Abstract
Despite significant advances in microbiology and molecular biology over the last decades, several infectious diseases remain global concerns, resulting in the death of millions of people worldwide each year. According to the Center for Disease Control (CDC) in 2012, there were 34 million people infected with HIV, 8.7 million new cases of tuberculosis, 500 million cases of hepatitis, and 50–100 million people infected with dengue. Several of these pathogens, despite high incidence, do not have reliable clinical detection methods. New or improved protocols have been generated to enhance detection and quantitation of several pathogens using high-end microscopy (light, confocal, and STORM microscopy) and imaging software. In the current manuscript, we discuss these approaches and the theories behind these methodologies. Thus, advances in imaging techniques will open new possibilities to discover therapeutic interventions to reduce or eliminate the devastating consequences of infectious diseases.
Keywords: HIV, tuberculosis, dengue, viruses
Introduction
Although significant advances have been made in the area of infectious diseases such as pathogenesis, therapy, and public awareness; the spread of highly contagious infectious disease remains a current and menacing prospect. The earth had once been divided into separate bio-geographic areas, but this division has been dissolved by the ease of worldwide transportation. Air travel in particular has enabled rapid spread of disease. The constant shuffle of human traffic and livestock trade among the continents has brought new species of pathogens and their vectors into previously unexposed regions. To counter this global health compromise, enormous efforts were made to institute a ‘Decade of Vaccines’ at the 2010 World Economic Forum as vaccines have served as the most successful protection for public health since Dr. Edward Jenner developed the vaccine for smallpox (D’Argenio & Wilson, 2010). However, the most lethal and debilitating infectious diseases remain public health challenges because of their abilities to elude host immune defenses, and to develop multidrug resistance, as well as the lack of information about their pathogenic mechanisms (Koff et al., 2013). To detect, quantitate, and analyze pathogens, classic microbiological approaches such as: plaque assay, culture, quantitation of colony-forming units (CFU), dye-based staining (e.g. Gram or acid fast/Ziehl Nelson), immune-based assays, and PCR are the standard means; however, several of those techniques are limited under certain microbial-induced conditions such as latency, undetectable replication, low infectivity, high-level adaption/mutation rates or encapsulation alterations. Additionally, application of these techniques has been limited in part due to biosafety concerns. To address several of these issues, the National Institute of Allergy and Infectious Diseases (NIAID) established two National Biocontainment Laboratories (NBL) and 13 Regional Biocontainment Laboratories (RBL). The main focus of the RBL, of the Public Health Research Institute (PHRI) at Rutgers The State University of New Jersey, is to understand the pathogenesis/virulence of several infectious diseases and to develop vaccines, diagnostics, and therapeutics (http://njms.rutgers.edu/research/rbl). Within the RBL of PHRI, our shared imaging facility integrates the advances in high-end microscopy with microbial research. The imaging facility is composed of several types of BSL2/3 imaging equipment including microscopy, full animal imaging, and imaging mass spectrometry—described in detail at http://phri.org/facilities/facil_imaging.asp. In the current manuscript, we will focus on key features of high-end microscopy and associated imaging equipment that allow us to generate new protocols for detection and quantitation of several pathogens. For a brief description of these critical improvements, refer to Table 1. These elements include the following: improved spectral detection systems; better cameras and detection systems; advanced concepts in 3D super-resolution; redevelopment of improved live cell imaging techniques; improved software; and new and/or improved detection and amplification techniques for optimal identification and quantitation of pathogens.
Table 1.
