Abstract
In eukaryotic cells, alternative cleavage of 3’ untranslated regions (UTRs) can affect transcript stability, transport and translation. For polyadenylated (poly(A)) transcripts, cleavage sites can be characterized with short-read sequencing using specialized library construction methods. However, for large-scale cohort studies as well as for clinical sequencing applications, it is desirable to characterize such events using RNA-seq data, as the latter are already widely applied to identify other relevant information, such as mutations, alternative splicing and chimeric transcripts. Here we describe KLEAT, an analysis tool that uses de novo assembly of RNA-seq data to characterize cleavage sites on 3’ UTRs. We demonstrate the performance of KLEAT on three cell line RNA-seq libraries constructed and sequenced by the ENCODE project, and assembled using Trans-ABySS. Validating the KLEAT predictions with matched ENCODE RNA-seq and RNA-PET libraries, we show that the tool has over 90% positive predictive value when there are at least three RNA-seq reads supporting a poly(A) tail and requiring at least three RNA-PET reads mapping within 100 nucleotides as validation. We also compare the performance of KLEAT with other popular RNA-seq analysis pipelines that reconstruct 3’ UTR ends, and show that it performs favourably, based on an ROC-like curve.
1. Introduction
The section of an mRNA transcript that is translated into protein sequence is flanked by 5’ and 3’ untranslated regions (UTRs). These UTRs play a number of important biological roles. The 3’ end of an mRNA molecule (the 3' UTR) helps to regulate its stability and localization, hence the amount of corresponding protein that is produced [1–4]. Over 50% of human genes produce two or more transcript isoforms via alternative polyadenylation (APA) of the 3’ UTRs [5]. APA is recognized as playing a role in cancer biology [6–9].
A number of direct sequencing protocols have been developed for characterizing polyadenylated (poly(A)) tails of 3’ UTRs and APA [9–15]. A cost-effective alternative to these direct sequencing protocols would be high throughput transcriptome sequencing (RNA-seq) [16], coupled with a validated bioinformatics pipeline to detect 3’ UTR cleavage sites (CS).
RNA-seq is a central data type for many studies, including the ENCODE (ENCyclopedia Of DNA Elements) project, whose goal is to identify all functional elements in the human genome sequence [17]. Using various sequencing protocols, an ENCODE study [18] identified over 100,000 transcripts, about 60,000 of which were protein coding, and reported that transcript expression levels span six orders of magnitude. This is remarkable, as it speaks to the sensitivity of the RNA-seq technology. The lower range of the reported expression levels of 10−2 RPKM in that study implies that RNA-seq can detect a transcript expressed by 1 in 100 cells [16]. This resolution of RNA-seq data can be leveraged to identify 3’ UTR ends of transcripts. An earlier study [19] inferred 3’ UTR switching using sudden changes in expression profiles near cleavage sites, but did not utilize the direct evidence of observed poly(A) sequences.
In this report, we introduce KLEAT, a post-processing tool for characterizing 3’ UTRs in assembled RNA-seq data through direct observation of poly(A) tails. While we developed KLEAT as an extension to the Trans-ABySS analysis pipeline [20, 21], it can also accept contigs from other transcriptome assembly tools, as we demonstrate below. It analyses the structures of assembled transcripts for poly(A) tails, filters 3’ UTR cleavage site (CS) candidates using several evidence types within RNA-seq reads, and gathers and reports metrics that can be used in downstream post-processing, such as for filtering calls by their levels of read support.
2. Methods
The key technology KLEAT uses in detecting 3’ UTR ends is de novo transcriptome assemblies. Compared to genome assembly, a successful transcriptome assembly has to address some particular challenges. These include robust assembly of transcripts from a wide range of transcript abundance levels, and resolution of transcripts from alternative isoforms and gene families. There are several specialized de novo assembly tools, including Trans-ABySS [21], Trinity [22] and Oases [23] that successfully address these challenges.
