique reagent required for MALDI-TOF MS, must be stored at +4°C when purchased or prepared on the day of use and stored at room temperature. The room containing the MALDI-TOF MS instrument must be thermally isolated, air-conditioned, and protected from insects and dust. Electricity must be supplied continuously. The instrument as well as three computers must each be equipped with inverters in case of voltage dips or brief power outages. To prevent power failure, an electric generator must be provided. The MALDI-TOF MS unit was supplied with several spare parts. Maintenance should be performed one time per year; the first annual maintenance was performed by an engineer of bioMérieux, and it was supported by bioMérieux.
Human constraints.
Local personnel operating the MALDI-TOF MS instrument should be specifically trained to use the instrument. Before the initiation of the project, a local operator (B.S.-B.), a PhD student, was trained to prepare target slides, use an MALDI-TOF MS instrument (Bruker Daltonik, Wissembourg, France), and analyze results during a 6-month period in the IHU Méditerranée Infection. In July of 2012, B.S.-B. (recruited then by HPD) and other local operators (B.F., C.I.L., and M.A.-L.) were trained to use the VITEK MS instrument specifically in a 4-day course in Dakar given by two engineers from bioMérieux. After this training course, all were capable of performing MALDI-TOF MS autonomously. A follow-up review of this training was conducted in November of 2012.
Identification of microorganisms using MALDI-TOF MS in Africa.
Bacterial and fungal isolates.
Fresh isolates were obtained from 2,640 specimens in the course of routine clinical work in the HPD clinical laboratory and tested over 10 months during the study period (August of 2012 to May of 2013). All isolates recovered from blood, cerebrospinal fluid, pus, biopsies, the respiratory tract, the urogenital tract, wounds, stool specimens, and devices were prospectively included in the study. The isolates were recovered after the inoculation of clinical specimens on 5% horse blood Mueller–Hinton agar, trypticase soy agar, and MacConkey agar media. Sabouraud agar media was inoculated when required. All media were prepared in the laboratory. In all cases, the cultures were incubated under standard conditions for a minimum of 18 hours at 35–37°C in ambient air with either CO2 enrichment or in anaerobic atmospheres.
To assess the accuracy of the VITEK MS RUO system for routine bacterial identification, we evaluated 93 strains, including all of the most current detected bacteria in a clinical microbiology laboratory; these bacteria had previously been isolated from patients and identified using a Bruker BioTyper in our laboratory in Marseille, France (Supplemental Table 1). A 100% concordance in identification was observed, allowing us to use the VITEK MS RUO for diagnostic purposes.
MALDI-TOF MS analysis.
The isolated colonies were deposited in a single well of a disposable, barcode-labeled target slide (VITEK MS-DS) using a 1.0-μL loop, then overlaid with 1.0 μL of a saturated solution of α-cyano-4-hydroxycinnamic acid matrix (VITEK MS-CHCA; bioMérieux), and air-dried. If the presence of fungi was suspected, 1 μL formic acid solution (VITEK MS-FA; bioMérieux) was first added. Two spots were prepared for each isolated colony. For instrument calibration, an Escherichia coli reference strain (Lyfocults Escherichia coli ATCC 8739; bioMérieux) was transferred to designated wells on the target slide using the procedure described above. For quality control purposes, positive controls (E. coli strains) were analyzed in each assay.
The Biotyper software compared the protein profile of the microorganisms obtained from the Saramis database, version 4.0 (bioMérieux). The Saramis software color-codes identification results (by default) according to confidence levels as follows: 99.9%, dark green; 99.8–90.0%, light green; 89.9–85.0%, yellow; and 84.9–70.0%, white. Identification results obtained between 70.0% and 99.9% confidence were considered to be correct identifications at the genus and species levels.
Results
Overall, 2,429 bacteria and fungi were isolated from 2,640 specimens received in the laboratory (Table 1) and directly tested using MALDI-TOF MS, leading to the identification of 2,082 bacteria (85.7%) and 206 fungi (8.5%) at the species level, 109 bacteria (4.5%) at the genus level, and 16 bacteria (0.75%) at the family level. Sixteen isolates were not identified (16 of 2,429; 0.65%).
Table 1.
Distribution of 2,640 specimens analyzed at the Principal Hospital, Dakar
| Specimens | Number | Percentage |
|---|---|---|
| Urine | 979 | 37.1 |
| Pus | 471 | 17.9 |
| Vagina | 381 | 14.4 |
| Blood | 289 | 10.9 |
| Respiratory tract | 194 | 7.4 |
| Stomach | 113 | 4.3 |
| Peripheral devices (catheters, probes, and others) | 72 | 2.7 |
| Feces | 61 | 2.3 |
| Genitalia (other than vagina) | 38 | 1.5 |
| Puncture fluid | 13 | 0.5 |
| Other | 29 | 1 |
Accurate identification at the species level.
