Regulations for the selection of patients with metastatic colorectal cancer for anti-EGFR treatment changed at the end of 2013. A European external quality assessment scheme monitored the performance of laboratories and evaluated the implementation of the new regulations. The findings show great variety in the quality of cancer biomarker testing and should raise awareness about the importance of selecting reliable laboratories for biomarker testing in the context of targeted therapy decisions.
Keywords: Biological markers, Colorectal neoplasms, Quality assurance, Patient safety
Abstract
Background.
Regulations for the selection of patients with metastatic colorectal cancer for anti-EGFR treatment changed at the end of 2013. The set of mutations to be tested extended from KRAS codons 12 and 13 to KRAS and NRAS exons 2, 3, and 4. A European external quality assessment scheme monitored the performance of laboratories and evaluated the implementation of the new regulations.
Materials and Methods.
The 131 participating laboratories received 10 samples of formalin-fixed paraffin-embedded material, including RAS (exon 2, 3, 4) and BRAF mutations. Mock clinical data were provided for three cases. Using their routine methods, laboratories determined the genotypes and submitted three written reports. Assessors scored the results according to predefined evaluation criteria.
Results.
Half of the participants (49.3%) had completely implemented the new test requirements (codons 12, 13, 59, 61, 117, and 146 of KRAS and NRAS), and 96 laboratories (73.3%) made no genotype mistakes. Correct nomenclature, according to the Human Genome Variation Society, was used by 82 laboratories (62.6%).
Conclusion.
Although regulations were effective for several months, many laboratories were not ready for full RAS testing in the context of anti-EGFR therapy. Nevertheless, in each participating country, there are laboratories that provide complete and correct testing. External quality assessments can be used to monitor implementation of new test regulations and to stimulate the laboratories to improve their testing procedures. Because the results of this program are available on the website of the European Society of Pathology, patients and clinicians can refer test samples to a reliable laboratory.
Implications for Practice:
The European Medicines Agency recently extended the labels of panitumumab and cetuximab with an additional biomarker, NRAS, and additional regions of the previously defined KRAS biomarker. An external quality assessment organized by the European Society of Pathology monitored the performance of laboratories under this new regulation. This article summarizes the performance results to reflect the impact of the new regulations on the laboratories. The findings show great variety in the quality of cancer biomarker testing. Many laboratories offer incomplete or unreliable tests that may compromise patient safety. These findings should raise awareness among oncologists of the importance of selecting reliable laboratories for biomarker testing in the context of targeted therapy decisions.
Introduction
A personalized approach for treatment of cancer is becoming the new standard. Using a person’s biological information to adapt the treatment reduces the toxicity and increases the efficacy of cancer therapy [1].
The addition of anti-epidermal growth factor receptor (EGFR) therapy to standard chemotherapy has been shown to significantly improve the survival of patients with metastatic colorectal carcinoma (mCRC). These monoclonal antibodies were initially shown to be active only in patients without mutations in codons 12 or 13 of the KRAS gene. The public announcement of the outcome of phase III trials (PRIME and FIRE studies) demonstrated that patients carrying additional KRAS and NRAS mutations are also resistant to EGFR monoclonal antibodies. The treatment guidelines for EGFR therapy (cetuximab or panitumumab) for patients with mCRC are now more stringent. The European Medicine Agency (EMA) extended the labels with an additional biomarker (NRAS) and additional regions of the previously defined KRAS biomarker. Confirmation of the wild-type status of exons 2, 3 and 4 of both the KRAS and the NRAS gene is now required before EGFR therapy [2, 3]. This new label resulted in a significant amount of pressure on the laboratories to adjust their techniques within a very short time frame [4].
Correct biomarker test results are extremely important for the patient. False-positive or false-negative results can lead to denial of treatment to a patient who would actually benefit from it or to superfluous use of high-priced therapeutic agents and unnecessary side effects in patients who will have no benefit from the drug. Negative treatment effects occur on administration of panitumumab plus the FOLFOX4 regimen or cetuximab plus FOLFOX4 to patients with RAS mutant tumors [4, 5].
