Abstract
AIM: SMAD4 immunohistochemistry is considered a valuable prognostic marker in colorectal cancer, but individual studies have often been small and the results variable. A meta-analysis could potentially clarify these findings. METHODS: In September 2014, a Pubmed and Google Scholar search was conducted to find publications that reported the prognostic value of SMAD4 expression. A meta-analysis was performed to clarify the association between SMAD4 expression and survival outcomes. RESULTS: 137 studies were found, of which 13 were considered eligible. The studies consisted of a total of 3800 patients. Three different endpoints were taken into account, namely, overall survival (OS), disease-free survival (DFS), and cancer-specific survival (CSS). In addition, the studies were divided into univariate and multivariate analyses. The pooled hazard ratios were given as follows: univariate CSS = 1.75 [95% confidence interval (CI): 0.93-3.32; z= 1.69; P= .09]; multivariate CSS = 2.17 (95% CI: 1.56-3.01; z= 4.65; P= .000); univariate DFS = 2.11 (95% CI: 1.36-3.28; z= 3.32; P= .001); multivariate DFS = 2.15 (95% CI: 1.56-3.01; z= 4.65; P= .000); univariate OS and DFS = 2.30 (95% CI: 1.41-3.73; z= 3.36; P= .001); univariate OS = 2.28 (95% CI: 1.30-4.00; z= 2.89; P= .004). CONCLUSION: The results of the presented meta-analyses indicate that SMAD4 expression status using immunohistochemistry is a prognostic marker for patient survival.
Introduction
Colorectal cancer (CRC) is the third most commonly diagnosed cancer and the second leading cause of cancer death in men and women combined in the United States [1]. CRC is thought to result from the accumulation of genetic alterations, which give cells a survival advantage over surrounding cells. An important genetic change in CRC is mutation of SMAD4 leading to loss of SMAD4 protein expression. SMAD4 protein expression is lost in approximately 30% to 40% of the CRCs [2], [3], [4] and is associated with metastasis formation and poor response to chemotherapy [5], [6], [7], [8]. SMAD4 is the common mediator of the transforming growth factor–β and bone morphogenetic protein pathways and is located on chromosome 18q21. SMAD4 immunohistochemistry correlates very well with the genetic status as is shown in juvenile polyposis [9]. SMAD4 immunohistochemistry is therefore frequently used to ascertain the SMAD4 status of a tumor. Although the use of SMAD4 immunohistochemistry as a molecular marker has been studied in multiple studies, this has mostly been performed in small cohorts and in different subgroups using different survival endpoints. Most studies report an association between SMAD4 loss and a poor prognosis, but this is not consistent. To our knowledge, there has never been a comprehensive study combining all these results to truly establish the predictive value of SMAD4 immunohistochemistry for clinical use. Therefore, we conducted a meta-analysis to correlate the SMAD4 immunohistochemistry expression with survival outcomes.
Methods
Publication Selection
Pubmed and Google Scholar were used to search for potentially relevant literature. The search entry used was (“colorectal OR large intestine” OR “large bowel” OR “colon” OR “colonic” OR “rectal” OR “rectum”) AND (“cancer” OR “carcinoma” OR “tumor” OR “tumour” OR “neoplasm” OR “cancers”) AND (“SMAD4” OR “DPC4”) AND (“marker” OR “signature molecule” OR “molecular marker” OR “markers” OR “biomarkers” OR “biomarker” OR “marker” OR “prognosis” OR “predictive” OR “survival”). Additionally, reference lists of the studies found and of systematic reviews were also checked for potential articles. The search was performed in September 2014.
First, the abstracts were checked for relevance and full articles were retrieved when potentially eligible. To be included in the analysis, studies had to have been performed in resected colorectal carcinomas and immunohistochemistry for SMAD4 had to have been performed. One or more of the following endpoints had to be described: overall survival (OS), disease-free survival (DFS), or cancer-specific survival (CSS). Both univariate and multivariate analyses were taken into account, although separately analyzed. All selected studies were checked according to a 20-point quality control system developed previously (Table S1) [10], [11], [12].