Improvements in detection systems
| Key elements | Former techniques | Advanced techniques |
|---|---|---|
| Spectral detector system | Limited wavelength distinction Limited use of multiple colors/stains Excess autofluorescence |
Separation of wavelengths to 2.5 nm Simultaneous use of more colors/stains Minimize autofluorescence |
| Increased sensitivity, speed and resolution of new cameras on the market | Low quantum efficiency: 50–65% Less image area/pixel depth Low frames per second (fps) Limited cooling systems |
Efficiencies 60–94% Image area/pixel depth:13–24 µm, 17 mHz, 12 million pixels Up to 11 074 fps Improved cooling system (to −100 °C) Integrated software Extremely low read noise (to 1 e− read noise) |
| 3D super-resolution | Maximum resolution: 200 nm (diffraction barrier) -Propensity to photobleach -Limited XYZ axes |
Super-resolution: SIM: 8–300 nm, TIRF: 25–800 nm, STORM: 2–30 nm Improved XYZ axes Photo-switching fluorophore |
| Capacity for extended live cell imaging | Single laser beam; single pinhole Slow speed Photobleaching Rapid or heavy cell damage Short-term imaging |
- disk; scattered beam; multiple pinholes Fast speed of 5000–10 000 r.p.m., (1000–2000 fps) Reduced photobleaching Minimal cellular or pathogen photodamage Live cell imaging coupled with better cameras resulted in decreased photodamage over long-term imaging |
| Improved software for image acquisition and processing | Limited by equipment | Improved application potential of software with advanced equipment Better algorithms |
| Generation of novel or improved techniques for sample preparation (hydration, antigen recovery, tissue sectioning) | Limited to thin sections (5–10 µm) Excess background Sterilization of pathogens to increase biosafety results in compromised antigen viability |
Minimalized background due to better cameras, spectral detection, or less photobleaching Tissue sample thickness > 100 µm: preserves 3D structure and wide XYZ fields for better analysis Optimal sample preparation: better antigen recovery & staining techniques Better sterilization of BSL2/3 pathogen |
| New or improved staining techniques to amplify minimal signals for DNA, RNA, lipids, and proteins as well as small molecules | Detection limited by size and quantity of target | Single copy number and small molecules detectable Application of tyramide signal amplification technology Application of molecular beacons or similar technology |
Recent developments in microscopy that contribute to more reliable BSL2/3 pathogen detection and quantitation
We identified seven key elements to achieve better pathogen detection. First, a former setback with previous confocal systems was the inability to finely separate close emission wavelengths in fluorescence, thereby limiting protocols to the use of fewer fluorophores and working with high autofluorescence. The new confocal systems overcome this setback with our first key element: an improved spectral detector. The spectral detector is the mechanism responsible for emitting light through a high-efficiency gate into its individual components, similar to how a prism separates white light into its individual ‘rainbow’ components. The improved spectral detector allows for use of multiple lasers with extreme accuracy and thus enables data acquisition of 5–6 distinct colors in a sequential manner (http://www.microscopyu.com). Precise separation of emission wavelengths occurs as they pass through a 32-photomultiplier tube (PMT) array detector that enables distinction between wavelengths as small as 2.5 nm apart. This detection system allows the researcher to analyze specific wavelengths or eliminate autofluorescence. This technology allows us to identify signals that would otherwise be impossible to isolate with wide-field fluorescence or standard confocal microscopy. This key feature allows separation of signal from autofluorescence or from fluorophores that are extremely close together (e.g. GFP & Alexa488). The identification of the specific emission wavelength of an unknown fluorophore, drug, or a particular signature wavelength of a pathogen can be established. The advantages of simultaneous spectral detection are increased speed of acquisition, better precision despite vibration or sample movement, etc., and protection of biological samples from phototoxicity and photobleaching.
A second key element that allows for development of better protocols for detection of pathogens is the increased sensitivity (quantum efficiency, %), speed (frames per second, fps), and resolution of new cameras on the market. Normally, cameras have quantum efficiencies equal to or lower than 50–65%, but new cameras have detection efficiencies that range from 60% to over 94%. Thus, detection of dim signals is not an obstacle. While improved spectral detectors and PMTs are essential for decreasing/eliminating photobleaching and phototoxicity, camera efficiency is just as important for decreasing exposure time and increasing resolution capabilities. Therefore, higher quantum efficiency would overcome the problem for very dim samples that quickly undergo photobleaching or that have a very short fluorescence lifetime. To achieve high resolution and minimal exposure, the STORM, SIM, and spinning disk systems generally use Andor iXon or similar cameras, which capture images with a pixel size of 13–24 × 24 µm or 1024 × 1024 active pixels. In these cameras, capture rate reaches up to a maximal readout rate of 17 mHz with 56–11 074 fps. These cameras have excellent cooling ability and several systems of noise correction including low read noise (< 1 e− with EM gain). The image area pixel depth is around 180 000 e− and gain of 800 000 e− providing outstanding sensitivity and detection capabilities not previously possible. This vigorous speed is uniquely capable of protecting the sample from fluorescent phototoxicity and fluorescent decay, thereby allowing extraction of images from very dim samples despite limited photon exposure (http://www.microscopyu.com/). Because of this improved speed and sensitivity, the newer cameras cause less phototoxicity and enhance resolution. In consequence, a completely new and distinct technology has been innovated to surpass current applications.