Bacterial identification.
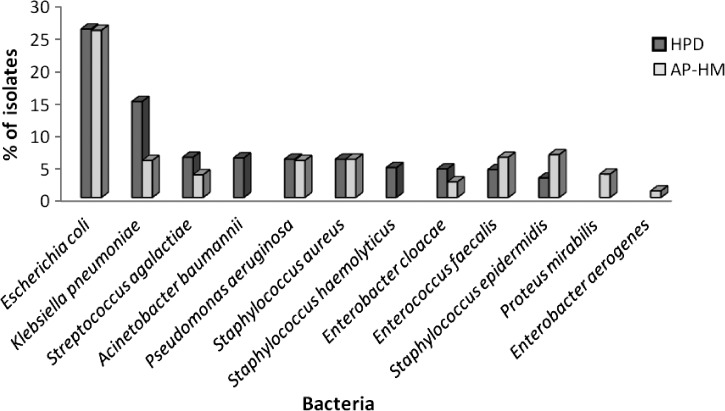
Ten bacteria were identified more than 50 times and together, represented 94.2% (1,962 of 2,083) of the bacterial isolates (Figure 1). E. coli was the most frequently identified bacterial species (538 of 2,083; 25.8%) followed by Klebsiella pneumoniae (308; 14.8%), S. agalactiae (130; 6.2%), Acinetobacter baumannii (128; 6.1%), Pseudomonas aeruginosa (124; 5.9%), S. aureus (124; 5.9%), S. haemolyticus (95; 4.6%), Enterobacter cloacae (92; 4.4%), Enterococcus faecalis (90; 4.3%), and S. epidermidis (63; 3%).
Figure 1.
Percentage of 10 bacteria most frequently identified using VITEK MALDI-TOF MS in Dakar, Senegal and Marseille, France. AP-HM = Assistance Publique - Hôpitaux de Marseille.
Ten bacteria were identified between 10 and 50 times: Morganella morganii (39; 1.9%), S. hominis (31; 1.5%), Proteus mirabilis (20; 1%), S. pyogenes (16; 0.8%), S. cohnii (14; 0.7%), S. saprophyticus (14; 0.7%), E. faecium (13; 0.6%), Stenotrophomonas maltophilia (13; 0.6%), S. anginosus (13; 0.6%), S. warneri (11; 0.5%), and P. putida (10; 0.48%).
Among 35 bacteria identified more than 1 time but less than 10 times (Table 2), Salmonella enterica (9), A. radioresistens (8), Citrobacter koseri (8), Providencia rettgeri (8), S. salivarius (8), A. junii (7), Bacillus cereus (7), P. stuartii (7), E. asburiae (6), K. oxytoca (6), P. stutzeri (6), and S. parasanguinis (6) were the most frequent.
Table 2.
Thirty-five bacteria identified at the species level from two to nine times
| Phylum, genus, and bacterial species | Number of isolates |
|---|---|
| Actinobacteria | |
| Corynebacterium | |
| C. amycolatum | 3 |
| C. striatum | 5 |
| C. aurimucosum | 3 |
| Firmicutes | |
| Aerococcus | |
| A. viridans | 4 |
| Bacillus | |
| B. cereus | 7 |
| B. pumilus | 3 |
| Lactobacillus | |
| L. delbrueckii | 2 |
| Micrococcus | |
| M. luteus | 2 |
| Staphylococcus | |
| S. auricularis | 3 |
| S. lugdunensis | 4 |
| S. sciuri | 2 |
| S. simulans | 2 |
| Streptococcus | |
| S. dysgalactiae | 2 |
| S. parasanguinis | 6 |
| S. pneumoniae | 5 |
| S. salivarius | 8 |
| Proteobacteria | |
| Achromobacter | |
| A. xylosoxidans | 5 |
| Acinetobacter | |
| A. junii | 7 |
| A. lwoffii | 2 |
| A. radioresistens | 8 |
| A. schindleri | 4 |
| Citrobacter | |
| C. freundii | 3 |
| C. koseri | 8 |
| Enterobacter | |
| E. asburiae | 6 |
| E. gergoviae | 5 |
| Haemophilus | |
| H. influenzae | 4 |
| Klebsiella | |
| K. oxytoca | 6 |
| Neisseria | |
| N. meningitidis | 2 |
| Plesiomonas | |
| P. shigelloides | 3 |
| Providencia | |
| P. rettgeri | 8 |
| P. stuartii | 7 |
| Pseudomonas | |
| P. fluorescens | 2 |
| P. stutzeri | 6 |
| Ralstonia | |
| R. pickettii | 2 |
| Salmonella | |
| S. enterica | 9 |
| Serratia | |
| S. marcescens | 4 |
Among 34 bacteria identified only one time (Table 3), emerging pathogens, such as Alloiococcus otitis, and rare pathogens, such as Arthrobacter cumminsii or S. australis, were detected.