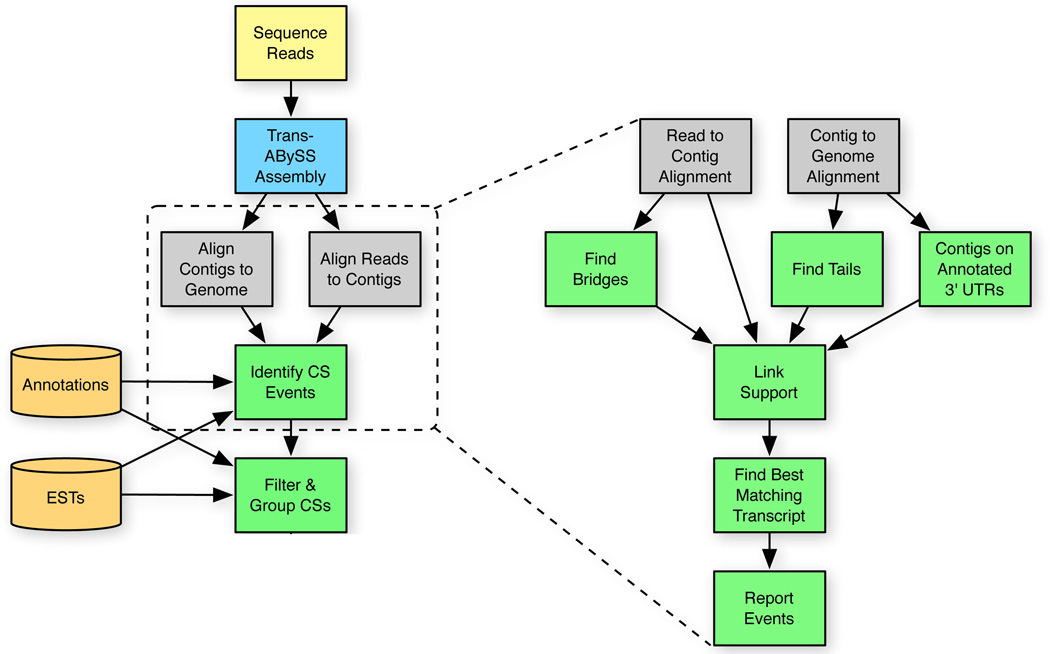
The KLEAT pipeline (Figure 1) uses Trans-ABySS by default. Using the raw reads and assembled contigs, it performs two levels of alignments in parallel: (1) reads to contigs; and (2) contigs to reference genome. It processes these alignment results to identify tail, bridge, and link evidence (Figure 2), and collates the evidence to predict cleavage sites.
Fig. 1.
Flowchart of the KLEAT pipeline. Two shades of yellow flowchart elements designate raw and external input to the pipeline; blue and grey indicate existing internal and external tools, respectively; green denotes new tools developed specifically for KLEAT.
Fig. 2.
Three types of support for detecting cleavage sites using RNA-seq data. The gene annotation (grey) indicates a single 3’ UTR isoform, while the sample expresses two APA (red) variants. RNA-seq data capture the presence of these two alternatives with reads that end in poly(A) sequence (red). Contigs with supporting evidence have either a poly(A) “tail”, an overhanging read that is “bridge” to a poly(A) sequence, or a read that has a “link” to a pair with poly(A) sequence.
2.1. Tail
Contig sequences that end in a poly(A) stretch represent high-confidence candidates. We filter these candidates to identify true poly(A) tails by aligning the flagged contigs to a reference genome. Accounting for the direction of transcription, we classify contigs with untemplated poly(A) sequence (a stretch of poly(A) sequence not observed in the reference genome) at their 3’ ends as tail type events. For a transcript that is sufficiently abundant, this would be the expected default event type.
2.2. Bridge
Expressed alternative long and short 3’ UTRs present alternative paths for contig extensions during de novo assembly. In such cases, if the graph indicates a branch that does not extend to a poly(A) sequence, the alternative branch with the poly(A) sequence is removed by an assembly quality assurance stage within ABySS, in an operation called trimming [24]. While this is a desirable behaviour in general, it creates a particular challenge in assembling contigs with poly(A) tails. However, the information removed during this step can be recovered later by aligning reads to contigs, then assessing the sequences of partial read alignments at the contig edges. When an overhanging read alignment represents an untemplated poly(A) sequence, we infer the presence of a cleavage site. We call such cases bridge type evidence.
2.3. Link
The sequence complexity of 3’ UTRs may drop substantially near their 3’ ends, where the region is dominated by AU-rich sequence [25], and this may affect contig extensions due to loss of specificity of read-to-read overlaps. When this happens near a cleavage site, the corresponding contig may fail to present a tail type evidence, and may terminate extension before a read with a poly(A) tail can bridge it to beyond the cleavage site. However, the 3’ UTR end may be within a typical sequencing fragment length, and if we identify read pairs linking the end of a contig to an untemplated poly(A) sequence, we classify the corresponding contig as having link type evidence.
Some cleavage sites may have supporting evidence from a combination of these evidence types, and even multiple observations from the same evidence type. The latter is partly due to the fuzzy definition of a cleavage site, where the end of a 3’ UTR may fluctuate by about ±30 nucleotides (nt) between mRNA molecules of the same transcript species [26]. Accordingly, we cluster cleavage sites predicted from multiple contigs if they fall within a certain window, label them as being representatives of the same cleavage site, and tally the counts presented by each evidence type in a given cluster to score the strength of our prediction.
We note that read-to-contig alignments performed in the pipeline have unique requirements. Although we demonstrate our results using BWA [27] – an established general-purpose sequence alignment tool – we recognize that detecting cleavage sites is most effective when reads are aligned to contigs with a tool that is capable of handling alignments with overhangs, that is, when a read aligns to the end of a contig with its sequence extending beyond the boundary(ies) of the contig. Because many high-throughput general-purpose sequence alignment tools are developed by the explicit or implicit assumption of a reference sequence that is composed of a small number of long contigs (i.e. chromosomes), they may suffer from accuracy and performance issues when the reference sequence is in many short pieces (as in an assembled transcriptome). Alignments near contig or scaffold edges are particularly challenging for general-purpose alignment software. We call this the edge effect, and address it by an FM-index based aligner within the ABySS genome assembly package [24], as an alternative. This aligner weighs edge alignments that are shorter but with fewer mismatches more favourably than longer alignments with more mismatches to provide local alignments.
