Abstract
BACKGROUND: We previously reported that the addition of erlotinib to gemcitabine and oxaliplatin (GEMOX) resulted in greater antitumor activity and might be a treatment option for patients with biliary tract cancers (BTCs). Molecular subgroup analysis of treatment outcomes in patients who had specimens available for analysis was undertaken. METHODS: Epidermal growth factor receptor (EGFR), KRAS, and PIK3CA mutations were evaluated using peptide nucleic acid–locked nucleic acid polymerase chain reaction clamp reactions. Survival and response rates (RRs) were analyzed according to the mutational status. Sixty-four patients (48.1%) were available for mutational analysis in the chemotherapy alone group and 61 (45.1%) in the chemotherapy plus erlotinib group. RESULTS: 1.6% (2/116) harbored an EGFR mutation (2 patients; exon 20), 9.6% (12/121) harbored a KRAS mutation (12 patients; exon 2), and 9.6% (12/118) harbored a PIK3CA mutation (10 patients, exon 9 and 2 patients, exon 20). The addition of erlotinib to GEMOX in patients with KRAS wild-type disease (n = 109) resulted in significant improvements in overall response compared with GEMOX alone (30.2% vs 12.5%, P = .024). In 95 patients with both wild-type KRAS and PIK3CA, there was evidence of a benefit associated with the addition of erlotinib to GEMOX with respect to RR as compared with GEMOX alone (P = .04). CONCLUSION: This study demonstrates that KRAS mutational status might be considered a predictive biomarker for the response to erlotinib in BTCs. Additionally, the mutation status of PIK3CA may be a determinant for adding erlotinib to chemotherapy in KRAS wild-type BTCs.
Introduction
In South Korea, biliary tract cancers (BTCs), including cholangiocarcinoma and gallbladder cancer (GBC), are not uncommon [1]. Due to the non-specific symptoms associated with BTCs, more than 75% of cases are unresectable because of the advanced stage of disease at diagnosis. Moreover, even after a complete resection, many patients experience a recurrence of disease. Patients with advanced or recurrent BTCs can be considered for palliative chemotherapy [2], [3]. Combination chemotherapy with gemcitabine and a platinum-based agent is regarded as a standard first-line chemotherapy for advanced BTC, based on the results of previous randomized phase II and III trials (ABC01 and 02) [4], [5]. Nevertheless, prognosis is still poor and overall survival (OS) is less than 12 months in patients with advanced or recurrent BTCs [5].
Molecularly targeted therapies, either given alone or in combination with chemotherapy, have emerged as the standard treatment for a variety of cancer types [6], [7]. These therapies have been applied to treatment of BTCs. Erlotinib is an orally active tyrosine kinase inhibitor of epidermal growth factor receptor (EGFR), which has been associated with improved outcomes in various cancers [8]. Erlotinib, alone or in combination, has shown promising results in phase II trials in patients with advanced BTC, with response rates (RRs) of 8% to 12%, median OS of 7.5 to 9.9 months, and median time to progression of 2.6 to 4.4 months [9], [10]. In our phase III trial (NCT01149122) of gemcitabine and oxaliplatin (GEMOX) with or without erlotinib, the median progression-free survival (PFS) was 4.2 months in the GEMOX group and 5.8 months in the GEMOX plus erlotinib group [11]. Results of this phase III trial suggested that the addition of erlotinib to GEMOX might be a new treatment option for patients with cholangiocarcinoma, although no significant difference in PFS was noted between the groups.
BTCs have a spectrum of mutations in EGFR and its downstream signaling pathways, which include EGFR, KRAS, and PIK3CA [12], [13], [14], [15]. The EGFR signaling pathway has been extensively explored for several years as a therapeutic target for cancer therapy. Dysregulation of EGFR signaling has been shown to stimulate cell proliferation, angiogenesis, and metastatic spread while inhibiting apoptosis. It is well known that oncogenic activation of EGFR and its downstream pathways, including KRAS, BRAF, PTEN, PIK3CA, and AKT, is correlated with responsiveness to anti-EGFR therapies [16], [17]. Given the evidence that KRAS mutations are associated with less efficient EGFR-directed targeted therapy in various cancer types [18], [19], we evaluated KRAS mutation status in 60 of 268 patients who were enrolled in our previous phase III trial [11]. However, the predictive value of KRAS mutation for response to erlotinib was limited by the small number of tissues we analyzed.
Herein, to clarify the roles of EGFR, KRAS, and PIK3CA as predictive biomarkers in patients with advanced BTC who received erlotinib, we investigated the mutational status of tumors from archival specimens in an expanded subset of patients enrolled on our study.
Patients and Methods
Patients and Samples
Eligibility criteria and study design have been previously described. This was an open-label, randomized, phase III trial comparing erlotinib plus GEMOX with GEMOX alone as a first-line treatment for advanced BTCs [11]. The primary endpoint was PFS and analyses were conducted by intention to treat (ITT). The study included 268 randomized patients and a separate written consent for the optional correlative study was obtained to allow participation in the exploratory biomarker study of archival tumor specimens. Specimens were labeled with site of origin and a unique patient identifier. Sixty-four patients (48.1%) were available for mutational analysis in the chemotherapy alone group (n = 131) and 61 (45.1%) in the chemotherapy plus erlotinib group (n = 135).