Table 3.
Thirty-four bacteria identified at the species level one time
| Phylum and genus | Bacterial species |
|---|---|
| Actinobacteria | |
| Arthrobacter | A. cumminsii |
| Corynebacterium | C. jeikeium |
| Nocardia | N. brasiliensis |
| Bacteroidetes | |
| Bacteroides | B. fragilis |
| Firmicutes | |
| Alloiococcus | A. otitis |
| Bacillus | B. megaterium |
| Bacillus | B. subtilis |
| Bacillus | B. weihenstephanensis |
| Enterococcus | E. avium |
| Enterococcus | E. hirae |
| Lactobacillus | L. jensenii |
| Lysinibacillus | L. fusiformis |
| Paenibacillus | P. durus |
| Staphylococcus | S. arlettae |
| Staphylococcus | S. caprae |
| Streptococcus | S. australis |
| Streptococcus | S. gallolyticus |
| Streptococcus | S. haemolyticus |
| Streptococcus | S. intermedius |
| Streptococcus | S. porcinus |
| Proteobacteria | |
| Acinetobacter | A. haemolyticus |
| Acinetobacter | A. johnsonii |
| Aggregatibacter | A. segnis |
| Alcaligenes | A. faecalis |
| Bordetella | B. bronchiseptica |
| Enterobacter | E. aerogenes |
| Escherichia | E. hermanii |
| Haemophilus | H. haemolyticus |
| Haemophilus | H. parainfluenzae |
| Kluyvera | K. ascorbata |
| Neisseria | N. subflava |
| Neisseria | N. elongata |
| Shewanella | S. putrifaciens |
| Brachyspira | B. pilosicoli |
Fungal identification.
Among 206 identified fungi (Table 4), 197 were from the Candida genus (95.6%). Only one fungus was identified more than 50 times: C. albicans (98; 47.6%). Three fungi were identified between 10 and 50 times: C. tropicalis (42; 20.4%), C. glabrata (30; 14.6%), and C. krusei (14; 6.8%). Five fungi were identified more than 1 time but less than 10 times: C. parapsilosis (6), Aspergillus niger (3), C. dubliniensis (2), Clavispora lusitaniae (2), and Kluyveromyces marxianus (2). Seven were identified only one time: A. flavus, Microsporum canis, Trichosporon asahii, Kodamaea ohmeri, C. africana (an emergent and rare pathogen described in 2001 for the first time),13 C. nivariensis (an emergent and rare pathogen described in 2005 for the first time),14 and C. utilis (an industrially important yeast that is rarely reported as a human pathogen, with approximately 10 reported cases found in a PubMed search on May 6, 2014).15
Table 4.
Two hundred six fungi identified at the species level
| Phylum, genus, and species | Number of isolates |
|---|---|
| Ascomycota | |
| Aspergillus | |
| A. niger | 3 |
| A. flavus | 1 |
| Candida | |
| C. albicans | 98 |
| C. tropicalis | 42 |
| C. glabrata | 30 |
| C. krusei | 14 |
| C. parapsilosis | 6 |
| C. dubliniensis | 2 |
| C. africana | 1 |
| C. nivariensis | 1 |
| C. utilis | 1 |
| Clavispora | |
| C. lusitaniae | 2 |
| Kluyveromyces | |
| K. marxianus | 2 |
| Kodamaea | |
| K. ohmeri | 1 |
| Microsporum | |
| M. canis | 1 |
| Basidiomycota | |
| Trichosporon | |
| T. asahii | 1 |
Imprecise identification at the genus or family levels.
Among 109 bacteria identified at the genus level (Table 5), most were from the genus Streptococcus (48 of 109; 44%), including 29 (26.6%) isolates with a misidentification between S. mitis, S. oralis, and S. pneumoniae. The other misidentifications primarily included bacteria from the Proteobacteria phylum, with difficulties occurring in identifying bacteria from the Citrobacter genus (10; 9.2%), Achromobacter genus (10; 9.2%), Burkholderia genus (9; 8.3%), and Aeromonas genus (7; 6.4%). In 16 cases, bacteria from the Enterobacteriaceae family were identified, but MALDI-TOF MS could not accurately distinguish between E. coli and Shigella, because they are likely pathovars belonging to the same species with similar ribosomal protein patterns.
Table 5.
