SUMMARY
Subterranean mammals spend their lives in dark, unventilated environments rich in carbon dioxide and ammonia, and low in oxygen. Many of these animals are also long-lived and exhibit reduced aging-associated diseases, such as neurodegenerative disorders and cancer. We sequenced the genome of the Damaraland mole rat (DMR, Fukomys damarensis) and improved the genome assembly of the naked mole rat (NMR, Heterocephalus glaber). Comparative genome analysis, along with transcriptomes of related subterranean rodents, reveal candidate molecular adaptations for subterranean life and longevity, including a divergent insulin peptide, expression of oxygen-carrying globins in the brain, prevention of high CO2-induced pain perception, and enhanced ammonia detoxification. Juxtaposition of the genomes of DMR and other more conventional animals with the genome of NMR revealed several truly exceptional NMR features: unusual thermogenesis, aberrant melatonin system, pain insensitivity, and novel processing of 28S rRNA. Together, the new genomes and transcriptomes extend our understanding of subterranean adaptations, stress resistance and longevity.
INTRODUCTION
Subterranean rodents comprise approximately 250 species that spend their lives in dark, unventilated environments, and are found on all continents except Australia and Antarctica (Begall et al., 2007). African mole rats (hystricognath rodent family Bathyergidae) are long-lived, strictly subterranean rodents that feed on underground roots and tubers. They are able to flourish in habitats poor in oxygen and rich in carbon dioxide and ammonia (Bennet and Faulkes, 2000), conditions that are harmful to mice and rats. It is hypothesized that the African Rift Valley acted as a geographical barrier shaping the adaptive radiation of African mole rats into Southern Africa, and from there to other regions (Faulkes et al., 2004). Until now, the only African mole rat genome has been that of the ~35g naked mole rat (NMR, Heterocephalus glaber), which is resident to North-East Africa, and is the most basal African mole rat lineage (Kim et al., 2011). The lack of genomic information for closely related species has thus far prevented detailed analysis of African mole rat traits. To better understand the molecular mechanisms underlying the traits of African mole rats, we developed an improved NMR genome assembly, determined the genome sequence of the related ~160g Damaraland mole rat (DMR, Fukomys damarensis) found in the arid regions of South-West Africa (Figures 1A and 1B), and sequenced the transcriptomes of additional subterranean rodents. Further analyses allowed us to decipher molecular adaptations consistent with subterranean life and shed light on unique traits of the most unusual mammal, the NMR.
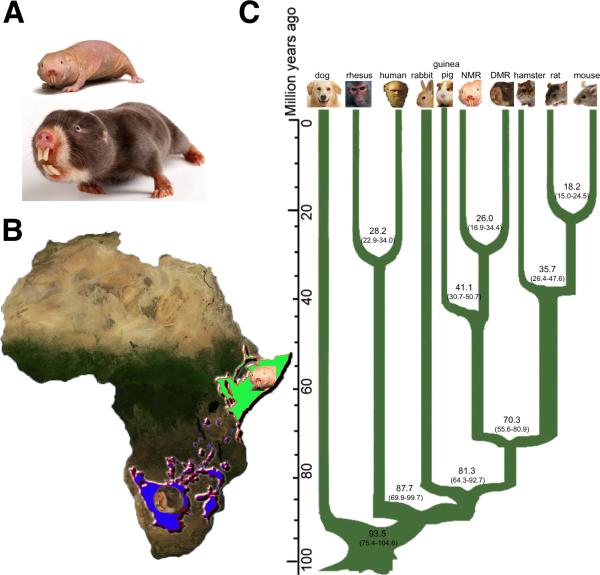
Figure 1. Relationship of NMR and DMR.
(A) The ~35 g NMR and the ~160 g DMR. (B) Species range map of African mole rats. DMR and NMR occurrence is shown in blue and green, respectively. (C) Phylogenetic tree constructed using fourfold degenerate sites from single-copy orthologs, with branch lengths scaled to estimated divergence time (with error range shown in parentheses). Distances are shown in millions of years (Myr). See also Figure S1.
RESULTS AND DISCUSSION
Genome assembly and gene content
The DMR genome yielded a 2.5 Gb sequence (~76-fold coverage) with a scaffold N50 size of 5 Mb (Table 1 and Figure S1A). The sequencing depth of 91% of the DMR assembly had more than 10-fold coverage (Figure S1B). We identified 1.3 million heterozygous single nucleotide polymorphisms (SNPs) and estimated a nucleotide diversity (heterozygosity) of 0.06%, which is comparable to the NMR, but lower than in rodents such as mouse and rat (Kim et al., 2011). The low level of nucleotide diversity in DMR and NMR may reflect their unique social system of a single breeding ‘queen’ per colony and the low effective size of their populations (Bennet and Faulkes, 2000). The number of repeat elements in the DMR genome was also lower (~28%) than in other mammals, but comparable to that of the NMR (Table 1) (Kim et al., 2011). We employed homology and de novo methods as well as RNA-seq data to predict 22,179 protein-coding genes in the DMR genome (Table 1 and Figure S1C), which is comparable to other mammals. Our analysis revealed that the common ancestor of the DMR and NMR lived approximately 26 million years ago (Figures 1C and S1D), similar to the distance between mice and rats, or between humans and macaques.
Table 1.
General features of the DMR (Fukomys damarensis) genome and the improved NMR (Heterocephalus glaber) genome assembly. See also Figures S1A and S1B.
| Sequencing | Fukomys damarensis | Heterocephalus glaber | ||||
|---|---|---|---|---|---|---|
| Insert size(bp) | Total data (Gb) | Sequence coverage (X) | Insert size(bp) | Total data (Gb) | Sequence coverage (X) | |
| Pair end library | 250~800 | 151.6 | 50.5 | 170~800 | 126.5 | 46.9 |
| 2~20×103 | 77.5 | 25.8 | 2~20×103 | 120.7 | 44.7 | |
| Total | 229.0 | 76.4 | Total | 247.2 | 91.6 | |
| Assembly | N50 (kb) | Longest (kb) | Size (Gb) | N50 (kb) | Longest (kb) | Size (Gb) |
|---|---|---|---|---|---|---|
| Contig | 22.9 | 229.6 | 2.46 | 19.3 | 178.9 | 2.45 |
| Scaffold | 4,996 | 22,231 | 2.51 | 21,307 | 80,826 | 2.75 |
| Annotation | Number | Total length (Mb) | Percentage of genome | Number | Total length (Mb) | Percentage of genome |
|---|---|---|---|---|---|---|
| Repeats | 4,585,125 | 717.6 | 28.6 | 3,090,116 | 666.7 | 24.2 |
| Genes | 22,179 | 745.6 | 29.7 | 22,561 | 722.3 | 26.3 |
| CDS | 187,627 | 33.0 | 1.31 | 181,641 | 32.5 | 1.18 |
We further prepared a new version of the NMR genome based on the original genome sequence (Kim et al., 2011), additional sequencing and the data generated by the Broad Institute (Table 1). The new NMR assembly had a genome size of 2.7 Gbp (92-fold coverage), with a scaffold N50 of 21 Mb, compared to 1.6 Mb in the assembly previously published (Kim et al., 2011). The resulting DMR and NMR genomes, gene models and transcriptome data for these and related rodents were used to reveal common and unique features of these animals. We primarily focused on genes likely to be involved in the ecophysiology and exceptional longevity of underground-dwelling African mole rats.
Sensory cues
Analyses of gene family contractions and expansions provide insights into evolutionary forces that have shaped genomes. Based on 19,839 gene families inferred to be present in the most recent common ancestor of mammals, we found that 212 gene families were gained, and 59 lost, from the DMR genome (Figure S1E, and Table S1). Over the same period, the NMR gained 378 gene families and lost 29. Gained gene families included olfaction (sense of smell) genes that likely play an important role in locating food and social interaction in complete darkness (Heth and Todrank, 2007). The NMR and DMR live exclusively in the dark and display small eyes and poor visual acuity (Bennet and Faulkes, 2000). However, their eyes could still serve to alert the colony to invasion by predators by detecting light entering tunnels (Nemec et al., 2008; Kott et al., 2010). The visual perception category was enriched in both DMR (Table S2) (GO: 0007601, P < 0.001, Fisher's exact test) and NMR pseudogene lists. We found that one visual perception gene (AOC2) was lost) and 13 were pseudogenized in the DMR (Table S2). Three visual genes were inactivated or missing in the DMR and the new NMR genome assembly: CRB1, GRK7, and GJA10 (Table S2). Positive selection of the rhodopsin gene (RHO), which enables dim light vision, was found in the lineage leading to the common ancestor of DMR and NMR (Table S3). This is consistent with accelerated evolution of African mole rat RHO, while preserving sites critical for spectral tuning (Zhao et al., 2009). Interestingly, we observed cataracts in all examined NMRs ranging from 4 to 20 years of age (Figure S2A). This phenotype may be a consequence of captive life under atmospheric oxygen levels, but could highlight inadequate antioxidant defence. Low glutathione peroxidase 1 (GPx1) levels may contribute to a decreased protection of the lens against oxidative stress (Kasaikina et al., 2011). A premature stop codon occurs in the gene encoding GPx1 (GPX1) in both NMR and DMR (Figure S2B), and knockout of this gene in mice results in cataract formation (Reddy et al., 2001; Wang et al., 2009; Wolf et al., 2005).
