Summary
Mitochondria are known to be functional organelles, but their role as a signaling unit is increasingly being appreciated. The identification of a short open reading frame (sORF) in the mitochondrial DNA (mtDNA) that encodes a signaling peptide, humanin, suggests the possible existence of additional sORFs in the mtDNA. Here we report a sORF within the mitochondrial 12S rRNA encoding a 16 amino acid peptide named MOTS-c (mitochondrial open-reading-frame of the twelve S rRNA -c) that regulates insulin sensitivity and metabolic homeostasis. Its primary target organ appears to be the skeletal muscle and its cellular actions inhibit the folate cycle and its tethered de novo purine biosynthesis, leading to AMPK activation. MOTS-c treatment in mice prevented age-dependent and high-fat diet-induced insulin resistance, as well as diet-induced obesity. These results suggest that mitochondria may actively regulate metabolic homeostasis at the cellular and organismal level via peptides encoded within their genome.
Introduction
The mitochondrion is largely viewed as a utilitarian organelle that provides various services to the cell, chiefly metabolism and energy production. The regulation of these pathways is often described to occur through vertical, rather than horizontal, communication where mitochondria are at the receiving end. Nonetheless, mitochondria also relay information via several known retrograde signaling molecules, such as reactive oxygen species (ROS), Ca2+, and cytochrome C (Goodwin et al., 2009; Houtkooper et al., 2011; Sethe et al., 2006). For instance, numerous studies suggest that mitochondrial ROS have evolved as a key communication method between the mitochondria and the cell to regulate homeostasis and normal cellular function (Sena and Chandel, 2012). Although essential, these are not of mitochondrial origin but are secondary metabolites, transient molecules, or nuclear-encoded proteins that reside in the mitochondria. Recent reports provide further evidence of mitochondrial signals transmitted as part of the mitochondrial unfolded protein response (mtUPR) that regulate critical processes such as aging, inflammation, and stress resistance (Durieux et al., 2011; Lee et al., 2013; Long et al., 2014; Nakahira et al., 2011; Yun and Finkel, 2014). Yet, humanin is currently the only reported mitochondrial-derived peptide (MDP) whose short open reading frame (sORF) maps to the mitochondrial DNA (Guo et al., 2003; Hashimoto et al., 2001; Ikonen et al., 2003; Lee et al., 2013).
The mitochondrial genome is traditionally described as a compact circular genetic system that encodes 13 proteins dedicated to energy production. Relatively little is understood of its unique genetic system that reflects its bacterial ancestry. The mitochondrial transcriptome, once considered relatively simple, is in fact a highly complex system with previously unknown characteristics including small RNAs encoded by the mtDNA (Mercer et al., 2011). Following on the identification of humanin, we set out to identify additional sORFs in the mitochondrial DNA (mtDNA). Notably, sORFs have recently been actively investigated in the nuclear genomes of Drosophila (Galindo et al., 2007; Kondo et al., 2010; Magny et al., 2013; Savard et al., 2006) and mammals (Frith et al., 2006; Slavoff et al., 2013), largely owing to technological advances that enable their detection and analysis. Here, we describe the identity of a sORF encoded within the mitochondrial 12S rRNA that yields a bioactive peptide involved in regulating metabolic homeostasis.
Results
Identification of a novel expressed short open reading frame (sORF) encoded within the mitochondrial genome
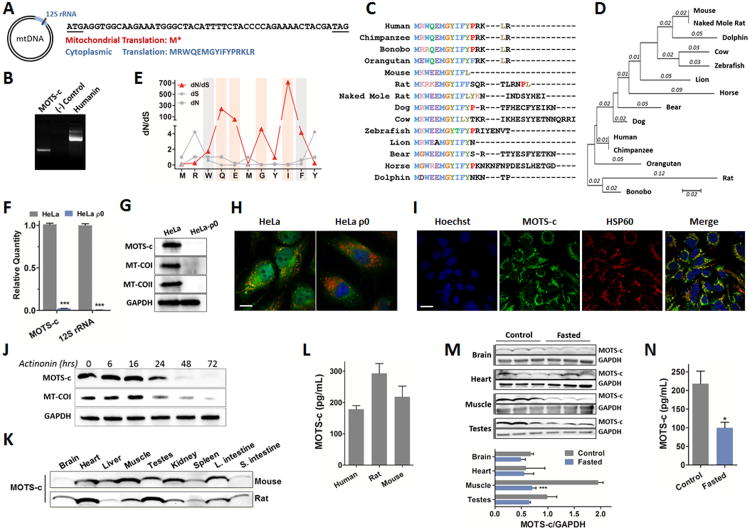
Previously, cDNAs mapping to the 12S rRNA region have been cloned from human myeloblasts after stimulation with interferon, but the exact sequences have not been identified (Tsuzuki et al., 1983). An in silico search for potential sORFs within the human 12S rRNA revealed one consisting of 51 base pairs with a strong Kozak sequence (Harhay et al., 2005) that translates into a 16 amino acid peptide subject to several putative post-translation modifications (Figures 1A and S1A-B). We have named this peptide MOTS-c (mitochondrial open-reading-frame of the twelve S rRNA type-c) (GenBank accession# KP715230). MOTS-c peptide translation obligatorily occurs in the cytoplasm using the standard genetic code because mitochondrial translation, using the mitochondria-specific genetic code, yields tandem start and stop codons (Figure 1A). This suggests that its polyadenylated transcript (Figure 1B) is exported from the mitochondria. Mitochondrial RNAs, both tRNA and rRNA, are exported out of the mitochondria via mechanisms that are still poorly understood (Amikura et al., 2001; Attardi and Attardi, 1968; Kashikawa et al., 2001; Kashikawa et al., 1999; Kobayashi et al., 1998; Kobayashi et al., 1993; Maniataki and Mourelatos, 2005; Nakamura et al., 1996). Multiple peptide sequence alignment of 14 species and its deduced phylogenetic tree suggest that MOTS-c is highly conserved, especially the first 11 residues (Figure 1C-D). To test if these 11 amino acids evolved over positive selection, we calculated ratios of non-synonymous (dN) to synonymous substitutions (dS), and their rate (dN/dS) for each residue in 14 species. We found that four residues were significantly undergoing positive selection (dN/dS > 1) at positions 4 (Q), 5 (E), 7 (G), and 9 (I), and two residues were undergoing purifying selection (dN/dS < 1) at positions 3 (W) and 10 (F) (Figure 1E). Positive selection was also detected by random effects likelihood (REL) analysis implemented in the HyPhy package with posterior probability greater than 0.95 and Bayes factor greater than 50 (Figure S1C) (Pond and Frost, 2005). All further studies were performed based on the human MOTS-c sequence.
Figure 1. Identification of a novel expressed short open reading frame (sORF) encoded within the mitochondrial genome.
(A) MOTSC is encoded as a 51-bp short open reading frame (sORF) in the mitochondrial 12S rRNA. The mitochondrial genetic code yields tandem start/stop codons, whereas the cytoplasmic genetic code yields a viable peptide. (B) 3′ rapid amplification of cDNA ends (3′ RACE) of polyadenylated MOTS-c and humanin transcripts. (C-E) Phylogenetic analysis of MOTS-c with (C) multiple peptide sequence alignment in 14 species, and (D) its phylogenetic tree and branch length (estimated number of substitutions per site), and (E) the rate of non-synonymous (dN) and synonymous substitutions (dS), and their ratio (dN/dS) for the first well-conserved 11 residues of MOTS-c. The red bars represent residues that evolved under significant positive selection and the grey bars those under significant purifying selection. (F-H) HeLa-ρ0 cells are devoid of mitochondrial DNA. HeLa cells vs HeLa-ρ0 La cells: (F) 12S rRNA and MOTS-c transcripts by qRT-PCR, (G) MOTS-c peptide and mitochondrial-encoded cytochrome C oxidase I and II (MT-COI/II) protein, and nuclear-encoded GAPDH by immunoblotting, (H) MOTS-c (green), mitochondrial resident HSP60 (red), and nucleus (blue) by immunocytochemistry (ICC). Scale bar, 100-μm. (I) MOTS-c (green), HSP60 (red), and nucleus (blue) immunostaining in HEK293 cells. Scale bar, 20-μm. (J) Time-course detection of MOTS-c and MT-COI after treating with actinonin (150μM), which causes mitochondrial RNA degradation. (K-L) MOTS-c is detected in (K) various tissues in mice and rats and (L) circulation (plasma) in humans and rodents. (M-N) Fasting alters expression of endogenous MOTS-c in (M) tissues and (N) plasma in mice. Data shown as mean ± SEM. Student's t-test. *P<0.05, **P<0.01, ***P<0.001. See also Figure S1 and Table S1 for additional information on MOTS-c and validation for Western blotting, ICC, and ELISA.