Statistical Analysis
Data were extracted by two independent researchers. When not reported, the log hazard ratio (HR) and confidence interval (CI) were extracted using previously published methods [13], [14]. The meta-analysis was performed in STATA12 using the metan package. Pooled HR and 95% CI were calculated for each endpoint (OS, DFS, and CSS) and presented in a forest plot. An HR > 1 implies a worse prognosis for SMAD4-negative CRCs. A P value of < .05 was considered significantly different. Heterogeneity was calculated and presented as X2 and I2. We chose to perform random effect models based on the fact that the studies differed in size, country, and most importantly in the method of scoring and cutoff values. The loge standard error of the HR and loge HR of each study were plotted in a funnel plot to assess potential publication bias.
Results
Search Results
The search resulted in 137 studies. On the basis of the abstracts, 19 studies were considered eligible, of which 6 were eventually excluded on the basis of the fact that the HR and CI could not be extracted. The 13 eligible studies consisted of a total of 3800 patients, ranging from 86 to 1381 per study. The main characteristics of the studies can be found in Table 1. We included OS, CSS, and DFS in this study but analyzed the different endpoints separately. We also made a distinction between univariate and multivariate analyses. Five studies only included colon cancer, and the other eight studies included both colon and rectal cancers. Six studies reported that a portion or all of the patients received adjuvant chemotherapy after resection. Three studies did not report the mean or median age and three studies did not report the mean or median follow-up period. Five studies reported CSS, five studies reported DFS, and seven studies reported OS. Eight studies described specifically that only nuclear staining of SMAD4 was considered positive. One study considered both cytoplasmic and nuclear staining as positive and four studies did not describe their scoring methods. All studies used SMAD4 antibodies produced by Santa Cruz Biotechnology Inc (Dallas, TX) except for Li et al. who used an antibody from Zhongshan Biotechnology Inc. All studies except Isaksson-Mettävainio [24] had dichotomized the scoring. For Isaksson-Mettävainio [24], we have compared the categories depicted as SMAD4highversus SMAD4loss and we have not considered the category SMAD4moderate. The mean and median positive SMAD4 score is 70.9% and 76.5%, respectively, with a range of 26% to 90.7%.
Table 1.
Studies included in meta-analysis
| Study | Year | Country | Stage | No. of Patients | Inclusion Period | AB | Dilution | Site | Age (year) | Follow-Up | Outcome | Cytoplasm Nuclear | Preserved SMAD4 (%) | Adjusted Therapy |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Alazzouzi [15] | 2005 | Finland | III | 86 | 1993-1997 | Santa Cruz Biotechnology Inc | 1:1000 | Colon/rectum | Mean, 70.1 | At least 6 years | DFS and OS | NA | 26 | No |
