Abstract
Background
Gene expression in archived newborn blood spots remaining from newborn screening may reflect pathophysiological disturbances useful in understanding the etiology of cerebral palsy (CP).
Methods
We quantified the expression of gene sets representing four physiological pathways hypothesized to contribute to CP in archived unfrozen residual newborn blood spot specimens from 53 children with CP and 53 age, gender, and gestational-age–matched controls. We selected four empirical and three canonical gene sets representing inflammatory, hypoxic, coagulative, and thyroidal pathways, and examined mRNA expression using an 8×60K oligonucleotide microarray. The log2 fold change of gene expression between matched cases and controls were analyzed using the Generally Applicable Gene Set Enrichment (GAGE) method.
Results
The empirical inflammatory and empirical hypoxic gene sets were significantly down-regulated in term-born CP cases (N = 33) as compared to matched controls (P = 0.0007 and 0.0009, respectively), while both gene sets were significantly up-regulated (P = 0.0055 and 0.0223, respectively) in preterm-born CP cases (N = 20). The empirical thyroidal gene set was significantly up-regulated in preterm-born CP (P = 0.0023).
Conclusion
The newborn blood spot transcriptome can serve as a platform for investigating distinctive gene expression patterns in children who later develop CP.
Introduction
Cerebral palsy (CP) is a neurological syndrome of onset in early childhood, characterized by impaired control of motor function (1), with a prevalence of about 2–3 per 1,000 live births in Western countries (2). The etiology of conditions, such as CP, that originate with exposures occurring in pregnancy and the perinatal period is difficult to study, because our capacity to interrogate such exposures once disease is manifest years later is limited. We here report on the assessment of biological processes occurring around birth, via examination of the transcriptome in residual filter paper blood spots archived after newborn genetic screening in 53 singleton children with CP and 53 age-, gender-, and gestational-age– matched controls.
Examination of the transcriptome has found little application thus far in etiologic epidemiology. Messenger RNA (mRNA) is hard to retrieve from serum samples, because ribonucleases and micro RNAs degrade mRNA quickly once transcriptional activity is complete. But while blood is ordinarily collected in glass or plastic tubes, permitting such enzymatic activity to continue, newborn blood is directly spotted from a heel stick incision onto filter paper, completely drying at room temperature within 3 hours (3,4). RNA degradation is an enzymatic process tightly controlled by ribonucleases (RNases), water-soluble enzymes that require one water molecule for each reaction (5). Drying of the sample interferes with this reaction and also limits access of RNases to RNA.
White blood cells share more than 80% of the transcriptome with at least 9 organs, including the brain (6); 82% of the 13,961 genes expressed in brain, according to the UNIgene database, are expressed in blood (7). We hypothesized that gene sets reflecting four pathophysiological pathways to CP (inflammatory, hypoxic, coagulative and thyroidal) would be dysregulated shortly after birth in children later diagnosed with CP, and that this dysregulation would be detectable in newborn blood spots. We also performed exploratory gene set analyses using a database of clinically relevant gene sets (8). The hypothesized gene sets were obtained by searching the literature, aiming for one canonical gene set (i.e. based on expert opinion (9) and one experimental gene set (i.e. based on experimental findings) for each pathway. For coagulation, we could only find a canonical gene set.
Results
Comparability of cases and controls
Cases and controls were similar in gestational age, a matching criterion, but fetal growth was somewhat reduced in cases. More cases were admitted to newborn intensive care and had lower Apgar scores. Mean day of blood spot collection did not differ between cases and controls. Cases and controls were broadly comparable in socio-economic and demographic characteristics (Table 1).
Table 1.
Characteristics of cases and controls at birth of child
| 37 weeks and above | Under 37 weeks | |||
|---|---|---|---|---|
| Cases (n =33) | Controls (n =33) | Cases (n =20) | Controls (n = 20) | |
| Biological/Clinical | ||||
| Mean Gestational Age | 39.5 wks (±1.2) | 39.6 wks (±1.1) | 30.2 wks (±3.6) | 31.8 wks (±3.7) |
| Mean Birthweight | 3,372 g (±495) | 3,562 g (±432) | 1,884 g (±889) | 2,106 g (±902) |
| Fetal Growth Ratioa | 0.99 | 1.04 | 0.95 | 1.01 |
| % Small for Gestational Agea | 9% | 6% | 10% | 5% |
| Mean 5 minute Apgar Score | 8.2 (± 1.6) | 8.8 (±0.6) | 7.5 (±1.4) | 8.5 (±0.7) |
| % Admitted to NICU | 36% | 9% | 95% | 78% |
| Age blood spot obtained | 1.7 days (±1.9) | 1.3 days (±0.5) | 1.5 days (±1.7) | 1.2 days (±0.9) |
| Socio-Demographic | ||||
| % Black | 12% | 3% | 10% | 20% |
| % Hispanic | 0 | 4% | 6% | 0 |
| % White or other | 88% | 93% | 84% | 80% |
| % Mother married | 82% | 82% | 71% | 82% |
| % Home ownership | 86% | 68% | 59% | 71% |
| % Medicaid support | 21% | 6% | 25% | 35% |
Ref. (38) was used as the source of fetal growth standards. (Fetal growth ratio = infant’s BW/median BW for gestational week.)