More predictive biomarkers for mCRC treatment are likely to become relevant in the near future [6, 7]. Consequently, new techniques will enter routine clinical practice, such as next-generation sequencing (NGS) [8]. This puts additional pressure not only on the laboratories but also on the suppliers of new assays.
Because of the increasing importance of biomarkers for therapy decisions, the European Society of Pathology (ESP) pushed the founding of a working group around external quality assessment (EQA) of colon biomarker testing [9]. The ESP Colon EQA scheme assesses nearly the whole analytical process and addresses both genotyping and reporting performance. It is coordinated according to accepted predefined standards of quality [10, 11]. The scheme monitors the performance of laboratories, allows interlaboratory comparison, and aims to educate and support the participating laboratories to reach accurate test results.
The ESP Colon EQA scheme for KRAS testing has been organized yearly since 2010, after two pilot schemes [12, 13]. Only in 2014 was there a need to include an additional biomarker (NRAS). A sample with a BRAF mutation was also added because it is seen as a possible prognostic and predictive marker [14, 15]. BRAF is not yet mandatory for testing but is often requested by medical oncologists for mCRC patients.
This article summarizes the results of the 2013 ESP Colon EQA scheme to reflect the impact of the new regulations on the performance of laboratories.
Materials and Methods
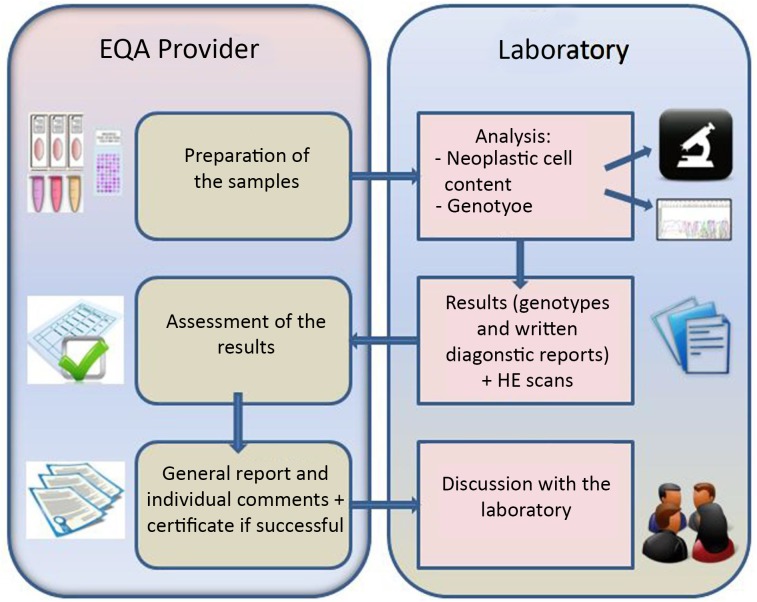
Figure 1 depicts the set-up of the ESP Colon EQA scheme. The participating laboratories received 10 samples with mock clinical information for three cases. The laboratories had to determine the 10 genotypes and submit written diagnostic reports for three cases using their routine methods of biomarker testing. In addition, information was requested regarding characteristics of the laboratory (e.g., DNA extraction method, mutation detection method, setting of the laboratory). All of this information could be completed on the participating laboratory’s page on the website of the ESP Colon EQA scheme [9]. The deadline for submission of the results was 14 calendar days after the receipt of the samples.
Figure 1.
The colon EQA scheme process.
Abbreviations: EQA, external quality assessment; HE, hematoxylin and eosin.
Samples
Eight samples consisted of three slides of formalin-fixed paraffin-embedded (FFPE) tissue; one of those was meant for staining with hematoxylin and eosin and for determination of neoplastic cell content, and two were intended for DNA extraction and mutation analysis. The other two samples were sections of artificial FFPE cell line material (Horizon Diagnostics, Cambridge, U.K., http://www.horizondx.com). The participants were informed beforehand that these samples mimicked a neoplastic cell content of 100%.