| Alhopuro [5] | 2005 | Finland | III | 75 | 1994-1998 | Santa Cruz Biotechnology Inc | 1:1000 | Colon/rectum | Mean, 59 | Mean, 8.7 years | DFS and OS | NA | 86.7 | Yes |
| Isaksson-Mettävainio [18] | 2006 | Sweden | I-III | 86 | 1987-1994 | Santa Cruz Biotechnology Inc | 1:50 | Colon/rectum | NA | NA | CSS | Nuclear | 90.7 | No |
| Bacman [17] | 2007 | Germany | II-III | 305 | 1991-2001 | Santa Cruz Biotechnology Inc | 1:50 | Colon | Median, 64 | Median, 91 months | CSS | Nuclear | 85.6 | Yes |
| Mesker [19] | 2009 | Netherlands | I-II | 118 | 1980-2001 | Santa Cruz Biotechnology Inc | 1:400 | Colon | Mean, 68.2 | Up to 25 years | DFS and OS | Nuclear | 76.5 | No |
| Gulubova [20] | 2010 | Bulgary | I-IV | 138 | 1997-2006 | Santa Cruz Biotechnology Inc | 1:50 | Colon/rectum | Median, 65 | Median, 37.6 months | OS | Nuclear | 88.4 | No |
| Li [21] | 2011 | China | I-IV | 147 | 2003-2004 | Zhongshan Biotechnology | 1:150 | Colon | NA | Up to 5 years | DFS and OS | Nuclear | 74.1 | No |
| Baraniskin [22] | 2011 | Germany | IV | 190 | NA | Santa Cruz Biotechnology Inc | 1:100 | Colon/rectum | Mean, 64.4 | NA | OS | Nuclear | 65.8 | Yes |
| Ahn [23] | 2011 | South Korea | I-IV | 429 | 1991-2000 | Santa Cruz Biotechnology Inc | 1:200 | Colon/rectum | Mean, 57 | Median, 56 months | DFS | NA | 47.3 | Yes |
| Isaksson-Mettävainio [24] | 2012 | Sweden | I-IV | 441 | 1995-2003 | Santa Cruz Biotechnology Inc | 1:100 | Colon/rectum | NA | NA | CSS | Nuclear | 80.3 | Yes |
| Lampropoulos [25] | 2012 | Greece | I-IV | 195 | 2005-2006 | Santa Cruz Biotechnology Inc | 1:100 | Colon/rectum | Mean, 68.6 | Median, 56 months | CSS | Both | 61.5 | No |
| Roth [26] | 2012 | Switzerland | II-III | 1381 | NA | Santa Cruz Biotechnology Inc | 0.2 mg/ml | Colon | Median, 60 | Median, 69 months | OS and DFS | NA | 78.8 | Yes |
| Voorneveld [16] | 2013 | Netherlands | I-IV | 209 | 1983-2004 | Santa Cruz Biotechnology Inc | 1:400 | Colon | Mean, 68.9 | Median, 65 months | CSS | Nuclear | 60 | No |
Cancer-Specific Survival
Three studies included a univariate CSS with a total of 600 patients (range 86-305). The pooled HR is 1.75, which is not significantly different (95% CI: 0.93-3.32; z= 1.69; P= .09; Figure 1). Four studies included a multivariate CSS with a total of 931 patients (range 86-441). The pooled HR is 2.17, which is significantly different (95% CI: 1.56-3.01; z= 4.65; P= .000), indicating that SMAD4 loss is associated with a worse CSS in this pooled multivariate analysis.
Figure 1.
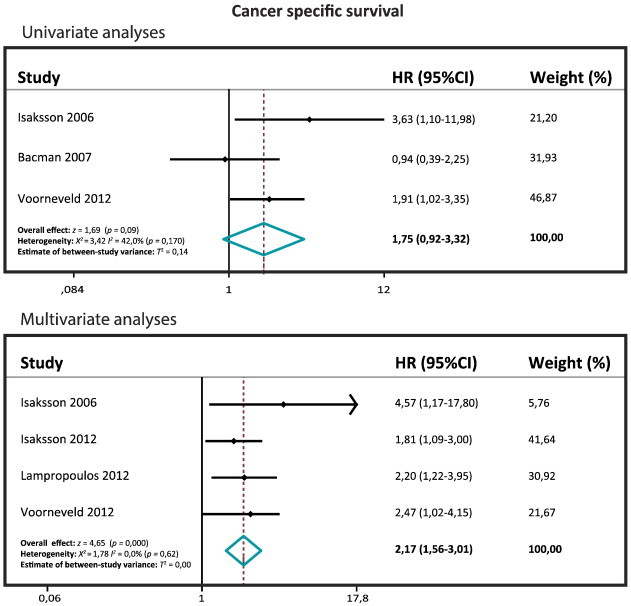
Forest plots of the CSS.
Disease-Free Survival
Six studies included a univariate DFS with a total of 2236 patients (range 75-1381), and three studies included a multivariate DFS with a total of 1646 patients (range 118-1381). The pooled HR for the univariate DFS is 2.11 (95% CI: 1.36-3.28; z= 3.32; P= .001), and the pooled HR for the multivariate DFS is 2.15 (95% CI: 1.56-3.01; z= 4.65; P= .000), which are both significantly different (Figure 2). The univariate and the multivariate DFS meta-analyses confirm that SMAD4 loss is associated with a poor DFS.
Figure 2.
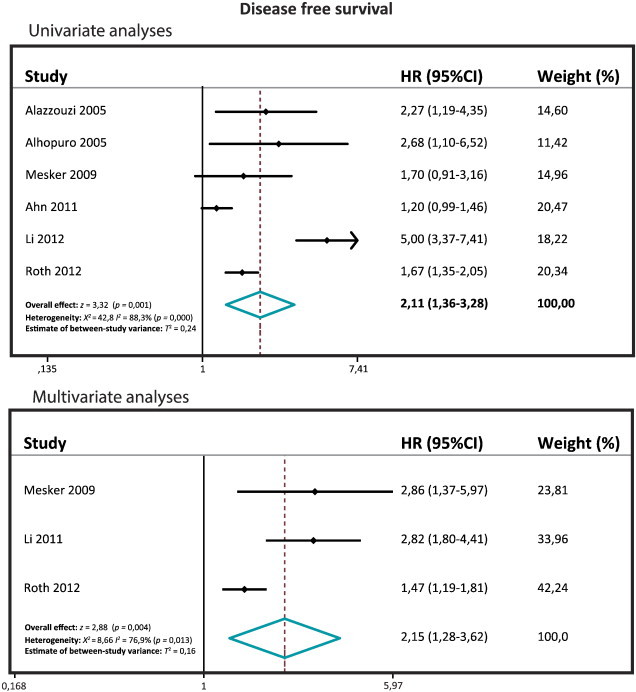
Forest plots of the DFS.