DNA Extraction and Mutation Analysis for EGFR, KRAS, and PIK3CA
DNA was extracted from five 10-μm formalin-fixed paraffin-embedded sections containing a representative portion of each tumor block, using the QIAamp DNA Mini Kit (Qiagen, Hilden, Germany). A pathologist (K.-T.J.) reviewed each slide and verified the presence of adequate tumor tissue with greater than 50% representative malignant cells.
Peptide nucleic acid (PNA)–locked nucleic acid polymerase chain reaction (PCR) clamp reactions were carried out using the PNA-Clamp EGFR, PNA-Clamp KRAS, and PNA-Clamp PIK3CA Mutation Detection kits (Panagene, Inc, Daejeon, Korea), as described previously [20], [21]. Briefly, this reaction consists of the following: all reactions were done in 20-μl volumes using 10 to 25 ng of template DNA, primer and PNA probe set, and SYBR Green PCR master mix. All necessary reagents are included with the kit. Real-time PCR reactions of PNA-mediated clamping PCR were performed using a CFX 96 System (Bio-Rad, Foster City, California). PCR cycling conditions were a 5-minute holding period at 94°C, followed by 40 cycles of 94°C for 30 seconds, 70°C for 20 seconds, 63°C for 30 seconds, and 72°C for 30 seconds. Detection of each of the 29 mutations in EGFR exons 18 to 21, 7 mutations in KRAS exon 2, and 3 mutations in PIK3CA exons 2 and 9 was possible using one-step PNA-mediated real-time PCR clamping.
Statistical Analysis
PFS, OS, and overall RR were evaluated with respect to tumor mutation status (KRAS and PIK3CA). PFS and OS were analyzed by the Kaplan-Meier method and stratified log-rank test. The predefined variables used to investigate the association of potential prognostic factors for PFS were age, sex, primary site, differentiation, disease status, liver-only metastasis, number of involved sites, the use of erlotinib, KRAS mutation status, PIK3CA mutation status, and any mutation of EGFR, KRAS, or PIK3CA. The Cox proportional hazards model was used for univariate and multivariate analyses to assess the independent effects of variables and to obtain their Hazard Ratio (HR estimates). P < .05 values were considered significant.
Results
Patients and Samples
In our previous study that included 268 patients randomly assigned to receive GEMOX plus erlotinib (N = 133) or GEMOX alone (N = 135), 227 events were observed over a median follow-up time of 15 months (range, 11.0-18.9 months) [11].
In the initial analysis, we were able to analyze either EGFR overexpression or KRAS mutation status from 60 tissue specimens (22.3% of the ITT population) that had sufficient DNA (Figure 1). In contrast, a total of 125 tissue specimens (mutation analysis population; 46.6% of the ITT population) was included for current analysis (Figure 1).
Figure 1.
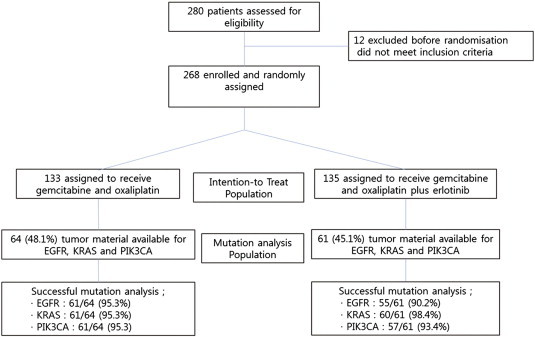
Consort.
The baseline characteristics were well balanced between the treatment groups in the ITT and mutation analysis population (Table 1). In the mutation analysis population (N = 125), 64 patients (51.2%) received GEMOX alone and 61 (48.9%) received GEMOX plus erlotinib.
Table 1.