The third key element is 3D super-resolution. Except for electron microscopy, previous light- and laser-based microscopes were incapable of resolving images beyond 200 nm, due to the diffraction limit. However, the advent of super-resolution has enabled us to resolve images near the level of electron microscopy. For example, the detection capacities of the N-SIM (structured illumination microscopy) is from 85 × 300 nm; the 2D Non-TIRF (total internal reflection fluorescence microscopy) is from 25 × 800 nm; and the 2D TIRF is from 25 × 100 nm. The detection of the STORM (stochastic optical reconstruction microscopy) is at a fine level with a lateral resolution of 2–30 nm and an axial resolution of 5–50 nm. This super-resolution concept requires cyanine family fluorophores (Alexa 647, Cy3, Cy5, Cy5.5, and Cy7) that are intrinsically capable of photo-switching between activated and deactivated statuses. The major notion of this is that alternative and random fluorophore activation and deactivation contrives an image of the specimen with nanometer localization accuracy. Cy5 in particular is able to be switched from fluorescent to dark state hundreds of times prior to the occurrence of photobleaching. Recovery time of a Cy family dye is enhanced by the immediacy of a switch to the nonfluorescent state. This photo-switching is achieved using a specific laser to drive labeled antibodies, in a reducing thiol buffer, back to a dark state. The random point scanning is executed to collect points on a Gaussian scale of photon detection along x, y, and z-axes. The image that is engineered from this method is constructed at a molecular level and thus allowing detection under the diffraction limit. STORM imaging is primarily designed for fixed cell imaging; however, N-SIM is available on the market for high-resolution analysis by live cell imaging (Rust et al., 2006; Bates et al., 2007; Huang et al., 2008) (see details at www.microscopyu.com). One current limitation of super-resolution is the selection of fluorophores and dyes; each of which has its limitations. Photo-switchable fluorophores are the most optimal, though there is a great necessity for the development of smaller sized fluorophores, as well as a broader family of them. Additionally, buffer optimizations are required to prevent any negative impact on the photo-switching fluorophores due to the presence of thiols and low levels of oxygen that cause them to stay in their dark state rather than their re-excited state. A benefit of photo-switching is that it protects the sample from photobleaching while maintaining a high quality photon exposure. Thus, better fluorophores and additional laser applications have resulted in large improvements in image resolution.
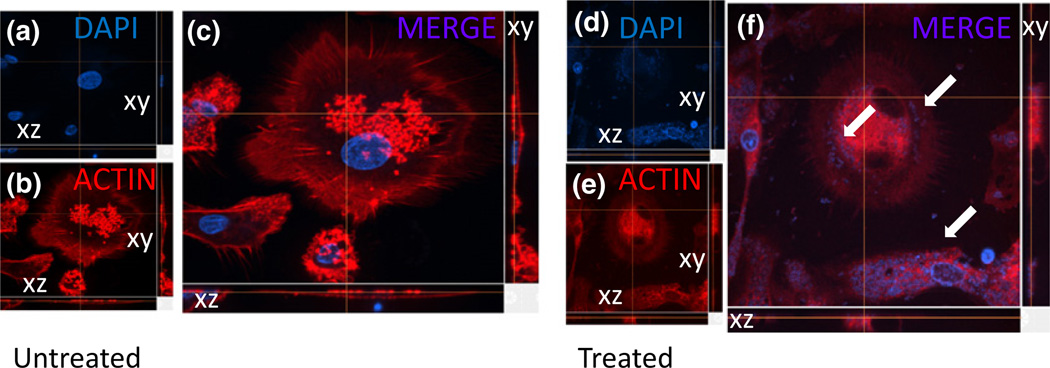
A fourth key element of improvement is the redevelopment of the spinning disk live cell imaging platforms. The spinning disk technology surmounts confocal microscopy with novel and improved focusing technology. Typically, a standard laser scanning microscope focuses a single beam on the specimen plane to sequentially point-scan a region of interest. This emission light is applied through a single pinhole that results in rejection of light from regions that are out of focus. However, this widely used system is extremely limited in image acquisition speed due to the photons emitted by the specimen during the pixel dwell time. In general, a regular confocal scans at the rate of 1 microsecond per pixel, thus, one image is reconstructed in around 2-s depending on the size and resolution used. In addition, most of the single-beam laser confocal microscopes produce significant damage to cells as well as photobleaching of fluorophores due to the constant stream of photons applied to the sample during the