Adaptations to hypoxia and a high carbon dioxide and ammonia environment
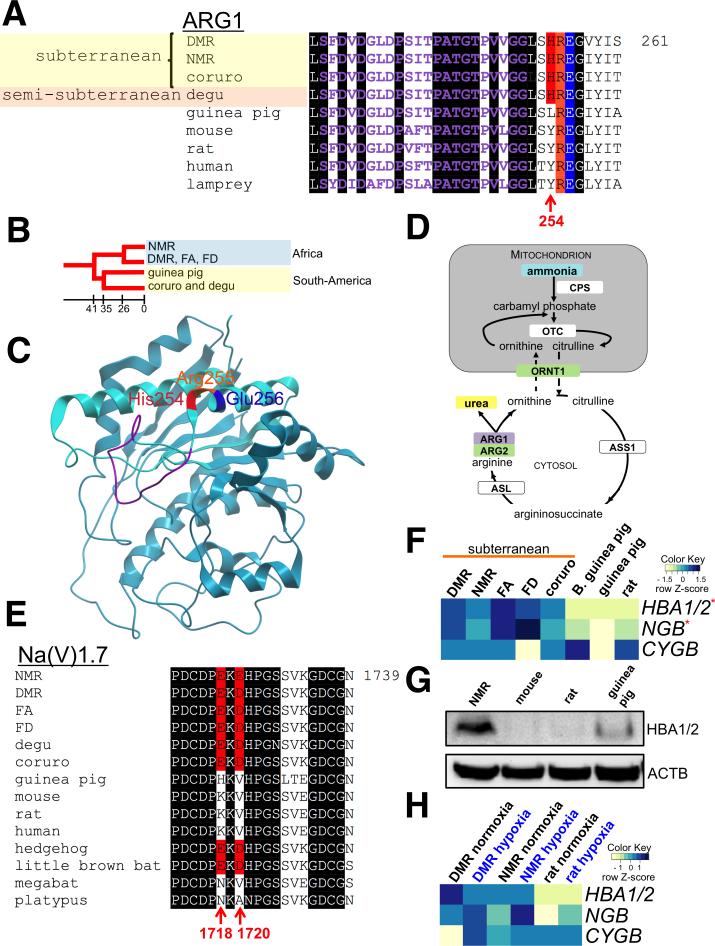
DMR, NMR and other subterranean rodents rest with conspecifics in deep, underground nests. These nests constitute environments low in oxygen and high in carbon dioxide and ammonia, conditions that would evoke cellular damage and behavioral stress responses in other mammals (Bennet and Faulkes, 2000). Ammonia is a potent irritant, and arises from nitrogen and methane accumulation in latrines and nest (Burda et al., 2007; Lavinka et al., 2009). We found that arginase 1 (ARG1), which catalyzes the final step of the hepatic urea cycle and removes ammonia from the body, has a radical residue change in both NMR and DMR: His254 replaces Leu/Tyr present in 38 other vertebrate species (Figure 2A). This amino acid change was also detected in the distantly related subterranean coruro (Spalacopus cyanus) and the semi-subterranean degu (Octodon degus) of South America. The common ancestor of Octodontoidea (coruro and degu) and Cavioidea (guinea pig) diverged approximately 35 Mya, while African and South American rodents diverged ~41 Mya (Antoine et al., 2011; Meredith et al., 2011) (Figure 2B). His254 is located immediately downstream of a conserved motif required for binding manganese and ARG1 function (Dowling et al., 2008) (Figure 2C). Moreover, ARG1 is a homotrimer, with the salt bridges formed by Arg255 and Glu256 critical for its assembly (Lavulo et al., 2001; Sabio et al., 2001). The charged residue flanking the ARG1 core may improve ammonia removal efficiency by interacting with the acidic Glu256 or by strengthening the Arg255-Glu256 salt bridge. In addition, several genes in the urea cycle were expressed at higher levels in NMR and DMR livers compared to mouse and rat (Table S4) (Figure 2D). This included arginase 2 (ARG2), the second arginase gene that is normally not expressed in rodent liver. Moreover, the expression of mitochondrial ornithine transporter ORNT1 (SLC25A15), essential for the urea cycle (Fiermonte et al., 2003), was elevated in NMR and DMR. Taken together, these data indicate that subterranean hystricognath rodents present enhanced ammonia detoxification.
Figure 2. Subterranean adaptations in hystricognath rodents.
(A) Subterranean rodents share a charged residue at position 254 of arginase 1 (ARG1). The manganese-binding site, residues critical for enzyme trimer assembly (Arg255 and Glu256), and unique His254 changes are highlighted in purple, orange, blue and red, respectively. Identical residues in vertebrates are shaded in black. (B) Phylogenetic relationship of hystricognath rodent lineages examined in this study. Approximate divergence times (Myr) are indicated. (C) Structural model of human ARG1 monomer. Residues are highlighted as in (A). (D) Schematic representation of the roles of components of the urea cycle with altered sequence (purple box) or expression in NMR and DRM (green boxes). CPS1, carbamoyl phosphate synthase 1; Ornithine transcarbamylase, OTC; ORNT1, ornithine transporter 1; ASS1, argininosuccinate synthase; ASL, argininosuccinate lyase; ARG1, arginase 1; ARG2, arginase 2. (E) Species of hypercapnic habitats share a negatively charged three-residue motif in the Na(V)1.7 sodium channel protein. Acidic amino acid residues in the motif, corresponding to amino acids 1718 and 1720 of the human sequence, are shown in red. Identical residues in vertebrates are shaded in black. (F). Heatmap of globin expression in normoxic rodent brains. Scaled log2 transformed normalized read counts, denoted as the row Z-score, is plotted in beige–blue color with blue indicating high expression and beige indicating low expression, respectively. B. guinea pig, Brazilian guinea pig; HBA1/2, hemoglobin α; NGB, neuroglobin; CYGB, cytoglobin. Red stars indicate differentially expressed genes in subterranean rodents. (G) Western blot of hemoglobin α in normoxic rodent brains with antibodies against the mouse protein. (H) Comparison of globin gene expression under normoxia (21% O2) and hypoxia (8% O2 over 8 hours). Annotated as in (F). See also Figure S2.
The buildup of CO2 in underground habitats evokes pain, as CO2 is converted into acid that stimulates pain receptors in the upper respiratory tract, nose and eyes (Brand et al., 2010). A recent study found that a negatively charged motif in the sodium channel Na(V)1.7 protein (SCN9A), which is highly expressed in nociceptor neurons, prevents acid-induced pain signaling to the NMR brain (Smith et al., 2011). We compared 44 vertebrate sequences and found that the motif is also present in the DMR, two African mole rats in the same genus as DMR (Ansell's mole rat, Fukomys anselli (FA) and Mashona mole rat, Fukomys darlingi (FD)), the South American subterranean coruro and semi-subterranean degu, the cave-roosting little brown bat (Myotis lucifugus), and the European hedgehog (Erinaceus europaeus). These animals are exposed to chronic hypoxia and hypercapnia in burrows or caves (Figure 2E), and suggests that convergent evolution has resulted in similar amino acid changes in Na(V)1.7 and adaptation to high CO2 levels.
Changes in both gene expression and gene sequences contribute to adaptive mechanisms in subterranean rodents (Avivi et al., 2010). We compared the normoxic brain transcriptomes of subterranean rodents to those of rodents living primarily ‘aboveground’ (surface-dwelling). In addition to NMR and DMR, the transcriptomes of three subterranean hystricognath rodents were generated: FA, FD and the coruro of South America. These were further compared to rat and two guinea pig subspecies (Table S5).