Characteristics of 125 Advanced BTC Patients Treated with GEMOX with or without Erlotinib
| ITT Population |
Mutation Analysis Population |
|||
|---|---|---|---|---|
| GEMOX (n = 133) | GEMOX Plus Erlotinib (n = 135) | GEMOX (n = 64) | GEMOX Plus Erlotinib (n = 61) | |
| Age, years | ||||
| Median | 61 (55-68) | 59 (54-66) | 61 (45-77) | 58 (39-77) |
| Sex | ||||
| Male | 79 (59%) | 91 (67%) | 42 (65.6%) | 39 (63.9%) |
| Female | 54 (41%) | 44 (33%) | 22 (34.4%) | 22 (36.1%) |
| Primary site | ||||
| Cholangiocarcinoma (intra-hepatic and extra-hepatic) | 84 (63%) | 96 (71%) | 43 (67.2%) | 44 (72.1%) |
| Gallbladder | 47 (35%) | 35 (26%) | 20 (31.3%) | 14 (23.0%) |
| AoV | 2 (2%) | 4 (3%) | 1 (1.6%) | 3 (4.9%) |
| Differentiation | ||||
| Well | 6 (5%) | 11 (8%) | 3 (4.7%) | 7 (11.5%) |
| Moderate | 92 (69%) | 91 (67%) | 36 (56.3%) | 28 (45.9%) |
| Poorly or unknown | 35 (26%) | 33 (24%) | 25 (39.1%) | 26 (42.6%) |
| EOCG performance status | ||||
| 0 | 20 (15%) | 26 (19%) | 5 (7.8%) | 7 (11.5%) |
| 1 | 100 (75%) | 104 (77%) | 56 (87.5%) | 54 (88.5%) |
| 2 | 13 (10%) | 5 (4%) | 1 (1.6%) | - |
| Disease status | ||||
| Recurrent | 23 (17%) | 31 (23%) | 16 (25.0%) | 22 (36.1%) |
| Primarily metastatic | 110 (83%) | 104 (77%) | 48 (75.0%) | 39 (63.9%) |
| Liver-only metastasis | ||||
| Yes | 46 (35%) | 42 (31%) | 13 (20.3%) | 10 (16.4%) |
| No | 87 (65%) | 93 (69%) | 51 (79.7%) | 51 (83.6%) |
| Number of metastatic sites | ||||
| 1 | 82 (62%) | 84 (62%) | 52 (81.3%) | 52 (85.2%) |
| 2 ≤ | 51 (38%) | 51 (38%) | 12 (18.8%) | 9 (14.7%) |
Frequency of EGFR, KRAS, and PIK3CA Mutations
The mutational status of EGFR, KRAS, and PIK3CA was identifiable in 116, 121, and 118 patients, respectively. EGFR mutations were found in 2 of 116 patients (1.7%). Two cholangiocarcinoma patients harbored EGFR T790M mutations and were treated with GEMOX alone. Following chemotherapy, one of these patients showed a complete response, while the second patient maintained stable disease. The incidence of KRAS mutations was 9.9% [12 of 121, 4 GBCs, 7 cholangiocarcinomas, and 1 Ampulla of Vater (AoV) cancer]. Most KRAS mutations occurred at codon 12 (11 of 12 patients, 91.6%). PIK3CA mutations, which were observed in 12 of 118 patients (10.1%), occurred mainly in exons 9 (10 patients) and 10 (2 patients). All patients with PIK3CA mutations had cholangiocarcinoma (Table 2). Mutations in both KRAS and PIK3CA were found in the tumor tissues from three patients.
Table 2.
Mutational Spectrum of EGFR, KRAS, and PIK3CA
| Gene | Gallbladder, n = 34, (%) | Cholangiocarcinoma, n = 87, (%) | AoV, n = 4, (%) |
|---|---|---|---|
| EGFR | 0 (0.0%) | 2 (2.3%) | 0 (0.0%) |
| T790M | 0 (0.0%) | 2 (2.3%) | 0 (0.0%) |
| KRAS | 4 (11.7%) | 7 (8.0%) | 1 (25%) |
| Codon 12 | 4 (11.7%) | 6 (6.9%) | 1 (25%) |
| Codon 13 | 0 (0.0%) | 1 (1.1%) | 0 (0.0%) |
| PIK3CA | 0 (0.0%) | 12 (13.8%) | 0 (0.0%) |
| E542 | 0 (0.0%) | 3 (3.5%) | 0 (0.0%) |
| E545 | 0 (0.0%) | 7 (8.0%) | 0 (0.0%) |
| H1047 | 0 (0.0%) | 2 (2.3%) | 0 (0.0%) |
Impact of Mutation of KRAS and PIK3CA on Clinical Outcomes
The statistical significances (P value) for RR, PFS, and OS in the ITT and mutational analysis populations were comparable (Table 3). Patients with wild-type KRAS tumors who received erlotinib plus GEMOX (GEMOXT) had significantly increased overall RR compared with those who received GEMOX alone (Table 3). However, there was no significant difference in RR in patients with KRAS mutations between the GEMOX with and without erlotinib treatment groups. In patients whose tumors carried wild-type PIK3CA, erlotinib plus GEMOX showed a favorable trend for overall response, but no statistically significance difference (P = .08). In 95 patients with both wild-type KRAS and PIK3CA, there was evidence of a benefit associated with the addition of erlotinib to GEMOX in relation to RR as compared with GEMOX alone (P = .04). No significant difference was observed in PFS and OS between the treatment groups, irrespective of KRAS or PIK3CA mutation status. Additionally, in 92 patients with wild-type EGFR, KRAS, and PIK3CA, the addition of erlotinib to GEMOX improved tumor responses (P = .03; Table 3).
Table 3.