In 2014, the EQA set consisted of four samples with KRAS mutations (c.34G > T p.Gly12Cys; c.35G > A p.Gly12Asp; c.38G > A p.Gly13Asp; c.436G > A p.Ala146Thr), one with an NRAS mutation (c.181C > A p.Gln61Lys), one with a BRAF mutation (c.1799T > A p.Val600Glu), and four wild-type samples.
Because of the large number of participants, the preparation and validation of the samples was done in collaboration with eight scheme organizers (SO). Each SO prepared, validated, and sent samples for approximately 16 participants (subschemes A–H). The samples originated from tissue blocks of leftover patient material that was obtained during routine care and testing for the treatment of metastatic colorectal cancer. Each SO signed a subcontractor agreement stating that the way in which the samples were obtained conformed to the national legal requirements for the use of patient samples. The samples were used for test validation and thus were excluded from research regulations requiring informed consent.
Before sample preparation and distribution, the department of pathology at Radboud University Medical Center (Radboud UMC) in Nijmegen, The Netherlands, revalidated the samples to ensure harmonization of genotypes and quality of the material across all subschemes. Moreover, after sample preparation, Radboud UMC also tested the last cut section of the tissue blocks. The percentage of neoplastic cells was assessed to have at least 30%, and samples were analyzed for KRAS, BRAF, NRAS, PIK3CA, EGFR, ERBB2, and AKT1 by NGS (Ion Torrent; ThermoFisher Scientific, Waltham, MA, http://www.lifetechnologies.com/us/en/home/brands/ion-torrent.html). The sensitivity threshold was set at 5% mutant alleles. The reference laboratory verified also the percentage of mutant alleles in the artificial samples by NGS.
Despite the harmonization procedure, in subschemes E and H, different tissue blocks were used that contained an extra BRAF mutation (c.1799T > A; p.Val600Glu) or no BRAF mutation, respectively.
Scoring
Assessment of the results was done in cooperation with two medical and two technical experts and a team of assessors with established knowledge and expertise in biomarker testing in mCRC (KU Leuven, Belgium; INT Fondazione “G. Pascale,” Naples, Italy; and Radboud UMC). Evaluation criteria were determined beforehand according to international guidelines for EQA [10]. At least two independent assessors evaluated the results of each participant. An assessment meeting was organized for discussion of the results.
Both the genotypes and the written reports were scored. For each sample, correct genotyping was awarded 2 points, and a test failure received 0.5 point. Correct genotyping was defined as a correct genotype based on the tests that were performed; for example, if a laboratory did not test for the NRAS codon 61 mutation, it obtained 2 points for a wild-type result in the sample with this mutation. The test reports of the three cases were evaluated on 28 elements, according to accepted standards [10]. This study focuses on the presence of a correct interpretation and the list of tested mutations.
The correct use of nomenclature in the written reports was scored according to the Human Genome Variation Society (HGVS) recommendations [16]. When wrong nomenclature was used in different reports, points were deducted only once.
At the time of the EQA scheme registration (July 2013), wild-type RAS confirmation was not yet mandatory for anti-EGFR therapy. Participants were grouped into three categories, according to the information in the written reports: those that performed RAS testing defined as KRAS exon 2, 3, and 4 and NRAS exon 2, 3, and 4 (complete RAS testing); those that tested KRAS exon 2, 3, and 4 and NRAS exon 2 and 3 (almost complete RAS testing); and those who tested even fewer exons (incomplete RAS testing).
Statistical Analysis
Statistical analysis was done with chi-square tests or with Fisher’s exact test in case the expected frequency was <5 in >20%. The p value was set at 5%, and a Bonferroni correction was applied as necessary.
Results
Participants
The 2013 ESP Colon EQA scheme included 133 participants, of which 131 submitted results. The laboratories originated from 30 different countries and were usually part of a hospital, including academic hospitals (70.2%). Other settings were private companies (13.7%) or university laboratories (7.6%). Fifty-two laboratories (39.7%) were accredited according to an international standard [17–19].
Of the 131 participants, 39 laboratories participated for the first time in an ESP Colon EQA scheme. The new participants were spread over 24 countries, with substantial proportions from Poland (17.9%), Hungary (15.4%), and Belgium (10.3%). The new participant settings were mostly hospitals (48.7%) or private companies (28.2%). Only a quarter (25.6%) were accredited.