period of time required to build an image. Whereas the high resolution of the conventional confocal microscope requires long exposures, the spinning disk confocal microscope applies a series of 1000 or more parallel excitation light beams through multiple pinholes thus decreasing the level required for photon exposure to capture a comparable image. Depending upon the configuration, the spinning disk mechanism reaches spinning speeds of 5000 or 10 000 r.p.m.; these speeds correspond to an image capture rate of 1000 or 2000 fps, respectively. This system allows us to perform long-term live cell imaging with minimal cellular damage of the sample analyzed. As a proof of principle, we analyzed internalization and intracellular accumulation of Staphylococcus aureus in a time-dependent manner, without significant loss of fluorescence or detection of phototoxicity. Figure 1 illustrates Nikon spinning disk live cell images of neutrophils untreated (Fig. 1a–c) or treated with Staphylococcus for up to 120 h (Fig. 1d–f). In this case, the numbers of internalized bacteria were quantified by fluorescence intensity and 3-D structure, at different time points, cells were fixed and counterstained with actin (red staining, Fig. 1b and e) and 4’, 6-diamidino-2-phenylindole (DAPI) (blue staining, Fig. 1a and d). Figure 1a–c corresponds to control cells without Staphylococcus. Figure 1f corresponds to cells with internalized bacteria after 120 h of incubation with laser exposure every 1 min for 120 h. The loss of fluorescence, 9.22%, was minimal during the time course examined. Thus, these cells after 5 days of imaging still maintained high survival, minimal cell death, and intact fluorescence. Additional analyses include autophagy, apoptosis, intercellular uptake or release, cell proliferation, cell death or damage, phagocytosis, and internalization. Using the approaches and techniques described above, we have successfully imaged tagged proteins and pathogens for up to 2 weeks without significant toxicity, evaporation of medium, loss of fluorescence, or photodamage. It is important to indicate that most commercially available spinning disk systems use the same scanning/optical head, manufactured from Yokogawa (Masashino-shi, Tokyo, Japan). Another high-resolution system adapted for live cell imaging is SIM which applies the moiré effect for capturing three-dimensional images (see description of STORM). The moiré effect, or superimposition, is due to the moiré fringes that develop from the overlaying of different frequencies that result in multiplication of emission intensities (Gustafsson, 2005). Five fluorescent colors, three phases, and three angles converge to create a full 3D image. Combined with a piezo z stage, the movement is very rapid at 1–2 s per frame, even if using thick sections of 20–30 µm. In summary, live cell imaging has been advanced to prolong the life and integrity of the sample and has been incorporated with 3D super-resolution.
Fig. 1.
Detection and quantitation of uptake of Staphylococcus aureus by neutrophils. Using a spinning disk system, we performed time lapse experiments up to 120 h postincubation with the bacteria. (a–c) represent untreated cells, stained for nuclei (DAPI, nuclei staining), actin (phalloidin conjugated to Texas red, red staining), and S. aureus was stained blue; none present in untreated cells. Due to the accelerated and improved acquisition of data using the spinning disk, a decay of only 9.22% of fluorescent was detected during the time course analyzed. On the side of each picture, the XY and XZ focal planes are shown. (d–f) corresponds to cells incubated for 120 h with S. aureus. Internalized bacteria become evident, and quantitation is highly accurate due to the stability of the fluorescence (see arrows).