One hundred nine identifications at the genus level
| Phylum and genus | MALDI-TOF MS identification | Number of isolates |
|---|---|---|
| Actinobacteria | ||
| Corynebacterium | Corynebacterium sp. | 1 |
| Firmicutes | ||
| Bacillus | Bacillus sp. | 2 |
| Enterococcus | Enterococcus sp. | 2 |
| Lactobacillus | Lactobacillus sp. | 6 |
| Streptococcus | S. mitis/oralis/pneumoniae | 29 |
| Streptococcus | Streptococcus sp. | 19 |
| Proteobacteria | ||
| Achromobacter | Achromobacter sp. | 10 |
| Acinetobacter | Acinetobacter sp. | 5 |
| Aeromonas | Aeromonas sp. | 7 |
| Burkholderia | Burkholderia sp. | 9 |
| Chryseobacterium | Chryseobacterium sp. | 1 |
| Citrobacter | Citrobacter sp. | 10 |
| Ochrobactrum | Ochrobactrum sp. | 1 |
| Proteus | P. penneri/vulgaris | 2 |
| Proteus | Proteus sp. | 1 |
| Pseudomonas | Pseudomonas sp. | 1 |
| Salmonella | Salmonella sp. | 3 |
Discussion
Several studies, including comparative and/or multicenter studies, have already been performed to evaluate and compare the performance of the most diffuse commercial systems of MALDI-TOF MS systems, such as the Bruker BioTyper and the bioMérieux VITEK MS (with both SARAMIS v4.09 and Knowledge Base v2.0 VITEK MS v2.0 systems), by checking the discrepancies through molecular methods and sequencing.11,16–19 Overall, MALDI-TOF MS identification has been found to be highly accurate for clinically relevant bacteria, including Gram-positive, Gram-negative, and fastidious anaerobic bacteria as well as fungi detected in routine microbiology.11,16,18,20 Herein, we confirm the power of MALDI-TOF MS in identifying bacteria and fungi, with 94.2% identification accuracy at the species level of isolates tested in a laboratory in tropical Africa (2,289 of 2,430 isolates). In the pioneering work performed in 2010 in our laboratory in Marseille, France, 84.1% of 1,660 tested isolates were accurately identified at the species level, and 11.3% of tested isolates were accurately identified at the genus level.5 In Dakar, the 10 most commonly identified bacteria also represented 94.2% of all bacteria that were accurately identified at the laboratory. MALDI-TOF MS has also enabled the detection of rare microorganisms, including bacteria, such as A. cumminsii and S. australis, as well as fungi, such as C. africana and C. nivariensis.
Most of the misidentifications were caused by the potential inability of MALDI-TOF MS (already highlighted in other studies) to accurately differentiate S. pneumoniae from the viridans group of streptococci, which was observed in our laboratory in France, or differentiate some strains of E. coli from Shigella.1,6,7,18,21 Thus, conventional biochemical techniques, such as S. pneumoniae latex agglutination and indole tests, are sometimes still necessary for accurate identification. Other misidentifications rarely observed but also previously reported were within the genera Achromobacter, Burkholderia, and Aeromonas.16 Most of these misidentifications can be attributed to an incomplete population of databases associated with the instrument.1,7,18,21,22 For instance, it has recently been reported that improvements to the database enable a more reliable distinction between S. pneumoniae and viridans group streptococci.23,24 Because the reference databases are not static and expanded regularly to fill in current gaps in identification, updates will continue to improve the performance of MALDI-TOF MS.18
To the best of our knowledge, this study represents the first implementation and use of MALDI-TOF MS for the identification of bacteria and fungi in a hospital in tropical Africa. MALDI-TOF MS has previously been implemented and used for research purposes in South Africa for protein identification and bacterial identification, but its use was restricted to a few bacterial species in environmental studies of plant pathology or river water.25–27 Sample preparation is simple (direct deposit of colonies onto the target slide followed by addition of ready-to-use matrix solution) and can be performed widely. Thus, personnel training requirements are minimal, and samples can be analyzed within minutes. Currently, the estimated wait for one bacterial identification is reported to be from 1 minute and 46 seconds to 2 minutes per sample.6,16 The use and necessity of this new system were quickly shown by the fact that traditional phenotypic systems were abandoned on the arrival of the MALDI-TOF MS in the laboratory.