Overall Survival
Seven studies included a univariate OS with a total of 2135 patients (range 75-1381), and four studies included a multivariate OS with a total of 1836 patients (range 118-1381). The pooled HR for the univariate OS is 2.30 (95% CI: 1.41-3.73; z= 3.36; P= .001), and the pooled HR for the multivariate OS is 2.28 (95% CI: 1.30-4.00; z= 2.89; P= .004) (Figure 3). Both the univariate and the multivariate OS meta-analyses showed that SMAD4 loss is associated with a poor OS.
Figure 3.
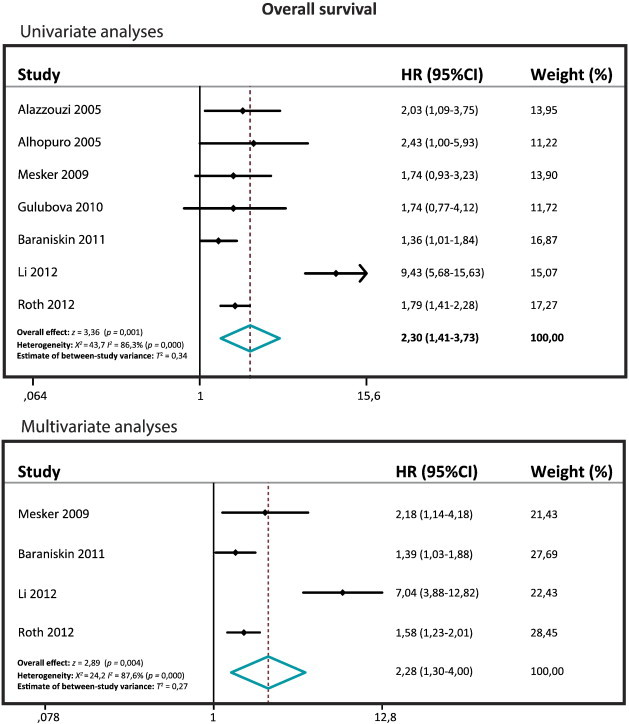
Forest plots of the OS.
Publication Bias
The funnel plots for the assessment of potential publication bias show one considerable outlier in the univariate and multivariate OS and the univariate DFS (Figure 4). Although in the univariate OS this does not result in a significant Egger test, we chose to exclude the outliers in all the meta-analyses and recalculate the pooled HRs. Exclusion of the outlier reduces the pooled HR of the univariate OS to 1.673 (95% CI: 1.42-1.96; z= 6.08; P= .000). The pooled HR of the multivariate OS is reduced to 1.55 (95% CI: 1.29-1.86; z= 4.68; P= .000), and the pooled HR of the univariate DFS is 1.6 (95% CI: 1.23-2.08; z= 3.53; P= .000).
Figure 4.
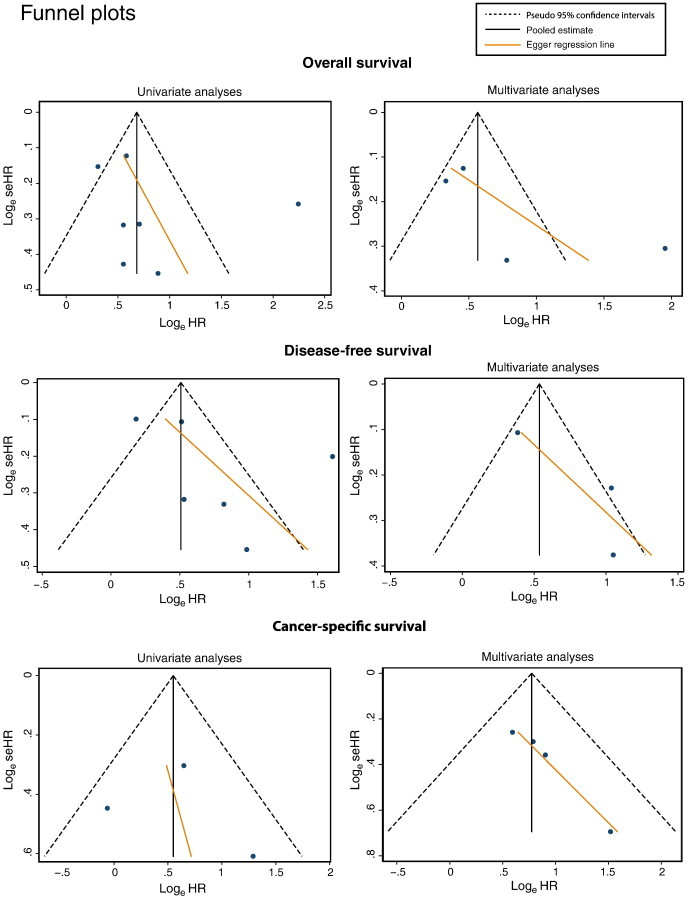
Funnel plots of all the meta-analyses.