Genotype Scoring
The laboratories tested 1,310 samples, of which 3.6% were incorrectly genotyped. Significantly more errors were made in mutation-positive samples compared with wild-type samples (5.0% vs. 1.5%; p = .001). A technical failure was reported in 2.7% of the samples. Table 1 summarizes the genotype mistakes and failures.
Table 1.
Subdivision of the scoring of different mutations

At the laboratory level, only 96 laboratories (73.3%) had no genotype mistakes for the 10 tested samples; 26 (19.9%) had one genotype mistake, and 9 laboratories (6.9%) had more than one genotype mistake. Incorrect HGVS nomenclature was used by 49 laboratories (37.4%) [16]. Table 2 summarizes these results in relation to different laboratory characteristics. Supplemental online Table 1 gives an overview of the performance of different laboratories by country.
Table 2.
Different laboratory characteristics versus errors and scoring

The synthetic samples used in this EQA program turned out to be inadequate for some laboratories, depending on the primer set used for amplification. An engineering scar produced by the introduction of an artificial mutation might interfere with amplification. Because of this problem, 14 laboratories could not be evaluated for the synthetic samples containing a KRAS exon 4 mutation, and 4 could not be evaluated for the synthetic samples containing an NRAS mutation.
Because NRAS exon 4 mutations are extremely rare and are absent in published articles, we divided the laboratories into three groups: complete RAS testing, almost complete RAS testing, and incomplete RAS testing. This characteristic is included in Table 2. There was only limited information on the codon level, which is provided in Table 3, from which it can be derived that only 37 laboratories (49.3%) tested all recommended codons (12, 13, 59, 61, 117, and 146). Laboratories using homebrew tests were more likely to obtain a maximal score (Table 2) and to use a complete or almost complete RAS test (83.3% vs. 42.3%; p = .000024 with α = .017) than laboratories using commercial kits.
Table 3.
Overview of codons of KRAS and NRAS that are considered relevant for RAS testing

The turnaround time (TAT) was calculated as the number of days between the receipt of the samples, as indicated by the participant, and the submission of the data in the online data sheet. The data show that 52.7% completed the genotype testing and the submission of the report within the required 14 calendar days. There was no indication that the use of homebrew assays or commercial kits was linked to a higher TAT. The minimum TAT was 7 days, and the maximum TAT was up to 31 days.
Report Scoring
The report score was based on a KRAS codon 12-positive sample, an NRAS codon 61-positive sample, and a wild-type sample. Of the 41 laboratories using a limited RAS test (mainly KRAS exon 2), 23 (56.1%) did not mention that the spectrum of tested mutations was incomplete according to the latest regulatory requirements, and 10 (24.4%) did not specify the tested nucleotide positions; 5 (12.2%) did not mention either item.
Discussion
Precision medicine is becoming an essential part of cancer treatment. In 2013, regulations for anti-EGFR treatment (cetuximab and panitumumab) for mCRC patients became more stringent. Wild-type RAS status (KRAS exons 2, 3, and 4 and NRAS exons 2, 3, and 4) must now be confirmed before treatment [2, 3]. We report the first EQA that monitored the performance of laboratories under this regulation.
There was a remarkable number of new participants in 2013 compared with the 18 new participants in the 2012 scheme. This shows the increasing interest of laboratories in EQA schemes. Laboratories understand the educational aspect of EQA and participate to test and improve their procedures. Surprisingly, at the time that the samples were sent—6 months after the change in regulation for panitumumab and 3 months after the change for cetuximab—not all laboratories offered full RAS testing. The reason could be the short time frame in which the laboratories had to change their routine process of biomarker testing. Laboratories using commercial tests are dependent on the availability of the tests that need to be verified in their own settings, whereas laboratories using homebrew assays need to expand and validate their test sets. Laboratories using homebrew assays seem to be more flexible because a significantly higher percentage had already implemented an (almost) complete RAS test.
Several laboratories did not provide NRAS exon 4 testing. The need for testing of this exon is debated because mutations were seldom identified in the different clinical trials leading to the additional rules for panitumumab or cetuximab [4, 20]. These mutations should be investigated because the label for EGFR monoclonal antibodies includes NRAS exon 4.