Several genes associated with DNA damage repair and responses to stress showed higher expression in subterranean rodents even during normoxia (Table S5 and Figure S2C). Hypoxia induces DNA damage, and in agreement with a recent report on the blind mole rat (Shams et al., 2013), our data suggest that improved DNA repair is an intrinsic mechanism of adaptation to an underground environment. The most obvious adaptation to a hypoxic subterranean environment is improved oxygen uptake to highly oxygen-demanding tissues, such as the brain. The globin family comprises proteins responsible for the delivery and storage of oxygen in cells and tissues. We found that hemoglobin α (HBA1 and HBA2; identical coding sequences) and neuroglobin (NGB) displayed elevated expression in the brains of subterranean rodents during normoxia (Figure 2F) (Table S5), and Western blot analysis verified higher hemoglobin α protein expression in the normoxic NMR brain compared to several surface-dwelling rodents (Figure 2G). We next compared the gene expression of NMR and DMR to the hypoxia-sensitive rat after 8 hours at oxygen levels (8% O2) comparable to NMR burrows (Bennet and Faulkes, 2000). As under normoxia, hemoglobin α and neuroglobin expression in the hypoxic brain was higher in the NMR and DMR than rat (Figure 2H). We observed a 3.4-fold decrease of hemoglobin α mRNA in DMR under hypoxia, whereas the expression in the NMR did not change significantly. Higher NGB expression was observed in the hypoxic rat and NMR brain, but not in the DMR, while there was a trend towards higher cytoglobin (CYGB) expression in the DMR. Species-specific expression of globins in response to hypoxia has previously been reported in subterranean blind mole rat species (family Spalacidae) distantly related to African mole rats (family Bathyergidae) (Avivi et al., 2010). NMR hemoglobin has a higher affinity for oxygen and is able to unload oxygen more efficiently than that of the mouse (Johansen et al., 1976), and hemoglobin α plays a neuroprotective role in the brain of rodents during hypoxia (Schelshorn et al., 2009). Neuroglobin and cytoglobin mRNA expression is elevated in two blind mole rat species compared to the rat under normoxia (Avivi et al., 2010). Importantly, neuroglobin is expressed in blind mole rat glial cells and neurons, while its expression is limited to neurons in surface-dwelling rodents, such as rat, indicating an organ-wide protective function. Glial globin expression has also been associated with improved oxygen delivery in hypoxia-tolerant seals (Schneuer et al., 2012) and shellfish (Kraus and Colacino, 1986), suggestive of a common adaptation with subterranean rodents. Overall, our data suggests that globins are constitutively and highly expressed in the brain of hypoxia-resistant rodents and play a major role in rodent adaptation to an oxygen-poor subterranean environment. Experiments are in progress to further dissect the expression and function of globins in African mole rats.
Potential longevity-associated adaptations in the NMR and DMR
Subterranean rodents have the highest maximum lifespans for their body weight, with species in both the Bathyergidae (e.g. NMR and DMR) and Spalacidae (e.g. the blind mole rat) families living for over 20 years (Dammann and Burda, 2007). The NMR is the longest-lived rodent known, with a lifespan exceeding 30 years, while the longest-lived DMRs in our laboratories survived for 20 years. These rodents have a similar longevity quotient to humans, and may show a comparable age-related disease pattern (Edrey et al., 2011).
We compared the transcriptomes of the liver, a relatively homogenous organ, of NMR and DMR (Hystricognathi) to those of short-lived rat and mouse (Muridae) (Table S4). Compared to mouse and rat, which spend considerable time above ground, NMR and DMR show differential expression and enrichment of several genes associated with oxidoreduction. Two out of six peroxiredoxins (PRDX2 and PRDX5) were expressed at lower levels (Table S4) in NMR and DMR livers, which, together with reduced GPx1 activity, may result in increased levels of reactive oxygen species (ROS). These observations are consistent with reports of oxidative stress in the NMR (Andziak and Buffenstein, 2006), and suggest that the long-lived NMR and DMR can thrive despite elevated oxidative stress.
Loss of FASTK, a sensor of mitochondrial stress, in African mole rats
We found that the Fas-activated serine/threonine kinase gene (FASTK) is inactivated in both NMR and DMR (Figure S2D). FASTK encodes a kinase that serves as a regulator of Fas-mediated apoptosis and is located at the inner mitochondrial membrane. FASTK is associated with cell survival and is overexpressed in tumors and immune-mediated inflammatory diseases, such as asthma and AIDS, where it can delay the onset of apoptosis and contribute to pathogenesis. Knockdown of this gene results in reduced lung inflammation in mice (Simarro et al., 2010) and reduced oncogenic potential of cultured human cancer cells (Zhi et al., 2013). Chronic inflammation, cancer, and cellular senescence are intertwined in the pathogenesis of premature aging (Campisi et al., 2011). Furthermore, knockdown of FASTK is also associated with improved neuron elongation and regeneration (Loh et al., 2008). The ability of neurons to regenerate and their rate of elongation decrease with age. Loss of FASTK may help maintain neuronal integrity in long-lived mole rats, keeping their brains ‘younger’. Taken together, the loss of FASTK in NMR and DMR suggests a role for FASTK in the aging phenotype of somatic cells, as well as in cancer resistance.
Divergent insulin in African mole rats
It has been reported that NMR insulin cannot be detected using rodent antibody-based assays, similar to the findings in the guinea pig several decades ago (Chan et al., 1984; Kramer and Buffenstein, 2004). We found that NMR, DMR and other hystricognath rodents harbor a divergent insulin β-chain sequence (Figure 3A). This finding is consistent with an observation that the South American hystricognath insulin is rapidly evolving (Opazo et al., 2005). In the guinea pig and other South American hystricognaths, the regions encoding the β-chain and, in particular, the β-chain, are highly divergent, with concomitant alterations in insulin structure and reduced activity compared to most other mammals, and possibly an alternative receptor (King et al., 1983; Opazo et al., 2004). Mutations in the human β-chain result in reduced insulin processing, misfolding, and less effective insulin (based on receptor binding) (Liu et al., 2010). Interestingly, residue 22 of the β-chain, whose mutation (Arg22Gln) is associated with misfolding of insulin and diabetes (Liu et al., 2010), is uniquely changed in both African and South American hystricognaths (Figure 3A). In the African crested porcupine, this residue has previously been linked to an altered insulin structure with reduced affinity for insulin receptors (Horuk et al., 1980). We hypothesize that NMR and DMR insulin exists as a monomer with low insulin receptor activity that targets alternative receptor(s) outside classic insulin-responsive tissues such as liver, muscle and adipose tissue.
Figure 3. Unique changes in the insulin sequence of hystricognath rodents.
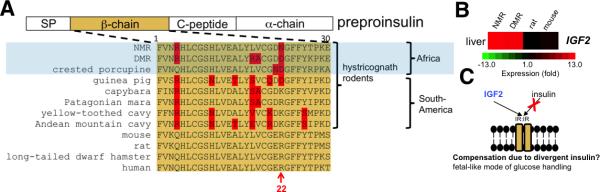
(A) Preproinsulin contains an amino-terminal signal peptide (SP) and is processed to yield two insulin peptide chains (α and β), as well as connecting peptide (C-peptide). The sequence of the β-chain is shown in orange. Residues that are uniquely altered in hystricognath rodents are highlighted in red. (B) Heatmap showing the expression (log2 fold-change) of IGF2 mRNA in the liver. (C) Schematic model, which applies to African mole rats and other hystricognaths, wherein IGF2 (normally only expressed in the fetus) signals via the insulin receptor and partly compensates for altered insulin sequence. See also Figure S2.
Surprisingly, NMR (Edrey et al., 2011) and South American hystricognaths (Opazo et al., 2004) are able to handle glucose in the absence of conventional insulin, suggesting that these animals have evolved compensatory mechanisms. In mammals, insulin is not secreted from the pancreas until after birth, and mice lacking insulin die a few days after birth due to acute diabetes mellitus (Duvillie et al., 1997). Until recently, it was unknown how glucose handling in the liver was achieved before birth. It has now been established that IGF2, which has high homology to insulin, is abundantly expressed in the fetal liver, and signals exclusively via the insulin receptor (IR) to maintain glycemia (Liang et al., 2010a). In most mammals, including mouse and rat, IGF2 expression is down-regulated after birth in liver; however, primates and guinea pig harbor residual IGF2 expression (Lui and Baron, 2013). We found that NMR and DMR also express IGF2 and its binding protein IGF2BP2 in liver (Figure 3B and Table S4). We hypothesize that autocrine/paracrine production of IGF2 in the liver substitutes for insulin, and may partly mediate a fetal-like mode of glucose handling in hystricognath rodents (Figure 3C).
Reduced levels of insulin are observed during calorie restriction and inhibition of the growth hormone/IGF1 axis, two manipulations that extend lifespan in various species (Blagosklonny, 2012). Interestingly, molecular innovations of this axis may contribute to the lifespan of the long-lived Brandt's bat Seim et al., 2013). In addition to induction of IGF2 expression in the NMR and DMR liver, we observed differential expression of genes associated with insulin signaling: decreased IGF1 and insulin induced gene 2 (INSIG2) and increased IGF1R and resistin (RETN) (Table S4). Taken together, a less bioactive insulin and altered downstream signaling may partly explain the enhanced longevity of African mole rats, and possibly other hystricognaths (e.g., porcupine, guinea pig). Our findings support the hypothesis that hystricognath rodents have evolved a distinct insulin peptide.
Cancer resistance
Studies of the NMR (Liang et al., 2010b; Manov et al., 2013; Seluanov et al., 2008; Seluanov et al., 2009), and the distantly (~70 million years) related blind mole rat (Gorbunova et al., 2012; Nasser et al., 2009; Manov et al., 2013), suggest that many species of long-lived mole rats are resistant to cancer and, even if they do develop pathology, present a milder phenotype in comparison to short-lived rodents (e.g. mouse) (Azpurua and Seluanov, 2012; de Magalhaes, 2013).