A fifth key element is the new and improved analysis software as compared to software limited to acquisition and basic analysis. Several software programs can be used for image acquisition and advanced analysis to accommodate the complexity of the confocal microscopic image. Some of the commercially available programs include axiovision/zen (Zeiss), cellsens (Olympus), and nis elements (Basic or Advance Research from Nikon). Additional proprietary software includes metamorph and volocity. image-j is one of the freely available and widely used types of the applied software. All of these programs allow the generation and adaptation of algorithms to particular applications that provide unique identification properties of proteins, lipids, dyes, DNA/RNA, or pathogens with extreme accuracy. Most of the BSL2/3 research described (see Figures) in this manuscript was generated using nis elements (Nikon) or zen (Zeiss). nis elements allow the capture, display, peripheral control, and data management for novel analysis of up to 6 dimensions (X, Y, Z, lambda (wavelength), T, and multipoint). It also offers sophisticated image processing features, such as an extremely powerful deconvolution module, exclusive one-click database capability, and extended depth of focus function. This software also interfaces with all Nikon microscopes and cameras, in contrast to many of the other manufacturers that have different software for confocal imaging vs. standard wide-field (camera-based) imaging, and some even utilize additional software for image analysis.
A sixth element is the modification of sample fixation for staining techniques in conjunction with the seventh element: the amplification of minimal signals of DNA, RNA, lipid, protein, and small molecule samples. Samples range from tissue sections/slices (fresh, culture, or fixed), cells, pathogens, or others (e.g. cell extract, purified DNA, RNA, proteins; indicated as samples). Preferably, thicker tissue sections are selected to allow for detection of pathogens in expansive areas of the XYZ axes. Stained or unstained samples are subjected to the following protocol with milder tissue/cell treatments for hydration, permeabilization, and antigen retrieval according to the bigger size and thickness of the tissue to reach the internal areas of the sample. In general, the protocol includes the following steps: (1) blocking endogenous biotin expression if needed; (2) maintaining the 3D structure of the thicker sections; (3) blocking nonspecific antibody reactivity using blocking solution; (4) applying primary antibody or probe (e.g. unconjugated, directly conjugated, biotinylated, and/or tyramine treated); (5) applying secondary antibody or amplification system to enhance the signal; and (6) analyzing fluorescent or individual wavelength signatures by light, confocal, or STORM microscopy. Moreover, improving blocking and elimination of autofluorescence using low-intensity lasers can reduce or eliminate particular wavelengths. Our laboratory has been developing these techniques in the last couple of years to identify low levels of several pathogens (including viruses, fungi, and bacteria) associated with several diseases.
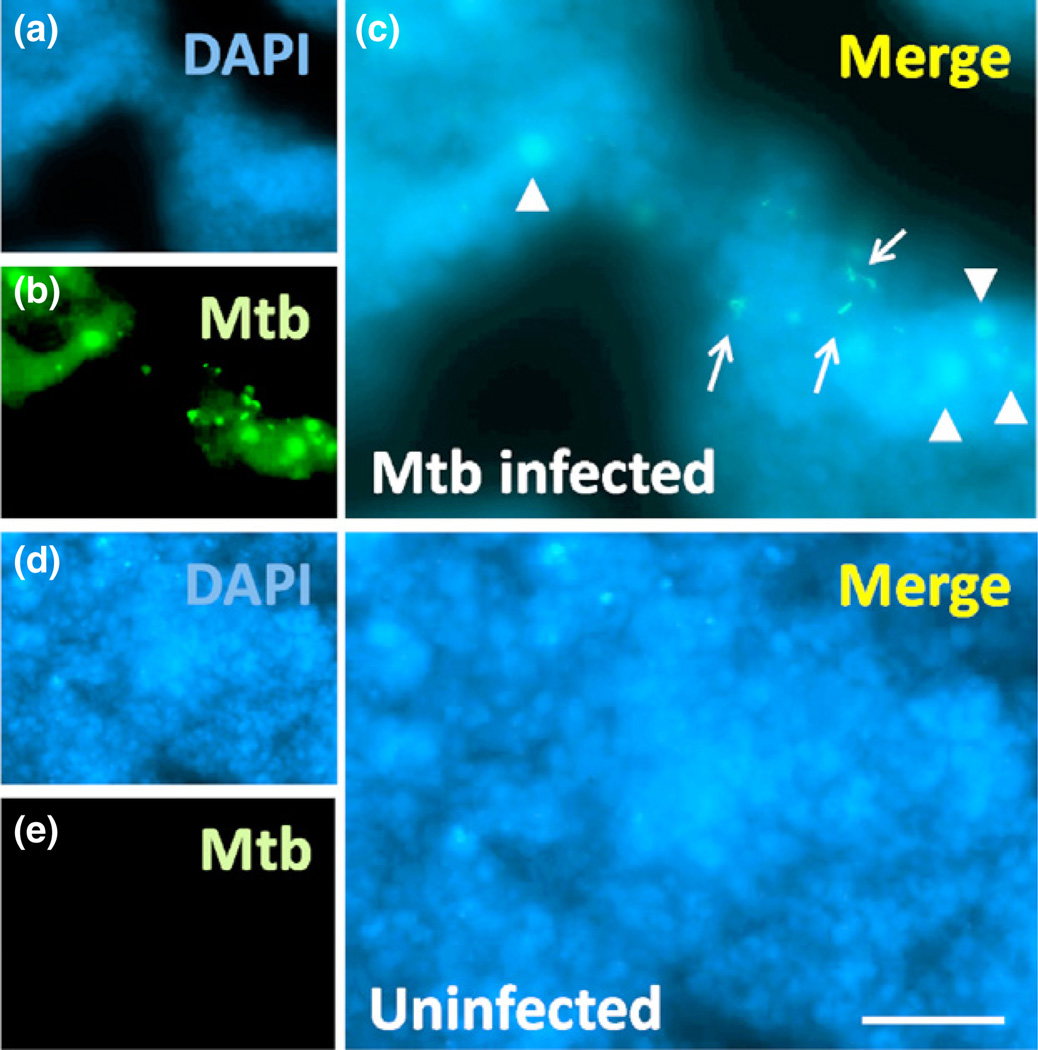
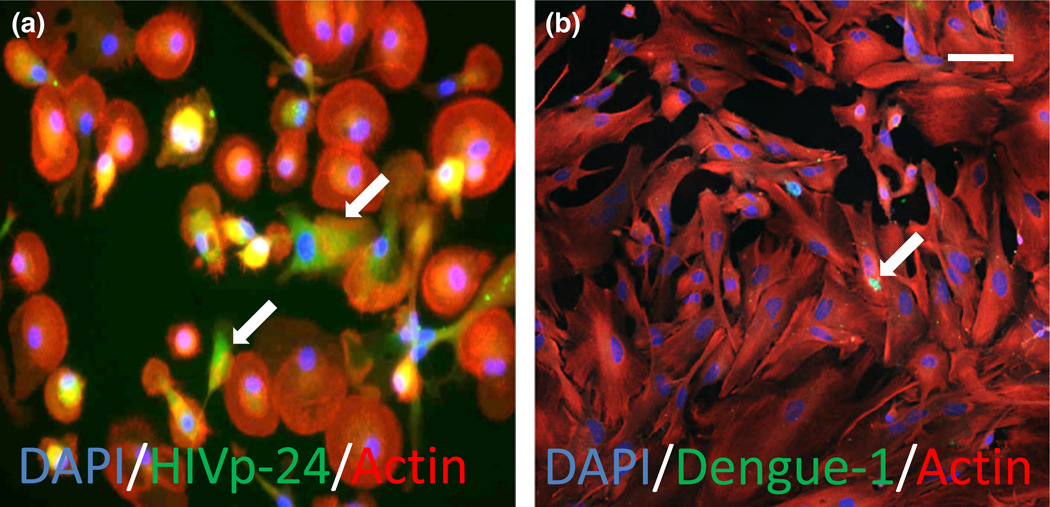