We recognized the possibility that MOTS-c could also be of nuclear origin due to a phenomenon known as nuclear mitochondrial DNA transfer (NUMT) (Ricchetti et al., 2004). Using the NCBI nucleotide basic local alignment search tool (BLASTN 2.2.29+) (Altschul et al., 1997), we found that none of the putative NUMT-derived peptides shared complete homology with mitochondrial encoded MOTS-c (Figure S1D). Furthermore, a BLAST search using the human expression sequence tag (EST) database revealed 101 hits whose mRNA sequences were all completely homologous to the mitochondrial 12S rRNA locus (Table S1). Interestingly, rats (Rattus Norvegicus) do not have any NUMT sequences for MOTS-c, making mitochondrial DNA its exclusive source. Because of the lack of methods for targeted knock-out or knock-down of mitochondrial genes, to further confirm its mitochondrial origin in humans, we selectively depleted mitochondrial DNA in HeLa cells (HeLa-ρ0) (Hashiguchi and Zhang-Akiyama, 2009) and show the elimination of both 12S rRNA and MOTS-c transcripts (Figure 1F). HeLa-ρ0 cells had undetectable levels of mitochondrial-encoded cytochrome oxidase I and II (MT-COI/II) and MOTS-c, measured using specific MOTS-c antibodies (Figure S1E), whereas nuclear-encoded GAPDH expression was unaltered (Figure 1G). HeLa and HEK293 cells showed a certain degree of mitochondrial co-localization (Figures 1H-I and S1F); HeLa-ρ0 cells showed loss of MOTS-c expression and mitochondrial co-localization (Figure 1H). Furthermore, we also selectively depleted mitochondrial RNA using actinonin (Richter et al., 2013), and show loss of MOTS-c and MT-COI expression in a time-dependent manner (Figures 1J and S1H). Therefore, using the ρ0 system and actinonin, we provide evidence of mitochondrial origin for MOTS-c at the DNA and RNA level, respectively. MOTS-c was detected in various tissues in mice and rats (Figure 1K), as well as in circulation in human and rodent plasma as determined with a MOTS-c specific ELISA (Figures 1L and S1G-H); again, NUMTs are absent in the rat making the mtDNA the only possible source of MOTS-c. Notably, fasting lowered endogenous expression of MOTS-c in certain metabolically active and mitochondria-rich tissues (skeletal muscle and testes) as well as in plasma, whereas homeostatic tissues (such as brain and heart) showed sustained levels (Figure 1M-N).
MOTS-c is a bioactive peptide that regulates gene expression and cellular metabolism
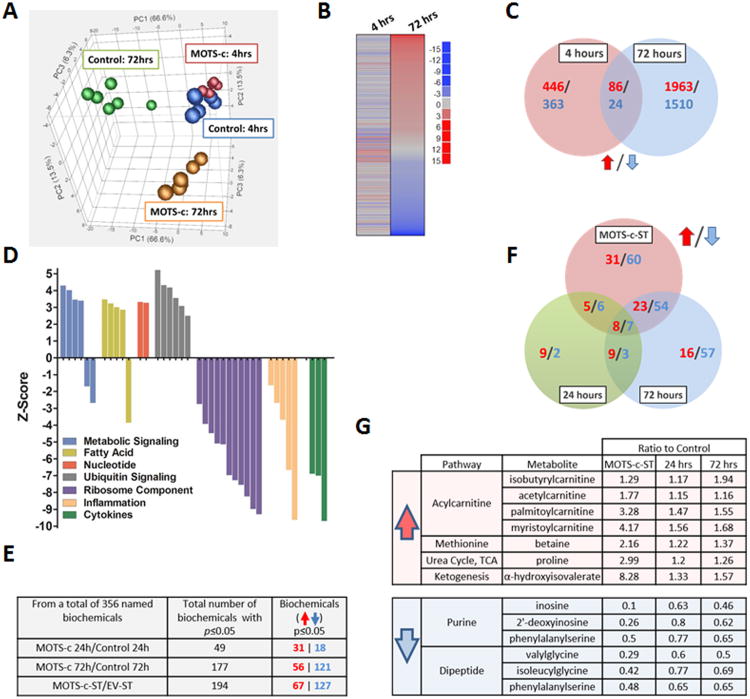
Microarray analyses performed on HEK293 cells treated with MOTS-c for 4 and 72 hours indicated that MOTS-c is a biologically active peptide (Figure 2A-D). Principal component analysis (PCA) showed a clear global gene expression profile shift by 72 hours after MOTS-c treatment (Figure 2A). To further highlight differences between functional pathways modified by MOTS-c, we employed parametric analysis of gene set enrichment (PAGE) to show that gene expression was significantly altered by 4 hr after MOTS-c treatment, which became remarkably distinct by 72 hr (Figure 2B). There was only a modest overlap between gene signatures at 4 and 72 hours, suggesting a time-dependent progression in response to MOTS-c treatment (Figure 2C). Notably, MOTS-c had a marked effect on expression of genes associated with cellular metabolism and inflammation (Figure 2D and Table S2). Since the central function of mitochondria involves metabolic processes, we next examined the effect of MOTS-c on cellular metabolism using unbiased global metabolomic profiling in vitro with a “gain-of-function” model using HEK293 cells either stably overexpressing MOTS-c (MOTS-c-ST) or treated exogenously with synthetic MOTS-c (10 μM) for 24 and 72 hours. Of the 356 named metabolites, 194 were significantly altered in MOTS-c-ST cells and 49 and 177 were significantly altered after 24 and 72 hours, respectively (Figure 2E). The pattern of metabolites also progressed with time and most biochemicals remained consistently altered both at 24 and 72 hours (Figure 2F). The majority of metabolites altered in MOTS-c-ST cells overlapped with those in cells treated with MOTS-c for 72 hours (Figure 2F). There were metabolites that were consistently changed in all 3 groups; those involved in purine metabolism and dipeptide metabolism were consistently reduced, whereas those involved in acylcarnitine metabolism and the methionine cycle were significantly increased (Figure 2G).
Figure 2. MOTS-c is a bioactive peptide that regulates gene expression and cellular metabolism.
(A to D) Microarray analyses on HEK293 cells treated with MOTS-c (10 μM) for 4- and 72-hours (N=6). (A) Principal component analysis (PCA), (B) Parametric analysis of gene set enrichment (PAGE) relative to control cells at the same time point, (C) Venn diagram depicting upregulated (Red) and downregulated (Blue) genes per time point. (D) Gene ontology analysis on HEK293 cells treated with MOTS-c for 72-hours. (E-G) Global unbiased metabolomics on HEK293 cells treated with MOTS-c (10 μM) for 24-and 72-hours or stably transfected with MOTS-c (MOTS-c-ST) or empty vector (EV-ST) (N=5). Welch's two sample t-test. (E) Total number of significantly or near-significantly changed metabolites. (F) Venn diagram depicting upregulated (Red) and downregulated (Blue) metabolites in MOTS-c-ST cells and HEK293 cells treated with MOTS-c for 24-and 72-hours compared to their controls. (G) List of metabolites that were consistently altered in all 3 groups. See also Table S2.