Bolded case-control contrasts, p <.05
Analysis of seven pre-selected gene sets
Each case-control pair received a score (expressed as a GAGE t-statistic) summarizing the case-control difference in expression of all genes (from 31–200 genes, depending on the gene set). This score can be interpreted as the difference between cases and controls expressed as fractions of a standard deviation. These score differences were then summarized across pairs and assessed for the statistical significance of the summarized difference, expressed as the global P-value for the contrast of cases and controls for each gene set.
Three gene sets, all empirical, showed significantly different gene regulation across the population of case-control pairs (Table 2). The empirical inflammatory and asphyxial gene sets were both significantly down-regulated in CP cases, compared to controls, with effect sizes of −0.19 SD units and −0.16 SD units respectively. The thyroidal gene set was significantly up-regulated by +0.13 SD units. These three differences were highly significant, either with (q value) or without (p value) correction for multiple testing.
Table 2.
Gamma GAGE analysis for seven gene sets representing four pre-hypothesized pathways
| Gene sets | Reference to the source of the gene set | Mean of GAGE t-statisticsa | P-values (q-values) for up-regulationb | P-values (q-values) for down regulationb | P-values (q-values) for Bi-directional regulationb |
|---|---|---|---|---|---|
| Coagulative | |||||
| Canonical (n=93; ne=92) | GO:0007596; blood clotting (28) | −0.08 | 0.9749 (>0.1) | 0.7737 (>0.1) | 0.6048 (>0.1) |
| Inflammatory | |||||
| Canonical (n=31; ne=31) | GO:0050727; regulation of inflammatory response (28) | −0.10 | 0.9870 (>0.1) | 0.7796 (>0.1) | 0.9439 (>0.1) |
| Empirical (n=36; ne=36) | Fetal inflammatory response (31) | −0.19 | 0.0791 (>0.1) | 0.0001 (0.0008) | 0.2139 (>0.1) |
| Asphyxial | |||||
| Canonical (n=37; ne=36) | Hypoxia (27) | 0.18 | 0.1656 (>0.1) | 0.9401 (>0.1) | 0.9620 (>0.1) |
| Empirical (n=127; ne=126) | Hypoxia (26) | −0.16 | 0.0983 (>0.1) | 0.0016 (0.0059) | 0.9749 (>0.1) |
| Thyroidal | |||||
| Canonical (n=200; ne=198) | V$T3R_Q6; TRE consensus (9) | −0.03 | 0.8183 (>0.1) | 0.7344 (>0.1) | 0.9993 (>0.1) |
| Empirical (n=140; ne=139) | Thyroid hormone (29) | 0.13 | 0.0039 (0.0273) | 0.2873 (>0.1) | 0.9767 (>0.1) |
Since the number of genes in all of the seven selected gene sets is >30, the GAGE t-statistic can be taken as a z-statistic. Thus, the mean of GAGE t-statistics, which can be expressed in terms of standard deviation (SD) units, is an approximation of effect size.
global P values of all pairs, with q-values in parentheses for the three significant findings calculated to adjust for multiple testing using q-values R package with FDR set at 0.05
n = Number of genes in the gene set. ne = Number of genes of the gene set that are found in the array used in this study
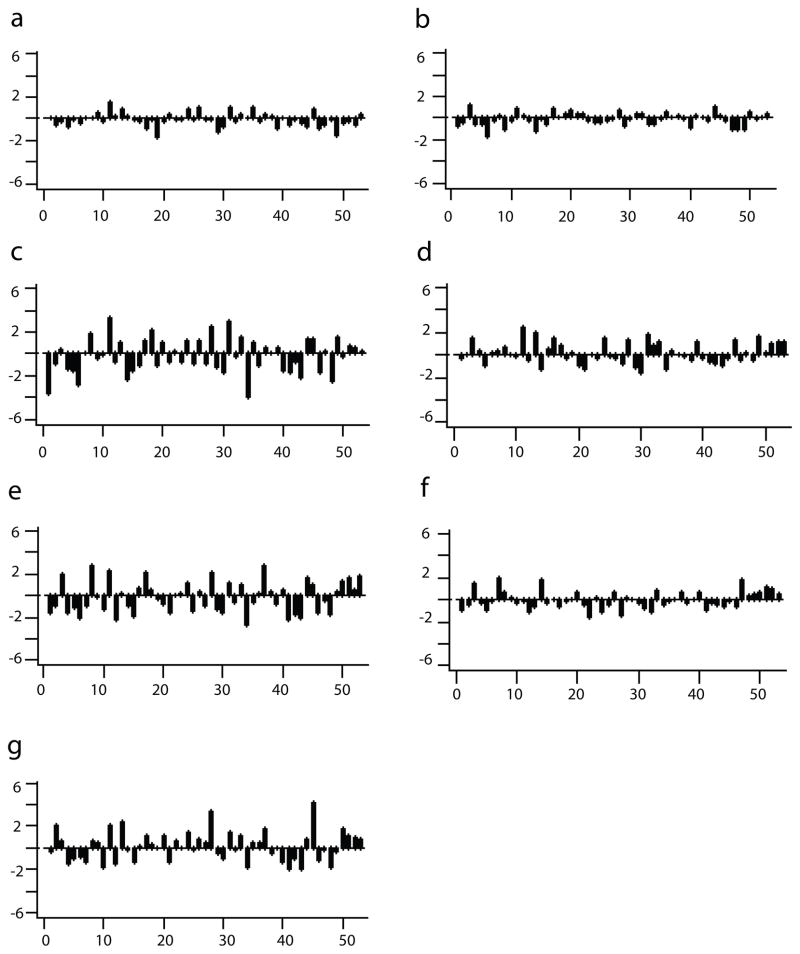
To illustrate gene set differences across case and control pairs, the distribution of GAGE t-statistics for each of the gene sets is shown in Figure 1. The three significant gene sets showed many pairs with large inter-pair differences in gene expression, while the canonical hypoxic and thyroidal gene sets showed modest inter-pair differences. The coagulation gene set and the canonical inflammatory gene set showed virtually no differences. Heterogeneity across pairs in the direction of differential expression (either up or down) is notable for all three statistically significant gene sets (Table 2, Figure 1).