A recent study suggests that one potential explanation for mole rats’ cancer resistance lies in the enzyme hyaluronan synthase 2 (HAS2) (Tian et al., 2013). Two amino acid residues in the HAS2 active site were reported to be unique to the NMR and hypothesized to result in the synthesis of high-molecular-mass hyaluronan (HMM-HA), an extracellular matrix polysaccharide. HMM-HA serves as an extracellular signal that results in induction of the tumor suppressor p16INK4A, early contact inhibition (ECI), and cancer resistance (Tian et al., 2013). We found that one of the unique amino acid changes in the NMR HAS2 sequence (Asn301Ser) is shared by the DMR, while Asn178Ser is unique to the NMR (Figure S2E). In contrast to residue 178, Asn301Ser is present in a highly conserved region. Interestingly, the blind mole also secretes HMM-HA (Tian et al., 2013). Taken together, these data suggest that the DMR and the blind mole rat produce high molecular-mass hyaluronan that confers cancer resistance. Surprisingly, a recent study found that HMM-HA does not influence the anti-cancer properties of blind mole rat fibroblasts (Manov et al., 2013), which supports the current evidence showing independent paths to cancer resistance in the blind mole rat (Azpurua and Seluanov, 2012). Future functional studies in mole rats are required, however, including sequencing blind mole rat HAS2.
Unique features of the naked mole rat
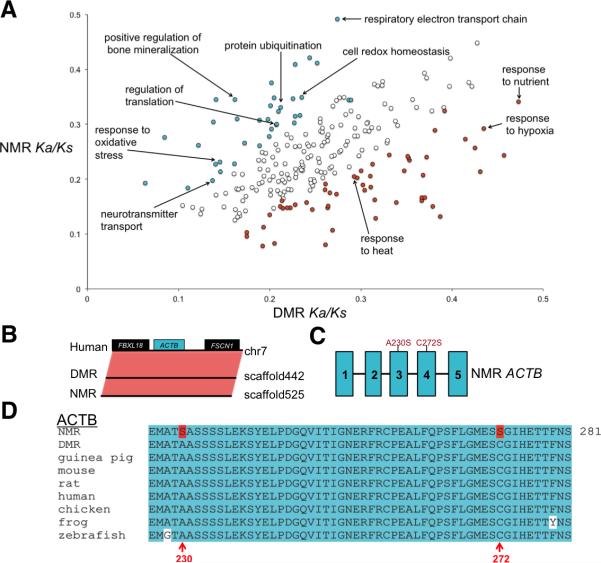
Although NMR and DMR share a relatively recent common ancestor ~26 million years ago, the NMR has several exceptional features and is considered the most unusual mammal. Accelerated gene evolution among lineages could indicate an association between genetic changes and the evolution of traits (Qiu et al., 2012). Analysis of nonsynonymous-to-synonymous substitution (Ka/Ks) ratios of 9,367 1:1 orthologs of 10 mammalian species revealed that the NMR was significantly enriched for Gene Ontology (GO) categories including the respiratory electron transport chain, cell redox homeostasis, and response to oxidative stress (Figure 4A and Table S6). To test if genes in the rapidly evolving GO categories were under positive selection, we used a branch likelihood ratio test to identify positively selected genes in the NMR and DMR lineages (Table S3).
Figure 4. Adaptive evolution in the NMR and DMR genomes.
(A) Accelerated evolution of NMR and DMR genomes. GO categories with putatively accelerated (P ≤ 0.05, binomial test) non-synonymous divergence in the NMR lineage (turquoise) and in the DMR lineage (orange) are highlighted. (B) Conserved gene synteny of the β-actin gene (ACTB) region between human, DMR and NMR. Boxes represent genes. (C) Schematic of the intron and exon structure of the ACTB, along with the location of amino acid changes found in the NMR ACTB protein. Turquoise boxes represent exons. (D) NMR has unique amino acid changes (highlighted in red) in the highly conserved (turquoise) ACTB, including the redox-sensitive Cys272 residue. See also Figure S3.
Body temperature regulation
Like other mammals, the DMR tightly controls body temperature (stable at 35°C). The NMR, in contrast, lacks an insulating layer of fur, and cannot maintain thermal homeostasis if housed on its own away from the warm confines of its humid burrows (Daly and Buffenstein, 1998). The NMR is effectively poikilothermic, taking on the temperature of its surroundings. To accomplish this, it employs non-shivering thermogenesis (NST), using large pads of brown adipose tissue interspersed between muscle (Hislop and Buffenstein, 1994). Thermogenin (uncoupling protein 1, UCP1) is the major protein used in this kind of heat generation. The NMR UCP1 harbors amino acid changes at the site regulated by fatty acids and nucleotides (Kim et al., 2011), whereas we find that the DMR sequence is typical of other mammals (Figure S3A). Thus, the altered UCP1 is an adaptation of the NMR rather than of mole rats in general, and it is strongly linked to the lack of thermogenesis.
Melatonin is a regulator of circadian rhythm and body temperature (Cagnacci et al., 1992). We found that the NMR is the only known ‘natural’ melatonin receptor 1a and 1b knockout animal (Figures S3B and S3C). In rodents, there are two high-affinity receptors for melatonin, MTNR1a and 1b. Both DMR and NMR lost MTNR1b, although the inactivating mutations are located in different positions (Figure S3B). MTNR1b is also a pseudogene in the distantly related Siberian hamster (Phodopus sungorus) (Prendergast, 2010) and the Syrian hamster (Mesocricetus auratus) (GenBank accession no. AY145849). However, MTNR1a alone is sufficient to maintain photoperiod and melatonin responses in the Siberian hamster (Prendergast, 2010). Interestingly, MTNR1a is intact in the DMR, but inactivated in the NMR (Figure S3C). The lack of cognate melatonin receptors could contribute to the inability of NMR to adequately respond to fluctuating temperature.
Pain insensitivity
C-fibers are small, unmyelinated axons associated with slow pain signaling in response to a range of external stimuli, which can be thermal, mechanical or chemical. The NMR has fewer C-fibers than other rodents, including the DMR (St John Smith et al., 2012), and the C-fibers of the NMR's skin, eyes and nose do not produce the pain-relaying neuropeptides substance P (SP) and calcitonin gene-related peptide (CGRP) (Park et al., 2003). It should be noted that there is not a complete loss of expression, as low levels of these neuropeptides can be found in internal organs (Park et al., 2003). It is currently not known how the expression of SP and CGRP is repressed in the sensory neurons of the NMR, but it was shown that NMRs receiving gene therapy with the SP-encoding preprotachykinin gene (TAC1) respond to pain induced by peripheral inflammation (Park et al., 2008). These observations suggest that the SP-encoding gene itself is altered in the NMR. The NMR TAC1 gene harbors an 8 base pair deletion in its proximal promoter (Kim et al., 2011). The presence of the 8 base pair region in the DMR (Figure S3D) suggests that this region is associated with pain insensitivity.
The calcitonin gene (CALCA) is responsible for the synthesis of two distinct preprohormones by means of alternative splicing (Rosenfeld et al., 1981). Splicing into exon 4 results in calcitonin (CT), while splicing into the exon 5 and the 3’ untranslated exon 6 encodes the sensory neuropeptide calcitonin-gene related peptide (CGRP) (Figure S3E). The splicing of CALCA is under tight endocrine control, and the regulation of CT and CGRP is complex and involves distal and proximal elements in the CALCA promoter, as well as a range of regulatory elements within and flanking exon 4 (van Oers et al., 1994). Our analysis revealed that there are unique deletions in NMR CALCA, including a conserved 6 base pair region, in the 3’ untranslated part of exon 4 (Figure S3F). Given that splicing factors are known to be composite- and context-dependent (Wang and Burge, 2008), and that elements within exon 4 of CALCA regulate CT and CGRP isoform switching, the deleted region in exon 4 may disrupt the mutually exclusive tissue specific splicing of CT exon 4 and CGRP exons 5-6, resulting in the observed lack of calcitonin-gene related peptide expression in sensory neurons of the NMR.
In addition to unique changes in the NMR genes encoding SP and CGRP, genes associated with neurotransmission of pain were under positive selection in the NMR. This included the NMDA receptor NR2B (GRIN2B), the TRP channel TRPC5, and proenkephalin (PENK) (Table S3).
β-actin may mediate enhanced oxidative stress resistance in the NMR
Actins are highly conserved proteins involved in cell structure, motility and integrity. The vertebrate actin family contains six genes, of which only the cytoplasmic actins, β-actin (ACTB) and γ-actin (ACTG1), are ubiquitously expressed (Herman, 1993). The β-actin protein is highly conserved throughout evolution (Vandekerckhove and Weber, 1978). During oxidative stress, cysteine residues of actins can be oxidized, which is associated with depolymerization and altered regulatory protein interactions (Terman and Kashina, 2013). Increased ROS levels and actin overoxidation is symptomatic of senescence and diseases such as Alzheimer's (Aksenov et al., 2001). We observed that β-actin (ACTB) is under positive selection in the NMR (Table S3). RNA-sequencing and synteny analysis (Figure 4B) confirmed the identity of NMR ACTB. We found that both Cys272 and Ala230 of β-actin are converted to serine in the NMR (Figures 4C and 4D). Cys272 is highly redox-sensitive and may serve as a ‘redox sensor’ Lassing et al., 007). These observations suggest that NMR β-actin is more resistant to oxidation and may contribute to the longevity of the NMR, which can live at least 10 years longer than the DMR, despite its exposure to high ROS levels and lower body mass (Lewis et al., 2012). The potential involvement of ACTB Cys272 in senescence and disease can now be evaluated more extensively in the NMR, an animal model that lacks this residue.