Discussion
Molecular profiling of individual tumors potentially allows a personalized approach to cancer treatment. Estimation of prognosis plays an important role in decisions about treatment, and this is currently almost entirely dependent on histopathologic staging. SMAD4, located on chromosome 18q21, has frequently been reported to be a useful prognostic marker. Several studies have reported the prognostic value of SMAD4 expression using immunohistochemistry [15], [16], although this has not been entirely consistent [17]. To our knowledge, this is the first comprehensive meta-analysis of the predictive value of SMAD4 expression using immunohistochemistry.
We included three different endpoints, namely OS, CSS, and DFS, and both univariate and multivariate analyses. All the meta-analyses, except the univariate CSS, showed a significant difference in HR implying that loss of SMAD4 expression as measured by immunohistochemistry is associated with a poor prognosis. Thirteen studies have investigated the prognostic value of SMAD4 loss using different types of endpoints resulting in relative few studies per single endpoint. The analyses of the univariate OS and DFS included the largest numbers of studies (seven each), which make these the most reliable results. To investigate the heterogeneity, I2 was calculated, which was more than 50% in most of the cases, except for the univariate and multivariate analyses of the CSS. Conventionally, when I2 is more than 50% and the distribution is significantly heterogeneous (P< .05), the fixed model cannot be used and the random effects model has to be applied, but prior to conducting the analysis, we had already decided to use the random effects model for all the meta-analyses because of the variation in the methodology of the studies included. One aspect that varies between the studies is the percentage of SMAD4 preservation, which is dependent on the staining and scoring method used. SMAD4 is the common mediator of the bone morphogenetic protein/transforming growth factor–β signaling pathways and complexes with phosphorylated R-SMADs, which then enter the nucleus to modulate gene transcription. Only nuclear localization shows active functional SMAD4. Not all studies used only nuclear staining and four studies did not report what they considered as positive. Another source of heterogeneity is the population that was used in each of the studies. Seven studies included all the stages and the other six included only one or two stages. Six studies included patients that had received chemotherapy, while in seven studies no adjuvant therapy was applied. These differences can affect the outcome of the individual studies and can increase the heterogeneity in the meta-analysis.
Despite the fact that the pooled studies of univariate CSS did not show a significant difference, we conclude, on the basis of the other five meta-analyses all showing statistically significant associations, that immunohistochemical analysis of SMAD4 expression is a useful prognostic marker in colorectal cancer. Immunohistochemistry is a relative easy technique to perform and is readily available in most hospital pathology departments. International recommendations to standardize SMAD4 scoring methodology are required.
The following is the supplementary data related to this article
20-point quality control checklist applied to all the studies used in the meta-analysis
Footnotes
This article refers to supplementary material, which is designated by Table S1 and is available online at www.transonc.com.
Conflict of interest statement: The authors declare no conflict of interest. Funding: P.W.V. and J.C.H.H. are funded by the Netherlands Digestive Diseases Foundation. L.L.K. is funded by the Dutch Cancer Society. Author contributions: Writing of the manuscript—P.W.V., L.L.K., and J.C.H.H.; supervision project—P.W.V. and J.C.H.H.; searching articles—P.W.V. and R.J.J.; extracting data—P.W.V. and R.J.J.; statistics—P.W.V.