MOTS-c targets the methionine-folate cycle, increases AICAR levels, and activates AMPK
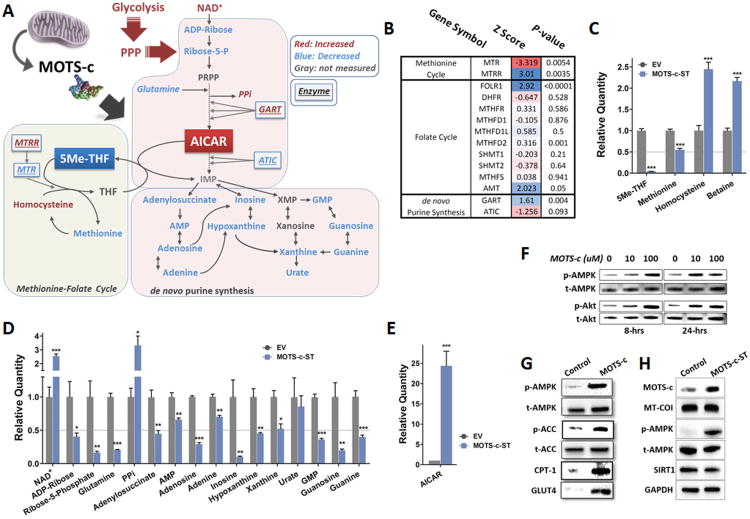
We identified a distinct metabolic signature from our global unbiased metabolomic profiling in MOTS-c-ST cells that strongly suggested its target of action as the folate-methionine cycle and the directly tethered de novo purine biosynthesis pathway (Figure 3A). We overlaid our microarray analysis with our metabolomics profiling and found that MOTS-c altered gene expression of enzymes involved in the folate-methionine cycle and de novo purine synthesis as early as 4 hours post-treatment (Figure 3A-B). We observed a concerted decrease in the levels of 5-methyl-tetrahydrofolate (5Me-THF), the most abundant form of activated folate, and methionine and elevated levels of homocysteine in MOTS-c-ST cells (Figure 3A, C). Notably, 5Me-THF levels declined prior to changes in methionine-homocysteine in HEK293 cells treated exogenously with MOTS-c, suggesting the regulation of the folate cycle to occur earlier (Figure S2A). Also, as expected, 5Me-THF depletion was coupled with the blockade of de novo purine biosynthesis (Figure 3A, D), resulting in an accumulation of endogenous AICAR (5-aminoimidazole-4-carboxamide ribonucleotide) to levels higher than 20-fold in MOTS-c-ST cells compared to control cells (Figure 3A, E), causing an expected significant decrease in purines (Chan and Cronstein, 2010); this was also observed, but to a lesser extent, 72 hours after exogenous MOTS-c treatment (Figure S2B). Because de novo purine synthesis is feedback-regulated by purine products, our data suggest accelerated de novo purine synthesis in MOTS-c-ST cells indicated by increased levels of its precursor metabolites NAD+, glycolysis, and the pentose phosphate pathway (PPP), as discussed below, simultaneously with reduced levels of ADP-ribose and R5P (Figures 3A, D and S2C).
Figure 3. MOTS-c targets the methionine-folate cycle, increases AICAR levels, and activates AMPK.
(A-B) The effect of ‘gain-of-function’ of MOTS-c on the folate-methionine cycle and de novo purine biosynthesis in MOTS-c-ST cells. (A) Metabolites were measured by mass spectrometry (N=5). (B) Enzymes altered 4-hours after MOTS-c treatment were determined by microarray (N=6). GART: phosphoribosylglycinamide formyltransferase (a trifunctional enzyme) ATIC: AICAR transformylase IMP carboxylase (a bifunctional enzyme), MTR: 5-methyltetrahydrofolate-homocysteine methyltransferase, MTRR: MTR reductase, 5Me-THF: 5-methyl-tetrahydrofolate, AICAR: 5-aminoimidazole-4-carboxamide ribonucleotide. (C) 5Me-THF and the methionine cycle intermediates in MOTS-c-ST cells (N=5). (D) Metabolic intermediates of the de novo purine biosynthesis pathway in MOTS-c-ST cells (N=5). (E) AICAR levels in MOTS-c-ST cells measured by mass spectrometry (N=5). (F) MOTS-c promotes AMPK (Thr172) and Akt (Ser473) phosphorylation in a time- and dose-dependent manner in HEK293 cells. (G) AMPK phosphorylation and its downstream pathways that control fatty acid oxidation (ACC and CPT-1) 72-hours after MOTS-c treatment (10 μM) in HEK293 cells. (H) MOTS-c-ST cells have higher levels of MOTS-c and phosphorylation of AMPK, but no changes in MT-COI, SIRT1, and GAPDH levels. Data shown as mean ± SEM. Student's t-test. *P<0.05, **P<0.01, ***P<0.001. See also Figure S2 for data on exogenous MOTS-c treatment and NAD+/NADH and adenine derivatives.
A well described role of AICAR is to activate AMPK and stimulate fatty acid oxidation via phosphorylation-induced inactivation of acetyl-CoA carboxylase (ACC) that consequently alleviates allosteric inhibition of carnitine palmitoyltransferase 1 (CPT-1), and also enhance glucose uptake in muscle (Steinberg and Kemp, 2009). MOTS-c treatment led to the phosphorylation of AMPKα (Thr172) and Akt (Ser473) in a time and dose-dependent manner (Figure 3F). Further, after 72 hours of MOTS-c treatment, increased phosphorylation of AMPKα2 (Thr172) and ACC (Ser79), and elevated CPT-1 protein levels were detected (Figure 3G). Notably AMPK activation occurred despite lower AMP levels and higher ADP and ATP levels (Figure S2D) similar to that achieved by salicylate (Hashiguchi and Zhang-Akiyama, 2009), leptin (Ohta et al., 2009), and metformin (Fujii et al., 2006). Consistent with this notion, the folate cycle, specifically 5Me-THF, has recently been shown to be a target of the AMPK-activating drug metformin (Cabreiro et al., 2013; Corominas-Faja et al., 2012). We also confirmed elevated levels of MOTS-c and phosphorylation of AMPKα2 (Thr172) in MOTS-c-ST cells (Figure 3H).
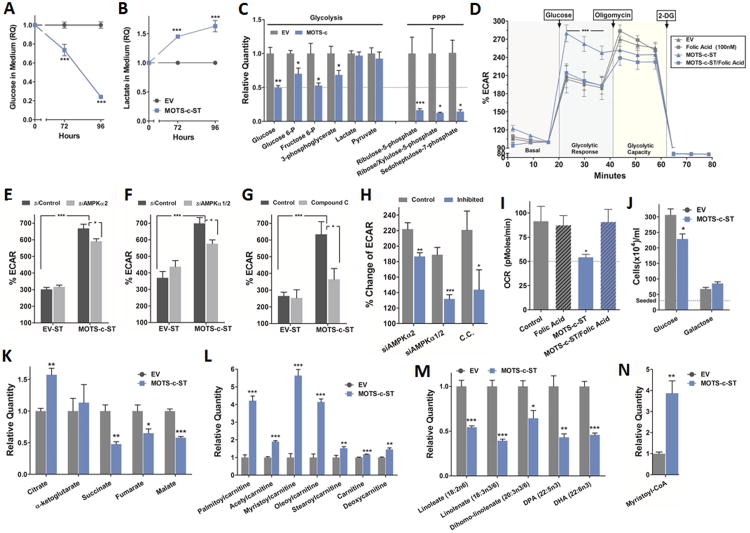
We next investigated the effect of MOTS-c on relevant cellular metabolism including glycolysis, mitochondrial function, and fatty acid oxidation. MOTS-c appeared to stimulate glucose utilization evidenced by increased glucose clearance and lactate accumulation in culture media (Figures 4A-B and S3A-C) coupled with decreased intracellular glucose levels, along with other glycolytic intermediates (Figure 4C). Similar findings were observed in HEK293 cells within 24 hours of MOTS-c treatment (Figure S3D). Notably, our data suggest increased routing of glucose to the pentose phosphate pathway (PPP), the anabolic branch of glycolysis that provides ribose-5-phosphate as a precursor for de novo purine synthesis, as indicated by reduced intermediates leading to the accumulation of AICAR in MOTS-c-ST cells and also in HEK293 cells after exogenous MOTS-c treatment (Figures 4C and S3D). To directly test glycolytic flux in real-time, we measured extracellular acidification rate (ECAR). Upon glucose stimulation MOTS-c-ST cells showed significantly enhanced glycolytic response, reaching maximum glycolytic capacity only typically achieved after oligomycin treatment (Figure 4D). Because MOTS-c targets the folate cycle and depletes intracellular 5Me-THF levels, we were able to reverse the enhanced glycolytic response in MOTS-c-ST cells by supplementing the culture media with folic acid (100nM) (Figure 4D); similar results were observed in HEK293 treated with MOTS-c (Figure S3E). To confirm the specificity of the MOTS-c sequence, we substituted 2 highly conserved residues to alanine and created null mutants that did not show enhanced glycolytic response to glucose stimulation in HEK293 cells. These mutants were glutamic acid in position 5 (E5A) and glycine in position 7 (G7A) (Figure S3F). Furthermore, we generated a randomly scrambled MOTS-c peptide sequence that did not have any primary sequence homology to known peptides/proteins (M3S1) and showed lack of enhanced glycolytic response to glucose stimulation in HEK293 cells (Figure S3G).
Figure 4. MOTS-c coordinates cellular glucose, mitochondrial, and fatty acid metabolism.