Figure 1.
GAGE t-statistics for the seven pre-hypothesized gene sets. (a) canonical coagulation, (b) canonical inflammation, (c) empirical inflammation, (d) canonical asphyxia, (e) empirical asphyxia, (f) canonical thyroid, (g) empirical thyroid gene sets respectively. For each graph: X-axis: matched pair (total 53 pairs); Y-axis: scale of GAGE t-statistic; each bar within each graph: the GAGE t-statistic of the gene set for each pair.
Individual Genes in the Fetal Inflammatory Response Syndrome (FIRS) Gene Set
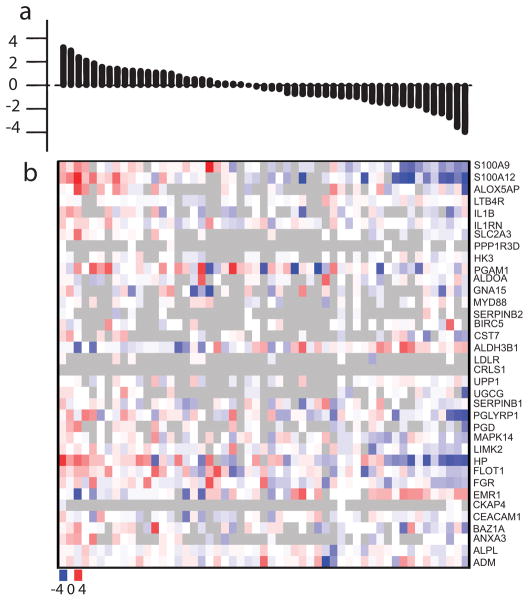
The largest case-control differences in gene expression (0.19 SD) were seen for the FIRS gene set. Differences in gene expression are often described as “fold changes”, that is, the ratio of the degree of gene expression in cases compared to controls, most conveniently expressed as the binary logarithm (log2 n = logarithm to the base 2) of the fold change. Figure 2 shows the heatmap of the log2 fold change of all genes in the FIRS gene set for all case-control pairs ordered by the magnitude of the GAGE t-statistics. The largest pair differences were seen in the following genes: S100A9 (S100 calcium binding protein A9), S100A12 (S100 calcium binding protein A12), ALOX5AP (arachidonate 5-lipoxygenase-activating protein), PGLYRP1 (peptidoglycan recognition protein 1), HP (haptoglobin), FLOT1 (flotillin 1), and FGR (Gardner-Rasheed feline sarcoma viral oncogene homolog) (Figure 2).
Figure 2.
Heatmap of FIRS gene set with pairs ordered by magnitude of GAGE t-statistics. (a) FIRS gene set in which the matched pairs are ordered by the values of the GAGE t-statistics of the pairs from most positive to most negative. (b) Heatmap: X-axis: matched pairs in the same order as the upper graph; Y-axis: gene names. Each small square represents log2 fold change of each of all genes of FIRS gene set of each of all pairs. Gradient scale for color from bluest (most negative log2 fold change or the gene expresses lowest in case vs. control) to white (log2 fold change is zero or the gene expresses equally in case vs. control) to reddest (positive log2 fold change or the gene expresses highest in case vs. control): −4 to 0 to + 4. Grey color: absence of data (missing values) due to unmet filtering criteria.
Assessing Significant Gene Set Findings in CP sub-sets
The heterogeneity of GAGE t-statistics across pairs suggests the need to stratify on covariates such as gestational age and motor type. Table 3 shows the findings for each gene set for children born at 37 weeks or later (N pairs = 33), before 37 weeks (N pairs = 20), with quadriplegia (N pairs = 24), diplegia (N pairs = 15) and hemiplegia (N pairs = 13).
Table 3.