References
- 1.Siegel R, DeSantis C, Virgo K, Stein K, Mariotto A, Smith T, Cooper D, Gansler T, Lerro C, Fedewa S. Cancer treatment and survivorship statistics, 2012. CA Cancer J Clin. 2012;62:220–241. doi: 10.3322/caac.21149. [DOI] [PubMed] [Google Scholar]
- 2.Riggins GJ, Kinzler KW, Vogelstein B, Thiagalingam S. Frequency of Smad gene mutations in human cancers. Cancer Res. 1997;57:2578–2580. [PubMed] [Google Scholar]
- 3.Fearon ER, Cho KR, Nigro JM, Kern SE, Simons JW, Ruppert JM, Hamilton SR, Preisinger AC, Thomas G, Kinzler KW. Identification of a chromosome 18q gene that is altered in colorectal cancers. Science. 1990;247:49–56. doi: 10.1126/science.2294591. [DOI] [PubMed] [Google Scholar]
- 4.Salovaara R, Roth S, Loukola A, Launonen V, Sistonen P, Avizienyte E, Kristo P, Järvinen H, Souchelnytskyi S, Sarlomo-Rikala M. Frequent loss of SMAD4/DPC4 protein in colorectal cancers. Gut. 2002;51:56–59. doi: 10.1136/gut.51.1.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alhopuro P, Alazzouzi H, Sammalkorpi H, Davalos V, Salovaara R, Hemminki A, Järvinen H, Mecklin JP, Schwartz S, Jr, Aaltonen LA. SMAD4 levels and response to 5-fluorouracil in colorectal cancer. Clin Cancer Res. 2005;11:6311–6316. doi: 10.1158/1078-0432.CCR-05-0244. [DOI] [PubMed] [Google Scholar]
- 6.Miyaki M, Iijima T, Konishi M, Sakai K, Ishii A, Yasuno M, Hishima T, Koike M, Shitara N, Iwama T. Higher frequency of Smad4 gene mutation in human colorectal cancer with distant metastasis. Oncogene. 1999;18:3098–3103. doi: 10.1038/sj.onc.1202642. [DOI] [PubMed] [Google Scholar]
- 7.Papageorgis P, Cheng K, Ozturk S, Gong Y, Lambert AW, Abdolmaleky HM, Zhou JR, Thiagalingam S. Smad4 inactivation promotes malignancy and drug resistance of colon cancer. Cancer Res. 2011;71:998–1008. doi: 10.1158/0008-5472.CAN-09-3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang B, Halder SK, Kashikar ND, Cho YJ, Datta A, Gorden DL, Datta PK. Antimetastatic role of Smad4 signaling in colorectal cancer. Gastroenterology. 2010;138:969–980. doi: 10.1053/j.gastro.2009.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Langeveld D, van Hattem WA, de Leng WW, Morsink FH, ten Kate FJ, Giardiello FM, Offerhaus GJ, Brosens LA. SMAD4 immunohistochemistry reflects genetic status in juvenile polyposis syndrome. Clin Cancer Res. 2010;16:4126–4134. doi: 10.1158/1078-0432.CCR-10-0168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hayden JA, Côté P, Bombardier C. Evaluation of the quality of prognosis studies in systematic reviews. Ann Intern Med. 2006;144:427–437. doi: 10.7326/0003-4819-144-6-200603210-00010. [DOI] [PubMed] [Google Scholar]
- 11.McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM. Reporting recommendations for tumor marker prognostic studies (remark) Exp Oncol. 2006;28:99–105. [PubMed] [Google Scholar]
- 12.Smith RA, Tang J, Tudur-Smith C, Neoptolemos JP, Ghaneh P. Meta-analysis of immunohistochemical prognostic markers in resected pancreatic cancer. Br J Cancer. 2011;104:1440–1451. doi: 10.1038/bjc.2011.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parmar MK, Torri V, Stewart L. Extracting summary statistics to perform meta-analyses of the published literature for survival endpoints. Stat Med. 1998;17:2815–2834. doi: 10.1002/(sici)1097-0258(19981230)17:24<2815::aid-sim110>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 14.Tierney JF, Stewart LA, Ghersi D, Burdett S, Sydes MR. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials. 2007;8:16. doi: 10.1186/1745-6215-8-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alazzouzi H, Alhopuro P, Salovaara R, Sammalkorpi H, Jarvinen H, Mecklin JP, Hemminki A, Schwartz S, Jr, Aaltonen LA, Arango D. SMAD4 as a prognostic marker in colorectal cancer. Clin Cancer Res. 2005;11:2606–2611. doi: 10.1158/1078-0432.CCR-04-1458. [DOI] [PubMed] [Google Scholar]