The use of patient samples in an EQA scheme is important; however, finding and validating the patient samples to optimize harmonization of the scheme is highly labor intensive. Consequently, two synthetic samples were added. During this colon EQA scheme, the limitations of the synthetic samples were elucidated. This prompted the manufacturer to openly communicate the limitations to the users and will enable laboratories to better evaluate unexpected negative results obtained with these samples.
At the laboratory level, 26.7% of the participating laboratories made one mistake or more. In comparison with previous schemes, the error rate is similar to the first full ESP Colon EQA scheme in 2010 (error rate: 30%) [12]. The error rate was 18.5% in 2011 and 26.7% in 2012, but this was mainly because of a sample with only 5% mutant alleles to challenge the laboratories [21]; without this sample, the error rate would have been only 12.4%. The high error rate of the present scheme indicates that many laboratories have difficulties extending their routine clinical testing. A new learning phase is probably ongoing, and the laboratories need to adjust to the new routine.
Half of the laboratories exceeded the turnaround time of 14 calendar days. Some delay in turnaround time might be due to the requirements of filling forms and uploading reports in a format and a system different from routine reports; however, in a clinical context, rapid availability of results is important to start the correct treatment [17].
The fact that only half of the participants performed complete RAS testing, in addition to the high error rate for the testing of the new NRAS biomarker, is alarming. Because the latter is based on only one NRAS mutated sample, additional independent sample sets will be needed to infer generalizability. Considering the consequences of an incomplete or incorrect test on therapy administration, faster implementation and validation of the techniques is needed to better guarantee patient safety. Nevertheless, in each country, laboratories are available that offer reliable testing. These laboratories are listed on the ESP website, which is available to patients and physicians.
The label of anti-EGFR therapies does not include a mutation-specific indication [2, 3]. Currently, only false-positive or false-negative results may influence therapy decisions. In the future, mutation-specific therapy decisions may be necessary. Several studies have already suggested that codon 13 mutated patients may benefit from anti-EGFR therapy, but the role of p.G13D, c.38G > A remains controversial [22, 23]. Currently, the ICE CREAM trial is recruiting patients with this mutation to determine whether cetuximab could help them.
The 2013 scheme was the first in which nomenclature was taken into account in the ESP RAS EQA for the scoring of the participants. Homogeneous nomenclature is essential for clear communication with the clinic, for avoiding misinterpretation, for the linkage of mutations in databases, and for additional clinical trials. It is recommended that laboratories follow the HGVS correctly [16, 21]. Not all laboratories perform mutation analysis according to the latest EMA decisions for RAS testing; therefore, especially in this transition state, it is important to state a clear interpretation of the results and to refer samples for further testing of the additional regions in case it is necessary. In addition, the list of tested codons is also of great importance. When this is not provided, the result can be misinterpreted by the physician.
The characteristics of the participating laboratories were also evaluated. Although there is growing interest for accreditation, no improvement was seen compared with previous years; only some of the participating laboratories were accredited according to a well-known international standard [17–19]. More efforts are needed to drive laboratories toward accreditation, even if it is not mandatory according to the national legal requirements. Accreditation provides the opportunity to externally verify whether laboratories perform well and whether they participate in an EQA program. The number of laboratories that make mistakes is scary and needs to be monitored to ensure the quality of health care.
The importance of participation in an EQA must be stressed. In this way, a longitudinal analysis can be performed to identify indicators related to a higher quality assurance of diagnostic biomarker testing. Working with a larger data set could help in obtaining significant results for this analysis.
Conclusion
With a high percentage of laboratories performing incomplete or unreliable tests, the results of this ESP Colon EQA scheme are concerning. It shows that many laboratories do not have enough experience with molecular testing to rapidly introduce new tests, although the techniques are considered to be straightforward and the number of mutations to be tested is still limited. This suggests that more extensive training in validation, implementation, and interpretation of molecular testing and frequent external quality assessments are pivotal in this era during which many different molecular tests are expected to be introduced in the clinic to stratify patients for optimal therapy. Clinicians prescribing anti-EGFR targeted for colorectal cancer patients are advised to select a reliable laboratory for testing. The ESP website can be a good source. Concerning future developments in the biomarker field, these results show the need for guidelines concerning the approval of medicines and the time frame for implementation of new regulations for both laboratories and industry.