Extracellular (A) glucose and (B) lactate in the culture medium of MOTS-c-ST cells (N=6). (C) Intracellular metabolites of glycolysis and the pentose phosphate pathway (PPP) in MOTS-c-ST cells (N=5). Cells were seeded and allowed to acclimate for 72 hours prior to collection, at which time they were 90-95% confluent. (D) Real-time glycolytic flux determined by extracellular acidification rate (ECAR) in MOTS-c-ST cells treated with folic acid (100nM) for 72 hours (N=6). (E-H) Glucose-stimulated real-time glycolytic measurements in MOTS-c-ST cells treated with (E) siRNA against AMPKα2 (48-hours), or (F) siRNA against AMPKα1/2 (48-hours), or (G) compound C (10μM) (24-hours) and (H) the relative percent changes of ECAR of each treated groups to their corresponding controls in (E-G). (I) Real-time oxygen consumption rate (OCR) in MOTS-c-ST cells treated with folic acid (100nM) for 72 hours (N=6). All values normalized against DNA content. (J) MOTS-c-ST cell proliferation after 72 hours under equimolar (25mM) glucose or galactose medium (N=6). (K) Tricarboxylic acid (TCA) cycle intermediates in MOTS-c-ST cells (N=5). (L) Acyl-carnitine shuttle levels, (M) essential fatty acid levels, and (N) the β-oxidation intermediate myristoyl-CoA levels in MOTS-c-ST cells (N=5). Data shown as mean ± SEM. Student's t-test. *P<0.05, **P<0.01, ***P<0.001. See also Figures S3 and S4 for data on exogenous MOTS-c treatment, null MOTS-c mutants and non-specific scrambled MOTS-c peptide, and the role of SIRT1 in mediating the glycolytic effects of MOTS-c.
Because treating and over-expressing MOTS-c increased AMPK activity (p-Thr172-AMPKα2), we tested the requirement of AMPK for the glycolytic effects of MOTS-c. We used siRNA to knock-down AMPKα2 or AMPKα1/2 (Figure S3I) as well as the AMPK inhibitor compound C in MOTS-c-ST cells. Knock-down of AMPKα2 alone and AMPKα1/2 showed a 16% and 30% decrease in glucose-stimulated glycolytic rate, respectively (Figure 4E, F, and H), and the inhibition of AMPK by compound C led to a 40% decrease (Figure 4G-H) compared to their controls. This suggests that AMPK activation plays a partial role in mediating the actions of MOTS-c. Furthermore, because of the high levels of NAD+ (Figures 3D and S2C) and the fact that SIRT1 has an essential role in resveratrol-dependent AMPK activation (Price et al., 2012), we tested the possible role of SIRT1 in mediating the glycolytic actions of MOTS-c. Again, we used siRNA to knock-down SIRT1 (Figure S3J) as well as the SIRT1 inhibitor EX527 (Price et al., 2012) in MOTS-c-ST cells. Knockdown of SIRT1 led to a 40% decrease in glucose-stimulated glycolytic rate, and the inhibition of SIRT1 by EX527 led to a 45% decrease (Figure S3K-M) in MOTS-c expressing cells compared to their controls suggesting that SIRT1 is partially necessary for some MOTS-c actions.
Consistent with the “Crabtree effect”, a phenomenon whereby cells exhibit suppressed respiration in response to high glucose uptake (Ibsen, 1961), MOTS-c-ST cells and HEK293 cells treated with MOTS-c showed the characteristic increased glucose uptake coupled with reduced basal oxygen consumption rate (OCR). Remarkably, co-treatment with folic acid fully reversed this effect in both cellular model systems (Figures 4I and S4A). Transient overexpression of multiple MOTS-c clones in HEK293 cells grown in complete media also decreased OCR (Figure S4B). Also, the scrambled MOTS-c peptide (M3S1) did not show decreased OCR (Figure S3H). To test whether MOTS-c targets mitochondrial metabolism per se, MOTS-c-ST cells were cultured in medium with either glucose or galactose as the main carbon source. In mammalian cells, galactose is largely metabolized by the mitochondria (Marroquin et al., 2007). MOTS-c reduced cellular proliferation under conditions of abundant glucose, but failed to affect cells cultured in galactose medium (Figure 4J). Reduced oxidative capacity was associated with tricarboxylic acid (TCA) cycle dysregulation in MOTS-c-ST cells and in HEK293 cells treated with exogenous MOTS-c (Figures 4K and S4C). These data suggest that MOTS-c-induced respiratory suppression is likely secondary to increased glucose uptake.
MOTS-c coordinates cellular glucose, mitochondrial, and fatty acid metabolism
As the AICAR-AMPK pathway increases lipid metabolism as discussed above, we assessed the effects of MOTS-c on fatty acid metabolism. MOTS-c-ST cells exhibited higher levels of carnitine shuttles (Figure 4L), reduced levels of essential fatty acids (Figure 4M), and increased levels of the β-oxidation intermediate myristoyl-CoA compared with control cells (Figure 4N). These findings were also observed, but to a lesser extent, in cells treated with exogenous MOTS-c (Figure S4D-F). Furthermore, other long-chain fatty acids were significantly reduced, supporting increased fatty acid utilization (Figure S4G-H). Notably, β-oxidation itself is not an aerobic reaction, and increased fatty acid oxidation can occur under reduced respiration by AICAR (Spangenburg et al., 2013) and metformin (Martin-Montalvo et al., 2013).
To test the effect of MOTS-c on whole body metabolism, outbred CD-1 male mice fed a normal diet were treated acutely with MOTS-c (5 mg/kg/day; BID, 4 days) or vehicle control. Modest reductions in body weight, food intake, and blood glucose levels were observed in MOTS-c treated mice (Figure S5A-C). Basal levels of circulating IL-6 and TNFα (Figure S5D), implicated in the pathogenesis of obesity and insulin resistance, were significantly reduced by MOTS-c treatment.
MOTS-c targets skeletal muscle and regulates insulin sensitivity in mice
As MOTS-c enhanced cellular glucose flux in vitro and acute treatment reduced glucose levels in mice fed a normal diet, we hypothesized its actions in vivo would be related to glucose handling and insulin sensitivity. We treated mice with intraperitoneal injections of MOTS-c for 7 days and then subjected them to a glucose tolerance test (GTT), and found significantly enhanced glucose clearance indicative of improved insulin sensitivity (Figure 5A). Next, we performed hyperinsulinemic-euglycemic clamp studies to quantify the effects of a 7-day MOTS-c treatment on whole body insulin sensitivity independent of changes in body weight that occur with extended treatment durations (Figure 5B-D and Table S3). MOTS-c improved whole body insulin sensitivity as reflected by a ∼30% increase in the exogenous glucose infusion rate (GIR) required to maintain euglycemia during insulin stimulation (Figure 5B). Insulin promotes glucose disposal into peripheral tissues and suppresses hepatic glucose production (HGP) to maintain homeostasis during periods of increased glucose availability (Kahn, 1994). Tritiated glucose was infused during the clamp to determine the tissue specificity of MOTS-c action on insulin sensitivity. We observed that MOTS-c treatment significantly enhanced the insulin-stimulated glucose disposal rate (IS-GDR) (Figure 5C), indicative of enhanced skeletal muscle insulin sensitivity, but the rate of hepatic glucose production (HGP) was comparable between the groups (Figure 5D). The insulin-stimulated skeletal muscles were collected at the end of the hyperinsulinemic-euglycemic clamp. MOTS-c treated mice showed enhanced ability of infused insulin to activate skeletal muscle Akt, concurrent with increased detection of MOTS-c (Figure 5E). Considering that 70-85% of insulin-stimulated glucose disposal is into skeletal muscle, MOTS-c actions to enhance insulin sensitivity and glucose homeostasis are likely mediated in this tissue. Because MOTS-c levels in skeletal muscle (Figure 5F) and in circulation (Figure 5G) decline concomitantly with the development of insulin resistance during aging in mice, we tested if MOTS-c could reverse age-dependent impairments in insulin action by measuring insulin-stimulated glucose (2-deoxyglucose) uptake into soleus muscles of middle-aged (12 months) and young (3 months) male C57BL/6 mice. Reduced insulin sensitivity becomes apparent around one year of age in C57BL/6 mice (Pearson et al., 2008). Muscles from older (12 months old) mice were more insulin resistant than younger (3 months old), but 7 days of MOTS-c treatment restored sensitivity in the old mice to levels comparable to young animals (Figure 5H).
Figure 5. MOTS-c targets skeletal muscle and regulates insulin sensitivity in mice.