The rapid and accurate identification of routinely encountered bacterial and fungal species as well as those that are rare and difficult to identify using phenotypic methods provides a promising way to improve the care of patients with infectious diseases in Africa. The greatest expenses are associated with purchasing the instrument as well as maintenance fees. The required reagents are not expensive and do not require specific storage conditions if they are prepared in the laboratory.1,6 Overall, it has been clearly shown that MALDI-TOF MS is less expensive than traditional methods, even when taking into account the costs of reagents, labor, performance measurements, waste disposal, microorganism prevalence, and instrument maintenance expenses as well as multiple runs and additional tests needed to maximize accuracy.6,18,21,22 Moreover, several studies have shown the clinical benefits of using MALDI-TOF MS. This technique can provide microorganism identification up to 30 hours faster than conventional phenotypic methods; such gains in the speed of identification can have a substantial impact on patient care and management.6,16,21 MALDI-TOF MS can also be used for the rapid and effective identification of microorganisms from positive blood cultures within 30–45 minutes after a positive signal is provided by a blood culture instrument, and such rapid identification can lead to earlier initiation of treatment with appropriate antimicrobial therapies and increase the chances of obtaining optimal clinical outcomes.28
The primary obstacle to the use of MALDI-TOF MS in Africa is the cost of the machine. In several countries, the consolidation of clinical microbiology laboratories into large core laboratories was intended to lower management costs.29 The feasibility of MALDI-TOF MS networking in a university hospital in Belgium has recently been shown.30 We suggest that a common MS platform be developed to be shared among several clinical microbiology laboratories within the city and in nearby areas. The cost of the acquisition can be supported and shared between several organizations, including research organizations, non-governmental organizations, or charity foundations, such as the Mérieux Foundation or the Bill & Melinda Gates Foundation, both of which are already involved in the implementation of new tools to prevent and treat deadly diseases in Africa. Additional costs to be considered relate to equipment maintenance because of the lack of trained personnel in Africa and the cost of spare parts and maintenance contracts.
MALDI-TOF MS also has the potential to identify microorganisms at the subspecies and serotype levels, type strains, and profile antibiotic resistance within minutes.5,18,31–38 Moreover, MALDI-TOF MS has enabled the rapid detection of tick and mosquito vectors without requiring previous expertise in entomology.39,40 Thus, this technique will be of use in the implementation of effective prevention measures for vector-borne diseases.39,40 In the future, it will be useful for tropical countries to evaluate whether MALDI-TOF MS may be used to distinguish between uninfected mosquitoes and those infected with sporozoites of Plasmodium spp. and determine potential vector resistance to insecticides.
In the future, an exhaustive repertory of the bacteria correctly identified using MALDI-TOF MS could become available. These data will enable the comparison of bacterial diversity across different areas of the world. They will enable the comparison of rarely observed bacterial species in addition to frequently detected bacteria. For example, comparisons between bacteria observed at the HPD in Dakar and in our laboratory in Marseille show that, during the same time period, among the 10 most frequently identified bacteria at each institution, 8 were found in common: E. coli, K. pneumoniae, S. aureus, S. epidermidis, S. agalactiae, E. cloacae, P. aeruginosa, and E. faecalis (Figure 1). However, the prevalence rates of these strains were found to be different; S. agalactiae was more frequently observed in Dakar, although it has recently become more prevalent in Marseille, whereas S. epidermidis was more frequently observed in Marseille.41 A. baumannii and S. haemolyticus were among the 10 most frequently identified bacteria only in Dakar, whereas two Enterobacteriaceae (P. mirabilis and E. aerogenes) were more prevalent in Marseille than Dakar.
MALDI-TOF MS is a single, rapid, robust, and simple-to-use system that has proven its broad applicability and robustness in tropical Africa through its ability to quickly identify a broad range of microorganisms. Despite the initial cost of the MS instrument, the MS technique is more cost-effective than current phenotypic methods, and it would be advantageous to expand the capabilities of the mass MS platform in Africa.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank Cédric Abat, Thierry Schmidgen, Virginie Giroudon, Dr. Hervé Tissot-Dupont, and the staff of the Hôpital Principal de Dakar (Senegal), especially Dr. Elimane Mbaye and Mr. Diene Bane, for their contributions.
Footnotes
Financial support: This study was supported by the French Ministry of Foreign Affairs, the Research Institute for Development, and the Institut Hospitalo-Universitaire (IHU) Méditerranée Infection.
Authors' addresses: Bécaye Fall, Bissoume Samb-Ba, Silman Diawara, Mamadou Wague Gueye, Kowry Sow, Yaya Diemé, and Boubacar Wade, Hôpital Principal, Dakar, Sénégal, E-mails: becayefall@gmail.com, bissamb@yahoo.fr, diawaraps@hotmail.com, wax304@hotmail.fr, kowryndiaye@yahoo.fr, yaya_dieme@hotmail.com, and bwade55@yahoo.com. Cheikh Ibrahima Lo, Maxence Aubadie-Ladrix, Oleg Mediannikov, Cheikh Sokhna, Didier Raoult, and Florence Fenollar, Aix Marseille Université, Unité des Rickettsies, Unité de Recherche sur les Maladies Infectieuses et Tropicales Emergentes (URMITE), UM63, CNRS 7278, Research Institute for Development 198, Inserm 1095, Marseille, France, and Campus commun UCAD Research Institute for Development, Hann, Dakar, Sénégal, E-mails: cibrahimalo@gmail.com, lamenacexr@yahoo.fr, olegusss1@gmail.com, cheikh.sokhna@ird.fr, didier.raoult@gmail.com, and florence.fenollar@univ-amu.fr. Nadine Perrot and Sonia Chatellier, Département Microbiologie Innovation, bioMérieux, La-Balme-Les-Grottes, France, E-mails: nadine.perrot@biomerieux.com and sonia.chatellier@biomerieux.com.