KLEAT compares putative cleavage sites to annotation and EST databases to characterize and annotate them with other supporting observations, if any. Again using the annotation and EST databases, KLEAT groups, classifies and filters the putative events.
For method validation, we used the RNA-PET protocol [10] as our gold standard. We quantified the concordance between the “putative” (KLEAT) and “real” (RNA-PET) cleavage sites (CS) using the following definitions:
| false positive | | A called CS not within a certain window of an RNA-PET cluster |
| true positive | | An RNA-PET cluster with at least one called CS in a window |
| false negative | | An RNA-PET cluster without a called CS in a window |
| true negative | | Cannot be defined |
One way to gauge the performance of a detection tool is to study its receiver-operator-characteristic (ROC) curve, where a stringency parameter is varied to plot the true positive rate, TPR (the ratio of the true positive count to the total number of events) versus the false positive rate, FPR (the ratio of the false positive count to the total number of negatives). Note that, because true negative is undefined in this context, FPR cannot be defined either. A common practice in such cases is to use the false discovery rate, FDR (defined as the ratio of the false positive count to the total number of calls) as surrogate for the FPR.
3. Results and Discussion
For validating our method, we employed experimental data collected by the ENCODE project [18]. The ENCODE consortium characterized transcript ends using the RNA-PET protocol [10], and generated RNA-seq data for some of the same samples. We considered three cell lines (H1-hESC, A549 and MCF-7) for which RNA-PET and RNA-seq data were available (Table 1).
Table 1.
Three ENCODE cell lines used for validation. All libraries were prepared using the long polyA+ RNA fraction protocol. RNA samples represent the whole cell transcriptome. RNA-PET reads were sequenced at 2×36 nt. H1-hESC cell line RNA-seq reads were sequenced at 2×78 nt, and A549 and MCF-7 cell lines at 2×76 nt. All data were generated and made publicly available by the ENCODE project [18].
| Cell Line | # RNA-PET Read Pairs (million) | # RNA-seq Read Pairs (million) |
|---|---|---|
| H1-hESC | 50.2 | 78.3 |
| A549 | 181.7 | 70.5 |
| MCF-7 | 174.0 | 87.4 |
For RNA-PET coverage data, we applied an expression level threshold (default, 3 reads), and clustered observations that occurred within a certain distance (default, 100 nt) of each other. We used the resulting clusters as ‘true’ events.
We used Trans-ABySS v1.4.7 [20, 21] to assemble the RNA-seq reads; aligned the assembled transcripts to the human genome reference hg19 using GMAP 2012-12-20 [28]; and aligned reads back to the assembled contigs using BWA-SW v0.6.2-r126 [27]. We processed the results to identify the tail, bridge and link evidence, and clustered the CS calls. We also used Trinity Release 2013-02-25 [22] for RNA-seq assembly, and used the same pipeline to identify CS calls. Table 2 summarizes the performance of KLEAT on the assembled transcriptomes (with Trans-ABySS and Trinity contigs) for the three cell lines.
Table 2.
Summary statistics on de novo assembly of the RNA-seq data and KLEAT calls. Assembly figures are for contigs longer than 500 nt in length. The total number of cleavage sites called by KLEAT, and the average number of alternative polyadenylation sites per gene, are shown in the last two columns. Two sub-columns for the number of cleavage sites and APA per gene represent total and true positive (TP) calls, at a support threshold of three reads.
| Cell Line | Assembler | # Contigs | N50 (nt) | Total Reconstruction (Mnt) |
# Cleavage Sites | APA per gene | ||
|---|---|---|---|---|---|---|---|---|
| All | TP | All | TP | |||||
| H1-hESC | Trans-ABySS | 879,277 | 1,049 | 309.1 | 11,975 | 8,998 | 2.54 | 2.49 |
| Trinity | 159,627 | 2,250 | 188.3 | 9,896 | 7,565 | 2.37 | 2.30 | |
| A549 | Trans-ABySS | 788,688 | 1,352 | 299.9 | 13,984 | 12,940 | 2.83 | 2.80 |
| Trinity | 149,880 | 2,791 | 197.4 | 12,249 | 11,369 | 2.62 | 2.57 | |
| MCF-7 | Trans-ABySS | 1,002,984 | 1,171 | 391.5 | 15,240 | 13,308 | 3.09 | 3.05 |
| Trinity | 237,875 | 2,925 | 323.6 | 12,845 | 11,387 | 2.82 | 2.76 | |
Using contigs from either transcriptome assembly tool as input, we observed the number of cleavage site calls to be the lowest for H1-hESC, and highest for MCF-7. The number of APA sites per gene also follows this pattern, ranging from roughly 2.5 to 3.0 APA isoforms, on average. Interestingly, while the fraction of true positive cleavage site calls range from 75 to 93%, the average number of APA isoforms per gene is insensitive to filtering for true positives.