(A) Intraperitoneal glucose tolerance test (1g/kg glucose) in male C57BL/6 mice after MOTS-c treatment (5 mg/kg/day; IP) for 7 days (N=7). (B to E) Euglycemic-hyperglycemic clamps on C57BL/6 mice fed a high-fat diet (60% by calories) and treated with MOTS-c (5 mg/kg/day; IP) for 7 days (N=6-8). (B) Glucose infusion rate (GIR), reflecting whole body insulin sensitivity, (C) insulin-stimulated glucose disposal rate (IS-GDR), primarily reflecting skeletal muscle insulin sensitivity, and (D) hepatic glucose production (HGP). (E) Akt activation (assessed by phosphorylation at Ser473) and MOTS-c levels were detected in the insulin-stimulated skeletal muscles obtained at the end of the hyperinsulinemic-euglycemic clamp. (F and G) MOTS-c levels in young (4-month) and aged (32-month) mice (N=3-4) decline in (F) skeletal muscle, and (G) circulation (serum). (H) Insulin-stimulated (60 μU/ml) 2-deoxyglucose uptake into soleus muscles of young (3-month) and middle-aged (12-month) male C57BL/6 mice after MOTS-c treatment (5 mg/kg/day; IP) for 7 days (N=6). (I-K) Differentiated mature rat L6 myotubes that stably over-express MOTS-c (L6-MOTS-c-ST) show (I) accelerated media glucose clearance, (J) enhanced glucose-stimulated glycolytic response, and (K) maximum glycolytic capacity estimated by oligomycin treatment. Data shown as mean ± SEM. Student's t-test. *P<0.05, **P<0.01, ***P<0.001. See also Table S3.
To examine the glycolytic effects of MOTS-c on muscle in vitro, we generated L6 rat myocytes that stably over-express MOTS-c (L6-MOTS-c-ST). Following differentiation to mature myotubes, we measured glucose levels in the media (steady-state) and glucose-stimulated and maximum glycolytic capacity (flux). Similar to the MOTS-c-ST (HEK293) cells, L6-MOTS-c-ST cells showed accelerated media glucose clearance (Figure 5I), and enhanced glucose-stimulated and maximum glycolytic rate (Figures 5J-K).
MOTS-c treatment prevents high fat diet-induced obesity and insulin resistance in mice
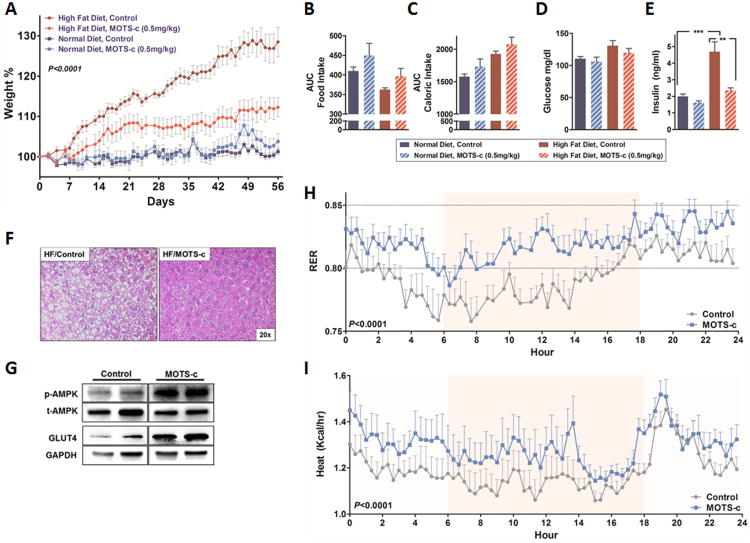
Given the role of MOTS-c in enhancing insulin sensitivity and glucose homeostasis, we next tested the effects of MOTS-c (0.5 mg/kg/day; IP) on outbred CD-1 mice fed a high-fat diet (HFD 60% by calories). Although MOTS-c treatment had no effect on body weight in mice fed a normal diet, it remarkably prevented obesity when administrated to mice fed a HFD (Figure 6A). This difference in body weight was not attributed to food intake, as caloric intake was identical between the groups (Figure 6B-C). Additionally, MOTS-c treatment prevented HFD-induced hyperinsulinemia, indicating improved glucose homeostasis (Figure 6D-E). Hepatic lipid accumulation was dramatically reduced in HFD-fed mice treated with MOTS-c (Figure 6F). Notably, MOTS-c promoted AMPK activation and GLUT4 expression in the skeletal muscles of HFD-fed mice (Figure 6G). This corroborates the in vitro actions of MOTS-c on AMPK activation and GLUT4 expression and also further supports the skeletal muscle as a major target organ. Mice fed a HFD and treated with MOTS-c (0.5 mg/kg/day) for 3 weeks showed increased respiratory exchange ratio (RER; CO2 exhaled/O2 inhaled) (Figure 6H), reflecting increased glucose utilization. Notably, MOTS-c treated mice also generated significantly more heat that may, in part, account for the increased energy expenditure (Figure 6I). Total activity was comparable between the 2 groups (Figure S6). MOTS-c was also able to prevent HFD-induced obesity and hyperinsulinemia independent of caloric intake in C57BL/6 mice (Figure S7). These studies suggest that MOTS-c prevents HFD-induced obesity by increasing energy expenditure, including heat production, and improving glucose utilization and insulin sensitivity. Reduced fat accumulation may be a result of robust carbohydrate usage that reduces fatty acid synthesis, but the possible involvement of increased fatty acid oxidation, as observed in vitro, would need detailed examination before being ruled out.
Figure 6. MOTS-c treatment prevents high fat diet-induced obesity and insulin resistance in mice.
8-week old male CD-1 mice fed a high fat diet (HFD, 60% by calories) or matched control diet (N=10) treated with MOTS-c daily (0.5 mg/kg/day; IP) for 8 weeks. (A) body weight, (B) food intake, (C) caloric intake, (D) glucose levels, (E) insulin levels determined at the time of euthanasia, (F) liver H&E staining, and (G) AMPK phosphorylation (Thr172) and GLUT4 levels in the skeletal muscles of HFD-fed mice treated with MOTS-c or vehicle control. (H-I) Respiratory exchange ratio (RER) and body heat production values of 8-week old male CD-1 mice fed a high fat diet (HFD, 60% by calories) or matched control diet treated with MOTS-c daily (0.5 mg/kg/day; IP) for 3 weeks (N=6). Data shown as mean ± SEM. (A, H, and I) Repeated measures two-way ANOVA and (B-E) Student's t-test. **P<0.01, ***P<0.001. See also Figure S6 for total activity and Figure S7 for similar data in C57BL/6 mice.
Discussion
Novel Peptides Derived from Mitochondrial sORFs as Signaling Molecules
Our understanding of the molecular mechanisms involved in the communication signals from the mitochondria of particular cells to the rest of those cells and to distant organs is rapidly evolving. Signals that are sent from the mitochondria to the cell are largely categorized as mitochondrial retrograde signals and have been thought to consist of nuclear-encoded proteins that reside in mitochondria, secondary and transient metabolites, and fragments of damaged mitochondrial DNA (mtDNA). Notably, in the nematode C. elegans, a stress-induced tissue-specific signal originating from mitochondria has been shown to communicate with distant organs and extend lifespan; the exact identity of this signal, termed ‘mitokine’, is unknown and currently being investigated (Durieux et al., 2011; Woo and Shadel, 2011). The demonstration of MOTS-c as a peptide encoded as a sORF in the mtDNA that is metabolically and developmentally regulated, which has endocrine like effects on muscle metabolism, insulin sensitivity, and weight regulation, establishes its role as a member of a new class of mitochondrial signals which can be called “MDPs” or mitochondrial-derived peptides (Lee et al., 2013). The MDP humanin similarly acts in a systemic fashion to protect neuronal and vascular systems from disease processes and toxic insults (Cohen, 2014; Lee et al., 2013).
Technological advances have unveiled previously unknown properties of mitochondrial genetics that suggest the existence of sORFs in the mtDNA (Mercer et al., 2011). MOTS-c may be a product of adaptation for effective bilateral communication between mitochondria in energetically sensitive tissues and cells of distant organs such as skeletal muscle. We provide here experimental evidence that MOTS-c, like humanin, is a mitochondrial signaling peptide encoded within the mtDNA that regulates global physiology (Lee et al., 2013).
Regulation of the Folate-AICAR-AMPK Pathway
The unique involvement of the interplay between the folate cycle, AICAR, and AMPK signaling in MOTS-c action (Figure 7) provide an interesting opportunity to expand our understanding of the role of mitochondria in regulating glucose homeostasis. Interestingly, MOTS-c shares many effects with the antifolate drug methotrexate: they both (1) target the folate cycle, (2) lead to the depletion of intracellular 5Me-THF, (3) increase levels of AICAR (Chan and Cronstein, 2010), (4) activate AMPK (O’Neill and Hardie, 2013) in the presence of ATP accumulation most likely resulting from increased glucose uptake (Diamanti-Kandarakis et al., 2010), and (5) decrease mitochondrial respiration (Sauter et al., 2003). Notably, methotrexate was initially developed to treat malignancy, but is currently widely used as an anti-inflammatory drug in rheumatic arthritis (Tennstedt et al., 2012). Because we observed reduced inflammatory markers in the microarray and following acute treatment in mice, future directions of MOTS-c investigation should involve its anti-inflammatory roles.