Efficacy of GEMOX with or without Erlotinib according to the Mutational Status
| ITT Population (n = 268) |
Mutation Analysis Population (n = 125) |
KRAS Mutation Status (n = 121) |
PIK3CA Mutation Status (n = 118) |
Overall Mutation Status (n = 114) |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| KRAS Wild (n = 109) |
KRAS Mutant (n = 12) |
PIK3CA Wild (n = 106) |
PIK3CA Mutant (n = 12) |
No Mutation⁎ (n = 92) |
Any Mutant† (n = 22) |
|||||||||||
| GEMOX | GEMOXT | GEMOX | GEMOXT | GEMOX | GEMOXT | GEMOX | GEMOXT | GEMOX | GEMOXT | GEMOX | GEMOXT | GEMOX | GEMOXT | GEMOX | GEMOXT | |
| RR | 16% | 30% | 15.6% | 31.1% | 12.5% | 30.2% | 60% | 28.6% | 14.8% | 28.8% | 14.3% | 60.0% | 12.7% | 31.1% | 25.0% | 40.0% |
| 0.005 | 0.040 | 0.024 | 0.558 | 0.080 | 0.222 | 0.033 | 0.452 | |||||||||
| DCR | 67% | 66% | 65.6% | 62.3% | 66.1% | 60.4% | 60% | 71.4% | 66.7% | 61.5% | 57.1% | 80.0% | 68.0% | 60.0% | 58.3% | 80.0% |
| - | 0.698 | 0.538 | 1.000 | 0.582 | 0.576 | 0.419 | 0.381 | |||||||||
| PFS | 4.2 months | 5.8 months | 2.7 months | 6.2 months | 2.7 months | 6.6 months | 11.5 months | 3.1 months | 2.9 months | 6.2 months | 2.1 months | 7.3 months | 4.0 months | 6.4 months | 2.1 months | 4.1 months |
| 0.087 | 0.172 | 0.125 | 0.501 | 0.180 | 0.858 | 0.296 | 0.735 | |||||||||
| OS | 9.5 months | 9.5 months | 8.0 months | 9.7 months | 8.0 months | 9.1 months | NA months | 10.7 months | 8.5 months | 9.7 months | 4.0 months | 10.7 months | 8.5 months | 8.5 months | 7.2 months | 10.7 months |
| 0.611 | 0.415 | 0.449 | 0.509 | 0.674 | 0.547 | 0.766 | 0.742 | |||||||||
DCR; Disease Conrol Rate.
Wild type for all EGFR, KRAS, and PIK3CA
Any mutant type for EGFR, KRAS, or PIK3C
In the mutation analysis population (n = 125), the estimated median PFS time was 4.8 months [95% confidence interval (CI), 3.2-6.4]. Median PFS was not significantly different in either treatment group [2.7 months (95% CI, 1.3-4.1) in GEMOX alone group and 6.2 months (95% CI, 4.5-7.8) in GEMOX plus erlotinib group, P = .172; Figure 2]. There was also no significant difference in PFS between the GEMOX with and without erlotinib groups in either patients with wild-type EGFR/KRAS/PIK3CA or with any mutation in EGFR or KRAS or PIK3CA (Figure 3). Univariate analysis revealed that PFS was not associated with the addition of erlotinib to chemotherapy, mutation status of KRAS and PIK3CA, and wild-type status of EGFR/KRAS/PIK3CA (Table 4).
Figure 2.
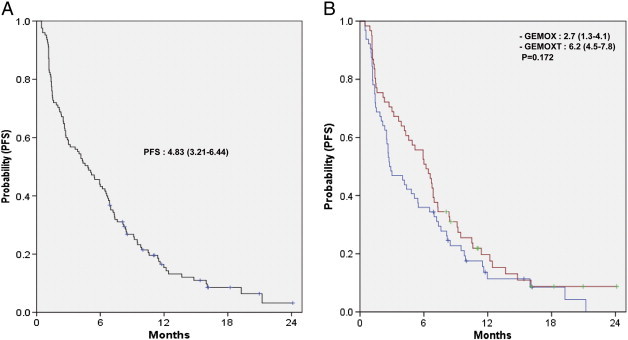
PFS. (A) Overall and (B) according to GEMOX with or without erlotinib.
Figure 3.
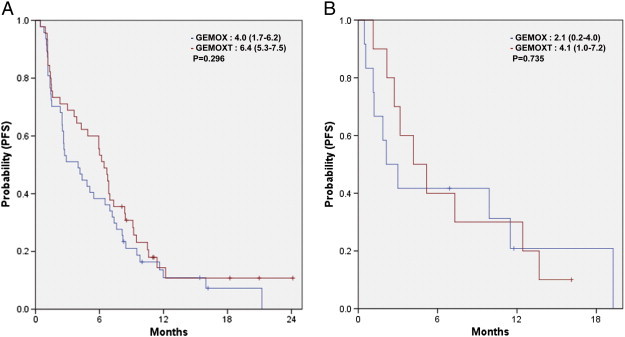
PFS according to GEMOX with or without erlotinib in patients with (A) EGFR/KRAS/PIK3CA wild type and (B) any mutation of EGFR or KRAS or PIK3CA.
Table 4.