28S ribosomal RNA processing in evolutionary and geographically distant hystricognath rodents
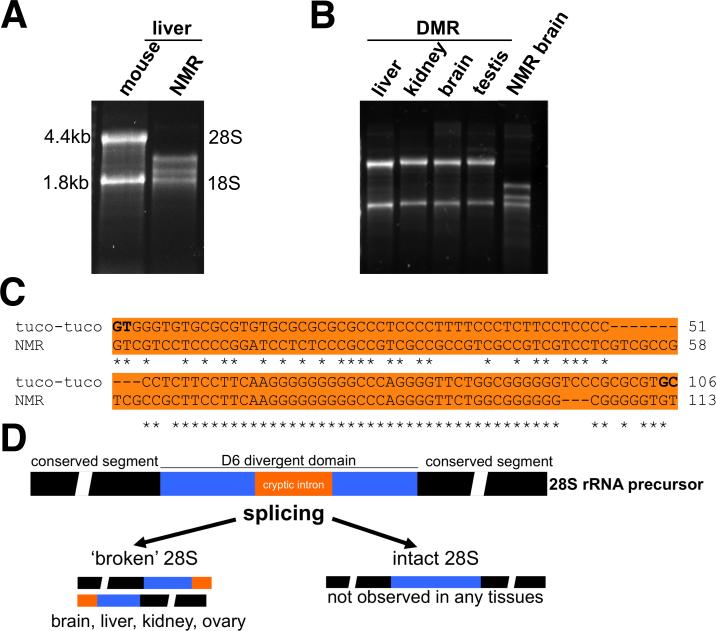
We discovered that NMR rRNA did not display the typical banding pattern, i.e., 28S at ~4.4 kb and 18S at ~1.8 kb, during denaturing gel electrophoresis (Figures 5A and 5B). In contrast, the DMR had the standard pattern (Figure 5B). The unusual NMR pattern occurred in every tissue tested (ovary, kidney, liver, brain), from separate animals and at any age tested (from 1 to 23 years old). A similar phenomenon, where 28S rRNA is split into two subunits held together as a single 28S rRNA molecule by hydrogen bonding under native conditions, has been described in insects and plants (Winnebeck et al., 2010). The only vertebrates reported to produce shorter S rRNA are South America's tuco-tucos (Ctenomys) and the degu (Octodontomys gliroides) (Melen et al., 1999). We found that the ‘break’ region is in the D6 domain of 28S. This NMR region corresponds to a cryptic GC- and simple repeat-rich intron in the Talas tuco-tuco (Ctenomys talarum) (Melen et al., 1999), and there is also a high degree of sequence conservation between these species (Figure 5C). In tuco-tuco, an unknown site within this cryptic intron results in ‘breakage’ of S rRNA molecules Melen et al., 1999). The cryptic intron in the NMR may explain the banding pattern observed (Figure 5D). Two other hystricognaths, the South American guinea pig and the African DMR do not harbor the cryptic intron (Figure S4). The data suggest that the cryptic intron and the resulting ‘broken’ S rRNA was present in a common ancestor prior to the cross-Atlantic migration of small African hystricognath rodents to South America ~41 million years ago (Antoine et al., 2012). This proposition is consistent with the predicted ancestor of NMR, DMR and guinea pig (Figure 1C). Following submission of our manuscript, a study by Azpurua and colleagues also found the unusual 28S processing in NMR tissues and proposed that the unique NMR 28S may result in improved protein synthesis fidelity and a more stable proteome (Azpurua et al., 2013). The NMR thus holds promise as a model organism to investigate the mechanism and function of ‘hidden breaks’ in ribosomal RNAs in a laboratory setting.
Figure 5. Novel 28S rRNA in the NMR.
Denaturing agarose gel electrophoresis of total RNA from (A) mouse liver and NMR liver; and (B) NMR brain and DMR brain, liver, kidney, testis. (C) A cryptic intron in the divergent region D6 of 28S rRNA is conserved between the NMR and tuco-tuco. Tuco-tuco splice donor and acceptor sites are indicated in bold. Dashes (–) indicate gaps and stars (*) identical nucleotides. (D) Proposed biogenesis of S rRNA subunits by ‘splicing’ of the cryptic intron in the D6 domain. Two 28S subunits are produced in NMR organs. No conventional 28S rRNA exists in the NMR. See also Figure S4.
Conclusions
We performed genome sequencing and de novo assembly of two related subterranean rodents, Damaraland mole rat and naked mole rat. Transcriptomes of five subterranean rodents were also sequenced. African mole rats share a unique ecology and physiology. These animals are also remarkably long-lived for their size, and are characterized by similar traits of cancer resistance, maintenance of neuronal integrity, altered insulin structure, and elevated brain globin. However, in many other traits, the use of the DMR genome pinpointed features responsible for the truly unusual characteristics of the NMR, including unusual thermogenesis, aberrant melatonin system, pain insensitivity, and processing of ribosomal RNA. The genomes and transcriptomes can be further mined to provide insights into the fascinating biology of these animals.
EXPERIMENTAL PROCEDURES
See Supplemental Experimental Procedures for additional protocols.
Animals
A breeding colony of Damaraland mole rats (DMRs; Fukomys damarensis) was housed at the University of Illinois-Chicago. DMR was known as Cryptomys damarensis prior to a recent sub-classification into a new genus, Fukomys (Kock et al., 2006). Animals were sacrificed and DNA and RNA isolated for subsequent sequencing. The genome sequenced was that of a 5-year-old male DMR. Liver and brain transcriptomes were obtained by sequencing individuals from the same colony. Animal experiments were approved by the University of Illinois at Chicago Institutional Animal Care and Use Committee. Species range maps were obtained and adapted from the World Wildlife Fund's WildFinder database (http://www.worldwildlife.org/WildFinder).
DMR genome sequencing and assembly
We employed a whole genome shotgun strategy and next-generation sequencing technologies, using Illumina HiSeq 2000 as platform, to sequence the genome of a captive male DMR. We constructed 16 paired-end libraries, with insert sizes of 250 base pairs (bp), 500 bp, 800 bp, 2 kbp, 5 kbp, 10 kbp and 20 kbp. In total, 229 Gbp (or 76×) high quality data, including 151 Gbp (or 50×) short insert size reads, were generated (Table 1). The genome was de novo assembled by SOAPdenovo (Li et al., 2010a). 151 Gpb (or 50×) data from short insert size libraries (250-800 bp) were split into 63-mers and contigs with unambiguous connections in de Bruijn graphs retained. All reads were aligned onto contigs for scaffold building using paired-end information. k-mer analysis (Li et al., 2010b) was used to estimate the genome size of DMR. In this study, K was 17, K_num was 61,533,145,821 and the K_depth was 22.5. The DMR genome size was, therefore, estimated to be 2.73 Gbp in size (Figure S1A).
Assembly of the NMR genome
To develop an improved assembly of the naked mole rat genome, we used lastz (Harris, 2007), with the parameter “M=60 Y= 400 T=2 --format=axt”, to align previously generated genome sequences (Kim et al., 2011), and additional sequence data to a recently-released genome assembly from the Broad Institute (GenBank accession no. AHKG00000000). ChainNet (Kent et al., 2003) was used to combine traditional alignments into larger structures. After obtaining the primary alignment, paired end (PE) reads with insert sizes from 2 kb to 20 kb were mapped to the ‘newly formed’ genome. A new NMR assembly with a scaffold N50 of 21.3 Mb was generated (Table 1).
Whole-genome heterozygosity analysis
We aligned all high-quality short insert size reads to the genome assembly using BWA (Li and Durbin, 2009) with parameters -I. Since the alignment results were stored in BAM/SAM format, SAMtools, which is based on the Bayesian model, was selected for variation analysis (Li et al., 2009). After sorting alignments by leftmost coordinates and removing potential PCR duplicates, we used SAMtools mpileup to call SNPs and short InDels. We rejected SNPs and InDels within reads with depth either much lower or much higher than expected, since large copy number variation might lead to miscalling of SNPs. The sequencing depth ranged from 4 to 100, and the upper limit was about triple the sequencing depth. SNP-miscalling caused by alignment around short InDels and low-quality sequences were removed. We applied samtools.pl varFilter, which can be found in the SAMtools package, as the filter tool with parameters -Q 20 -q 20 -d 4 -D 100 -S 20 -i 20 -N 5 -l 5 -W 5 -N 1.
Repeat annotation
RepeatProteinMask and RepeatMasker (Tarailo-Graovac and Chen, 2009) were used to identify and classify transposable elements (TEs) by aligning the DMR genome sequences against a library of known repeats, Repbase, with default parameters. The repeats obtained were combined together to form a list of non-redundant repeats of DMR. The same approach was used to identify repeats in related mammals, including the NMR.