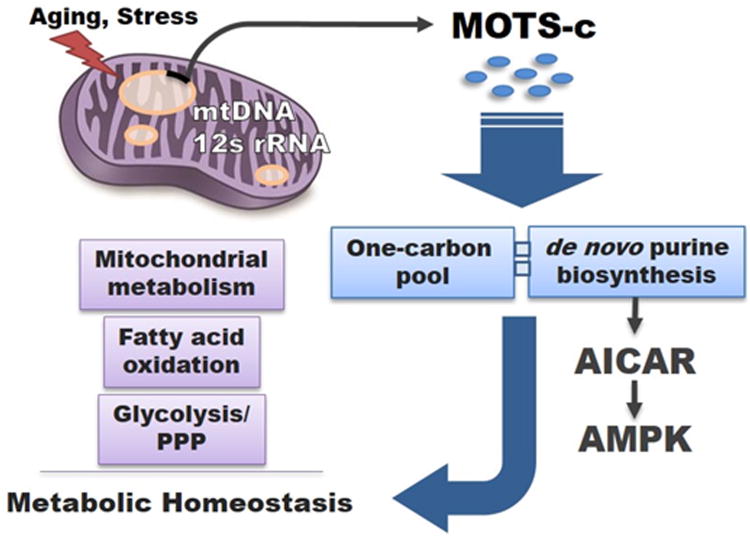
Figure 7. MOTS-c: mitochondrial-encoded regulator of metabolic homeostasis.
Proposed model of MOTS-c as a mitochondrial signaling peptide encoded in the mtDNA that regulates metabolic homeostasis. MOTS-c targets the skeletal muscle and acts on the folate cycle (one carbon pool) and inhibits the directly tethered de novo purine biosynthesis pathway. This leads to the accumulation of the de novo purine synthesis intermediate AICAR that is also a potent activator of the metabolic regulator AMPK, thus partially mediating the metabolic effects of MOTS-c.
Mitochondrial Regulation of Energy Metabolism and Insulin Sensitivity
Mitochondria are thought to sense the cellular energetic status and are well recognized to modulate carbohydrate metabolism through incompletely characterized mechanisms (Woo and Shadel, 2011). We propose MOTS-c as a novel key endocrine signal that originates from mitochondria and systemically regulates in vivo glucose metabolism and muscle insulin action. MOTS-c also shares some physiological similarities to the first-line diabetes drug metformin in terms of regulating glucose utilization, mitochondrial and fatty acid metabolism, and body weight (Ferguson et al., 2007) which is thought to occur by targeting the folate cycle (Cabreiro et al., 2013; Corominas-Faja et al., 2012) and signaling via cAMP (Miller et al., 2013) and AMPK (Shaw, 2013).
One key difference between the actions of metformin and MOTS-c is that MOTS-c appears to directly target skeletal muscle as the key site of its activity, while metformin is thought to act primarily on the liver (Diamanti-Kandarakis et al., 2010). Interestingly, the mitochondrial peptide humanin has been shown to regulate carbohydrate metabolism primarily by acting on hypothalamic and islet cells (Kuliawat et al., 2013; Muzumdar et al., 2009); this suggests a highly regulated, tissue and context specific targeted MDP effect on whole body metabolism.
Implications for Metabolic Diseases of Aging
Age-dependent accumulation of mtDNA mutations and consequent metabolic dysfunction are strongly implicated in aging (Bratic and Larsson, 2013; Wallace, 2011). We hypothesize these mitochondrial genetic alterations may underlie the age-dependent decline of MOTS-c and humanin levels (Muzumdar et al., 2009), thus adding another layer of mitochondrial contribution to aging via the emerging biology of MDPs. Thus, age-related mtDNA dysfunction could result in both a direct decline in mitochondrial function as well as progressive loss of MDP expression that will diminish their functions as regulatory peptides.
As aging is associated with worsening of mitochondrial function concomitantly with the development of aging-related diseases such as diabetes and metabolic syndrome, and since the tissue and circulating levels of humanin and MOTS-c fall with age, it is compelling to hypothesize that declining MDP levels are also related to age-related metabolic deterioration. The profound systemic effects of the administration of MOTS-c as a daily injection in aged mice suggest that derivatives of this molecule might be useful in ameliorating the abnormal metabolism associated with aging in humans.
Finally, the recognition of the mitochondrial-encoded peptides MOTS-c and humanin suggest the existence of additional functional sORFs in the mitochondrial genome and repositions the mitochondria from an “end-function” organelle to an active regulator of biological processes such as metabolism and weight homeostasis.
Methods
Cell Culture
HEK293 and HeLa cells were routinely cultured in Dulbecco's modified Eagle's medium (DMEM), and L6 myoblasts were cultured in minimum essential media (MEM) supplemented with 10% fetal bovine serum (FBS) at 37°C with 5% CO2. ρ0 cells, which are devoid of mitochondrial DNA, were generated by culturing HeLa cells in low doses of ethidium bromide (EtBr; 100ng/mL) as described before (Hashiguchi and Zhang-Akiyama, 2009). MOTS-c-ST and L6-MOTS-c-ST cells are HEK293 and L6 cells, respectively, stably overexpressing MOTS-c and were generated by transfecting MOTS-c expression clone (described below) followed by selection in and maintenance in G418 (500 μM; Sigma) in DMEM or MEM with 10% FBS. The control cells for MOTS-c-ST and L6-MOTS-c-ST cells were HEK293 or L6 cells, respectively, stably transfected with empty vector (EV) with the same selection and maintenance method. L6 myoblasts were differentiated to mature myotubes by culturing in MEM 2% media, once 80-90% confluence was reached, for 8-10 days (media replaced every 2-3 days).
Mice Care
All animal work was performed in accordance with the University of Southern California and University of California Los Angeles Institutional Animal Care and Use Committee. MOTS-c (Genscript, USA) was injected daily via intraperitoneal injections in all in vivo experiments. 8-week old CD-1 (ICR) mice were purchased from Harlan. C57BL/6 mice were purchased from Jackson Laboratories. Mice were fed a high-fat diet (60% by calories) and matching control diet purchased from Research Diets (Cat# D12492 and D12450J, respectively) for 8 weeks. Pellets were replaced twice weekly, and body weight and food consumption were recorded daily (N=10).
Microarray Analysis
RNA was isolated using the RNeasy kit (Qiagen, Valencia, CA) and then hybridized to BD-103-0603 Illumina Beadchips. Raw data were subjected to Z-normalization, as previously described (Cheadle et al., 2003). Principal component analysis, performed on the normalized Z-scores of all of the detectable probes in the samples, was performed by using the DIANE 6.0 software: (http://www.grc.nia.nih.gov/branches/rrb/dna/diane_software.pdf). Significant genes were selected by the z-test < 0.05, false discovery rate < 0.30, as well as z-ratio > 1.5 in both directions and ANOVA p value < 0.05. Parametric analysis of gene set enrichment (PAGE) was analyzed as previously described (Kim and Volsky, 2005). Gene regulatory network and canonic pathway analysis was performed using Ingenuity Pathways Analysis© (Ingenuity Systems; Redwood City, CA) (N=6). Microarray data are accessible in GEO database under accession code GSE65068.
Oxygen Consumption and Extracellular Acidification Rate
Real-time oxygen consumption rates (OCR) were measured using XF24/96 Extracellular Flux Analyzer (Seahorse Bioscience). ATP turnover and maximum respiratory capacity were estimated by challenging cells with oligomycin and FCCP (carbonyl cyanide 4-[trifluoromethoxy]phenylhydrazone) or DNP (2,4-dinitrophenol). Glycolytic rate was determined using extracellular acidification rate (ECAR) and individually reported relative to basal level in percentage. Cells were stimulated with glucose to determine active glycolytic rate, and with oligomycin to determine maximum glycolytic capacity, and with 2-DG to determine glycolytic capacity. All readings were normalized to total DNA content.
Supplementary Material
Acknowledgments
We would like to thank Y. Zhang, E. Lehrmann, W. Wood and K. Becker for microarray assistance and Brianna Manes for graphical abstract assistance. P.C. is a stockholder of and consultant for, and C.L. is a consultant for CohBar, Inc. This research was supported by UCSD-UCLA Diabetes Research Center grant (NIH DK063491), a Glenn Foundation Award and National Institute of Health Grants to P.C. (1R01AG 034430, 1R01GM 090311, 1R01ES 020812) and an Ellison Medical Foundation New Scholar Award, a SC-CTSI grant, Hanson Thorell Family Research Award, and a Zumberge award to C.L.