We compared the use of contigs from de novo transcriptome assembly to detect cleavage sites, with the use of transcripts reconstructed by aligning the reads to a reference genome. We ran the Cufflinks pipeline v2.1.1 [29] on the same dataset, and used the reconstructed 3’ UTR ends of predicted transcripts to measure its accuracy in detecting poly(A) tails. Cufflinks takes RNA-seq read alignments to a reference genome as input, and builds those alignments into a parsimonious set of transcripts with or without annotation support. We ran the pipeline with annotation support, allowing for transcript discovery.
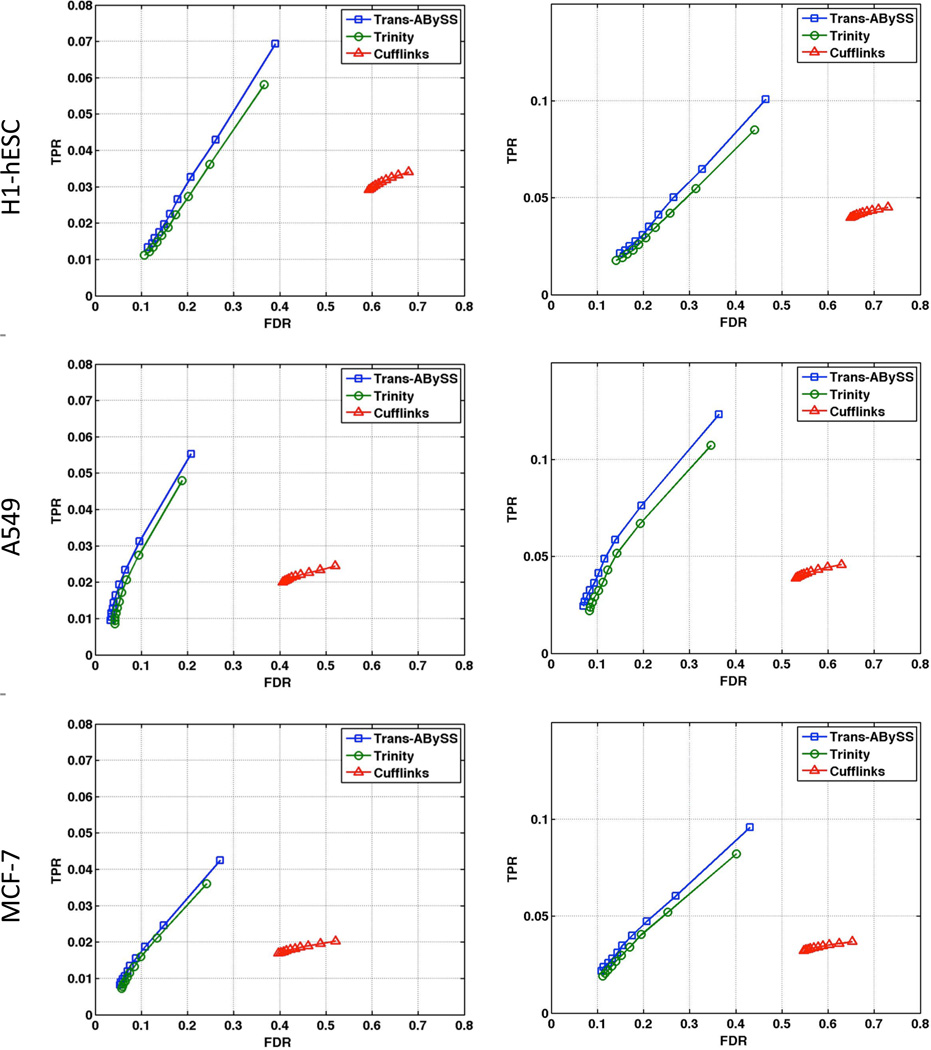
Figure 3 depicts the ROC curves for these two paradigms for the three cell lines used. Curves closer to the top-left corner indicate better performance. These results suggest that to identify 3’ UTR cleavage sites an assembly-first approach (using either Trans-ABySS or Trinity) may be preferred over the alignment-first approach implemented in the Cufflinks pipeline. We note that the Trans-ABySS and Trinity results are similar, while the Trans-ABySS assemblies perform marginally yet consistently better. This may be due to the difference between the total reconstruction figures of the two tools (Table 2).
Fig. 1.
Performance of KLEAT on three ENCODE cell lines. Curves represent the true positive rate (TPR) as a function of the false discovery rate (FDR), for KLEAT with Trans-ABySS (blue) and KLEAT with Trinity (green) and Cufflinks (red). RNA-PET evidence is considered as the gold standard at two support cutoffs: left column: 3 or more, right column: 10 or more RNA-PET reads.
Although the magnitudes of the reported TPR figures are low (<10%), we note that this reflects our simple but relaxed definition of the ground truth, with no distinction between 3’ and 5’ UTR ends, and applying none of the filtering suggested in the ENCODE report. These choices would inflate the denominator of TPR, and lead to underestimates in the reported figures. However, neither of these would change the relative performance of the analysis tools we present.