Tuberculosis, a BSL3 pathogen, is a diagnostically challenging pathogen that can undergo latency, in which it essentially hides in a host cell; thereby, evading easy detection (Helb et al., 2010; Antonenka et al., 2013). The combination of the spectral detector, evolved camera, and integrated imaging software generates an advanced capacity for background elimination, thus creating the opportunity for 3D images from thicker tissue sections. This is exemplified in the detection of a single viable TB bacterium, active or dormant, from within large 300-µm-thick tissue sections (Fig. 2), despite no detection of CFU and acid fast staining negativity. Using similar approaches, we are able to detect latent HIV infection as well as low-load dengue infection. Latent HIV infections are not readily detectable. After HIV infection for 35 days, a state of latency occurred with minimal to undetectable HIV production by HIV-p24 ELISA, or conventional HIV-p24 staining in human primary macrophages. Despite labeling intracellular HIV with biotin-conjugated antibodies against HIV p24 (Fig 3a), or Nef (not shown), and amplifying with streptavidin, the final signal remained low, but using a high-resolution camera, we were able to detect a significant amount of viral proteins in these latently infected macrophages (King et al., 2010; Eugenin et al., 2011). Using this staining regimen and technologies devised to improve image resolution, we have been successful in the detection of virus in vitro, in HIV-infected cultured primary macrophages (Fig. 3a), as well as in vivo, in SIV-infected brain tissue and arteries (not shown) (Eugenin et al., 2008,2011). We have been able to detect single dengue-infected cells as shown in Fig. 3b. Dengue was stained using primary antibodies directly conjugated to FITC, with labeled actin as a counter stain, and DAPI to identify nuclei (Fig. 3b). Thus, utilizing the improved laser-based systems, cameras, and software; these advancements allow examination of latency, reactivation, and pathogenesis at a level of sensitivity not possible before. Thus, the combination of all seven elements described above results in an outstanding degree of detection, despite minimal expression of cellular and pathogen markers (see Figs 1–3).
Fig. 2.
Detection of latent Mycobacterium tuberculosis (TB) in infected lung sections. Three-hundred micrometer tissue sections of lungs were stained for mTB and nuclei. (a)–(c) corresponds to tissue sections containing latent TB with no CFU detected. (d)–(f) corresponds to uninfected lungs where we observe no non-specific staining. Using the spectral detection system and antibody-based staining as well as an A1 Nikon confocal microscope with a high-resolution camera; we are able to detect one viable bacterium in each lung (see arrows) despite a negative result by CFU assay. In addition, we detect bacteria engulfed by macrophages (arrowheads). Bar: 2.5 µm.
Fig. 3.
Detection of latent HIV or dengue virus at low levels in macrophages and astrocytes, respectively. Merged images of HIV (a) or dengue-1 (b) staining (FITC, green staining) and actin staining (phalloidin conjugated to Texas red, red staining) demonstrate low to undetectable levels of infection. Numbers of positive infected cells were calculated using nuclear staining, DAPI. The merge of these three colors (Merge of the two cytoplasmic markers, HIV-p24 and actin, yellow plus nuclear staining) is also illustrated. In the case of HIV, HIV-p24 production was not detected by ELISA. In dengue infection, only 3% of human astrocytes were infected. These results were obtained using spinning disk and A1 confocal microscopy, respectively. Bar: 50 µm