- 16.Voorneveld PW, Jacobs RJ, de Miranda NF, Morreau H, van Noesel CJ, Offerhaus GJ, Kodach LL, Hardwick JC. Evaluation of the prognostic value of pSMAD immunohistochemistry in colorectal cancer. Eur J Cancer Prev. 2013;22:420–424. doi: 10.1097/CEJ.0b013e32835e88e2. [DOI] [PubMed] [Google Scholar]
- 17.Bacman D, Merkel S, Croner R, Papadopoulos T, Brueckl W, Dimmler A. TGF-beta receptor 2 downregulation in tumour-associated stroma worsens prognosis and high-grade tumours show more tumour-associated macrophages and lower TGF-beta1 expression in colon carcinoma: a retrospective study. BMC Cancer. 2007;7:156. doi: 10.1186/1471-2407-7-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Isaksson-Mettävainio M, Palmqvist R, Forssell J, Stenling R, Oberg A. SMAD4/DPC4 expression and prognosis in human colorectal cancer. Anticancer Res. 2006;26:507–510. [PubMed] [Google Scholar]
- 19.Mesker WE, Liefers GJ, Junggeburt JM, van Pelt GW, Alberici P, Kuppen PJ, de Miranda NF, van Leeuwen KA, Morreau H, Szuhai K. Presence of a high amount of stroma and downregulation of SMAD4 predict for worse survival for stage I-II colon cancer patients. Cell Oncol. 2009;31:169–178. doi: 10.3233/CLO-2009-0478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gulubova M, Manolova I, Ananiev J, Julianov A, Yovchev Y, Peeva K. Role of TGF-β1, its receptor TGFβRII, and Smad proteins in the progression of colorectal cancer. Int J Colorectal Dis. 2010;25:591–599. doi: 10.1007/s00384-010-0906-9. [DOI] [PubMed] [Google Scholar]
- 21.Li X, Liu B, Xiao J, Yuan Y, Ma J, Zhang Y. Roles of VEGF-C and Smad4 in the lymphangiogenesis, lymphatic metastasis, and prognosis in colon cancer. J Gastrointest Surg. 2011;15:2001–2010. doi: 10.1007/s11605-011-1627-2. [DOI] [PubMed] [Google Scholar]
- 22.Baraniskin A, Munding J, Schulmann K, Meier D, Porschen R, Arkenau HT, Graeven U, Schmiegel W, Tannapfel A, Reinacher-Schick A. Prognostic value of reduced SMAD4 expression in patients with metastatic colorectal cancer under oxaliplatin-containing chemotherapy: a translational study of the AIO colorectal study group. Clin Colorectal Cancer. 2011;10:24–29. doi: 10.3816/CCC.2011.n.003. [DOI] [PubMed] [Google Scholar]
- 23.Ahn BK, Jang SH, Paik SS, Lee KH. Smad4 may help to identify a subset of colorectal cancer patients with early recurrence after curative therapy. Hepatogastroenterology. 2011;58:1933–1936. doi: 10.5754/hge11186. [DOI] [PubMed] [Google Scholar]
- 24.Isaksson-Mettävainio M, Palmqvist R, Dahlin AM, van Guelpen B, Rutegård J, Oberg A, Henriksson ML. High SMAD4 levels appear in microsatellite instability and hypermethylated colon cancers, and indicate a better prognosis. Int J Cancer. 2012;131:779–788. doi: 10.1002/ijc.26473. [DOI] [PubMed] [Google Scholar]
- 25.Lampropoulos P, Zizi-Sermpetzoglou A, Rizos S, Kostakis A, Nikiteas N, Papavassiliou AG. Prognostic significance of transforming growth factor beta (TGF-β) signaling axis molecules and E-cadherin in colorectal cancer. Tumour Biol. 2012;33:1005–1014. doi: 10.1007/s13277-012-0333-3. [DOI] [PubMed] [Google Scholar]
- 26.Roth AD, Delorenzi M, Tejpar S, Yan P, Klingbiel D, Fiocca R, d'Ario G, Cisar L, Labianca R, Cunningham D. Integrated analysis of molecular and clinical prognostic factors in stage II/III colon cancer. J Natl Cancer Inst. 2012;104:1635–1646. doi: 10.1093/jnci/djs427. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
20-point quality control checklist applied to all the studies used in the meta-analysis