References
- 1.Seng P, Drancourt M, Gouriet F, La Scola B, Fournier PE, Rolain JM, Raoult D. Ongoing revolution in bacteriology: routine identification of bacteria by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin Infect Dis. 2009;49:543–551. doi: 10.1086/600885. [DOI] [PubMed] [Google Scholar]
- 2.Cherkaoui A, Hibbs J, Emonet S, Tangomo M, Girard M, Francois P, Schrenzel P. Comparison of two matrix-assisted laser desorption ionization-time of flight mass spectrometry methods with conventional phenotypic identification for routine identification of bacteria to the species level. J Clin Microbiol. 2010;48:1169–1175. doi: 10.1128/JCM.01881-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stevenson LG, Drake SK, Shea YR, Zelazny AM, Murray PR. Evaluation of matrix-assisted laser desorption ionization-time of flight mass spectrometry for identification of clinically important yeast species. J Clin Microbiol. 2010;48:3482–3486. doi: 10.1128/JCM.00687-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Patel R. Matrix-assisted laser desorption ionization-time of flight mass spectrometry in clinical microbiology. Clin Infect Dis. 2013;57:564–572. doi: 10.1093/cid/cit247. [DOI] [PubMed] [Google Scholar]
- 5.Seng P, Rolain JM, Fournier PE, La Scola B, Drancourt M, Raoult D. MALDI-TOF-mass spectrometry applications in clinical microbiology. Future Microbiol. 2010;5:1733–1754. doi: 10.2217/fmb.10.127. [DOI] [PubMed] [Google Scholar]
- 6.Seng P, Abat C, Rolain JM, Colson P, Gouriet F, Fournier PE, Drancourt M, La Scola B, Raoult D. Emergence of rare pathogenic bacteria in a clinical microbiology laboratory: impact of MALDI-TOF mass spectrometry. J Clin Microbiol. 2013;51:2182–2194. doi: 10.1128/JCM.00492-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bizzini A, Durussel C, Bille J, Greub G, Prod'hom G. Performance of matrix-assisted laser desorption ionization-time of flight mass spectrometry for identification of bacterial strains routinely isolated in a clinical microbiology laboratory. J Clin Microbiol. 2010;48:1549–1554. doi: 10.1128/JCM.01794-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen JH, Yam WC, Ngan AH, Fung AM, Woo WL, Yan MK, Choi GK, Ho PL, Cheng VC, Yuen KY. Advantages of using MALDI-TOF mass spectrometry as a rapid diagnostic tool for yeast and mycobacteria identification in clinical microbiological laboratory. J Clin Microbiol. 2013;51:3981–3987. doi: 10.1128/JCM.01437-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Itoh E, Watari T, Azuma Y, Watanabe N, Tomoda Y, Akasaka K, Kino S. Performance of two matrix-assisted laser desorption/ionization time of flight mass spectrometry (MALDI-TOF MS) models for identification of bacteria isolated from blood culture. Rinsho Byori. 2013;61:382–389. [PubMed] [Google Scholar]
- 10.Panda A, Kurapati S, Samantaray JC, Myneedu VP, Verma A, Srinivasan A, Ahmad H, Behera D, Singh UB. Rapid identification of clinical mycobacterial isolates by protein profiling using matrix assisted laser desorption ionization-time of flight mass spectrometry. Indian J Med Microbiol. 2013;31:117–122. doi: 10.4103/0255-0857.115217. [DOI] [PubMed] [Google Scholar]
- 11.Moon HW, Lee SH, Chung HS, Lee M, Lee K. Performance of the Vitek MS matrix-assisted laser desorption ionization time-of-flight mass spectrometry system for identification of Gram-positive cocci routinely isolated in clinical microbiology laboratories. J Med Microbiol. 2013;62:1301–1306. doi: 10.1099/jmm.0.062950-0. [DOI] [PubMed] [Google Scholar]
- 12.Nori P, Ostrowsky B, Dorokhova O, Gialanella P, Moy M, Muggia V, Grossberg R, Kornblum J, Lin Y, Levi MH. Use of matrix-assisted laser desorption ionization-time of flight mass spectrometry to resolve complex clinical cases of patients with recurrent bacteremias. J Clin Microbiol. 2013;51:1983–1986. doi: 10.1128/JCM.00083-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tietz HJ, Hopp M, Schmalreck A, Sterry W, Czaika V. Candida africana sp. nov., a new human pathogen or a variant of Candida albicans? Mycoses. 2001;44:437–445. doi: 10.1046/j.1439-0507.2001.00707.x. [DOI] [PubMed] [Google Scholar]