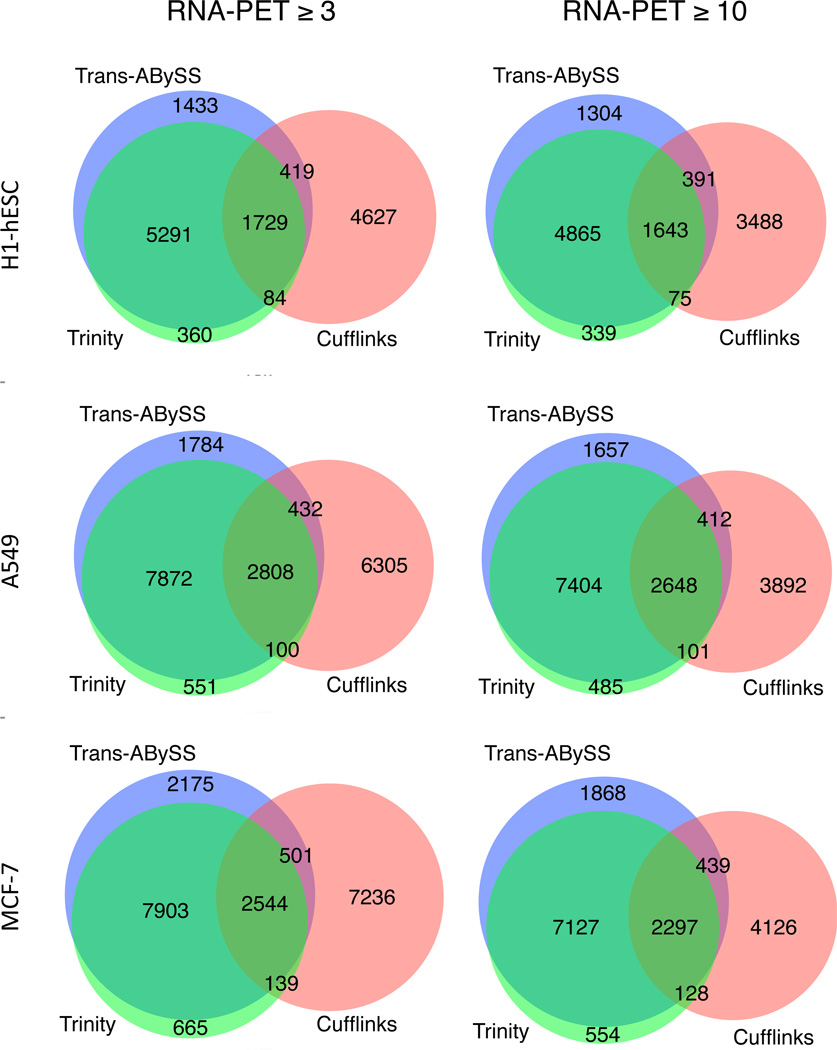
We also compared the concordance between these three sets of results (Figure 4). Our analysis indicates that Trans-ABySS and Trinity contigs are largely concordant in their reconstruction of cleavage sites, identifying roughly 9,000 to 13,000 and 7,000 to 11,000 true positive calls, respectively (for an RNA-PET threshold of 3 reads) and agreeing on about 80 to 90% of the calls. In contrast, Cufflinks would identify 7,000 to 10,000 true positive calls with the same RNA-PET threshold, agreeing with the assembly-first results only about 25 to 30% of the time, meaning that it identifies a smaller yet largely distinct set of events.
Fig. 4.
Concordance between three methods. Blue, green and red sets indicate events detected by KLEAT/Trans-ABySS, KLEAT/Trinity and Cufflinks, respectively, that are supported by at least 3 or 10 RNA-PET reads, as indicated.
We note that in this dataset the Cufflinks pipeline is more sensitive for detecting weakly expressed transcripts. This observation is supported by the performance metrics of the three tools when we increased the RNA-PET threshold from three to 10 reads. At the increased threshold, KLEAT with Trans-ABySS and Trinity loses about 6 to 10% of its true positive calls, while Cufflinks loses about 10 to 33% of such calls.