Prognostic Factors for PFS in Univariate Analysis
| Characteristics | Median PFS (95% CI), Months | Univariate |
|
|---|---|---|---|
| HR (95% CI) | P | ||
| Age | 0.880 (0.606-1.278) | .502 | |
| ≤ 60 | 5.93 (3.57-8.28) | ||
| > 60 | 4.00 (1.62-6.37) | ||
| Sex | 0.942 (0.775-1.144) | .546 | |
| Male | 4.17 (2.30-6.03) | ||
| Female | 5.93 (3.90-7.95) | ||
| Primary sites | 1.003 (0.666-1.510) | .989 | |
| Cholangiocarcinoma | 4.90 (3.13-6.66) | ||
| GBC/AoV cancer | 3.63 (0.05-7.20) | ||
| Differentiation | 1.114 (0.763-1.626) | .578 | |
| Well/moderate | 4.57 (1.61-7.52) | ||
| Poorly | 4.90 (2.43-7.36) | ||
| Disease status | 1.406 (0.928-2.130) | .108 | |
| Recurrent | 6.60 (3.77-9.42) | ||
| Primarily metastatic | 4.17 (1.93-6.40) | ||
| Liver-only metastasis | 1.239 (0.761-2.018) | .388 | |
| Yes | 3.87 (1.72-6.01) | ||
| No | 4.90 (3.09-6.70) | ||
| Number of metastatic sites | 0.969 (0.600-1.562) | .896 | |
| 1 | 4.30 (2.38-6.21) | ||
| 2 ≤ | 6.00 (3.45-8.54) | ||
| GEMOX | 0.772 (0.532-1.122) | .175 | |
| Without erlotinib | 2.77 (1.34-4.19) | ||
| With erlotinib | 6.20 (4.52-7.87) | ||
| KRAS | 0.846 (0.427-1.626) | .631 | |
| Wild type | 4.90 (3.04-6.76) | ||
| Mutant type | 3.17 (0.00-7.31) | ||
| PIK3CA | 0.974 (0.519-1.828) | .934 | |
| Wild type | 5.10 (3.25-6.95) | ||
| Mutant type | 3.00 (0.00-6.46) | ||
| Mutation status | 1.095 (0.662-1.812) | .723 | |
| EGFR/KRAS/PIK3CA wild | 5.10 (3.19-7.00) | ||
| Any mutation | 3.17 (0.36-5.97) | ||
Discussion
Emerging evidence has implicated the EGFR pathway as a potential therapeutic target in BTC [22], [23]. Oncogenic activation of the EGFR pathway, including EGFR-RAS-RAF or EGFR-PIK3CA signaling, is a key mechanism in the efficacy of EGFR-directed therapy [16]. Presently, we do not know the biologic significance of mutations in these genes or to what extent they confer sensitivity to currently available small-molecule inhibitors, including erlotinib, in BTCs. We have previously analyzed the efficacy of chemotherapy plus erlotinib with respect to KRAS status using data from our previous phase III trial for BTC. Clinical samples from which tumor DNA could be successfully analyzed were initially available for a subset of 22.3% of the ITT population. Subsequently, through the use of improved ascertainment, the total number of patients from whom tumor KRAS mutation status could be assessed was approximately doubled to 46.6% of the ITT population (mutation analysis population). Moreover, mutation status for EGFR and PIK3CA was analyzed in this mutation analysis population. Herein, we report the impact of the mutation status of EGFR, KRAS, and PIK3CA on the clinical outcomes of patients treated with a small-molecule inhibitor of the EGFR kinase domain in an expanded patient cohort.
This analysis suggested that KRAS mutation might be a predictor of resistance to the small-molecule inhibitor erlotinib in BTCs. It is yet unknown if KRAS mutation in BTCs is a negative predictor of the effectiveness of anti-EGFR treatment. In colorectal cancer, it has been established that KRAS mutation precludes any therapeutic benefit from anti-EGFR antibodies (cetuximab and panitumumab) [24], [25]. In addition, we have also published that KRAS mutations might be negative predictive biomarkers in advanced pancreatic cancer patients who have been treated with a gemcitabine-erlotinib combination [26]. In contrast, this concept has not yet been confirmed in other types of cancer including BTCs. Gruenberger et al. investigated the efficacy of gemcitabine, oxaliplatin, and cetuximab combinations in patients with advanced BTCs and reported that two of three patients harboring KRAS mutations showed a partial response to cetuximab plus chemotherapy [27]. We also analyzed KRAS mutations in the 60 specimens in our study and showed that three of six patients with KRAS mutations responded to erlotinib [11]. These findings suggest that anti-EGFR therapies might be beneficial, irrespective of KRAS mutational status, in BTCs. However, our sample size was too small to allow us to draw any significant conclusions. There is a discrepancy between our current findings and previous reports, which may have resulted from our small sample size, geographic differences, different testing techniques, and heterogeneity of the patient populations between studies [11], [27]. Although our present analysis used a larger sample set as compared with previous analyses, the sample size of this study was still too small.