- 14.Alcoba-Florez J, Mendez-Alvarez S, Cano J, Guarro J, Perez-Roth E, del Pilar AM. Phenotypic and molecular characterization of Candida nivariensis sp. nov., a possible new opportunistic fungus. J Clin Microbiol. 2005;43:4107–4111. doi: 10.1128/JCM.43.8.4107-4111.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eddouzi J, Lohberger A, Vogne C, Manai M, Sanglard D. Identification and antifungal susceptibility of a large collection of yeast strains isolated in Tunisian hospitals. Med Mycol. 2013;51:737–746. doi: 10.3109/13693786.2013.800239. [DOI] [PubMed] [Google Scholar]
- 16.Manji R, Bythrow M, Branda JA, Burnham CA, Ferraro MJ, Garner OB, Jennemann R, Lewinski MA, Mochon AB, Procop GW, Richter SS, Rychert JA, Sercia L, Westblade LF, Ginocchio CC. Multi-center evaluation of the VITEK(R) MS system for mass spectrometric identification of non-enterobacteriaceae gram-negative bacilli. Eur J Clin Microbiol Infect Dis. 2014;33:337–346. doi: 10.1007/s10096-013-1961-2. [DOI] [PubMed] [Google Scholar]
- 17.Alby K, Gilligan PH, Miller MB. Comparison of MALDI-TOF mass spectrometry platforms for the identification of gram negative rods from cystic fibrosis patients. J Clin Microbiol. 2013;51:3852–3854. doi: 10.1128/JCM.01618-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clark AE, Kaleta EJ, Arora A, Wolk DM. Matrix-assisted laser desorption ionization-time of flight mass spectrometry: a fundamental shift in the routine practice of clinical microbiology. Clin Microbiol Rev. 2013;26:547–603. doi: 10.1128/CMR.00072-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martiny D, Busson L, Wybo I, El Haj RA, Dediste A, Vandenberg O. Comparison of the Microflex LT and Vitek MS systems for routine identification of bacteria by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol. 2012;50:1313–1325. doi: 10.1128/JCM.05971-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garner O, Mochon A, Branda J, Burnham CA, Bythrow M, Ferraro M, Ginocchio C, Jennemann R, Manji R, Procop GW, Richter S, Rychert J, Sercia L, Westblade L, Lewinski M. Multi-centre evaluation of mass spectrometric identification of anaerobic bacteria using the VITEK MS system. Clin Microbiol Infect. 2014;20:335–339. doi: 10.1111/1469-0691.12317. [DOI] [PubMed] [Google Scholar]
- 21.Tan KE, Ellis BC, Lee R, Stamper PD, Zhang SX, Carroll KC. Prospective evaluation of a matrix-assisted laser desorption ionization-time of flight mass spectrometry system in a hospital clinical microbiology laboratory for identification of bacteria and yeasts: a bench-by-bench study for assessing the impact on time to identification and cost-effectiveness. J Clin Microbiol. 2012;50:3301–3308. doi: 10.1128/JCM.01405-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gaillot O, Blondiaux N, Loiez C, Wallet F, Lemaitre N, Herwegh S, Courcol RJ. Cost-effectiveness of switch to matrix-assisted laser desorption ionization-time of flight mass spectrometry for routine bacterial identification. J Clin Microbiol. 2011;49:4412. doi: 10.1128/JCM.05429-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Werno AM, Christner M, Anderson TP, Murdoch DR. Differentiation of Streptococcus pneumoniae from nonpneumococcal streptococci of the Streptococcus mitis group by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol. 2012;50:2863–2867. doi: 10.1128/JCM.00508-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dubois D, Segonds C, Prere MF, Marty N, Oswald E. Identification of clinical Streptococcus pneumoniae isolates among other alpha and nonhemolytic streptococci by use of the Vitek MS matrix-assisted laser desorption ionization-time of flight mass spectrometry system. J Clin Microbiol. 2013;51:1861–1867. doi: 10.1128/JCM.03069-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Standing TA, du Plessis E, Duvenage S, Korsten L. Internalisation potential of Escherichia coli O157:H7, Listeria monocytogenes, Salmonella enterica subsp. enterica serovar Typhimurium and Staphylococcus aureus in lettuce seedlings and mature plants. J Water Health. 2013;11:210–223. doi: 10.2166/wh.2013.164. [DOI] [PubMed] [Google Scholar]
- 26.Gemmell ME, Schmidt S. Is the microbiological quality of the Msunduzi River (KwaZulu-Natal, South Africa) suitable for domestic, recreational, and agricultural purposes? Environ Sci Pollut Res Int. 2013;20:6551–6562. doi: 10.1007/s11356-013-1710-1. [DOI] [PubMed] [Google Scholar]