ACCESSION NUMBERS
The GenBank accession number for the MOTS-c (mitochondrial open reading frame of the 12S rRNA type-c) peptide reported in this paper is KP715230. Microarray data are accessible in the Gene Expression Omnibus database under accession code GSE65068.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic acids research. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amikura R, Kashikawa M, Nakamura A, Kobayashi S. Presence of mitochondria-type ribosomes outside mitochondria in germ plasm of Drosophila embryos. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:9133–9138. doi: 10.1073/pnas.171286998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attardi G, Attardi B. Mitochondrial origin of membrane-associated heterogeneous RNA in HeLa cells. Proceedings of the National Academy of Sciences of the United States of America. 1968;61:261–268. doi: 10.1073/pnas.61.1.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bratic A, Larsson NG. The role of mitochondria in aging. The Journal of clinical investigation. 2013;123:951–957. doi: 10.1172/JCI64125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabreiro F, Au C, Leung KY, Vergara-Irigaray N, Cocheme HM, Noori T, Weinkove D, Schuster E, Greene ND, Gems D. Metformin retards aging in C. elegans by altering microbial folate and methionine metabolism. Cell. 2013;153:228–239. doi: 10.1016/j.cell.2013.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan ES, Cronstein BN. Methotrexate--how does it really work? Nature reviews Rheumatology. 2010;6:175–178. doi: 10.1038/nrrheum.2010.5. [DOI] [PubMed] [Google Scholar]
- Cheadle C, Vawter MP, Freed WJ, Becker KG. Analysis of microarray data using Z score transformation. The Journal of molecular diagnostics : JMD. 2003;5:73–81. doi: 10.1016/S1525-1578(10)60455-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen P. A Potential New Role for the Mitochondrial Peptide Humanin as a Protective Agent Against Chemotherapy-Induced Side Effects. Jounal of the National Cancer Institute. 2014 doi: 10.1093/jnci/dju006. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corominas-Faja B, Quirantes-Pine R, Oliveras-Ferraros C, Vazquez-Martin A, Cufi S, Martin-Castillo B, Micol V, Joven J, Segura-Carretero A, Menendez JA. Metabolomic fingerprint reveals that metformin impairs one-carbon metabolism in a manner similar to the antifolate class of chemotherapy drugs. Aging. 2012;4:480–498. doi: 10.18632/aging.100472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamanti-Kandarakis E, Christakou CD, Kandaraki E, Economou FN. Metformin: an old medication of new fashion: evolving new molecular mechanisms and clinical implications in polycystic ovary syndrome. European journal of endocrinology / European Federation of Endocrine Societies. 2010;162:193–212. doi: 10.1530/EJE-09-0733. [DOI] [PubMed] [Google Scholar]
- Durieux J, Wolff S, Dillin A. The cell-non-autonomous nature of electron transport chain-mediated longevity. Cell. 2011;144:79–91. doi: 10.1016/j.cell.2010.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson M, Sohal BH, Forster MJ, Sohal RS. Effect of long-term caloric restriction on oxygen consumption and body temperature in two different strains of mice. Mechanisms of ageing and development. 2007;128:539–545. doi: 10.1016/j.mad.2007.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frith MC, Forrest AR, Nourbakhsh E, Pang KC, Kai C, Kawai J, Carninci P, Hayashizaki Y, Bailey TL, Grimmond SM. The abundance of short proteins in the mammalian proteome. PLoS genetics. 2006;2:e52. doi: 10.1371/journal.pgen.0020052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii N, Jessen N, Goodyear LJ. AMP-activated protein kinase and the regulation of glucose transport. American journal of physiology Endocrinology and metabolism. 2006;291:E867–877. doi: 10.1152/ajpendo.00207.2006. [DOI] [PubMed] [Google Scholar]
- Galindo MI, Pueyo JI, Fouix S, Bishop SA, Couso JP. Peptides encoded by short ORFs control development and define a new eukaryotic gene family. PLoS biology. 2007;5:e106. doi: 10.1371/journal.pbio.0050106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin PJ, Ligibel JA, Stambolic V. Metformin in breast cancer: time for action. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2009;27:3271–3273. doi: 10.1200/JCO.2009.22.1630. [DOI] [PubMed] [Google Scholar]
- Guo B, Zhai D, Cabezas E, Welsh K, Nouraini S, Satterthwait AC, Reed JC. Humanin peptide suppresses apoptosis by interfering with Bax activation. Nature. 2003;423:456–461. doi: 10.1038/nature01627. [DOI] [PubMed] [Google Scholar]
- Harhay GP, Sonstegard TS, Keele JW, Heaton MP, Clawson ML, Snelling WM, Wiedmann RT, Van Tassell CP, Smith TP. Characterization of 954 bovine full-CDS cDNA sequences. BMC genomics. 2005;6:166. doi: 10.1186/1471-2164-6-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashiguchi K, Zhang-Akiyama QM. Establishment of human cell lines lacking mitochondrial DNA. Methods in molecular biology. 2009;554:383–391. doi: 10.1007/978-1-59745-521-3_23. [DOI] [PubMed] [Google Scholar]
- Hashimoto Y, Niikura T, Tajima H, Yasukawa T, Sudo H, Ito Y, Kita Y, Kawasumi M, Kouyama K, Doyu M, et al. A rescue factor abolishing neuronal cell death by a wide spectrum of familial Alzheimer's disease genes and Abeta. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:6336–6341. doi: 10.1073/pnas.101133498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houtkooper RH, Argmann C, Houten SM, Canto C, Jeninga EH, Andreux PA, Thomas C, Doenlen R, Schoonjans K, Auwerx J. The metabolic footprint of aging in mice. Scientific reports. 2011;1:134. doi: 10.1038/srep00134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibsen KH. The Crabtree effect: a review. Cancer research. 1961;21:829–841. [PubMed] [Google Scholar]
- Ikonen M, Liu B, Hashimoto Y, Ma L, Lee KW, Niikura T, Nishimoto I, Cohen P. Interaction between the Alzheimer's survival peptide humanin and insulin-like growth factor-binding protein 3 regulates cell survival and apoptosis. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:13042–13047. doi: 10.1073/pnas.2135111100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn CR. Banting Lecture. Insulin action, diabetogenes, and the cause of type II diabetes. Diabetes. 1994;43:1066–1084. doi: 10.2337/diab.43.8.1066. [DOI] [PubMed] [Google Scholar]
- Kashikawa M, Amikura R, Kobayashi S. Mitochondrial small ribosomal RNA is a component of germinal granules in Xenopus embryos. Mechanisms of development. 2001;101:71–77. doi: 10.1016/s0925-4773(00)00553-0. [DOI] [PubMed] [Google Scholar]
- Kashikawa M, Amikura R, Nakamura A, Kobayashi S. Mitochondrial small ribosomal RNA is present on polar granules in early cleavage embryos of Drosophila melanogaster. Development, growth & differentiation. 1999;41:495–502. doi: 10.1046/j.1440-169x.1999.00451.x. [DOI] [PubMed] [Google Scholar]
- Kim SY, Volsky DJ. PAGE: parametric analysis of gene set enrichment. BMC bioinformatics. 2005;6:144. doi: 10.1186/1471-2105-6-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi S, Amikura R, Mukai M. Localization of mitochondrial large ribosomal RNA in germ plasm of Xenopus embryos. Current biology : CB. 1998;8:1117–1120. doi: 10.1016/s0960-9822(98)70466-x. [DOI] [PubMed] [Google Scholar]
- Kobayashi S, Amikura R, Okada M. Presence of mitochondrial large ribosomal RNA outside mitochondria in germ plasm of Drosophila melanogaster. Science. 1993;260:1521–1524. doi: 10.1126/science.7684857. [DOI] [PubMed] [Google Scholar]
- Kondo T, Plaza S, Zanet J, Benrabah E, Valenti P, Hashimoto Y, Kobayashi S, Payre F, Kageyama Y. Small peptides switch the transcriptional activity of Shavenbaby during Drosophila embryogenesis. Science. 2010;329:336–339. doi: 10.1126/science.1188158. [DOI] [PubMed] [Google Scholar]