Additionally, we found that PIK3CA mutation was also associated with a lack of erlotinib activity in BTCs that retain wild-type KRAS. PIK3CA mutations, which are commonly found in colon, breast, gastric, and brain cancers, are rarely found in BTC [15]. PIK3CA mutations mainly occur in exons 9 (E542K, E545K) and 20 (H1047R) [18], [28], [29]. In our analysis, 12 patients had PIK3CA mutations, with 10 having a mutation in exon 9 and 2 in exon 20. Of the 12 patients with PIK3CA mutation, 3 patients also had a KRAS mutation. In the 95 patients with both wild-type KRAS and PIK3CA, there was evidence of a benefit associated with the addition of erlotinib to GEMOX, as compared with GEMOX alone, with respect to RR (P = .04). However, in 9 patients with wild-type KRAS and PIK3CA mutations (8 in exon 9 and 1 in exon 20), the addition of erlotinib to chemotherapy did not result in an improved response (P = .08). The effect of the mutation status of PIK3CA on anti-EGFR therapies has been studied in KRAS wild-type colorectal cancers [13], [18], [30], [31]. To the best of our knowledge, this is the first report to evaluate the correlation between the mutation status of PIK3CA and the activity of the anti-EGFR therapy erlotinib in BTCs. Clearly, however, the role of KRAS and PIK3CA mutations in response to treatment with anti-EGFR agents needs further analysis in a larger homogeneous population.
EGFR mutations occur in a significant minority of patients (13-15%) with BTC; however, the results of mutation analyses from two studies were quite different [13], [14]. In our study, EGFR mutation was found in 2 of 116 patients (1.7%). These two patients harbored an EGFR T790M mutation that is known to confer resistance to currently available small-molecule inhibitors. This EGFR T790M mutation was not described in previous studies [13], [14].
Our analysis revealed that KRAS mutational status might be considered a negative predictive biomarker for response to erlotinib in BTCs. Additionally, the mutation status of PIK3CA may be a determinant in the decision as to whether to add erlotinib to chemotherapy in KRAS wild-type BTCs. However, tissue availability was a potential limitation of the current retrospective mutational analysis. This is as critical an issue in BTC research as it is in pancreatic cancer. In the current study, about 20% of patients in the ITT population underwent prior surgery, as did about 30% of patients in the mutation analysis population, thereby effectively limiting the availability and adequacy of surgical specimens for biomarker analysis. Therefore, possible selection bias in the current study may make definitive conclusions difficult and may have influenced the prognostic and predictive results of KRAS and PIK3CA status, which need to be interpreted with caution and validated. Further prospective studies are needed for defining predictive or prognostic biomarkers. The rarity of BTC hinders clinicians from conducting definitive trials and from producing rigorous scientific data. Thus, coordination of trials among institutions and cooperative groups, both nationally and internationally, will be the key to improving treatment outcomes in BTCs. Additionally, because of the heterogeneity of BTCs, appropriate stratification using clinical or molecular factors will help to define more clearly outcomes of the research.
Acknowledgements
This study was supported by a grant from the Korean Health Technology R&D Project, Ministry of Health and Welfare, Republic of Korea to J.O.P. (HI11C1416) and by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology to J.O.P. (NRF-2013R1A1A2013441). Disclosure of potential conflicts of interest: The authors have declared no conflict of interest.
References
- 1.Won Y.J., Sung J., Jung K.W., Kong H.J., Park S., Shin H.R., Park E.C., Ahn Y.O., Hwang I.K., Lee D.H. Nationwide cancer incidence in Korea, 2003–2005. Cancer Res Treat. 2009;41:122–131. doi: 10.4143/crt.2009.41.3.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Glimelius B., Hoffman K., Sjoden P.O., Jacobsson G., Sellström H., Enander L.K., Linné T., Svensson C. Chemotherapy improves survival and quality of life in advanced pancreatic and biliary cancer. Ann Oncol. 1996;7:593–600. doi: 10.1093/oxfordjournals.annonc.a010676. [DOI] [PubMed] [Google Scholar]
- 3.Thongprasert S. The role of chemotherapy in cholangiocarcinoma. Ann Oncol. 2005;16(Suppl. 2):ii93–ii96. doi: 10.1093/annonc/mdi712. [DOI] [PubMed] [Google Scholar]