Gene expression findings for three gene sets stratified by GA and CP type
| Gene sets | Empirical inflammatory gene set | Empirical asphyxial gene set | Empirical thyroidal gene set | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Mean GAGE t-stat | P-values up | P-values down | Mean GAGE t-stat | P-values up | P-values down | Mean GAGE t-stat | P-values up | P-values down | |
| Gestational age | |||||||||
| >=37 weeks (n=33) | −0.42 | 0.6567 | 0.00007 | −0.36 | 0.5014 | 0.00089 | −0.04 | 0.1345 | 0.1204 |
| <37 weeks (n=20) | 0.19 | 0.0055 | 0.1537 | 0.16 | 0.0223 | 0.2381 | 0.42 | 0.0023 | 0.7353 |
| CP typea | |||||||||
| Quadriplegia (n=24) | −0.26 | 0.60869 | 0.02830 | −0.18 | 0.11721 | 0.01024 | 0.08 | 0.01865 | 0.16745 |
| Diplegia (n=15) | 0.16 | 0.03753 | 0.23559 | 0.03 | 0.27462 | 0.30122 | 0.32 | 0.10591 | 0.84832 |
| Hemiplegia (n=13) | −0.45 | 0.11210 | 0.00009 | −0.32 | 0.30896 | 0.01476 | 0.01 | 0.09332 | 0.19450 |
CP type missing for 1 case
FIRS showed a clear interaction with gestational age; among 20 preterm pairs, FIRS was significantly up-regulated in CP cases, whereas among 33 term-born pairs, FIRS was significantly down-regulated among cases. The FIRS up-regulation seen in preterm cases was paralleled by up-regulation in diplegic cases, who are dominantly preterm. In parallel, the strongest contribution to down-regulation of inflammation came from hemiplegic cases (N = 13) who are nearly all born at term. Quadriplegia also showed down-regulation of FIRS, but not as strongly as hemiplegia.
The empirical hypoxic gene set also showed significant up-regulation in preterm cases and the opposite with term cases. Down-regulation of the hypoxic gene set was seen in hemiplegia and quadriplegia. The thyroidal up-regulation signal was derived entirely from preterms and from children with quadriplegia. The sample size precluded consideration of interactions between gestational age and motor typology (Table 3).
Kyoto Encyclopedia of Genes and Genomes (KEGG) gene sets
We explored case-control differences for the 205 gene sets archived by KEGG (2009 version) (8). The five most up-regulated gene sets in CP cases compared to controls were: Ribosome, Systemic lupus erythematosus (SLE), Olfactory transduction (OT), Cell cycle (CC) and Oxidative phosphorylation (OP). Down-regulation was seen most strongly for three of the above five - Ribosome, CC and SLE, and for leukocyte transendothelial migration (LTM) and Regulation of actin cytoskeleton (RAC) gene sets. Using the approach of Storey to control for the false discovery rate (FDR) (10, 11), Ribosome, SLE and OT were significantly up-regulated, Ribosome, LTM and RAC were significantly down-regulated, and the Ribosome gene set was significantly bi-directionally regulated.
The analysis for all individual genes available in the arrays reveals that no individual gene was significantly differentially expressed between cases and controls, after adjusting for multiple testing. The lack of single gene expression differences confirms the value of gene set analysis for aggregating coordinated expression signals from related genes in gene sets in exploring pathophysiological pathways to disease (Table 4).
Table 4.
Most up-regulated and most down-regulated KEGG gene sets in cases compared to controls
| Most up-regulated gene sets | Most down-regulated gene sets | Most bi-directionally regulated gene sets | ||||||
|---|---|---|---|---|---|---|---|---|
| Gene sets | P-values | q-valuesa | Gene sets | P-values | q-valuesa | Gene sets | P-values | q-valuesa |
| Ribosome | 4.4e-40 | 9.1e-38 | Ribosome | 4.1e-42 | 8.3e-40 | Ribosome | 1.9e-14 | 3.9e-12 |
| SLE | 1.7e-06 | 1.8e-04 | LTM | 4.2e-04 | 3.5e-02 | BTF | 3.3e-03 | >0.1 |
| OT | 5.8e-05 | 3.9e-03 | RAC | 5.1e-04 | 3.5e-02 | APM | 4.9e-02 | >0.1 |
| CC | 2.4e-03 | >0.1 | CC | 1.8e-03 | 9.5e-02 | PD | 5.0e-02 | >0.1 |
| OP | 5.6e-03 | >0.1 | SLE | 9.2e-03 | >0.1 | MODY | 5.1e-02 | >0.1 |
q-values were calculated using q-values with FDR level set at 0.05.
SLE: Systemic lupus erythematosus, OT: Olfactory transduction, CC: Cell cycle, OP: Oxidative phosphorylation, LTM: Leukocyte transendothelial migration, RAC: Regulation of actin cytoskeleton, BTF: Basal transcription factors, APM: Aminophosphonate metabolism, PD: Parkinson’s disease, MODY: Maturity onset diabetes of the young.
Quantitative polymerase chain reaction (qPCR) validation of mRNA data
To validate our microarray findings, we used qPCR techniques to examine the housekeeping genes, ACTB (beta actin) and PPIA (peptidylprolyl isomerase A), both commonly used in the literature to validate microarray findings. To validate genes differentially expressed, we selected FCGR2A (Fc fragment of IgG, low affinity IIa receptor), a representative gene of the Lupus pathway from the KEGG database.