Supplementary Material
ACKNOWLEDGEMENTS
We acknowledge financial support from NIH (AG047745, AG047200, AG038004, AG021518 and GM061603), the WCU Program (R31-2008-000-10010-0), and National Natural Science Foundation of China (31171190). NMR and DMR photos by Joel Sartore/joelsartore.com.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
V.N.G. coordinated the study. T.J.P. collected and prepared DMR, NMR and rat samples. R.S. collected and prepared FA, FD and coruro samples. A.A.T., S.H.Y. and R.T.B. prepared and analyzed biological samples. X.F., Z.H., Z.X., Y.Z., X.Y., D.F. and L.Y. performed genome sequencing, assembly and annotation. X.F., X.Z., G.Z., J.W. supervised genome sequencing, assembly and annotation. X.F, I.S., Z.H., M.V.G., Z.X., Y.Z., A.V.L., D.F., X.Y., S.M., L.Y., S.G.L., E.B.K., R.B., X.Z., A.K., T.J.P. and V.N.G. performed genome and transcriptome analyses. All authors contributed to data interpretation. I.S. and V.N.G. wrote the paper with significant contributions from X.F., Z.X, Z.H. and input from all authors.
ACCESSION NUMBERS
The DMR whole genome shotgun project has been deposited at DDBJ/EMBL/GenBank under the accession code ANXX00000000. The version described in this paper is the first version, ANXX01000000. All short read data have been deposited into the Short Read Archive under the accession code SRA000000. Raw sequencing data of the transcriptome have been deposited in Gene Expression Omnibus with the accession code GSX00000.
SUPPLEMENTAL INFORMATION
Supplemental Information includes Extended Experimental Procedures, 4 Figures, and 6 Tables and can be found with this article online at doi: xyz.
REFERENCES
- Aksenov MY, Aksenova MV, Butterfield DA, Geddes JW, Markesbery WR. Protein oxidation in the brain in Alzheimer's disease. Neuroscience. 2001;103:373–383. doi: 10.1016/s0306-4522(00)00580-7. [DOI] [PubMed] [Google Scholar]
- Andziak B, Buffenstein R. Disparate patterns of age-related changes in lipid peroxidation in long-lived naked mole-rats and shorter-lived mice. Aging Cell. 2006;5:525–532. doi: 10.1111/j.1474-9726.2006.00246.x. [DOI] [PubMed] [Google Scholar]
- Antoine PO, Marivaux L, Croft DA, Billet G, Ganerod M, Jaramillo C, Martin T, Orliac MJ, Tejada J, Altamirano AJ, et al. Middle Eocene rodents from Peruvian Amazonia reveal the pattern and timing of caviomorph origins and biogeography. Proc. Biol. Sci. 2012;279:1319–1326. doi: 10.1098/rspb.2011.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avivi A, Gerlach F, Joel A, Reuss S, Burmester T, Nevo E, Hankeln T. Neuroglobin, cytoglobin, and myoglobin contribute to hypoxia adaptation of the subterranean mole rat Spalax. Proc. Natl. Acad. Sci. U.S.A. 2010;107:21570–21575. doi: 10.1073/pnas.1015379107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azpurua J, Seluanov A. Long-lived cancer-resistant rodents as new model species for cancer research. Front. Genet. 2012;3:319. doi: 10.3389/fgene.2012.00319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azpurua J, Ke Z, Chen IX, Zhang Q, Ermolenko DN, Zhang ZD, Gorbunova V, Seluanov A. Naked mole-rat has increased translational fidelity compared with the mouse, as well as a unique 28S ribosomal RNA cleavage. Proc. Natl. Acad. Sci. U.S.A. 2013;110:17350–17355. doi: 10.1073/pnas.1313473110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker DJ, Perez-Terzic C, Jin F, Pitel KS, Niederlander NJ, Jeganathan K, Yamada S, Reyes S, Rowe L, Hiddinga HJ, et al. Opposing roles for p16Ink4a and p19Arf in senescence and ageing caused by BubR1 insufficiency. Nature Cell Biol. 2008;10:825–836. doi: 10.1038/ncb1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker DJ, Wijshake T, Tchkonia T, LeBrasseur NK, Childs BG, van de Sluis B, Kirkland JL, van Deursen JM. Clearance of p16Ink4a-positive senescent cells delays ageing-associated disorders. Nature. 2011;479:232–236. doi: 10.1038/nature10600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begall S, Burda H, Schleich CE. Subterranean Rodents: News from Underground. In: Begall S, Burda H, Schleich CE, editors. Subterranean Rodents. Springer; Berlin, Germany: 2007. pp. 3–9. [Google Scholar]
- Bennett NC, Faulkes CG. African mole-rats: ecology and eusociality. Cambridge University Press; Cambridge, UK: 2000. [Google Scholar]
- Blagosklonny MV. Once again on rapamycin-induced insulin resistance and longevity: despite of or owing to. Aging. 2012;4:350–358. doi: 10.18632/aging.100461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand A, Smith ES, Lewin GR, Park TJ. Functional neurokinin and NMDA receptor activity in an animal naturally lacking substance P: the naked mole-rat. PLoS ONE. 2010;5:e15162. doi: 10.1371/journal.pone.0015162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burda H, Šumbera R, Begall S. Microclimate in Burrows of Subterranean Rodents – Revisited. In: Begall S, Burda H, Schleich CE, editors. Subterranean Rodents: News from Underground. Subterranean Rodents. Springer; Berlin, Germany: 2007. pp. 21–31. [Google Scholar]
- Cagnacci A, Elliott JA, Yen SS. Melatonin: a major regulator of the circadian rhythm of core temperature in humans. J. Clin. Endocrinol. Metab. 1992;75:447–452. doi: 10.1210/jcem.75.2.1639946. [DOI] [PubMed] [Google Scholar]
- Campisi J, Andersen JK, Kapahi P, Melov S. Cellular senescence: a link between cancer and age-related degenerative disease? Semin. Cancer Biol. 2011;21:354–359. doi: 10.1016/j.semcancer.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SJ, Episkopou V, Zeitlin S, Karathanasis SK, MacKrell A, Steiner DF, Efstratiadis A. Guinea pig preproinsulin gene: an evolutionary compromise? Proc. Natl. Acad. Sci. U.S.A. 1984;81:5046–5050. doi: 10.1073/pnas.81.16.5046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daly TJ, Buffenstein R. Skin morphology and its role in thermoregulation in mole-rats, Heterocephalus glaber and Cryptomys hottentotus. J. Anat. 1998;193(Pt 4):495–502. doi: 10.1046/j.1469-7580.1998.19340495.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dammann P, Burda H. Senescence Patterns in African Mole-rats (Bathyergidae, Rodentia). In: Begall S, Burda H, Schleich CE, editors. Subterranean Rodents. Springer; Berlin, Germany: 2007. pp. 251–262. [Google Scholar]
- de Magalhaes JP. How ageing processes influence cancer. Nature Rev. Cancer. 2013;13:357–365. doi: 10.1038/nrc3497. [DOI] [PubMed] [Google Scholar]
- Dowling DP, Di Costanzo L, Gennadios HA, Christianson DW. Evolution of the arginase fold and functional diversity. Cell. Mol. Life Sci. 2008;65:2039–2055. doi: 10.1007/s00018-008-7554-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvillie B, Cordonnier N, Deltour L, Dandoy-Dron F, Itier JM, Monthioux E, Jami J, Joshi RL, Bucchini D. Phenotypic alterations in insulin-deficient mutant mice. Proc. Natl. Acad. Sci. U.S.A. 1997;94:5137–5140. doi: 10.1073/pnas.94.10.5137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebensperger LA, Hurtado MJ, Soto-Gamboa M, Lacey EA, Chang AT. Communal nesting and kinship in degus (Octodon degus). Naturwissenschaften. 2004;91:391–395. doi: 10.1007/s00114-004-0545-5. [DOI] [PubMed] [Google Scholar]
- Edrey YH, Park TJ, Kang H, Biney A, Buffenstein R. Endocrine function and neurobiology of the longest-living rodent, the naked mole-rat. Exp. Gerontol. 2011;46:116–123. doi: 10.1016/j.exger.2010.09.005. [DOI] [PubMed] [Google Scholar]
- Faulkes CG, Verheyen E, Verheyen W, Jarvis JU, Bennett NC. Phylogeographical patterns of genetic divergence and speciation in African mole-rats (Family: Bathyergidae). Mol. Ecol. 2004;13:613–629. doi: 10.1046/j.1365-294x.2004.02099.x. [DOI] [PubMed] [Google Scholar]
- Fiermonte G, Dolce V, David L, Santorelli FM, Dionisi-Vici C, Palmieri F, Walker JE. The mitochondrial ornithine transporter. Bacterial expression, reconstitution, functional characterization, and tissue distribution of two human isoforms. J. Biol. Chem. 2003;278:32778–32783. doi: 10.1074/jbc.M302317200. [DOI] [PubMed] [Google Scholar]