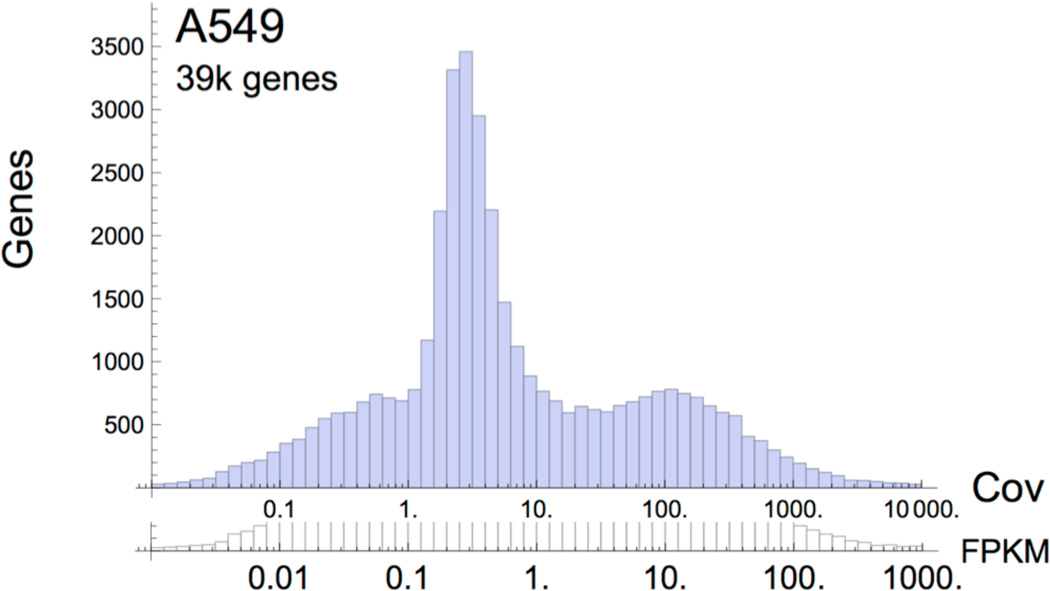
Further supporting this observation, a coverage histogram of the expressed genes in the A549 cell line, as detected by Cufflinks, is depicted in Figure 5. Here the two x-axes represent expression levels in units of average coverage and FPKM on logarithmic scales. Cufflinks reports a major peak for transcripts represented at 1- to 10-fold average coverage, also reconstructing a large number of transcripts with less than 1-fold coverage, some of which would report cleavage sites also observed by RNA-PET data. In contrast, de novo assembly methods would typically reconstruct transcripts over 10-fold coverage. This apparent difference in target expression levels for transcript reconstruction explains the lack of concordance between KLEAT (in conjunction with Trans-ABySS or Trinity) and Cufflinks, as reported in Figures 3 and 4.
Fig. 5.
Gene level coverage histogram for the A549 cell line RNA-seq data, as reported by Cufflinks. The histogram is presented with two logarithmic scales on the x-axis, average coverage (Cov) and FPKM, to show the correspondence between the two units for the sequencing depth of the experimental data.
4. Conclusions
In this study, we introduced KLEAT as an analysis tool for detecting 3’ UTR cleavage sites using de novo assembled RNA-seq reads. We validated our method using data from the ENCODE project [30]. We measured the accuracy of KLEAT using two transcriptome assembly tools (Trans-ABySS [20, 21] and Trinity [22]), and compared its performance to results from an alignment-based analysis tool (Cufflinks [29], the method of choice in the ENCODE study).
Our results demonstrate that one can reliably detect around 10,000 poly(A) tails per sample using RNA-seq data, at a sequencing depth of 70 million read pairs. The depth of sequencing data will certainly affect the number of transcripts observed, hence the number of poly(A) tails detected. Therefore, although we suggest that detecting on the order of 10,000 features will already provide important biological insights for highly expressed transcripts, if one wants to observe more features, one approach might be to sequence a library to greater depth (albeit with diminishing returns). With sequencing throughput on the Illumina platform pushing beyond 250 million read pairs per lane, experimental design (such as pooling multiple samples per lane) reflects a balance between cost and value, and that balance is determined by the particular experimental goals and the budget of a study.
We also note that overlapping sense/anti-sense gene annotations can potentially confuse the poly(A) tail calls. Using a strand-specific RNA-seq protocol should help mitigate this issue.
Surveying 15 human cell lines, the ENCODE study reports a total of 128,824 poly(A) sites mapping within annotated Gencode transcripts [30]. This observation puts the average number of polydenylation sites in this dataset to 2.5 per gene. Interestingly, before this landmark publication, the APA multiplicity was estimated to be around 1.1. Our analysis of RNA-seq data from three ENCODE cell lines (APA per gene statistics in Table 2) are in agreement with this ENCODE estimate.
There is a growing appreciation of 3’ UTRs, their molecular assembly, mechanistic roles and variants [9–15]. Many of these studies developed novel wet lab techniques to build specialized sequencing libraries, and applied them to interrogate a particular biological condition.
KLEAT offers an alternative analysis method to characterize 3’ UTRs and APA from RNA-seq data at a nucleotide scale resolution. We anticipate the tool to be an enabling technology for many applications, including large-scale disease studies and clinical genomics, and to provide added value to the large volume of sequencing data already generated using the data type. KLEAT is available at www.bcgsc.ca/platform/bioinfo/software, and is offered free for academic use.
Acknowledgments
The authors thank, Canadian Institutes of Health Research, Genome Canada, Genome British Columbia and British Columbia Cancer Foundation for their generous support. The work is also partially funded by the National Institutes of Health under Award Number R01HG007182. The content of this work is solely the responsibility of the authors and does not necessarily represent the official views of any of our funding agencies. We also thank the ENCODE project for enabling the validation work presented by making their datasets publicly available.