For ACTB and PPIA genes, the correlation coefficient between the log2 intensity of microarray data and mean CT (cycle threshold value) of qPCR was −0.52 (P < 0.0001). For FCGR2A, the correlation coefficient between the log2 intensity of microarray data and mean CT of qPCR is −0.43 (P < 0.0001) and the correlation coefficient between log2 fold change of microarray data and −log2 fold change of qPCR data was 0.38 (P = 0.0197).
Discussion
Principal findings and their direction
We found three gene sets, each reflecting a separate pathophysiological pathway, to be significantly dysregulated in newborns who later developed CP. The direction of the signals was not entirely as expected. While the up-regulation of inflammation noted among prematures later developing CP is consistent with several previous studies (12, 13, 14), the significant down-regulation of inflammation in term CP, and up-regulation of thyroidal function in premature CP seem opposite to what might be expected based on the literature (15, 16). However, dysregulation of a molecular pathway, as measured by changes in gene expression, can result from the influences of several types of cellular control processes, not all in the same direction. For example, mitochondrial defects leading to decreased energy output activate a transcriptional response to increase energy output, registered as increased expression of mitochondrial genes (17). It is thus more useful to consider dysregulation in either direction as indicating some perturbation of cellular control systems involved in the pathophysiologic pathway of interest.
Validity Studies of Archived Newborn Blood Spots
In Michigan, spots are normally collected on day one or two of life, allowed to dry in ambient conditions for four hours, and sent to the Michigan Department of Community Health (MDCH), where, after testing, they have been stored in boxes at room temperature. Since 2009, with the establishment of the Michigan Biobank for Health (18), all new specimens are placed in −80°C storage. None of the spots used in this study were frozen.
Four sources of evidence support the validity of the mRNA data obtained from our samples:
Published research by others. Several studies have shown that even after 20 years of unfrozen storage, messenger RNA can be recovered from unfrozen blood spots (19, 20, 21, 22).
Our own published work. We have published two papers demonstrating the reliability and validity of the archived unfrozen newborn blood spots we use in this study. Haak et al have demonstrated the replicability (R > .90) of microarray gene expression (23), and Resau et al have shown that sex-specific genes such as XIST and KDM5D are distinguishable in male and female archived unfrozen neonatal blood spots (24). A third paper shows the high sensitivity of gene detection in blood spotted on filter paper with the Agilent 4 × 44K microarray system used in this study (Agilent Technologies, Santa Clara, CA) (25).
qPCR confirmation of microarray findings: In our earlier study (23), we showed that using qPCR we could identify two housekeeping genes (HKI and MRPLII) and two inflammatory genes (ITGAX and NFKB-1) in archived unfrozen blood spots. As shown above, in our present study we move beyond qPCR identification, and show the strong correlation between the amount of mRNA detected by microarray and qPCR for another three genes.
Unpublished pilot data comparing frozen and unfrozen newborn blood spots. Microarray was performed in our laboratory on paired spots from the same newborns studied ten years ago. Frozen (−80°C) blood spots obtained from 5 Michigan newborns as part of a research study were compared to the blood spots archived by the state after genetic screening obtained at about the same age. The average Pearson correlation coefficient in total gene expression between frozen and unfrozen spots was 0.94.
Exploratory gene set analyses
Our exploratory analysis of the KEGG gene set reinforced the importance of inflammatory processes in CP, since several gene sets related to such processes were significantly dysregulated. The lupus gene set, for example, contains genes such as T cell and B cell receptor signaling pathways and cytokine-cytokine receptor interaction. Several KEGG inflammatory gene sets that differed in case-control expression showed the same interaction with gestational age as FIRS, with up-regulation in preterm-born CP and down-regulation in term-born CP.
Sensitivity analyses
To determine whether values resulting from filtering procedures affect the results, we repeated the analyses with imputation for missing values and found nearly identical gene set findings. We also performed permutations on randomly selected gene sets of different sizes to evaluate the false positive rate of the GAGE method on our actual microarray data. The probability of false positive findings of the GAGE method is <0.05 when the significance level for the test is set at <0.05. (A detailed description is in the supplementary materials online.)
In conclusion, we find that archived unfrozen filter paper blood spots, even after many years of storage, retain enough mRNA to describe a distinct gene expression profile in neonates who later develop CP. The pathophysiological pathways we identify get us closer to understanding the changes that take place in the interval between prenatal or perinatal brain insult and the clinical manifestation of the CP syndrome. Our results also underline the scientific promise of using the large collections of state-archived newborn blood spots to develop compendia of expression profiles in disease states characterized by perturbations in gene expression that can be captured shortly after birth.