See http://www.TheOncologist.com for supplemental material available online.
Supplementary Material
Acknowledgment
Funding from AMGEN was received for validation of the samples with next-generation sequencing.
Author Contributions
Conception/Design: Véronique Tack, Marjolijn J.L. Ligtenberg, Nicola Normanno, J. Han van Krieken, Elisabeth M.C. Dequeker
Provision of study material or patients: Marjolijn J.L. Ligtenberg, Sara Vander Borght, J. Han van Krieken
Collection and/or assembly of data: Véronique Tack, Lien Tembuyser
Data analysis and interpretation: Véronique Tack, Marjolijn J.L. Ligtenberg, Lien Tembuyser, Nicola Normanno, Sara Vander Borght, J. Han van Krieken, Elisabeth M.C. Dequeker
Manuscript writing: Véronique Tack, Marjolijn J.L. Ligtenberg, Lien Tembuyser, Nicola Normanno, Sara Vander Borght, J. Han van Krieken, Elisabeth M.C. Dequeker
Final approval of manuscript: Véronique Tack, Marjolijn J.L. Ligtenberg, Lien Tembuyser, Nicola Normanno, Sara Vander Borght, J. Han van Krieken, Elisabeth M.C. Dequeker
Disclosures
Marjolijn J.L. Ligtenberg: Amgen (RF); Nicola Normanno: Qiagen, Roche (C/A); Amgen, Merck (H); J. Han van Krieken: Amgen, Merck-Serrono (RF, H); Elisabeth M.C. Dequeker: Pfizer, Amgen (RF); Johnson & Johnson (OI). The other authors indicated no financial relationships.
(C/A) Consulting/advisory relationship; (RF) Research funding; (E) Employment; (ET) Expert testimony; (H) Honoraria received; (OI) Ownership interests; (IP) Intellectual property rights/inventor/patent holder; (SAB) Scientific advisory board
References
- 1.Heinemann V, Douillard JY, Ducreux M, et al. Targeted therapy in metastatic colorectal cancer -- an example of personalised medicine in action. Cancer Treat Rev. 2013;39:592–601. doi: 10.1016/j.ctrv.2012.12.011. [DOI] [PubMed] [Google Scholar]
- 2.Erbitux (cetuximab) [European Public Assessment Report]. Available at http://www.ema.europa.eu/docs/en_GB/document_library/EPAR_-_Summary_for_the_public/human/000558/WC500029111.pdf. Accessed September 25, 2014.
- 3.Vectibix (panitumumab) [European Public Assessment Report]. Available at http://www.ema.europa.eu/docs/en_GB/document_library/EPAR_-_Summary_for_the_public/human/000741/WC500047704.pdf. Accessed September 25, 2014.
- 4.Douillard JY, Oliner KS, Siena S, et al. Panitumumab-FOLFOX4 treatment and RAS mutations in colorectal cancer. N Engl J Med. 2013;369:1023–1034. doi: 10.1056/NEJMoa1305275. [DOI] [PubMed] [Google Scholar]
- 5.Bokemeyer C, Bondarenko I, Hartmann JT, et al. Efficacy according to biomarker status of cetuximab plus FOLFOX-4 as first-line treatment for metastatic colorectal cancer: The OPUS study. Ann Oncol. 2011;22:1535–1546. doi: 10.1093/annonc/mdq632. [DOI] [PubMed] [Google Scholar]
- 6.Cathomas G. PIK3CA in colorectal cancer. Front Oncol. 2014;4:35. doi: 10.3389/fonc.2014.00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peeters M, Price TJ, Cervantes A, et al. Randomized phase III study of panitumumab with fluorouracil, leucovorin, and irinotecan (FOLFIRI) compared with FOLFIRI alone as second-line treatment in patients with metastatic colorectal cancer. J Clin Oncol. 2010;28:4706–4713. doi: 10.1200/JCO.2009.27.6055. [DOI] [PubMed] [Google Scholar]
- 8.Peeters M, Oliner KS, Parker A, et al. Massively parallel tumor multigene sequencing to evaluate response to panitumumab in a randomized phase III study of metastatic colorectal cancer. Clin Cancer Res. 2013;19:1902–1912. doi: 10.1158/1078-0432.CCR-12-1913. [DOI] [PubMed] [Google Scholar]
- 9.Colon external quality assessment scheme. Available at http://kras.eqascheme.org. Accessed September 25, 2014.