Footnotes
This work is supported by Canadian Institutes of Health Research, Genome Canada, Genome British Columbia, British Columbia Cancer Foundation and the National Institutes of Health under Award Number R01HG007182. The content of this work is solely the responsibility of the authors and does not necessarily represent the official views of any of our funding agencies.
References
- 1.Keene JD. RNA regulons: coordination of post-transcriptional events. Nature reviews. Genetics. 2007 Jul;8(7):533–543. doi: 10.1038/nrg2111. [DOI] [PubMed] [Google Scholar]
- 2.To KK, Zhan Z, Litman T, Bates SE. Regulation of ABCG2 expression at the 3' untranslated region of its mRNA through modulation of transcript stability and protein translation by a putative microRNA in the S1 colon cancer cell line. Molecular and cellular biology. 2008 Sep;28(17):5147–5161. doi: 10.1128/MCB.00331-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ji Z, Lee JY, Pan Z, Jiang B, Tian B. Progressive lengthening of 3' untranslated regions of mRNAs by alternative polyadenylation during mouse embryonic development. Proceedings of the National Academy of Sciences of the United States of America. 2009 Apr 28;106(17):7028–7033. doi: 10.1073/pnas.0900028106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Muller S, Rycak L, Afonso-Grunz F, Winter P, Zawada AM, Damrath E, Scheider J, Schmah J, Koch I, Kahl G, Rotter B. APADB: a database for alternative polyadenylation and microRNA regulation events. Database (Oxford) 2014;2014 doi: 10.1093/database/bau076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tian B, Hu J, Zhang H, Lutz CS. A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Res. 2005;33(1):201–212. doi: 10.1093/nar/gki158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Singh P, Alley TL, Wright SM, Kamdar S, Schott W, Wilpan RY, Mills KD, Graber JH. Global changes in processing of mRNA 3' untranslated regions characterize clinically distinct cancer subtypes. Cancer Res. 2009 Dec;69(24):9422–9430. doi: 10.1158/0008-5472.CAN-09-2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hamaya Y, Kuriyama S, Takai T, Yoshida K, Yamada T, Sugimoto M, Osawa S, Sugimoto K, Miyajima H, Kanaoka S. A distinct expression pattern of the long 3'-untranslated region dicer mRNA and its implications for posttranscriptional regulation in colorectal cancer. Clin Transl Gastroenterol. 2012;3:e17. doi: 10.1038/ctg.2012.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Devany E, Zhang X, Park JY, Tian B, Kleiman FE. Positive and negative feedback loops in the p53 and mRNA 3' processing pathways. Proc Natl Acad Sci U S A. 2013 Feb;110(9):3351–3356. doi: 10.1073/pnas.1212533110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fu Y, Sun Y, Li Y, Li J, Rao X, Chen C, Xu A. Differential genome-wide profiling of tandem 3' UTRs among human breast cancer and normal cells by high-throughput sequencing. Genome Res. 2011 May;21(5):741–747. doi: 10.1101/gr.115295.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ng P, Wei CL, Sung WK, Chiu KP, Lipovich L, Ang CC, Gupta S, Shahab A, Ridwan A, Wong CH, Liu ET, Ruan Y. Gene identification signature (GIS) analysis for transcriptome characterization and genome annotation. Nat Methods. 2005 Feb;2(2):105–111. doi: 10.1038/nmeth733. [DOI] [PubMed] [Google Scholar]
- 11.Ruan Y, Ooi HS, Choo SW, Chiu KP, Zhao XD, Srinivasan KG, Yao F, Choo CY, Liu J, Ariyaratne P, Bin WG, Kuznetsov VA, Shahab A, Sung WK, Bourque G, Palanisamy N, Wei CL. Fusion transcripts and transcribed retrotransposed loci discovered through comprehensive transcriptome analysis using Paired-End diTags (PETs) Genome Res. 2007 Jun;17(6):828–838. doi: 10.1101/gr.6018607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shepard PJ, Choi EA, Lu J, Flanagan LA, Hertel KJ, Shi Y. Complex and dynamic landscape of RNA polyadenylation revealed by PAS-Seq. RNA. 2011 Apr;17(4):761–772. doi: 10.1261/rna.2581711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ruan X, Ruan Y. Genome wide full-length transcript analysis using 5' and 3' paired-end-tag next generation sequencing (RNA-PET) Methods Mol Biol. 2012;809:535–562. doi: 10.1007/978-1-61779-376-9_35. [DOI] [PubMed] [Google Scholar]