- Kuliawat R, Klein L, Gong Z, Nicoletta-Gentile M, Nemkal A, Cui L, Bastie C, Su K, Huffman D, Surana M, et al. Potent humanin analog increases glucose-stimulated insulin secretion through enhanced metabolism in the beta cell. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2013;27:4890–4898. doi: 10.1096/fj.13-231092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Yen K, Cohen P. Humanin: a harbinger of mitochondrial-derived peptides? Trends in endocrinology and metabolism: TEM. 2013 doi: 10.1016/j.tem.2013.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long YC, Tan TM, Takao I, Tang BL. The biochemistry and cell biology of aging: metabolic regulation through mitochondrial signaling. American journal of physiology Endocrinology and metabolism. 2014;306:E581–591. doi: 10.1152/ajpendo.00665.2013. [DOI] [PubMed] [Google Scholar]
- Magny EG, Pueyo JI, Pearl FM, Cespedes MA, Niven JE, Bishop SA, Couso JP. Conserved regulation of cardiac calcium uptake by peptides encoded in small open reading frames. Science. 2013;341:1116–1120. doi: 10.1126/science.1238802. [DOI] [PubMed] [Google Scholar]
- Maniataki E, Mourelatos Z. Human mitochondrial tRNAMet is exported to the cytoplasm and associates with the Argonaute 2 protein. RNA. 2005;11:849–852. doi: 10.1261/rna.2210805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marroquin LD, Hynes J, Dykens JA, Jamieson JD, Will Y. Circumventing the Crabtree effect: replacing media glucose with galactose increases susceptibility of HepG2 cells to mitochondrial toxicants. Toxicological sciences : an official journal of the Society of Toxicology. 2007;97:539–547. doi: 10.1093/toxsci/kfm052. [DOI] [PubMed] [Google Scholar]
- Martin-Montalvo A, Mercken EM, Mitchell SJ, Palacios HH, Mote PL, Scheibye-Knudsen M, Gomes AP, Ward TM, Minor RK, Blouin MJ, et al. Metformin improves healthspan and lifespan in mice. Nature communications. 2013;4:2192. doi: 10.1038/ncomms3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer TR, Neph S, Dinger ME, Crawford J, Smith MA, Shearwood AM, Haugen E, Bracken CP, Rackham O, Stamatoyannopoulos JA, et al. The human mitochondrial transcriptome. Cell. 2011;146:645–658. doi: 10.1016/j.cell.2011.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller RA, Chu Q, Xie J, Foretz M, Viollet B, Birnbaum MJ. Biguanides suppress hepatic glucagon signalling by decreasing production of cyclic AMP. Nature. 2013;494:256–260. doi: 10.1038/nature11808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzumdar RH, Huffman DM, Atzmon G, Buettner C, Cobb LJ, Fishman S, Budagov T, Cui L, Einstein FH, Poduval A, et al. Humanin: a novel central regulator of peripheral insulin action. PloS one. 2009;4:e6334. doi: 10.1371/journal.pone.0006334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahira K, Haspel JA, Rathinam VA, Lee SJ, Dolinay T, Lam HC, Englert JA, Rabinovitch M, Cernadas M, Kim HP, et al. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nature immunology. 2011;12:222–230. doi: 10.1038/ni.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura A, Amikura R, Mukai M, Kobayashi S, Lasko PF. Requirement for a noncoding RNA in Drosophila polar granules for germ cell establishment. Science. 1996;274:2075–2079. doi: 10.1126/science.274.5295.2075. [DOI] [PubMed] [Google Scholar]
- O'Neill LA, Hardie DG. Metabolism of inflammation limited by AMPK and pseudo-starvation. Nature. 2013;493:346–355. doi: 10.1038/nature11862. [DOI] [PubMed] [Google Scholar]
- Ohta T, Masutomi N, Tsutsui N, Sakairi T, Mitchell M, Milburn MV, Ryals JA, Beebe KD, Guo L. Untargeted metabolomic profiling as an evaluative tool of fenofibrate-induced toxicology in Fischer 344 male rats. Toxicologic pathology. 2009;37:521–535. doi: 10.1177/0192623309336152. [DOI] [PubMed] [Google Scholar]
- Pearson KJ, Baur JA, Lewis KN, Peshkin L, Price NL, Labinskyy N, Swindell WR, Kamara D, Minor RK, Perez E, et al. Resveratrol delays age-related deterioration and mimics transcriptional aspects of dietary restriction without extending life span. Cell metabolism. 2008;8:157–168. doi: 10.1016/j.cmet.2008.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pond SLK, Frost SD. Not so different after all: a comparison of methods for detecting amino acid sites under selection. Molecular biology and evolution. 2005;22:1208–1222. doi: 10.1093/molbev/msi105. [DOI] [PubMed] [Google Scholar]
- Price NL, Gomes AP, Ling AJ, Duarte FV, Martin-Montalvo A, North BJ, Agarwal B, Ye L, Ramadori G, Teodoro JS, et al. SIRT1 is required for AMPK activation and the beneficial effects of resveratrol on mitochondrial function. Cell metabolism. 2012;15:675–690. doi: 10.1016/j.cmet.2012.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricchetti M, Tekaia F, Dujon B. Continued colonization of the human genome by mitochondrial DNA. PLoS biology. 2004;2:E273. doi: 10.1371/journal.pbio.0020273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter U, Lahtinen T, Marttinen P, Myohanen M, Greco D, Cannino G, Jacobs HT, Lietzen N, Nyman TA, Battersby BJ. A mitochondrial ribosomal and RNA decay pathway blocks cell proliferation. Current biology : CB. 2013;23:535–541. doi: 10.1016/j.cub.2013.02.019. [DOI] [PubMed] [Google Scholar]
- Sauter G, Simon R, Hillan K. Tissue microarrays in drug discovery. Nature reviews Drug discovery. 2003;2:962–972. doi: 10.1038/nrd1254. [DOI] [PubMed] [Google Scholar]
- Savard J, Marques-Souza H, Aranda M, Tautz D. A segmentation gene in tribolium produces a polycistronic mRNA that codes for multiple conserved peptides. Cell. 2006;126:559–569. doi: 10.1016/j.cell.2006.05.053. [DOI] [PubMed] [Google Scholar]
- Sena LA, Chandel NS. Physiological roles of mitochondrial reactive oxygen species. Molecular cell. 2012;48:158–167. doi: 10.1016/j.molcel.2012.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethe S, Scutt A, Stolzing A. Aging of mesenchymal stem cells. Ageing research reviews. 2006;5:91–116. doi: 10.1016/j.arr.2005.10.001. [DOI] [PubMed] [Google Scholar]
- Shaw RJ. Metformin trims fats to restore insulin sensitivity. Nature medicine. 2013;19:1570–1572. doi: 10.1038/nm.3414. [DOI] [PubMed] [Google Scholar]
- Slavoff SA, Mitchell AJ, Schwaid AG, Cabili MN, Ma J, Levin JZ, Karger AD, Budnik BA, Rinn JL, Saghatelian A. Peptidomic discovery of short open reading frame-encoded peptides in human cells. Nature chemical biology. 2013;9:59–64. doi: 10.1038/nchembio.1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spangenburg EE, Jackson KC, Schuh RA. AICAR inhibits oxygen consumption by intact skeletal muscle cells in culture. Journal of physiology and biochemistry. 2013 doi: 10.1007/s13105-013-0269-0. [DOI] [PubMed] [Google Scholar]
- Steinberg GR, Kemp BE. AMPK in Health and Disease. Physiological reviews. 2009;89:1025–1078. doi: 10.1152/physrev.00011.2008. [DOI] [PubMed] [Google Scholar]
- Tennstedt P, Koster P, Bruchmann A, Mirlacher M, Haese A, Steuber T, Sauter G, Huland H, Graefen M, Schlomm T, et al. The impact of the number of cores on tissue microarray studies investigating prostate cancer biomarkers. International journal of oncology. 2012;40:261–268. doi: 10.3892/ijo.2011.1216. [DOI] [PubMed] [Google Scholar]
- Tsuzuki T, Nomiyama H, Setoyama C, Maeda S, Shimada K, Pestka S. The majority of cDNA clones with strong positive signals for the interferon-induction-specific sequences resemble mitochondrial ribosomal RNA genes. Biochemical and biophysical research communications. 1983;114:670–676. doi: 10.1016/0006-291x(83)90833-1. [DOI] [PubMed] [Google Scholar]
- Wallace DC. Bioenergetic origins of complexity and disease. Cold Spring Harbor symposia on quantitative biology. 2011;76:1–16. doi: 10.1101/sqb.2011.76.010462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo DK, Shadel GS. Mitochondrial stress signals revise an old aging theory. Cell. 2011;144:11–12. doi: 10.1016/j.cell.2010.12.023. [DOI] [PubMed] [Google Scholar]
- Yun J, Finkel T. Mitohormesis. Cell metabolism. 2014;19:757–766. doi: 10.1016/j.cmet.2014.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.