Methods
Study subjects
Children with clinically verified CP aged 2–15 (N = 53) were recruited in specialty clinics in Lansing, Ann Arbor, and Grand Rapids, MI, if they had GMFCS scores >1, and did not have a recognized malformation syndrome accounting for the CP. Control subjects were recruited in primary care settings in Michigan and were individually matched on year of birth, sex, and gestational age in four categories (<28 weeks, 28–32 weeks, 33–37 weeks, >37 weeks) and were free of major brain disorders. Birth certificates, maternal and infant hospital discharge abstracts and residual newborn blood specimens were retrieved from MDCH, after obtaining written informed consent from the mother or guardian of the study subject. Case and control mothers were interviewed by telephone about reproductive exposures. Approval for this study was obtained from the Institutional Review Boards of all participating institutions.
RNA isolation and cDNA preparation
RNA isolation, cDNA synthesis, and microarray on blood spot samples was performed at the Laboratory of Microarray Technology at VARI as per our published protocol (25). Three 3-mm blood spot punches were homogenized using a TissueLyser (Qiagen, Valencia, CA) before isolation using the illustra RNAspin Mini kit (GE Healthcare, Buckinghamshire, UK). DNase digestion was carried out during RNA isolation to remove any contaminating DNA in RNA. Total RNA was then concentrated with an RNA Clean & Concentrator-5 Kit (Zymo Research, Orange, CA). RNA quality and quantity were evaluated using an RNA Pico Lab Chip on the Agilent BioAnalyzer. The average RIN (RNA Integrity Number) was 2.3±0.71. The WT-Ovation Pico RNA Amplification System (NuGEN Technologies, San Carlos, CA) based on the Ribo-SPIA technology was used to amplify total RNA and synthesize first and second-stranded cDNA. Since this amplification system is initiated both at the 3′ end and randomly throughout the whole transcriptome, it has the advantage of amplifying non-poly A transcripts and compromised RNA.
Gene expression microarray assays
For gene expression microarray, cDNA was labeled with Alexa Fluor 3 fluorescent dye from the BioPrime Total Genomic Labeling System (Invitrogen Life Technologies, Carlsbad, CA) before purification using the PureLink PCR Purification System (Invitrogen). Purified labeled product was then applied onto a 8×60K whole human genome gene expression microarray (Agilent). Each array contains 60,000 oligonucleotide probes (60bp probe) covering 27,958 Entrez gene RNAs and 7,419 long intergenic non-coding RNAs. The arrays were hybridized for 17 hours at 65 °C and 10 rpm rotation speed, then washed for 2 min each with wash buffer 1 and 2 and scanned with an Agilent G3 high-resolution scanner. Probe features were extracted from the microarray scan data using Feature Extraction software v. 10.7.3.1 (Agilent).
Microarray confirmation by qPCR
Masked cDNA samples synthesized from neonatal blood spot RNA were used for qPCR analysis. Specific optimized Taqman probes and primers were obtained from Applied Biosystems by Life Technologies (Carlsbad, CA) and qPCR was performed using Applied Biosystems 7500 Fast Real-Time PCR System. We examined the correlation between log2 intensity of microarray data and mean CT of qPCR data for all three genes, and for the FCGR2A gene, we also assessed the correlation between log2 fold change of microarray data and −log2 fold change of qPCR.
Selection of 7 gene sets representing 4 hypothesized pathways to CP
The empirical Hypoxic gene set is based on responses of cells in tissue culture exposed to hypoxemia compared to normoxemia (26) while the canonical Hypoxic gene set is based on the view that hypoxia-inducible transcription factor (HIF) binds a consensus DNA sequence termed the hypoxia-responsive element (HRE) (27). A similar approach was used to construct the canonical thyroid responsive element (TRE) gene set (28). The experimentally derived thyroid gene set was isolated following human exposure to thyroid hormone (29). The canonical inflammation pathway is GO:005072; inflammatory response (30), while the empirical gene set is from an experiment in prematures with and without evidence of fetal inflammation and infection (31). The coagulation cascade was represented by GO:0007596; (blood clotting biological processes) (30).
Statistical methods
Data processing and analysis were performed using statistical software R (version 2.13.2). Unqualified spots were filtered using the method of Patterson et al, where expression data were removed whenever the gProcessed signal was less than twice the gProcessed signal error (32). Gene expression data were normalized using a between-array quantile normalization method (33) and further aggregated to the gene level using the mean of the expression signal of all probes of each gene. Differential expression of individual genes was examined with the moderated paired t-test (appropriate for matched pairs) of the linear model and an empirical Bayes method implemented in R package limma (34). The significance of gene expression was corrected for multiple testing, setting the false discovery rate (FDR) at 0.05 (10). We chose the Generally Applicable Gene Set Enrichment (GAGE) method (35) the only published method applicable to a matched case-control study among several methods for gene set analysis (36,37). A fuller description of the GAGE method is provided in the supplementary materials (online). For case-control comparisons of clinical data, exact conditional logistic regression and paired t-tests were used for categorical variables and continuous variables, respectively.