- 14.Hafez D, Ni T, Mukherjee S, Zhu J, Ohler U. Genome-wide identification and predictive modeling of tissue-specific alternative polyadenylation. Bioinformatics. 2013 Jul;29(13):i108–i116. doi: 10.1093/bioinformatics/btt233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Minasaki R, Rudel D, Eckmann CR. Increased sensitivity and accuracy of a single-stranded DNA splint-mediated ligation assay (sPAT) reveals poly(A) tail length dynamics of developmentally regulated mRNAs. RNA Biol. 2014 Feb;11(2) doi: 10.4161/rna.27992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008 Jul;5(7):621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 17.ENCODE Project Consortium. The ENCODE (ENCyclopedia Of DNA Elements) Project. Science. 2004 Oct;306(5696):636–640. doi: 10.1126/science.1105136. [DOI] [PubMed] [Google Scholar]
- 18.Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, Xue C, Marinov GK, Khatun J, Williams BA, Zaleski C, Rozowsky J, Roder M, Kokocinski F, Abdelhamid RF, Alioto T, Antoshechkin I, Baer MT, Bar NS, Batut P, Bell K, Bell I, Chakrabortty S, Chen X, Chrast J, Curado J, Derrien T, Drenkow J, Dumais E, Dumais J, Duttagupta R, Falconnet E, Fastuca M, Fejes-Toth K, Ferreira P, Foissac S, Fullwood MJ, Gao H, Gonzalez D, Gordon A, Gunawardena H, Howald C, Jha S, Johnson R, Kapranov P, King B, Kingswood C, Luo OJ, Park E, Persaud K, Preall JB, Ribeca P, Risk B, Robyr D, Sammeth M, Schaffer L, See LH, Shahab A, Skancke J, Suzuki AM, Takahashi H, Tilgner H, Trout D, Walters N, Wang H, Wrobel J, Yu Y, Ruan X, Hayashizaki Y, Harrow J, Gerstein M, Hubbard T, Reymond A, Antonarakis SE, Hannon G, Giddings MC, Ruan Y, Wold B, Carninci P, Guigo R, Gingeras TR. Landscape of transcription in human cells. Nature. 2012 Sep 6;489(7414):101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang W, Wei Z, Li H. A change-point model for identifying 3'UTR switching by next-generation RNA sequencing. Bioinformatics. 2014 Aug 1;30(15):2162–2170. doi: 10.1093/bioinformatics/btu189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Birol I, Jackman SD, Nielsen CB, Qian JQ, Varhol R, Stazyk G, Morin RD, Zhao Y, Hirst M, Schein JE, Horsman DE, Connors JM, Gascoyne RD, Marra MA, Jones SJM. De novo transcriptome assembly with ABySS. Bioinformatics. 2009 Nov 1;25(21):2872–2877. doi: 10.1093/bioinformatics/btp367. [DOI] [PubMed] [Google Scholar]
- 21.Robertson G, Schein J, Chiu R, Corbett R, Field M, Jackman SD, Mungall K, Lee S, Okada HM, Qian JQ, Griffith M, Raymond A, Thiessen N, Cezard T, Butterfield YS, Newsome R, Chan SK, She R, Varhol R, Kamoh, A.-L. Prabhu B, Tam A, Zhao Y, Moore RA, Hirst M, Marra MA, Jones SJM, Hoodless PA, Birol I. De novo assembly and analysis of RNA-seq data. Nature Methods. 2010 Nov;7(11) doi: 10.1038/nmeth.1517. pp. 909-U62, 2010. [DOI] [PubMed] [Google Scholar]
- 22.Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, Chen Z, Mauceli E, Hacohen N, Gnirke A, Rhind N, di Palma F, Birren BW, Nusbaum C, Lindblad-Toh K, Friedman N, Regev A. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011 Jul;29(7):644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schulz MH, Zerbino DR, Vingron M, Birney E. Oases: robust de novo RNA-seq assembly across the dynamic range of expression levels. Bioinformatics. 2012 Apr 15;28(8):1086–1092. doi: 10.1093/bioinformatics/bts094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Simpson JT, Wong K, Jackman SD, Schein JE, Jones SJM, Birol I. ABySS: A parallel assembler for short read sequence data. Genome Research. 2009 Jun;19(6):1117–1123. doi: 10.1101/gr.089532.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang T, Kruys V, Huez G, Gueydan C. AU-rich element-mediated translational control: complexity and multiple activities of trans-activating factors. Biochem Soc Trans. 2002 Nov;30(Pt 6):952–958. doi: 10.1042/bst0300952. [DOI] [PubMed] [Google Scholar]
- 26.Beaudoing E, Freier S, Wyatt JR, Claverie JM, Gautheret D. Patterns of variant polyadenylation signal usage in human genes. Genome Res. 2000 Jul;10(7):1001–1010. doi: 10.1101/gr.10.7.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009 Jul 15;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu TD, Watanabe CK. GMAP: a genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics. 2005 May 1;21(9):1859–1875. doi: 10.1093/bioinformatics/bti310. [DOI] [PubMed] [Google Scholar]
- 29.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010 May;28(5):511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, Aken BL, Barrell D, Zadissa A, Searle S, Barnes I, Bignell A, Boychenko V, Hunt T, Kay M, Mukherjee G, Rajan J, Despacio-Reyes G, Saunders G, Steward C, Harte R, Lin M, Howald C, Tanzer A, Derrien T, Chrast J, Walters N, Balasubramanian S, Pei B, Tress M, Rodriguez JM, Ezkurdia I, van Baren J, Brent M, Haussler D, Kellis M, Valencia A, Reymond A, Gerstein M, Guigó R, Hubbard TJ. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012 Sep;22(9):1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]