- 4.Valle J.W., Wasan H., Johnson P., Jones E., Dixon L., Swindell R., Baka S., Maraveyas A., Corrie P., Falk S. Gemcitabine alone or in combination with cisplatin in patients with advanced or metastatic cholangiocarcinomas or other biliary tract tumours: a multicentre randomised phase II study – The UK ABC-01 Study. Br J Cancer. 2009;101:621–627. doi: 10.1038/sj.bjc.6605211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Valle J., Wasan H., Palmer D.H., Cunningham D., Anthoney A., Maraveyas A., Madhusudan S., Iveson T., Hughes S., Pereira S.P. Cisplatin plus gemcitabine versus gemcitabine for biliary tract cancer. N Engl J Med. 2010;362:1273–1281. doi: 10.1056/NEJMoa0908721. [DOI] [PubMed] [Google Scholar]
- 6.Llovet J.M., Ricci S., Mazzaferro V., Hilgard P., Gane E., Blanc J.F., de Oliveira A.C., Santoro A., Raoul J.L., Forner A. Sorafenib in advanced hepatocellular carcinoma. N Engl J Med. 2008;359:378–390. doi: 10.1056/NEJMoa0708857. [DOI] [PubMed] [Google Scholar]
- 7.Hurwitz H., Fehrenbacher L., Novotny W., Cartwright T., Hainsworth J., Heim W., Berlin J., Baron A., Griffing S., Holmgren E. Bevacizumab plus irinotecan, fluorouracil, and leucovorin for metastatic colorectal cancer. N Engl J Med. 2004;350:2335–2342. doi: 10.1056/NEJMoa032691. [DOI] [PubMed] [Google Scholar]
- 8.Moore M.J., Goldstein D., Hamm J., Figer A., Hecht J.R., Gallinger S., Au H.J., Murawa P., Walde D., Wolff R.A. Erlotinib plus gemcitabine compared with gemcitabine alone in patients with advanced pancreatic cancer: a phase III trial of the National Cancer Institute of Canada Clinical Trials Group. J Clin Oncol. 2007;25:1960–1966. doi: 10.1200/JCO.2006.07.9525. [DOI] [PubMed] [Google Scholar]
- 9.Philip P.A., Mahoney M.R., Allmer C., Thomas J., Pitot H.C., Kim G., Donehower R.C., Fitch T., Picus J., Erlichman C. Phase II study of erlotinib in patients with advanced biliary cancer. J Clin Oncol. 2006;24:3069–3074. doi: 10.1200/JCO.2005.05.3579. [DOI] [PubMed] [Google Scholar]
- 10.Lubner S.J., Mahoney M.R., Kolesar J.L., Loconte N.K., Kim G.P., Pitot H.C., Philip P.A., Picus J., Yong W.P., Horvath L. Report of a multicenter phase II trial testing a combination of biweekly bevacizumab and daily erlotinib in patients with unresectable biliary cancer: a phase II Consortium study. J Clin Oncol. 2010;28:3491–3497. doi: 10.1200/JCO.2010.28.4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee J., Park S.H., Chang H.M., Kim J.S., Choi H.J., Lee M.A., Jang J.S., Jeung H.C., Kang J.H., Lee H.W. Gemcitabine and oxaliplatin with or without erlotinib in advanced biliary-tract cancer: a multicentre, open-label, randomised, phase 3 study. Lancet Oncol. 2012;13:181–188. doi: 10.1016/S1470-2045(11)70301-1. [DOI] [PubMed] [Google Scholar]
- 12.Hanada K., Tsuchida A., Iwao T., Eguchi N., Sasaki T., Morinaka K., Matsubara K., Kawasaki Y., Yamamoto S., Kajiyama G. Gene mutations of K-ras in gallbladder mucosae and gallbladder carcinoma with an anomalous junction of the pancreaticobiliary duct. Am J Gastroenterol. 1999;94:1638–1642. doi: 10.1111/j.1572-0241.1999.01155.x. [DOI] [PubMed] [Google Scholar]
- 13.Leone F., Cavalloni G., Pignochino Y., Sarotto I., Ferraris R., Piacibello W., Venesio T., Capussotti L., Risio M., Aglietta M. Somatic mutations of epidermal growth factor receptor in bile duct and gallbladder carcinoma. Clin Cancer Res. 2006;12:1680–1685. doi: 10.1158/1078-0432.CCR-05-1692. [DOI] [PubMed] [Google Scholar]
- 14.Gwak G.Y., Yoon J.H., Shin C.M., Ahn Y.J., Chung J.K., Kim Y.A., Kim T.Y., Lee H.S. Detection of response-predicting mutations in the kinase domain of the epidermal growth factor receptor gene in cholangiocarcinomas. J Cancer Res Clin Oncol. 2005;131:649–652. doi: 10.1007/s00432-005-0016-1. [DOI] [PubMed] [Google Scholar]
- 15.Riener M.O., Bawohl M., Clavien P.A., Jochum W. Rare PIK3CA hotspot mutations in carcinomas of the biliary tract. Genes Chromosomes Cancer. 2008;47:363–367. doi: 10.1002/gcc.20540. [DOI] [PubMed] [Google Scholar]
- 16.Ciardiello F., Tortora G. EGFR antagonists in cancer treatment. N Engl J Med. 2008;358:1160–1174. doi: 10.1056/NEJMra0707704. [DOI] [PubMed] [Google Scholar]
- 17.Marino D., Leone F., Cavalloni G., Cagnazzo C., Aglietta M. Biliary tract carcinomas: from chemotherapy to targeted therapy. Crit Rev Oncol Hematol. 2013;85:136–148. doi: 10.1016/j.critrevonc.2012.06.006. [DOI] [PubMed] [Google Scholar]