- 10.van Krieken JH, Normanno N, Blackhall F, et al. Guideline on the requirements of external quality assessment programs in molecular pathology. Virchows Arch. 2013;462:27–37. doi: 10.1007/s00428-012-1354-4. [DOI] [PubMed] [Google Scholar]
- 11.ISO . IEC 17043:2010 Conformity assessment -- general requirements for proficiency testing. Geneva, Switzerland: International Organization for Standardization; 2010. [Google Scholar]
- 12.Bellon E, Ligtenberg MJ, Tejpar S, et al. External quality assessment for KRAS testing is needed: Setup of a European program and report of the first joined regional quality assessment rounds. The Oncologist. 2011;16:467–478. doi: 10.1634/theoncologist.2010-0429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dequeker E, Ligtenberg MJ, Vander Borght S, et al. Mutation analysis of KRAS prior to targeted therapy in colorectal cancer: Development and evaluation of quality by a European external quality assessment scheme. Virchows Arch. 2011;459:155–160. doi: 10.1007/s00428-011-1094-x. [DOI] [PubMed] [Google Scholar]
- 14.Reimers MS, Zeestraten EC, Kuppen PJ, et al. Biomarkers in precision therapy in colorectal cancer. Gastroenterol Rep (Oxf) 2013;1:166–183. doi: 10.1093/gastro/got022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu Q, Xu AT, Zhu MM, et al. Predictive and prognostic roles of BRAF mutation in patients with metastatic colorectal cancer treated with anti-epidermal growth factor receptor monoclonal antibodies: A meta-analysis. J Dig Dis. 2013;14:409–416. doi: 10.1111/1751-2980.12063. [DOI] [PubMed] [Google Scholar]
- 16.Guidelines and recommendations. Available at http://www.hgvs.org/content/guidelines. Accessed September 25, 2014.
- 17.Commission on Laboratory Accreditation. Laboratory Accreditation Program. Molecular pathology checklist. CAP 2014. College of American Pathologists.
- 18.ISO 15189:2012 Medical laboratories -- Requirements for quality and competence. Geneva, Switzerland: International Organization for Standardization, 2012.
- 19.Coordination commission for the promotion of quality assurance in laboratory research in health care. CCKL code of practice. Available at http://www.cckl.nl. Accessed September 25, 2014.
- 20.Lièvre A. Des mutations de KRAS aux mutations de RAS: Vers une meilleure définition de la réponse aux anticorps anti-EGFR dans le cancer colorectal métastatique. Oncologie. 2014;16:120. [in French] [Google Scholar]
- 21.Tembuyser L, Ligtenberg MJ, Normanno N, et al. Higher quality of molecular testing, an unfulfilled priority: Results from external quality assessment for KRAS mutation testing in colorectal cancer. J Mol Diagn. 2014;16:371–377. doi: 10.1016/j.jmoldx.2014.01.003. [DOI] [PubMed] [Google Scholar]
- 22.Chen CC, Er TK, Liu YY, et al. Computational analysis of KRAS mutations: Implications for different effects on the KRAS p.G12D and p.G13D mutations. PLoS One. 2013;8:e55793. doi: 10.1371/journal.pone.0055793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tejpar S, Celik I, Schlichting M, et al. Association of KRAS G13D tumor mutations with outcome in patients with metastatic colorectal cancer treated with first-line chemotherapy with or without cetuximab. J Clin Oncol. 2012;30:3570–3577. doi: 10.1200/JCO.2012.42.2592. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.