Supplementary Material
Acknowledgments
We appreciate the work of the membership of the OWL (Outcome, Wellness and Life-history in cerebral palsy) study team, including Dr. Mary Jo Cooley Hidecker, Dr. Jaime Slaughter, Suzette Baez Vanderbeek, Joseph Bonner, Ariel Brovont, Mary Crawford, Matt Francis, Alice Stanley and Dr. Sainan Wei; referring physicians Drs. Zachary Dyme, Steven DeRoos, and Andrea Kuldanek; and collaborators Harry Hawkins and Glenn Copeland of the Michigan Department of Community Health. All authors participated in manuscript preparation and editing. In addition, Dr. Ho conceived of and performed the data analyses; Dr. Furge chose the gene sets and advised on analysis; Dr. Fu and Dr. Lu advised on statistical analysis; Dr. Busik performed the qPCR analyses and advised on laboratory aspects of the study; Dr. Khoo performed the microarray analyses and advised on laboratory aspects of the study; Ms. Lenski, Dr. Hurvitz, Dr. Wirth and Dr. Dodge supervised and organized data collection in clinical settings, advised on clinical aspects of the study, and participated in discussions of findings and manuscript reviews; Dr. Resau supervised the microarray work; and Dr. Paneth conceived of the study, obtained its funding, and serves as principal investigator of the grant.
Statement of financial support: Funded by National Institutes of Health grant R01 NS055101 and the Van Andel Research Institute. Nhan Thi Ho is partially funded by Vietnam Education Foundation.
Footnotes
Conflicts of interest: None
References
- 1.Rosenbaum P, Dan B, Fabiola R, et al. Proposed definition and classification of cerebral palsy, April 2005. The definition of cerebral palsy. Dev Med Child Neurol. 2005;47:571–6. doi: 10.1017/s001216220500112x. [DOI] [PubMed] [Google Scholar]
- 2.Paneth N, Hong T, Korzeniewski S. The descriptive epidemiology of cerebral palsy. Clin Perinatol. 2006;33:251–67. doi: 10.1016/j.clp.2006.03.011. [DOI] [PubMed] [Google Scholar]
- 3.Doh.sd.gov [Internet] Approved standard. 4. Vol. 23. National Committee for Clinical Laboratory Standards; 2003. Blood collection on filter paper for neonatal screening programs; p. 21. NCCL Document LA4-A4. Available from: http://doh.sd.gov/NewbornScreening/Manual/bloodcollection.pdf. [Google Scholar]
- 4.Whatman.com [Internet] Neonatal screening. General Electric Company; Blood specimen collection and handling procedure. [updated Mar 2010; cited Jul 9 2012]. Available from: http://www.whatman.com/References/Neonatal%20Screening%20Cue%20Card%20LR%20FINAL%203.2.10.pdf. [Google Scholar]
- 5.Roberts GCK, Dennis AD, Meadows DH, et al. The mechanism of action of ribonuclease. Proc Natl Acad Sci. 1969;62:1151–1158. doi: 10.1073/pnas.62.4.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liew CC, Ma J, Tang HC, Zheng R, Dempsey AA. The peripheral blood transcriptome dynamically reflects system wide biology: a potential diagnostic tool. J Lab Clin Med. 2006;147:126–132. doi: 10.1016/j.lab.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 7.Mohr S, Liew CC. The peripheral-blood transcriptome: new insights into disease and risk assessment. Trends Mol Med. 2007;13:422–432. doi: 10.1016/j.molmed.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 8.Genome.jp [Internet] KEGG: Kyoto Encyclopedia of Genes and Genomes. [cited 2009 Nov 30]. Available at: www.genome.jp/kegg/
- 9.Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Storey JD. A direct approach to false discovery rates. J R Statist Soc B. 2002;64:479–498. [Google Scholar]
- 11.Storey JD. The positive false discovery rate: a Bayesian interpretation and the q-value. Ann Statist. 2003;6 :2013–2035. [Google Scholar]
- 12.Leviton A, Allred EN, Kuban KC, et al. Microbiologic and histologic characteristics of the extremely preterm infant’s placenta predict white matter damage and later cerebral palsy. Pediatr Res. 2010;67:95–101. doi: 10.1203/PDR.0b013e3181bf5fab. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yoon BH, Jun JK, Romero R, et al. Amniotic fluid inflammatory cytokines (interleukin-6, interleukin-1β, and tumor necrosis factor-α), neonatal brain white matter lesions, and cerebral palsy. Am J Obstet Gynecol. 1997;177:19–26. doi: 10.1016/s0002-9378(97)70432-0. [DOI] [PubMed] [Google Scholar]
- 14.Huang HC, Wang CL, Huang LT, et al. Association of cord blood cytokines with prematurity and cerebral palsy. Early Hum Dev. 2004;77:29–36. doi: 10.1016/j.earlhumdev.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 15.Wu YW, Escobar GJ, Grether JK, et al. Chorioamnionitis and cerebral palsy in term and near-term infants. JAMA. 2003;290:2677–84. doi: 10.1001/jama.290.20.2677. [DOI] [PubMed] [Google Scholar]