- 18.De Roock W., Claes B., Bernasconi D., De Schutter J., Biesmans B., Fountzilas G., Kalogeras K.T., Kotoula V., Papamichael D., Laurent-Puig P. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis. Lancet Oncol. 2010;11:753–762. doi: 10.1016/S1470-2045(10)70130-3. [DOI] [PubMed] [Google Scholar]
- 19.Massarelli E., Varella-Garcia M., Tang X., Xavier A.C., Ozburn N.C., Liu D.D., Bekele B.N., Herbst R.S., Wistuba I.I. KRAS mutation is an important predictor of resistance to therapy with epidermal growth factor receptor tyrosine kinase inhibitors in non-small-cell lung cancer. Clin Cancer Res. 2007;13:2890–2896. doi: 10.1158/1078-0432.CCR-06-3043. [DOI] [PubMed] [Google Scholar]
- 20.Kwon M.J., Lee S.E., Kang S.Y., Choi Y.L. Frequency of KRAS, BRAF, and PIK3CA mutations in advanced colorectal cancers: Comparison of peptide nucleic acid-mediated PCR clamping and direct sequencing in formalin-fixed, paraffin-embedded tissue. Pathol Res Pract. 2011;207:762–768. doi: 10.1016/j.prp.2011.10.002. [DOI] [PubMed] [Google Scholar]
- 21.Kobunai T., Watanabe T., Yamamoto Y., Eshima K. The frequency of KRAS mutation detection in human colon carcinoma is influenced by the sensitivity of assay methodology: a comparison between direct sequencing and real-time PCR. Biochem Biophys Res Commun. 2010;395:158–162. doi: 10.1016/j.bbrc.2010.03.167. [DOI] [PubMed] [Google Scholar]
- 22.Hezel A.F., Deshpande V., Zhu A.X. Genetics of biliary tract cancers and emerging targeted therapies. J Clin Oncol. 2010;28:3531–3540. doi: 10.1200/JCO.2009.27.4787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu A.X., Hezel A.F. Development of molecularly targeted therapies in biliary tract cancers: reassessing the challenges and opportunities. Hepatology. 2011;53:695–704. doi: 10.1002/hep.24145. [DOI] [PubMed] [Google Scholar]
- 24.Amado R.G., Wolf M., Peeters M., Van Cutsem E., Siena S., Freeman D.J., Juan T., Sikorski R., Suggs S., Radinsky R. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26:1626–1634. doi: 10.1200/JCO.2007.14.7116. [DOI] [PubMed] [Google Scholar]
- 25.Van Cutsem E., Köhne C.H., Láng I., Folprecht G., Nowacki M.P., Cascinu S., Shchepotin I., Maurel J., Cunningham D., Tejpar S. Cetuximab plus irinotecan, fluorouracil, and leucovorin as first-line treatment for metastatic colorectal cancer: updated analysis of overall survival according to tumor KRAS and BRAF mutation status. J Clin Oncol. 2011;29:2011–2019. doi: 10.1200/JCO.2010.33.5091. [DOI] [PubMed] [Google Scholar]
- 26.Kim S.T., Lim do H., Jang K.T., Lim T., Lee J., Choi Y.L., Jang H.L., Yi J.H., Baek K.K., Park S.H. Impact of KRAS mutations on clinical outcomes in pancreatic cancer patients treated with first-line gemcitabine-based chemotherapy. Mol Cancer Ther. 2011;10:1993–1999. doi: 10.1158/1535-7163.MCT-11-0269. [DOI] [PubMed] [Google Scholar]
- 27.Gruenberger B., Schueller J., Heubrandtner U., Wrba F., Tamandl D., Kaczirek K., Roka R., Freimann-Pircher S., Gruenberger T. Cetuximab, gemcitabine, and oxaliplatin in patients with unresectable advanced or metastatic biliary tract cancer: a phase 2 study. Lancet Oncol. 2010;11:1142–1148. doi: 10.1016/S1470-2045(10)70247-3. [DOI] [PubMed] [Google Scholar]
- 28.Moroni M., Veronese S., Benvenuti S., Marrapese G., Sartore-Bianchi A., Di Nicolantonio F., Gambacorta M., Siena S., Bardelli A. Gene copy number for epidermal growth factor receptor (EGFR) and clinical response to antiEGFR treatment in colorectal cancer: a cohort study. Lancet Oncol. 2005;6:279–286. doi: 10.1016/S1470-2045(05)70102-9. [DOI] [PubMed] [Google Scholar]
- 29.Lièvre A., Bachet J.B., Le Corre D., Boige V., Landi B., Emile J.F., Côté J.F., Tomasic G., Penna C., Ducreux M. KRAS mutation status is predictive of response to cetuximab therapy in colorectal cancer. Cancer Res. 2006;66:3992–3995. doi: 10.1158/0008-5472.CAN-06-0191. [DOI] [PubMed] [Google Scholar]
- 30.Prenen H., De Schutter J., Jacobs B., De Roock W., Biesmans B., Claes B., Lambrechts D., Van Cutsem E., Tejpar S. PIK3CA mutations are not a major determinant of resistance to the epidermal growth factor receptor inhibitor cetuximab in metastatic colorectal cancer. Clin Cancer Res. 2009;15:3184–3188. doi: 10.1158/1078-0432.CCR-08-2961. [DOI] [PubMed] [Google Scholar]
- 31.Ogino S., Nosho K., Kirkner G.J., Shima K., Irahara N., Kure S., Chan A.T., Engelman J.A., Kraft P., Cantley L.C. PIK3CA mutation is associated with poor prognosis among patients with curatively resected colon cancer. J Clin Oncol. 2009;27:1477–1484. doi: 10.1200/JCO.2008.18.6544. [DOI] [PMC free article] [PubMed] [Google Scholar]