- Gorbunova V, Hine C, Tian X, Ablaeva J, Gudkov AV, Nevo E, Seluanov A. Cancer resistance in the blind mole rat is mediated by concerted necrotic cell death mechanism. Proc. Natl. Acad. Sci. U.S.A. 2012;109:19392–19396. doi: 10.1073/pnas.1217211109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris RS. PhD thesis. Pennsylvania State University; University Park, Pennsylvania, PA: 2007. Improved Pairwise Alignment of Genomic DNA. [Google Scholar]
- Herman IM. Actin isoforms. Curr. Opin. Cell. Biol. 1993;5:48–55. doi: 10.1016/s0955-0674(05)80007-9. [DOI] [PubMed] [Google Scholar]
- Heth G, Todrank J. Using Odors Underground. In: Begall S, Burda H, Schleich CE, editors. Subterranean Rodents: News from Underground. Subterranean Rodents. Springer; Berlin, Germany: 2007. pp. 85–96. [Google Scholar]
- Hislop MS, Buffenstein R. Noradrenaline induces nonshivering thermogenesis in both the naked mole-rat (Heterocephalus glaber) and the Damara mole-rat (Cryptomys damarensis) despite very different modes of thermoregulation. J. Therm. Biol. 1994;19:25–32. [Google Scholar]
- Horuk R, Blundell TL, Lazarus NR, Neville RW, Stone D, Wollmer A. A monomeric insulin from the porcupine (Hystrix cristata), an Old World hystricomorph. Nature. 1980;286:822–824. doi: 10.1038/286822a0. [DOI] [PubMed] [Google Scholar]
- Johansen K, Lykkeboe G, Weber RE, Maloiy GM. Blood respiratory properties in the naked mole rat Heterocephalus glaber, a mammal of low body temperature. Respir Physiol. 1976;28:303–314. doi: 10.1016/0034-5687(76)90025-6. [DOI] [PubMed] [Google Scholar]
- Kasaikina MV, Lobanov AV, Malinouski MY, Lee BC, Seravalli J, Fomenko DE, Turanov AA, Finney L, Vogt S, Park TJ, et al. Reduced utilization of selenium by naked mole rats due to a specific defect in GPx1 expression. J. Biol. Chem. 2011;286:17005–17014. doi: 10.1074/jbc.M110.216267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Baertsch R, Hinrichs A, Miller W, Haussler D. Evolution's cauldron: duplication, deletion, and rearrangement in the mouse and human genomes. Proc. Natl. Acad. Sci. U.S.A. 2003;100:11484–11489. doi: 10.1073/pnas.1932072100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King GL, Kahn CR, Heldin CH. Sharing of biological effect and receptors between guinea pig insulin and platelet-derived growth factor. Proc. Natl. Acad. Sci. U.S.A. 1983;80:1308–1312. doi: 10.1073/pnas.80.5.1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim EB, Fang X, Fushan AA, Huang Z, Lobanov AV, Han L, Marino SM, Sun X, Turanov AA, Yang P, et al. Genome sequencing reveals insights into physiology and longevity of the naked mole rat. Nature. 2011;479:223–227. doi: 10.1038/nature10533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kock D, Ingram C, Frabotta L, Honeycutt R, Burda H. On the nomenclature of Bathyergidae and Fukomys n. gen. Mammalia: Rodentia). Zootaxa. 2006;1142:51–55. [Google Scholar]
- Kott O, Sumbera R, Nemec P. Light perception in two strictly subterranean rodents: life in the dark or blue? PLoS ONE. 2010;5:e11810. doi: 10.1371/journal.pone.0011810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer B, Buffenstein R. The pancreas of the naked mole-rat (Heterocephalus glaber): an ultrastructural and immunocytochemical study of the endocrine component of thermoneutral and cold acclimated animals. Gen. Comp. Endocrinol. 2004;139:206–214. doi: 10.1016/j.ygcen.2004.09.006. [DOI] [PubMed] [Google Scholar]
- Kraus DW, Colacino JM. Extended Oxygen Delivery from the Nerve Hemoglobin of Tellina alternata (Bivalvia). Science. 1986;232:90–92. doi: 10.1126/science.232.4746.90. [DOI] [PubMed] [Google Scholar]
- Lassing I, Schmitzberger F, Bjornstedt M, Holmgren A, Nordlund P, Schutt CE, Lindberg U. Molecular and structural basis for redox regulation of beta-actin. J. Mol. Biol. 2007;370:331–348. doi: 10.1016/j.jmb.2007.04.056. [DOI] [PubMed] [Google Scholar]
- LaVinka PC, Brand A, Landau VJ, Wirtshafter D, Park TJ. Extreme tolerance to ammonia fumes in African naked mole-rats: animals that naturally lack neuropeptides from trigeminal chemosensory nerve fibers. J. Comp. Physiol. A Neuroethol. Sens. Neural Behav. Physiol. 2009;195:419–427. doi: 10.1007/s00359-009-0420-0. [DOI] [PubMed] [Google Scholar]
- Lavulo LT, Sossong TM, Jr., Brigham-Burke MR, Doyle ML, Cox JD, Christianson DW, Ash DE. Subunit-subunit interactions in trimeric arginase. Generation of active monomers by mutation of a single amino acid. J. Biol. Chem. 2001;276:14242–14248. doi: 10.1074/jbc.M010575200. [DOI] [PubMed] [Google Scholar]
- Lewis KN, Andziak B, Yang T, Buffenstein R. The Naked Mole-Rat Response to Oxidative Stress: Just Deal with It. Antioxid. Redox. Signal. 2013;19:1388–1399. doi: 10.1089/ars.2012.4911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Zhu H, Ruan J, Qian W, Fang X, Shi Z, Li Y, Li S, Shan G, Kristiansen K, et al. De novo assembly of human genomes with massively parallel short read sequencing. Genome Res. 2010a;20:265–272. doi: 10.1101/gr.097261.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Fan W, Tian G, Zhu H, He L, Cai J, Huang Q, Cai Q, Li B, Bai Y, et al. The sequence and de novo assembly of the giant panda genome. Nature. 2010b;463:311–317. doi: 10.1038/nature08696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang L, Guo WH, Esquiliano DR, Asai M, Rodriguez S, Giraud J, Kushner JA, White MF, Lopez MF. Insulin-like growth factor 2 and the insulin receptor, but not insulin, regulate fetal hepatic glycogen synthesis. Endocrinology. 2010a;151:741–747. doi: 10.1210/en.2009-0705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang S, Mele J, Wu Y, Buffenstein R, Hornsby PJ. Resistance to experimental tumorigenesis in cells of a long-lived mammal, the naked mole-rat (Heterocephalus glaber). Aging Cell. 2010b;9:626–635. doi: 10.1111/j.1474-9726.2010.00588.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Hodish I, Haataja L, Lara-Lemus R, Rajpal G, Wright J, Arvan P. Proinsulin misfolding and diabetes: mutant INS gene-induced diabetes of youth. Trends Endocrinol. Metab. 2010;21:652–659. doi: 10.1016/j.tem.2010.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh SH, Francescut L, Lingor P, Bahr M, Nicotera P. Identification of new kinase clusters required for neurite outgrowth and retraction by a loss-of-function RNA interference screen. Cell Death Differ. 2008;15:283–298. doi: 10.1038/sj.cdd.4402258. [DOI] [PubMed] [Google Scholar]
- Lui JC, Baron J. Evidence that Igf2 down-regulation in postnatal tissues and up-regulation in malignancies is driven by transcription factor E2f3. Proc. Natl. Acad. Sci. U.S.A. 2013;110:6181–6186. doi: 10.1073/pnas.1219079110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manov I, Hirsh M, Iancu TC, Malik A, Sotnichenko N, Band M, Avivi A, Shams I. Pronounced cancer resistance in a subterranean rodent, the , Spalax: in vivo and in vitro evidence. BMC Biol. 2013;11:91. doi: 10.1186/1741-7007-11-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melen GJ, Pesce CG, Rossi MS, Kornblihtt AR. Novel processing in a mammalian nuclear 28S pre-rRNA: tissue-specific elimination of an ‘intron’ bearing a hidden break site. EMBO J. 1999;18:3107–3118. doi: 10.1093/emboj/18.11.3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meredith RW, Janecka JE, Gatesy J, Ryder OA, Fisher CA, Teeling EC, Goodbla A, Eizirik E, Simao TL, Stadler T, et al. Impacts of the Cretaceous Terrestrial Revolution and KPg extinction on mammal diversification. Science. 2011;334:521–524. doi: 10.1126/science.1211028. [DOI] [PubMed] [Google Scholar]
- Nasser NJ, Avivi A, Shafat I, Edovitsky E, Zcharia E, Ilan N, Vlodavsky I, Nevo E. Alternatively spliced Spalax heparanase inhibits extracellular matrix degradation, tumor growth, and metastasis. Proc. Natl. Acad. Sci. U.S.A. 2009;106:2253–2258. doi: 10.1073/pnas.0812846106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemec P, Cvekova P, Benada O, Wielkopolska E, Olkowicz S, Turlejski K, Burda H, Bennett NC, Peichl L. The visual system in subterranean African mole-rats (Rodentia, Bathyergidae): retina, subcortical visual nuclei and primary visual cortex. Brain Res. Bull. 2008;75:356–364. doi: 10.1016/j.brainresbull.2007.10.055. [DOI] [PubMed] [Google Scholar]