Discussion
Safety concerns present the greatest limitation to investigations of infectious pathogens in their natural physiologic states and environments. The infectious pathogens must be rendered innocuous, usually inactivated by fixation, so that it no longer present as contaminating biohazards for researchers. Thus, to examine these pathogens ‘live’ and unaltered, dedicated imaging equipment, highly trained personnel, and excellent techniques of biosafety are required. The Rutgers University Biocontainment Laboratory provide a safe and secure environment to support research work with live infectious pathogens. In the current review, we discuss several microscopy technologies currently found in several point scanners, spinning disk, N-SIM and STORM systems that facilitate detection of pathogens like never before. These new technologies allow the detection and quantitation of pathogens/drugs/cellular products at specific wavelengths with outstanding resolution and accuracy. These technologies will lead to the development of novel clinical approaches and detection systems, thus enhancing our understanding of the pathogenesis of several infectious diseases. For example, human tissue or cells can be explanted for detection of several pathogens that are harbored within different host cell reservoirs, and results may be obtained in a few hours or within a day, vs. previous techniques that take weeks. The development of new technology, improved protocols, several improvements in sample preparation, DNA/mRNA/protein staining, and imaging detection systems creates unique systems capable of detection and quantitation of latent and actively replicating pathogens, including TB, HCV, HBV, HIV, and dengue. We have reached new heights in microbial research of BSL2/3 pathogens by applying the seven components described above to achieve: (1) high sensitivity to detect one pathogen in large portions of tissue or among millions of negative cells (the only limitation is the speed of the microscope); (2) high accuracy, with no amplification in negative samples or cross-identification of other pathogens; (3) high reproducibility; (4) simultaneous use of multiple markers of pathogen activation, such as DNA, mRNA and protein in the same sample; (5) faster thus more cost effective results as compared to culture or determination of CFU; (6) compatibility with a variety of samples types including cells and tissue samples; and (7) an easily scalable method for clinical analysis. In summary, new and improved imaging-based technologies enable us to identify, quantitate, and characterize viral reservoirs in multiple samples with high accuracy and reliability. In order to find a treatment for an infectious disease, we must first be able to detect the pathogen. The equipment and technologies described here promote the generation of new and highly precise protocols for the detection and quantitation of pathogens. New methods of pathogen detection open new avenues of research and of drug discovery.