- 27.Haarburger D, Bergstrom J, Pillay TS. Serum proteome changes following human immunodeficiency virus infection. Clin Lab. 2013;59:639–646. doi: 10.7754/clin.lab.2012.120419. [DOI] [PubMed] [Google Scholar]
- 28.Foster AG. Rapid identification of microbes in positive blood cultures by matrix-assisted laser desorption/ionisation time-of-flight (Maldi-Tof) mass spectrometry- (Vitek MS–bioMerieux.) J Clin Microbiol. 2013;51:3717–3719. doi: 10.1128/JCM.01679-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fournier PE, Drancourt M, Colson P, Rolain JM, La Scola B, Raoult D. Modern clinical microbiology: new challenges and solutions. Nat Rev Microbiol. 2013;11:574–585. doi: 10.1038/nrmicro3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martiny D, Cremagnani P, Gaillard A, Miendje Deyi VY, Mascart G, Ebraert A, Attalibi S, Dediste A, Vandenberg O. Feasibility of matrix-assisted laser desorption/ionisation time-of-flight mass spectrometry (MALDI-TOF MS) networking in university hospitals in Brussels. Eur J Clin Microbiol Infect Dis. 2014;33:745–754. doi: 10.1007/s10096-013-2006-6. [DOI] [PubMed] [Google Scholar]
- 31.Hrabak J, Walkova R, Studentova V, Chudackova E, Bergerova T. Carbapenemase activity detection by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol. 2011;49:3222–3227. doi: 10.1128/JCM.00984-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Walker J, Fox AJ, Edwards-Jones V, Gordon DB. Intact cell mass spectrometry (ICMS) used to type methicillin-resistant Staphylococcus aureus: media effects and inter-laboratory reproducibility. J Microbiol Methods. 2002;48:117–126. doi: 10.1016/s0167-7012(01)00316-5. [DOI] [PubMed] [Google Scholar]
- 33.Jackson KA, Edwards-Jones V, Sutton CW, Fox AJ. Optimisation of intact cell MALDI method for fingerprinting of methicillin-resistant Staphylococcus aureus. J Microbiol Methods. 2005;62:273–284. doi: 10.1016/j.mimet.2005.04.015. [DOI] [PubMed] [Google Scholar]
- 34.Du Z, Yang R, Guo Z, Song Y, Wang J. Identification of Staphylococcus aureus and determination of its methicillin resistance by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Anal Chem. 2002;74:5487–5491. doi: 10.1021/ac020109k. [DOI] [PubMed] [Google Scholar]
- 35.Majcherczyk PA, McKenna T, Moreillon P, Vaudaux P. The discriminatory power of MALDI-TOF mass spectrometry to differentiate between isogenic teicoplanin-susceptible and teicoplanin-resistant strains of methicillin-resistant Staphylococcus aureus. FEMS Microbiol Lett. 2006;255:233–239. doi: 10.1111/j.1574-6968.2005.00060.x. [DOI] [PubMed] [Google Scholar]
- 36.Camara JE, Hays FA. Discrimination between wild-type and ampicillin-resistant Escherichia coli by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Anal Bioanal Chem. 2007;389:1633–1638. doi: 10.1007/s00216-007-1558-7. [DOI] [PubMed] [Google Scholar]
- 37.Sparbier K, Schubert S, Weller U, Boogen C, Kostrzewa M. Matrix-assisted laser desorption ionization-time of flight mass spectrometry-based functional assay for rapid detection of resistance against beta-lactam antibiotics. J Clin Microbiol. 2012;50:927–937. doi: 10.1128/JCM.05737-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kostrzewa M, Sparbier K, Maier T, Schubert S. MALDI-TOF MS: an upcoming tool for rapid detection of antibiotic resistance in microorganisms. Proteomics Clin Appl. 2013;7:767–778. doi: 10.1002/prca.201300042. [DOI] [PubMed] [Google Scholar]
- 39.Yssouf A, Socolovschi C, Flaudrops C, Ndiath MO, Sougoufara S, Dehecq JS, Lacour G, Berenger JM, Sokhna CS, Raoult D, Parola P. Matrix-assisted laser desorption ionization—time of flight mass spectrometry: an emerging tool for the rapid identification of mosquito vectors. PLoS ONE. 2013;8:e72380. doi: 10.1371/journal.pone.0072380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yssouf A, Flaudrops C, Drali R, Kernif T, Socolovschi C, Berenger JM, Raoult D, Parola P. Matrix-assisted laser desorption ionization-time of flight mass spectrometry for rapid identification of tick vectors. J Clin Microbiol. 2013;51:522–528. doi: 10.1128/JCM.02665-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Abat C, Chaudet H, Raoult D, Colson P. Increasing trend of invasive group B streptococcal infections, Marseille, France. Clin Infect Dis. 2014;58:750–751. doi: 10.1093/cid/cit777. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.