- Opazo JC, Soto-Gamboa M, Bozinovic F. Blood glucose concentration in caviomorph rodents. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2004;137:57–64. doi: 10.1016/j.cbpb.2003.09.007. [DOI] [PubMed] [Google Scholar]
- Opazo JC, Palma RE, Melo F, Lessa EP. Adaptive evolution of the insulin gene in caviomorph rodents. Mol. Biol. Evol. 2005;22:1290–1298. doi: 10.1093/molbev/msi117. [DOI] [PubMed] [Google Scholar]
- Park TJ, Comer C, Carol A, Lu Y, Hong HS, Rice FL. Somatosensory organization and behavior in naked mole-rats: II. Peripheral structures, innervation, and selective lack of neuropeptides associated with thermoregulation and pain. J. Comp. Neurol. 2003;465:104–120. doi: 10.1002/cne.10824. [DOI] [PubMed] [Google Scholar]
- Park TJ, Lu Y, Juttner R, Smith ES, Hu J, Brand A, Wetzel C, Milenkovic N, Erdmann B, Heppenstall PA, et al. Selective inflammatory pain insensitivity in the African naked mole-rat (Heterocephalus glaber). PLoS Biol. 2008;6:e13. doi: 10.1371/journal.pbio.0060013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prendergast BJ. MT1 melatonin receptors mediate somatic, behavioral, and reproductive neuroendocrine responses to photoperiod and melatonin in Siberian hamsters (Phodopus sungorus). Endocrinology. 2010;151:714–721. doi: 10.1210/en.2009-0710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Q, Zhang G, Ma T, Qian W, Wang J, Ye Z, Cao C, Hu Q, Kim J, Larkin DM, et al. The yak genome and adaptation to life at high altitude. Nature Genet. 2012;44:946–949. doi: 10.1038/ng.2343. [DOI] [PubMed] [Google Scholar]
- Reddy VN, Giblin FJ, Lin LR, Dang L, Unakar NJ, Musch DC, Boyle DL, Takemoto LJ, Ho YS, Knoernschild T, et al. Glutathione peroxidase-1 deficiency leads to increased nuclear light scattering, membrane damage, and cataract formation in gene-knockout mice. Invest. Ophthalmol. Vis. Sci. 2001;42:3247–3255. [PubMed] [Google Scholar]
- Rosenfeld MG, Amara SG, Roos BA, Ong ES, Evans RM. Altered expression of the calcitonin gene associated with RNA polymorphism. Nature. 1981;290:63–65. doi: 10.1038/290063a0. [DOI] [PubMed] [Google Scholar]
- Sabio G, Mora A, Rangel MA, Quesada A, Marcos CF, Alonso JC, Soler G, Centeno F. Glu-256 is a main structural determinant for oligomerisation of human arginase I. FEBS Lett. 2001;501:161–165. doi: 10.1016/s0014-5793(01)02650-3. [DOI] [PubMed] [Google Scholar]
- Schelshorn DW, Schneider A, Kuschinsky W, Weber D, Kruger C, Dittgen T, Burgers HF, Sabouri F, Gassler N, Bach A, et al. Expression of hemoglobin in rodent neurons. J. Cereb. Blood Flow Metab. 2009;29:585–595. doi: 10.1038/jcbfm.2008.152. [DOI] [PubMed] [Google Scholar]
- Schneuer M, Flachsbarth S, Czech-Damal NU, Folkow LP, Siebert U, Burmester T. Neuroglobin of seals and whales: evidence for a divergent role in the diving brain. Neuroscience. 2012;223:35–44. doi: 10.1016/j.neuroscience.2012.07.052. [DOI] [PubMed] [Google Scholar]
- Seim I, Fang X, Xiong Z, Lobanov AV, Huang Z, Ma S, Feng Y, Turanov AA, Zhu Y, Lenz TL, et al. Genome analysis reveals insights into physiology and longevity of the Brandt's bat Myotis brandtii. Nat. Commun. 2013;4:2212. doi: 10.1038/ncomms3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seluanov A, Hine C, Bozzella M, Hall A, Sasahara TH, Ribeiro AA, Catania KC, Presgraves DC, Gorbunova V. Distinct tumor suppressor mechanisms evolve in rodent species that differ in size and lifespan. Aging Cell. 2008;7:813–823. doi: 10.1111/j.1474-9726.2008.00431.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seluanov A, Hine C, Azpurua J, Feigenson M, Bozzella M, Mao Z, Catania KC, Gorbunova V. Hypersensitivity to contact inhibition provides a clue to cancer resistance of naked mole-rat. Proc. Natl. Acad. Sci. U.S.A. 2009;106:19352–19357. doi: 10.1073/pnas.0905252106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shams I, Malik A, Manov I, Joel A, Band M, Avivi A. Transcription pattern of p53-targeted DNA repair genes in the hypoxia-tolerant subterranean Spalax. J. Mol. Biol. 2013;425:1111–1118. doi: 10.1016/j.jmb.2013.01.007. [DOI] [PubMed] [Google Scholar]
- Simarro M, Giannattasio G, De la Fuente MA, Benarafa C, Subramanian KK, Ishizawar R, Balestrieri B, Andersson EM, Luo HR, Orduna A, et al. Fas-activated serine/threonine phosphoprotein promotes immune-mediated pulmonary inflammation. J. Immunol. 2010;184:5325–5332. doi: 10.4049/jimmunol.1000104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St John Smith E, Purfurst B, Grigoryan T, Park TJ, Bennett NC, Lewin GR. Specific paucity of unmyelinated C-fibers in cutaneous peripheral nerves of the African naked-: comparative analysis using six species of Bathyergidae. The J. Comp. Neurol. 2012;520:2785–2803. doi: 10.1002/cne.23133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ES, Omerbasic D, Lechner SG, Anirudhan G, Lapatsina L, Lewin GR. The molecular basis of acid insensitivity in the African naked mole-rat. Science. 2011;334:1557–1560. doi: 10.1126/science.1213760. [DOI] [PubMed] [Google Scholar]
- Tarailo-Graovac M, Chen N. Using RepeatMasker to identify repetitive elements in genomic sequences. In: Baxevanis AD, editor. Current Protocols in Bioinformatics. John Wiley & Sons; Hoboken, NJ: 2009. pp. 4.10.1–4.10.14. [DOI] [PubMed] [Google Scholar]
- Terman JR, Kashina A. Post-translational modification and regulation of actin. Curr. Opin. Cell Biol. 2013;25:30–38. doi: 10.1016/j.ceb.2012.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian X, Azpurua J, Hine C, Vaidya A, Myakishev-Rempel M, Ablaeva J, Mao Z, Nevo E, Gorbunova V, Seluanov A. High-molecular-mass hyaluronan mediates the cancer resistance of the naked. Nature. 2013;499:346–349. doi: 10.1038/nature12234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandekerckhove J, Weber K. At least six different actins are expressed in a higher mammal: an analysis based on the amino acid sequence of the amino-terminal tryptic peptide. J. Mol. Biol. 1978;126:783–802. doi: 10.1016/0022-2836(78)90020-7. [DOI] [PubMed] [Google Scholar]
- van Oers CC, Adema GJ, Zandberg H, Moen TC, Baas PD. Two different sequence elements within exon 4 are necessary for calcitonin-specific splicing of the human calcitonin/calcitonin gene-related peptide I pre-mRNA. Mol. Cell. Biol. 1994;14:951–960. doi: 10.1128/mcb.14.2.951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Burge CB. Splicing regulation: from a parts list of regulatory elements to an integrated splicing code. RNA. 2008;14:802–813. doi: 10.1261/rna.876308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Gao J, Sun X, Martinez-Wittinghan FJ, Li L, Varadaraj K, Farrell M, Reddy VN, White TW, Mathias RT. The effects of GPX-1 knockout on membrane transport and intracellular homeostasis in the lens. J. Membr. Biol. 2009;227:25–37. doi: 10.1007/s00232-008-9141-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winnebeck EC, Millar CD, Warman GR. Why does insect RNA look degraded? J. Insect Sci. 2010;10:159. doi: 10.1673/031.010.14119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf N, Penn P, Pendergrass W, Van Remmen H, Bartke A, Rabinovitch P, Martin GM. Age-related cataract progression in five mouse models for anti-oxidant protection or hormonal influence. Exp. Eye Res. 2005;81:276–285. doi: 10.1016/j.exer.2005.01.024. [DOI] [PubMed] [Google Scholar]
- Zhao H, Ru B, Teeling EC, Faulkes CG, Zhang S, Rossiter SJ. Rhodopsin molecular evolution in mammals inhabiting low light environments. PLoS ONE. 2009;4:e8326. doi: 10.1371/journal.pone.0008326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhi F, Zhou G, Shao N, Xia X, Shi Y, Wang Q, Zhang Y, Wang R, Xue L, Wang S, et al. miR-106a-5p inhibits the proliferation and migration of astrocytoma cells and promotes apoptosis by targeting FASTK. PLoS ONE. 2013;8:e72390. doi: 10.1371/journal.pone.0072390. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.