- 16.Reuss ML, Paneth N, Pinto-Martin JA, Lorenz JM, Susser M. The relation of transient hypothyroxinemia in preterm infants to neurologic development at two years of age. N Engl J Med. 1996;28:821–7. doi: 10.1056/NEJM199603283341303. [DOI] [PubMed] [Google Scholar]
- 17.Klomp JA, Petillo D, Niemi NM, et al. Birt-Hogg-Dubé renal tumors are genetically distinct from other renal neoplasias and are associated with up-regulation of mitochondrial gene expression. BMC Med Genomics. 2010 Dec 16;3:59. doi: 10.1186/1755-8794-3-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Michigan.gov [Internet] Michigan Department of Community Health; [cited 2012 Mar 30]. Available at: http://www.michigan.gov/mdch/0,1607,7-132-2942_4911_4916_53246-209738--,00.html. [Google Scholar]
- 19.Karlsson H, Guthenberg C, von Dobeln U, Kristenssson K. Extraction of RNA from dried blood on filter papers after long-term storage. Clin Chem. 2003;49:979–81. doi: 10.1373/49.6.979. [DOI] [PubMed] [Google Scholar]
- 20.Zhang YH, McCabe ER. RNA analysis from newborn screening dried blood specimens. Hum Genet. 1992;89:311–4. doi: 10.1007/BF00220548. [DOI] [PubMed] [Google Scholar]
- 21.Matsubara Y, Ikeda H, Endo H, Narisawa K. Dried blood spot on filter-paper as a source of messenger-RNA. Nucleic Acids Res. 1992;20:1998. doi: 10.1093/nar/20.8.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gauffin F, Nordgren A, Barbany G, et al. Quantitation of RNA decay in dried blood spots during 20 years of storage. Clin Chem Lab Med. 2009;47:1467–1469. doi: 10.1515/CCLM.2009.351. [DOI] [PubMed] [Google Scholar]
- 23.Haak PT, Busik JV, Kort EJ, et al. Archived unfrozen neonatal blood spots are amenable to quantitative gene expression analysis. Neonatology. 2009;95:210–216. doi: 10.1159/000155652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Resau JH, Ho NT, Dykema K, et al. Evaluation of sex-specific gene expression in archived dried blood spots (DBS) Int J Mol Sci. 2012;13:9599–9608. doi: 10.3390/ijms13089599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khoo SK, Dykema K, Vadlapatla NM, et al. Acquiring genome-wide gene expression profiles in Guthrie card blood spots using microarrays. Pathol Int. 2011;61:1–6. doi: 10.1111/j.1440-1827.2010.02611.x. [DOI] [PubMed] [Google Scholar]
- 26.Chi JT, Wang Z, Nuyten DS, et al. Gene expression programs in response to hypoxia: Cell type specificity and prognostic significance in human cancers. PLoS Med. 2006;3:e47. doi: 10.1371/journal.pmed.0030047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maynard MA, Ohh M. von Hippel-Lindau tumor suppressor protein and hypoxia-inducible factor in kidney cancer. Am J Nephrol. 2004;24:1–13. doi: 10.1159/000075346. [DOI] [PubMed] [Google Scholar]
- 28.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102;43:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Clement K, Viguerie N, Diehn M, et al. In vivo regulation of human skeletal muscle gene expression by thyroid hormone. Genome Res. 2002;12:281–91. doi: 10.1101/gr.207702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Geneontology.org [Internet] [cited 2010 April 30]. Available at: http://www.geneontology.org.
- 31.Madsen-Bouterse SA, Romero R, Tarca AL, et al. The transcriptome of the fetal inflammatory response syndrome. Am J Reprod Immunol. 2010;63:73–92. doi: 10.1111/j.1600-0897.2009.00791.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Patterson TA, Lobenhofer EK, Fulmer-Smentek SB, et al. Performance comparison of one-color and two-color platforms within the MicroArray Quality Control (MAQC) project. Nat Biotechnol. 2006;24:1140–50. doi: 10.1038/nbt1242. [DOI] [PubMed] [Google Scholar]
- 33.Bolstad BM, Irizarry RA, Astrand M, et al. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–93. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 34.Smyth GK. Limma: linear models for microarray data. In: Gentleman R, Carey V, Dudoit S, Irizarry R, Huber W, editors. Bioinformatics and Computational Biology Solutions using R and Bioconductor. Springer; New York: 2005. pp. 397–420. [Google Scholar]
- 35.Luo W, Friedman MS, Shedden K, Hankenson KD, Woolf PJ. GAGE: generally applicable gene set enrichment for pathway analysis. BMC Bioinformatics. 2009;10:161. doi: 10.1186/1471-2105-10-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Curtis RK, Oresic M, Vidal-Puig A. Pathways to the analysis of microarray data. Trends Biotechnol. 2005;23:429–35. doi: 10.1016/j.tibtech.2005.05.011. [DOI] [PubMed] [Google Scholar]
- 37.Goeman JJ, Buhlmann P. Analyzing gene expression data in terms of gene sets: methodological issues. Bioinformatics. 2007;23:980–7. doi: 10.1093/bioinformatics/btm051. [DOI] [PubMed] [Google Scholar]
- 38.Alexander GR, Himes JH, Kaufman RB, Mor J, Kogan M. A United States national reference for fetal growth. Obstet Gynecol. 1996;87:163–8. doi: 10.1016/0029-7844(95)00386-X. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.