Acknowledgements
This work was supported by the National Institutes of Mental Health grants, MH096625 (to E.A.E), and internal funding from the Public Health Research Institute (PHRI). We would like to thank the Analytical Imaging Facility at PHRI for microscopy support (http://www.phri.org/facilities/facil_imaging.asp). We thank Nikon instruments for its commitment to develop new techniques and technologies in the area of infectious diseases (Richard W. Francis and Brian T. Kain, Nikon Bioscience Specialists).
Footnotes
The authors have no financial interest or conflict of interest, but some of the protocols are under the patent No. 61/944,122 entitled ‘Methods for the identification and quantification of pathogens using imaging and microscopy techniques’.
References
- Antonenka U, Hofmann-Thiel S, Turaev L, et al. Comparison of Xpert MTB/RIF with ProbeTec ET DTB and COBAS TaqMan MTB for direct detection of M. tuberculosis complex in respiratory specimens. BMC Infect Dis. 2013;13:280. doi: 10.1186/1471-2334-13-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates M, Huang B, Dempsey GT, Zhuang X. Multicolor super-resolution imaging with photo-switchable fluorescent probes. Science. 2007;317:1749–1753. doi: 10.1126/science.1146598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Argenio DA, Wilson CB. A decade of vaccines: integrating immunology and vaccinology for rational vaccine design. Immunity. 2010;33:437–440. doi: 10.1016/j.immuni.2010.10.011. [DOI] [PubMed] [Google Scholar]
- Eugenin EA, Morgello S, Klotman ME, Mosoian A, Lento PA, Berman JW, Schecter AD. Human immunodeficiency virus (HIV) infects human arterial smooth muscle cells in vivo and in vitro: implications for the pathogenesis of HIV-mediated vascular disease. Am J Pathol. 2008;172:1100–1111. doi: 10.2353/ajpath.2008.070457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eugenin EA, Clements JE, Zink MC, Berman JW. Human immunodeficiency virus infection of human astrocytes disrupts blood-brain barrier integrity by a gap junction-dependent mechanism. J Neurosci. 2011;31:9456–9465. doi: 10.1523/JNEUROSCI.1460-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafsson MG. Nonlinear structured-illumination microscopy: wide-field fluorescence imaging with theoretically unlimited resolution. P Natl Acad Sci USA. 2005;102:13081–13086. doi: 10.1073/pnas.0406877102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helb D, Jones M, Story E, et al. Rapid detection of Mycobacterium tuberculosis and rifampin resistance by use of on-demand, near-patient technology. J Clin Microbiol. 2010;48:229–237. doi: 10.1128/JCM.01463-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B, Wang W, Bates M, huang X. Three-dimensional super-resolution imaging by stochastic optical reconstruction microscopy. Science. 2008;319:810–813. doi: 10.1126/science.1153529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King JE, Eugenin EA, Hazleton JE, Morgello S, Berman JW. Mechanisms of HIV-tat-induced phosphorylation of N-methyl-D-aspartate receptor subunit 2A in human primary neurons: implications for neuroAIDS pathogenesis. Am J Pathol. 2010;176:2819–2830. doi: 10.2353/ajpath.2010.090642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koff WC, Burton DR, Johnson PR, et al. Accelerating next-generation vaccine development for global disease prevention. Science. 2013;340:1232910. doi: 10.1126/science.1232910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rust MJ, Bates M, Zhuang X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM) Nat Methods. 2006;3:793–795. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]